Introduction
Colorectal cancer (CRC) is the third most commonly
diagnosed cancer in males, and the second most common in females
worldwide (1). The highest
incidence rates are mainly in developed countries and the incidence
is increasing in developing countries, which is partly attributable
to lipid metabolism (1–3). Despite an enormous amount of effort
spent in the development of therapies to treat CRC, the 5-year
survival rate has increased slowly in the USA, from 60% during the
1980s to 66% during 2005 to 2011 (4). To a large extent, this discouraging
observation is largely due to the fact that patients with CRC
demonstrate resistance to both new and older anticancer drugs
(3,5).
Three-dimensional (3D) cell culture models provide
an optimal experimental system to study the mechanisms of
anticancer drug resistance, as they represent a suitable in
vivo approximation of solid tumor tissue microenvironment,
including cell adhesion (3,6,7). The
formation and stability of cell adhesion, including cell-cell
adhesion and cell-extracellular matrix (ECM) adhesion, rely on cell
adhesion molecules, particularly the integrin family (6,8,9).
Conversely, cell adhesion triggers certain integrin signaling
cascades, and influences tumor cell biological behavior, including
progression, proliferation, survival and chemosensitivity (6,9–11).
Accumulating evidence has demonstrated that the expression of
integrin is negatively correlated with prognosis in multiple cancer
types (9,10,12).
However, the underlying mechanism remains unclear (9,10,12).
In the present study, 3D cultures were used to
explore the role of integrin β4 in the response of human CRC cells
to platinum. Our data demonstrated that integrin β4 reduced DNA
damage-induced p53 activation in CRC.
Materials and methods
Cell lines and cell culture
HCT116 and LoVo, two human CRC cell lines that
contain stabilized wild-type p53 protein (3), and the cell line 293T were obtained
from the Cell Bank of the Chinese Academy of Sciences (Shanghai,
China).
Two-dimensional (2D) cultures were grown and
passaged routinely as previously described (3). In brief, HCT116 cells were grown in
McCoy's 5A (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA,
USA, F12K (Gibco Thermo Fisher Scientific, Inc.) for LoVo cells or
DMEM (Gibco) for 293T cells, respectively supplemented with 100
ml/l newborn calf serum (Gibco Thermo Fisher Scientific, Inc.),
100,000 IU/l penicillin and 100 µg/ml streptomycin (Gibco Thermo
Fisher Scientific, Inc.) under a humidified atmosphere of 5%
CO2 at 37°C.
Subsequently, 3D cultures were prepared without the
use of any extracellular components as previously described
(3,6,13). In
brief, plates were coated with poly-2-hydroxyethylmethacrylate
(Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) as previously
described (13). Exponentially
growing CRC cells were inoculated into the plates. The plates were
then gently horizontally swirled for 2 min every 4 h for the first
24 h, then 6 min every 8 h (3,6). The
cells were incubated under a humidified atmosphere of 5%
CO2 at 37°C. Half of the medium was replaced every
day.
Preparation of slides for scanning
electron microscopy
Sample slides were routinely prepared. In brief, 3D
cultures were fixed in 2.5% glutaraldehyde. Samples were then sent
to The Medical Research Center of The Third Military Medical
University for preparation. The sections were imaged using a
scanning electron microscope (S3400N II; Hitachi, Tokugawa,
Japan).
Hematoxylin and eosin (H&E)
staining
HCT116 3D cultures were fixed in 4%
paraformaldehyde, and OCT-embedded samples were sectioned at a
thickness of 10 µm. Sample slides were routinely stained with
H&E (3).
Preparation of slides for transmission
electron microscopy
Sample slides were routinely prepared as previously
described (3). In brief, 3D
cultures were fixed in 2.5% glutaraldehyde and then in 1% osmium
tetroxide. Samples were dehydrated and ultrathin sections were
generated. The sections were stained with uranium acetate and lead
citrate and were observed using a transmission electron microscope
(Tecnai 10; Philips, Amsterdam, The Netherlands).
Western blot analysis
Western blotting was performed as previously
described (3,6). Cells were washed with PBS and lysed in
2X SDS loading buffer [0.1 M Tris-HCl (pH 6.8), 0.2 M DTT, 4% SDS,
20% glycerol and 0.2% bromophenol blue] with Protease Inhibitor
Cocktail, Phosphatase Inhibitor Cocktail 2 and Phosphatase
Inhibitor Cocktail 3 (all from Sigma-Aldrich; Merck KGaA) for 5 min
on ice, inverting the tube. Following sonication, protein was
quantitated using the RC DC protein assay (Bio-Rad Laboratories,
Hercules, CA, USA) according to the manufacturer's
instructions.
The protein was resolved by SDS/PAGE and blotted on
nitrocellulose membranes (Bio-Rad Laboratories). The nitrocellulose
membranes were incubated with specific primary antibodies overnight
at 4°C. Following incubation with secondary antibodies for 90 min
at 37°C, immunoreactive proteins were visualized using the Enhanced
Chemiluminescent Substrate (Thermo Fisher Scientific, Inc.).
Primary antibodies against integrin β4 (1:500; cat.
no. 14803), p53 (1:1,000; cat. no. 2524), phospho-p53 (Ser15)
(p-p53; 1:500; cat. no. 9286), glyceraldehyde 3-phosphate
dehydrogenase (GAPDH) (1:1,000; cat. no. 5174) and HRP-linked
secondary antibodies (anti-mouse IgG; 1:5,000; cat. no. 7076;
anti-rabbit IgG; 1:5,000; cat. no. 7074) were purchased from Cell
Signaling Technology, Inc. (Danvers, MA, USA).
Lentiviral delivery of small hairpin
(sh)RNA
Integrin β4 and p53 were knocked down by lentiviral
vector-mediated shRNA interference using The RNAi Consortium system
(Open Biosystems, Inc., Huntsville, AL, USA) according to the
manufacturer's instructions (3). In
brief, an integrin β4 or p53-targeting shRNA-pLKO.1 vector or a
control shRNA-pLKO.1 vector, with the packaging plasmid pCMV-Dr8.91
and the enveloping plasmid pCMV-VSV-G, was co-transfected into 293T
cells using Lipofectamine® 2000 (Invitrogen; Thermo
Fisher Scientific, Inc.) according to the manufacturer's
instructions (3). Virus-containing
medium was collected at 48 and 72 h post-transfection and was
filtered using a 0.22-µm filter (EMD Millipore, Billerica, MA,
USA). Cells were infected with the lentivirus, and then were
selected using puromycin (Sigma-Aldrich; Merck KGaA). Control shRNA
(shcontrol) was targeted against green fluorescent protein, and the
sense sequence of this shRNA is TACAACAGCCACAACGTCTAT (3). Sense sequences of shRNAs targeting
specific genes were GAGGGTGTCATCACCATTGAA (shβ4-1) (14) or ACGATGACAACCGACCTATTG (shβ4-2) for
integrin β4, and GACTCCAGTGGTAATCTACT for p53 (3). Knockdown efficiency was confirmed by
western blotting.
Immunohistochemical staining
HCT116 3D cultures were fixed in 4%
paraformaldehyde, and OCT-embedded samples were sectioned at a
thickness of 10 µm. Immunohistochemistry was performed according to
the protocol of the SPlink Detection kits (ZSGB-Bio, Beijing,
China), as previously described (3).
Immunofluorescence staining
Immunofluorescence staining was performed as
previously described (7). HCT116
cells were grown as 2D cultures on cover slides, and then treated
with 2.5 µg/ml cisplatin (CDDP; Sigma-Aldrich; Merck KGaA) for 24
h. Following fixation in 4% paraformaldehyde for 30 min, cells were
incubated in 0.2% Triton X-100 in 2% BSA/PBS for 30 min.
Subsequently, cells were incubated in p53 (Cell Signaling
Technology, Inc.; 1:500) antibody solution and Alexa
Fluor® 555 goat anti-mouse (Invitrogen; Thermo Fisher
Scientific, Inc.; 1:1,000) solution for 2 h and 30 min,
respectively. The nuclei were stained by incubating with
4′,6-diamidino-2-phenylindole (DAPI, 1 µg/ml) (Sigma-Aldrich; Merck
KGaA) for 30 min.
