Introduction
Protein diversity is essential for yielding the
remarkable regulatory and functional multiformity observed in human
cells. A prevalent mechanism for protein diversity is the
alternative processing and modification of precursor mRNA (1). Alternative mRNA splicing (AS) is a
regulated process that occurs in >95% of multi-exon human genes
(2,3), resulting in an enormous amount of
flexibility in the regulation of gene function and protein
diversity. Recent extensive genomic and functional studies have
firmly established the critical role of AS in cancer (4–6). The
major mechanism may be the involvement of AS in a full spectrum of
oncogenic processes, including cell proliferation, apoptosis,
hypoxia, angiogenesis, immune escape and metastasis (6,7). In
addition, a transcriptome-wide change in AS programming during
epithelial-mesenchymal transition also serves an important role in
cancer cell invasion and metastasis (8,9).
Gastric cancer (GC) is a major global health threat
and the third leading cause of cancer-associated mortality
worldwide (10). In addition to DNA
sequence alterations (11,12), epigenetic alterations have also been
extensively studied as major players in GC pathogenesis, including
DNA methylation (13), histone
modifications (acetylation and methylation) (14), expression of GC-associated microRNAs
(15) and long non-coding RNAs
(16). However, there have been
relatively limited efforts to explore the mechanism of AS involved
in the pathogenesis and progression of GC.
The Cancer Genome Atlas (TCGA) project provides
abundant sources for the investigation of AS patterns in cancer,
including data on the exon, splice and transcript isoform levels,
available via the Genomic Data Commons (GDC; http://gdc.cancer.gov/). To date, systematic analyses
have been performed to reveal the associations of the copy number
variation, DNA methylation, gene expression and microRNA expression
profiles with the survival of cancer patient (17,18).
However, the majority of RNA sequencing studies focus on
identifying cancer-specific AS events (19,20). A
recent analysis of TCGA RNA sequencing data identified 163
cancer-specific AS events for three cancer types, among which five
were found to be potentially associated with survival in breast
cancer (21). There has been a lack
of studies, however, that comprehensively analyze systematic cancer
survival associated with AS at individual exon resolution,
particularly in GC.
Recently, Armero et al (22) provided the first comprehensive
portrait of global modifications in the cellular RNA splicing
signatures that occur in Epstein-Barr virus (EBV)-associated GC.
Since EBV infection is associated with only 10% of all GC cases
reported worldwide (23), the
present study aimed to systematically analyze the unclassified
GC-specific AS events in the TCGA-stomach adenocarcinoma (STAD)
cohort. Furthermore, survival-associated AS events were identified,
and gene models with clinical parameters were constructed as
prognosis predictors for GC patients.
Materials and methods
TCGA RNA sequencing data and
processing
RNA sequencing read counts and medical information
of GC patients, namely the TCGA-STAD cohort, were downloaded from
TCGA data portal (https://tcga-data.nci.nih.gov/). The TCGA dataset
provided the number of RNA sequencing read counts on the splice
junctions as part of the level 3 RNA sequencing data. As shown in
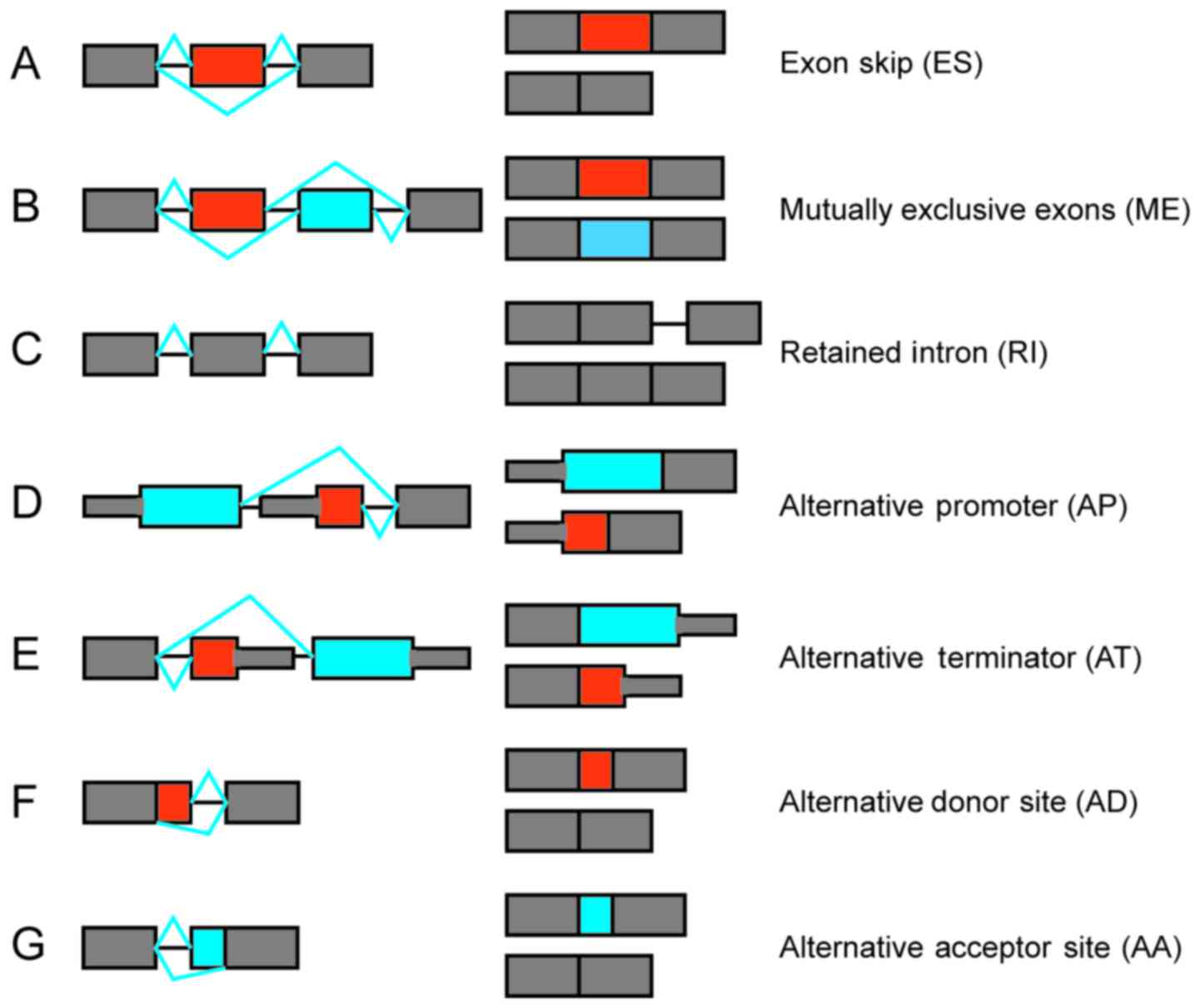
Fig. 1, there are seven types of
splice events, as follows: Exon skipping (ES), mutually exclusive
exons (ME), retained intron (RI), alternative promoter (AP),
alternative terminator (AT), alternative donor site (AD) and
alternative acceptor site (AA). For each ES event defined in the
Ensembl annotations, the read counts were identified for
exon-inclusion and skipping splice junctions.
Cancer specific mRNA splice variations
analysis
SpliceSeq tool (version 2.0) (http://bioinformatics.mdanderson.org/main/SpliceSeq:Methods)
was applied to analyze the mRNA splicing patterns of TCGA-STAD
samples (24), which started with a
reference model for each gene constructed from all the
protein-coding transcripts in the Ensembl database. The percent
spliced in (PSI) index indicates how efficiently sequences of
interest are spliced into transcripts, with values ranging between
0 and 1 (25). The PSI values were
calculated for the seven types of AS events.
