Introduction
Oral cancer is a serious and increasingly prevalent
disease in several regions worldwide. Oral and pharyngeal cancer,
grouped together, constitute the sixth most common type of cancer
globally. The estimated annual incidence is ~275,000 for oral
cancer, and two thirds of these cases occur in developing countries
(1). In China, ~8/100,000 of the
population experience oral cancer per year according to the World
Health Organization (2). Of these
tumors, >90% are oral squamous cell carcinoma (OSCC). Despite
advances in diagnosis and treatment, the 5-year survival rate of
patients with OSCC is no more than 50% (3). In the late stages of tongue cancer,
the 5-year survival rate has markedly decreased to 5% [National
Cancer Institute's Surveillance, Epidemiology, and End Results
program (2003–2009)] (https://seer.cancer.gov/archive/csr).
The histologic examination of routine hematoxylin
and eosin (H&E)-stained sections is the current standard used
in the diagnosis of OSCC; however, the discrimination of normal
tissue, pre-cancerous lesions, cancerous lesions, and
micrometastasis in lymph nodes based on H&E-stained sections of
the oral epithelium can be difficult (4,5). It is
well established that the development of oral cancer consists of
two steps, the initial presence of a precursor lesion and its
subsequent development into cancer (6). Although certain morphological changes
may underlie the process of pre-cancerous lesions developing into
cancer and thus provide certain clues for assessing the risk of
malignancy of lesions, pathological decisions based on such
observations can be subjective and even controversial. For example,
opinions vary among pathologists regarding the interpretation of
the extent of dysplasia in the same lesion. Therefore, the
identification of relatively objective biomarkers that can be used
to identify oral malignancy or micrometastasis in lymph nodes will
benefit the evaluation and diagnosis of patients with OSCC
(7).
To identify proteins capable of differentiating OSCC
tissues from para-cancerous tissues in the same patient, the
present study performed assays directly on diseased tissues using
stringent protocols and a well-designed workflow. First, a vast
archive of biopsied and excised specimens, which had been processed
for histological diagnosis, formalin-fixed, and paraffin-embedded
(FFPE), were accessed. These specimens represent a unique source of
morphologically defined and disease-specific samples, containing
substantial protein information and the accompanying patient
clinical information, and have already been applied in the
assessment of novel and classical biomarkers (8–10).
Inevitably, investigations based on FFPE tissues
must account for the loss of protein in FFPE samples, as the
process of fixation and long duration of storage may induce
degradation and alteration of proteins in FFPE tissues. Therefore,
the present study used a commercially available protein extraction
kit, which enabled the extraction of as many protein/peptides as
possible from relatively newly-preserved FFPE tissues (within 6
months). Mass spectrometry technologies have been widely used for
biomarker screening in human diseases (11). Another important technical aspect in
the biomarker identification process in the present study was the
use of laser capture microdissection (LCM), which allows the
identification and capture of targeted cells from complex clinical
specimens without interference from the tumor stroma (12). Using LCM, oral cancer cells,
pre-cancerous cells, and normal mucosal epithelial cells were
obtained from FFPE specimens derived from the same individual, on
which proteomic analysis using liquid chromatography-tandem mass
spectrometry (LC/MS) was performed. This facilitated the
high-throughput proteomic analysis of differentially expressed
proteins (10).
The approach used in the present study to quantify
proteome changes in oral cancer tissues allowed for the
high-throughput and unbiased identification of novel proteomic
changes in OSCC. This LCM-FFPE-proteomic and LC/MS analysis
approach, followed by IHC validation in formalin-fixed tissues,
serves as a reliable platform for the identification of diagnostic
biomarkers for OSCC.
Materials and methods
Clinical samples and histological
analysis
Archived FFPE tissue samples, which had been
pathologically diagnosed within the prior 6 months, were classified
into two groups: Group 1 consisted of three paired tissue samples.
Each paired sample was derived from the same individual and
consisted of OSCC lesions and their corresponding adjacent normal
mucosal lesions. H&E staining was used to determine the tumor
tissue boundaries. The distance from the normal tissues to
cancerous tissues was ~1–2 mm.
Group 2 also consisted of three paired cases
(leukoplakia carcinogenesis tissue), and each was derived from the
same individual, including paired normal oral mucosa, oral
leukoplakia with mild-moderate dysplasia, and OSCC lesions. H&E
staining was used to determine the boundaries of the tumor tissues
or leukoplakia with mild-moderate dysplastic tissues. The distance
from the normal tissues to leukoplakia with mild-moderate dysplasia
tissues, or leukoplakia with mild-moderate dysplasia tissues to
cancerous tissues was also ~1–2 mm. For cytokeratin (CK)
immunohistochemistry (IHC) analysis and validation, three groups of
samples were used: Group 1: 20 FFPE specimens diagnosed as OSCC
within the most recent 5 years, which included normal,
pre-cancerous, and cancerous areas; Group 2: 20 neck dissection
specimens with lymph node micro-metastases; and Group 3:10 samples
diagnosed with non-metastatic lymph nodes.
The histological analysis and diagnosis were
performed by a pathologist (Dr Ning Geng) and then confirmed by a
second pathologist (Dr Yu Chen) of the State Key Laboratory of Oral
Diseases, National Clinical Research Center for Oral Diseases, West
China Hospital of Stomatology (Sichuan, China). All tissue samples
were obtained from the Oral Pathology Archives of West China
Hospital of Stomatology, and were fixed and paraffin-embedded using
standard procedures. The Institutional Review Board of the West
China College of Stomatology, Sichuan University approved the
protocol.
LCM using FFPE sections
The FFPE oral cancer tissue sections (5-µM thick) on
Arcturus® PEN membrane glass slides (Thermo Fisher
Scientific, Inc., Waltham, MA, USA) were deparaffinized in xylene,
hydrated and stained in hematoxylin, followed by dehydration. For
LCM, the stained uncovered Arcturus® PEN membrane glass
slides were air dried and ~10,000 cells were captured onto
CapSure® HS LCM caps (Thermo Fisher Scientific, Inc.)
using an ArcturusXT™ microdissection system (Thermo Fisher
Scientific, Inc.). The caps were transferred to a 0.5-ml sterile
Eppendorf tube for protein extraction.