Water-soluble tetrazolium salt (WST)
assay
3D cultures were treated with 10 µg/ml CDDP or 5
µg/ml oxaliplatin (L-OHP) (Sigma-Aldrich; Merck KGaA) for 48 h.
Control cultures received 10 µl PBS only. Cell viability was
assayed using WST
[2-(2-methoxy-4-nitrophenyl)-3-(4-nitrophenyl)-5-(2,4-disulfophenyl)-2H-tetrazolium,
monosodium salt], as previously described (3,7). The
WST assay was performed using Cell Counting Kit-8 (CCK-8; Dojindo
Laboratories, Kumamoto, Japan) according to the manufacturer's
instructions (3,7). In brief, 3D cultures detached using
accutase (Non-enzyme Cell Detach Solution; Applygen Technologies,
Beijing, China) were incubated with WST/media for 3–4 h, after
which the absorbance at 450 nm was determined using a microplate
reader with a reference wavelength of 650 nm. Cell viability was
normalized to the control.
Clonogenic assay
Clonogenic assay was performed as previously
described (3,7). In brief, 3D cultures with the same
numbers of cells were treated with 10 µg/ml CDDP for 48 h. Then,
the 3D cultures were detached as single-cell suspensions using
accutase, and the same ratio was inoculated into 24-well plates.
The cells were grown for 7 days and were subsequently stained with
crystal violet and counted using a stereomicroscope and an
automatic ‘counting colony counter pen’.
Statistical analysis
The data shown represent the mean ± standard error.
Statistical differences between groups were analyzed by Student's
t-test or one-way ANOVA. P<0.05 was considered to indicate a
statistically significant difference.
Results
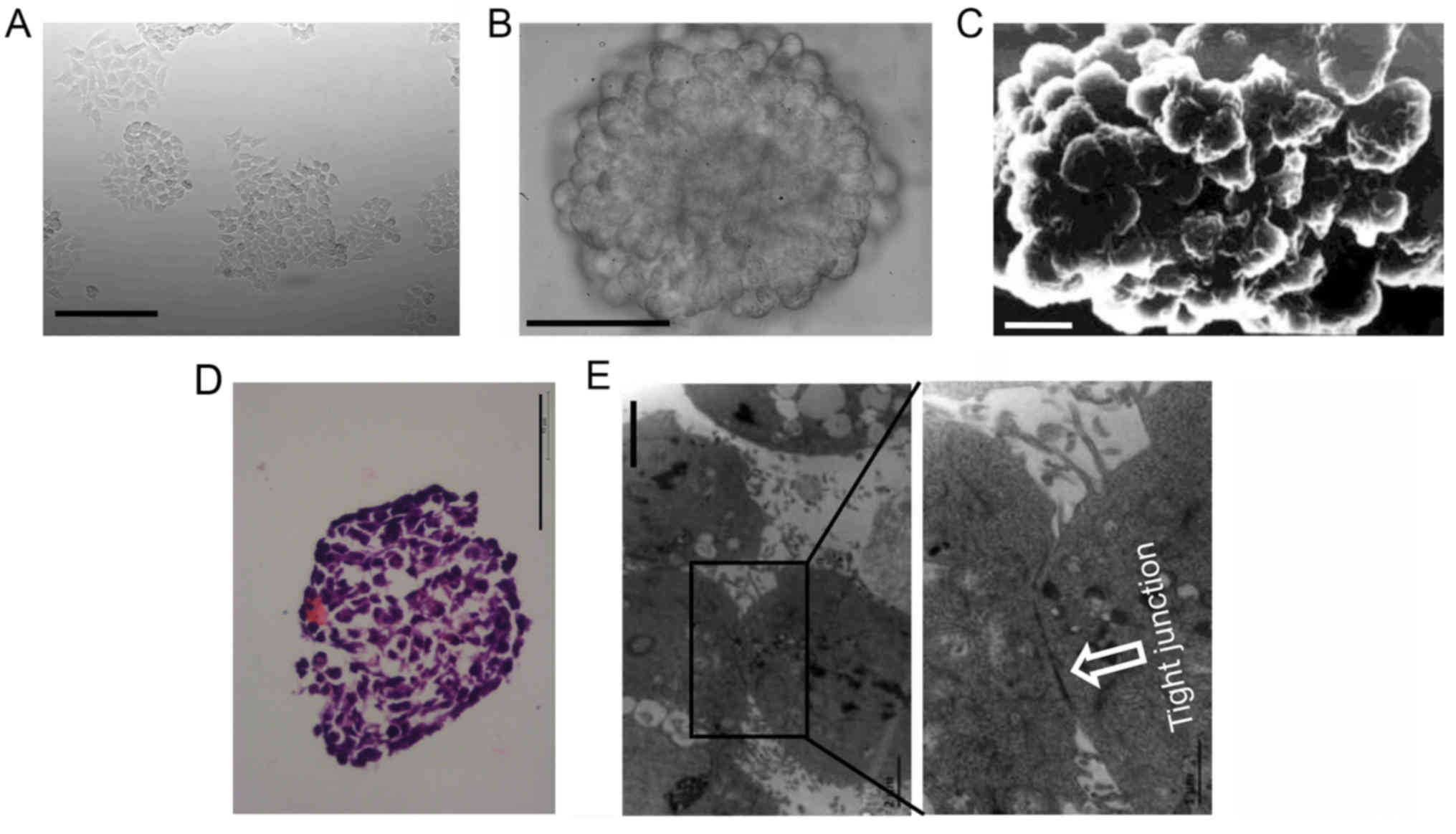
Histopathology of 3D cultures
HCT116 cells grew as a layer of cells under 2D
condition (Fig. 1A). Under 3D
condition, dispersed cells aggregated automatically and formed 3D
cultures (multicellular spheroids). The surface of 3D cultures was
observed using an inverted microscope (Fig. 1B) or a scanning electron microscope
(Fig. 1C). The cells in the
outermost layer of 3D cultures were uniformly spherical.
HCT116 3D cultures were stained with H&E to
further analyze the inner structure. 3D cultures consisted of
layers of cells packed tightly. The cells in the inner layers were
heteromorphic, not uniformly spherical as in the outermost layer
(Fig. 1D). These structures
mimicked colorectal tumors at an avascular stage or avascular
regions of colorectal tumors in vivo (2,7). The
ultrastructure of 3D cultures was observed using a transmission
electron microscope. Cells adhered with each other in 3D cultures,
and cell junctions were commonly found (Fig. 1E).
Association between integrin β4 and 3D
cultures
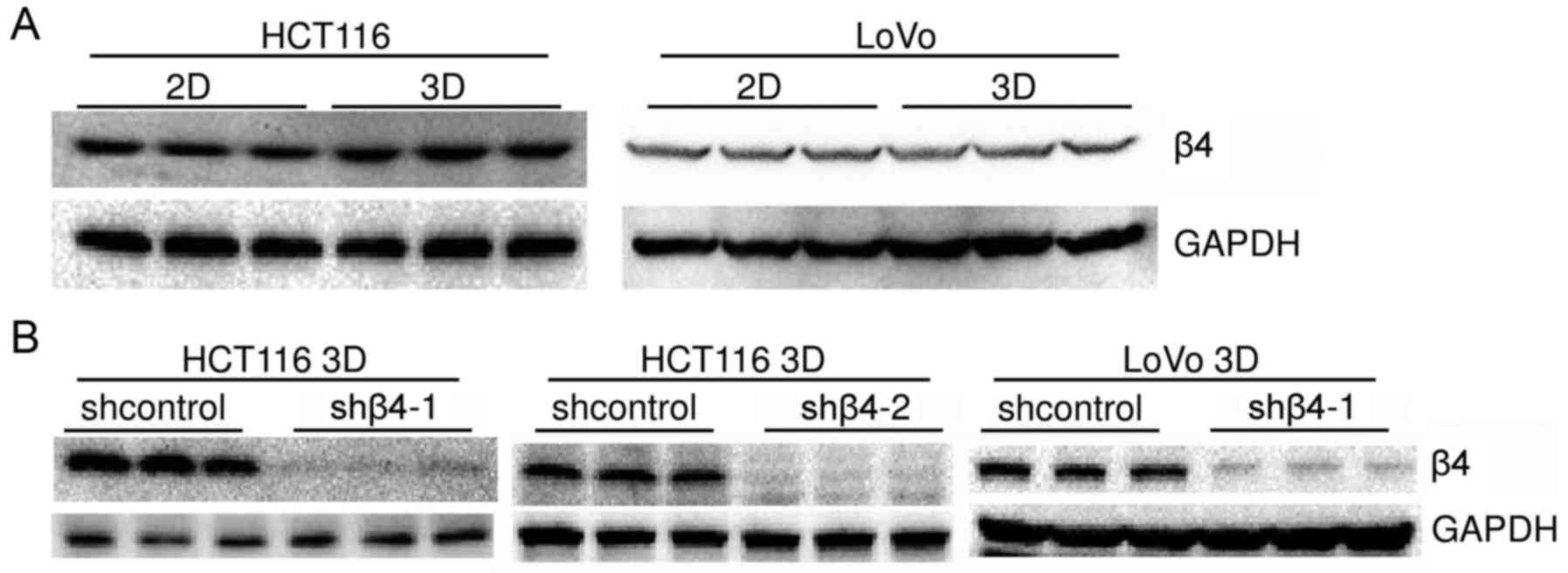
Western blot analysis revealed that the integrin β4
expression level in HCT116 3D cultures was slightly higher than in
2D cultures. The integrin β4 expression level in LoVo 3D cultures
was similar to or mildly lower than in 2D cultures (Fig. 2A). These results indicated that the
function of cell adhesion in integrin β4 expression may be
cell-type specific.