To identify possibly abnormal AS events in GC,
comparisons of the PSI values were conducted between GC and normal
samples (415 STAD tumors vs. 37 normal controls). Student's t-test,
followed by an adjustment of the P-value (Benjamini-Hochberg method
for multiple comparisons), was applied, and P<0.001 was
considered to denote a statistically significant difference.
UpSet plot, a novel visualization technique for the
quantitative analysis of interactive sets (26), was also used to analyze the
intersections between seven types of AS in GC.
Survival analysis
Only 373 GC patients with an overall survival (OS)
of ≥30 days were enrolled in the survival analysis. Clinical
covariates contained age, sex and the pathologic T stage, N stage,
M stage and TNM stage (27), as
characterized by TCGA consortium (summarized in Table I). For each AS events, GC patients
were divided into two groups using the median as a cutoff value.
The association between AS events and OS were evaluated by
univariate Cox regression analysis. In addition, multivariate Cox
regression was conducted to remove any genes that may not be
independent factors in the prognostic models. Following the
construction of each multivariate model, a high-risk status for
GC-related death was defined when the right side of the Cox
regression equation was >0, while low risk was defined when the
value in the right side of the equation was <0. Based on the
definition of high and low risk, survival curves were constructed
to evaluate the performance of prognostic models.
 | Table I.Clinical parameters of patients in
the stomach adenocarcinoma cohort obtained from The Cancer Genome
Atlas (n=373). |
Table I.
Clinical parameters of patients in
the stomach adenocarcinoma cohort obtained from The Cancer Genome
Atlas (n=373).
| Parameter | Value |
|---|
| Age
(years)a | 67 (30–90) |
| Sex (male) | 245 |
| T stage |
|
| T1 | 18 |
| T2 | 82 |
| T3 | 169 |
| T4 | 100 |
| TX | 4 |
| N
stageb |
|
| N0 | 111 |
| N1 | 105 |
| N2 | 72 |
| N3 | 75 |
| NX | 9 |
| M stage |
|
| M0 | 338 |
| M1 | 22 |
| MX | 13 |
| TNM
stagec |
|
| I | 50 |
| II | 118 |
|
III | 157 |
| IV | 36 |
Gene Ontology (GO) analysis
GO enrichment analysis (https://david.ncifcrf.gov/) was conducted to explore
the functions of differentially spliced genes identified in the
present study. Furthermore, the Kyoto Encyclopedia of Genes and
Genomes (KEGG) database (http://www.genome.ad.jp/kegg/) was used to assess the
potential functions of these target genes in pathways. Fisher's
exact test was used to determine the GO category and GO annotation
list.
Statistical analysis
All statistical analyses were performed using
R/Bioconductor software (version 3.4.1; http://www.bioconductor.org/). All statistical tests
were two-sided, and P=0.05 was considered as the threshold for a
statistically significant difference, unless otherwise stated.
Results
Integrated mRNA splice variant profile
in the TCGA-STAD cohort
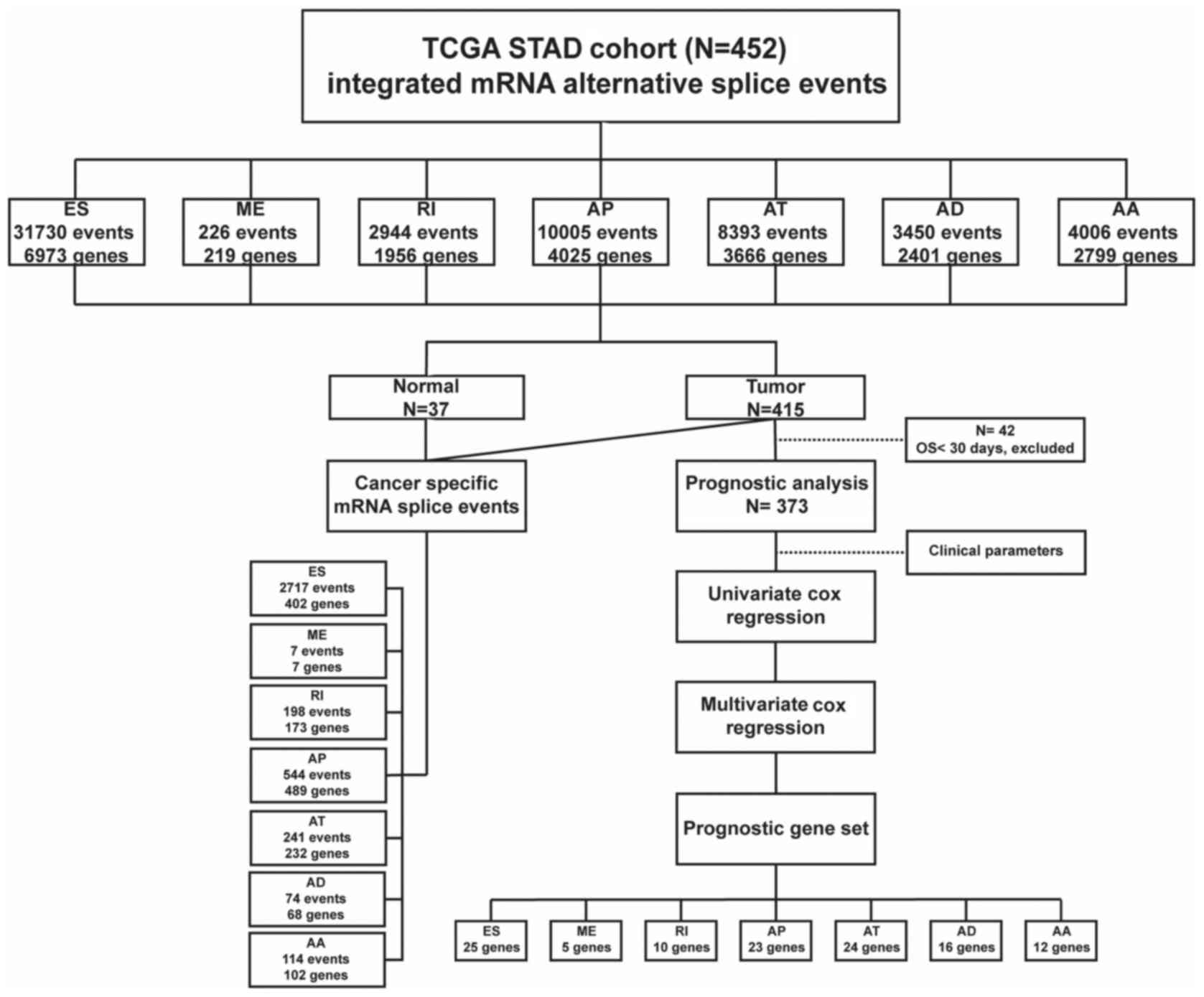
Integrated mRNA splice variant profiles were
explored in depth for 452 tissues in the STAD cohort. In total,
60,754 mRNA splice variants were detected in 10,611 genes,
comprised of 31,730 ES events in 6,973 genes, 226 ME events in 219
genes, 2,944 RI events in 1,956 genes, 10,005 AP events in 4,025
genes, 8,393 AT events in 3,666 genes, 3,450 AD events in 2,401
genes and 4,006 AA events in 2,799 genes (Fig. 2). The findings of the current study
indicated that AS may be universal in human multi-exon genes.