Protein extraction from captured cells
derived from FFPE tissues
The protein lysates were prepared according to the
manufacturer's protocol (Liquid Tissue kit; Expression Pathology,
Inc., Gaithersburg, MD, USA). The procured cells (~10,000 cells for
each set of tissues) were suspended in 20 µl of proprietary Liquid
Tissue buffer (Expression Pathology, Inc.) and heated at 95°C for
90 min. Following cooling for 2 min on ice, 1 µl of trypsin reagent
was added and incubated at 37°C for 1 h with vigorous shaking for 1
min at 20 min intervals. The samples were then incubated at 37°C
for 18 h followed by heating at 95°C for 5 min, and then stored at
−20°C prior to use for LC/MS analysis.
Proteomic analysis by LC/MS
The FFPE-extracted samples were processed,
quantified, and analyzed in a Thermo LTQ 7000 high-performance
liquid chromatography (HPLC)-mass spectrophotometer (LTQ; Thermo
Fisher Scientific, Inc.). Protein separation was performed on a
Thermo Scientific Surveyor HPLC system with Hypersil GOLD columns
(Thermo Fisher Scientific, Inc.). The spectra were searched against
the NCBI Protein Bank human protein database (https://www.ncbi.nlm.nih.gov/protein/)
from the European Bioinformatics Institute using SEQUEST 3.2
(Thermo Fisher Scientific, Inc.). The results were further filtered
using software developed in-house to determine unique peptides and
proteins, which has a predicted error of <1.5%.
IHC analysis
Archived FFPE tissue blocks from the Department of
Oral Pathology, West China College of Stomatology, Sichuan
University were used for histopathologic and immunohistochemical
analysis. EnVision/HRP, a two-step IHC staining technique, was used
for IHC staining (EnVision Detection system, Peroxidase/DAB,
rabbit/mouse; code no. 500750; Gene Tech Co., Ltd., Shanghai,
China). Sections (4-µM) were prepared from the FFPE blocks on
adhesive glass slides. Antigen retrieval was achieved by boiling
the sections in 0.01 M citrate buffer (pH 6.0) in a high-pressure
cooker for 3 min. Primary antibodies used in this procedure were as
follows: CK4 (rabbit monoclonal; 1:200; cat. no. M07410; Boster
Biological Technology, Pleasanton CA, USA); CK14 (rabbit
monoclonal; 1:200; cat. no. ZA-0540; ZSGB-BIO, Beijing, China);
CK10/13 (mouse monoclonal; 1:30; cat. no. ZM-0314; ZSGB-BIO); CK17
(rabbit monoclonal; 1:200; cat. no. ZA-0551; ZSGB-BIO); and Pan CK
(mouse monoclonal; 1:200; cat. no. ZM-0069; ZSGB-BIO). Duplicate
sections were incubated overnight in 4°C with primary antibody.
Lymph node metastasis mouse model
Female BALB/c-nu/nu nude mice (5–6 weeks old)
weighing 18–20 g (n=6) were used in the present study. Six mice
were housed in a specific pathogen free class laboratory animal
room, which was maintained at 22±2°C on a regular light-dark cycle.
All animals were provided with food and water ad libitum.
The use of animals in the experiments was approved by the State Key
Laboratory of Oral Diseases Research Ethics Committee.
HSC-3 cells were purchased from the Cell Bank of
Japanese Collection of Research Bioresource (JCRB; Shinjuku,
Japan). This cell was cultured in DMEM (Sigma-Aldrich; Merck KGaA)
with 10% fetal bovine serum (FBS) in a humidified atmosphere with
5% CO2. The cells were collected separately in the
