Introduction
Thyroid cancer (TC) is the most common endocrine
malignancy, accounting for approximately 3% of all malignant tumors
(1,2). Moreover, it has a higher incidence in
women, and is the most common cancer of the head and neck (3,4). In
recent years, the incidence of TC has significantly increased. A
better understanding of the underlying mechanism of TC would
provide novel insights for the treatment of TC.
ETS-domain containing protein (Elk1), a
transcription factor belonging to the ETS oncogene family,
regulates the oncogene c-fos by phosphorylation through activation
of the PKC/ERK pathways (5–8). Studies have reported that Elk1 has
roles in cell proliferation, the cell cycle, apoptosis and
tumorigenesis (9,10). It has been demonstrated that Elk1
expression is upregulated, and promotes cell viability in bladder
cancer (10). Elk1 has also been
shown to be induced, and to play a crucial role in
hormone-resistant or metastatic prostate cancers (11) and is reported to play an important
role in breast cancer and ovarian cancer (7,12–14).
However, the molecular mechanism of Elk1 in TC remains unknown. In
our study, we investigated the role of Elk1 in cell proliferation
and apoptosis in TC. A previous study has shown that in SH-SY5Y
neuroblastomas Elk1 represses the expression of Egr-1, which is
implicated in different cellular processes containing cell
proliferation, differentiation and apoptosis (15).
Early growth response-1 (Egr-1), also called Zif268,
NGF1-A, and Krox24, is a transcription factor containing a
zinc-finger DNA binding domain, and is known as an important
immediate-early gene (IEG) (16–21).
Egr-1 promotes quiescent cells to enter the proliferative phase,
regulating cell growth and differentiation (20,22,23).
Egr-1 is found in eukaryotic genomes, and is highly conserved
evolutionarily (24). Many factors
can activate Egr-1, and activated Egr-1 regulates target gene
transcription by interacting with the binding sites of target
genes. The biological function of Egr-1 is realized by upregulating
or downregulating target gene expression. Egr-1 is considered to be
a class II tumor suppressor gene (25). It has been demonstrated that the
expression of Egr-1 is decreased in breast cancer, non-small cell
lung cancer (NSCLC) and glioma (26,27).
Nevertheless, other studies have demonstrated that Egr-1 expression
is increased in prostate cancer, lymphoma and Wilms' tumor, among
others (28,29). It has been reported that Egr-1 could
directly regulate the transcription of the phosphatase and tensin
homolog deleted on chromosome ten (PTEN) (30).
PTEN is also recognized as a tumor suppressor
(31,32), and it was observed that PTEN is
inactivated or inhibited in multiple types of cancer including
thyroid carcinoma (33). Research
over the past few year has shown the mechanism by which loss of
PTEN function contributes to tumor development (34). It has been reported that PTEN
inhibition induces cell survival and cisplatin resistance in human
ovarian cancer, and promotes the risk of breast and endometrial
cancers and leukemia (35,36). Furthermore, it has been demonstrated
that PTEN suppression causes thyroid cancer development,
progression and invasion, authenticating PTEN as a crucial tumor
suppressor in thyroid carcinogenesis (37). Moreover, transient ectopic
expression of PTEN promotes cell cycle arrest and cell death in
thyroid cancer cell lines (38).
In the present study, we investigated the molecular
mechanism underlying Elk1 action in thyroid cancer progression
in vitro. We found that Elk1 expression was upregulated in
thyroid cancer cell lines and tissues. Loss of Elk1 function
significantly inhibited the proliferation and induced apoptosis in
thyroid cancer cell lines. Furthermore, the results also
demonstrated that Elk1 inhibition induced PTEN expression by
upregulating Egr-1. Therefore, this study proposes the potential
role of Elk1 in preventing thyroid cancer, providing a potential
novel target for treatment.
Materials and methods
Cell lines and tissues
Human thyroid cancer cell lines FTC-133 and TPC-1
were purchased from the Protection Agency Culture Collections
(HPACC, Salisbury, Wiltshire, UK). The cells were cultured in
Dulbecco's modified Eagle's medium (DMEM, Sigma-Aldrich, Merck
KGaA, Darmstadt, Germany) containing 10% fetal bovine serum (FBS,
HyClone; GE Healthcare Life Sciences, Logan, UT, USA). The normal
thyroid cell line FRTL-5 was obtained and cultured according to a
previously described protocol (39). The cell lines were incubated at 37°C
with a 5% CO2 atmosphere. Additionally, 10 pairs (sample
collection: From February 2017 to October 2017) of tumor tissue
samples and matched adjacent normal tissues were obtained from
Xinxiang Central Hospital along with written informed consent of
patients (4 males and 6 females; 39–50 years old), and were
immediately stored in liquefied ammonia. The study was approved by
the Xinxiang Central Hospital Ethics Board.
Cell transfection
The cells (FTC-133 and TPC-1) were separately seeded
in 12-well plates and incubated in a humid atmosphere with 5%
CO2 at 37°C until 80% fusion was achieved. The
transfection procedure was performed according to the
manufacturer's instructions. Elk1 siRNA
(5′-AACCACCCGCCACTCTTCCT-3′), Egr-1 siRNA
(5′-GTAGGTTGCTGTCGTCAGGGTAAAT-3′), and non-specific siRNA were
separately diluted in FBS-free DMEM medium (200 µl) with 6 µl
TurboFect (Thermo Fisher Scientific, Inc., Waltham, MA, USA), and
the mixtures were added in the well. The cells were then cultured
under conditions of 5% CO2 at 37°C for 24 h.
Cell growth and viability
Cell growth and viability were measured using the
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT)
assay, and the assay was performed in accordance with standard
protocols. The cells (1×05 cells/well) were cultured in
96-well plates with DMEM containing 10% FBS. The medium in each
well was then replaced by 18 µl MTT (5 g/l) diluted in
phosphate-buffered saline (PBS) followed by incubation at 37°C for
5 h. Subsequently, a total of 150 µl dimethyl sulfoxide was added
per well, in order to dissolve the crystals. Finally, the result
was read using a microplate reader (Thermo Fisher Scientific) at
490 nm. The analysis was repeated three times.
Bromodeoxyuridine (BrdU) assay
The BrdU cell proliferation assay kit (Cell
Signaling Technology, Danvers, MA, USA) was used to detect cell
proliferation based on the manufacturer's protocol. Briefly, the
cells were plated in 96-well plates and incubated with BrdU
solution (10 µl per well) for 1.5 h. A total of 150 µl denaturing
solution was added per well to replace the medium followed by
culturing for 30 min, and cells were incubated with anti-BrdU
conjugated with peroxidase. After addition of the substrate and
incubation for 20 min, the optical density at 450 nm was determined
at room temperature using a SpectroFluor Plus multiwell plate
reader (Tecan, Research Triangle Park, NC, USA). The experiment was
repeated three times.
Caspase-3 activity detection
The caspase-3 activity assay was performed as per
the manufacturer's instructions using the caspase-3 activity assay
kit (Beyotime Institute of Biotechnology, Nantong, China). Briefly,
cells were lysed on ice for 15 min, and 10-µl cell lysate per
sample in 90 µl reaction buffer [1% NP-40, 20 mM Tris-HCl (pH 7.5),
137 mM Nad and 10% glycerol] containing 12 µl caspase-3 substrate
(Ac-DEVD-pNA) (2 mM) were added into 96-well microtitre plates.
Lysates were incubated at 37°C for 2 h. The results were measured
with an ELISA reader (Tecan) at an absorbance of 405 nm.