Furthermore, the results demonstrated that over half of the mRNA
splice variants were ES variants.
Analysis of cancer-specific mRNA
splice variants
To discern any differences in mRNA splice variants,
the PSI values of mRNA splice variants in STAD tumor tissues were
compared with those in normal tissues (Fig. 2). Since only 7 ME variants in 7
genes were detected to be differentially spliced in GC tissues
compared with normal tissues, the study focused on the other six
modes of AS for a more detailed analysis. For instance, 2,717 ES
variants that belong to 402 genes were found to be differentially
spliced (P<0.001; fold change >2). Thus, on average, a gene
may have more than one cancer-specific ES event, indicating a
diverse involvement of ES in GC.
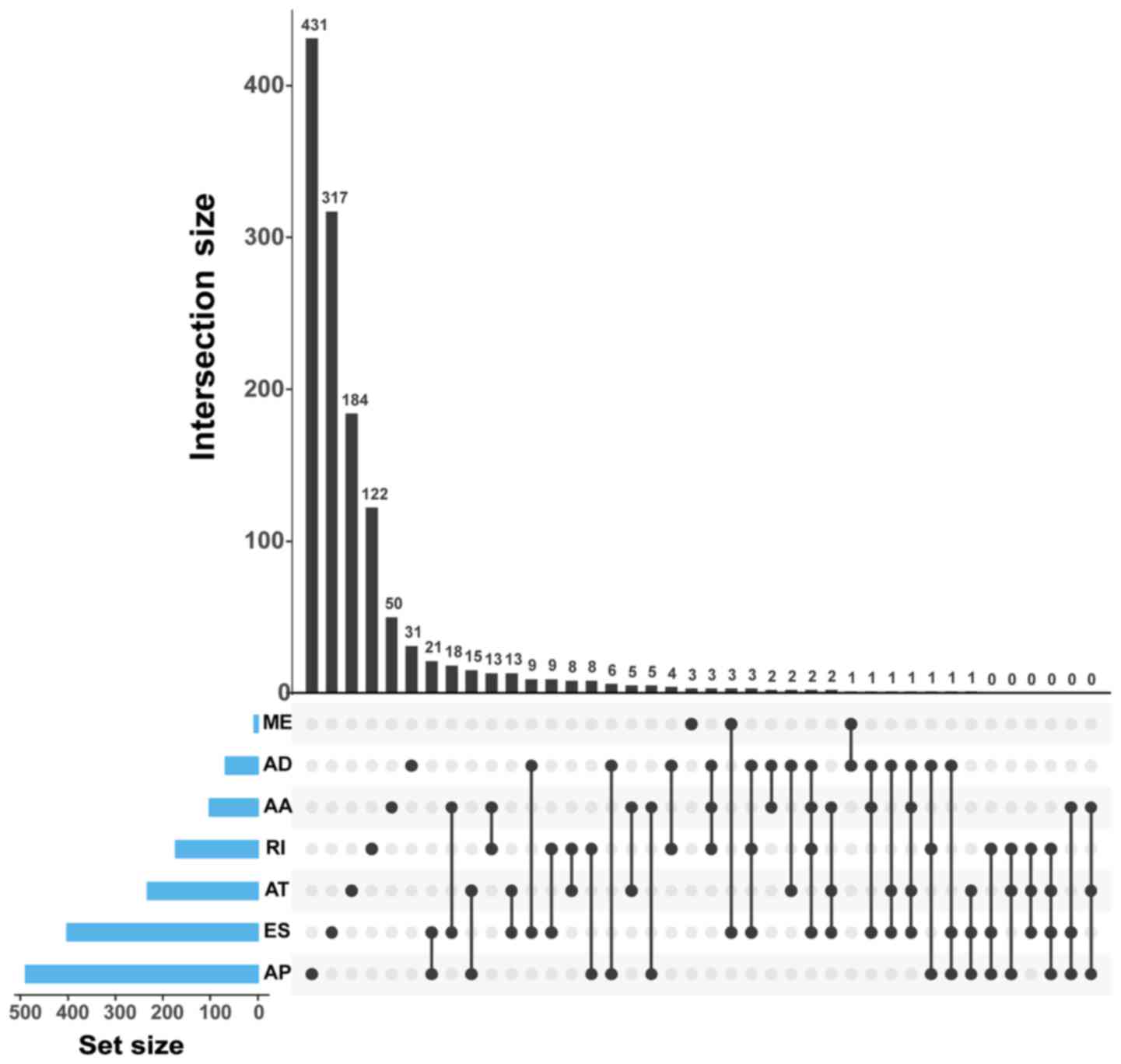
The top ranked GC-specific splice genes were labeled
in the volcano plots, with the exception of ME with only 7 variants
(Fig. 3). Furthermore, it was
observed that there were a few genes that had more than one type of
mRNA splice variant and were differentially spliced in GC. The
intersection of gene sets is visualized in Fig. 4. For instance, RPS6 and UBE2C had
four types of variants (ES, RI, AD and AA) that were differentially
spliced in GC. In addition, the ES, AT, AD and AA variants in gene
RPS3A were also differentially spliced.
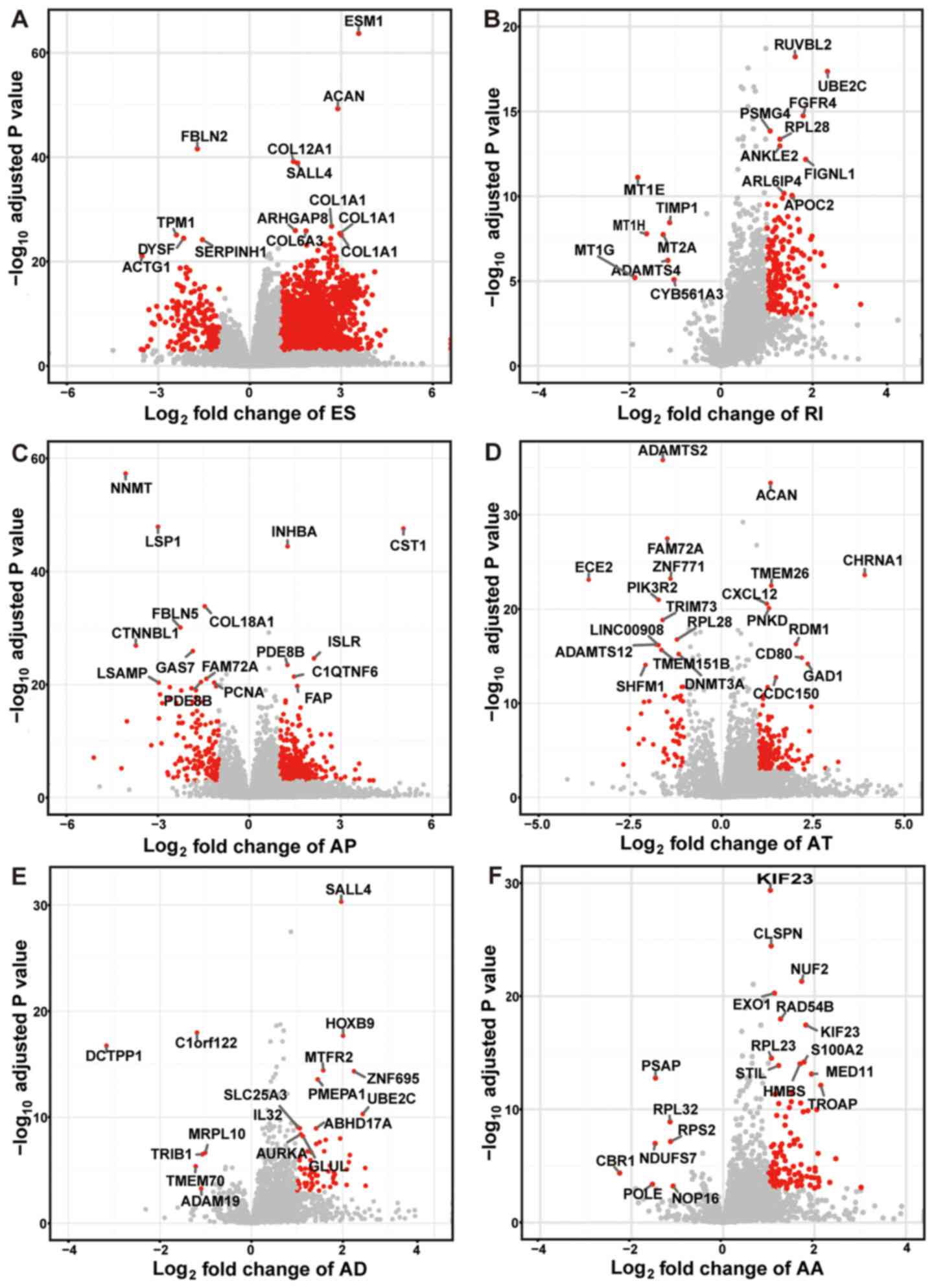 | Figure 3.Volcano plots for the six types of
alternative splicing with top ranked cancer-specific splice genes.
