Introduction
The bone marrow microenvironment consists of a
specialized population of cells that play essential roles in
regulation, self-renewal and differentiation of adult stem cells
(1–4). The microenvironment supports
maturation of hematopoietic stem cells (HSCs) and hematopoietic
progenitor cells (HPCs) and their release into the vascular system
(5–7). Human bone marrow-derived stromal cell
lines HS5 and HS27A, co-cultured with myeloid cells, are frequently
used in studies of crosstalk between cells in the bone marrow
microenvironment and hematopoietic cells (8–13). For
example, HS5 and HS27A have been used to establish a
xenotransplantation murine model of myelodysplastic syndromes
(MDS), a group of clonal hematopoietic disorders characterized by
dysregulation of programmed cell death (apoptosis) and ineffective
hematopoiesis in both normal and clonal (transformed) hematopoietic
cells (12,14). Kerbauy et al (14) observed engraftment of distinct
clonal MDS-derived hematopoietic precursors when stromal cells (HS5
and HS27A combined) were co-injected via an intramedullary route.
Our previous study found that intravenous co-administration of
HS27A cells (but not HS5 cells) with HPCs from MDS patients
facilitated engraftment of clonal CD34+ cells of any
karyotype (12), indicating that
HS27A cells were more effective than HS5 in supporting primitive
clonal MDS precursors (12).
Glycans, a major category of biomolecules, are
commonly attached to proteins and lipids to form glycoproteins,
glycolipids, glycosaminoglycans and other types of glycoconjugates
(15–17). Glycans play key roles in a variety
of biological processes, including cell adhesion, molecular
trafficking, receptor activation and signal transduction (17,18).
They are also involved in malignant hematopoiesis. CoCl2
(hypoxia mimic) induced expression of cell surface glycans
recognized by both β-galactoside- and GlcNAc-binding lectins in
HL-60 cells (19). Galectin-3 is
developmentally expressed in human myeloid cells, and is strongly
upregulated on the surface of late mature myeloid cells (20). Conversely, it also promotes
proliferation and angiogenesis of endothelial cells differentiated
from bone marrow mesenchymal stem cells (21). An important characteristic of the
HSC microenvironment is that low oxygen (O2) tension and
hypoxia (defined as O2 tension <2%) is essential for
HSC function (22–25). In vitro BM cells are always
cultured under hypoxic conditions in order to maintain their
primitive phenotype and self-renewal ability (26). In addition, the key regulator of
cellular adaptation to hypoxic stress, hypoxia-inducible factor 1 α
(HIF-1α) can be stabilized under hypoxia (25,26).
No study to date has included global analysis of the expression of
N-glycan in hematopoietic cells co-cultured with vs. without
stromal cells under hypoxia.
An in vitro model in which KG1a hematopoietic
cells were co-cultured with HS27A cells under hypoxia conditions
(1% O2) was used in the present study. We examined the
apoptosis rate of the KG1a cells, and the related apoptotic
pathway. N-glycans linked to glycoproteins were released by
PNGase F and analyzed by MALDI-TOF/TOF-MS, and selectively altered
N-glycans were further analyzed by lectin staining. By
presenting global analysis of patterns of altered N-glycans
in hematopoietic cells before and after stroma contact under
hypoxia, the present study provides valuable information for future
investigations of the bone marrow microenvironment.
Materials and methods
Cell culture
KG1a cells (derived from acute myeloid leukemia) and
bone marrow-derived stromal cell line HS27A are generous gifts from
Professor H. Joachim Deeg (Fred-Hutchinson Cancer Research Center,
Seattle, WA, USA). KG1a and HS27A were grown, propagated and
subjected to experiments between passages 8 and 24, as previously
described (27). Multiple aliquots
from early passages were cryopreserved for later use.
Total protein extraction
Cells were harvested and lysed with T-PER Tissue
Protein Extraction reagent (Thermo Fisher Scientific, Hudson, NH,
USA) according to the manufacturer's protocol. In brief, cells were
trypsinized, resuspended in phosphate-buffered saline (PBS), added
with T-PER reagent containing 1% protease inhibitor cocktail
(Biotool Ltd., Houston, TX, USA), incubated on ice for 30 min and
homogenized. The sample was centrifuged for 15 min at 13,000 × g
(4°C), and the supernatant was collected and stored at −80°C.
Enzyme-linked immunosorbent assay
(ELISA)
Concentrations of secreted tumor necrosis factor α
(TNFα) in culture supernatants were determined (in triplicate)
using an ELISA kit (R&D Systems, Minneapolis, MN, USA) for
human TNFα (detection limit 0.5 pg/ml). Color intensity of the
chromogenic reaction was determined at 490 nm by a plate reader
(Bio-Rad Laboratories, Hercules, CA, USA).
Western blot analysis
Proteins from each sample were separated by 10%
SDS-PAGE and transferred onto polyvinylidene fluoride (PVDF)
membranes using the Trans-Blot Turbo Transfer System (Bio-Rad
Laboratories). Membranes were soaked in 5% (w/v) skim milk in TBST
(20 mM Tris-HCl, 150 mM NaCl, 0.05% Tween-20, pH 8.0) for 2 h at
37°C, probed with primary antibodies against HIF-1α (1:1,000; cat.
no. 3716; Cell Signaling Technology, Inc., Beverly, MA, USA), p53
(1:1,000; cat. no. 2524; Cell Signaling Technology, Inc.), Bax
(1:1,000; cat. no. 2774; Cell Signaling Technology, Inc.), Bcl-2
(1:1,000; 2872; Cell Signaling Technology, Inc.), caspase-3
(1:1,000; cat. no. 9662; Cell Signaling Technology, Inc.),
caspase-8 (1:1,000; cat. no. 9746; Cell Signaling Technology,
Inc.), caspase-9 (1:1,000; cat. no. 9502; Cell Signaling
Technology, Inc.), NF-κB (1:1,000; cat. no. 8242; Cell Signaling
Technology, Inc.), phospho-NF-κB (1:1,000; cat. no. 3031; Cell
Signaling Technology, Inc.) and β-tubulin (1:5,000; cat. no. T7816;
Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) overnight at 4°C,
and incubated with the appropriate HRP-conjugated secondary
antibody (1:5,000; cat. nos. A0208 and A0216; Beyotime Institute of
Biotechnology, Shanghai, China). Bands were visualized using
enhanced chemiluminescence detection kit Westar Nova (Cyanagen Srl,
Bologna, Italy) with imaging by ChemiDoc™ XRS+ (Bio-Rad
Laboratories).
Cell sorting
KG1a cells were co-cultured with HS27A cells for 24
h and added together with CD45+ magnetic beads. KG1a
cells were isolated by magnetic-activated cell sorting (Miltenyi
Biotec, Bergisch Gladbach, Germany) according to the manufacturer's
protocol. Cell purity was assessed by staining with
fluorescent-tagged anti-CD45 antibody and quantified by flow
cytometry with the purity of sorted cells reaching >95%.
Apoptosis was assessed by flow cytometry.
Caspase activity assay
Caspase activity assay was performed according to
the manufacturer's instructions (Beyotime Institute of
Biotechnology). Briefly, after treatment, isolated KG1a cells were
lysed with lysis buffer and the supernatants were incubated with
Ac-DEVD-pNA (200 µM) substrate for caspase-3 and Ac-IETD-pNA (200
µM) substrate for caspase-8, respectively. The reaction was
assessed with a plate reader at 405 nm wavelength.
Quantitative real-time PCR
Total RNA was extracted as previously described
(28). Primers were designed using
the Primer-BLAST program (https://www.ncbi.nlm.nih.gov/tools/primer-blast/).
First-strand cDNA was synthesized from total RNA using a OneScript
Plus cDNA Synthesis kit (Abm Canada, Milton, ON, Canada).
