Introduction
Lung cancer is one of the most common cancers in the
world (1), and non-small cell lung
cancer (including adenocarcinoma and squamous cell carcinoma)
accounts for most of the diagnosed cases of lung cancer (2). Tumor progression mechanisms and
therapeutic strategies remain crucial subjects for investigation.
Dysregulation of metabolic pathways such as the tricarboxylic acid
cycle and the metabolic pathways of serine, glycine, and glutamine
play a critical role in tumor progression (3–5). Lipid
metabolic pathways are involved in various physiological functions.
Multiple lipid metabolic enzymes regulate the behavior of lung
cancer cells. High expression of fatty acid synthase is associated
with relatively high risk of lung carcinoma recurrence (6). In lung adenocarcinoma, high expression
of stearoyl CoA desaturase 1 (SCD) leads to enhanced cell migration
and invasion capacity in lung cancer cells and is associated with
poor prognosis in patients (7).
Furthermore, the plasma levels of some saturated and unsaturated
fatty acids, including arachidonic, palmitic, linoleic, and oleic
acids, in patients with lung adenocarcinoma are higher than those
in healthy people (8–10). These findings suggest that lipid
metabolic enzymes and fatty acid transporters are affected in the
progression of lung cancer.
The endoplasmic reticulum (ER) is an organelle that
performs protein folding and posttranslational modification and is
involved in the processes of energy metabolism, lipid biosynthesis,
and homeostasis of intracellular calcium ions (Ca2+)
(11). Certain physiological
conditions interfere with protein folding, including calcium
depletion, nutrient deprivation and DNA damage. Accumulation of
unfolded and misfolded proteins in ER lumen causes ER stress
(12). In a tumor microenvironment,
hypoxia, nutrient deprivation, and calcium dysregulation induce ER
stress in tumor cells (12).
Generally, unresolved ER stress leads to apoptosis (13). Unfolded protein responses (UPRs) can
protect cells from apoptosis. Thus, UPR activation serves as a
prognostic marker for several human cancers (14). UPR signaling affects sterol
regulatory element binding proteins (SREBPs), which are upstream
regulators of lipid biosynthesis (15). In liver cells, activation of UPR
signaling results in increased lipid accumulation (16). Furthermore, silencing of SREBPs
decreases the desaturation of fatty acids and increases
accumulation of reactive oxygen species (ROS) (17). Thus, ER stress and lipid metabolism
are highly regulated in human physiology.
Fatty acyl-CoA is a critical metabolite in the lipid
metabolic pathway and is involved in various physiological
processes, including β-oxidation and triacylglycerol and
phospholipid synthesis (18). Fatty
acyl-CoA formation and degradation respectively occur in two enzyme
families: acyl-CoA synthetases (ACSs) and acyl-CoA thioesterases
(ACOTs) (19). Human physiology
involves more than 26 ACS enzymes and 12 ACOT enzymes (19). Recent studies have demonstrated the
critical role of these enzymes in lung cancer. An increased risk of
lymph node metastasis in lung adenocarcinoma is associated with
high ACOT8 expression (20). Our
previous study revealed that high expression of ACOT11 and ACOT13
in patients with lung adenocarcinoma was associated with cell
proliferation and poor prognosis (21). Regulation of other ACS and ACOT
enzymes under the condition of ER stress is not fully understood.
The present study investigated these phenomena with a view to
identifying the relevant mechanisms through bioinformatic and
experimental approaches.
Materials and methods
Bioinformatic analysis
Cancer gene expression and clinical outcomes for
each type of lung cancer were evaluated using PREdiction of
Clinical Outcomes from Genomic Profiles (PRECOG; http://precog.stanford.edu) (22). Z-scores were obtained from the
PRECOG website and a heatmap was drawn using Morpheus (https://software.broadinstitute.org/morpheus/).
Overall survival in lung cancer patients was evaluated using the
Kaplan-Meier (KM) plotter (http://kmplot.com/; 23). For KM plotter analysis, the
studied lung cancer patients were divided into high- and
low-expression groups according to median gene expression. To
evaluate gene expression in lung cancer cell line A549 under ER
stress, a microarray dataset was obtained from the National Center
for Biotechnology Information Gene Expression Omnibus (GEO;
accession number: GSE76515; 24). The log2 gene expression value was
obtained from the GEO2R interface (http://www.ncbi.nlm.nih.gov/geo/geo2r/). Gene
expression with log2 fold changes of >1 or ≤1 under ER stress
were chosen for analysis with a biological pathway assay and
functional enrichment analysis (FunRich) software (version 3.1.3;
25,26). Gene transcription factors were evaluated using MotifMap
(http://motifmap.ics.uci.edu) with the
human hg18 multiz28way_placental background, and analyses were
conducted within −1,000 to 1,000 of the transcription start sites
(27).
Cell culture
Human lung adenocarcinoma CL1-0 cell line was
provided by Dr Pan-Chyr Yang (Department of Internal Medicine,
National Taiwan University Hospital, Taipei, Taiwan (28). Human lung carcinoma A549 cells were
purchased from the American Type Culture Collection (ATCC;
Manassas, VA, USA). The CL1-0 and A549 cells were respectively
maintained in RPMI-1640 medium and Ham's F12K medium. Both media
consisted of 10% fetal bovine serum and 1%
penicillin-streptomycin-amphotericin B (Lonza, Walkersville, MD,
USA), and the cultures were stored in an incubator at 37°C with a
5% CO2 environment.
Western blot analysis
The cells were lysed in radioimmunoprecipitation
lysis buffer (RIPA) with a protease inhibitor cocktail at a 100:1
ratio at 24 h after vehicle control [dimethyl sulfoxide (DMSO)] or
thapsigargin (Tg; dissolved in DMSO; Sigma-Aldrich; Merck KGaA,
Darmstadt, Germany) treatment. A bicinchoninic acid (BCA) protein
assay kit (Thermo Fisher Scientific, Inc., Rockford, IL, USA) was
used to determine protein concentration. Equal amounts (30 µg) of
protein were loaded and separated using 12% sodium dodecyl
sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). After
electrophoresis, the proteins were transferred onto polyvinylidene
difluoride (PVDF) membranes (EMD Millipore, Billerica, MA, USA).
After 1 h of blocking in 5% skim milk and Tris-buffered saline
solution with Tween-20 (TBST) buffer, the PVDF membranes were
incubated with the following primary antibodies: anti-ACOT11
(1:3,000; cat. no. ab153835; Abcam, Cambridge, UK), anti-ACSL1
(1:1,000, cat. no. 4047; Cell Signaling Technology, Danvers, MA,
USA), anti-ACSL4 (1:1,000, cat. no. ab155282; Abcam) and anti-GAPDH
(dilution, 1:5,000; cat. no. MAB374; EMD Millipore). After TBST
washing, the membranes were incubated with secondary antibodies,
including peroxidase-conjugated goat anti-rabbit immunoglobulin G
(IgG; 1:5,000; cat. no. AP132P; EMD Millipore) and
peroxidase-conjugated goat anti-mouse IgG (1:5,000; cat. no.