(A) ES, (B) RI, (C) AP, (D) AT, (E) AD and (F) AA event plots are
shown. The y-axis represents the negative log10
P-values, while the x-axis represents the log2 of the
fold change for every type of alternative splicing. The red dots
indicate that alternative splicing variants were found to be
significantly differentially spliced between GC tissues and normal
tissues. The gene names of top ranked cancer-specific splice
variants are shown. AA, alternative acceptor site; AD, alternative
donor site; AP, alternative promoter; AT, alternative terminator;
ES, exon skipping; RI, retained intron. |
GO and KEGG pathway analysis of
cancer-specific differentially spliced genes
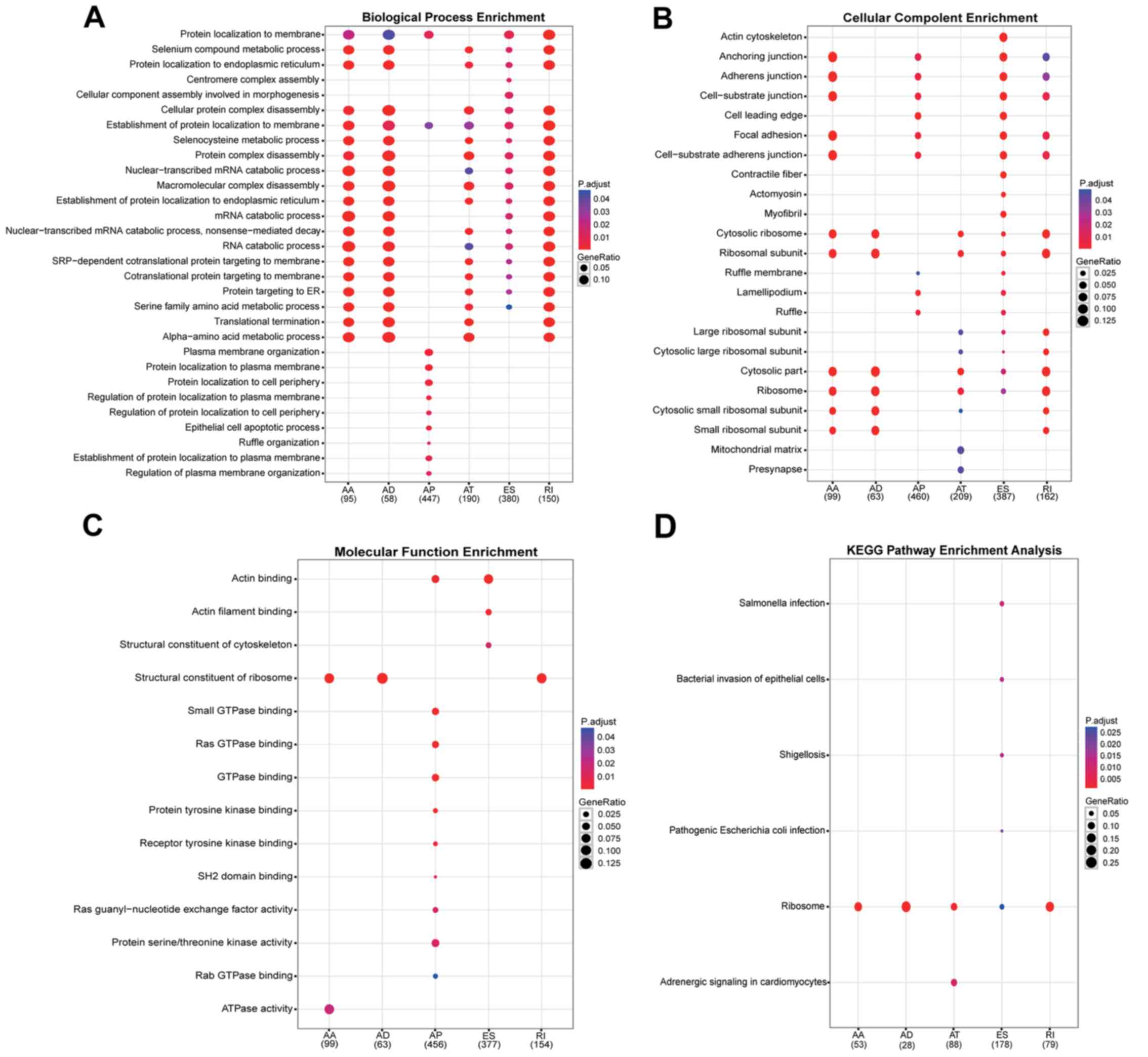
Through GO analysis, genes were organized into
hierarchical categories to uncover gene regulatory networks on the
basis of biological process (excluding the ME events). It was
observed that, among these differentially spliced genes, 1,320 are
involved in biological processes, 1,380 in cellular components and
1,149 in molecular functions (Fig
5A-C).
To further specify and identify target mRNAs
(excluding the ME events), significant pathways of differentially
spliced mRNAs were analyzed using the KEGG database (Fig. 5D). This analysis revealed that
GC-specific ES genes were focused in six pathways, including
Salmonella infection, bacterial invasion of epithelial
cells, shigellosis, pathogenic Escherichia coli infection,
ribosome and adrenergic signaling in cardiomyocytes. Ribosome was
the only significantly enriched network for AA, AD, AT, ES and RI
variants.
Association of OS with mRNA splice
variants in the TCGA-STAD cohort
Univariate survival tests were first conducted to
assess the correlation between clinical parameters and OS in the
STAD cohort. The age, T stage, N stage, M stage and TNM stage were
significantly associated with OS. Next, univariate survival
analysis for OS was conducted using the integrated mRNA splice
variants profiles in the STAD cohort. The univariate survival
analyses identified that 1,168 ES, 16 ME, 138 RI, 675 AP, 434 AT,
186 AD and 170 AA events were statistically associated with OS. The
top 20 genes that were most significantly associated with survival
in the six types of AS are presented in Fig. 6 (excluding the ME events).