logarithmic phase, washed twice in 1X PBS, and added to fresh
medium without serum, cells were counted and the cell concentration
was adjusted to 2×107 cells/ml. On the inside of the
oral buccal mucosa of the nude mice, the cells were inoculated in a
50-µl cell suspension (lx106 cells/mouse). The mice were
sacrificed at 45 days and the neck lymph nodes were collected, for
the production of paraffinized sections for H&E staining and
immunohistochemical staining.
Statistical analysis
The statistical analysis was conducted using SPSS
version 19 (IBM Corp., Armonk, NY, USA). The rate of accuracy and
95% confidence intervals (CIs) were used to evaluate the effects of
CKs in differentiating OSCC from para-cancerous tissues, and the
95% CI of was performed by using Bootstrap test.
Results
Proteomic profiling based on LCM of
FFPE tissues represents a reliable approach for biomarker
identification
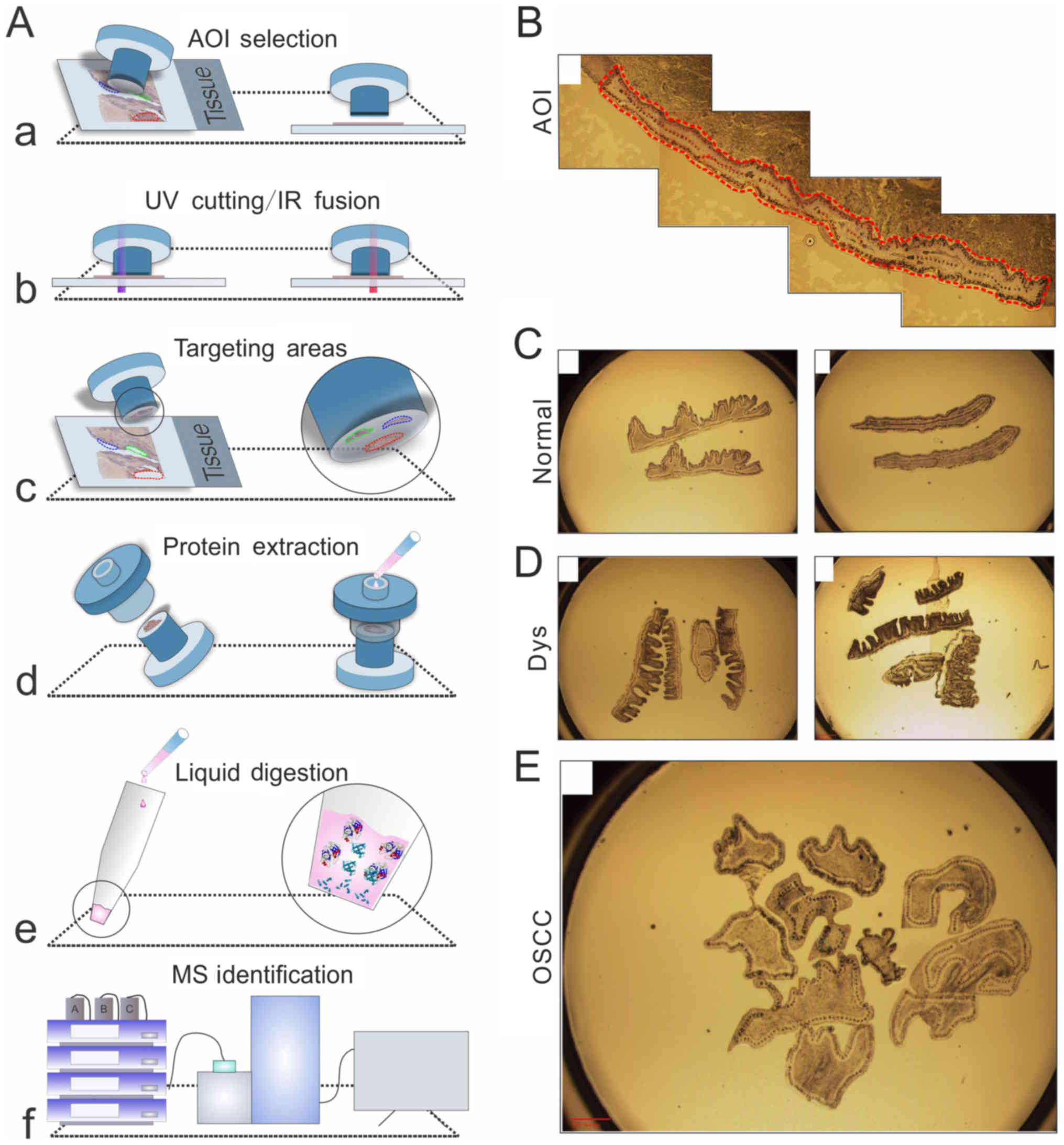
In the present study, the Arcturus LCM platform was
used, which features three functions: Cut, capture, and the
combination of the two. During the procedure, the platform was used
together with CapSure® HS LCM caps. An elevated ring
surrounds the cap, and the captured cells adhere to this. Proteins
extracted from these FFPE samples are amenable to LC/MS analysis.
The detailed experimental procedures are shown in Fig. 1A. For the FFPE samples,
~10,000–20,000 (two caps) cells were obtained from each type of
tissue, although in one case of mucosal epithelial cells,
6,000–10,000 cells were obtained. The prepared OSCC tissues/cells
micro-dissected for further analysis using LCM are shown in
Fig. 1B-E.
Detection of differentially expressed
proteins between OSCC tissues and tumor-adjacent normal
tissues
To obtain insights into the proteins distinctively
expressed in OSCC tissues and adjacent normal tissues, paired OSCC
and para-cancerous normal mucosal tissues were analyzed. Each
paired sample was derived from the same individual, and the group
consisted of three paired tissue samples (Fig. 2A). The LC/MS proteomics platform
used in this analysis enabled the identification of 143 total
proteins in OSCC tumor tissues and 133 adjacent normal tissues,
representing a diverse array of molecular functions, including
cytoskeleton proteins; a membrane protein; desmosomal proteins; and
proteins involved in cell differentiation, cell metabolism,
cell-cycle regulation, cell damage and repair, genetic regulation,
a signaling pathway, and transportation (Table I). Among these, 82 proteins were
shared across normal and OSCC tissues (Fig. 2B), whereas tumor-adjacent tissues
exhibited high levels of CK4, 13 and 10.
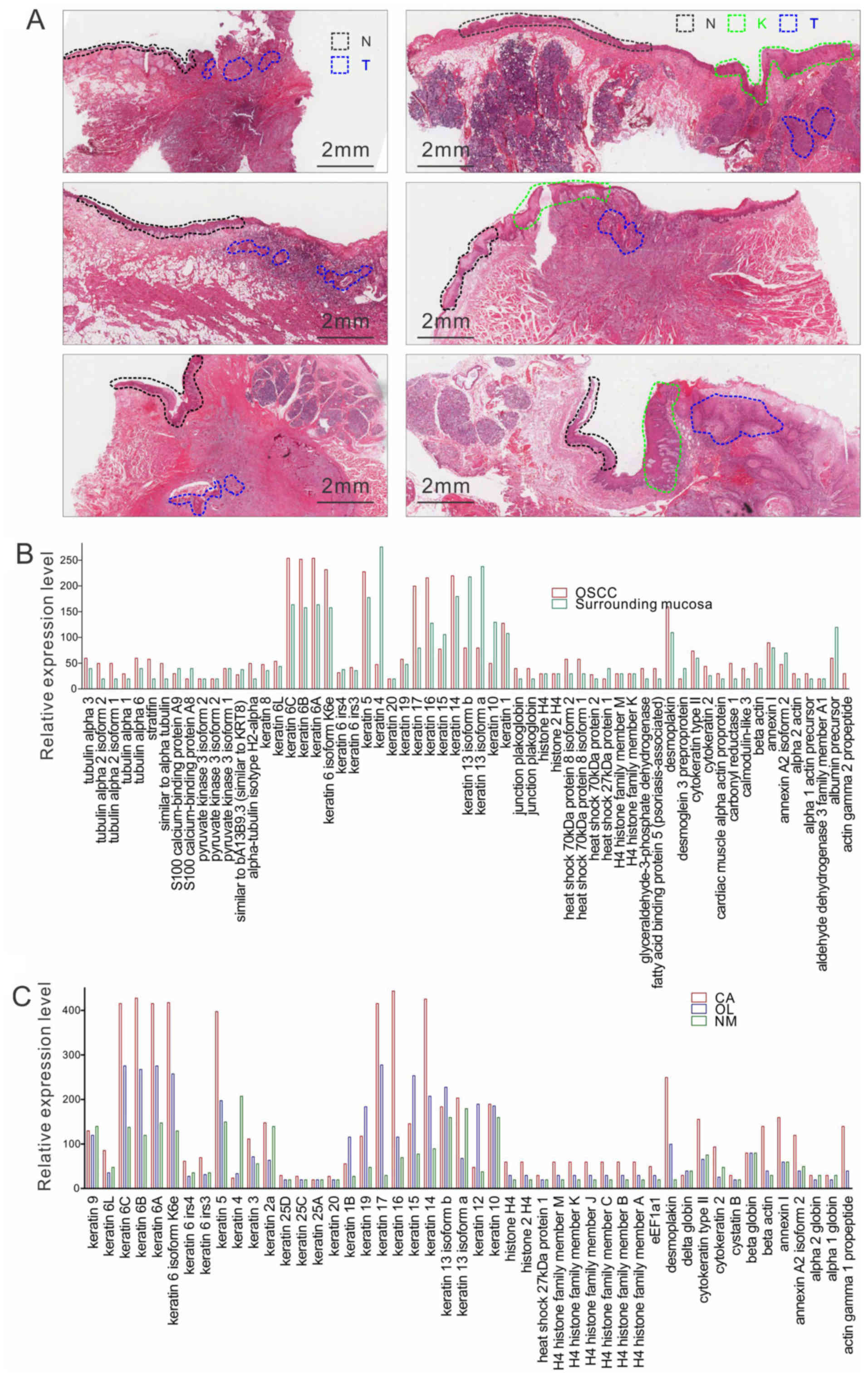 | Figure 2.Panoramic view of H&E staining and
protein expression in clinical samples analyzed with LC/MS. (A)
Representative H&E staining images of adjacent N, T, and/or K
regions in six cases used for LC/MS analysis. Group 1 cases are on
the left; Group 2 cases are on the right. Scale bar, 2 mm. (B)
LC/MS analysis of proteins shared by OSCC tissues and adjacent
normal surrounding mucosa tissues. y-axis represents the relative
level of the proteins. (C) LC/MS analysis of proteins shared by
OSCC (CA), pre-cancerous lesions (OLK) and normal (NM) tissues.