To explore the role of integrin β4 in the
architectural formation of 3D cultures, integrin β4 was knocked
down by lentiviral delivery of shRNA. The efficiency was confirmed
by western blot analysis (Fig. 2B).
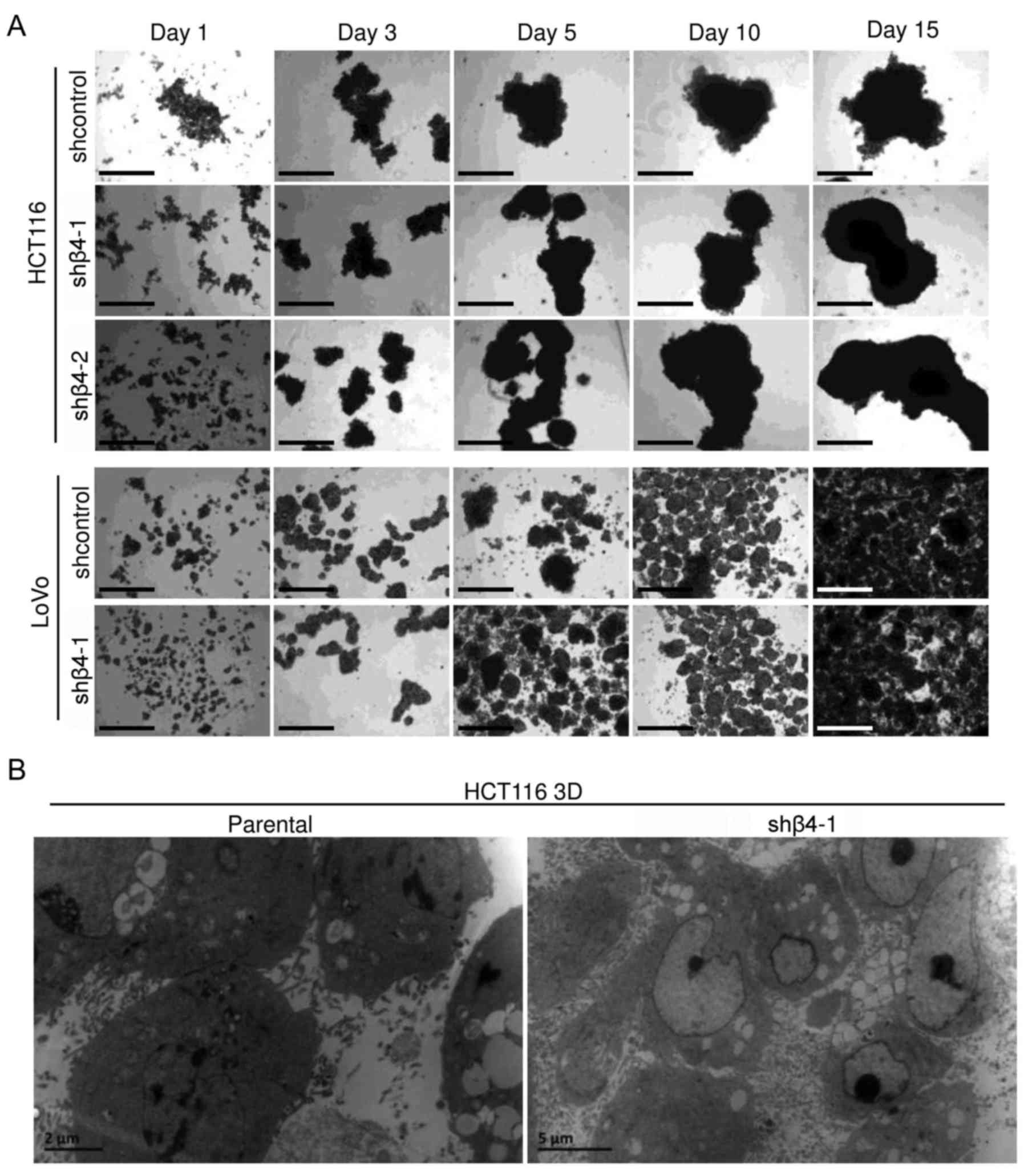
Cells with integrin β4-knockdown (shβ4) or cells transfected with
control lentivirus (shcontrol) were cultured under 3D condition for
15 days and were observed under an inverted microscope (Fig. 3A). LoVo 3D cultures were smaller and
more uniform than HCT116 3D cultures. Cells with integrin β4
knockdown grew as multicellular spheroids, similar to the
respective shcontrol. No differences between shβ4 and the
respective shcontrol were observed. The ultrastructures of parental
HCT116 3D cultures and of shβ4 HCT116 3D cultures were observed
using a transmission electron microscope (Fig. 3B). Integrin β4 knockdown did not
detectably change cell adhesion, and no significant difference was
observed. These results demonstrated that integrin β4 knockdown did
not detectably change the architecture of 3D cultures.