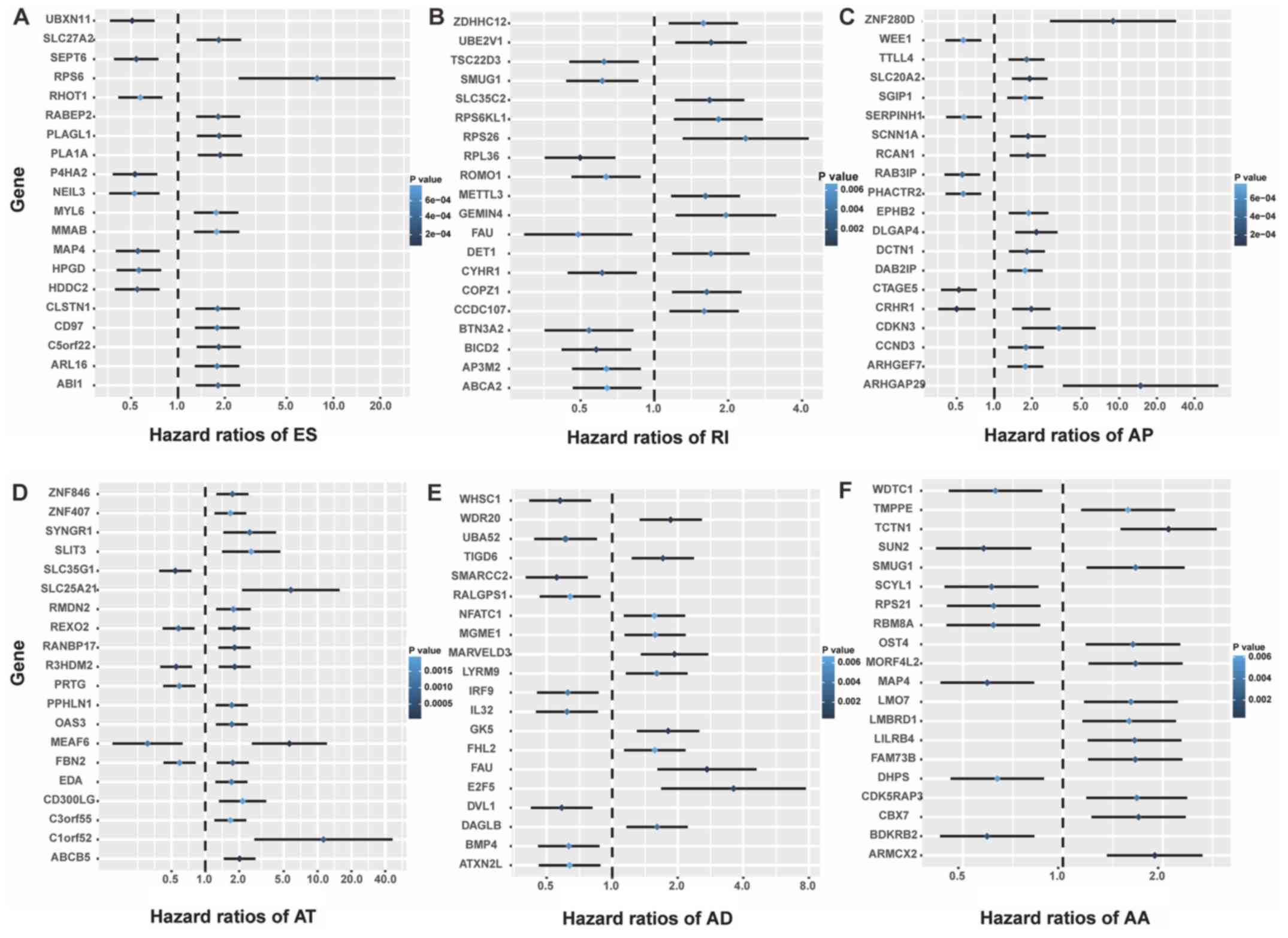 | Figure 6.Forrest plots of hazard ratios for
survival-associated alternative events in stomach adenocarcinoma.
Hazard ratios of the top 20 genes with survival-associated (A) ES,
(B) RI, (C) AP, (D) AT, (E) AD and (F) AA events. AA, alternative
acceptor site; AD, alternative donor site; AP, alternative
promoter; AT, alternative terminator; ES, exon skipping; ME,
mutually exclusive exons; RI, retained intron. |
Integrated STAD risk predictor:
Prognostic gene sets
Subsequent to determining the candidate genes across
the seven types of mRNA splice variants, multivariate Cox
regression was separately applied to the candidate gene sets for
each of the six types of AS (Fig.
2). The ES variants in 25 genes were found to be independent
prognostic factors in the survival of STAD patients. Notably,
MAPKBP1 was one of the most significant independent prognostic
factors [hazard ratio (HR), 116.77; 95% confidence interval (95%
CI), 15.66–870.87; P<0.001]. The age, T stage, N stage and M
stage were also independent prognostic factors in the models.
Similarly, numerous genes were identified as independent prognostic
factors in the other six types of mRNA splice variants. The
specific AS events of genes involved in the final models are listed
in Table II.
 | Table II.List of survival-associated splicing
factors of genes in multivariate Cox analysis. |
Table II.
List of survival-associated splicing
factors of genes in multivariate Cox analysis.
| Alternative
splicing | Genes |
|---|
| AA | CBX7, DHPS, DYNLL1,
LMO7, MORF4L2, RPS21, SCYL1, SHQ1, SMUG1, SUN2, TCTN1, TNIP1 |
| AD | BMP4, C1QC, DAGLB,
E2F5, GK5, IRF9, LYRM9, MARVELD3, MGME1, MT1F, NFATC1, NOB1,
RALGPS1, TAF1D, TXNDC9, WDR20 |
| AP | ALDOA, CCDC64B,
CCND3, CLIP3, CTBP2, DALRD3, DCTN1, HOXB3, IL1R1, KDM2B, KIAA1671,
MICAL2, NEDD1, NFATC2, NNT, NR2F2, PKM, PLAGL1, RTN4, SGIP1, TCF4,
TNFAIP8L1, ZNF544 |
| AT | ABCB5, ABCC5,
ACAD9, C1orf52, CD300LG, CPED1, DYNLL1, EDA, GHR, IL7R, MRPL30,
NOX4, PRTG, REXO2, RMDN2, SLC35G1, SMG8, SPINK5, TSTD2, UQCC1,
WNT9B, ZBTB8OS, ZNF407, ZNF680, ZNF846 |
| ES | ABI1, ARHGAP4,
C14orf80, CD44, EML2, FSTL1, GGA3, GRIPAP1, HPGD, LDB2, MAP4,
MAPKBP1, MEF2B, NCOA7, NFATC3, P4HA2, PLAGL1, RHOT1, RQCD1, SAE1,
SEC16A, SEC31A, SLC27A2, UBA52, WWP2 |
| ME | G3BP1, GRB10,
H2AFY, ZDHHC16, ZNF140 |
| RI | BICD2, BRWD1,
C18orf21, COPZ1, CYHR1, FAU, FOS, METTL3, SMUG1, UBE2V1 |
Based on the aforementioned prognostic models,
Kaplan-Meier survival plots were generated to test the performance
of these models as shown in Fig. 7
(excluding ME). All six mRNA splice variant prognostic models were
significantly associated with the OS of GC patients (all
P<0.05). Furthermore, the prediction models constructed for the
splice variants ES, AP, AT, AD and AA were good, since evident gaps
between the curves of high-risk and low-risk patients were
observed.
