Introduction
Thyroid cancer, the most common endocrine malignancy
worldwide, has increased 3-fold in the past 3 decades (1,2).
Well-differentiated thyroid cancer (WDTC), including papillary and
follicular thyroid cancer, represents >90% of all thyroid
cancers (3), and patients with WDTC
have a 10-year survival of 80–95%. Many studies have profiled WDTC
genes and rearrangements of RET/PTC and PAX8/PPARγ
(4), and frequent (70%) point
mutations of BRAF and RAS genes, which alter the
mitogen-activated protein kinase (MAPK) signaling pathway (5–7) were
documented in WDTC. Several genes, such as MUC1, PD-L1,
DPP4, and mutations of the BRAFV600E and
TERT promoter, have been reported as prognostic markers
(8–11).
For WDTC, 10-year survival is reduced to 14–40% when
distant metastasis (DM) occurs (12–14).
For patients with metastases, radioactive iodine (131I)
has been the mainstay of treatment, selectively combined with
surgical intervention, external beam radiation and chemotherapy
(15). Metastatic lesions are
sometimes resistant to 131I, so treatment options are
limited and survival is poor (10-year survival, 10%) (14,16).
Recurrence also greatly contributes to the morbidity of WDTC
(17,18), and recurrence at 30 years is 35%,
most being local with 32% being DM (17). Thus, we must identify potential
targets to improve therapeutic strategies to prevent metastasis and
recurrence.
MicroRNAs (miRNAs) are a class of endogenous small
(18–22 nucleotide) non-coding RNAs. Gene expression regulation by
miRNAs is of interest because many miRNAs in WDTC, such as miR-221,
miR-222 and miR-146 (19–21), have been reported to have critical
roles in the regulation of apoptosis, proliferation, the cell cycle
and epithelial-mesenchymal transition by negatively regulating
mRNAs post-transcriptionally. Other studies on mRNA and miRNA
expression indicate a miRNA-mRNA regulatory network that may
provide clues for genetic deregulation in WDTC (22,23).
Although many studies have focused on the tumorigenesis of WDTC,
few studies have identified the mechanism of metastasis and
recurrence in WDTC. This is due to the fact that tissues must be
taken from metastatic sites, yet metastasis-prone sites of WDTC,
such as the brain and bones, are challenging to sample properly.
Second, the incidence of metastasis and recurrence is relatively
low, and as such, WDTC patients have better outcomes than other
malignancies.
In the present study, we collected tissue samples
from four separate stages of WDTC: Normal, cancerous, metastatic
and recurrent stages and we performed mRNA and miRNA microarray
analysis. By integrating our study with gene expression data from
The Cancer Genome Atlas (TCGA) (21), we revealed novel prognostic markers
and key genes related to the progression of WDTC and possible
pathways involved in WDTC recurrence.
Materials and methods
Pipeline and workflow
We observed changes in the expression of mRNA and
miRNA in normal, cancerous, metastatic and recurrent stages of
WDTC. To study tumorigenesis and metastasis, we overlapped our
results with differentially expressed mRNAs (DEmRNAs) and miRNAs
(DEmiRNAs) from TCGA sequencing data, which contained more than 500
cancerous samples of papillary thyroid cancer, 59 normal samples, 8
lymph node metastatic (LNM) samples and corresponding clinical
data. We performed survival analysis to identify prognostic
influence, enrichment analysis to reveal relevant pathways and
ontology of gene sets and miRNA target prediction to depict
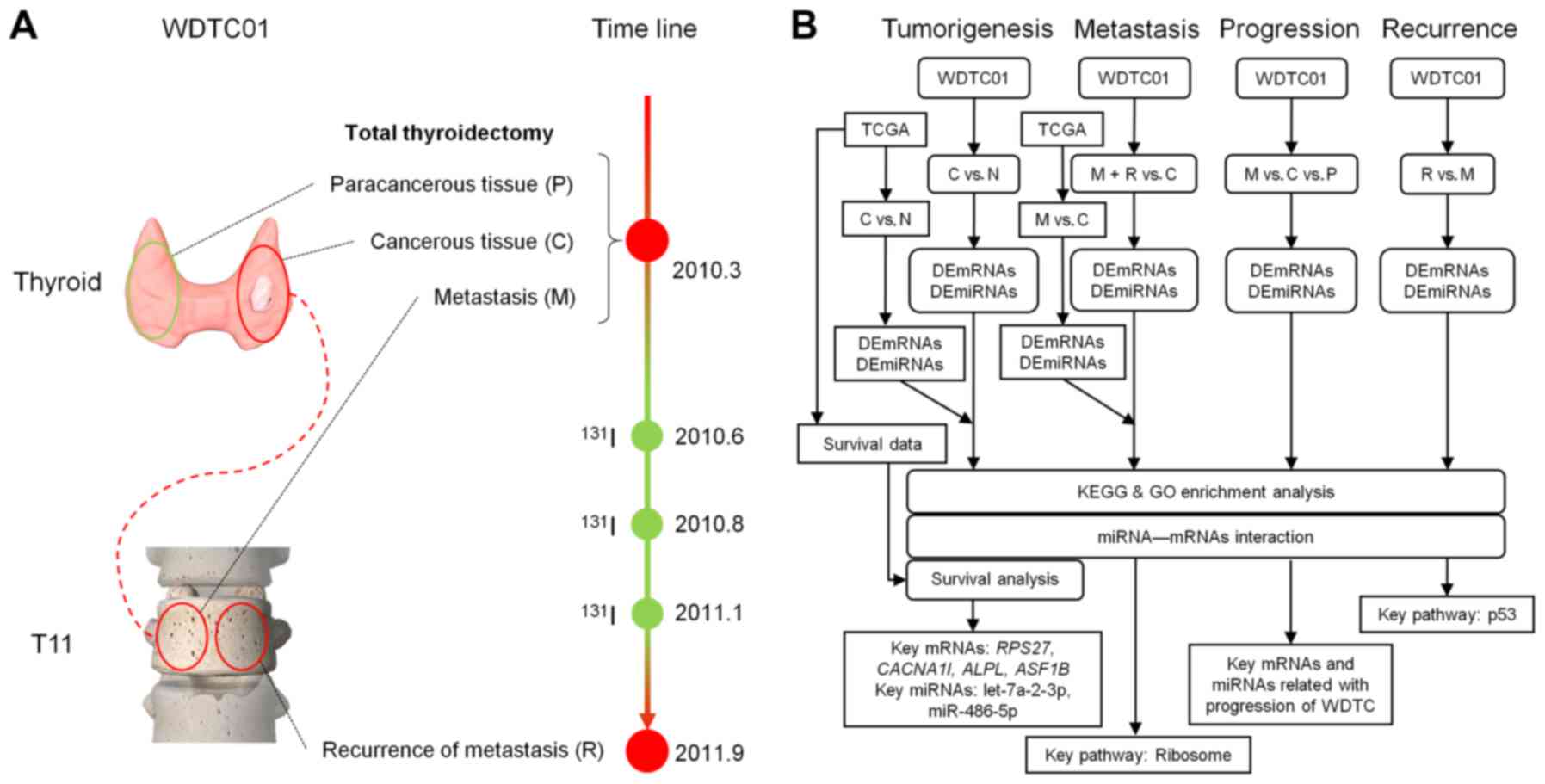
possible miRNA-mRNA interactions in WDTC (Fig. 1).