Quantitative real-time PCR was performed by SYBR-Green I dye
detection with BrightGreen Express 2× qPCR Master Mix (Abm Canada).
Gene expression was quantified by the 2−ΔΔCq method
(29). All primer sequences are
provided in Table I.
 | Table I.Quantitative real-time PCR primer
sequences. |
Table I.
Quantitative real-time PCR primer
sequences.
| Genes | Primer
sequences |
|---|
| FUT8 | F:
5′-TGTCCTGTACTTCATGCGCT-3′ |
|
| R:
3′-TCCATGACCCTAATGGTCTTTT-5′ |
| MGAT2 | F:
5′-AAAGAACACCTGCAGAACCG-3′ |
|
| R:
3′-GGAATTGACAACGTCCTCGT-5′ |
| MGAT3 | F:
5′-AGGAAGGAGATGAGGCACAG-3′ |
|
| R:
3′-TTGCTGAGACCCAGCGG-5′ |
| MGAT4B | F:
5′-TCACTGCCGAAGTGTACTGTG-3′ |
|
| R:
3′-CTGACACTCTGCACTCGCTC-5′ |
| MGAT5 | F:
5′-GTGAGGGTAGCCGTCCATAG-3′ |
|
| R:
3′-CAGCTTGGTTGCACTTGAGA-5′ |
Apoptosis assay
Apoptosis of hematopoietic cells was assayed by
Annexin V staining. KG1a cells were cultured under hypoxia for 24 h
with or without HS27A stroma contact, on the basis of our previous
studies (9,11). Twelve-well plates were centrifuged
at 1,200 rpm (300 × g), the supernatant was discarded, and cells in
each well were washed with cold PBS containing 2% bovine serum
albumin (BSA) and assayed using an Annexin V-fluorescein
isothiocyanate (FITC) apoptosis kit (Beyotime Institute of
Biotechnology). Cells were then labeled with Allophycocyanin
(APC)-conjugated anti-CD45 antibody (1:20; cat. no. 368512;
BioLegend, San Diego, CA, USA) to distinguish hematopoietic cells
(myeloid cells, CD45+) from stromal (CD45−),
and apoptosis was assayed by flow cytometry (model BD Accuri C6; BD
Biosciences, San Jose, CA, USA) using Annexin V-FITC and propidium
iodide (PI).
N-glycan mass profiling
N-glycans were separated as described in our
previous study (30). In brief, we
used a size-exclusion spin ultrafiltration unit (Amicon Ultra-0.5
10 KD; EMD Millipore; Merck KGaA, Darmstadt, Germany) to
concentrate 2 mg total proteins. Proteins were denatured,
centrifuged and then further digested with PNGase F (New England
BioLabs, Ipswich, MA, USA) overnight at 37°C. Released
N-glycans were collected and desalted using Sepharose 4B
(Sigma-Aldrich; Merck KGaA, Darmstadt, Germany), as previously
described (31), then characterized
by MALDI-TOF/TOF-MS (ultrafleXtreme; Bruker Daltonik GmbH, Bremen,
Germany) in positive-ion mode. Data were analyzed using the
FlexAnalysis software program (Bruker Daltonik GmbH) and annotated
using the GlycoWorkbench software program (http://code.google.com/p/glycoworkbench/). The
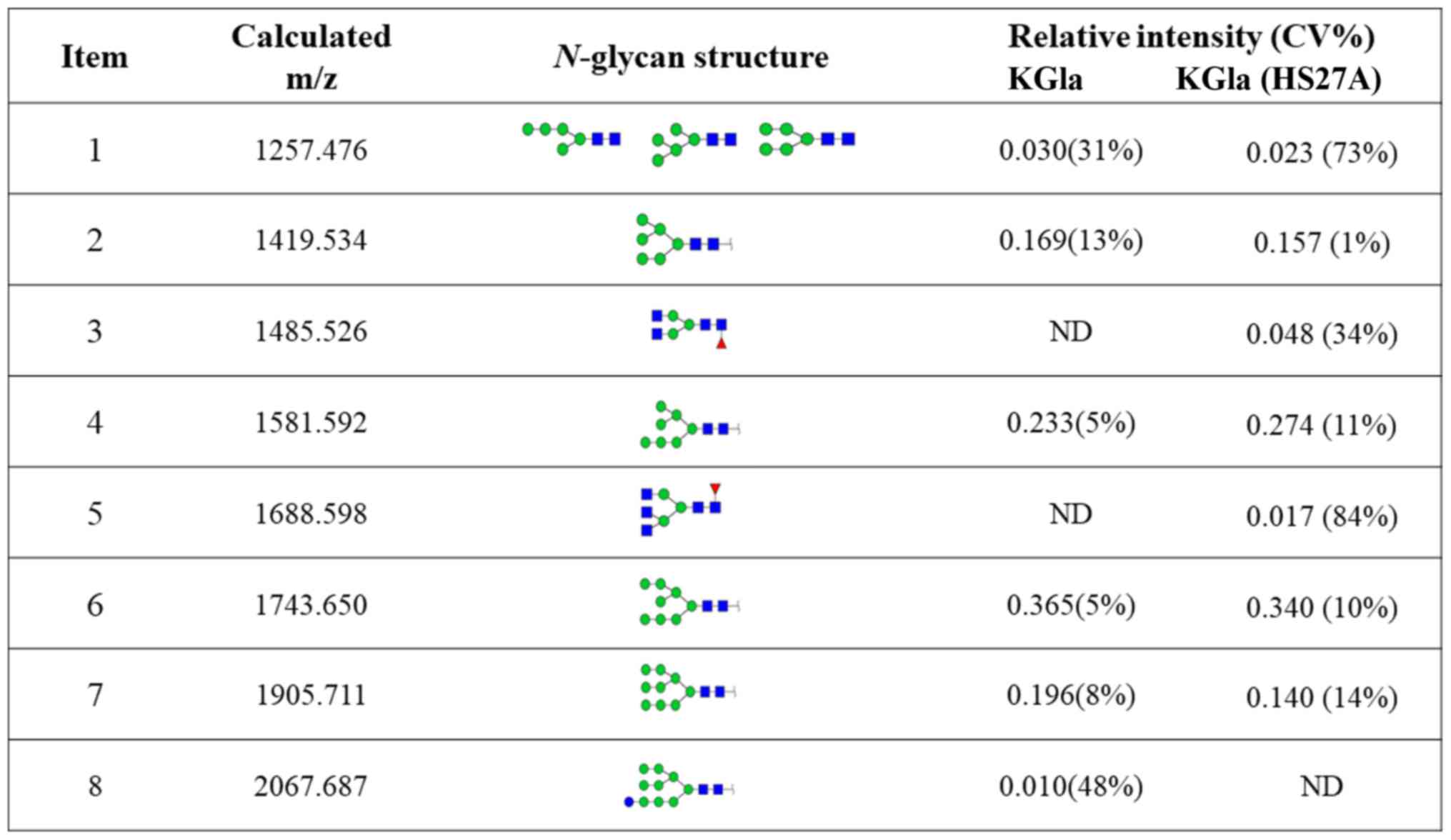
stability of MS was estimated using coefficient of variation (CV)
percentages based on relative intensity values. The relative
variation was calculated by dividing the amount of a given type of
N-glycan by the total N-glycan amount (30).
Flow cytometric (FACS) analysis of
lectin binding affinity
Cells were harvested to a single cell suspension
with trypsin or 0.2% EDTA, and washed three times with PBS.
Aliquots (106 cells) were resuspended in 100 µl diluting
Cy3-conjugated lectin (cat. no. L-1000, L-1020, L-1040 and L-1120;
Vector Labs, Peterborough, UK) and APC-conjugated anti-human CD45
antibody (1:20; cat. no. 368512; BioLegend) in 0.1% BSA/PBS, and
incubated on ice for 30 min in the dark. Lectins were used at a
final concentration 100 µg/ml. Cells were washed twice with 1 ml
PBS and analyzed by flow cytometry as above-mentioned.