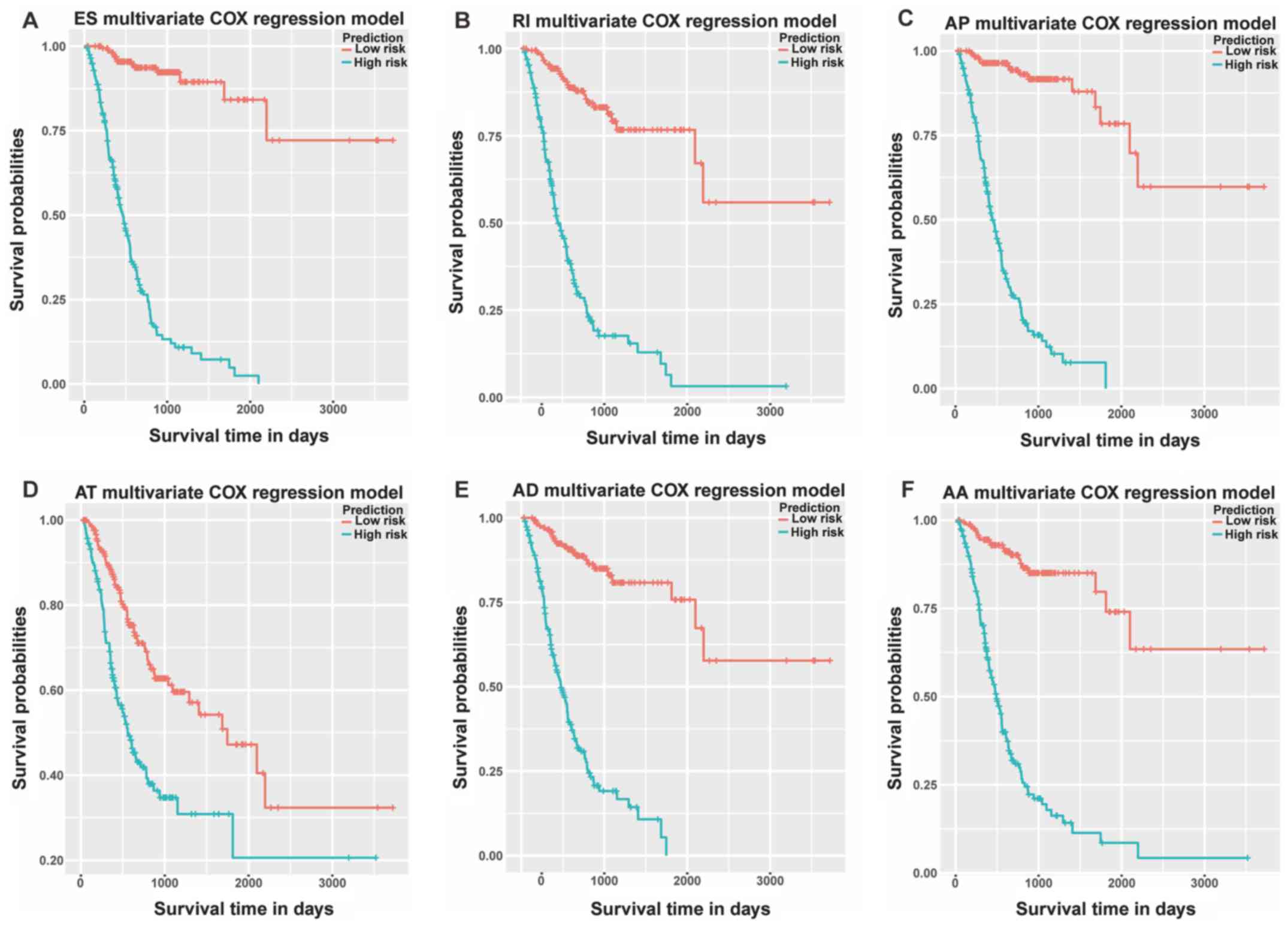 | Figure 7.Kaplan-Meier prognostic predictors
for gastric cancer. Prognostic predictor models were constructed
according to the (A) ES, (B) RI, (C) AP, (D) AT, (E) AD and (F) AA
events in stomach adenocarcinoma. AA, alternative acceptor site;
AD, alternative donor site; AP, alternative promoter; AT,
alternative terminator; ES, exon skipping; ME, mutually exclusive
exons; RI, retained intron. |
Discussion
AS generally allows cells to generate substantial
mRNA and protein isoforms with diverse regulatory and functional
properties, which may also facilitate the survival, proliferation
and metastasis of cancer cells (6,7). The
present study reported a comprehensive transcriptome-wide analysis
of the AS profiling landscape for GC, and identified a significant
association between AS and the GC clinical outcome. Through an
analysis of the RNA-Seq data of individuals in the TCGA-STAD
cohort, a total of 60,754 mRNA splice variants were detected in
10,611 genes. Recently, Tsai et al (21) reported that there were individually
65,152, 70,342 and 70,637 AS events in breast, liver and lung
cancer, respectively, all of which have a slightly higher number of
AS events compared with that reported for GC in the present
study.
Systematic identification and analysis of
cancer-specific AS events was also performed in the current study,
using TCGA-STAD data. The results revealed that there are thousands
of AS events significantly associated with GC, involving multiple
genes whose splicing is known to serve a critical role in cancer
development, such as CD44 (28).
Several studies have demonstrated that polymorphisms in CD44 are
associated with a risk for several types of cancer, including
breast cancer (29), colorectal
cancer (30) and GC (31). In the present study, CD44 was
differentially spliced between GC and normal tissues, and the main
splicing modes were ES and AA. It was also observed that one type
of ES events for CD44 was negatively associated with the survival
in GC. However, the AS mechanisms for CD44 have yet to be
explicitly defined, and the role of different CD44 spliced isoforms
in the development and metastasis of GC should be more clearly
elucidated in future studies. Conversely, several AS events of
genes with well-known roles in cancer development were not
identified by our procedure, possibly due to the highly stringent
inclusion criteria applied in the present study. For instance,
GC-specific AS genes in the present study did not include Bcl-xL,
whose splicing is known to control cell apoptosis in multiple
tumors (32). In addition, CDH1 was
also not identified as a GC-specific AS gene, yet CDH1 exon 11
skipping has been reported in leukemia cells, and head and neck
cancer cells (33,34), while a fragment lacking the final 83
base pairs of exon 8 in CDH1 was frequently detected in Chinese GC
patients (35). The fold change of
Bcl-xL and CDH1 was not, however, large enough to pass the
significance thresholds in the present analysis. It is expected
that additional cancer-specific genes could be identified if these
criteria are relaxed. Furthermore, the P-value of AS events was
required to be <0.001, which may cause certain uncommon events
to be omitted in the current study.
Notably, RPS6 and UBE2C were found to have four
splicing modes (ES, RI, AD and AA) of variants that were
differentially spliced in GC. RPS6, a component of the 40S
ribosomal subunit, is regarded to be involved in the translation of
specific mRNAs, as well as a regulator of cellular metabolisms,
survival and proliferation (36).
Several studies have reported the overexpression and activation of
RPS6 in esophageal squamous cell carcinoma (37), lung cancer (38) and breast cancer (39). Furthermore, the present study
demonstrated that increased ES events in RPS6 were associated with
poor prognosis. UBE2C encodes a member of the E2
ubiquitin-conjugating enzyme family, which is involved in the
destruction of mitotic cyclins and cell cycle progression (40). Overexpression of UBE2C is associated
with worse clinical outcomes in a number of cancer types, including
GC (41). MAPKBP1, acting as a
scaffold protein, facilitates the polyubiquitination of
TNF-receptor associated factor 2, leading to the TGF-β-activated
kinase 1-mediated activation of nuclear factor-κB (42,43).