Patient and sample preparation
The present study was approved by the Institutional
Review Board and Ethics Committee of The Shanghai Tenth People's
Hospital, Tongji University, Shanghai, China
(SHSY-IEC-KY-4.0/17-13/01). In March 2010, case WDTC01 was a
40-year-old male patient, diagnosed with follicular thyroid cancer
and distant bone metastasis (BM) in the 11th thoracic vertebrae.
Total thyroidectomy was performed at Zhongshan Hospital, Shanghai,
China. Paracancerous and cancerous tissues were collected during
surgery. To relieve neurological symptoms of the lower extremity
caused by BM, the metastatic lesion was debulked, collected and
internally fixed. After three 131I radiation ablations,
local recurrence was observed at the 11th thoracic vertebrae on
September 2011 and the recurred lesion was collected during surgery
(Fig. 1). All samples were
immediately stored at −80°C until use. Total RNA was harvested
using TRIzol (Invitrogen; Thermo Fisher Scientific, Inc., Waltham,
MA, USA) and an miRNeasy mini kit (Qiagen GmbH, Hilden, Germany)
according to the manufacturer's instructions.
Genome-wide transcriptional profiling
with Agilent microarray and microRNA microarray expression
profiling
RNA was immediately shipped on dry ice to KangChen
Bio-Tech (Shanghai, China) for mRNA and miRNA microarray assay. For
analysis via the Agilent Whole Human Genome Oligo Microarray
platform, total RNA from each sample was amplified and transcribed
into fluorescent cRNA using the manufacturer's instructions
(Agilent's Quick Amp Labeling protocol, version 5.7; Agilent
Technologies, Santa Clara, CA, USA). Labeled cRNAs were hybridized
onto the Whole Human Genome Oligo Microarray (4×44 K; Agilent
Technologies). After washing the slides, arrays were scanned using
the Agilent Scanner G2505C. Agilent Feature Extraction software
(version 11.0.1.1) was used to analyze the acquired array images.
Quantile normalization and subsequent data processing were
performed using the GeneSpring GX v11.5.1 software package (Agilent
Technologies).
For the miRNA microarray, samples were labeled using
the miRCURY Hy3/Hy5 Power labeling kit and hybridized with a
miRCURY LNA Array (v.16.0) (Exiqon, Vedbaek, Denmark). Following
washing, the slides were scanned using the Axon GenePix 4000B
microarray scanner. Scanned images were then imported into GenePix
Pro 6.0 software (Axon Instruments; Molecular Devices, LLC,
Sunnyvale, CA, USA) for grid alignment and data extraction.
Replicated miRNAs were averaged and miRNAs in which intensities
were ≥50 in all samples were chosen for calculating a normalization
factor.
Targeting prediction and enrichment
analysis
The online prediction tool MiRWalk 2.0 (24) was used to predict interactions
between miRNAs and mRNAs. In addition, miRanda (25) and TargetScan (26) were selected to generate an
intersecting gene list. When an mRNA was predicted by all three
databases, it was considered a target for the miRNA. Biological
pathways defined by the Kyoto Encyclopedia of Genes and Genomes
(KEGG) (27) and Gene Ontology (GO)
(28) were analyzed using the
Database for Annotation, Visualization and Integrated Discovery
(DAVID) (29). DAVID provides a set
of functional annotations for many genes. In the present study,
KEGG pathway and GO terms were selected with a threshold of
P<0.05.
Statistical analysis
For WDTC01, mRNAs and miRNAs with fold-changes (FC)
>1.5 times were defined as differentially expressed and
considered for analysis. RNA sequencing data with 505 samples of
papillary thyroid cancer (PTC), 59 normal control and eight LNM,
with the −3 miRNA sequencing data of 502 samples of PTC, 59 normal
control and eight LNM from TCGA thyroid carcinoma (THCA), and
survival data were downloaded from the TCGA analytical tool UCSC
Xena (http://xena.ucsc.edu). However,
recurrent sites were not available. The unit for the mRNA
sequencing data was log2 (RSEM+1) and the unit for the miRNA
sequencing data was log2 (RPM+1). To screen differentially
expressed mRNAs and miRNAs with a median expression >0, we used
the linear model of the limma package (30) of R-3.4.1. Benjamin and Hochberg's
(31) correction was applied to
ensure a false discovery rate (FDR). For comparisons between cancer
and normal samples, DEmRNAs and DEmiRNAs were identified when FC
>1.5, P<0.05 and FDR <0.05. For comparisons between 8 LNM
and cancer samples, criteria were FC >1.5 and P<0.05.
To depict expression differences, a scatter plot was
used to describe averages and standard deviations of expression. To
address overall survival (OS) and recurrence-free survival (RFS)
for patients, we retrieved follow-up states for patients in the
clinical data matrix from TCGA and merged this with the expression
matrix and performed Kaplan-Meier analysis for each mRNA and miRNA
with survival of R. OS and RFS were considered significant when
log-rank test was P<0.05.