Lectin staining
Lectin staining was performed as previously
described (30). Cells were spun
onto a microscope slide for 5 min at 300 × g, immobilized with 2%
fresh paraformaldehyde for 15 min at room temperature, and blocked
with 5% BSA in 1× PBS for 1 h at 37°C. Fixed cells were incubated
with 20 µg/ml Cy3-conjugated lectin (LCA, PHA-E) in 5% BSA for 3 h
in the dark, washed with 1× PBS, stained with 20 µg/ml DAPI in 1×
PBS for 10 min, washed again with 1× PBS, and images were captured
with a fluorescence microscope (model Eclipse E600; Nikon, Tokyo,
Japan) at the same exposure time and gain factor.
Statistical analysis
Data are presented as the mean ± SEM. Statistical
significance of differences between the means of two groups was
evaluated by Student's t-test. Multiple group comparisons were
evaluated by ANOVA with Bonferroni's post hoc test.
Results
Co-culture with stromal cells enhances
susceptibility of KG1a cells to apoptosis under a hypoxic
condition
Hematopoietic cell line KG1a is resistant to
apoptosis (32,33). However, apoptosis of KG1a was
significantly increased when they were co-cultured with HS27A
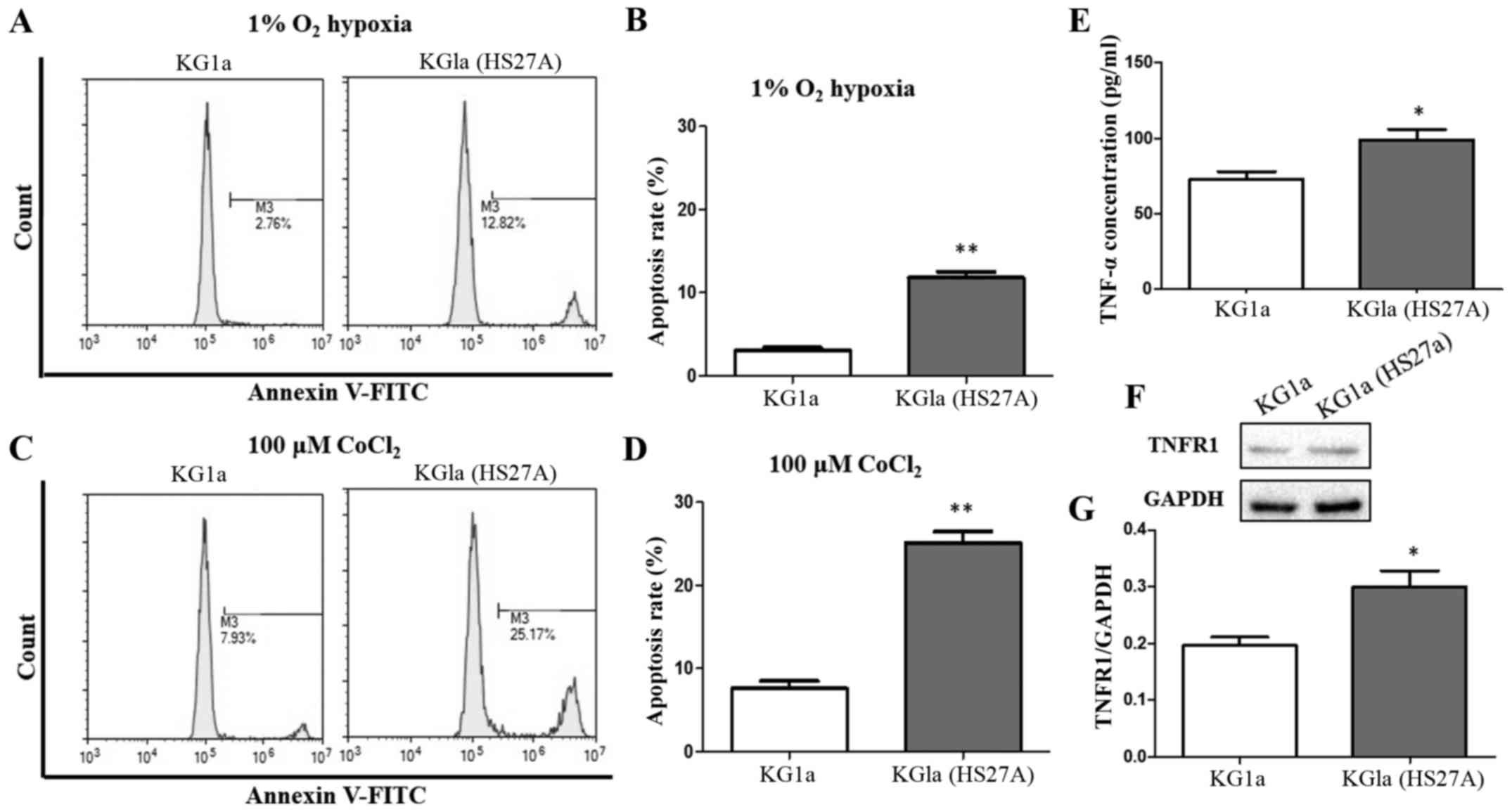
stromal cells under a hypoxic condition (Fig. 1A and B). We investigated the
apoptosis of KG1a cells before and after co-culture with HS27A in
the presence of 100 µM CoCl2. Treatment with
CoCl2 mimics hypoxic conditions by inhibiting
prolyl-4-hydroxylases involved in the degradation of HIF-1α
(34,35). CoCl2 was used as a
positive control in the present study. Apoptosis of KG1a cells
co-cultured with HS27A cells was also increased by CoCl2
treatment (Fig. 1C and D).
Concentration of TNFα, another factor that enhances susceptibility
of KG1a cells to apoptosis, was increased in the co-culture system
under hypoxic conditions (Fig. 1E).
TNF receptor I (TNFR1) was also increased in KG1a cells following
co-culture with stromal cells under hypoxic conditions (Fig. 1F and G).
Hypoxia induces the p53-dependent
pathway in co-cultured KG1a cells
Our previous studies demonstrated upregulation of
p53 levels in hematopoietic cells and primary MDS marrow cells in
stromal contact culture (8,9). To investigate the possible involvement
of a p53-dependent pathway in hypoxia-induced apoptosis in
co-cultured KG1a cells, we examined expression of p53, caspase-3,
caspase-8, caspase-9 and NF-κB in KG1a cells after co-culture with
HS27A cells under hypoxic conditions and CoCl2
treatment. In co-cultured KG1a cells, p53, caspase-8, caspase-9 and
phosphorylated NF-κB levels were elevated and caspase-3 and
caspase-8 were cleaved and active (Fig.
2A and B). We further observed that the activities of caspase-3
and caspase-8 were also elevated in co-cultured KG1a cells, which
appeared to be relevant with the increase in TNF-α and TNFR1
(Fig. 2C). Bax and Bcl-2 levels
were respectively upregulated and downregulated in the co-cultured
cells, as expected since their expression was affected by p53
through transcriptional regulation (36,37).