The results of the current study also revealed that MAPKBP1 was one
of the most significant independent prognostic factors. Fu et
al (44) reported the
prognostic relevance of MAPKBP1 expression in cytogenetically
normal acute myeloid leukemia. This evidence probably suggests the
functional and clinical relevance of these genes in the
survival-associated AS events. While certain of these events may
reflect a correlation but may not have a causal effect on the
survival of GC patients, others may serve an active role in
regulating the cancer cell phenotypes, cancer development and
cancer prognosis. Therefore, AS events for these marked genes
should be investigated further to elucidate their impact on the
development and prognosis of GC.
Currently, AS has been considered to be generally
regulated by multiple cis-acting splicing regulatory elements that
are specifically bound by trans-acting splicing factors to
enhance or inhibit the use of nearby splice sites (45,46).
AS events can respond not only to various signaling pathways that
target splicing mechanisms, but can also be influenced by
transcription factors and chromatin structure (47). Small interfering RNAs have been
reported to trigger AS events in human cells through
transcriptional gene silencing pathways (48). MicroRNAs can also serve an important
role in the regulation of AS. For instance, miR-124 promotes
neuronal differentiation by targeting the PTBP1 splicing repressor
(49). Shukla et al
(50) reported that DNA methylation
can regulate AS in CD45 exon 5. Furthermore, the expression of
numerous cellular splicing factors has been reported to be
dysregulated in various human diseases. Armero et al
(22) detected several changes in
the expression levels and/or splicing patterns of cellular splicing
factors in EBV-negative GC tissues, including CDK10, MBNL1 and
RBFOX2. Notably, recent data demonstrated a key role for RBFOX2 and
MBNL1, which seem to account for numerous splicing alterations in
breast, lung and prostate cancer (51). The current study also identified
that ES events in RBFOX2 and MBNL1 were significantly different in
GC samples. Although a few known factors regulating AS events can
be listed, it is likely that there are numerous other regulators
awaiting identification and validation.
In conclusion, the present study generated a common
set of cancer-specific and survival-associated AS events in GC.
Furthermore, the results demonstrated that a core set of AS events
can be potentially applied to build predictive models for the
survival of GC patients with a higher accuracy. This further
demonstrates the potential biological relevance and clinical
utility of these identified AS events. Ultimately, certain RNA
splicing isoforms may eventually be used as diagnostic or
prognostic biomarkers, and even as novel therapeutic targets.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used or analysed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
YS and GF were involved in the design of the study.
YS, JG, SW and HG conducted the statistical analysis and compiled
the figures. ZC collected the original data. All authors read and
approved the final manuscript and agreed to be accountable for all
aspects of the work in ensuring that questions related to the
accuracy or integrity of any part of the work are appropriately
investigated and resolved.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no conflict of
interest.
Glossary
Abbreviations
Abbreviations:
|
AA
|
alternative acceptor site
|
|
AD
|
alternative donor site
|
|
AP
|
alternative promoter
|
|
AS
|
alternative mRNA splicing
|
|
AT
|
alternative terminator
|
|
ES
|
exon skipping
|
|
GC
|
gastric cancer
|
|
GO
|
Gene Ontology
|
|
ME
|
mutually exclusive exons
|
|
OS
|
overall survival
|
|
PSI
|
percent spliced in
|
|
RI
|
retained intron
|
|
STAD
|
stomach adenocarcinoma
|
|
TCGA
|
The Cancer Genome Atlas
|
References
|
1
|
Nilsen TW and Graveley BR: Expansion of
the eukaryotic proteome by alternative splicing. Nature.
463:457–463. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wang ET, Sandberg R, Luo S, Khrebtukova I,
Zhang L, Mayr C, Kingsmore SF, Schroth GP and Burge CB: Alternative
isoform regulation in human tissue transcriptomes. Nature.
456:470–476. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Pan Q, Shai O, Lee LJ, Frey BJ and
Blencowe BJ: Deep surveying of alternative splicing complexity in
the human transcriptome by high-throughput sequencing. Nat Genet.
40:1413–1415. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
4
|
Venables JP: Aberrant and alternative
splicing in cancer. Cancer Res. 64:7647–7654. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kim E, Goren A and Ast G: Insights into
the connection between cancer and alternative splicing. Trends
Genet. 24:7–10. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Oltean S and Bates DO: Hallmarks of
alternative splicing in cancer. Oncogene. 33:5311–5318. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Liu S and Cheng C: Alternative RNA
splicing and cancer. Wiley Interdiscip Rev RNA. 4:547–566. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
De Craene B and Berx G: Regulatory
networks defining EMT during cancer initiation and progression. Nat
Rev Cancer. 13:97–110. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Warzecha CC, Jiang P, Amirikian K, Dittmar
KA, Lu H, Shen S, Guo W, Xing Y and Carstens RP: An ESRP-regulated
splicing programme is abrogated during the epithelial-mesenchymal
transition. EMBO J. 29:3286–3300. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hudler P: Genetic aspects of gastric
cancer instability. Sci World J. 2012:7619092012. View Article : Google Scholar
|
|
12
|
Tao J, Deng NT, Ramnarayanan K, Huang B,
Oh HK, Leong SH, Lim SS, Tan IB, Ooi CH, Wu J, et al: CD44-SLC1A2
gene fusions in gastric cancer. Sci Transl Med. 3:77ra302011.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zouridis H, Deng N, Ivanova T, Zhu Y, Wong
B, Huang D, Wu YH, Wu Y, Tan IB, Liem N, et al: Methylation
subtypes and large-scale epigenetic alterations in gastric cancer.
Sci Transl Med. 4:156ra1402012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Calcagno DQ, Gigek CO, Chen ES, Burbano RR
and Smith MA: DNA and histone methylation in gastric
carcinogenesis. World J Gastroenterol. 19:1182–1192. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wu WK, Lee CW, Cho CH, Fan D, Wu K, Yu J
and Sung JJ: MicroRNA dysregulation in gastric cancer: A new player
enters the game. Oncogene. 29:5761–5771. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Li T, Mo X, Fu L, Xiao B and Guo J:
Molecular mechanisms of long noncoding RNAs on gastric cancer.
Oncotarget. 7:8601–8612. 2016.PubMed/NCBI
|
|
17
|
Ludwig JA and Weinstein JN: Biomarkers in
cancer staging, prognosis and treatment selection. Nat Rev Cancer.