Results
Upregulated ASF1B and ALPL and
downregulated RPS27, CACNA1I, let-7a-2-3p and miR-486-5p are
related to the prognosis of WDTC
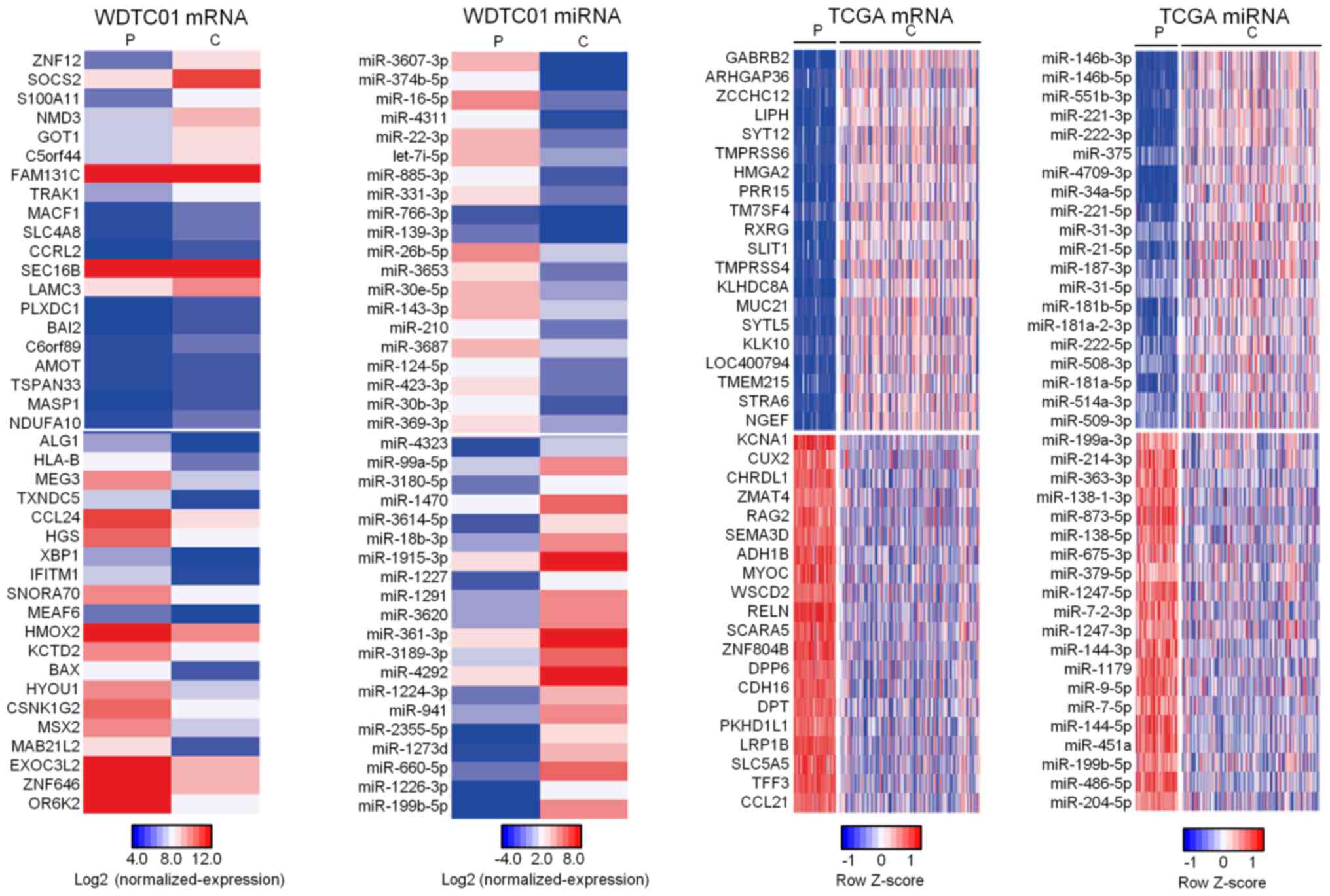
We observed 444 upregulation and 740 downregulated
DEmRNAs and 114 upregulation and 105 downregulated DEmiRNAs in
cancerous tissue for WDTC01 (Fig.
2). To validate commonly expressed DEmRNAs and miRNAs in WDTC,
DEmRNAs and miRNAs were compared with TCGA data. We identified 49
upregulated and 53 downregulated DEmRNAs, along with 2 upregulated
and 10 downregulated DEmiRNAs, which were commonly deregulated
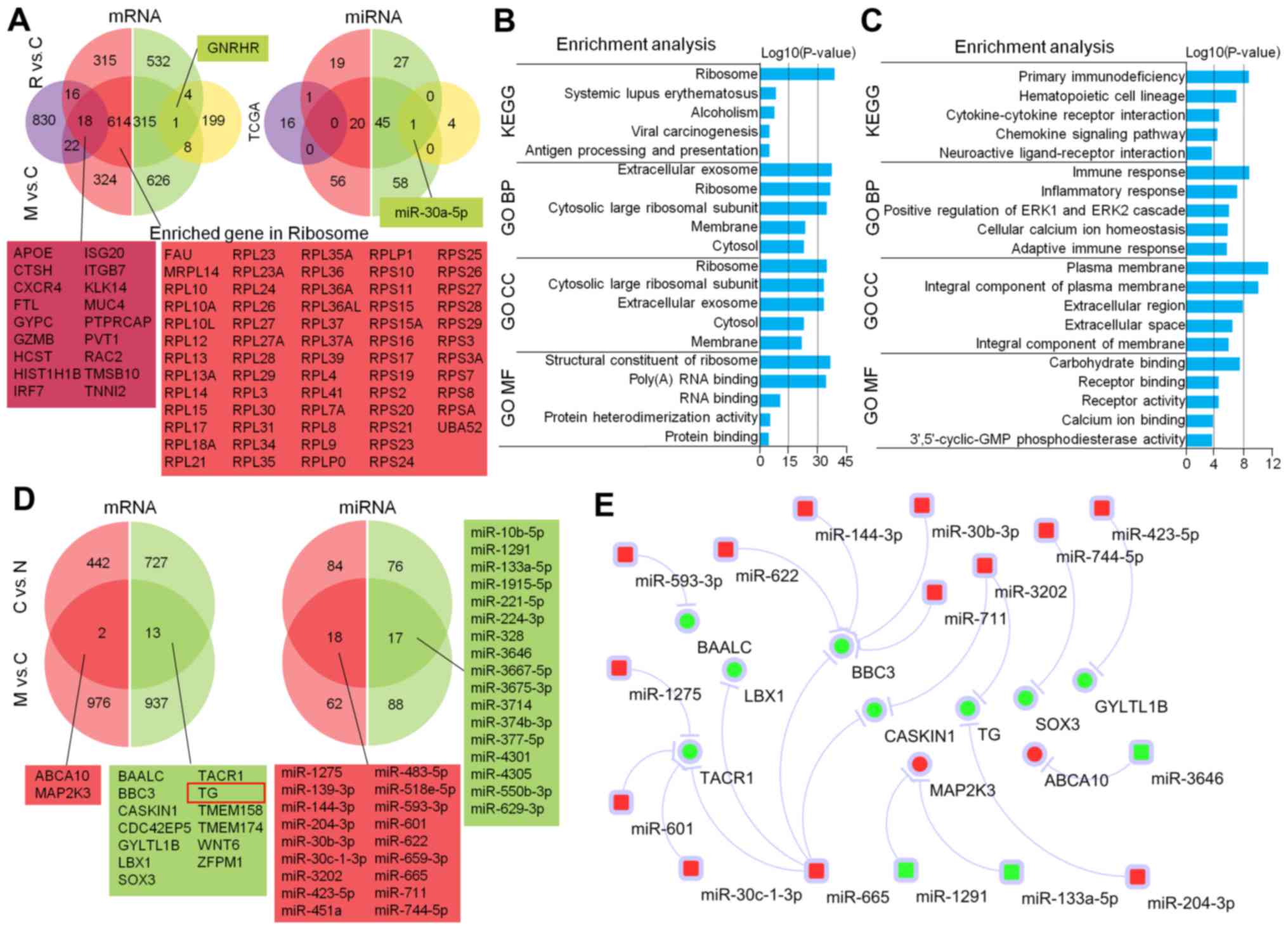
(Fig. 3A). We selected overlapping
molecules to perform survival analysis and construct a miRNA-mRNA
network using online miRNA-target predicting tools. We found 10
overlapping mRNAs that interacted with 21 commonly deregulated
mRNAs (Fig. 3B).
Among commonly expressed DEmRNAs and miRNAs, high
expression of alkaline phosphatase, liver/bone/kidney
(ALPL), low expression of ribosomal protein S27
(RPS27) and calcium voltage-gated channel subunit alpha1 I
(CACNA1I) were associated with overall survival of WCDT.
High expression of anti-silencing function 1B histone chaperone
(ASF1B) and low expression of let-7a-2-3p and miR-486-3p
were associated with RFS for WDTC (Fig.
3C).
Expression patterns are related to
metastatic variations of WDTC
We identified 632 upregulation and 316 downregulated
DEmRNAs and 20 upregulation and 46 downregulated DEmiRNAs in DM and
recurrent lesions of WDTC01. To identify evolutionary signatures
related to metastasis of WDTC, we investigated common expression
patterns shared with BM from WDTC01 and LNM from TCGA. DEmRNAs and
miRNAs from both metastatic sources were different. We found 18
upregulated DEmRNAs, such as apolipoprotein E (ApoE),
cathepsin H (CTSH), C-X-C motif chemokine receptor 4
(CXCR4) and mucin 4 (MUC4) and one downregulated
DEmRNA, gonadotropin releasing hormone receptor (GNRHR) in
BM and LNM. For miRNAs, downregulation of miR-30a-5p occurred in BM