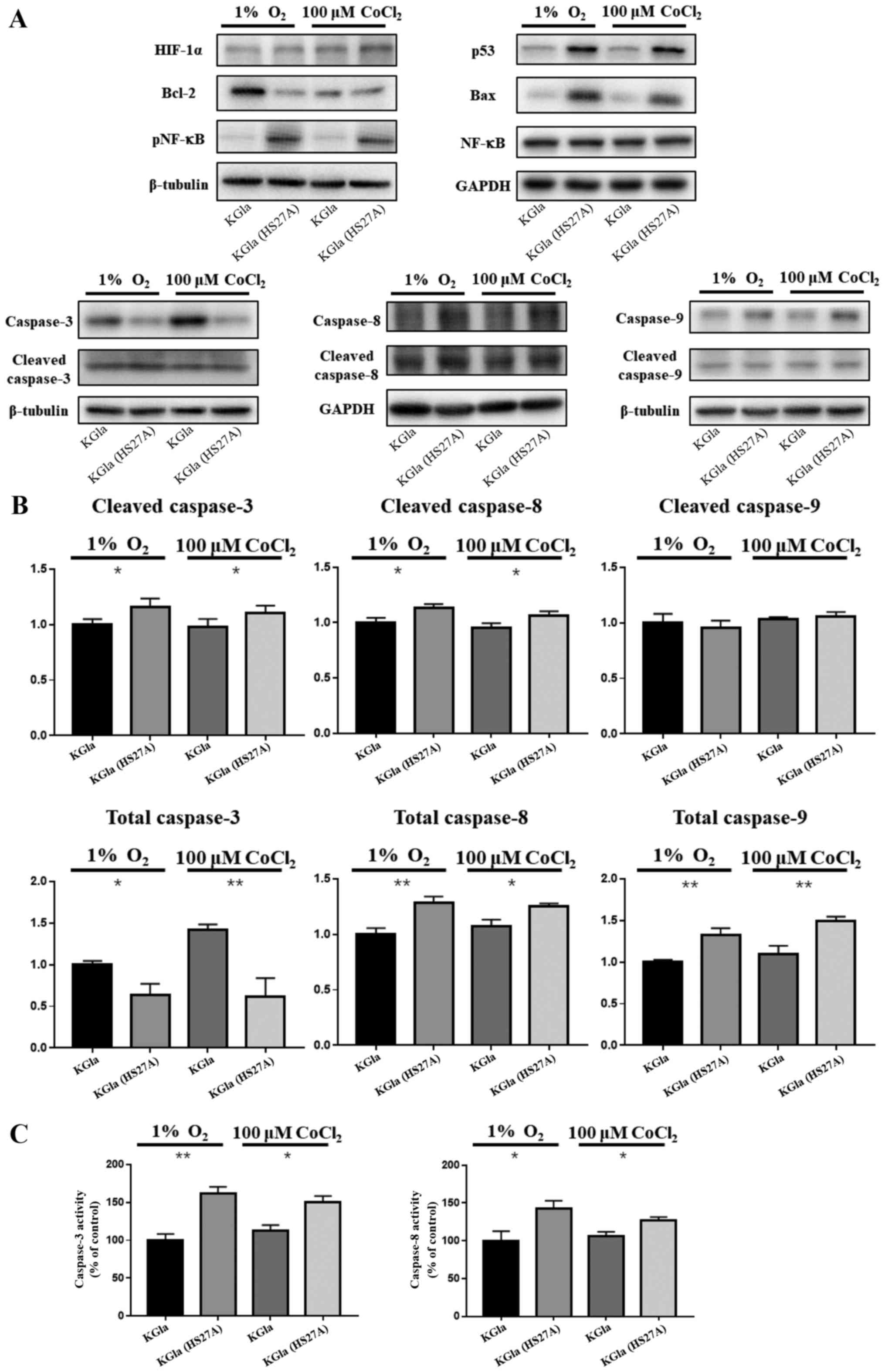 | Figure 2.Protein expression in KG1a cells and
co-cultured KG1a cells under hypoxia. (A) Protein lysates from KG1a
cells and co-cultured KG1a cells under 1% O2 hypoxia or
100 µM CoCl2 conditions were separated on 4 to 12%
Bis-Tris gels, and immunoblotted with antibodies against HIF-1α,
p53, Bax, Bcl-2, caspase-3, caspase-8, caspase-9, NF-κB, and
phospho-NF-κB. β-tubulin was used as loading control. (B)
Quantification of caspase-3, caspase-8 and caspase-9. (C) Activity
assay of caspase-3 and caspase-8. The data shown are representative
of three independent experiments. *P<0.05, **P<0.01. KG1a
(HS27A), co-cultured cells. |
MALDI-TOF/TOF-MS profiles of N-glycans
in KG1a cells before and after co-culture under hypoxic
conditions
N-glycans are involved in a variety of
biological processes, including cell-cell and cell-matrix
interactions, receptor-mediated functions and specific protein
functions. Many types of tumor cells display aberrant
N-glycosylation patterns, and cancer-associated
N-glycans are potential biomarkers for early detection and
diagnosis of cancer (38). We
compared MALDI-TOF/TOF-MS profiles of total N-glycans in
KG1a cells before vs. after co-culture with HS27A cells under
hypoxic conditions. MALDI-TOF/TOF-MS spectra of N-glycans
with signal-to-noise ratios >5 from total glycoproteins were
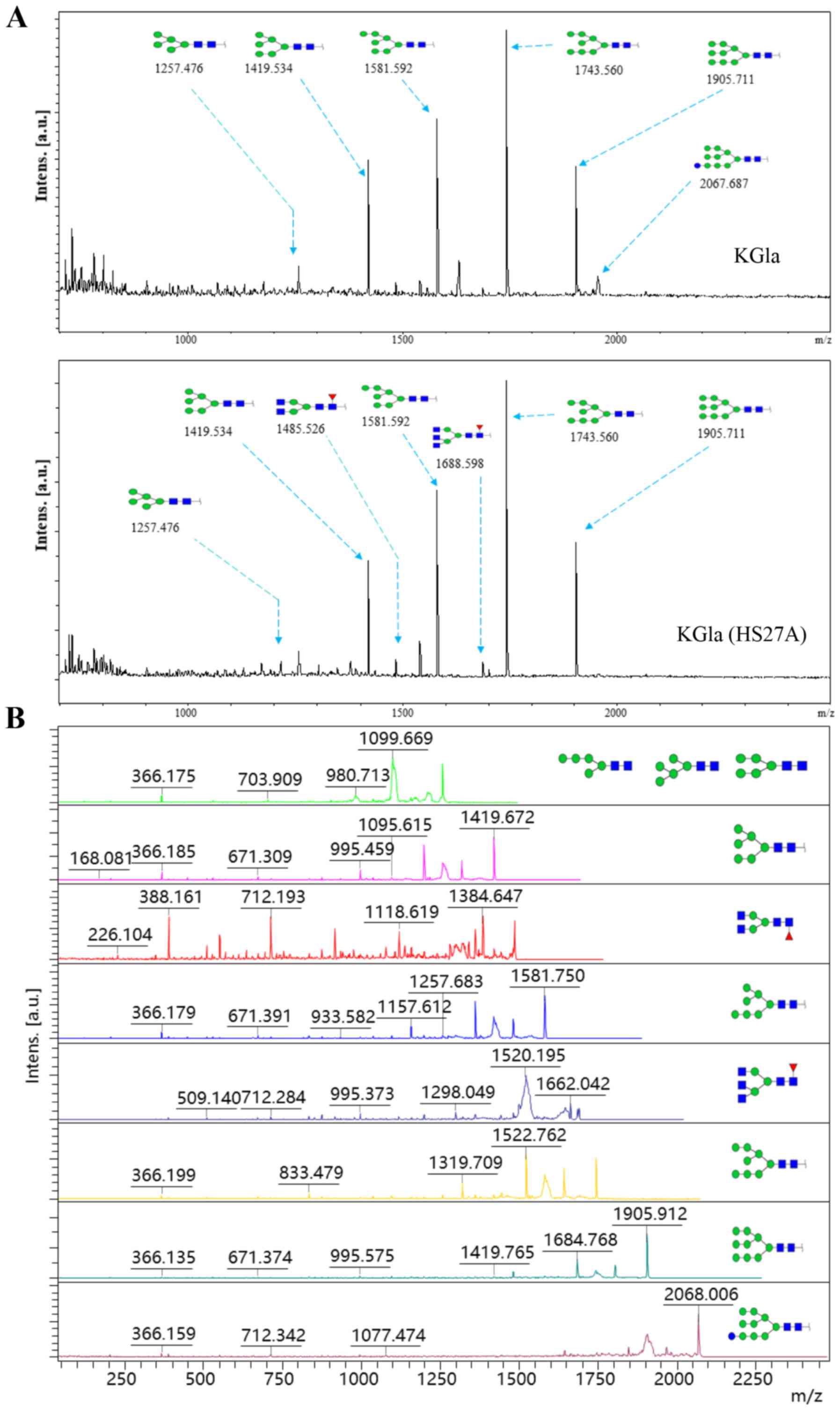
annotated using the GlycoWorkbench program (Fig. 3). Eight distinctive N-glycans
were observed in KG1a cells before vs. after co-culture, at m/z
1257.476, 1419.534, 1485.526, 1581.592, 1688.598, 1743.650,
1905.711 and 2067.687, corresponding to eight proposed
N-glycan structures including high-mannose, bi-antennary and
tri-antennary types (Fig. 4). There
were eight N-glycan structures found in both KG1a cells and
co-cultured KG1a cells, two structures unique to co-cultured KG1a
cells, and one structure unique to KG1a cells. Co-cultured KG1a
cells showed enhancement of two complex type N-glycan
structures with fucosylation.