Integrin β4 reduces DNA damage-induced
p53 activation in 3D cultures
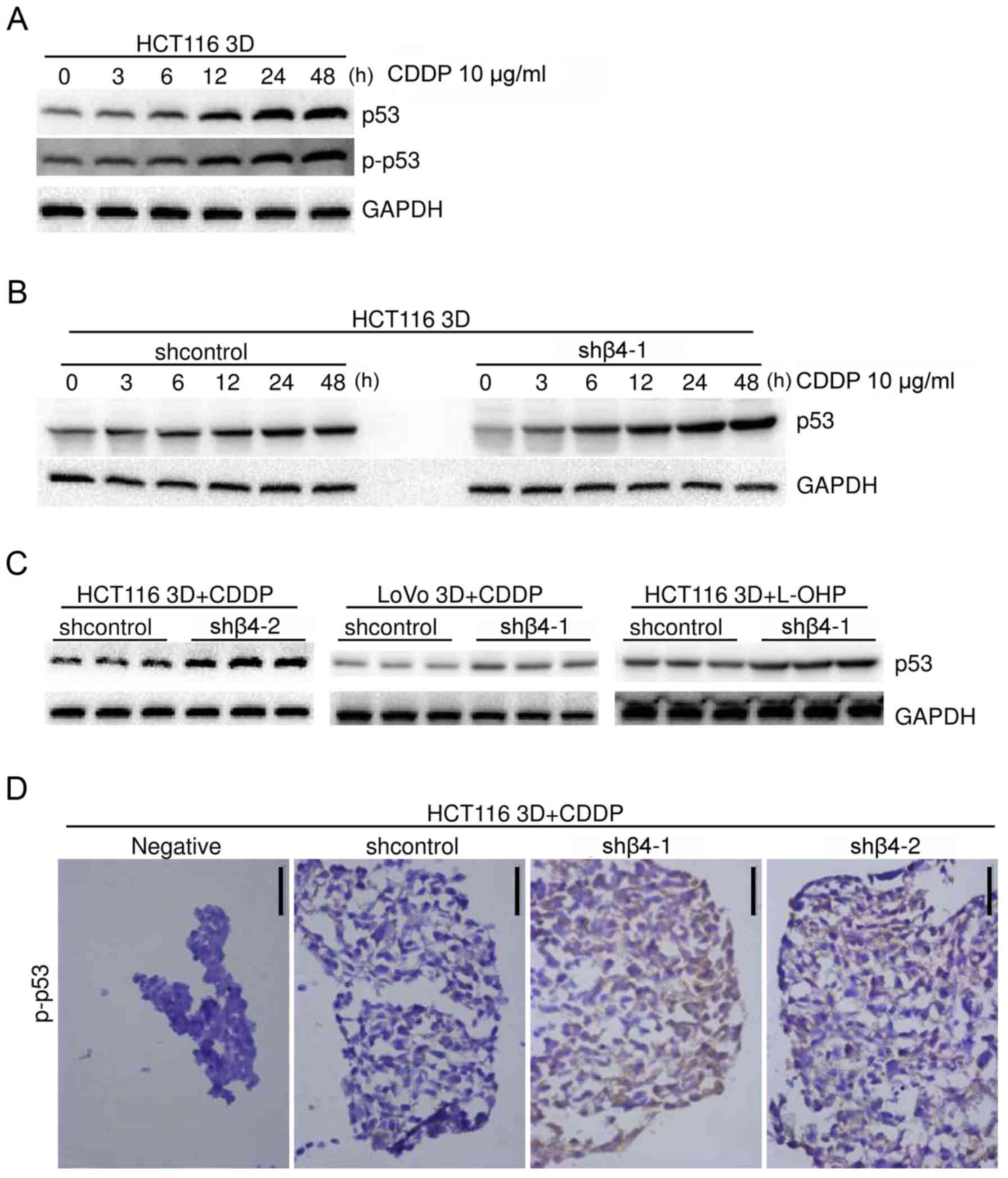
Platinum and irradiation kills cells by damaging
DNA, and p53 plays a key role in the DNA damage response (3,15,16).
CDDP caused p53 protein accumulation in HCT116 3D cultures in a
time-dependent manner (Fig. 4A).
DNA damage induces the phosphorylation of p53 at ser15 (16,17).
Western blot analysis revealed that CDDP induced p-p53 (ser15)
protein accumulation in a time-dependent manner (Fig. 4A). These results are consistent with
those of previous studies (3,15,17,18)
and, collectively, these results demonstrated that platinum caused
DNA damage to induce p53 activation in a time-dependent manner.
HCT116 3D cultures were treated with 10 µg/ml CDDP
in a time-gradient manner and the p53 protein level was assayed
using western blot analysis. Knockdown of integrin β4 did not
detectably change the basal p53 protein level but increased the
CDDP-induced p53 protein accumulation (Fig. 4B). HCT116 and LoVo 3D cultures were
treated with 10 µg/ml CDDP or 5 µg/ml L-OHP for 48 h, respectively.
Western blot analysis revealed that integrin β4 knockdown increased
platinum-induced p53 protein accumulation (Fig. 4C), but did not detectably change the
basal p53 level (data not shown). The effect of integrin β4
knockdown on p-p53 was explored. HCT116 3D cultures were treated
with 10 µg/ml CDDP for 48 h, and p-p53 was evaluated using
immunohistochemical staining. Integrin β4 knockdown increased the
CDDP-induced p-p53 accumulation that arose from DNA damage
(16,17) (Fig.
4D). These results unanimously supported the conclusion that
integrin β4 reduced p53 activation from platinum-induced DNA damage
in 3D cultures.
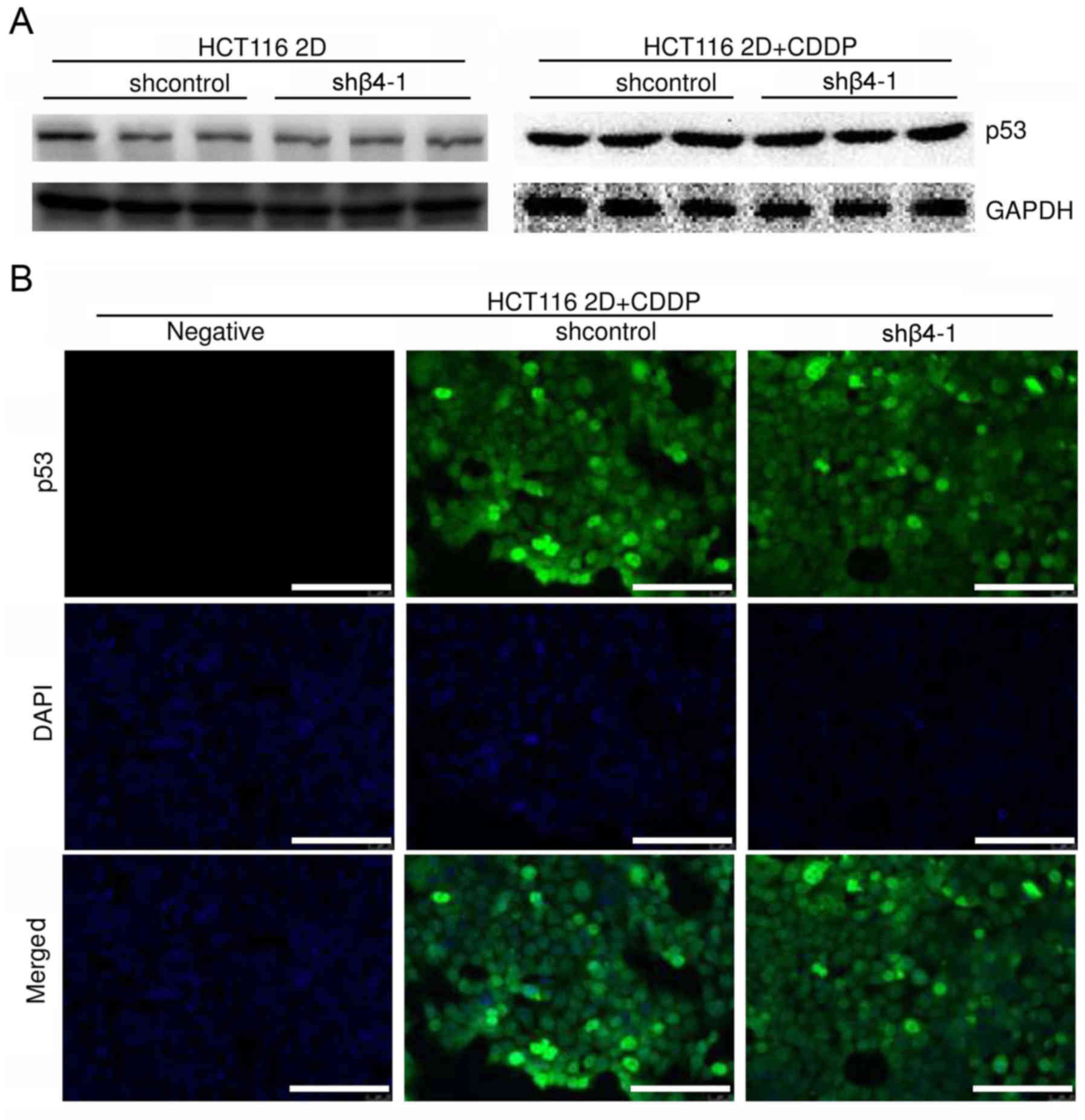
The effect of integrin β4 on p53 in HCT116 2D
cultures was studied. Since 2D cultures are more sensitive to
chemotherapy and radiotherapy than 3D cultures (3,8,19), 2D
cultures were treated with a lower concentration of CDDP. Results
of western blot analysis revealed that integrin β4 knockdown
(shβ4-1) did not markedly change the basal p53 protein level, or
the CDDP-induced (2.5 µg/ml, 48 h) p53 protein level (Fig. 5A). Results of immunofluorescence
staining were consistent with those of western blot analysis.