Annexin V fluorescein isothiocyanate
conjugate and propidium iodide (Annexin V FITC/PI)
Apoptosis was measured using the BD Pharmingen™
Annexin V FITC/PI apoptosis detection kit (BD Biosciences, Franklin
Lakes, NJ, USA) following the standard protocol. In brief, cells
were precooled in cold PBS and suspended in binding buffer. Then,
Annexin V FITC solution (10 µl) was added followed by incubation
for 23 min. Afterwards, 10 µl PI was added and the reaction was
allowed to proceed for 7 min. Cellular apoptosis was measured using
a FACS analyzer (Thermo Fisher Scientific, Inc.).
Real-time quantitative polymerase
chain reaction (RT-qPCR)
Total RNA was extracted from cells using TRIzol
(Thermo Fisher Scientific, Inc.). Total RNA extraction from tissues
was performed according to a previously published method (40). RNA (5 µg) was then synthesized into
cDNA using the Revert Aid First Strand cDNA Synthesis kit (Thermo
Fisher Scientific, Inc.) according to the manufacturer's
instructions. RT-qPCR was performed in 20 µl reaction volumes
containing 10 µl Applied Biosystems® SYBR®
Green PCR Master Mix (Thermo Fisher Scientific, Inc.). The genes
were normalized to GAPDH. The primers used were as follows: Elk1
sense primer, 5′-CCTTGCGGTACTACTATGAC-3′ and antisense primer,
5′-GGCTGCGGCTGCAGAGACTGG-3′; Egr-1 sense primer,
5′-TTTGCCAGGAGCGATGAAC-3′ and antisense primer,
5′-CCGAAGAGGCCACAACACTT-3′; GAPDH sense primer,
5′-CGTCTTCACCACCATGGAGA-3′ and antisense primer,
5′-CGGCCATCACGCCACAGTTT-3′. The protocol: 94°C for 30 sec; 35
cycles of 95°C for 30 sec, 58°C (Elk1) or 60°C (Egr-1) for 30 sec
and 72°C for 30 sec; 72°C for 10 min. The relative gene expression
levels were estimated using the 2−ΔΔCt method.
Immunoblotting analysis
Proteins were extracted from the cells treated with
the lysate (Beyotime Institute of Biotechnology), and quantified
using the BCA kit (Beyotime). A total of 25 µg of protein was
separated using 12% sodium dodecyl sulfate polyacrylamide gel
electrophoresis (SDS-PAGE), and transferred to a nitrocellulose
membrane using a semi-dry blotting apparatus (Bio-Rad Laboratories,
Inc., Hercules, CA, USA). The membranes containing the proteins
were incubated in Tris-buffered saline (TBS) containing 2% non-fat
dry milk at room temperature for 2 h followed by washing with TBS.
The nitrocellulose membranes were then incubated overnight at 4°C
with the primary antibodies against Elk1 (1:500; cat. no.
ab131465), Egr1 (1:500; cat. no. ab182624), PTEN (1:800; cat. no.
ab31392) and GAPDH (1:1,000; cat. no. ab37168; all from Abcam Inc.,
Cambridge, MA, USA), and then incubated with a horseradish
peroxidase conjugated secondary antibody (1:1,000; cat. no.
ab205718; Abcam) for 1 h at room temperature. Finally, proteins
were visualized using Pierce enhanced chemiluminescence (Thermo
Fisher Scientific, Inc.) in a Bio-Rad ChemiDoc apparatus.
Statistical analysis
Data are expressed as mean ± standard deviation
(SD). Statistical significance was determined by Student's t-test
for two groups or by one-way ANOVA for multiple groups. A P-value
of <0.05 was considered statistically significant.
Results
Induction of Elk1 in thyroid
cancer
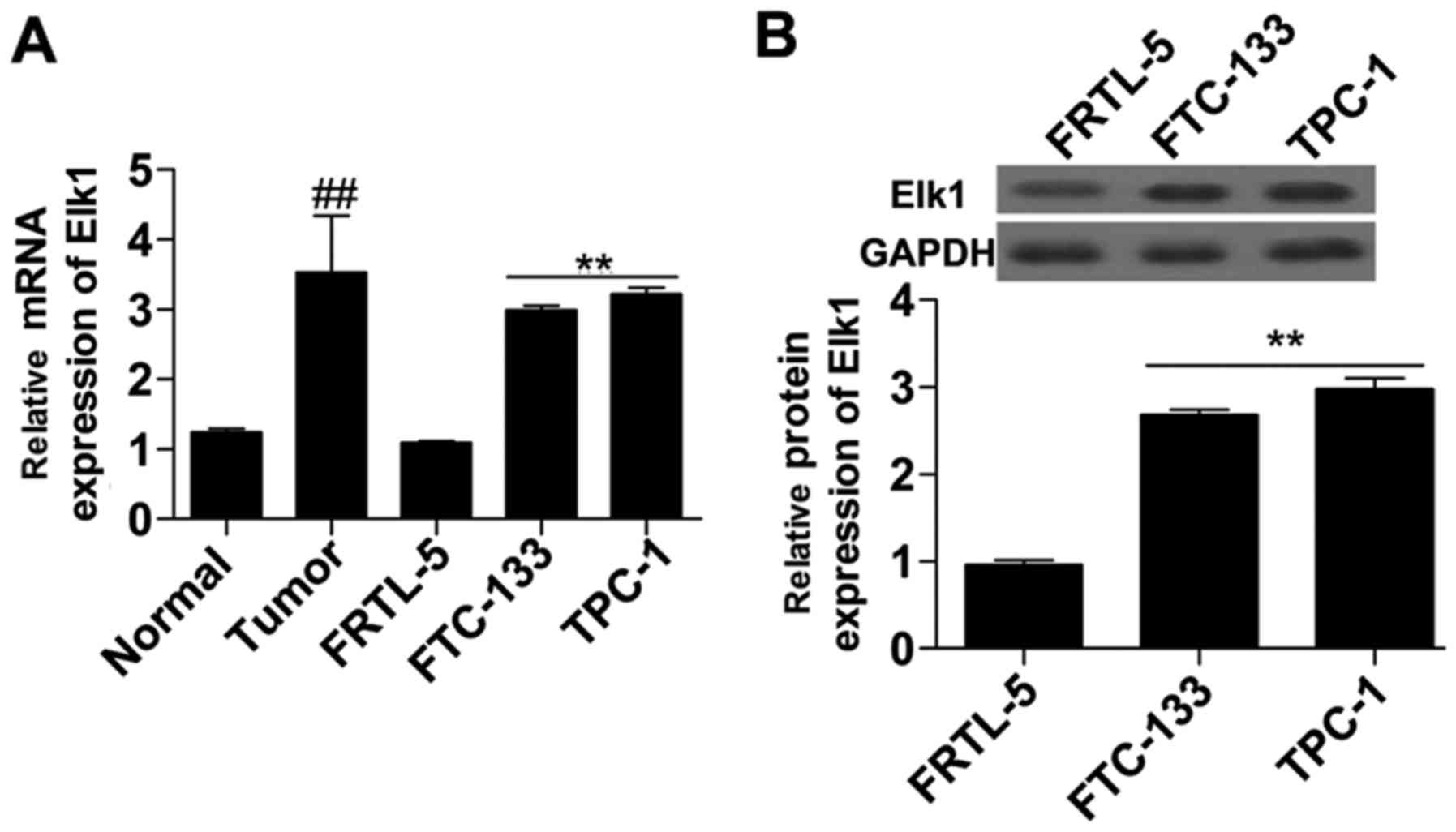
To investigate the expression of Elk1 in thyroid
cancer, we detected the expression of Elk1 in thyroid cancer
tissues and thyroid cancer cells. The results showed that the mRNA
(Fig. 1A) expression level of Elk1
in the thyroid cancer tissue was significantly higher than that in
the normal tissue. Additionally, mRNA (Fig. 1A) and protein (Fig. 1B) were both significantly increased
in the thyroid cancer cells (FTC-133 and TPC-1) compared with that
in the normal thyroid cells (FRTL-5). Thus, Elk1 expression was
upregulated in thyroid cancer.