Validation by real-time PCR of
N-glycan biosynthesis-related genes
On the basis of our MS data, we examined the
expression of N-glycan biosynthesis-related genes by
real-time PCR. The glycosyltransferase genes involved in the
synthesis of the N-glycan structures listed in Fig. 4 are α1,6-fucosyltransferase
(FUT8) and β1,4-N-acetylglucosaminyltransferases 2, 3, 4A,
4B and 5 (MGAT2, MGAT3, MGAT4A, MGAT4B and MGAT5)
(Fig. 5A). Among these six genes,
FUT8, MGAT3 and MGAT5 showed increased expression in
co-cultured KG1a cells, whereas the expression of MGAT2 and
MGAT4B was not notably altered. These findings were
consistent with our MS results. We then applied flow cytometric
(FACS) analysis with specific lectins to further examine
differentially expressed N-glycans in KG1a cells co-cultured
with HS27A cells under hypoxic conditions. Co-cultured cells showed
increased expression of N-glycan structures recognized by
PHA-E (i.e., bisecting GlcNAc structures), LCA (core
Fucα1-6GlcNAc) and WGA (multivalent Sia and (GlcNAc)n
structures), and reduced expression of structures recognized by
ConA (terminal mannose) (Fig.
5B-D).
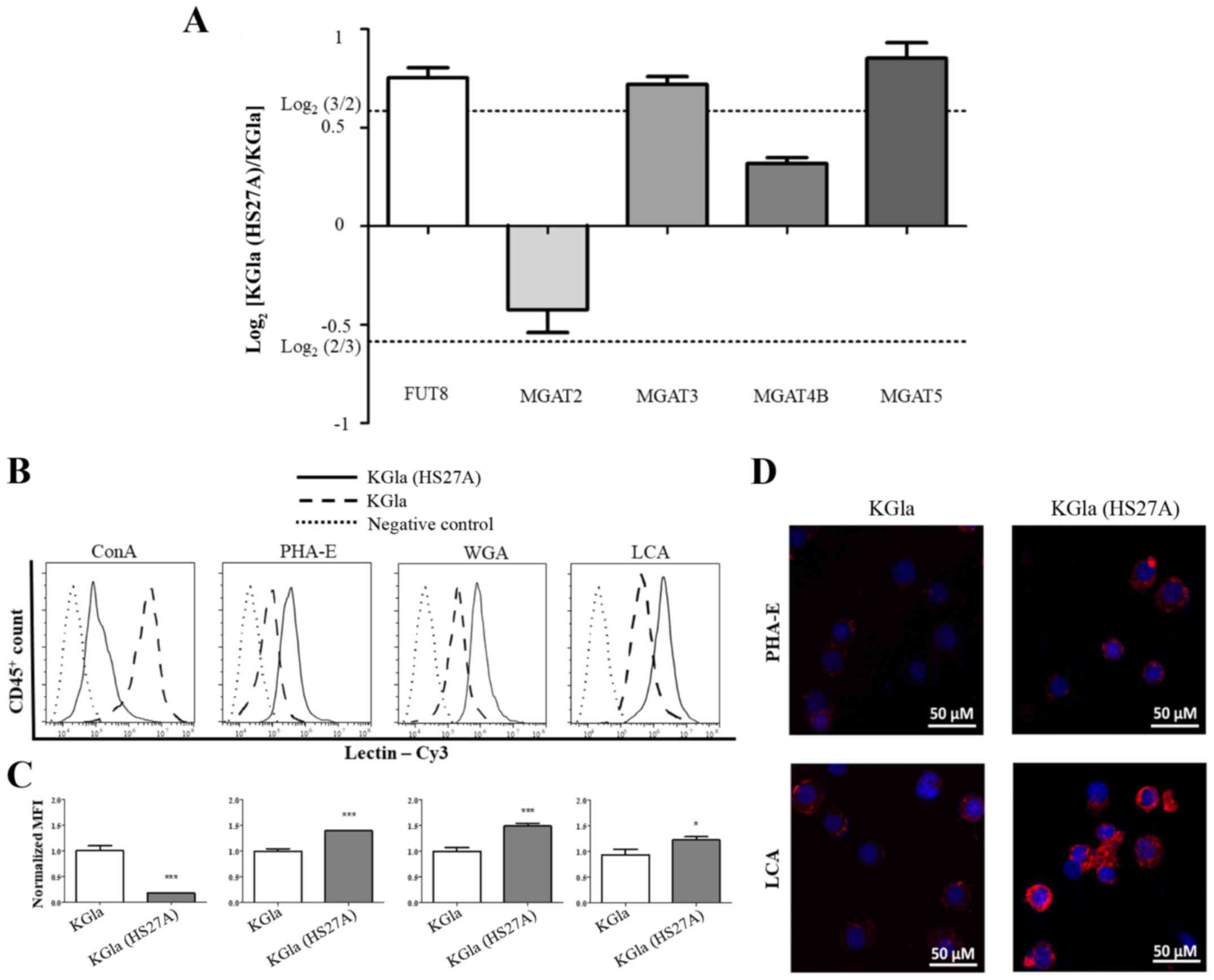 | Figure 5.Comparative glycogene expression and
differential glycopatterns of KG1a cells vs. co-cultured KG1a
cells. (A) Gene expression of FUT8, MGAT2, MGAT3, MGAT4B and
MGAT5 was analyzed by real-time PCR in triplicate
experiments. Relative expression was analyzed by the
2−ΔΔCq method and presented as Log2 relative
expression for KG1a vs. co-cultured KG1a cells, with
Log2(3/2) and Log2(2/3) as threshold values.
Values above Log2(3/2) and below Log2(2/3)
indicate significant upregulation and downregulation, respectively.