AP124P; EMD Millipore) at room temperature for 1 h. The results
were acquired using an imaging capturing system (Alpha Innotech
FluorChem FC2 imaging system, ProteinSimple; Bio-Techne,
Minneapolis, MN, USA). Protein expression was quantified using
ImageJ software (version 1.51; National Institutes of Health,
Bethesda, MD, USA).
Fatty acid uptake assay
Fatty acid uptake was determined using the Free
Fatty Acid Uptake Assay Kit (Fluorometric) according to the
manufacturer's instructions (cat. no. ab176768; Abcam). Before
fatty acid uptake assay, 2×104 A549 and CL1-0 cells were
seeded on a 96-well plate overnight. The cells were then treated
with Tg or vehicle control (DMSO; Sigma-Aldrich; Merck KGaA) for 24
h. After being washed with phosphate-buffered saline, the cells
were preincubated in serum-free media for 1 h and then incubated in
a fluorescent fatty acid mixture for 30 min. Fluorescence signals
were measured using a microplate fluorescence reader at 485/528 nm
(FLx800; BioTek Instruments Inc., Winooski, VT, USA). The
fluorescence signals from wells containing assay mix without cells
were used as the background. For relative quantification, vehicle
control (DMSO treatment) for CL1-0 and A549 was set to 100%.
ROS detection
Intracellular ROS was detected using the DCFDA
Cellular Reactive Oxygen Species Detection Assay kit (cat. no.
ab113851; Abcam) according to the manufacturer's instructions.
Briefly, 2×104 A549 and CL1-0 cells were seeded on a
96-well plate overnight. The cells were stained with 20 µM of DCFDA
solution for 30 min at 37°C and then treated with vehicle control
or Tg for 6 h. The fluorescence signal was detected using a
microplate fluorescence reader at 485/528 nm (FLx800; BioTek
Instruments Inc.) and the fluorescence signals from the cells
without DCFDA staining were used as the background.
Prediction of targeted gene
function
To predict the interaction between genes with
induced ER stress and transcription factors on each ACS and ACOT
enzyme, the GeneMANIA database was used (http://genemania.org) (29). The genes with induced ER stress are
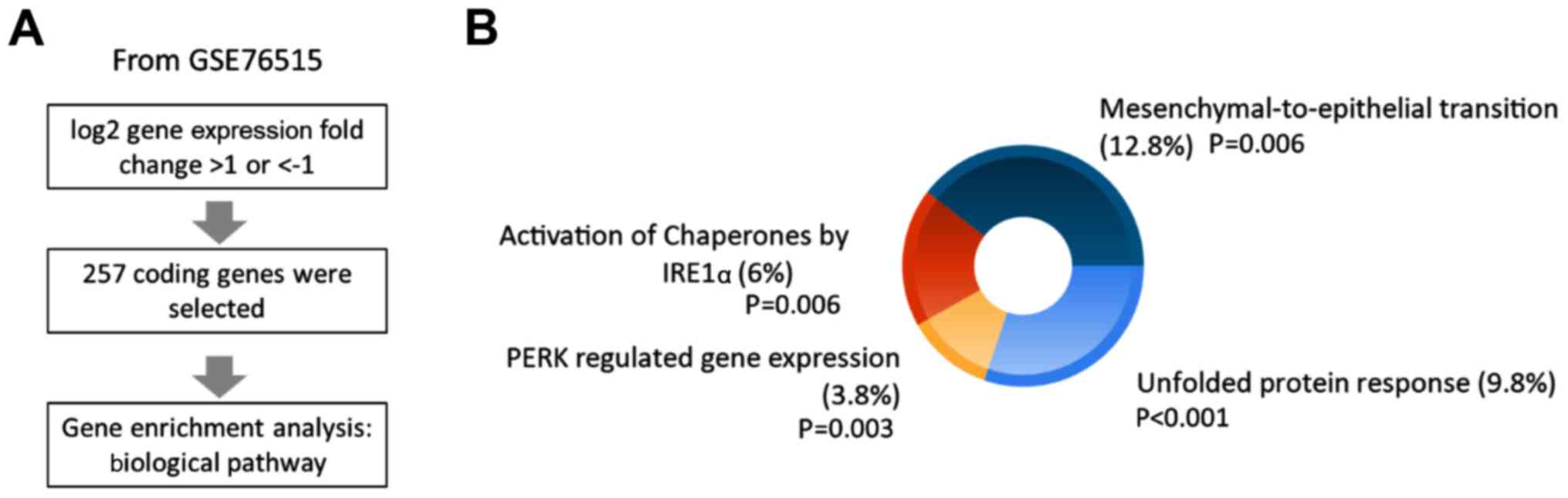
listed in Table I according to
‘Unfolded Protein Response’, ‘Activation of Chaperones by IRE1α’
and ‘PERK-regulated gene expression’. The potential transcription
factors of ACSL3, ACSL4, ACOT13 and SLC27A2 promoter are listed in
Table II.
 | Table I.Gene enrichment analysis for
biological pathways. |
Table I.
Gene enrichment analysis for
biological pathways.
| Biological
pathway | Enriched genes
(gene symbols are shown) |
|---|
| Unfolded protein
response | ASNS, CALR,
DDIT3, EDEM1, ERN1, FKBP14, GFPT1, HERPUD1, HSPA5, HYOU1, IGFBP1,
SYVN1, WIPI1 |
| Activation of
chaperones by IRE1α | EDEM1, ERN1,
FKBP14, GFPT1, HSPA5, HYOU1, SYVN1, WIPI1 |
| PERK-regulated gene
expression | ASNS, DDIT3,
HERPUD1, HSPA5, IGFBP1 |
|
Mesenchymal-to-epithelial transition | ANXA4, AREG,
CD24, CEACAM1, CXADR, DHCR24, ERBB3, ESRP1, EXPH5, GPX2, HES1,
HPGD, MALL, MTUS1, OAS1, SLPI |
 | Table II.Potential transcription factors on
the promoter region. |
Table II.
Potential transcription factors on
the promoter region.
| Gene (gene
symbol) | Transcription
factor (gene symbol) |
|---|
| ACOT4 | ETS2 |
| ACOT7 | GTF2IRD1,
TFAP2A |
| ACOT11 | ATF6, CTCF, CTF1,
ESR1, ESR2, ESRRA, ESRRB, HAND1, MAFA, MAFB, MYF5, MYOD1, MZF1,
NEUROD1, NF1, NFE2, NR1H4, PEBP1, PURA, SF1, TAL1, TCF12, TCF3,
TFAP2A, TFE3, TLX1 ETS2, MAFB, NEUROD1, NHLH1, NRF1 |
| ACSL1 | CLOCK, ETS2,
GTF2IRD1, MAFB, MAX, MYC, MYCN, NEUROD1, NFATC1, USF1 |
| ACSL3 | AHR, ARNT, CTCF,
ETS2, HSF1, MAFB, MZF1, NFKB1, NFYA, NR6A1, NR1H2, NRF1, NRIH3,
RXRA, TFAP2A, TOPORS |
| ACSL4 | ATF1, EGR1, EGR2,
EGR3, ELF1, ELK1, ETS2, GLI1, GLI2, GLI3, MZF1, NEUROD1, NFATC1,
NR1H4, REL, TAL1, ZEB1 |
| ACSM2B | NEUROD1 |
| SLC27A2 | CTCF, EGR2, ETS2,
MAFB, MYC, SMAD4, TAL1, TCF3 |
| SLC27A4 | ESRRA, NR1H4,
PPARG, RORA |
| SLC27A5 | ETS2, HNF4A, MAFB,
ZEB1 |
| SLC27A6 | MZF1 |
Statistical analysis
GraphPad Prism 7 software (GraphPad Software, Inc.,
La Jolla, CA, USA) was used to construct all graphs and perform all
statistical Differences between two groups and among more than
three groups were determined using the Student's t-test and a
one-way analysis of variance (ANOVA) with Tukey's multiple
comparison test, respectively. P<0.05 was considered to indicate
a statistically significant difference.