Suppression of Elk1 induces the
expression of Egr-1 and PTEN
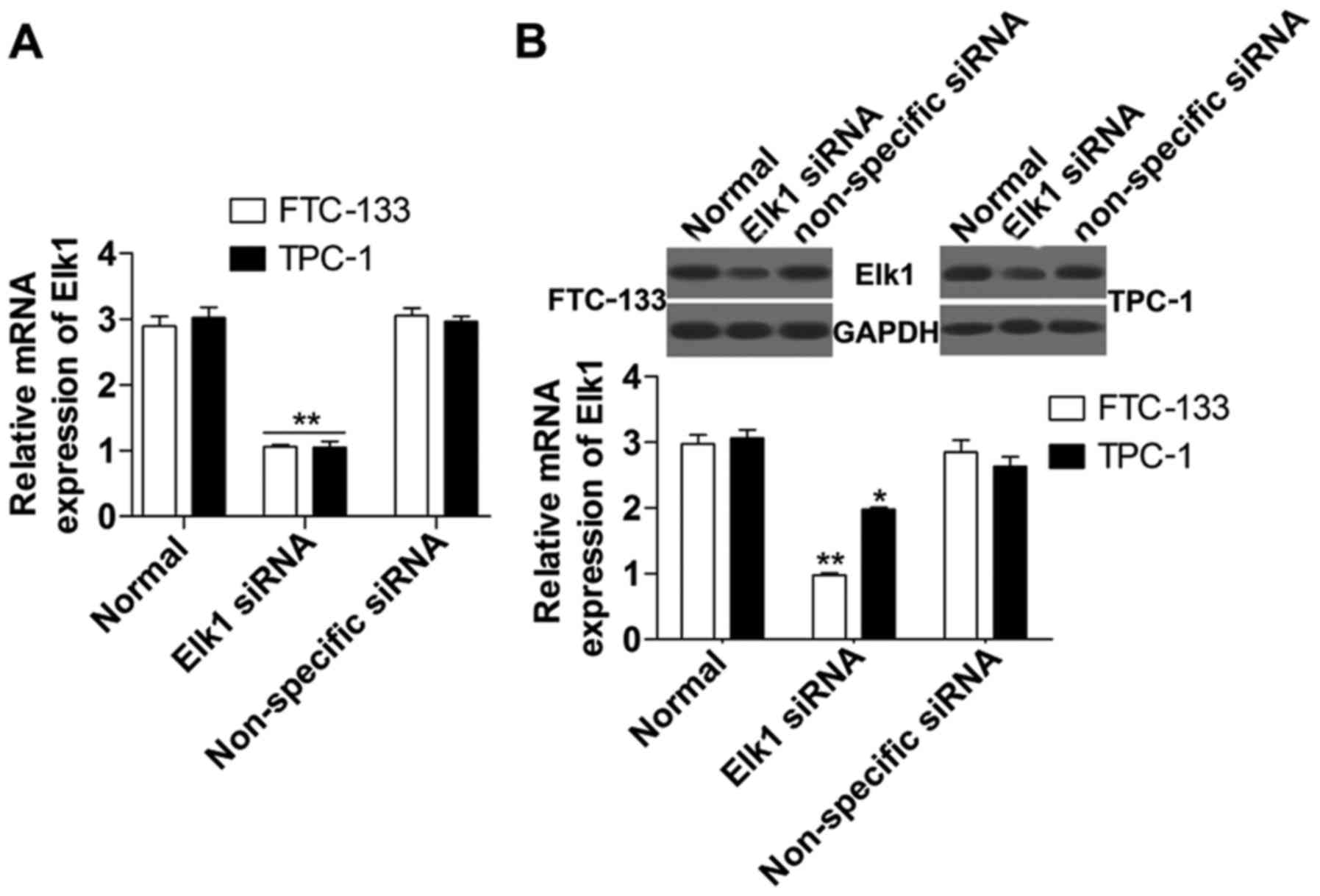
To detect the role of Elk1 in the regulation of
Egr-1 and PTEN, we inhibited Elk1 expression using cell
transfection in FTC-133 and TPC-1 cells. The results indicated that
the loss-of-function experiment was successful with a significant
reduction of Elk1 mRNA (Fig. 2A)
and protein (Fig. 2B) expression in
the Elk1 siRNA group compared with the non-specific siRNA group.
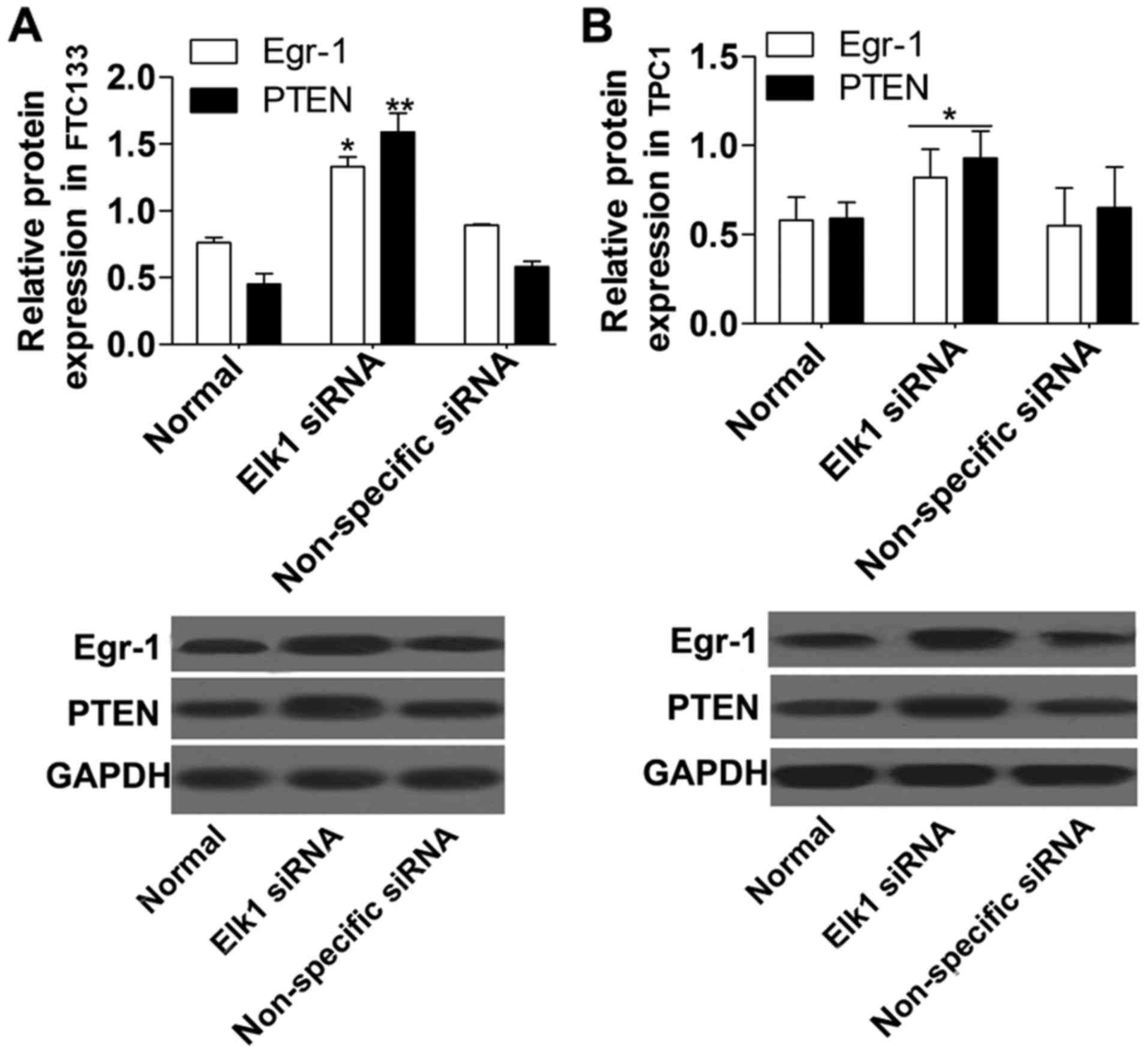
Furthermore, the protein (Fig. 3)
expression levels of Egr-1 and PTEN were obviously increased in the
Elk1 siRNA group compared with the non-specific siRNA group.