(B-D) Differential glycopatterns in KG1a and co-cultured KG1a. The
four indicated lectins, conjugated with Cy3, were used. (B) Flow
cytometric analysis. (C) Quantification of intensity. (D) Lectin
staining was performed. The data shown are representative of three
independent experiments. *P<0.05, ***P<0.001. Error bars,
SEM. KG1a (HS27A), co-cultured cells. |
Validation by flow cytometry of
N-glycan expression differences in co-cultured KG1a cells under
hypoxic conditions
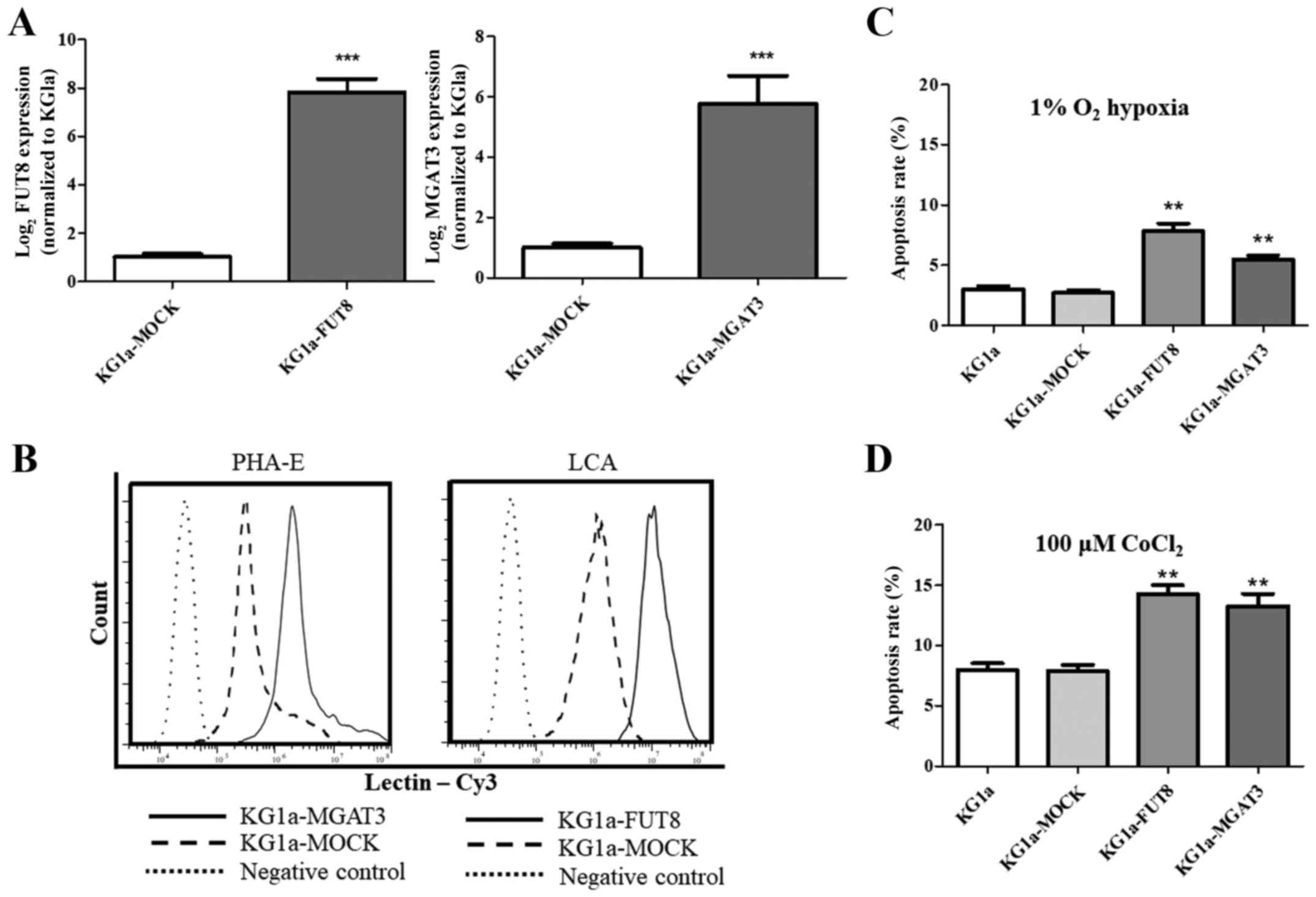
To verify the function of FUT8 and
MGAT3, we overexpressed these two genes in KG1a cells
(Fig. 6A). Flow cytometric data
revealed that the corresponding bisecting N-glycan or core
fucosylation in KG1a cells that overexpressed MGAT3 or
FUT8 was enhanced (Fig. 6B).
Our data showed an enhanced apoptosis rate in KG1a cells that
overexpressed MGAT3 or FUT8 under both hypoxic
conditions and CoCl2 treatment (Fig. 6C and D). We suggest that modulation
of glycogenes may play a role in regulating the apoptosis in KG1a
cells.
Discussion
Tumor/stromal cell interactions are frequently part
of cell social networks, play essential roles in tumor development
and growth, and provide useful therapeutic targets. Our 2010
studies were focused on an in vitro co-culture system
involving two stromal cell lines (HS5 and HS27A) derived from a
healthy bone marrow donor. Cells from patients with advanced MDS
showed increased susceptibility to TNFα-induced apoptosis following
stromal contact (8,9). Our 2013 in vivo study, based on
the in vitro observations, involved injection of HS5 or
HS27A cells, together with primary MDS marrow cells, into
NOD.Cg-Prkdcscid Il2rgtm1Wjl (NSG) mice
(12). HS27A cells, but not HS5
cells, facilitated the engraftment of clonal MDS cells in
vivo. The present study focused on the crosstalk that occurs
between hematopoietic and HS27A cells (but not HS5). Among the
numerous factors present in the tumor microenvironment, hypoxia is
a common feature of myeloid malignancies and has been shown to
affect such HSC behaviors as apoptosis, proliferation,
differentiation and resistance to chemotherapy. The role of hypoxia
and its downstream signaling in hematopoiesis is controversial,
since both promoting and inhibitory effects have been reported
(39). Velasco-Hernandez et
al found that deletion of HIF-1α in mouse hematopoietic cells
promoted acute myeloid leukemia progression (40). Other studies indicated that hypoxia
promoted leukemogenic niche metabolism and cytokine secretion
(41,42). In the present study, we compared the
apoptosis rates of hematopoietic KG1a cells alone or co-cultured
with HS27A cells under hypoxic conditions. Our data clearly
indicated that elevated caspase 8 activities were found in
co-cultured KG1a cells. However, cleaved caspase-9 had no
significant changes in co-cultured KG1a cells, even though the
total amount of caspase-9 was increased, indicating that the
co-culture may trigger the expression of total caspase-9 which
possibly make the co-cultured KG1a cells becoming more sensitive to
cell death signals (43–45). Increased levels of p53 in the
co-cultured cells revealed that the apoptotic pathway was
p53-dependent.
Functional proteins in both normal and malignant
cells are maintained by post-translational modifications (PTMs),
which include phosphorylation, ubiquitination, methylation,
N-acetylation and glycosylation (30). Glycosylation, the most common PTM,
is involved in a wide variety of biological processes.
Glycosylation of proteins (giving rise to glycoproteins) in
eukaryotic cells is classified on the basis of various linkages of
glycans to protein core regions e.g., N-linked glycan
(GlcNAc linkage to Asn), O-linked glycan (O-GalNAc linkage
to Ser/Thr), and other O-linked glycan varieties (e.g.,
O-linked mannose, O-linked GlcNAc) (30). Biological processes mediated by
glycans include signal transduction, inflammation,
virus/bacteria-host interactions, cell-cell interactions and cancer
development and progression. Our MS analyses of N-glycan
profiles of co-cultured KG1a cells under hypoxia revealed increases
in bi- and tri-antennary type, GlcNAc type and fucose type
N-glycans.