Integrin β4 knockdown did not markedly change the p53 protein level
in HCT116 2D cultures treated with CDDP (2.5 µg/ml, 24 h) (Fig. 5B). In summary, these results
indicated that integrin β4 reduced DNA damage-induced p53
activation in 3D cultures, but not in HCT116 2D cultures.
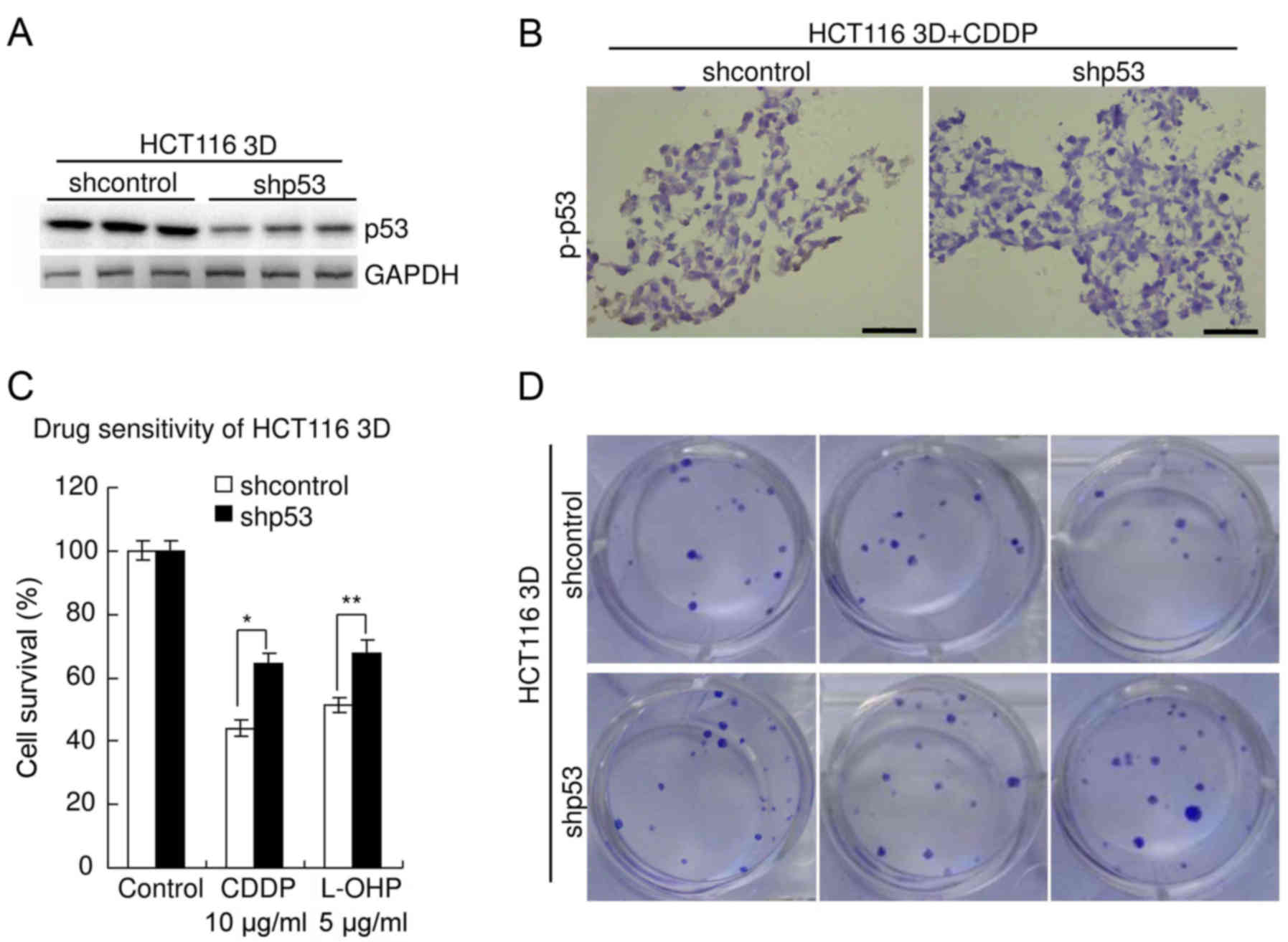
Knockdown of wild-type p53 decreases
sensitivity to platinum in CRC
The role of p53 in the sensitivity of HCT116
(containing stabilized wild-type p53 protein (3)) to platinum was explored. p53 was
knocked down by lentiviral delivery of shRNA. The efficiency was
confirmed by western blot analysis (Fig. 6A). The specificity was previously
verified (3,20). Immunohistochemistry revealed that
knockdown of p53 significantly decreased p-p53 level in HCT116 3D
cultures treated with 10 µg/ml CDDP for 48 h (Fig. 6B). Results of the WST assay revealed
that knockdown of p53 significantly decreased chemosensitivity of
HCT116 3D cultures to platinum (Fig.
6C). The viability of shcontrol HCT116 cells treated with CDDP
(10 µg/ml, 48 h) was 44.1±2.6%, whereas that of HCT116 cells with
p53 knockdown was 64.6±3.2% (P<0.01). The viability of shcontrol
HCT116 cells treated with L-OHP (5 µg/ml, 48 h) was 51.4±2.4%,
whereas that of HCT116 cells with p53 knockdown was 67.7±4.2%
(P<0.05). There was no significant difference in viability
between the parental HCT116 3D cell cultures and the shcontrol
(P≥0.05) (data not shown). The results of the clonogenic assay were
consistent with these of the WST assay. The clonogenicity of the
shp53 HCT116 cells was higher than that of the shcontrol (Fig. 6D).
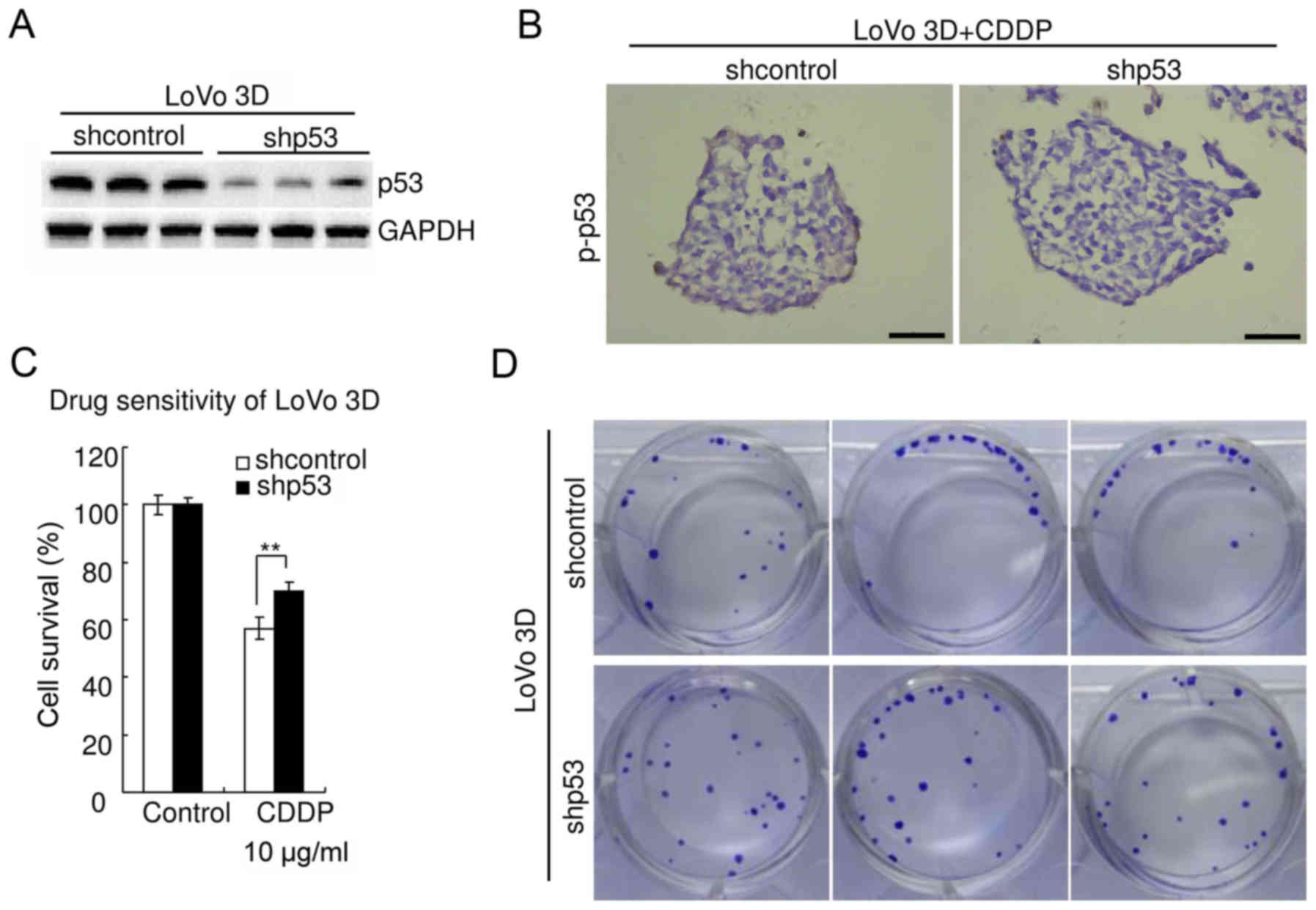
The aforementioned experiments were also performed
in LoVo cells [containing stabilized wild-type p53 protein
(3)]. The results were consistent
with those observed in HCT116 cells. Western blot analysis
confirmed the efficiency of p53-knockdown in LoVo 3D cultures
(Fig. 7A). Immunohistochemistry
revealed that knockdown of p53 significantly decreased p-p53 in
LoVo 3D cultures treated with 10 µg/ml CDDP for 48 h (Fig. 7B). WST assay revealed that knockdown
of p53 significantly decreased chemosensitivity of LoVo 3D cultures
to CDDP (P<0.05) (Fig. 7C). The
cell viability of shcontrol LoVo treated with CDDP (10 µg/ml, 48 h)
was 57.0±3.7%, whereas that of LoVo with p53 knockdown was
69.9±3.1% (P<0.05). The clonogenicity of the shp53 LoVo 3D
cultures was higher than that of the shcontrol (Fig. 7D). There was no significant
difference in viability between parental LoVo 3D cultures and
shcontrol (P≥0.05) (data not shown).