Results
Expression of fatty acyl-CoA-related
enzymes in lung cancer and related clinical outcomes for evaluated
patients
Our previous study indicated that expression of
ACOT11 and ACOT13 in lung adenocarcinoma cells was significantly
higher than that in cells from noncancerous tissues (21). In the present study, we further
evaluated expression and clinical outcomes for enzymes involved in
acyl-CoA formation [including short-chain ACS (ACSS), medium-chain
ACS (ACSM), long-chain ACS (ACSL), and very-long-chain ACS (SLC27A,
also called ‘ACSVL’)] and hydrolysis (ACOTs) by using the PRECOG
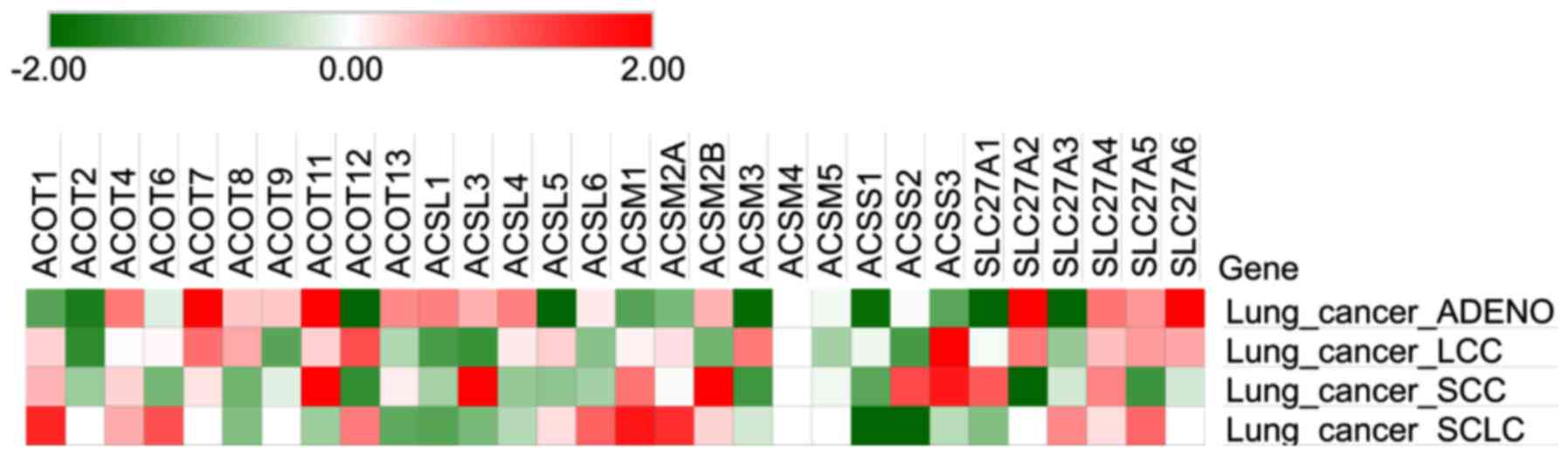
database. A higher Z-score was indicative of a relatively poor
prognosis. As presented in Fig. 1,
relatively high Z-scores for ACOT4, ACOT7, ACOT11, ACOT13, ACSL1,
ACSL3, ACSL4, ACSM2B, SLC27A2, SLC27A4, SLC27A5 and SLC27A6 in lung
adenocarcinoma and ACOT4, ACOT7, ACOT11, ACOT13, SLC27A4 and
SLC27A5 in other types of lung cancer were observed. Associations
between gene expression and clinical outcomes were further
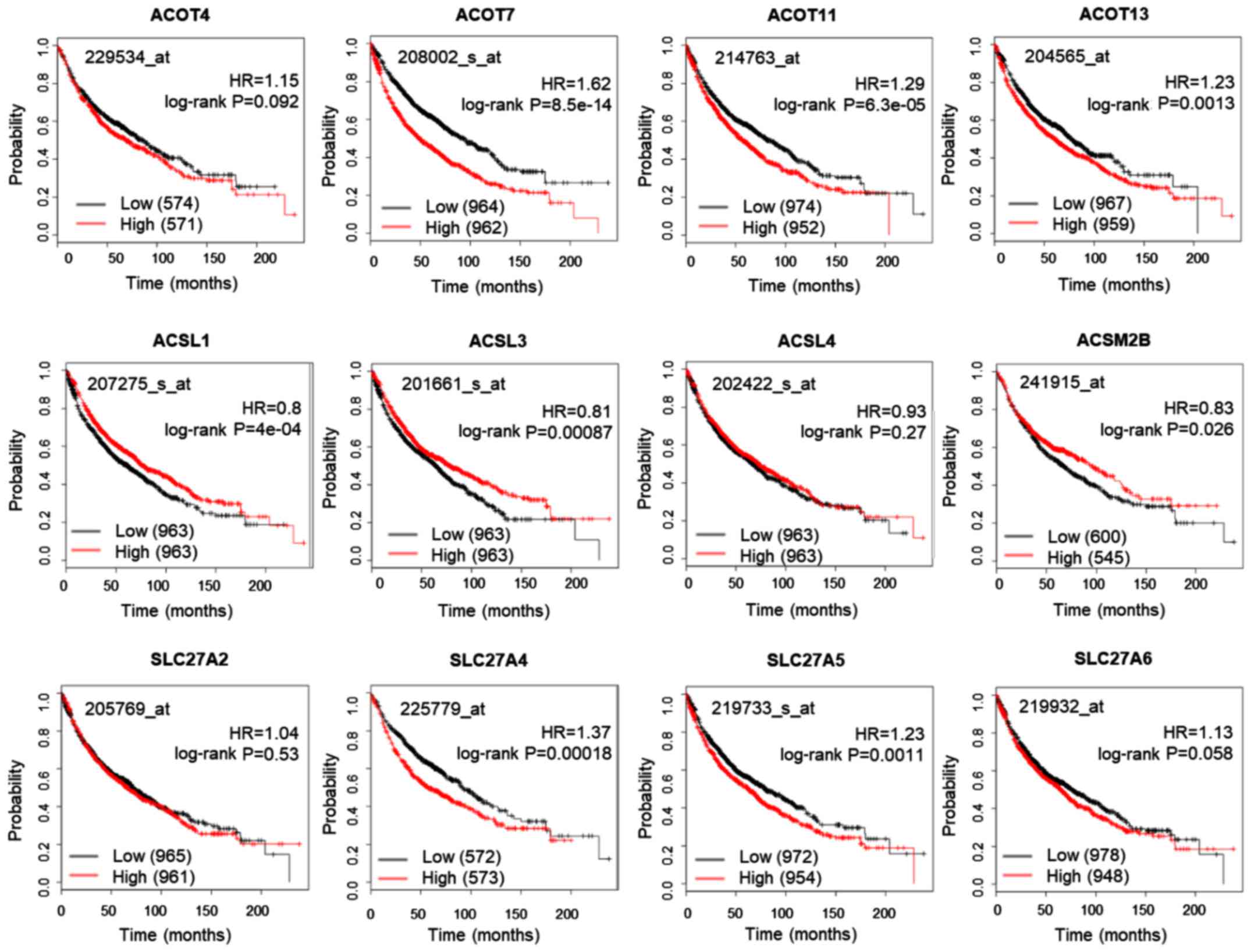
evaluated using the KM plotter database (Fig. 2). High expression of ACSL1, ACSL3
and ACSL5 was significantly associated with favorable prognosis,
whereas high expression of ACOT7, ACOT11, ACOT13, SLC27A4 and
SLC27A5 was significantly associated with poor prognosis in
patients with lung cancer. Thus, ACOT7, ACOT11, ACOT13, SLC27A4 and
SLC27A5 expression is generally associated with poor outcomes for
patients with lung cancer and may contribute to oncogenic
progression.