Synthesis of N-glycans is catalyzed by
various enzymes termed glycosyltransferases. Fucosylation is
catalyzed by FUT8, which transfers a fucose (Fuc) moiety from
GDP-β-L-Fuc to the innermost GlcNAc residue of an N-glycan,
resulting in α1,6-Fuc residue as core Fuc. FUT8 activity is
upregulated during epithelial-mesenchymal transition (EMT) through
transactivation of β-catenin/lymphoid enhancer-binding factor-1
(LEF-1) (46). The
glycosyltransferase MGAT3 catalyzes transfer of GlcNAc to the core
β-mannose residue of N-glycans having β1,4-linkage. Addition
of bisecting GlcNAc residue to core β-mannose by MGAT3 inhibits
activities of mannosidase II and other GlcNAc transferases (MGAT2,
MGAT4). Li et al and Xu et al reported reduced levels
of MGAT3 and its product (bisecting N-glycans) in a
TGF-β1-induced EMT model (16,47).
Takahashi et al observed that MGAT3 overexpression reduced
the amount of poly-N-acetyllactosamine on N-glycans of EGFR,
indicating that specific terminal structures of N-glycans
regulated EGFR endocytosis through interaction with carbohydrate
recognition molecules (48). MGAT3
exerts a stabilizing effect on E-cadherin at the cell membrane by
inducing a delay in the turnover rate of the protein, thereby
contributing to stable and functional adhesion, and blocking
clathrin-dependent E-cadherin endocytosis (49). Remodeling of glycans by MGAT3
contributes to the retention of E-cadherin in membrane by
mesalamine (5-ASA) (50). In
contrast, MGAT5 promotes destabilization of E-cadherin, leading to
its mislocalization, formation of unstable junctions and impairment
of cell-cell adhesion (49).
Inhibition of MGAT5 activity reduced liver fibrosis and suppressed
TGF-β1-induced EMT in hepatocytes, as evidenced by reversal of EMT
markers. Our model (co-cultured KG1a under hypoxia) showed
increased expression of FUT8, MGAT3 and MGAT5, but no
notable change of MGAT2 or MGAT4B. Increased binding
affinity of lectins PHA-E and WGA in our model revealed elevated
levels of bisecting-GlcNAc and (GlcNAc)n structures, in
striking contrast to N-glycan structures in co-cultured KG1a
under normoxia (data not shown). We then overexpressed FUT8
and MGAT3 genes in KG1a cells to ascertain whether modified
glycogenes could impact the cell apoptosis of KG1a cells under
hypoxia. Our data suggested that modified glycogenes of both
FUT8 and MGAT3 could enhance cell apoptosis in KG1a
cells. Our further studies will focus on the functional role of
FUT8 and MGAT3 in primary cells, as well as the
microenvironmental niche.
In conclusion, the present study provided the first
demonstration of hypoxia-induced apoptosis in KG1a hematopoietic
cells following contact with HS27A stromal cells, and
N-glycan profiles of KG1a cells before and after stromal
contact under hypoxia. Our findings are useful references for
future studies based on co-culture models, and may help elucidate
the roles of hematopoietic and stromal cells in bone-related
diseases. Our ongoing studies will focus on the functions of
N-glycans in co-culture systems and the clinical relevance
of these N-glycans.
Acknowledgements
The authors are grateful to Dr S. Anderson for the
English language editing of the manuscript.
Funding
The present study was supported in part by grants
from the National Natural Science Foundation of China (nos.
81470294, 31400691 and 81770123), the Natural Science Foundation of
Jiangsu Province (BK20140169), the Fundamental Research Funds for
the Central Universities (JUSRP51619B) and the 13115 Key Projects
of Scientific and Technical Innovation of Shaan'xi Province
(2010ZDKG-53).
Availability of data and materials
The datasets used during the present study are
available from the corresponding author upon reasonable
request.
Authors' contributions
XL and YW designed the study and wrote the
manuscript. XP, SZ, XL, ZT and JG performed the experiments. FG
critiqued the manuscript and was also involved in the conception
and design of the study. All authors have read and approved the
final manuscript. All authors agreed to be accountable for all
aspects of the work in ensuring that questions related to the
accuracy or integrity of any part of the work are appropriately
investigated and resolved.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
HSCs
|
hematopoietic stem cells
|
|
HPCs
|
hematopoietic progenitor cells
|
|
MDS
|
myelodysplastic syndrome
|
|
HIF-1α
|
hypoxia-inducible factor 1α
|
|
TNFα
|
tumor necrosis factor α
|
|
PTMs
|
post-translational modifications
|
|
EMT
|
epithelial-mesenchymal transition
|
|
LEF-1
|
lymphoid enhancer-binding factor-1
|
References
|
1
|
Trentin JJ: Determination of bone marrow
stem cell differentiation by stromal hemopoietic inductive
microenvironments (HIM). Am J Pathol. 65:621–628. 1971.PubMed/NCBI
|
|
2
|
Bennett M and Kumar V: 89Sr-induced bone
marrow aplasia: Effects on seed (stem cells) and soil (inductive
microenvironment). Lab Invest. 49:235–236. 1983.PubMed/NCBI
|
|
3
|
Morrison SJ and Scadden DT: The bone
marrow niche for haematopoietic stem cells. Nature. 505:327–334.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Simmons PJ and Torok-Storb B:
Identification of stromal cell precursors in human bone marrow by a
novel monoclonal antibody, STRO-1. Blood. 78:55–62. 1991.PubMed/NCBI
|
|
5
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Manabe A, Coustan-Smith E, Behm FG,
Raimondi SC and Campana D: Bone marrow-derived stromal cells
prevent apoptotic cell death in B-lineage acute lymphoblastic
leukemia. Blood. 79:2370–2377. 1992.PubMed/NCBI
|
|
7
|
Ayala F, Dewar R, Kieran M and Kalluri R:
Contribution of bone microenvironment to leukemogenesis and
leukemia progression. Leukemia. 23:2233–2241. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Marcondes AM, Li X, Gooley TA, Milless B
and Deeg HJ: Identification of DJ-1/PARK-7 as a determinant of
stroma-dependent and TNF-alpha-induced apoptosis in MDS using mass
spectrometry and phosphopeptide analysis. Blood. 115:1993–2002.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Li X, Marcondes AM, Gooley TA and Deeg HJ:
The helix-loop-helix transcription factor TWIST is dysregulated in
myelodysplastic syndromes. Blood. 116:2304–2314. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Marcondes AM, Li X, Tabellini L,
Bartenstein M, Kabacka J, Sale GE, Hansen JA, Dinarello CA and Deeg
HJ: Inhibition of IL-32 activation by α-1 antitrypsin suppresses
alloreactivity and increases survival in an allogeneic murine
marrow transplantation model. Blood. 118:5031–5039. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Li X, Xu F, Chang C, Byon J,
Papayannopoulou T, Deeg HJ and Marcondes AM: Transcriptional
regulation of miR-10a/b by TWIST-1 in myelodysplastic syndromes.
Haematologica. 98:414–419. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Li X, Marcondes AM, Ragoczy T, Telling A
and Deeg HJ: Effect of intravenous coadministration of human stroma
cell lines on engraftment of long-term repopulating clonal
myelodysplastic syndrome cells in immunodeficient mice. Blood
Cancer J. 3:e1132013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Li X and Deeg HJ: Murine xenogeneic models
of myelodysplastic syndrome: An essential role for stroma cells.
Exp Hematol. 42:4–10. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kerbauy DM, Lesnikov V, Torok-Storb B,
Bryant E and Deeg HJ: Engraftment of distinct clonal MDS-derived
hematopoietic precursors in NOD/SCID-beta2-microglobulin-deficient
mice after intramedullary transplantation of hematopoietic and
stromal cells. Blood. 104:2202–2203. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Li M, Song L and Qin X: Glycan changes:
Cancer metastasis and anti-cancer vaccines. J Biosci. 35:665–673.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Li X, Wang X, Tan Z, Chen S and Guan F:
Role of glycans in cancer cells undergoing epithelial-mesenchymal
transition. Front Oncol. 6:332016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Dube DH and Bertozzi CR: Glycans in cancer
and inflammation-potential for therapeutics and diagnostics. Nat
Rev Drug Discov. 4:477–488. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Groux-Degroote S, Guérardel Y and Delannoy
P: Gangliosides: Structures, biosynthesis, analysis, and roles in
cancer. Chembiochem. 18:1146–1154. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Timoshenko AV, Lanteigne J and Kozak K:
Extracellular stress stimuli alter galectin expression profiles and
adhesion characteristics of HL-60 cells. Mol Cell Biochem.