and LNM (Fig. 4A).
Hyper-activation of the ribosome
pathway contributes to DM of WDTC as indicated by enrichment
analysis
To analyze features of BM and LNM of WDTC, we
performed enrichment analysis with DEmRNAs that did not overlap
between the two metastatic sources. Ribosomes, first listed in KEGG
analysis and cell component (CC) in the GO analysis, were
involved.
Significant biological process (BP) and molecular
function (MF) terms were related to the translational process. Our
analysis focused on the ribosome pathway in which 63 enriched genes
were upregulated (Fig. 4B). For
LNM, KEGG terms were related to immune-related pathways. BP terms
were related to the ERK1/ERK2 signaling pathway and cellular
calcium ion homeostasis. CC terms were related to extracellular
activity and MF terms were associated with regulation of ion and
protein binding (Fig. 4C).
Comparison among different stages of
WDTC reveals that upregulation and downregulated DEmRNAs and
DEmiRNAs are associated with tumor progression
To investigate DEmRNAs and miRNAs associated with
tumor progression, we analyzed three stages of WDTC01 (normal,
cancerous and metastatic tissues) before 131I treatment.
Upregulated DEmRNAs included ATP-binding cassette subfamily A
member 10 (ABCA10) and Mitogen-Activated Protein Kinase
Kinase 3 (MAP2K3) and 13 downregulated DEmRNAs, such as BCL2
binding component 3 (BBC3) and thyroglobulin (TG), as
well as 18 upregulation and 17 downregulated DEmiRNAs that were
constantly deregulated with tumor progression (Fig. 4D).
Potential interactions among DEmRNAs and miRNAs
using the aforementioned algorithm suggested that downregulation of
miR-1291, miR-133a-5p and miR-3646 may increase the expression of
MAP2K3 and ABCA10. Downregulation of the
pro-apoptotic gene BBC3 may be caused by increases in 5
different progression-related miRNAs. The lineage upregulation of
miR-204-3p and miR-3202 may interfere with the expression of
TG, an important marker for thyroid cancer cell stemness
(Fig. 4E).