Expression of enzymes under ER
stress
To determine whether expression of the ACS and ACOT
enzymes is altered under ER stress, we first accessed the public
microarray database (GEO) and obtained a dataset that contained the
messenger RNA (mRNA) expression of A549 following control treatment
and tunicamycin treatment (accession number: GSE76515). Treatment
with tunicamycin blocks the biosynthesis of glycoproteins and
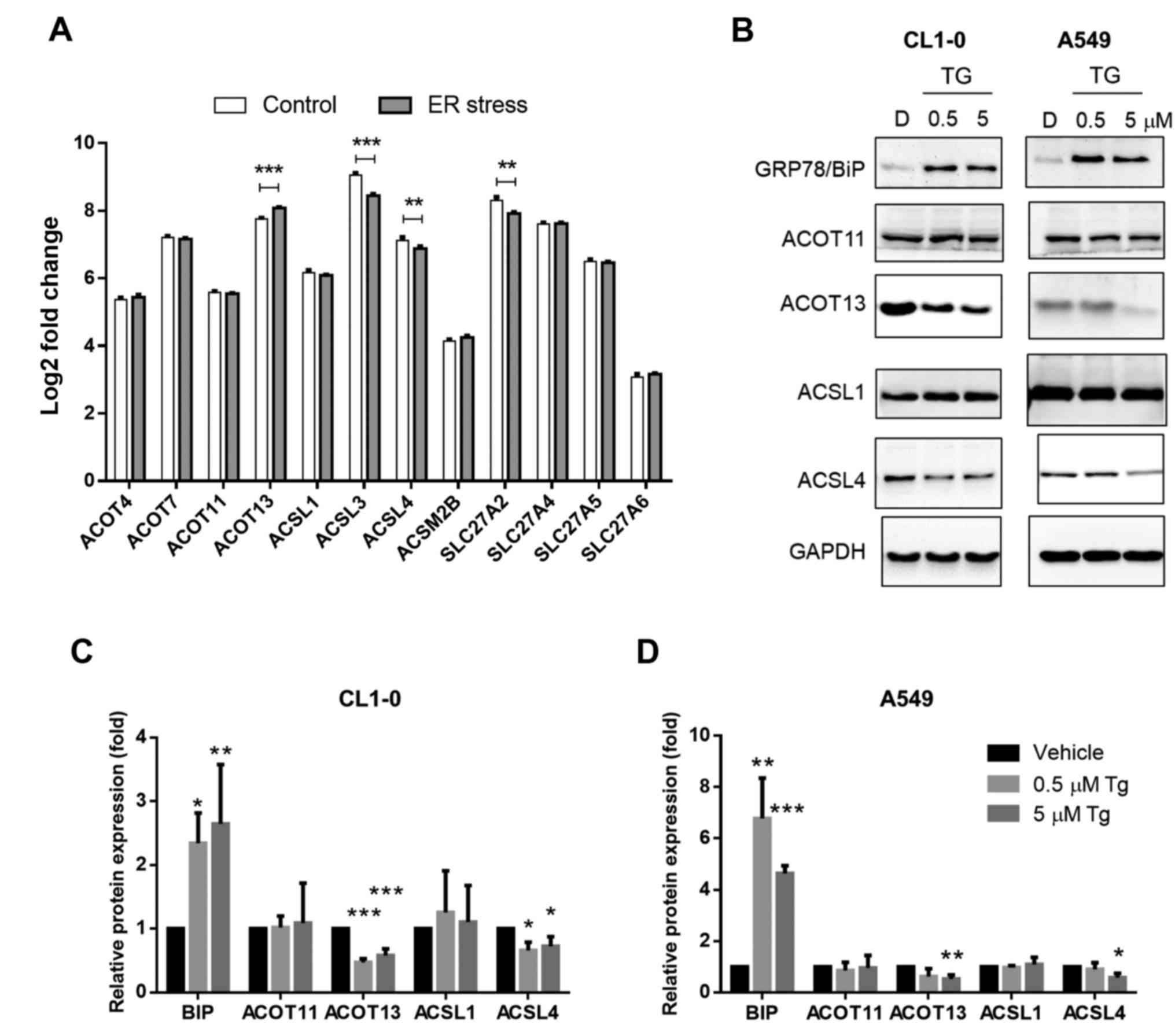
induces ER stress. Decreased levels of ACSL3, ACSL4 and SLC27A2 and
an increased level of ACOT13 were observed after ER stress
induction (Fig. 3A). We further
identified protein expression levels in lung adenocarcinoma cell
lines CL1-0 and A549. Tg, which inhibits sarcoplasmic and
endoplasmic reticulum Ca2+ ATPases, was used to induce
ER stress. As presented in Fig.
3B-D, expression of ER stress marker Bip/GRP78 was
significantly induced by Tg treatment. In addition, the protein
expression patterns of ACOT11, ACSL1 and ACSL4 were similar to the
mRNA expression pattern illustrated in Fig. 3A. By contrast, the expression of
ACOT13 was decreased in the Tg-treated groups (Fig. 3B-D). The results indicated that
expression levels of not all potential oncogenic ACS and ACOT
enzymes were altered under ER stress. In addition, the expression
of ACSL3 and SLC27A2, which was not associated with poor prognosis,
decreased after induction of ER stress.