Suppression of Elk1 promotes thyroid
cancer cell apoptosis
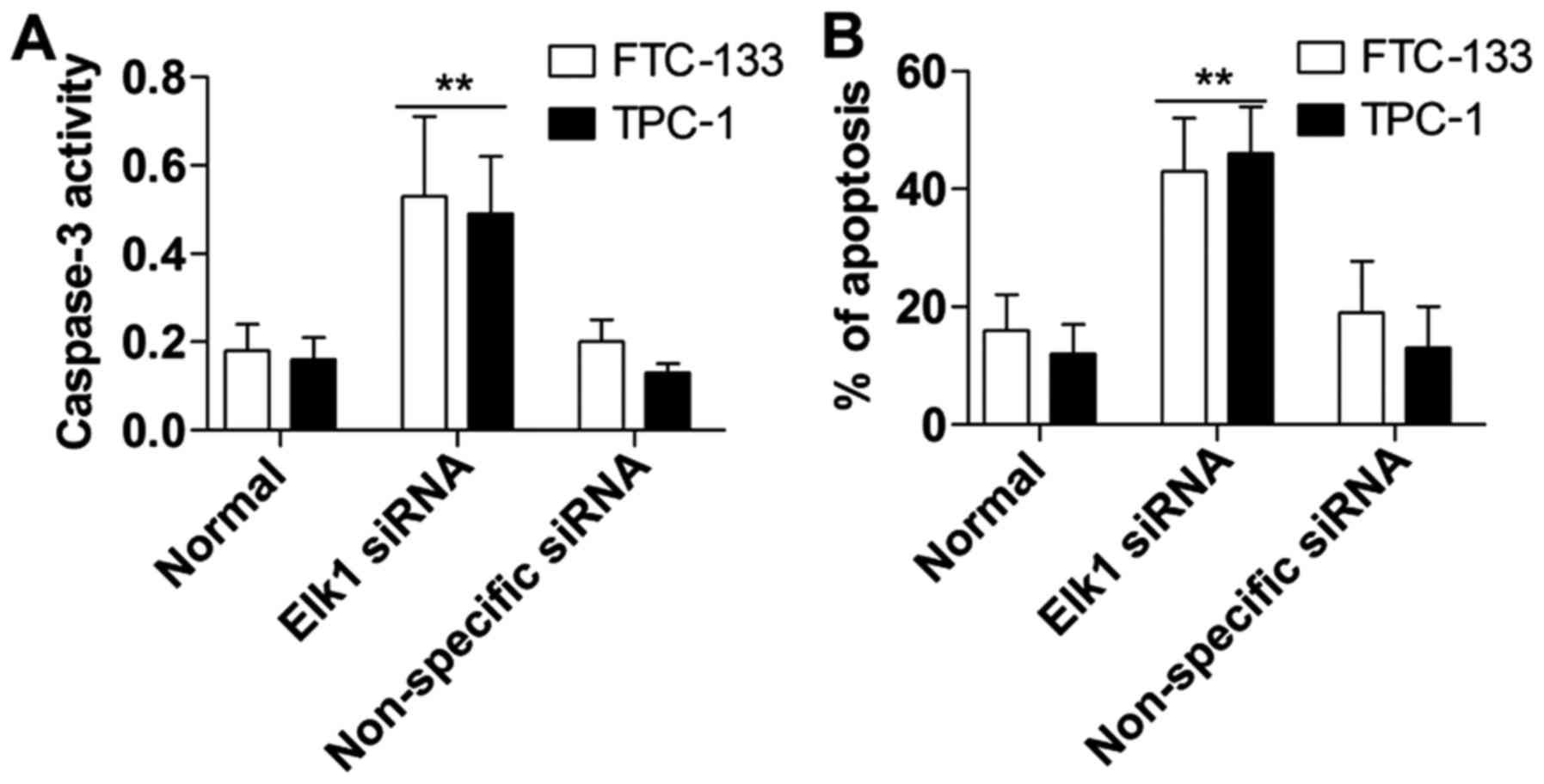
Annexin V FITC/PI and caspase-3 activity detection
were used to assess the role of Elk1 in the downregulation of
thyroid cancer cell apoptosis. The results demonstrated that the
caspase-3 activity in the Elk1 siRNA group was also obviously
induced compared with that noted in the non-specific siRNA group
(Fig. 4A). Furthermore, cellular
apoptosis in the Elk1 siRNA group was markedly increased compared
with the non-specific siRNA group as determined using the Annexin V
FITC/PI assay (Fig. 4B).