Regained TG expression and impaired
p53 downstream in recurrent lesions
To understand the underlying molecular mechanisms
for recurrence following three 131I treatments, we
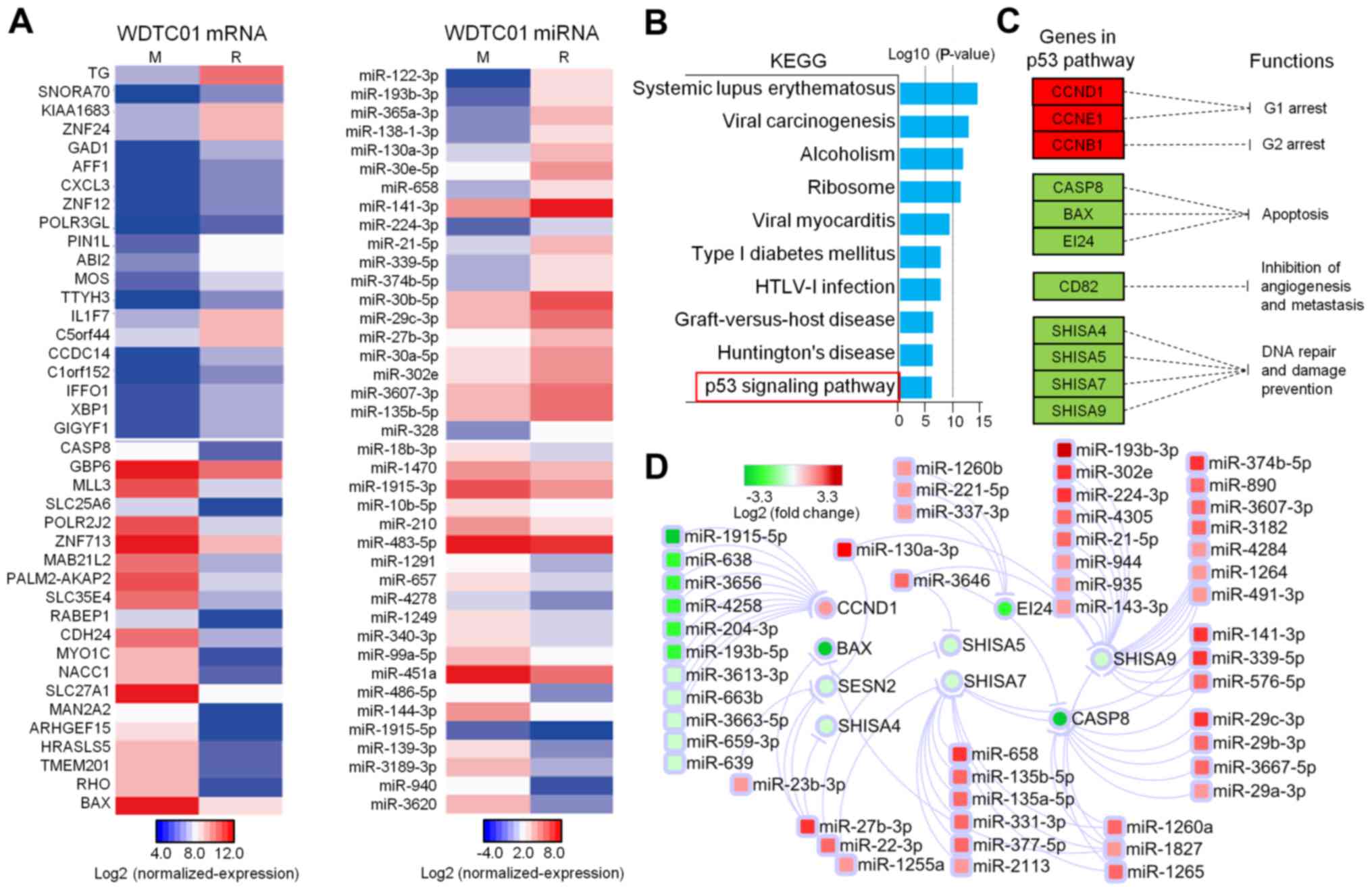
compared recurrence and metastasis and revealed that there were 740
upregulation and 754 downregulated mRNAs and 88 upregulation and 60
downregulated miRNAs. TG was the most upregulated gene
(10-fold) in recurrent lesions. Pro-apoptotic genes BAX
(FC=23.6) and caspase-8 (FC=9.2) were among the top 20
downregulated genes (Fig. 5A).
KEGG pathway enrichment analysis was applied to
1,494 DEmRNAs and the p53 signaling pathway was of interest
(Fig. 5B). Downregulation of BCL2
associated X, apoptosis regulator (BAX) and caspase-8
(CASP8) and upregulation of cyclin B1 (CCB1), D1
(CCD1) and E1 (CCE1) indicated attenuated downstream
effects of the p53 pathway, such as cell cycle arrest and apoptosis
(Fig. 5C). MiRNA-mRNA interaction
analysis revealed potential DEmiRNAs in recurrent lesions, which
regulated these mRNAs downstream of the p53 pathway. Among
apoptotic-inducing genes, BAX and CASP8 were targeted
by several miRNAs, and miR-1827 interacted with BAX and
CASP8; EI24 was targeted by miR-211-5p, a well-known
oncogene in WDTC (Fig. 5D).
Discussion
Despite increasing WDTC globally, few studies have
investigated metastatic sites of WDTC from a genomic perspective.
RNA-Seq analysis of metastatic sites, including two LNM and one
pleural metastasis from a single patient, revealed intratumor
heterogeneity and clonal evolution (32). Another miRNA study analyzed miRNA
transcriptomes of PTC with four LNM and noted that downregulation
of miRNAs was a common feature in PTC tumorigenesis (33). However, there has been no study to
investigate the underlying mechanisms of recurrent WDTC using
recurrent lesions and genomic profiling due to the difficulty of
collecting tissue. We used one paired bone metastatic and one
paired recurrent metastatic lesion and performed mRNA and miRNA
microarray analysis. Integrated with TCGA data, we found common
prognostic factors as key expression changes related to metastasis
and distinct features of LMN and BM. We also suggested a pattern
for recurrence from a genomic perspective, and we provided evidence
for future prevention and therapy for metastasis and relapse of
WDTC.