Effects of ER stress on biological
enzyme functions
ER stress-inducing UPR signaling pathways affect ACS
and ACOT enzyme expression; thus, the effect on enzyme function was
further investigated. Expression of several ACS enzymes (ACSL3,
ACSL4, and SLC27A2) was decreased under ER stress, and the function
of ACS is linked to β-oxidation, which contributes to increases in
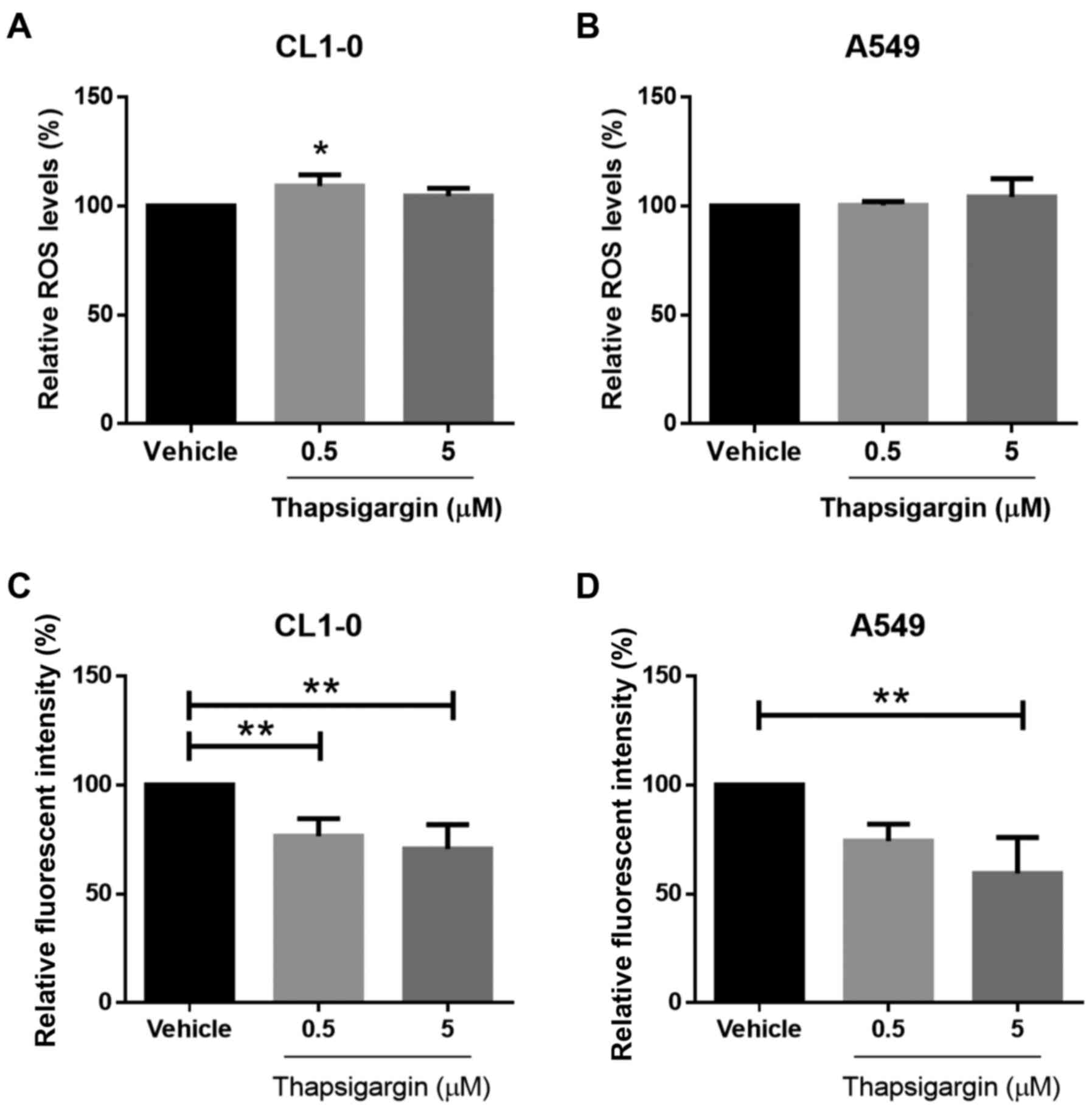
intracellular ROS (18). Therefore,
we evaluated the total levels of ROS in the CL1-0 and A549 lines.
However, the ROS levels did not differ significantly between the
groups (Fig. 4A and B). ACSL and
SLC27A enzymes can serve as fatty acid transporters (30,31).
As shown in Fig. 4C and D, capacity
for fatty acid uptake was significantly weakened after ER stress
induction. Our results suggested that fatty acid uptake capacity
was attenuated after Tg treatment. The causal relationship between
decreasing ACSL3, ACSL4 and SLC27A2 expression and fatty acid
uptake capacity requires further investigation.
Potential regulatory molecules and the
interaction network of ACSs and ACOTs under ER stress
Bioinformatic analysis and our results indicated
that the expression levels of ACOT13, ACSL3, ACSL4, and SCL27A2
were altered after ER stress induction. However, the regulatory
mechanism remained unclear. To screen for potential gene regulators
in lung cancer cells, data for genes with log2 fold changes of
>1 or ≤1 under ER stress were collected from the GSE76515
dataset and gene enrichment analysis for biological pathways was
conducted using FunRich software (Fig.
5A). The results revealed four biological pathways related to
ER stress and mesenchymal-to-epithelial transition, including UPR,
protein kinase RNA (PKR)-like ER kinase (PERK)-regulated gene
expression, and activation of chaperones by IRE1α (Fig. 5B). The studied genes are listed in
Table I. Previous studies have
demonstrated that these ER stress-induced pathways activate
transcription factors SREBP-1c and SREBP-2 as well as other lipid
metabolic enzymes such as SCD, acetyl-CoA carboxylase, and fatty
acid synthase (15). To further
investigate whether SREBPs regulate expression of ACS and ACOT
enzymes, potential regulatory elements in the promoters of these
genes were evaluated using the MotifMap website. The results listed
in Table II indicate that SREBPs
may not directly bind to the promoter regions of these genes after
induction of ER stress. The potential interaction network between
ER stress-induced biological pathways and ACS and ACOT enzymes was
further investigated using the GeneMania database. As illustrated
in Fig. 6, our results revealed
that ETS proto-oncogene 2 (ETS2) in the promoter region of ACSL4
and ETS2 and MAF BZIP transcription factor B (MAFB), which were
potential transcription factors in the promoter regions of ACSL3,
ACOT13 and SLC27A2, interacted with ER stress-induced UPR
genes.