Suppression of Elk1 constrains thyroid
cancer cell proliferation
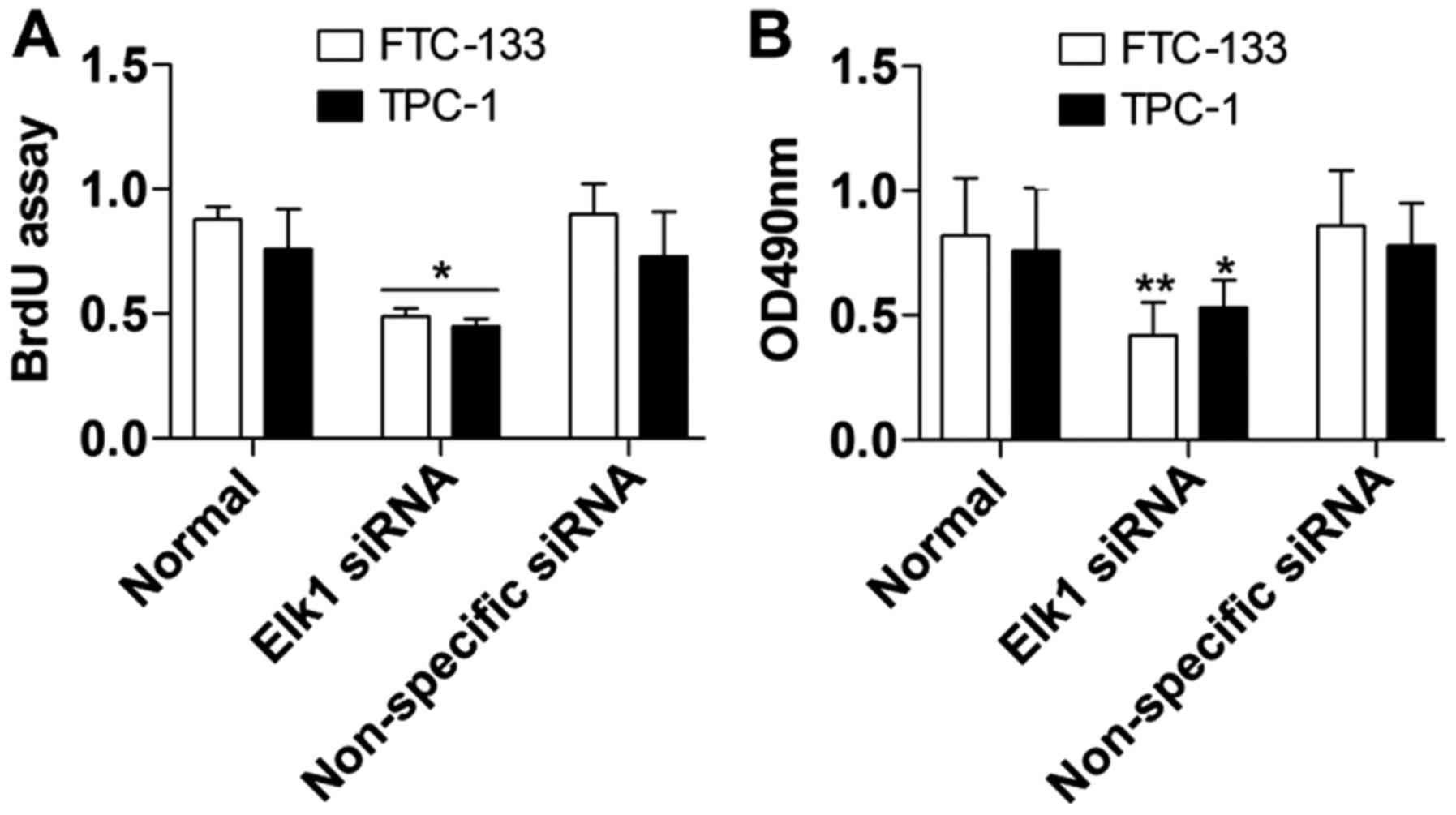
Cell proliferation was assessed using the BrdU and
MTT assays to further detect the biological effect of Elk1
inhibition on thyroid cancer cells. In the BrdU assay, the results
demonstrated that FTC-133 and TPC-1 cell proliferation (Fig. 5A) was markedly inhibited in the Elk1
siRNA group compared with the non-specific siRNA group. In the MTT
assay, FTC-133 and TPC-1 growth and viability (Fig. 5B) were both constrained in the Elk1
siRNA group compared with the non-specific siRNA group, and the
differences were significant.
Elk1 inhibition upregulates PTEN via
increased Egr-1 expression
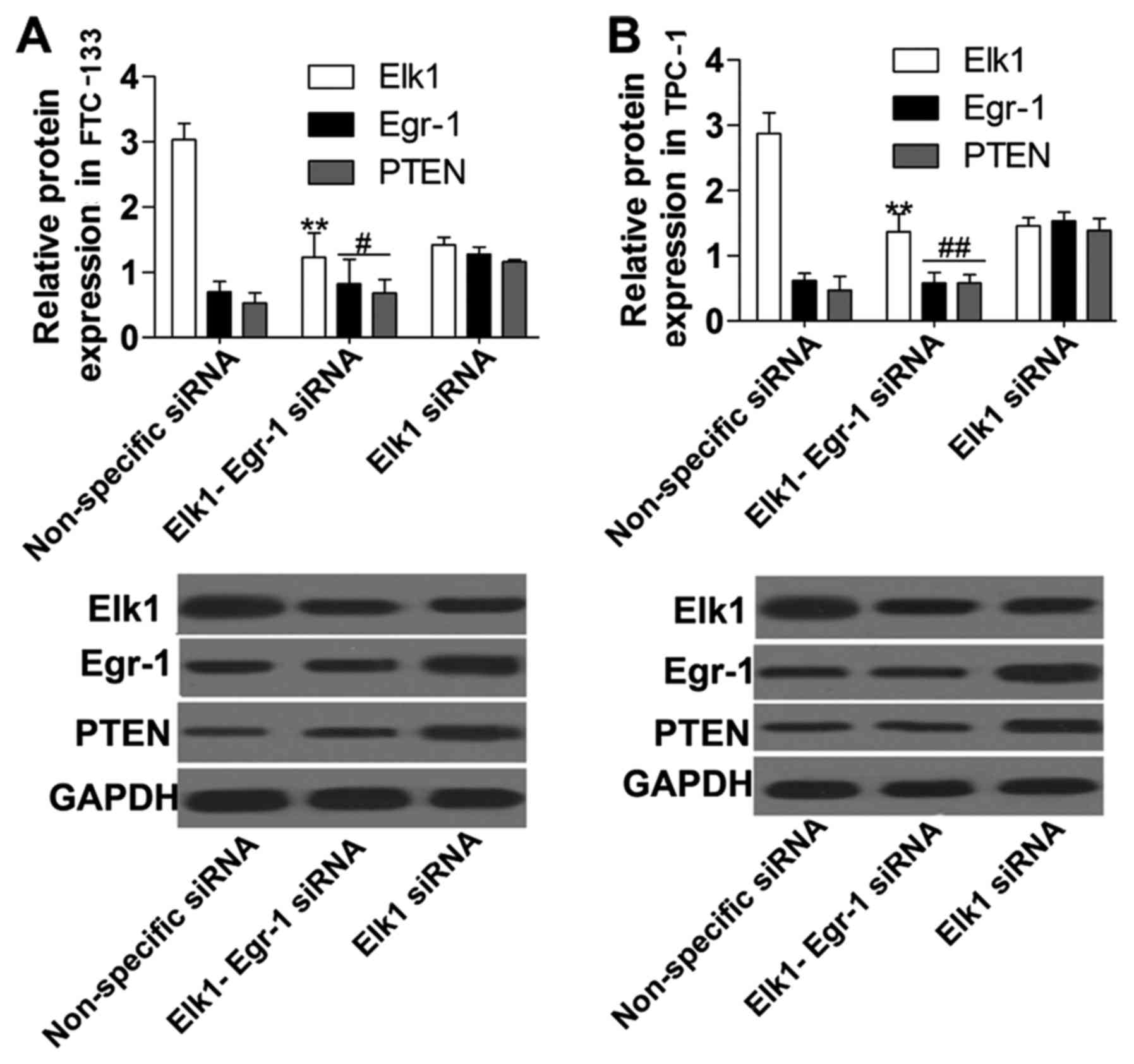
To explore the mechanism of Elk1 regulation of PTEN
and Egr-1, we performed co-transfection of Elk1 siRNA and Egr-1
siRNA into FTC-133 and TPC-1 cells, and the results showed that the
expression of Elk1 and Egr-1 protein (Fig. 6) was significantly suppressed in the
Elk1-Egr-1 siRNA group compared with the non-specific siRNA group
and with the Elk1 siRNA group, respectively. Furthermore, PTEN
protein expression (Fig. 6) was
significantly downregulated in the Elk1-Egr-1 siRNA group compared
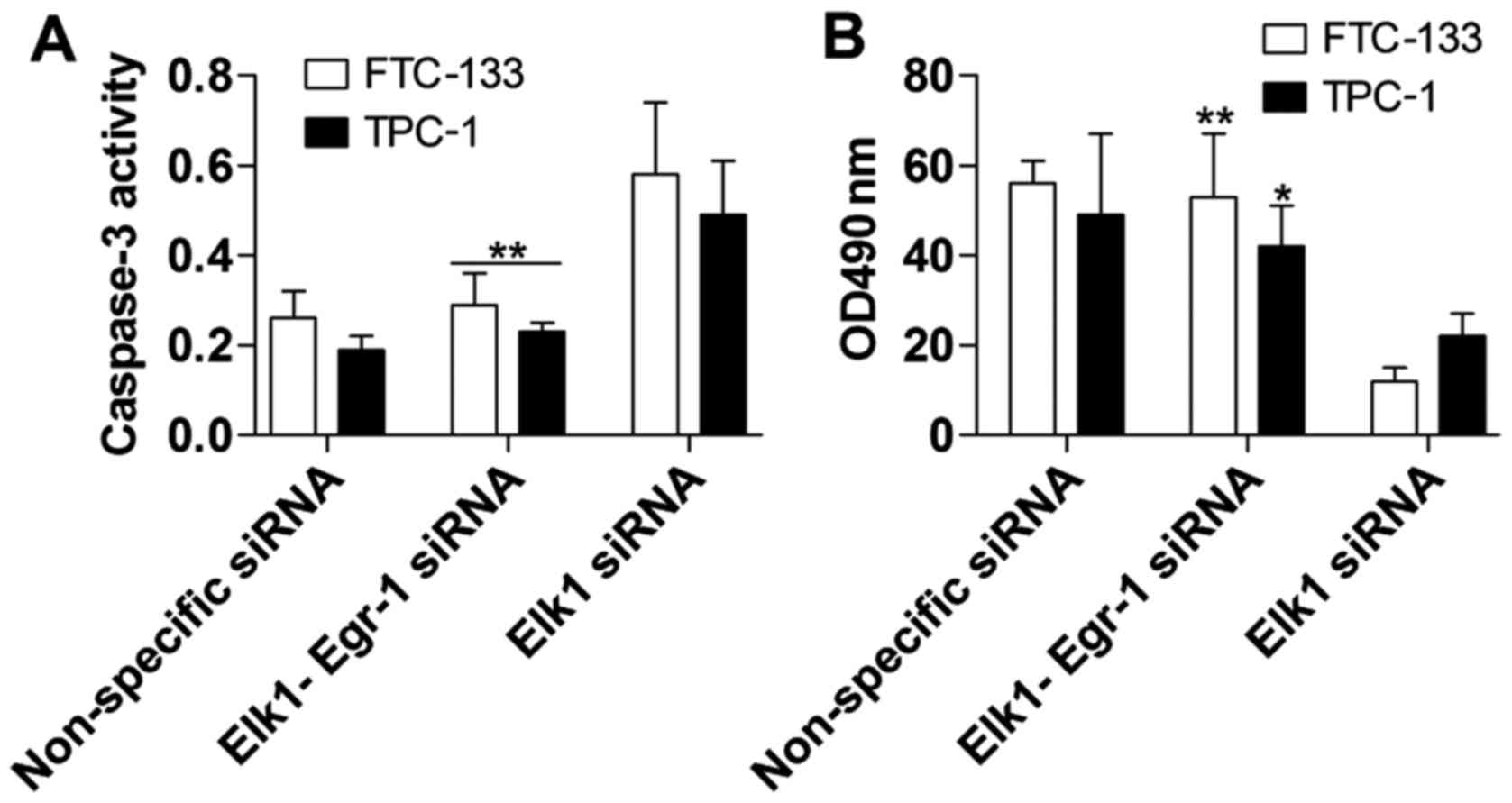
with the Elk1 siRNA group. The promotion of apoptosis (Fig. 7A) and inhibition of cell
proliferation (Fig. 7B) caused by
the suppression of Elk1 were markedly overcome by Egr-1
inhibition.
Discussion
Thyroid cancer (TC) has different histological and
biological types, and the clinically significant human thyroid
cancers are papillary and follicular carcinomas (41). Surgery-based treatment is the
primary clinical treatment. However, specific targets for drugs to
treat TC are still lacking, and the molecular mechanisms of TC
remain unclear. Elk1 is reported to be a transcriptional factor
that forms part of the ternary complex factor (TCF), and it can be
phosphorylated by the MAPK cascade (42). Elk1 regulates different factors
related to cell proliferation, differentiation and even
tumorigenesis (43). Studies have
demonstrated that Elk1 plays an important role in cancer
progression. Kawahara et al found that Elk1 is induced in
prostate cancer and promotes tumor development, whereas Elk1
inhibition suppresses tumor growth (44). Additionally, Elk1 is upregulated and
promotes cell proliferation in bladder cancer and non-small cell
lung cancer (NSCLC) (10). In this
study, we found that Elk1 expression was significantly upregulated
in TC tissues and cells. Moreover, the results showed that TC cell