The epithelial-mesenchymal transition is a
well-established model explaining thyroid cancer development
(34). TG is thought to be a
differentiation marker for WDTC (35), and lower TG expression was
revealed to be correlated with thyroid cancer cell stemness
(34). For WDTC01, TG
expression gradually decreased in normal tissue to metastatic stage
tissue (Fig. 4D). This may be
explained by the fact that tumor cells with lower TG
expression migrating from the primary tumor are prone to BM, which
is supported by the fact that poorly differentiated thyroid cancer
has more DM (36). Additionally,
significant repression of TG was absent in 8 LNM from TCGA,
indicating that TG suppression is specifically related to BM
and not LNM. Next, the microenvironment of the vertebrae may induce
an adaptive response to suppress expression of TG. According
to the miRNA-mRNA analysis, TG may be suppressed by aberrant
upregulation of miR-3202 and miR-204-3p (Fig. 4D). Along with TG, the
pro-apoptotic BBC3 gene was gradually downregulated during
lineage dedifferentiation. The putative miRNA-mRNA network revealed
that continuously increasing expression of miR-622, miR-144-3p,
miR-30b-3p, miR-711 and miR-665 may be the trigger of dysregulation
of BBC3.
High expression of ALPL and low expression of
miR-486-5p were related to a prognosis of WDTC, and these markers
were associated with cancer cell stemness. High expression of
ALPL, as a pluripotent stem cell marker (37), has been revealed to be associated
with poor OS for glioblastoma and prostate cancers (38,39).
For lung and prostate cancer, downregulation of miR-486-5p was
reported to be associated with poor survival (40–42).
Cellular function data revealed that loss of miR-486-5p
dedifferentiated cancer cells via epithelial-mesenchymal transition
(42).
To find the common molecules related to BM and LNM
in WDTC, we overlapped expression changes in both and among the 19
overlapped DEmRNAs, and CXCR4 was key to the
CXCL12/CXCR4 pathway, a critical and well-studied pathway in
BM (43). CXCR4 was reported
to be significantly upregulated and related to BRAF mutations and
neoplastic infiltration, resulting in more aggressive WDTC
(44). Another two molecules
involved in WDTC progression were also related to cancer cell
stemness. As reported in ovarian carcinoma (45), MUC4 can induce
epithelial-mesenchymal transition, and it can enhance the invasion
of tumors in pancreatic and breast cancers (46,47).
miR-30a-5p, the only overlapped miRNA in metastatic lesions, is
reported to be associated with cancer metastasis. The loss of
miR-30a-5p can lead to invasion and migration of breast cancer
(48) and colorectal cancer cells
(49) and epithelial-mesenchymal
transition (50).
In addition, BM and LNM had distinct features with
respect to enrichment analysis. Notably, all KEGG and GO terms of
BM for WDTC01 were associated with ribosomal activity, and all
genes enriched in this pathway were upregulated, indicating that
efficient ribosome translational machineries contribute to BM in
WDCT01. Although individual ribosomal proteins, such as
RPL17 and RPS27L, were reported to be tumor
suppressors, ribosome biogenesis and translation were directly
associated with increased cell growth and proliferation (51). In contrast, no sign of overturned
ribosomal activity was observed in the 8 LNM, perhaps due to the
different microenvironment between BM and LNM.
The most common reason for WDTC recurrence was due
to poor uptake of 131I in the poorly differentiated
variant of WDTC. For WDTC01, gradually decreased expression of TG
indicated a more dedifferentiated type of WDTC, so inefficient
uptake was considered to contribute to cancer relapse (52). However, following 131I
treatments, expression differed greatly from former metastatic
lesions, indicating heterogeneity of the recurrent lesion.
Well-differentiated neoplasms arise from residual cancer stem cells
(CSCs) following surgical debulking. TG expression was again
recovered to its pre-metastatic state because CSCs give rise to
differentiated progeny (53).
According to enrichment analysis, several critical genes of the p53
pathway were altered, and apoptotic resistance and a promoted cell
cycle should increase proliferation of the recurrent lesion.
In summary, we offered an integrated analysis of
mRNAs and miRNAs involved in the metastasis, progression and
recurrence of WDTC and identified numerous mRNA and miRNA related
to stemness, which contributed to the malignancy and recurrence of
WDTC. The ribosome pathway was hyper-activated in BM, and the p53
pathway was correlated with relapse in our case. This information
may provide useful regulatory targets and pathways for future
development of predictive tools and therapies of WDTC.
Acknowledgements
We thank the study subject for his participation. We
appreciate the experimental support of the Central Laboratory for
Medical Research, Shanghai Tenth People's Hospital.
Funding
The present study was supported partly by grants
from the National Natural Science Foundation of China (81472202,
81772932, 81201535, 81302065, 81301993, 81472501, 81372175,
81472209 and 81702243), The Fundamental Research Funds For the
Central Universities (22120170212 and 22120170117), The Shanghai
Natural Science Foundation (12ZR1436000 and 16ZR1428900) and the
Shanghai Municipal Commission of Health and Family Planning
(201440398 and 201540228). The funder has no role in the study
design, data collection and analysis, decision to publish or
preparation of the manuscript.
Availability of data and materials
The datasets used during the present study are
available from the corresponding author upon reasonable
request.
Authors' contributions
YJZ, YSM, QX, DF and XFW designed the study. YJZ,
YSM, QX, FY, ZWL, CYJ, DF and XFW performed the experiments. YJZ,
YSM, DF and XFW performed the statistical analyses and interpreted
the data. QX, FY, ZWL, CYJ, GXL, XQY and XFW are involved in the
patient recruitment. FY, ZWL, CYJ, XXJ, LZ, YCS, WTX, GXL, XQY, PZ,
DF and XFW contributed to the study materials and consumables. YJZ,
YSM, DF and XFW wrote the manuscript. YJZ, YSM and QX contributed
equally to this work. All authors read and approved the manuscript
and agree to be accountable for all aspects of the research in
ensuring that the accuracy or integrity of any part of the work are
appropriately investigated and resolved.
Ethics approval and consent to
participate
The present study was approved by the Institutional
Review Board and Ethics Committee of The Shanghai Tenth People's
Hospital, Tongji University, Shanghai, China
(SHSY-IEC-KY-4.0/17-13/01).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Kilfoy BA, Zheng T, Holford TR, Han X,
Ward MH, Sjodin A, Zhang Y, Bai Y, Zhu C, Guo GL, et al:
International patterns and trends in thyroid cancer incidence,
1973–2002. Cancer Causes Control. 20:525–531. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chen AY, Jemal A and Ward EM: Increasing
incidence of differentiated thyroid cancer in the United States,
1988–2005. Cancer. 115:3801–3807. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sherman SI: Thyroid carcinoma. Lancet.