y-axis represents the relative level of the proteins. H&E,
hematoxylin and eosin; LC/MS, liquid chromatography-tandem mass
spectrometry; OSCC, oral squamous cell carcinoma; N, normal; T,
tumor; K, pre-cancerous lesions. |
 | Table I.Representative proteins identified
using liquid chromatography-tandem mass spectrometry analysis. |
Table I.
Representative proteins identified
using liquid chromatography-tandem mass spectrometry analysis.
| Classification | Representative
proteins |
|---|
| Cell structure |
|
|
Cytoskeletal proteins | CK16, CK6, CK14,
CK17, CK1, Actin, Tubulin, Plectin-1 |
|
Desmosomal proteins | Desmoplakin,
Plakophilin, Desmoglein |
| Membrane-related
proteins | Annexin I, Annexin
A2 |
| Cell
differentiation | Involucrin, Small
proline-rich protein 1B, Small proline-rich protein 1A, Small
proline-rich protein 3, Cornifin A, Cornifin B |
| Cell
metabolism | Pyruvate kinase 3,
Glyceraldehyde-3-phosphate dehydrogenase, ATP synthase, Enolase,
Tyr3/Trp5-mono-oxygenase activation protein, Calgranulin B,
Calgranulin A. |
| Cell-cycle
regulation | Eukaryotic
translation elongation factor 1 |
| Cell damage and
repair | Heat shock protein
27, Heat shock protein 70, Heat shock protein 90. |
| Genetic
regulation | H4 histone family,
MYC promoter-binding protein |
| Signaling
pathway | Calmodulin-like 3,
S100 Calcium-binding protein A8, S100 Calcium-binding protein
A9 |
| Transportation | Serum albumin
precursor |
The proteins detected in the normal oral squamous
epithelium included CK12 and cornulin, also known as tumor-related
protein, which is a cytoplasmic protein considered to be involved
in epidermal differentiation and the epithelial immune response.
Several studies have shown that its downregulation is linked to
tumor progression (13,14). The proteins expressed uniquely in
normal tissues indicated that their biological functions may
include maintaining normal epithelial differentiation and
suppressing tumors.
Proteins differentially expressed in
normal, pre-cancerous, and cancerous oral mucosal epithelial
tissues may be effective biomarkers for OSCC diagnosis
To obtain insights into the proteins differentially
expressed at different stages of oral carcinogenesis, including
normal epithelium, pre-cancerous tissues (OLK), and OSCC tissues,
paired normal oral mucosa and oral leukoplakia with mild-moderate
dysplasia were included in the analysis. A total of 53 proteins
were expressed in all three tissue sets, including CKs and
desmosomal proteins (Fig. 2C;
Table I). Notably, CK4 was the most
abundant in normal tissues, and its expression level was
significantly lower in the OLK and OSCC tissues. By contrast, the
protein expression levels of CK6, 14, 16 and 17; desmoplakin; actin
γ1; eukaryotic translation elongation factor 1; and H4 histone were
found to be higher in the tumor samples than in the OLK samples,
and their levels continually decreased from the OLK samples to the
normal samples. This shift in the distribution pattern may be
indicative of the importance of these proteins in oral
carcinogenesis.
A total of 13 proteins were found to be expressed at
high levels specifically in OSCC and OLK tissues. The majority of
CKs, including CK3, 5, 6, 14 and 16, had higher expression scores
in OSCC tissues than in adjacent OLK tissues or normal tissues.
Notably, the expression of CK17 was 10-fold higher in the OSCC
tissues than in the normal tissues. In addition, a group of
proteins was detected only in OSCC tissues, including myosin and
CK18. Certain proteins were detected preferentially in normal
tissues, including CK5. Of note, this pattern was in accordance
with the previous analysis of OSCC tissues and tumor-adjacent
normal tissues.