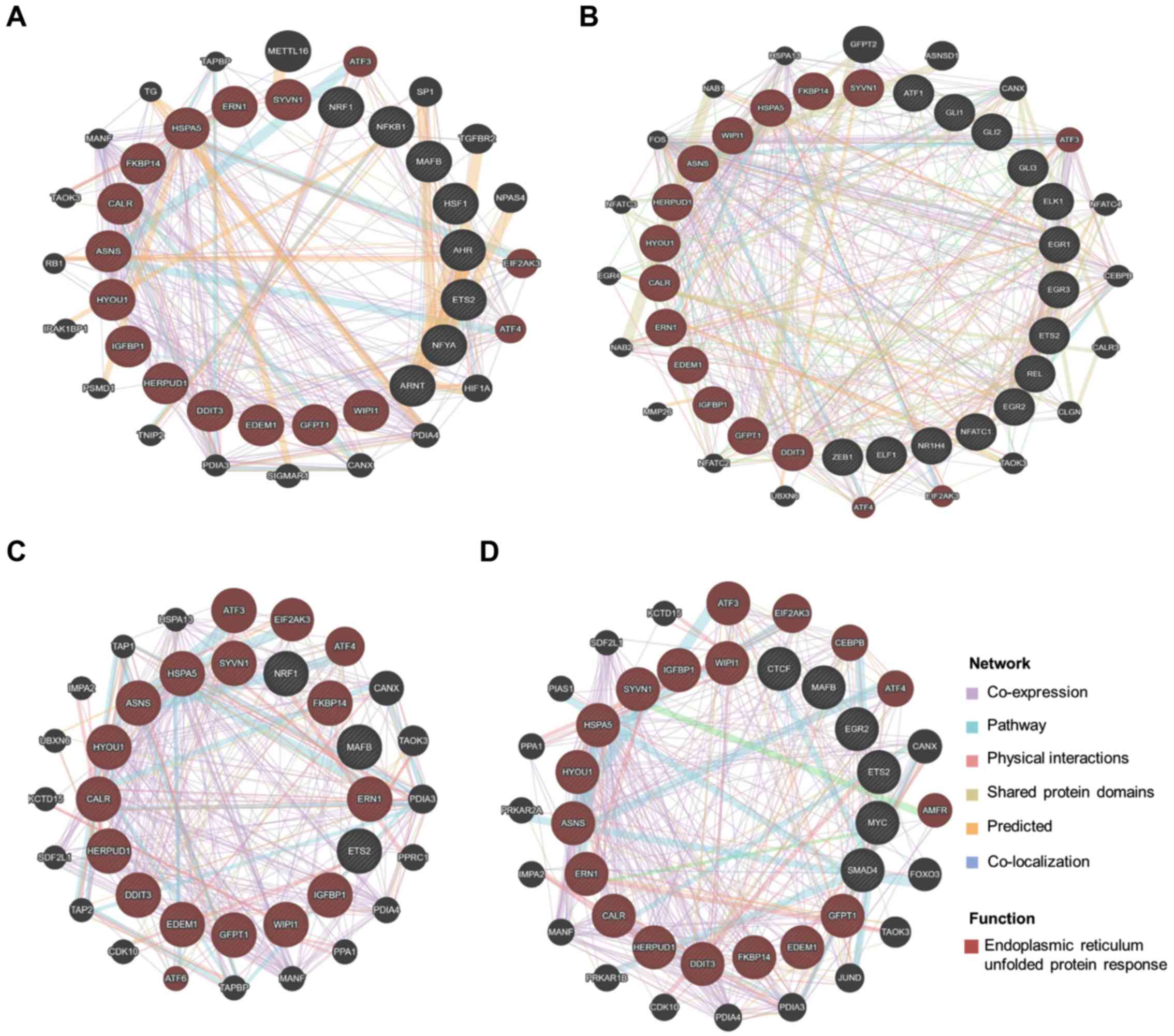 | Figure 6.Functional protein association
network. The interaction network of genes with induced ER stress
and transcription factors in promoter regions of ACSL3, ACSL4,
ACOT13, and SLC27A2 was predicted using the GeneMANIA database. The
input genes are displayed in striped circles within the inner
circle. The protein with the function ‘endoplasmic reticulum
unfolded protein response’ is marked by a red circle. The gene
interaction network was created using the terms ‘co-expression’,
‘pathway’, ‘physical interactions’, ‘shared protein domains’,
‘predicted’ and ‘co-localization’, and each interaction network is
marked by a differently colored line. The predicted interaction
network of (A) ACSL3, (B) ACSL4, (C) ACOT13 and (D) SLC27A2 is
displayed. The transcription factors that did not interact with
‘endoplasmic reticulum unfolded protein response’ genes are not
shown. |
Discussion
Studies have identified the role of acyl-CoA
metabolic enzymes in multiple types of cancer. Low expression of
ACSL5 serves as a prognostic factor for early recurrence of
colorectal carcinoma (32). The
ACSL1/ACSL4/SCD lipid network enhances migration and invasion
capacity by inducing epithelial-mesenchymal transition in
colorectal cancer, and microRNA-19b-1 is a key regulator in this
oncogenic axis (33,34). Additionally, high expression of
ACSL5 may be used as a biomarker for predicting relatively
favorable clinical outcomes in patients with breast cancer
(35). In hepatocellular carcinoma,
silencing of ACOT8 inhibits tumor cell proliferation (36). ACOT7 is involved in the development
of breast and lung cancer through regulation of the cell cycle via
p53/p21 signaling (37). Our
previous study indicated that proliferation of lung adenocarcinoma
was suppressed after ACOT11 and ACOT13 had been silenced (21). In the present study, we used the
PRECOG website and KM plotter to evaluate the association between
expression of ACOT and ACS enzymes and clinical outcomes in human
lung cancer patients. The analysis results revealed that some of
these enzymes may be linked to tumor progression. High expression
levels of ACOT7, ACOT11, ACOT13, SLC27A4 and SLC27A5 were observed
in most types of lung cancer and were associated with poor
outcomes. These findings confirmed that ACOT7, ACOT11, ACOT13,
SLC27A4 and SLC27A5 may play oncogenic roles in the development of
lung cancers. Other genes require further investigation.
In a tumor microenvironment, various stimuli can
induce ER stress (12). In the
present study, we evaluated gene expression levels after treatment
with two types of ER stress inducers. We expected the protein
expression levels in most enzymes to decrease after treatment with
UPR as UPR triggers ER-associated degradation response. However, of
the 12 enzymes, only ACOT13, ACSL3, ACSL4 and SLC27A2 were affected
after glycoprotein biosynthesis had been blocked by tunicamycin
treatment (Fig. 3A). Additionally,
we observed that the protein expression patterns of ACOT11, ACSL1,
and ACSL4 in Tg-treated CL1-0 and A549 were similar to those of
mRNA expression (Fig. 3B). These
results suggest that at the very least, ACOT11, ACSL1 and ACSL4 may
have activated the same regulatory mechanism during tunicamycin and
Tg treatment. ACOT13 expression was not consistent in mRNA and
protein expression; this indicated that Tg treatment and
tunicamycin treatment may have affected ACOT13 expression via
different regulatory pathways. ACSL3, ACSL4, and SLC27A2 are all
ACS enzymes, the biological function of which is connected to
β-oxidation, phospholipid and triglyceride synthesis, and fatty
acid uptake (18,30,31).
Thus, the weakened capacity for fatty acid uptake observed in both
lung cancer cell lines may have been due to decreased expression of
ACSL3, ACSL4, and SLC27A2 (Fig. 4C and
D). By contrast, the expression levels of other ACS enzymes
were not significantly affected by ER stress, regardless of whether
they were associated with favorable or poor prognosis.