413:137–143. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Le Marer N: GALECTIN-3 expression in
differentiating human myeloid cells. Cell Biol Int. 24:245–251.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wan SY, Zhang TF and Ding Y: Galectin-3
enhances proliferation and angiogenesis of endothelial cells
differentiated from bone marrow mesenchymal stem cells. Transplant
Proc. 43:3933–3938. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Mohyeldin A, Garzón-Muvdi T and
Quiñones-Hinojosa A: Oxygen in stem cell biology: A critical
component of the stem cell niche. Cell Stem Cell. 7:150–161. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wenger RH: Cellular adaptation to hypoxia:
O2-sensing protein hydroxylases, hypoxia-inducible transcription
factors, and O2-regulated gene expression. FASEB J. 16:1151–1162.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Semenza GL: Intratumoral hypoxia,
radiation resistance, and HIF-1. Cancer Cell. 5:405–406. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Semenza GL: Targeting HIF-1 for cancer
therapy. Nat Rev Cancer. 3:721–732. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Danet GH, Pan Y, Luongo JL, Bonnet DA and
Simon MC: Expansion of human SCID-repopulating cells under hypoxic
conditions. J Clin Invest. 112:126–135. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Li X, Wan T, Zhang S, Li D and Han X:
Quantitative proteomic analysis and comparison of two bone marrow
stromal cell lines using the SILAC method. Exp Hematol.
44:1059–1071. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Li X, Li D, Pang X, Yang G, Deeg HJ and
Guan F: Quantitative analysis of glycans, related genes, and
proteins in two human bone marrow stromal cell lines using an
integrated strategy. Exp Hematol. 43:760–769.e7. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2−ΔΔCT method. Methods. 25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Tan Z, Lu W, Li X, Yang G, Guo J, Yu H, Li
Z and Guan F: Altered N-Glycan expression profile in
epithelial-to-mesenchymal transition of NMuMG cells revealed by an
integrated strategy using mass spectrometry and glycogene and
lectin microarray analysis. J Proteome Res. 13:2783–2795. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Guan F, Tan Z, Li X, Pang X, Zhu Y, Li D
and Yang G: A lectin-based isolation/enrichment strategy for
improved coverage of N-glycan analysis. Carbohydr Res. 416:7–13.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Ying SX, Seal S, Abbassi N, Hockenbery DM,
Kiem HP, Li X, Pagel JM, Gopal AK and Deeg HJ: Differential effects
of bexarotene on intrinsic and extrinsic pathways in TRAIL-induced
apoptosis in two myeloid leukemia cell lines. Leuk Lymphoma.
48:1003–1014. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
de Thonel A, Bettaïeb A, Jean C, Laurent G
and Quillet-Mary A: Role of protein kinase C zeta isoform in Fas
resistance of immature myeloid KG1a leukemic cells. Blood.
98:3770–3777. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Lopez-Sánchez LM, Jimenez C, Valverde A,
Hernandez V, Peñarando J, Martinez A, Lopez-Pedrera C,
Muñoz-Castañeda JR, De la Haba-Rodríguez JR, Aranda E and
Rodriguez-Ariza A: CoCl2, a mimic of hypoxia, induces formation of
polyploid giant cells with stem characteristics in colon cancer.
PLoS One. 9:e991432014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Guan F, Schaffer L, Handa K and Hakomori
SI: Functional role of gangliotetraosylceramide in
epithelial-to-mesenchymal transition process induced by hypoxia and
by TGF-{beta}. FASEB J. 24:4889–4903. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Ohtsuka T, Ryu H, Minamishima YA, Macip S,
Sagara J, Nakayama KI, Aaronson SA and Lee SW: ASC is a Bax adaptor
and regulates the p53-Bax mitochondrial apoptosis pathway. Nat Cell
Biol. 6:121–128. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Marchenko ND and Moll UM: Mitochondrial
death functions of p53. Mol Cell Oncol. 1:e9559952014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Etxebarria J and Reichardt NC: Methods for
the absolute quantification of N-glycan biomarkers. Biochim Biophys
Acta. 1860:1676–1687. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Korn C and Méndez-Ferrer S: Myeloid
malignancies and the microenvironment. Blood. 129:811–822. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Velasco-Hernandez T, Hyrenius-Wittsten A,
Rehn M, Bryder D and Cammenga J: HIF-1α can act as a tumor
suppressor gene in murine acute myeloid leukemia. Blood.
124:3597–3607. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Drolle H, Wagner M, Vasold J, Kütt A,
Deniffel C, Sotlar K, Sironi S, Herold T, Rieger C and Fiegl M:
Hypoxia regulates proliferation of acute myeloid leukemia and
sensitivity against chemotherapy. Leuk Res. 39:779–785. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Kuschel A, Simon P and Tug S: Functional
regulation of HIF-1α under normoxia-is there more than
post-translational regulation? J Cell Physiol. 227:514–524. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Liedtke C, Gröger N, Manns MP and
Trautwein C: Interferon-alpha enhances TRAIL-mediated apoptosis by
up-regulating caspase-8 transcription in human hepatoma cells. J
Hepatol. 44:342–349. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Ruiz-Ruiz C, Ruiz de Almodóvar C,
Rodríguez A, Ortiz-Ferrón G, Redondo JM and López-Rivas A: The
up-regulation of human caspase-8 by interferon-gamma in breast
tumor cells requires the induction and action of the transcription
factor interferon regulatory factor-1. J Biol Chem.
279:19712–19720. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Gomyo Y, Sasaki J, Branch C, Roth JA and
Mukhopadhyay T: 5-aza-2′-deoxycytidine upregulates caspase-9
expression cooperating with p53-induced apoptosis in human lung
cancer cells. Oncogene. 23:6779–6787. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Chen CY, Jan YH, Juan YH, Yang CJ, Huang
MS, Yu CJ, Yang PC, Hsiao M, Hsu TL and Wong CH: Fucosyltransferase
8 as a functional regulator of nonsmall cell lung cancer. Proc Natl
Acad Sci USA. 110:630–635. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Xu Q, Isaji T, Lu Y, Gu W, Kondo M, Fukuda
T, Du Y and Gu J: Roles of N-acetylglucosaminyltransferase III in
epithelial-to-mesenchymal transition induced by transforming growth
factor β1 (TGF- β1) in epithelial cell lines. J Biol Chem.
287:16563–16574. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Takahashi M, Kizuka Y, Ohtsubo K, Gu J and
Taniguchi N: Disease-associated glycans on cell surface proteins.
Mol Aspects Med. 51:56–70. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Pinho SS, Figueiredo J, Cabral J, Carvalho
S, Dourado J, Magalhães A, Gärtner F, Mendonfa AM, Isaji T, Gu J,
et al: E-cadherin and adherens-junctions stability in gastric
carcinoma: Functional implications of glycosyltransferases
involving N-glycan branching biosynthesis,
N-acetylglucosaminyltransferases III and V. Biochim Biophys Acta.
1830:2690–2700. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Khare V, Lang M, Dammann K, Campregher C,
Lyakhovich A and Gasche C: Modulation of N-glycosylation by
mesalamine facilitates membranous E-cadherin expression in colon
epithelial cells. Biochem Pharmacol. 87:312–320. 2014. View Article : Google Scholar : PubMed/NCBI
|