Expression levels of CK10/13 and 4 or
CK14 and 17 are potential molecular markers for the extent of the
surgical resection of OSCC
Surgical resection beyond contrast-enhancing
boundaries may represent a promising strategy to improve outcome
and quality of life for patients with OSCC. Therefore, a specific
molecular marker to guide the determination of surgical margins is
urgently required. Using LC/MS proteomics analysis, the present
study found significant differences in the expression of CK14,
CK17, CK10/13 and CK4 in OSCC tissues, compared with that in normal
oral tissues/para-cancerous tissues. To identify molecular markers
that may be used to determine the extent of the surgical resection
of OSCC tissue, IHC of CK 10/13, 4, 14 and 17 was performed in 20
FFPE OSCC tissue samples. The expression levels of CK 10/13, 4, 14
and 17 in two representative cases are shown in Fig. 3A. CK4 was detected at high levels in
epithelial cell layers of tumor-adjacent tissues but was not
present in OSCC tissues. CK10/13 showed similar distribution
patterns in normal and OSCC tissues, but they were also expressed
in the basal layers of OSCC tissues. A high level of
plasma-positive staining of CK14 and 17 was observed in OSCC
tissues, whereas their distribution was confined to the basal layer
in the adjacent normal epithelium or in moderate dysplasia. The
observation that CK14 and CK17 made it possible to distinguish
between OSCC tissues and adjacent normal tissues/moderate
dysplastic tissues indicated a shift in intermediate filament
protein expression, which may be a potential diagnostic biomarker
for OSCC.
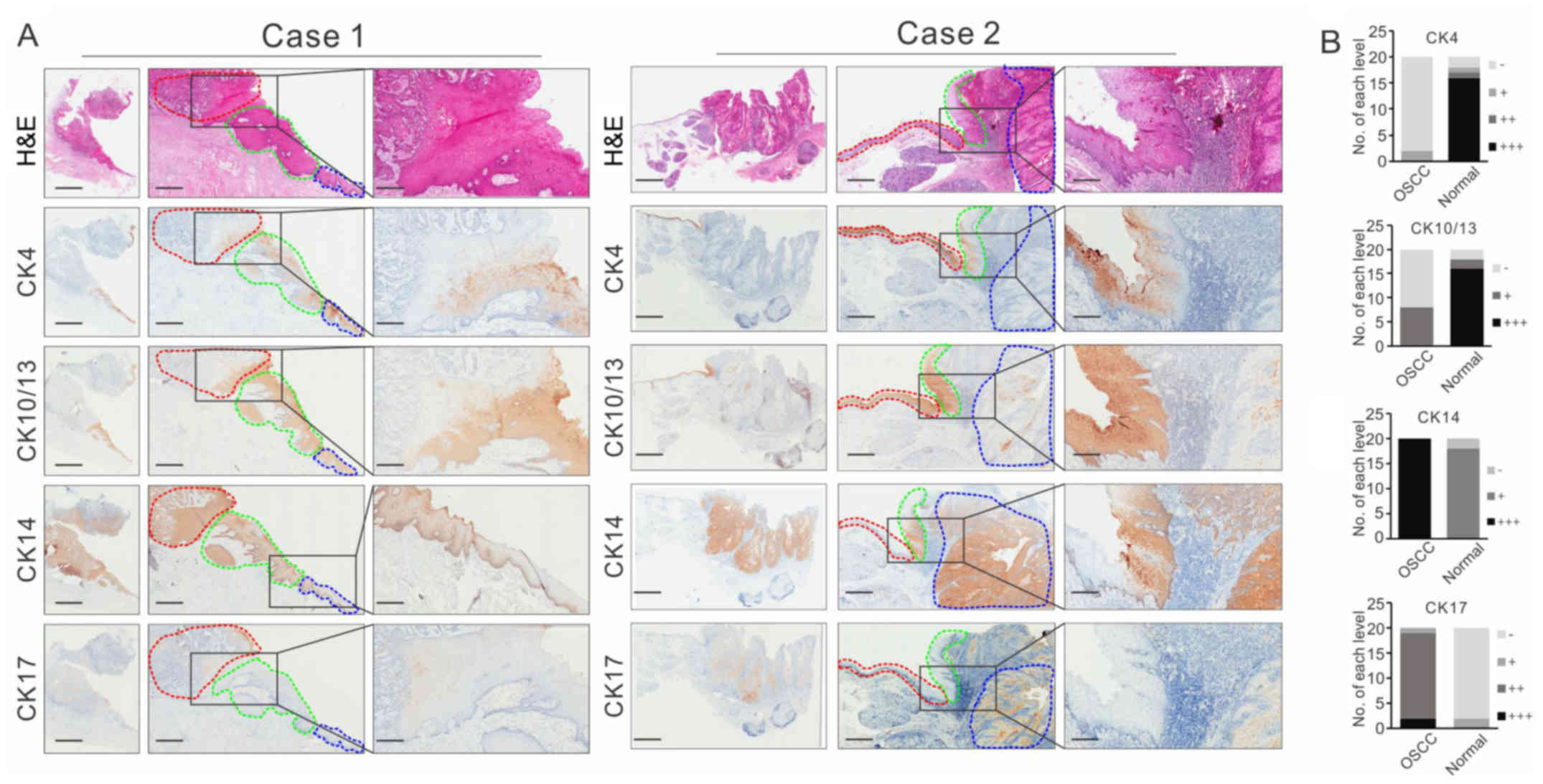 | Figure 3.Immunohistochemistry analysis of the
identified proteins in clinical tissues. (A) Representative images
of CK4, CK10/13, CK14 and CK17 immunostaining of OSCC tissues
accompanied by those of adjacent normal/pre-cancerous lesions. The
regions were identified by the pathologist: Adjacent normal regions
are shown in blue circles, pre-cancerous lesions are shown in green
circles and OSCC regions are shown in red circle. Scale bar, 2,000
µm (left panels), 1,000 µm (middle panels) and 250 µm (right
panels). (B) Semi-quantitative scores of CK-positive staining in
OSCC tissues and adjacent normal tissues of 20 cases. OSCC, oral
squamous cell carcinoma; H&E, hematoxylin and eosin; CK,
cytokeratin. |
The relative expression levels were evaluated and
analyzed (Fig. 3B), and the results
showed that negative staining of CK4 and CK10/13 clearly
distinguished between cancerous tissue and para-cancerous tissue,
with an accuracy of 90% (95% CI, 0.68–0.99) and 75% (95% CI,
0.51–0.91), respectively, whereas the positive staining of CK14 and
CK17 clearly distinguished between cancerous lesions and
para-cancerous lesions with an accuracy of 100% (95% CI, 83–100%)
and 90% (95% CI, 0.68–0.99), respectively. These results suggested
that CK4 and CK10/13 or CK14 and CK17 may be useful molecular
boundary markers for guidance in the surgical resection of
OSCC.