proliferation and apoptosis were constrained and promoted,
respectively, after experimental downregulation of Elk1 by
siRNA.
Egr-1 is an important immediate-early gene, and is
also a tumor suppressor related to different cancers (45), as well as being implicated in cell
proliferation and apoptosis (22).
Additionally, studies have demonstrated that Egr-1 expression is
down-regulated or absent in NSCLC and breast cancer, while it is
upregulated in prostate cancer and lymphadenoma (26,28).
Research has shown that Egr-1 can be regulated via Elk1, and Demir
and Kurnaz demonstrated that Egr-1 could be repressed by Elk-1
expression in SH-SY5Y neuroblastomas (15). In our study, Elk1 inhibition
markedly increased Egr-1 expression in the TC cell lines FTC-133
and TPC-1. Furthermore, it was previously indicated that Egr-1
positively regulates PTEN expression, and that loss of Egr-1
restrains the expression of PTEN. Thus, we investigated the role of
the Elk1/Egr-1 pathway in TC.
PTEN is accepted as a tumor suppressor, and its
suppression function is realized by inhibiting the activity of P13K
(32). PTEN induction is reported
to facilitate cell apoptosis in bladder cancer, lung squamous
carcinoma and ovarian cancer (46).
PTEN mutations are extremely common in melanoma cell lines,
advanced prostate cancers, and endometrial carcinomas, and PTEN
deficiency was found to accelerate the proliferation and invasion
of a range of cancers such as gastric cancer, pancreatic cancer,
prostate cancer, among other (47–49).
Most importantly, many correlative data suggest that inhibition of
PTEN leads to TC in vivo, and Guigon et al
demonstrated that suppression of PTEN facilitates TC tumor
development in a mouse model (50).
In our study, PTEN expression was induced by Elk1 inhibition,
decreasing TC cell proliferation and increasing apoptosis.
Additionally, loss of Egr-1 significantly reversed the effect of
Elk1 inhibition on PTEN expression, TC cell proliferation and
apoptosis. Thus, the results showed that Elk1 inhibition could
induce the expression of PTEN via upregulation of Egr-1.
In summary, this study revealed that Elk1 is induced
in TC tissues and cell lines. The loss of Elk1 can markedly
increase the expression of PTEN, promoting TC cell apoptosis and
inhibiting proliferation. Furthermore, Elk1 suppression upregulates
PTEN expression via increased Egr-1 expression, providing a novel
target for the treatment of TC.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used during the present study are
available from the corresponding author upon reasonable
request.