361:501–511. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Xing M, Haugen BR and Schlumberger M:
Progress in molecular-based management of differentiated thyroid
cancer. Lancet. 381:1058–1069. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kimura ET, Nikiforova MN, Zhu Z, Knauf JA,
Nikiforov YE and Fagin JA: High prevalence of BRAF mutations in
thyroid cancer: Genetic evidence for constitutive activation of the
RET/PTC-RAS-BRAF signaling pathway in papillary thyroid carcinoma.
Cancer Res. 63:1454–1457. 2003.PubMed/NCBI
|
|
6
|
Cohen Y, Xing M, Mambo E, Guo Z, Wu G,
Trink B, Beller U, Westra WH, Ladenson PW and Sidransky D: BRAF
mutation in papillary thyroid carcinoma. J Natl Cancer Inst.
95:625–627. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ciampi R, Knauf JA, Kerler R, Gandhi M,
Zhu Z, Nikiforova MN, Rabes HM, Fagin JA and Nikiforov YE:
Oncogenic AKAP9-BRAF fusion is a novel mechanism of MAPK pathway
activation in thyroid cancer. J Clin Invest. 115:94–101. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wreesmann VB, Sieczka EM, Socci ND, Hezel
M, Belbin TJ, Childs G, Patel SG, Patel KN, Tallini G, Prystowsky
M, et al: Genome-wide profiling of papillary thyroid cancer
identifies MUC1 as an independent prognostic marker. Cancer Res.
64:3780–3789. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Moon S, Song YS, Kim YA, Lim JA, Cho SW,
Moon JH, Hahn S, Park DJ and Park YJ: Effects of coexistent
BRAFV600E and TERT promoter mutations on poor clinical
outcomes in papillary thyroid cancer: A meta-analysis. Thyroid.
27:651–660. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lee JJ, Wang TY, Liu CL, Chien MN, Chen
MJ, Hsu YC, Leung CH and Cheng SP: Dipeptidyl peptidase IV as a
prognostic marker and therapeutic target in papillary thyroid
carcinoma. J Clin Endocrinol Metab. 102:2930–2940. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chowdhury S, Veyhl J, Jessa F, Polyakova
O, Alenzi A, MacMillan C, Ralhan R and Walfish PG: Programmed
death-ligand 1 overexpression is a prognostic marker for aggressive
papillary thyroid cancer and its variants. Oncotarget.
7:32318–32328. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Samaan NA, Schultz PN, Hickey RC, Goepfert
H, Haynie TP, Johnston DA and Ordonez NG: The results of various
modalities of treatment of well differentiated thyroid carcinomas:
A retrospective review of 1599 patients. J Clin Endocrinol Metab.
75:714–720. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Schlumberger MJ: Papillary and follicular
thyroid carcinoma. N Engl J Med. 338:297–306. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Durante C, Haddy N, Baudin E, Leboulleux
S, Hartl D, Travagli JP, Caillou B, Ricard M, Lumbroso JD, De
Vathaire F, et al: Long-term outcome of 444 patients with distant
metastases from papillary and follicular thyroid carcinoma:
Benefits and limits of radioiodine therapy. J Clin Endocrinol
Metab. 91:2892–2899. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Haugen BR: 2015 American Thyroid
Association management guidelines for adult patients with thyroid
nodules and differentiated thyroid cancer: What is new and what has
changed? Cancer. 123:372–381. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
O'Neill CJ, Oucharek J, Learoyd D and
Sidhu SB: Standard and emerging therapies for metastatic
differentiated thyroid cancer. Oncologist. 15:146–156. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Mazzaferri EL and Kloos RT: Clinical
review 128: Current approaches to primary therapy for papillary and
follicular thyroid cancer. J Clin Endocrinol Metab. 86:1447–1463.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Mazzaferri EL and Jhiang SM:
Differentiated thyroid cancer long-term impact of initial therapy.
Trans Am Clin Climatol Assoc. 106(151–168): discussion 168–170.
1995.PubMed/NCBI
|
|
19
|
Visone R, Russo L, Pallante P, De Martino
I, Ferraro A, Leone V, Borbone E, Petrocca F, Alder H, Croce CM and
Fusco A: MicroRNAs (miR)-221 and miR-222, both overexpressed in
human thyroid papillary carcinomas, regulate p27Kip1 protein levels
and cell cycle. Endocr Relat Cancer. 14:791–798. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Mancikova V, Castelblanco E, Pineiro-Yanez
E, Perales-Paton J, de Cubas AA, Inglada-Perez L, Matias-Guiu X,
Capel I, Bella M, Lerma E, et al: MicroRNA deep-sequencing reveals
master regulators of follicular and papillary thyroid tumors. Mod
Pathol. 28:748–757. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Cancer Genome Atlas Research Network:
Integrated genomic characterization of papillary thyroid carcinoma.
Cell. 159:676–690. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhao M, Wang KJ, Tan Z, Zheng CM, Liang Z
and Zhao JQ: Identification of potential therapeutic targets for
papillary thyroid carcinoma by bioinformatics analysis. Oncol Lett.
11:51–58. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Geraldo MV and Kimura ET: Integrated
analysis of thyroid cancer public datasets reveals role of
post-transcriptional regulation on tumor progression by targeting
of immune system mediators. PLoS One. 10:e01417262015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Dweep H and Gretz N: miRWalk2.0: A
comprehensive atlas of microRNA-target interactions. Nat Methods.
12:6972015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Betel D, Wilson M, Gabow A, Marks DS and
Sander C: The microRNA.org resource: Targets and expression.
Nucleic Acids Res. 36:D149–D153. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Agarwal V, Bell GW, Nam JW and Bartel DP:
Predicting effective microRNA target sites in mammalian mRNAs.