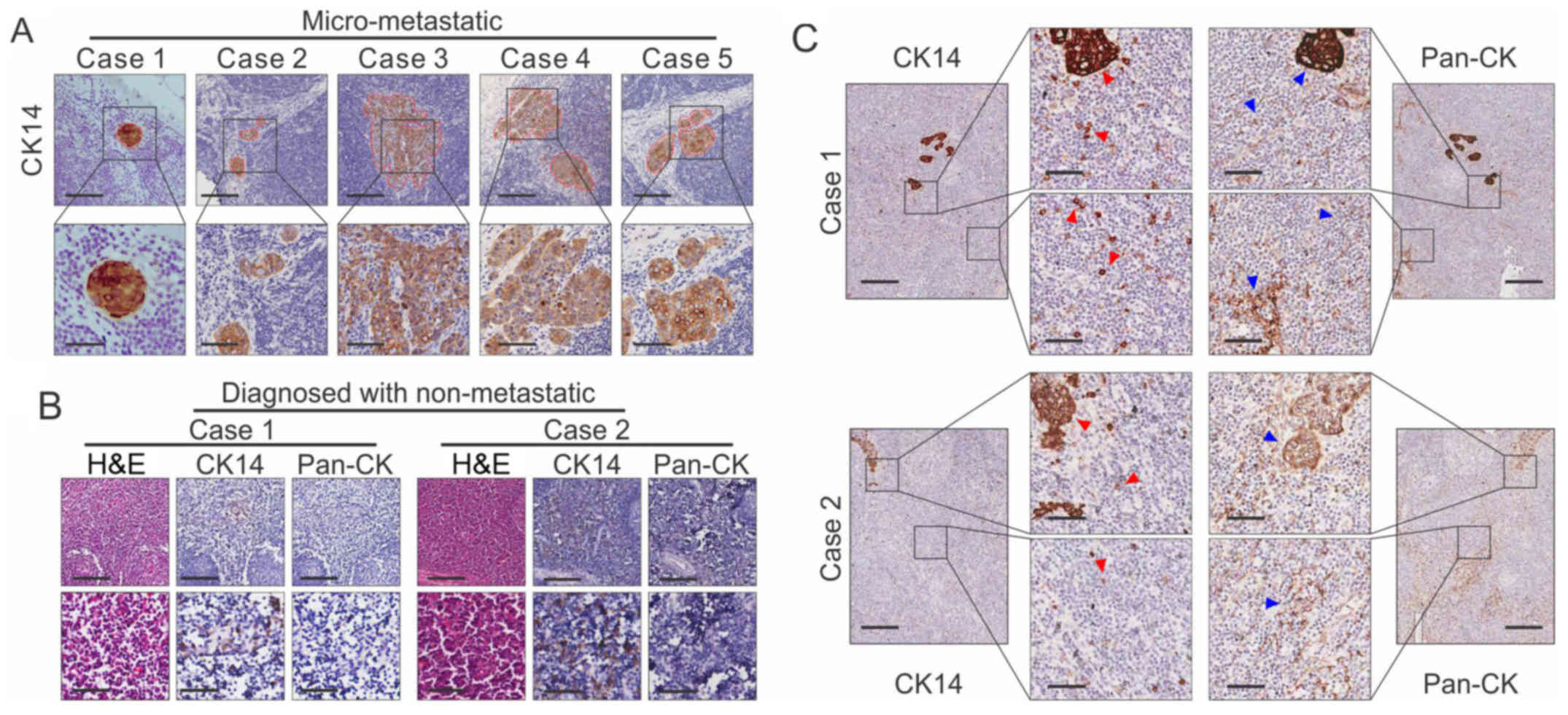
CK14 is a superior predictor of OSCC
lymph node micro-metastases compared with Pan-CK
A previous study indicated that CK14 was upregulated
in oral cancer tissues (15).
Therefore, the present study further evaluated the potential use of
CK14 for the detection of lymph node metastasis. As shown in
Fig. 4A, CK14 was specifically
positive for micrometastasis in all five cases. Pan-CK, an
important predictor of lymph node metastasis in OSCC, was used as a
control. An expanded set of 20 samples was further examined. The
IHC of CK14 was specifically positive for micrometastasis in all
cases and was more specific than Pan-CK (Fig. 4B). In addition, 10 samples diagnosed
as non-metastatic lymph nodes were examined, and it was found that
seven cases were CK14-positive. One of the seven CK14-positive
cases showed recurrence 1 year later (Fig. 4C). Collectively, the results from
the lymph node metastasis mouse model data were consistent with the
clinical results (Fig. 5). These
results indicated the potential value of CK14 as a biomarker for
the detection of lymph node micrometastasis in OSCC.
Discussion
The discrimination of OSCC tissues from
para-cancerous tissues in H&E sections can be difficult due to
the subjectivity of the microscopic analyses, particularly when the
histologic structure is irregularly distributed. The majority of
surgeons use a 0.5–1 cm empirical boundary to expand the excision
of OSCC (16,17). However, in maxillofacial surgery,
this can lead to the irreversible destruction of the facial
appearance, impacting the quality of life of the patients.
Therefore, identifying specific molecular markers to guide the
determination of surgical margins is urgently required (18,19).
A wealth of information has been provided by
intensive research efforts in cancer biomarker identification based
on the LCM-FFPE-proteomic analysis. In previous studies that
focused on the utility of proteomic analysis based on FFPE tissue,
it was suggested that this platform was an effective approach for
biomarker identification in cancer. In the present study, the
expression profiles of a large number of proteins were determined,
revealing significant shifts in expression between normal mucosal
epithelial tissues and pre-cancerous and cancer tissues. A total of
13 proteins were identified with altered expression levels based on
LCM-FFPE-proteomic and LC/MS analyses. Of these, the expression
patterns of CK14, 17, 4 and 10/13 were found to have unique
patterns that may serve as biomarkers for the histologic evaluation
of oral mucosa, particularly when histologic structures are
unclear. The results of the IHC and H&E staining were
consistent, and CK14, 17, 4 and 10/13 staining made it possible to
distinguish between early phenotype changes in tumor cells and
adjacent cells, which are difficult to detect by conventional
H&E staining.