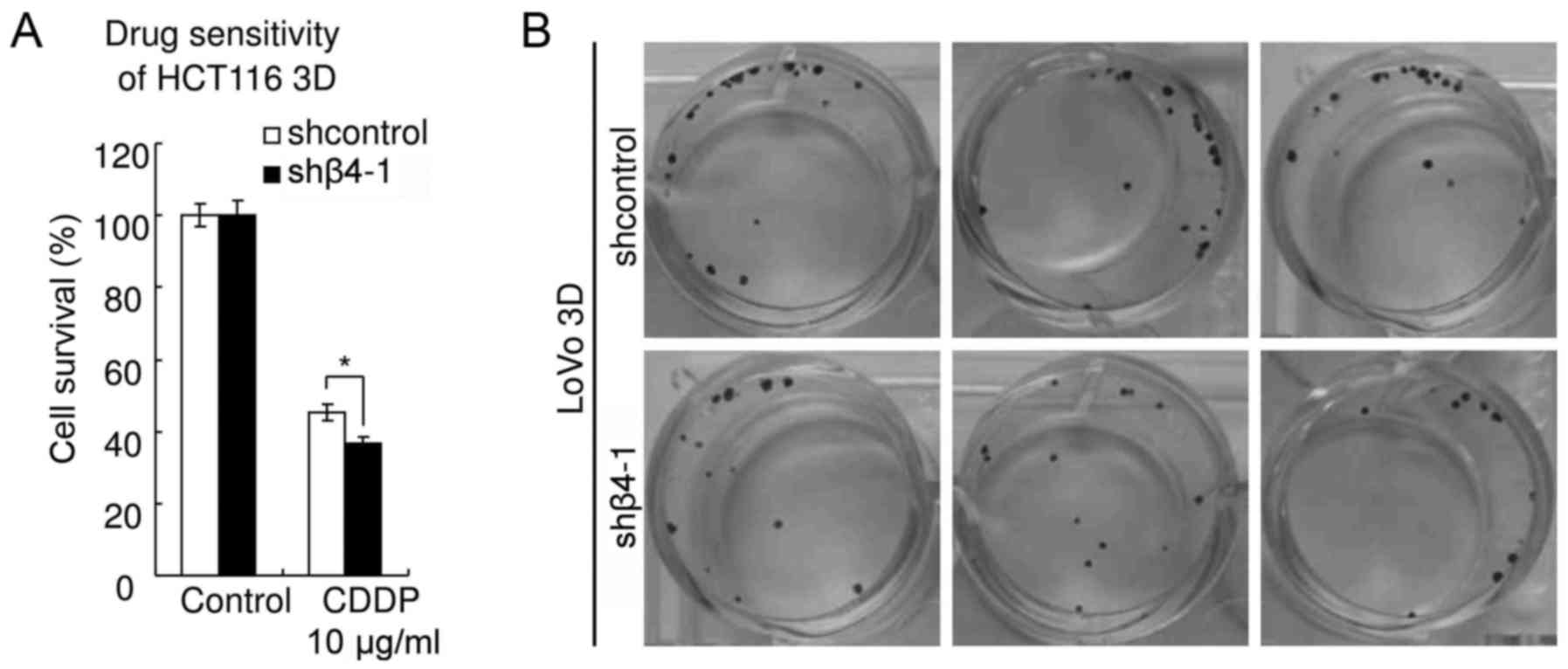
Knockdown of integrin β4 increases
sensitivity to CDDP in CRC
As aforementioned, integrin β4 reduced DNA
damage-induced p53 activation (Fig.
4) and knockdown of wild-type p53 decreased sensitivity to
platinum (Figs. 6 and 7). These results led to the hypothesis
that knockdown of integrin β4 may increase sensitivity to platinum
of CRC. Results of WST assay revealed that knockdown of integrin β4
significantly increased the chemosensitivity of HCT116 3D cultures
to CDDP (Fig. 8A). The viability of
shcontrol HCT116 cells treated with CDDP (10 µg/ml, 48 h) was
45.3±2.4%, whereas that of HCT116 cells with integrin β4 knockdown
(shβ4-1) was 36.5±2.1% (P<0.01). The results of the clonogenic
assay revealed that the clonogenicity of the shβ4-1 LoVo 3D
cultures was lower than that of the shcontrol (Fig. 8B). In summary, these results
indicated that integrin β4 reduces DNA damage-induced p53
activation to decrease CRC chemosensitivity to platinum.
Knockdown of p53 does not markedly
change integrin β4 protein levels in HCT116 cells
It was reported that p53 regulated integrin β4
expression in several cells types, including tumor cells (21–23).