Elife. 4:e050052015. View Article : Google Scholar :
|
|
27
|
Kanehisa M, Sato Y, Kawashima M, Furumichi
M and Tanabe M: KEGG as a reference resource for gene and protein
annotation. Nucleic Acids Res. 44:D457–D462. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ashburner M, Ball CA, Blake JA, Botstein
D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT,
et al: Gene ontology: Tool for the unification of biology. The gene
ontology consortium. Nat Genet. 25:25–29. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
da Huang W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using david
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW,
Shi W and Smyth GK: Limma powers differential expression analyses
for rna-sequencing and microarray studies. Nucleic Acids Res.
43:e472015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Benjamini Y and Hochberg Y: Controlling
the false discovery rate-a practical and powerful approach to
multiple testing. J R Stat Soc Ser B-Methodol. 57:289–300.
1995.
|
|
32
|
Le Pennec S, Konopka T, Gacquer D,
Fimereli D, Tarabichi M, Tomas G, Savagner F, Decaussin-Petrucci M,
Tresallet C, Andry G, et al: Intratumor heterogeneity and clonal
evolution in an aggressive papillary thyroid cancer and matched
metastases. Endocr Relat Cancer. 22:205–216. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Saiselet M, Gacquer D, Spinette A, Craciun
L, Decaussin-Petrucci M, Andry G, Detours V and Maenhaut C: New
global analysis of the microRNA transcriptome of primary tumors and
lymph node metastases of papillary thyroid cancer. BMC Genomics.
16:8282015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Lin RY: Thyroid cancer stem cells. Nat Rev
Endocrinol. 7:609–616. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Takano T, Miyauchi A, Yokozawa T,
Matsuzuka F, Maeda I, Kuma K and Amino N: Preoperative diagnosis of
thyroid papillary and anaplastic carcinomas by real-time
quantitative reverse transcription-polymerase chain reaction of
oncofetal fibronectin messenger RNA. Cancer Res. 59:4542–4545.
1999.PubMed/NCBI
|
|
36
|
Are C and Shaha AR: Anaplastic thyroid
carcinoma: Biology, pathogenesis, prognostic factors, and treatment
approaches. Ann Surg Oncol. 13:453–464. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Hou Z, Meyer S, Propson NE, Nie J, Jiang
P, Stewart R and Thomson JA: Characterization and target
identification of a DNA aptamer that labels pluripotent stem cells.
Cell Res. 25:390–393. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Rao SR, Snaith AE, Marino D, Cheng X, Lwin
ST, Orriss IR, Hamdy FC and Edwards CM: Tumour-derived alkaline
phosphatase regulates tumour growth, epithelial plasticity and
disease-free survival in metastatic prostate cancer. Br J Cancer.
116:227–236. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Iwadate Y, Suganami A, Tamura Y, Matsutani
T, Hirono S, Shinozaki N, Hiwasa T, Takiguchi M and Saeki N: The
pluripotent stem-cell marker alkaline phosphatase is highly
expressed in refractory glioblastoma with DNA hypomethylation.
Neurosurgery. 80:248–256. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Vosa U, Vooder T, Kolde R, Vilo J,
Metspalu A and Annilo T: Meta-analysis of microRNA expression in
lung cancer. Int J Cancer. 132:2884–2893. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Tan X, Qin W, Zhang L, Hang J, Li B, Zhang
C, Wan J, Zhou F, Shao K, Sun Y, et al: A 5-microRNA signature for
lung squamous cell carcinoma diagnosis and hsa-miR-31 for
prognosis. Clin Cancer Res. 17:6802–6811. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Zhang X, Zhang T, Yang K, Zhang M and Wang
K: miR-486-5p suppresses prostate cancer metastasis by targeting
Snail and regulating epithelial-mesenchymal transition. Onco
Targets Ther. 9:6909–6914. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Burton A: Regulation of RANKL might reduce
bone metastases. Lancet Oncol. 7:3672006. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Torregrossa L, Giannini R, Borrelli N,
Sensi E, Melillo RM, Leocata P, Materazzi G, Miccoli P, Santoro M
and Basolo F: Cxcr4 expression correlates with the degree of tumor
infiltration and braf status in papillary thyroid carcinomas. Mod
Pathol. 25:46–55. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Ponnusamy MP, Lakshmanan I, Jain M, Das S,
Chakraborty S, Dey P and Batra SK: Muc4 mucin-induced epithelial to
mesenchymal transition: A novel mechanism for metastasis of human
ovarian cancer cells. Oncogene. 29:5741–5754. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Singh AP, Moniaux N, Chauhan SC, Meza JL
and Batra SK: Inhibition of muc4 expression suppresses pancreatic
tumor cell growth and metastasis. Cancer Res. 64:622–630. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Rowson-Hodel AR, Wald JH, Hatakeyama J,
O'Neal WK, Stonebraker JR, VanderVorst K, Saldana MJ, Borowsky AD,
Sweeney C and Carraway KL III: Membrane mucin muc4 promotes blood
cell association with tumor cells and mediates efficient metastasis
in a mouse model of breast cancer. Oncogene. 37:197–207. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Li W, Liu C, Zhao C, Zhai L and Lv S:
Downregulation of β3 integrin by miR-30a-5p modulates cell adhesion
and invasion by interrupting Erk/Ets-1 network in triple-negative
breast cancer. Int J Oncol. 48:1155–1164. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Wei W, Yang Y, Cai J, Cui K, Li RX, Wang
H, Shang X and Wei D: miR-30a-5p suppresses tumor metastasis of
human colorectal cancer by targeting ITGB3. Cell Physiol Biochem.
39:1165–1176. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Chung YH, Li SC, Kao YH, Luo HL, Cheng YT,
Lin PR, Tai MH and Chiang PH: miR-30a-5p inhibits
epithelial-to-mesenchymal transition and upregulates expression of
tight junction protein claudin-5 in human upper tract urothelial
carcinoma cells. Int J Mol Sci. 18:E18262017. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Ruggero D and Pandolfi PP: Does the
ribosome translate cancer? Nat Rev Cancer. 3:179–192. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Ma ZF and Skeaff SA: Thyroglobulin as a
biomarker of iodine deficiency: A review. Thyroid. 24:1195–1209.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Vermeulen L, Sprick MR, Kemper K, Stassi G
and Medema JP: Cancer stem cells-old concepts, new insights. Cell
Death Differ. 15:947–958. 2008. View Article : Google Scholar : PubMed/NCBI
|