CKs are often used as a source for the
diagnosis/differential diagnosis of epithelial-derived tumors
(20). Several studies have
reported that these expression patterns correlate with the presence
of OSCC and oral epithelial dysplasia. Previous reports have
suggested that the downregulated expression of CK4 and CK13 in OSCC
may cause morphological alterations in oral carcinogenesis and are
relevant biomarkers of OSCC (8,21). The
expression patterns of CK13, 4, 14, and 17 during the development
of OSCC from oral precancerous lesions have also been investigated
(22–24). The results of the present study were
consistent with reports. In previous studies, CK14 was often
expressed in myoepithelial cells and other epithelial cells,
whereas CK10/13 was mainly expressed in the stratified squamous
epithelium (25,26). The results of the statistical
analysis in the present study confirmed the results of these
previous studies, demonstrating that CK14, 17, 4 and 10/13 staining
can distinguish between early phenotypic changes in tumor cells and
adjacent cells, which are difficult to detect by conventional
H&E staining.
H&E staining is the current standard for the
diagnosis of OSCC. However, the discrimination of micrometastasis
in lymph nodes based on H&E-stained sections of OSCC is
difficult. Therefore, the identification of relatively objective
biomarkers that can be used to identify micrometastasis in lymph
nodes is likely to benefit patients with OSCC. The expression of
CK14 is altered in several diseases, including human papillomavirus
infections, lichen planus, psoriasis, inflammation, and metaplasia
(27–30). The data from the clinical samples
and a mouse model in the present study showed for the first time,
to the best of our knowledge, that CK14 has potential value as a
biomarker for the detection of lymph node micrometastasis in OSCC.
Additionally, signals for CK14 were found in a high proportion of
lymph nodes diagnosed as non-metastatic from patients with OSCC who
developed micro-metastases. One of the CK14-positive cases showed
recurrence 1 year later, which may partly explain the significant
correlations between locoregional recurrence and the number of
resected lymph nodes and lymph node ratio of patients with oral
cavity cancer (31). The number of
resected lymph nodes is a prognosticator of multiple types of
cancer, including vulvar cancer (32), gastric cancer (33) and non-small cell lung cancer
(34). The data in the present
study are in agreement with those described previously. Larger
investigations with larger clinical cohorts are required prior to
CK14 being used as a clinical micrometastasis biomarker in the
future.
In conclusion, the data obtained in the present
study indicated that the expression levels of CK10/13/4 or CK14/17
may be considered molecular markers to guide the extent of surgical
resection of OSCC. The major finding of the present study was that
CK14 is a superior predictor of lymph node micro-metastases in
OSCC, performing better than Pan-CK. Taken together, the data
obtained indicated that the platform based on LCM-FFPE-proteomic
analysis is an effective approach for biomarker identification in
OSCC.
Acknowledgements
The authors would like to thank Dr Vyomesh Patel
(National Institutes of Health, National Institute of Dental and
Craniofacial Research, Bethesda, MD, USA), Dr Li-hong Yao, Dr
Ming-zhong Yang and Dr Gao Wu (University of Science and Technology
of China, Anhui, China) for their technical assistance and
advice.
Funding
The present study was supported by the National
Natural Science Foundation of China (grant nos. 81621062, 81321002,
81621062, 81672675 and 81472533), the 111 Project of MOE China
(grant no. B14038), the National Natural Science Foundation of
China-Canadian Institutes of Health Research (grant no. 81010066)
and the Open Foundation of State Key Laboratory of Oral Diseases,
Sichuan University (grant nos. SKLOD201701 and SKLOD201714).
Availability of data and materials
The datasets used during the present study are
available from the corresponding author upon reasonable
request.
Authors' contributions
RW, YZ and NG performed the LCM and the proteomic
analysis; RW, DZ, NJ, YZ, MZ, YY and JL performed the IHC
experiments. XL, QC, NG, XZ, YY and JL conceived, designed,
analyzed and interpreted the data; LZ, YC and NG performed the
pathological diagnosis of the patients. XZ and MZ were responsible
for the tissue collection and management. YZ, LZ and QC wrote and
revised the manuscript. All authors read and approved the
manuscript and agree to be accountable for all aspects of the
research in ensuring that the accuracy or integrity of any part of
the work are appropriately investigated and resolved.
Ethics approval and consent to
participate
The use of animals in the experiments was approved
by the State Key Laboratory of Oral Diseases Research Ethics
Committee. The Institutional Review Board of the West China College
of Stomatology, Sichuan University approved the protocol.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Warnakulasuriya S: Global epidemiology of
oral and oropharyngeal cancer. Oral Oncol. 45:309–316. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Panzarella V, Pizzo G, Calvino F,
Compilato D, Colella G and Campisi G: Diagnostic delay in oral
squamous cell carcinoma: The role of cognitive and psychological
variables. Int J Oral Sci. 6:39–45. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Perez-Ordoñez B, Beauchemin M and Jordan
RC: Molecular biology of squamous cell carcinoma of the head and
neck. J Clin Pathol. 59:445–453. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Messadi DV: Diagnostic aids for detection
of oral precancerous conditions. Int J Oral Sci. 5:59–65. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Warnakulasuriya S, Johnson NW and van der
Waal I: Nomenclature and classification of potentially malignant
disorders of the oral mucosa. J Oral Pathol Med. 36:575–580. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wulfkuhle JD, Liotta LA and Petricoin EF:
Proteomic applications for the early detection of cancer. Nat Rev
Cancer. 3:267–275. 2003. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Schaaij-Visser TB, Bremmer JF, Braakhuis
BJ, Heck AJ, Slijper M, van der Waal I and Brakenhoff RH:
Evaluation of cornulin, keratin 4, keratin 13 expression and grade
of dysplasia for predicting malignant progression of oral
leukoplakia. Oral Oncol. 46:123–127. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hood BL, Conrads TP and Veenstra TD: Mass
spectrometric analysis of formalin-fixed paraffin-embedded tissue:
Unlocking the proteome within. Proteomics. 6:4106–4114. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Sprung RW Jr, Brock JW, Tanksley JP, Li M,
Washington MK, Slebos RJ and Liebler DC: Equivalence of protein
inventories obtained from formalin-fixed paraffin-embedded and
frozen tissue in multidimensional liquid chromatography-tandem mass
spectrometry shotgun proteomic analysis. Mol Cell Proteomics.