5:845–856. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yuan Y, Van Allen EM, Omberg L, Wagle N,
Amin-Mansour A, Sokolov A, Byers LA, Xu Y, Hess KR, Diao L, et al:
Assessing the clinical utility of cancer genomic and proteomic data
across tumor types. Nat Biotechnol. 32:644–652. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li XW, Shi BY, Yang QL, Wu J, Wu HM, Wang
YF, Wu ZJ, Fan YM and Wang YP: Epigenetic regulation of CDH1 exon 8
alternative splicing in gastric cancer. BMC Cancer. 15:9542015.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Hafezi N, Ajami A, Farazmandfar T,
Hosseini V, Alizadeh-Navaei R and Tehrani M: Increased expression
of two alternative spliced variants of CD1d molecule in human
gastric cancer. Iran J Immunol. 12:129–140. 2015.PubMed/NCBI
|
|
21
|
Tsai YS, Dominguez D, Gomez SM and Wang Z:
Transcriptome-wide identification and study of cancer-specific
splicing events across multiple tumors. Oncotarget. 6:6825–6839.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Armero VE, Tremblay MP, Allaire A,
Boudreault S, Martenon-Brodeur C, Duval C, Durand M, Lapointe E,
Thibault P, Tremblay-Létourneau M, et al: Transcriptome-wide
analysis of alternative RNA splicing events in Epstein-Barr
virus-associated gastric carcinomas. PLoS One. 12:e01768802017.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Uozaki H and Fukayama M: Epstein-Barr
virus and gastric carcinoma - viral carcinogenesis through
epigenetic mechanisms. Int J Clin Exp Pathol. 1:198–216.
2008.PubMed/NCBI
|
|
24
|
Ryan M, Wong WC, Brown R, Akbani R, Su X,
Broom B, Melott J and Weinstein J: TCGASpliceSeq a compendium of
alternative mRNA splicing in cancer. Nucleic Acids Res.
44:D1018–D1022. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ryan MC, Cleland J, Kim R, Wong WC and
Weinstein JN: SpliceSeq: A resource for analysis and visualization
of RNA-Seq data on alternative splicing and its functional impacts.
Bioinformatics. 28:2385–2387. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lex A, Gehlenborg N, Strobelt H, Vuillemot
R and Pfister H: UpSet: Visualization of intersecting sets. IEEE
Trans Vis Comput Graph. 20:1983–1992. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Sobin LH, Gospodarowicz MK and Wittekind
C: TNM classification of malignant tumours. 7th edition.
Wiley-Blackwell; Oxford: pp. 3102009
|
|
28
|
Brown RL, Reinke LM, Damerow MS, Perez D,
Chodosh LA, Yang J and Cheng C: CD44 splice isoform switching in
human and mouse epithelium is essential for epithelial-mesenchymal
transition and breast cancer progression. J Clin Invest.
121:1064–1074. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
29
|
Jiang L, Deng J, Zhu X, Zheng J, You Y, Li
N, Wu H, Lu J and Zhou Y: CD44 rs13347 C>T polymorphism predicts
breast cancer risk and prognosis in Chinese populations. Breast
Cancer Res. 14:R1052012. View
Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wu XM, Yang HG, Zheng BA, Cao HF, Hu ZM
and Wu WD: Functional genetic variations at the microRNA
binding-site in the CD44 gene are associated with risk of
colorectal cancer in Chinese populations. PLoS One.
10:e01275572015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Suenaga M, Yamada S, Fuchs BC, Fujii T,
Kanda M, Tanaka C, Kobayashi D, Fujiwara M, Tanabe KK and Kodera Y:
CD44 single nucleotide polymorphism and isoform switching may
predict gastric cancer recurrence. J Surg Oncol. 112:622–628. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Adams JM and Cory S: The Bcl-2 apoptotic
switch in cancer development and therapy. Oncogene. 26:1324–1337.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Sharma S and Lichtenstein A: Aberrant
splicing of the E-cadherin transcript is a novel mechanism of gene
silencing in chronic lymphocytic leukemia cells. Blood.
114:4179–4185. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Sharma S, Liao W, Zhou X, Wong DT and
Lichtenstein A: Exon 11 skipping of E-cadherin RNA downregulates
its expression in head and neck cancer cells. Mol Cancer Ther.
10:1751–1759. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Li X, Gao Y, Pan Y, Pan Y, Wang L, Xiao N,
He Q, Fan Y and Wang Y: Mutation screen and RNA analysis disclose
the changed splicing of the E-cadherin transcription in gastric
cancer. Fam Cancer. 12:547–554. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Ruvinsky I and Meyuhas O: Ribosomal
protein S6 phosphorylation: From protein synthesis to cell size.
Trends Biochem Sci. 31:342–348. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Kim SH, Jang YH, Chau GC, Pyo S and Um SH:
Prognostic significance and function of phosphorylated ribosomal
protein S6 in esophageal squamous cell carcinoma. Mod Pathol.
26:327–335. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Chen B, Tan Z, Gao J, Wu W, Liu L, Jin W,
Cao Y, Zhao S, Zhang W, Qiu Z, et al: Hyperphosphorylation of
ribosomal protein S6 predicts unfavorable clinical survival in
non-small cell lung cancer. J Exp Clin Cancer Res. 34:1262015.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Akar U, Ozpolat B, Mehta K,
Lopez-Berestein G, Zhang D, Ueno NT, Hortobagyi GN and Arun B:
Targeting p70S6K prevented lung metastasis in a breast cancer
xenograft model. Mol Cancer Ther. 9:1180–1187. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Pagano M: Cell cycle regulation by the
ubiquitin pathway. FASEB J. 11:1067–1075. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Xie C, Powell C, Yao M, Wu J and Dong Q:
Ubiquitin-conjugating enzyme E2C: A potential cancer biomarker. Int
J Biochem Cell Biol. 47:113–117. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Lee SW, Han SI, Kim HH and Lee ZH:
TAK1-dependent activation of AP-1 and c-Jun N-terminal kinase by
receptor activator of NF-kappaB. J Biochem Mol Biol. 35:371–376.
2002.PubMed/NCBI
|
|
43
|
Yamaguchi T, Miyashita C, Koyano S, Kanda
H, Yoshioka K, Shiba T, Takamatsu N and Ito M: JNK-binding protein
1 regulates NF-kappaB activation through TRAF2 and TAK1. Cell Biol
Int. 33:364–368. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Fu L, Shi J, Hu K, Wang J, Wang W and Ke
X: Mitogen-activated protein kinase binding protein 1 (MAPKBP1) is
an unfavorable prognostic biomarker in cytogenetically normal acute
myeloid leukemia. Oncotarget. 6:8144–8154. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Matera AG and Wang Z: A day in the life of
the spliceosome. Nat Rev Mol Cell Biol. 15:108–121. 2014.
View Article : Google Scholar :
|
|
46
|
Matlin AJ, Clark F and Smith CW:
Understanding alternative splicing: Towards a cellular code. Nat
Rev Mol Cell Biol. 6:386–398. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Naftelberg S, Schor IE, Ast G and
Kornblihtt AR: Regulation of alternative splicing through coupling
with transcription and chromatin structure. Annu Rev Biochem.
84:165–198. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Alló M, Buggiano V, Fededa JP, Petrillo E,
Schor I, de la Mata M, Agirre E, Plass M, Eyras E, Elela SA, et al:
Control of alternative splicing through siRNA-mediated
transcriptional gene silencing. Nat Struct Mol Biol. 16:717–724.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Makeyev EV, Zhang J, Carrasco MA and
Maniatis T: The MicroRNA miR-124 promotes neuronal differentiation
by triggering brain-specific alternative pre-mRNA splicing. Mol
Cell. 27:435–448. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Shukla S, Kavak E, Gregory M, Imashimizu
M, Shutinoski B, Kashlev M, Oberdoerffer P, Sandberg R and
Oberdoerffer S: CTCF-promoted RNA polymerase II pausing links DNA
methylation to splicing. Nature. 479:74–79. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Danan-Gotthold M, Golan-Gerstl R,
Eisenberg E, Meir K, Karni R and Levanon EY: Identification of
recurrent regulated alternative splicing events across human solid
tumors. Nucleic Acids Res. 43:5130–5144. 2015. View Article : Google Scholar : PubMed/NCBI
|