Authors' contributions
All authors participated in the design,
interpretation of the studies and analysis of the data and review
of the manuscript; YK and XG designed and prepared the experiments;
YK performed the experiments; JY, YF, YC and YZ contributed to the
reagents/materials/analysis tools; YK wrote the manuscript; XG
modified and revised the manuscript. All authors read and approved
the manuscript and agree to be accountable for all aspects of the
research in ensuring that the accuracy or integrity of any part of
the work are appropriately investigated and resolved.
Ethics approval and consent to
participate
The present study was approved by the Xinxiang
Central Hospital Ethics Board and written informed consent obtained
from the patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbrevations
Abbreviations:
|
Elk1
|
ETS-domain containing protein
|
|
Egr-1
|
early growth response-1
|
|
TC
|
thyroid cancer
|
|
DMEM
|
Dulbecco's modified Eagle's nedium
|
|
FBS
|
fetal bovine serum
|
|
MTT
|
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
|
|
PBS
|
phosphate-buffered saline
|
|
BrdU
|
bromodeoxyuridine
|
|
Annexin V FITC/PI
|
Annexin V fluorescein isothiocyanate
conjugate/propidium iodide
|
|
RT-qPCR
|
real-time quantitative polymerase
chain reaction
|
|
TBS
|
Tris-buffered saline
|
References
|
1
|
Xing M, Haugen BR and Schlumberger M:
Progress in molecular-based management of differentiated thyroid
cancer. Lancet. 381:1058–1069. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kelly LM, Barila G, Liu P, Evdokimova VN,
Trivedi S, Panebianco F, Gandhi M, Carty SE, Hodak SP, Luo J, et
al: Identification of the transforming STRN-ALK fusion as a
potential therapeutic target in the aggressive forms of thyroid
cancer. Proc Natl Acad Sci USA. 111:4233–4238. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
McLeod DS, Sawka AM and Cooper DS:
Controversies in primary treatment of low-risk papillary thyroid
cancer. Lancet. 381:1046–1057. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Van Fossen VL, Wilhelm SM, Eaton JL and
McHenry CR: Association of thyroid, breast and renal cell cancer: A
population-based study of the prevalence of second malignancies.
Ann Surg Oncol. 20:1341–1347. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Tatler AL, Habgood A, Porte J, John AE,
Stavrou A, Hodge E, Kerama-Likoko C, Violette SM, Weinreb PH, Knox
AJ, et al: Reduced Ets Domain-containing protein Elk1 promotes
pulmonary fibrosis via increased integrin αvβ6 expression. J Biol
Chem. 291:9540–9553. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Buffet C, Catelli MG, Hecale-Perlemoine K,
Bricaire L, Garcia C, Gallet-Dierick A, Rodriguez S, Cormier F and
Groussin L: Dual specificity phosphatase 5, a specific negative
regulator of ERK signaling, is induced by serum response factor and
Elk-1 transcription factor. PLoS One. 10:e01454842015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Morris JF, Sul JY, Kim MS, Klein-Szanto
AJ, Schochet T, Rustgi A and Eberwine JH: Elk-1 phosphorylated at
threonine-417 is present in diverse cancers and correlates with
differentiation grade of colonic adenocarcinoma. Hum Pathol.
44:766–776. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Glidewell-Kenney CA, Trang C, Shao PP,
Gutierrez-Reed N, Uzo-Okereke AM, Coss D and Mellon PL: Neurokinin
B induces c-fos transcription via protein kinase C and activation
of serum response factor and Elk-1 in immortalized GnRH neurons.
Endocrinology. 155:3909–3919. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Chen YS, Aubee J, DiVito KA, Zhou H, Zhang
W, Chou FP, Simbulan-Rosenthal CM and Rosenthal DS: Id3 induces an
Elk-1-caspase-8-dependent apoptotic pathway in squamous carcinoma
cells. Cancer Med. 4:914–924. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kawahara T, Shareef HK, Aljarah AK, Ide H,
Li Y, Kashiwagi E, Netto GJ, Zheng Y and Miyamoto H: ELK1 is
up-regulated by androgen in bladder cancer cells and promotes tumor
progression. Oncotarget. 6:29860–29876. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Patki M, Chari V, Sivakumaran S, Gonit M,
Trumbly R and Ratnam M: The ETS domain transcription factor ELK1
directs a critical component of growth signaling by the androgen
receptor in prostate cancer cells. J Biol Chem. 288:11047–11065.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kim HR, Lee HN, Lim K, Surh YJ and Na HK:
15-Deoxy-Δ12,14-prostaglandin J2 induces expression of
15-hydroxyprostaglandin dehydrogenase through Elk-1 activation in
human breast cancer MDA-MB-231 cells. Mutat Res. 768:6–15. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hsu YL, Hou MF, Kuo PL, Huang YF and Tsai
EM: Breast tumor-associated osteoblast-derived CXCL5 increases
cancer progression by ERK/MSK1/Elk-1/snail signaling pathway.
Oncogene. 32:4436–4447. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Goncharenko-Khaider N, Matte I, Lane D,
Rancourt C and Piche A: Ovarian cancer ascites increase Mcl-1
expression in tumor cells through ERK1/2-Elk-1 signaling to
attenuate TRAIL-induced apoptosis. Mol Cancer. 11:842012.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Demir O and Kurnaz IA: Wildtype Elk-1, but
not a SUMOylation mutant, represses egr-1 expression in SH-SY5Y
neuroblastomas. Neurosci Lett. 437:20–24. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Pacini L, Suffredini S, Ponti D, Coppini
R, Frati G, Ragona G, Cerbai E and Calogero A: Altered calcium
regulation in isolated cardiomyocytes from Egr-1 knock-out mice.
Can J Physiol Pharmacol. 91:1135–1142. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zalcman G, Federman N, de la Fuente V and
Romano A: Nuclear factor kappa B-dependent Zif268 expression in
hippocampus is required for recognition memory in mice. Neurobiol
Learn Mem. 119:10–17. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Fan YY, Ye GH, Lin KZ, Yu LS, Wu SZ, Dong
MW, Han JG, Feng XP and Li XB: Time-dependent expression and
distribution of Egr-1 during skeletal muscle wound healing in rats.
J Mol Histol. 44:75–81. 2013.PubMed/NCBI
|
|
19
|
Klenke S, Rump K, Buschkamp K, Engler A,
Peters J, Siffert W and Frey UH: Characterization of the PLCB1
promoter and regulation by early growth response transcription
factor EGR-1. Eur J Pharmacol. 742:8–14. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Bhattacharyya S, Fang F, Tourtellotte W
and Varga J: Egr-1: New conductor for the tissue repair orchestra
directs harmony (regeneration) or cacophony (fibrosis). J Pathol.
229:286–297. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Witham EA, Meadows JD, Hoffmann HM,
Shojaei S, Coss D, Kauffman AS and Mellon PL: Kisspeptin regulates
gonadotropin genes via immediate early gene induction in pituitary
gonadotropes. Mol Endocrinol. 27:1283–1294. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Sysol JR, Natarajan V and Machado RF: PDGF
induces SphK1 expression via Egr-1 to promote pulmonary artery
smooth muscle cell proliferation. Am J Physiol Cell Physiol.