8:1988–1998. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yuan Y, Hou X, Feng H, Liu R, Xu H, Gong
W, Deng J, Sun C, Gao Y, Peng J, et al: Proteomic identification of
cyclophilin A as a potential biomarker and therapeutic target in
oral submucous fibrosis. Oncotarget. 7:60348–60365. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Espina V, Wulfkuhle JD, Calvert VS,
VanMeter A, Zhou W, Coukos G, Geho DH, Petricoin EF III and Liotta
LA: Laser-capture microdissection. Nat Protoc. 1:586–603. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Merkley MA, Weinberger PM, Jackson LL,
Podolsky RH, Lee JR and Dynan WS: 2D-DIGE proteomic
characterization of head and neck squamous cell carcinoma.
Otolaryngol Head Neck Surg. 141:626–632. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Pawar H, Maharudraiah J, Kashyap MK,
Sharma J, Srikanth SM, Choudhary R, Chavan S, Sathe G, Manju HC,
Kumar KV, et al: Downregulation of cornulin in esophageal squamous
cell carcinoma. Acta Histochem. 115:89–99. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Shores CG, Yin X, Funkhouser W and
Yarbrough W: Clinical evaluation of a new molecular method for
detection of micrometastases in head and neck squamous cell
carcinoma. Arch Otolaryngol Head Neck Surg. 130:937–942. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Loree TR and Strong EW: Significance of
positive margins in oral cavity squamous carcinoma. Am J Surg.
160:410–414. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Cheng A, Cox D and Schmidt BL: Oral
squamous cell carcinoma margin discrepancy after resection and
pathologic processing. J Oral Maxillofac Surg. 66:523–529. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hosseinpour S, Mashhadiabbas F and Ahsaie
MG: Diagnostic biomarkers in oral verrucous carcinoma: A systematic
review. Pathol Oncol Res. 23:19–32. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Tirelli G, Zacchigna S, Biasotto M and
Piovesana M: Open questions and novel concepts in oral cancer
surgery. Eur Arch Otorhinolaryngol. 273:1975–1985. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Khanom R, Sakamoto K, Pal SK, Shimada Y,
Morita K, Omura K, Miki Y and Yamaguchi A: Expression of basal cell
keratin 15 and keratin 19 in oral squamous neoplasms represents
diverse pathophysiologies. Histol Histopathol. 27:949–959.
2012.PubMed/NCBI
|
|
21
|
Sakamoto K, Aragaki T, Morita K, Kawachi
H, Kayamori K, Nakanishi S, Omura K, Miki Y, Okada N, Katsube K, et
al: Down-regulation of keratin 4 and keratin 13 expression in oral
squamous cell carcinoma and epithelial dysplasia: A clue for
histopathogenesis. Histopathology. 58:531–542. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ohkura S, Kondoh N, Hada A, Arai M,
Yamazaki Y, Sindoh M, Takahashi M, Matsumoto I and Yamamoto M:
Differential expression of the keratin-4, −13, −14, −17 and
transglutaminase 3 genes during the development of oral squamous
cell carcinoma from leukoplakia. Oral Oncol. 41:607–613. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Nobusawa A, Sano T, Negishi A, Yokoo S and
Oyama T: Immunohistochemical staining patterns of cytokeratins 13,
14, and 17 in oral epithelial dysplasia including orthokeratotic
dysplasia. Pathol Int. 64:20–27. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Valach J, Foltan R, Vlk M, Szabo P and
Smetana K Jr: Phenotypic characterization of oral mucosa: What is
normal? J Oral Pathol Med. 46:834–839. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Hansson A, Bloor BK, Haig Y, Morgan PR,
Ekstrand J and Grafström RC: Expression of keratins in normal,
immortalized and malignant oral epithelia in organotypic culture.
Oral Oncol. 37:419–430. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ming XY, Fu L, Zhang LY, Qin YR, Cao TT,
Chan KW, Ma S, Xie D and Guan XY: Integrin α7 is a functional
cancer stem cell surface marker in oesophageal squamous cell
carcinoma. Nat Commun. 7:135682016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Jacques CM, Pereira AL, Maia V, Cuzzi T
and Ramos-e-Silva M: Expression of cytokeratins 10, 13, 14 and 19
in oral lichen planus. J Oral Sci. 51:355–365. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Fay J, Kelehan P, Lambkin H and Schwartz
S: Increased expression of cellular RNA-binding proteins in
HPV-induced neoplasia and cervical cancer. J Med Virol. 81:897–907.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Bech R, Otkjaer K, Birkelund S,
Vorup-Jensen T, Agger R, Johansen C, Iversen L, Kragballe K and
Rømer J: Interleukin 20 protein locates to distinct mononuclear
cells in psoriatic skin. Exp Dermatol. 23:349–352. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Linskey KR, Gimbel DC, Zukerberg LR,
Duncan LM, Sadow PM and Nazarian RM: BerEp4, cytokeratin 14, and
cytokeratin 17 immunohistochemical staining aid in differentiation
of basaloid squamous cell carcinoma from basal cell carcinoma with
squamous metaplasia. Arch Pathol Lab Med. 137:1591–1598. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Safi AF, Grandoch A, Nickenig HJ, Zöller
JE and Kreppel M: The importance of lymph node ratio for
locoregional recurrence of squamous cell carcinoma of the tongue. J
Craniomaxillofac Surg. 45:1058–1061. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Polterauer S, Schwameis R, Grimm C, Macuks
R, Iacoponi S, Zalewski K and Zapardiel I: VULCAN Study
Collaborative Group: Prognostic value of lymph node ratio and
number of positive inguinal nodes in patients with vulvar cancer.
Gynecol Oncol. 147:92–97. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Gholami S, Janson L, Worhunsky DJ, Tran
TB, Squires MH III, Jin LX, Spolverato G, Votanopoulos KI, Schmidt
C, Weber SM, et al: Number of lymph nodes removed and survival
after gastric cancer resection: An analysis from the US gastric
cancer collaborative. J Am Coll Surg. 221:291–299. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Liang W, He J, Shen Y, Shen J, He Q, Zhang
J, Jiang G, Wang Q, Liu L, Gao S, et al: Impact of examined lymph
node count on precise staging and long-term survival of resected
non-small-cell lung cancer: A population study of the us seer
database and a chinese multi-institutional registry. J Clin Oncol.
35:1162–1170. 2017. View Article : Google Scholar : PubMed/NCBI
|