Additionally, the expression levels of ACOT4, ACOT7, and ACOT11
were not significantly affected by ER stress. ACOT enzymes regulate
intracellular fatty acyl-CoAs and are involved in various metabolic
processes such as energy expenditure and hepatic and neuronal
functions (38). The role of ACOT
enzymes in lung cancer is not completely understood. The results of
our present and previous study (21) indicate that the functions of ACOT4,
ACOT7 and ACOT11 may be associated with tumor progression. Based on
the finding that the expression of these enzymes was not
significantly altered by ER stress, the metabolic functions of
ACOT4, ACOT7 and ACOT11 may be essential for resolving ER stress.
In addition, the substrates of human ACOT4 are short-chain
dicarboxylyl-CoAs and medium- to long-chain acyl-CoAs (39); those of ACOT7 are long-chain
acyl-CoAs (40); and those of
ACOT11 range from acetyl-CoAs to long-chain acyl-CoAs (41). The roles of the metabolites of these
ACOT enzymes in ER stress-mediated lipid metabolic pathways warrant
further study.
With the exception of SREBPs, induction of ER stress
activates downstream pathways, including the PERK, IRE and ATF6
pathways (42). However, no direct
evidence has confirmed that these pathways are linked to ACS and
ACOT regulation under ER stress. Through investigation of the
potential regulatory molecules of ACS and ACOT in lung cancer, we
identified significant gene enrichment on four biological pathways
(Table I), three of which were
related to UPR pathways, and the other of which was a
mesenchymal-to-epithelial transition pathway. Therefore, PERK, IRE,
and ATF6 pathways may regulate the gene expression of ACOT13,
ACSL3, ACSL4 and SLC27A2. Table II
lists the possible regulators of gene promoters. ACOT11 may be
directly regulated by transcription factor ATF6; the other genes
require further investigation. The mesenchymal-to-epithelial
transition pathway was enriched after induction of ER stress. The
ACSL1/ACSL4/SCD lipid network can promote the
epithelial-mesenchymal transition pathway (33); therefore, a decreased number of ACS
enzymes or weakened fatty acid uptake may have been involved in
activation of the mesenchymal-to-epithelial transition pathway.
Furthermore, the proposed interaction network indicated that ETS2
and MAFB may be regulators of ACSL3, ACOT13, SLC27A2 and ACSL4. The
expression levels of ACOT4, ACOT11 and ACSL1 were not affected by
ER stress; however, ETS2 and MAFB are potential transcription
factors of these genes (Table II).
The details of the proposed regulatory network require further
investigation.
Our study evaluated the effect of ER stress on
oncogenic ACS and ACOT enzymes. The PRECOG and KM plotter databases
were used for evaluation, and the results suggested that high
expression of ACOT7, ACOT11, ACOT13, SLC27A4 and SLC27A5 was
associated with poor clinical outcomes. Additionally, ER stress
affected expression of some enzymes and attenuated fatty acid
uptake capacity but did not affect the expression levels of
oncogenic ACOT7, ACOT11, SLC27A4 and SLC27A5. Bioinformatic
analysis revealed potential regulatory molecules and a regulatory
network. In summary, our findings indicated potential oncogenic
acyl-CoA metabolic enzymes, the biological effects of decreased
enzyme levels, and possible regulatory elements under ER stress in
lung cancer.
Acknowledgements
The authors thank the Center for Research Resources
and Development of Kaohsiung Medical University.
Funding
The present study was supported by grants from the
Ministry of Science and Technology (MOST 107-2320-B-037-011-MY3),
the Kaohsiung Medical University Hospital Research Foundation
(KMUH106-6M58), the Kaohsiung Medical University Research
Foundation (105KMUOR05) and the Kaohsiung Medical University
(KMU-DK 108003).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
KTL and PLK designed the study; SKC and MCY
performed the experiments for the study; KTL, IJY, SKC, MCY and PLK
analyzed the data and interpreted the results; KTL, MCY and PLK
wrote the manuscript. All authors read and approved the manuscript
and agree to be accountable for all aspects of the research in
ensuring that the accuracy or integrity of any part of the work are
appropriately investigated and resolved.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2017. CA Cancer J Clin. 67:7–30. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Stewart B and Wild CP: World Cancer Report
2014International Agency for Research on Cancer. World Health
Organization; 2014, View Article : Google Scholar
|
|
3
|
Chen JQ and Russo J: Dysregulation of
glucose transport, glycolysis, TCA cycle and glutaminolysis by
oncogenes and tumor suppressors in cancer cells. Biochim Biophys
Acta. 1826:370–384. 2012.PubMed/NCBI
|
|
4
|
Locasale JW: Serine, glycine and
one-carbon units: Cancer metabolism in full circle. Nat Rev Cancer.
13:572–583. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hensley CT, Wasti AT and DeBerardinis RJ:
Glutamine and cancer: Cell biology, physiology, and clinical
opportunities. J Clin Invest. 123:3678–3684. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Visca P, Sebastiani V, Botti C, Diodoro
MG, Lasagni RP, Romagnoli F, Brenna A, De Joannon BC, Donnorso RP,
Lombardi G, et al: Fatty acid synthase (FAS) is a marker of
increased risk of recurrence in lung carcinoma. Anticancer Res.
24:4169–4173. 2004.PubMed/NCBI
|
|
7
|
Huang J, Fan XX, He J, Pan H, Li RZ, Huang
L, Jiang Z, Yao XJ, Liu L, Leung EL, et al: SCD1 is associated with
tumor promotion, late stage and poor survival in lung
adenocarcinoma. Oncotarget. 7:39970–39979. 2016.PubMed/NCBI
|
|
8
|
Wen T, Gao L, Wen Z, Wu C, Tan CS, Toh WZ
and Ong CN: Exploratory investigation of plasma metabolomics in
human lung adenocarcinoma. Mol Biosyst. 9:2370–2378. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Liu J, Mazzone PJ, Cata JP, Kurz A, Bauer
M, Mascha EJ and Sessler DI: Serum free fatty acid biomarkers of
lung cancer. Chest. 146:670–679. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhang Y, He C, Qiu L, Wang Y, Zhang L, Qin
X, Liu Y, Zhang D and Li Z: Serum unsaturated free fatty acids:
Potential biomarkers for early detection and disease progression
monitoring of non-small cell lung cancer. J Cancer. 5:706–714.
2014. View
Article : Google Scholar : PubMed/NCBI
|
|
11
|
Cao SS and Kaufman RJ: Endoplasmic
reticulum stress and oxidative stress in cell fate decision and
human disease. Antioxid Redox Signal. 21:396–413. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yadav RK, Chae SW, Kim HR and Chae HJ:
Endoplasmic reticulum stress and cancer. J Cancer Prev. 19:75–88.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Tabas I and Ron D: Integrating the
mechanisms of apoptosis induced by endoplasmic reticulum stress.
Nat Cell Biol. 13:184–190. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Corazzari M, Gagliardi M, Fimia GM and
Piacentini M: Endoplasmic reticulum stress, unfolded protein
response, and cancer cell fate. Front Oncol. 7:782017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Han J and Kaufman RJ: The role of ER
stress in lipid metabolism and lipotoxicity. J Lipid Res.