310:C983–C992. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Liu QF, Yu HW, You L, Liu MX, Li KY and
Tao GZ: Apelin-13-induced proliferation and migration induced of
rat vascular smooth muscle cells is mediated by the upregulation of
Egr-1. Biochem Biophys Res Commun. 439:235–240. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Fang F, Shangguan AJ, Kelly K, Wei J,
Gruner K, Ye B, Wang W, Bhattacharyya S, Hinchcliff ME,
Tourtellotte WG and Varga J: Early growth response 3 (Egr-3) is
induced by transforming growth factor-β and regulates fibrogenic
responses. Am J Pathol. 183:1197–1208. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kim K, Jutooru I, Chadalapaka G, Johnson
G, Frank J, Burghardt R, Kim S and Safe S: HOTAIR is a negative
prognostic factor and exhibits pro-oncogenic activity in pancreatic
cancer. Oncogene. 32:1616–1625. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kim JH, Choi DS, Lee OH, Oh SH, Lippman SM
and Lee HY: Antiangiogenic antitumor activities of IGFBP-3 are
mediated by IGF-independent suppression of Erk1/2 activation and
Egr-1-mediated transcriptional events. Blood. 118:2622–2631. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Calogero A, Arcella A, De Gregorio G,
Porcellini A, Mercola D, Liu C, Lombari V, Zani M, Giannini G,
Gagliardi FM, et al: The early growth response gene EGR-1 behaves
as a suppressor gene that is down-regulated independent of ARF/Mdm2
but not p53 alterations in fresh human gliomas. Clin Cancer Res.
7:2788–2796. 2001.PubMed/NCBI
|
|
28
|
Vockerodt M, Wei W, Nagy E, Prouzova Z,
Schrader A, Kube D, Rowe M, Woodman CB and Murray PG: Suppression
of the LMP2A target gene, EGR-1, protects Hodgkin's lymphoma cells
from entry to the EBV lytic cycle. J Pathol. 230:399–409. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Scharnhorst V, Menke AL, Attema J,
Haneveld JK, Riteco N, van Steenbrugge GJ, van der Eb AJ and
Jochemsen AG: EGR-1 enhances tumor growth and modulates the effect
of the Wilms' tumor 1 gene products on tumorigenicity. Oncogene.
19:791–800. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Virolle T, Adamson ED, Baron V, Birle D,
Mercola D, Mustelin T and de Belle I: The Egr-1 transcription
factor directly activates PTEN during irradiation-induced
signalling. Nat Cell Biol. 3:1124–1128. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Salmena L: PTEN: History of a tumor
suppressor. Methods Mol Biol. 1388:3–11. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Jing X, Cheng W, Wang S, Li P and He L:
Resveratrol induces cell cycle arrest in human gastric cancer
MGC803 cells via the PTEN-regulated PI3K/Akt signaling pathway.
Oncol Rep. 35:472–478. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Jolly LA, Novitskiy S, Owens P, Massoll N,
Cheng N, Fang W, Moses HL and Franco AT: Fibroblast-mediated
collagen remodeling within the tumor microenvironment facilitates
progression of thyroid cancers driven by BrafV600E and pten loss.
Cancer Res. 76:1804–1813. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Yang Z, Yuan XG, Chen J, Luo SW, Luo ZJ
and Lu NH: Reduced expression of PTEN and increased PTEN
phosphorylation at residue Ser380 in gastric cancer tissues: A
novel mechanism of PTEN inactivation. Clin Res Hepatol
Gastroenterol. 37:72–79. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Govatati S, Kodati VL, Deenadayal M,
Chakravarty B, Shivaji S and Bhanoori M: Mutations in the PTEN
tumor gene and risk of endometriosis: A case-control study. Hum
Reprod. 29:324–336. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Fortin J, Bassi C and Mak TW: PTEN enables
the development of pre-B acute lymphoblastic leukemia. Nat Med.
22:339–340. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Herranz D, Maraver A, Cañamero M,
Gómez-López G, Inglada-Pérez L, Robledo M, Castelblanco E,
Matias-Guiu X and Serrano M: SIRT1 promotes thyroid carcinogenesis
driven by PTEN deficiency. Oncogene. 32:4052–4056. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Weng LP, Gimm O, Kum JB, Smith WM, Zhou
XP, Wynford-Thomas D, Leone G and Eng C: Transient ectopic
expression of PTEN in thyroid cancer cell lines induces cell cycle
arrest and cell type-dependent cell death. Hum Mol Genet.
10:251–258. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Curcio F, Ambesi-Impiombato FS, Perrella G
and Coon HG: Long-term culture and functional characterization of
follicular cells from adult normal human thyroids. Proc Natl Acad
Sci USA. 91:9004–9008. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Welsh JB, Sapinoso LM, Su AI, Kern SG,
Wang-Rodriguez J, Moskaluk CA, Frierson HF Jr and Hampton GM:
Analysis of gene expression identifies candidate markers and
pharmacological targets in prostate cancer. Cancer Res.
61:5974–5978. 2001.PubMed/NCBI
|
|
41
|
Jankovic B, Le KT and Hershman JM:
Clinical review: Hashimoto's thyroiditis and papillary thyroid
carcinoma: Is there a correlation? J Clin Endocrinol Metab.
98:474–482. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Wozniak MA, Cheng CQ, Shen CJ, Gao L,
Olarerin-George AO, Won KJ, Hogenesch JB and Chen CS: Adhesion
regulates MAP kinase/ternary complex factor exchange to control a
proliferative transcriptional switch. Curr Biol. 22:2017–2026.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Doma E, Rupp C, Varga A, Kern F, Riegler B
and Baccarini M: Skin tumorigenesis stimulated by Raf inhibitors
relies upon Raf functions that are dependent and independent of
ERK. Cancer Res. 73:6926–6937. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Kawahara T, Aljarah AK, Shareef HK, Inoue
S, Ide H, Patterson JD, Kashiwagi E, Han B, Li Y, Zheng Y and
Miyamoto H: Silodosin inhibits prostate cancer cell growth via ELK1
inactivation and enhances the cytotoxic activity of gemcitabine.
Prostate. 76:744–756. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Chakraborty T, Asok A, Stanton ME and
Rosen JB: Variants of contextual fear conditioning induce
differential patterns of Egr-1 activity within the young adult
prefrontal cortex. Behav Brain Res. 302:122–130. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Peralta-Zaragoza O, Deas J, Meneses-Acosta
A, De la O-Gómez F, Fernández-Tilapa G, Gómez-Cerón C,
Benítez-Boijseauneau O, Burguete-García A, Torres-Poveda K,
Bermúdez-Morales VH, et al: Relevance of miR-21 in regulation of
tumor suppressor gene PTEN in human cervical cancer cells. BMC
Cancer. 16:2152016. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Yang SM, Huang C, Li XF, Yu MZ, He Y and
Li J: miR-21 confers cisplatin resistance in gastric cancer cells
by regulating PTEN. Toxicology. 306:162–168. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Soubani O, Ali AS, Logna F, Ali S, Philip
PA and Sarkar FH: Re-expression of miR-200 by novel approaches
regulates the expression of PTEN and MT1-MMP in pancreatic cancer.
Carcinogenesis. 33:1563–1571. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Wu Z, He B, He J and Mao X: Upregulation
of miR-153 promotes cell proliferation via downregulation of the
PTEN tumor suppressor gene in human prostate cancer. Prostate.
73:596–604. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Guigon CJ, Zhao L, Willingham MC and Cheng
SY: PTEN deficiency accelerates tumour progression in a mouse model
of thyroid cancer. Oncogene. 28:509–517. 2009. View Article : Google Scholar : PubMed/NCBI
|