The effect of p53 on integrin β4 expression in HCT116 3D cultures
was investigated. HCT116 3D cultures were treated with or without
10 µg/ml CDDP for 24 or 48 h, respectively. Integrin β4 expression
levels were assayed using western blot analysis. No detectable
difference in integrin β4 expression levels was observed between
shcontrol HCT116 cells and the respective shp53 HCT116 cells
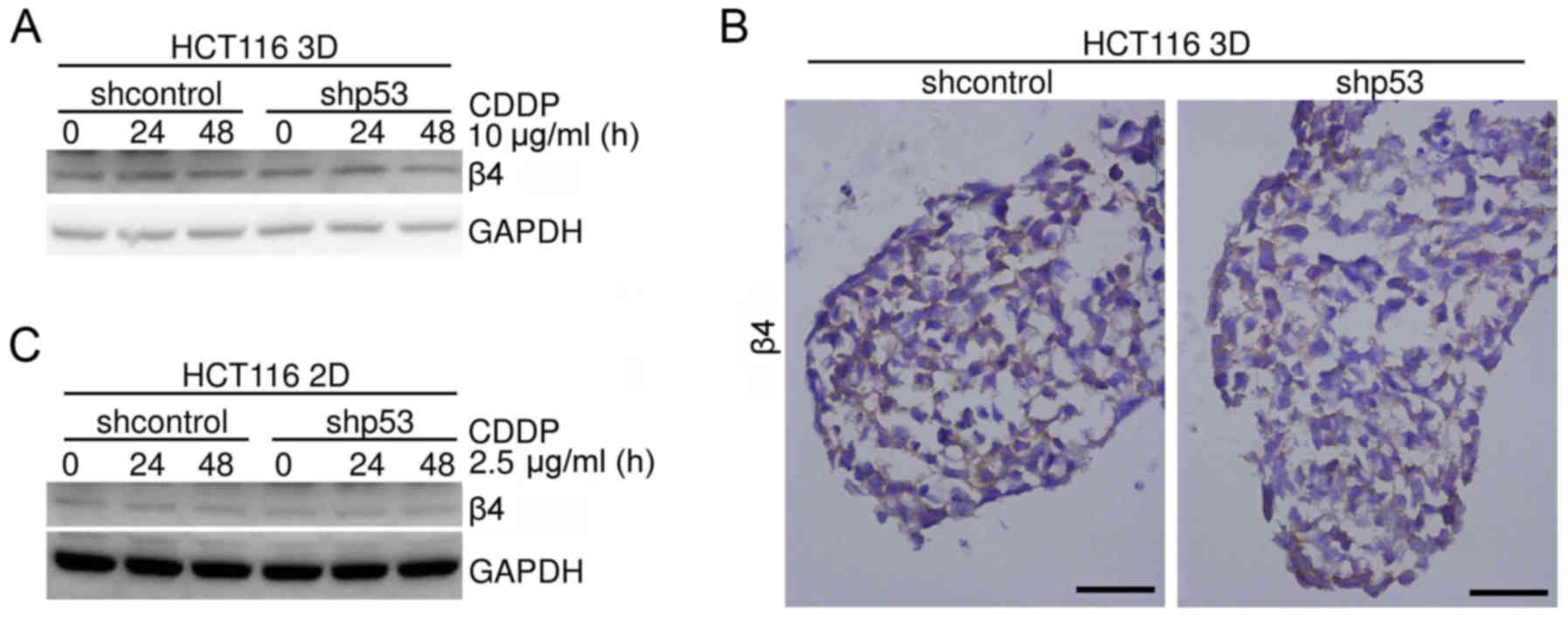
(Fig. 9A). Immunohistochemical
staining was employed to evaluate the integrin β4 protein level of
HCT116 3D cultures without any treatment. Knockdown of p53 (shp53)
did not detectably change the integrin β4 protein level (Fig. 9B). The effect of p53 on integrin β4
in HCT116 2D cultures was studied. HCT116 2D cultures were treated
with or without 2.5 µg/ml CDDP for 24 or 48 h, respectively.
Integrin β4 expression levels were assayed using western blot
analysis. No detectable difference in integrin β4 expression levels
was observed (Fig. 9C). These
results indicated that knockdown of p53 may not detectably change
the expression of integrin β4 under these specific conditions.
Discussion
Solid tumor cells, including CRC cells, proliferate,
survive and response to stimuli in a specific tissue
microenvironment in vivo (3,7,8). In
addition, 3D cell culture models provide an optimal experimental
system for mimicking solid tumor tissue microenvironments,
particularly cell adhesion in vivo (Fig. 1) (3,7,8). In 3D
cultures, cells adhered to each other within layers of cells, and
cell junctions were commonly found (Fig. 1B-E) (7). Their structures were more similar to
these of tumors at avascular stage and avascular tumor regions
(3,7,8).
The formation and stability of cell adhesion rely on
cell adhesion molecules, particularly E-cadherin and the integrin
family (6–11). Integrins, consisting of α and β
subunits, are a group of transmembrane heterodimeric cell surface
receptors that enhance cell anchorage to the ECM and cell-cell
interaction (6,9–11). It
has been reported that 24 definite integrin heterodimers are
established by the amalgamation of 18 α subunits and 8 β subunits
(9,10). In the present study, integrin β4
knockdown did not prevent suspended CRC cells from forming 3D
cultures (Fig. 3A). This indicated
that integrin β4 may not be essential for architectural formation
of 3D cultures or very low expression of integrin β4 may be
sufficient. Integrin β4 is different from other integrin β subunits
for its exceptionally large cytoplasmic domain and its upregulation
correlates with changes in cell biology (10,12,23,24).
High expression of integrin β4 has been reported in human CRC, and
it was also reported that cell adhesion affects cell adhesion
molecules expression levels (8,14). In
the present study, the integrin β4 expression level in HCT116 3D
cultures was higher than in 2D cultures, while in LoVo 3D cultures,
the integrin β4 expression level was similar to or slightly lower
than in 2D cultures (Fig. 2A).
These results indicated that the function of cell adhesion in
integrin β4 expression may be cell-type specific in CRC. Another
possible explanation is that the difference may be caused by the
difference in the microenvironment between HCT116 3D cultures and
LoVo 3D cultures since the two cell lines form different shapes of
multicellular spheroids (Fig.
3B).
Integrin contributes to the maintenance of cell
adhesion (6,9,10,23,25).
On the other hand, mounting evidence has indicated that integrin
couples intra- and extra-cellular signals, since it rapidly
undergoes conformational switches transduced via cytoplasmic
changes (‘inside-out’ signaling) and simultaneous ligand-induced
rearrangements (‘outside-in’ signaling) (6,9,25).
Inactive integrins are compact and bent, with their genu folded and
the headpiece ~5 nm from the membrane. Separation of the α and β
subunit legs destabilizes their interface with the headpiece,
converting the bent structure to an overall extended conformation
and relieving constraints on headpiece activation (6,9,10,21,25).
Several factors, such as ECM-cell adhesion, cell-cell adhesion and
force, were reported to activate integrins (6,9–11,25).
Furthermore, 3D cultures represent the solid tumor tissue
microenvironment, particularly cell adhesion in vivo
(Fig. 1B-E) (3,6–8). Under
3D condition, integrin β4 exhibited the ability to reduce DNA
damage-induced p53 acivation (Fig.
4B-D). This effect was not observed in HCT116 2D cultures
(Fig. 5A and B). These results
indicated that integrin β4 reduced DNA damage-induced p53
activation in 3D cultures and this may be due to integrin β4
activation. Due to a myriad of difference in the microenvironment
between 3D cultures and 2D cultures (3,6,8), it is
unclear what causes integrin β4 activation under the specific
condition.
Platinum and irradiation damages DNA by binding to
and causing crosslink of DNA to kill cells (3,15,16,26).
Mounting evidence has demonstrated that the activation of p53 plays
a key role in the DNA damage response, and loss of wild-type p53
leads to resistance to DNA damage-induced cell death (3,15,18,20,27,28).
In the present study, integrin β4 reduced DNA damage-induced p53
activation, and knockdown of integrin β4 increased sensitivity to
CDDP (Figs. 4, 5 and 8).
Knockdown of wild-type p53 [both HCT116 and LoVo contain a
stabilized wild-type p53 protein (3)] decreased sensitivity to platinum
(Figs. 6 and 7). In summary, these findings indicated
that integrin β4 reduced DNA damage-induced p53 activation. This
may contribute to explain the phenomenon that the integrin
expression level negatively correlated with prognosis in multiple
cancer types (10,12,14,23).
Furthermore, mutated p53 or loss of p53 is frequently
observed in cancer (17,21,27)
and p53 mutations can also inactivate the protein normal
function (18,22,28).
These results may also contribute to explain that loss of p53 or
mutated p53 often acquired drug resistance and that loss of
p53 or mutated p53 negatively correlated with survival of
cancer patients, including CRC patients (3,15,20,23,26–28).
In the present study, knockdown of integrin β4
appeared to increase chemosensitivity to a lesser degree than that
caused by knockdown of p53 (Figs.
6–8). Integrin β4 triggers
numerous signaling cascades, and p53 can be regulated by a variety
of factors (3,10,12,14,17,20–25,27).
It is reasonable to hypothesize that other signaling cascades,
besides integrin β4-p53 pathway, may be involved in the mechanism
underlying the development of CRC resistance to DNA damage.
In summary, 3D cultures consist of layers of cells
that preserve cell adhesive systems. These present a good model to
deciphering the function of cell adhesive systems in cancer. Cell
adhesion triggers certain integrin signaling cascades and
influences tumor cell biological behavior, including
chemosensitivity (6,9–11).