57:1329–1338. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lauressergues E, Bert E, Duriez P, Hum D,
Majd Z, Staels B and Cussac D: Does endoplasmic reticulum stress
participate in APD-induced hepatic metabolic dysregulation?
Neuropharmacology. 62:784–796. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Griffiths B, Lewis CA, Bensaad K, Ros S,
Zhang Q, Ferber EC, Konisti S, Peck B, Miess H, East P, et al:
Sterol regulatory element binding protein-dependent regulation of
lipid synthesis supports cell survival and tumor growth. Cancer
Metab. 1:32013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Faergeman NJ and Knudsen J: Role of
long-chain fatty acyl-CoA esters in the regulation of metabolism
and in cell signalling. Biochem J. 323:1–12. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ellis JM, Bowman CE and Wolfgang MJ:
Metabolic and tissue-specific regulation of acyl-CoA metabolism.
PLoS One. 10:e01165872015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Jung WY, Kim YH, Ryu YJ, Kim BH, Shin BK,
Kim A and Kim HK: Acyl-CoA thioesterase 8 is a specific protein
related to nodal metastasis and prognosis of lung adenocarcinoma.
Pathol Res Pract. 209:276–283. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hung JY, Chiang SR, Liu KT, Tsai MJ, Huang
MS, Shieh JM, Yen MC and Hsu YL: Overexpression and proliferation
dependence of acyl-CoA thioesterase 11 and 13 in lung
adenocarcinoma. Oncol Lett. 14:3647–3656. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Gentles AJ, Newman AM, Liu CL, Bratman SV,
Feng W, Kim D, Nair VS, Xu Y, Khuong A, Hoang CD, et al: The
prognostic landscape of genes and infiltrating immune cells across
human cancers. Nat Med. 21:938–945. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Gyorffy B, Surowiak P, Budczies J and
Lanczky A: Online survival analysis software to assess the
prognostic value of biomarkers using transcriptomic data in
non-small-cell lung cancer. PLoS One. 8:e822412013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Barrett T, Wilhite SE, Ledoux P,
Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH,
Sherman PM, Holko M, et al: NCBI GEO: Archive for functional
genomics data sets-update. Nucleic Acids Res. 41:D991–D995. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Pathan M, Keerthikumar S, Chisanga D,
Alessandro R, Ang CS, Askenase P, Batagov AO, Benito-Martin A,
Camussi G, Clayton A, et al: A novel community driven software for
functional enrichment analysis of extracellular vesicles data. J
Extracell Vesicles. 6:13214552017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Pathan M, Keerthikumar S, Ang CS, Gangoda
L, Quek CY, Williamson NA, Mouradov D, Sieber OM, Simpson RJ, Salim
A, et al: FunRich: An open access standalone functional enrichment
and interaction network analysis tool. Proteomics. 15:2597–2601.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Daily K, Patel VR, Rigor P, Xie X and
Baldi P: MotifMap: Integrative genome-wide maps of regulatory motif
sites for model species. BMC Bioinformatics. 12:4952011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Chu YW, Yang PC, Yang SC, Shyu YC, Hendrix
MJ, Wu R and Wu CW: Selection of invasive and metastatic
subpopulations from a human lung adenocarcinoma cell line. Am J
Respir Cell Mol Biol. 17:353–360. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Warde-Farley D, Donaldson SL, Comes O,
Zuberi K, Badrawi R, Chao P, Franz M, Grouios C, Kazi F, Lopes CT,
et al: The GeneMANIA prediction server: Biological network
integration for gene prioritization and predicting gene function.
Nucleic Acids Res. 38:W214–W220. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Watkins PA: Very-long-chain acyl-CoA
synthetases. J Biol Chem. 283:1773–1777. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Digel M, Ehehalt R, Stremmel W and
Fullekrug J: Acyl-CoA synthetases: Fatty acid uptake and metabolic
channeling. Mol Cell Biochem. 326:23–28. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Hartmann F, Sparla D, Tute E, Tamm M,
Schneider U, Jeon MK, Kasperk R, Gassler N and Kaemmerer E: Low
acyl-CoA synthetase 5 expression in colorectal carcinomas is
prognostic for early tumour recurrence. Pathol Res Pract.
213:261–266. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Sanchez-Martinez R, Cruz-Gil S, de Cedron
Gomez M, Álvarez-Fernández M, Vargas T, Molina S, García B, Herranz
J, Moreno-Rubio J, Reglero G, et al: A link between lipid
metabolism and epithelial-mesenchymal transition provides a target
for colon cancer therapy. Oncotarget. 6:38719–38736. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Cruz-Gil S, Sanchez-Martinez R, de Cedron
Gomez M, Martin-Hernandez R, Vargas T, Molina S, Herranz J, Davalos
A, Reglero G and de Molina Ramirez A: Targeting the lipid metabolic
axis ACSL/SCD in colorectal cancer progression by therapeutic
miRNAs: miR-19b-1 role. J Lipid Res. 59:14–24. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Yen MC, Kan JY, Hsieh CJ, Kuo PL, Hou MF
and Hsu YL: Association of long-chain acyl-coenzyme A synthetase 5
expression in human breast cancer by estrogen receptor status and
its clinical significance. Oncol Rep. 37:3253–3260. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Hung YH, Chan YS, Chang YS, Lee KT, Hsu
HP, Yen MC, Chen WC, Wang CY and Lai MD: Fatty acid metabolic
enzyme acyl-CoA thioesterase 8 promotes the development of
hepatocellular carcinoma. Oncol Rep. 31:2797–2803. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Jung SH, Lee HC, Hwang HJ, Park HA, Moon
YA, Kim BC, Lee HM, Kim KP, Kim YN, Lee BL, et al: Acyl-CoA
thioesterase 7 is involved in cell cycle progression via regulation
of PKCzeta-p53-p21 signaling pathway. Cell Death Dis. 8:e27932017.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Tillander V, Alexson SEH and Cohen DE:
Deactivating fatty acids: Acyl-coa thioesterase-mediated control of
lipid metabolism. Trends Endocrinol Metab. 28:473–484. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Hunt MC, Rautanen A, Westin MA, Svensson
LT and Alexson SE: Analysis of the mouse and human acyl-CoA
thioesterase (ACOT) gene clusters shows that convergent, functional
evolution results in a reduced number of human peroxisomal ACOTs.
FASEB J. 20:1855–1864. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Forwood JK, Thakur AS, Guncar G, Marfori
M, Mouradov D, Meng W, Robinson J, Huber T, Kellie S, Martin JL, et
al: Structural basis for recruitment of tandem hotdog domains in
acyl-CoA thioesterase 7 and its role in inflammation. Proc Natl
Acad Sci U S A. 104:10382–10387. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Han S and Cohen DE: Functional
characterization of thioesterase superfamily member 1/Acyl-CoA
thioesterase 11: Implications for metabolic regulation. J Lipid
Res. 53:2620–2631. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Basseri S and Austin RC: Endoplasmic
reticulum stress and lipid metabolism: Mechanisms and therapeutic
potential. Biochem Res Int. 2012:8413622012. View Article : Google Scholar : PubMed/NCBI
|