Data in the present study indicated that integrin β4 reduced DNA
damage-induced p53 activation to decrease DNA damage-induced cell
death; this may be due to integrin β4 activation in 3D cultures.
However, the mechanism of adhesion-associated CRC cell resistance
to DNA damage in vivo is complex and deserves further
investigation.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used during the present study are
available from the corresponding author upon reasonable
request.
Authors' contributions
JW, RZ and JL performed the experiments; JW and BL
analyzed the data, designed the research and wrote the paper. All
authors read and approved the manuscript and agree to be
accountable for all aspects of the research in ensuring that the
accuracy or integrity of any part of the work are appropriately
investigated and resolved.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
CRC
|
colorectal cancer
|
|
3D
|
three-dimensional
|
|
ECM
|
extracellular matrix
|
|
2D
|
two-dimensional
|
|
AOM
|
azoxymethane
|
|
DSS
|
dextran sodium sulfate
|
|
H&E
|
hematoxylin and eosin
|
|
PBS
|
phosphate-buffered saline
|
|
p-p53
|
phospho-p53 (Ser15)
|
|
GAPDH
|
glyceraldehyde 3-phosphate
dehydrogenase
|
|
sh
|
small hairpin
|
|
shcontrol
|
control shRNA
|
|
CDDP
|
cisplatin
|
|
DAPI
|
4′,6-diamidino-2-phenylindole
|
|
WST
|
water-soluble tetrazolium salt
|
|
L-OHP
|
oxaliplatin
|
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
He J, Shin H, Wei X, Kadegowda AK, Chen R
and Xie SK: NPC1L1 knockout protects against colitis-associated
tumorigenesis in mice. BMC Cancer. 15:1892015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
He J, Liang X, Luo F, Chen X, Xu X, Wang F
and Zhang Z: P53 is involved in a three-dimensional
architecture-mediated decrease in chemosensitivity in colon cancer.
J Cancer. 7:900–909. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2016. CA Cancer J Clin. 66:7–30. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
He J, Pei L, Jiang H, Yang W, Chen J and
Liang H: Chemoresistance of colorectal cancer to 5-fluorouracil is
associated with silencing of the BNIP3 gene through aberrant
methylation. J Cancer. 8:1187–1196. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
He JM, Wang FC, Qi HB, Li Y and Liang HJ:
Down-regulation of alphav integrin by retroviral delivery of small
interfering RNA reduces multicellular resistance of HT29. Cancer
Lett. 284:182–188. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Liang X, Xu X, Wang F, Chen X, Li N, Wang
C and He J: E-cadherin knockdown increases β-catenin reducing
colorectal cancer chemosensitivity only in three-dimensional
cultures. Int J Oncol. 47:1517–1527. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Liang X, Xu X, Wang F, Li N and He J:
E-cadherin increasing multidrug resistance protein 1 via
hypoxia-inducible factor-1alpha contributes to multicellular
resistance in colorectal cancer. Tumour Biol. 37:425–435. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Longmate W and DiPersio CM: Beyond
adhesion: Emerging roles for integrins in control of the tumor
microenvironment. F1000Res. 6:16122017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Das V, Kalyan G, Hazra S and Pal M:
Understanding the role of structural integrity and differential
expression of integrin profiling to identify potential therapeutic
targets in breast cancer. J Cell Physiol. 233:168–185. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zeltz C and Gullberg D: The
integrin-collagen connection-a glue for tissue repair? J Cell Sci.
129:653–664. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Bierie B, Pierce SE, Kroeger C, Stover DG,
Pattabiraman DR, Thiru P, Donaher Liu J, Reinhardt F, Chaffer CL,
Keckesova Z, et al: Integrin-β4 identifies cancer stem
cell-enriched populations of partially mesenchymal carcinoma cells.
Proc Natl Acad Sci USA. 114:E2337–E2346. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Phung YT, Barbone D, Broaddus VC and Ho M:
Rapid generation of in vitro multicellular spheroids for the study
of monoclonal antibody therapy. J Cancer. 2:507–514. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Tai YL, Lai IR, Peng YJ, Ding ST and Shen
TL: Activation of focal adhesion kinase through an interaction with
beta4 integrin contributes to tumorigenicity of colon cancer. FEBS
Lett. 590:1826–1837. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ye X, Zhang C, Chen Y and Zhou T:
Upregulation of acetylcholinesterase mediated by p53 contributes to
cisplatin-induced apoptosis in human breast cancer cell. J Cancer.
6:48–53. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Valentine JM, Kumar S and Moumen A: A
p53-independent role for the MDM2 antagonist Nutlin-3 in DNA damage
response initiation. BMC Cancer. 11:792011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
He J, Wang F, Luo F, Chen X, Liang X,
Jiang W, Huang Z, Lei J, Shan F and Xu X: Effects of long term low-
and high-dose sodium arsenite exposure in human transitional cells.
Am J Transl Res. 9:416–428. 2017.PubMed/NCBI
|
|
18
|
Yogev O, Barker K, Sikka A, Almeida GS,
Hallsworth A, Smith LM, Jamin Y, Ruddle R, Koers A, Webber HT, et
al: p53 loss in MYC-driven neuroblastoma leads to metabolic
adaptations supporting radioresistance. Cancer Res. 76:3025–3035.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Verjans ET, Doijen J, Luyten W, Landuyt B
and Schoofs L: Three-dimensional cell culture models for anticancer
drug screening: Worth the effort? J Cell Physiol. 233:2993–3003.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Godar S, Ince TA, Bell GW, Feldser D,
Donaher JL, Bergh J, Liu A, Miu K, Watnick RS, Reinhardt F, et al:
Growth-inhibitory and tumor-suppressive functions of p53 depend on
its repression of CD44 expression. Cell. 134:62–73. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yoshioka T, Otero J, Chen Y, Kim YM,
Koutcher JA, Satagopan J, Reuter V, Carver B, de Stanchina E,
Enomoto K, et al: β4 Integrin signaling induces expansion of
prostate tumor progenitors. J Clin Invest. 123:682–699.
2013.PubMed/NCBI
|
|
22
|
Bon G, Di Carlo SE, Folgiero V, Avetrani
P, Lazzari C, D'Orazi G, Brizzi MF, Sacchi A, Soddu S, Blandino G,
et al: Negative regulation of beta4 integrin transcription by
homeodomain-interacting protein kinase 2 and p53 impairs tumor
progression. Cancer Res. 69:5978–5986. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lakshmanan I, Rachagani S, Hauke R, Krishn
SR, Paknikar S, Seshacharyulu P, Karmakar S, Nimmakayala RK,
Kaushik G, Johansson SL, et al: MUC5AC interactions with integrin
β4 enhances the migration of lung cancer cells through FAK
signaling. Oncogene. 35:4112–4121. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Chao C, Lotz MM, Clarke AC and Mercurio
AM: A function for the integrin alpha6beta4 in the invasive
properties of colorectal carcinoma cells. Cancer Res. 56:4811–4819.
1996.PubMed/NCBI
|
|
25
|
Alon R and Dustin ML: Force as a
facilitator of integrin conformational changes during leukocyte
arrest on blood vessels and antigen-presenting cells. Immunity.
26:17–27. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Sarasqueta AF, Forte G, Corver WE, de
Miranda NF, Ruano D, van Eijk R, Oosting J, Tollenaar RA, van Wezel
T and Morreau H: Integral analysis of p53 and its value as
prognostic factor in sporadic colon cancer. BMC Cancer. 13:2772013.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Li H, Rokavec M, Jiang L, Horst D and
Hermeking H: Antagonistic effects of p53 and HIF1A on microRNA-34a
regulation of PPP1R11 and STAT3 and hypoxia-induced epithelial to
mesenchymal transition in colorectal cancer cells.
Gastroenterology. 153:505–520. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Tran TQ, Lowman XH, Reid MA,
Mendez-Dorantes C, Pan M, Yang Y and Kong M: Tumor-associated
mutant p53 promotes cancer cell survival upon glutamine deprivation
through p21 induction. Oncogene. 36:1991–2001. 2017. View Article : Google Scholar : PubMed/NCBI
|