Introduction
The intercellular communication and passage of small
molecules between neighbouring cells is mediated by gap junctions,
which play a variety of important roles in various biological
processes, including cell differentiation, homeostasis,
morphogenesis and growth control (1–3).
Connexins have been considered as tumor suppressors in many types
of cancer (4–6). However, recent evidence indicates the
involvement of connexins in promoting cell proliferation (7), regulating apoptosis (8), and promoting chemoresistance (9), migration (10), regulation of epithelial-mesenchymal
transition (EMT) (11), tumor
differentiation and angiogenesis (12–15).
Gap Junction β-2 (GJB2, also known as connexin 26)
is one of the members of 21 connexin family genes that form the gap
junction channels, which is abundantly expressed in the skin, liver
and breasts. A previous study indicated an association of the
mutations in the GJB2 gene with neurosensory deafness (16). Previous studies have also revealed
the overexpression of GJB2 in several carcinomas, including head
and neck, colon, prostate and skin tumors (17–21).
In breast cancer, GJB2 has been indicated to be a tumor suppressor
due to its very low expression in breast tumor tissues, perhaps due
to hypermethylation (22). By
contrast, there are also studies indicating the overexpression of
GJB2 in breast cancer tissues and its involvement in metastasis to
the lymph node and its association with a poor prognosis (23,24). A
high expression of GJB2 has also been detected in >50% of
invasive breast cancer tissues when compared to normal human breast
tissues. In addition, the expression of GJB2 has shown to be
associated with a poor prognosis of lung and esophagus carcinoma
(25–27). Collectively, these studies support a
pro-tumorigenic role for GJB2.
In view of these findings, we hypothesized that GJB2
may be associated with the hormonal status of breast tumors, which
may be one of the reasons for the discrepancy in the reported role
of GJB2 in breast cancer. In the present study, we examined the
expression of GJB2 in breast cancer tissues of various hormonal
statuses and investigated the role of GJB2 by knocking down its
expression in breast cancer cells followed by in vitro and
in vivo experiments. Our data revealed a negative
association of GJB2 protein with the ER status of breast tumor
tissues. Furthermore, GJB2 was found to be involved in the growth
of breast tumors.
Materials and methods
Patients and tumor samples
Breast tumor tissue biopsy samples as well as
adjescent normal breast tissue biopsy samples were obtained from
breast cancer patients who presented at the Kidwai Memorial
Institute of Oncology (KMIO) Bangalore from June, 2009 to March,
2012. For the present study, a total of 99 breast tumor samples and
46 tumor adjescent normal samples were collected. All patients
provided informed consent following the approval of the Kidwai
Memorial Instititue of Oncology (KMIO) Medical Ethics Committee.
The status of estrogen receptor (ER), progesterone receptor (PR),
HER2/neu and pathological data, including tumor grade, size and
lymph node status were obtained from the pathology records of the
respective patients
Cell lines and cell culture
The breast cancer cell lines, BT-474 and MCF7
(ER-positive), were obtained from the American Type Culture
Collection (ATCC, Manassas, VA, USA) and were cultured in
Dulbecco's modified Eagle's medium (DMEM, high glucose;
Sigma-Aldrich, St. Louis, MO, USA) supplemented with 10% fetal
bovine serum (FBS; Invitrogen/Thermo Fisher Scientific, Inc.,
Waltham, MA, USA), penicillin (1 kU/ml) and streptomycin (0.1
mg/ml). The cells were maintained under standard culture conditions
at 37°C, 5% CO2 in a humidified incubator (Thermo Fisher
Scientific, Inc.).
Reverse transcription-quantitative PCR
(RT-qPCR)
RNA was extracted from the tissues/cells using
TRIzol reagent (Sigma-Aldrich) according to manufacturer's
instructions. For quantitative (real-time) PCR analysis, total RNA
was reverse transcribed using a cDNA synthesis kit (Applied
Biosystems, Foster City, CA, USA). qPCR was performed in
triplicates using Dynamo™ SYBR-Green 2X mix (Finnzymes; Thermo
Fisher Scientific, Inc.) on an ABI Prism 7900HT sequence detection
system and analyzed with SDS 2.1 software (both from Applied
Biosystems). The expression of TBP was used for normalization. The
expression of TBP was consistent across the breast cancer tissues
and adjacent normal tissues in the microarray experiments. For the
normalization of gene expression in the cell lines, RPL35A was
used. The analysis was performed using SDS 2.1 software (Applied
Biosystems). The fold change in the levels of the mRNA was
calculated from the data using the 2−ΔΔCt method
(28). For analyzing the gene
expression of GJB2 in breast tissue samples, a set of 50 invasive
breast cancer tissue samples and 25 adjacent normal samples were
used.
The sequences of the primers used for PCR in the
present study were as follows: GJB2 forward,
5′-AGCGCAGAGACCCCAAC-3′ and reverse, 5′-GGTGGAGTGTTTGTTCAC-3′; TBP
forward, 5′-GATCAGAACAACAACAGCCTGCC-3′ and reverse,
5′-TTCTGAATAGGCTGTGGGGT; RPL35A forward, 5′-GGGTACAGCATCACTCGGA-3′
and reverse, 5′-ACGCCCGAGATGAAACAG-3′.
Immunohistochemistry
For histological analysis, a different set of breast
tumor tissues were obtained from blocks archived at the Department
of Pathology at the Kidwai Memorial Institute of Oncology (KMIO).
Tissue sections (5-µm-thick) from the paraffin-embedded tumor
specimens were collected on silane-coated slides and
immunohistochemistry for GJB2 was performed on 49 tumor tissue
sections (27 ER-negative and 22 ER-positive) and 21 adjacent normal
tissue sections. Antigen retrieval was performed by heat treatment
of the deparaffinised sections in citrate buffer (10 mM; pH 6.0).
Following the initial processing steps, the sections were incubated
overnight with goat polyclonal anti-GJB2 antibody (1:100 dilution;
ab59020; Abcam, Cambridge, MA, USA), at 4°C. This was followed by
incubation with the supersensitive non-biotin horseradish
peroxidase detection system (QD440-XAK, Biogenex, Fermont, CA,
USA). In addition, 3,3′-diaminobenzidine (Sigma-Aldrich) was used
as the chromogenic substrate.
Evaluation of immunohistochemistry
results
The immunohistochemistry scoring for the expression
of GJB2 was based on a semi-quantitative scoring method as
previously described from our laboratory (29), where both the intensity and
percentage of cells with positive staining were analyzed by
experienced pathologists and a combined score was obtained. The
combined score was calculated by the multiplication product of both
scores. The scores were as follows: i) percentage of cells: no
staining, 0; ≤10% or less of cells stained, 1; 11–50% of cells
stained, 2; and ≥50% or more of cells stained, 3; and ii)
intensity: no staining, 0; weak staining, 1; moderate staining, 2;
and strong staining, 3. Thus, the combined scores ranged from 0–9.
Only scores from 4–9 were considered positive for staining.
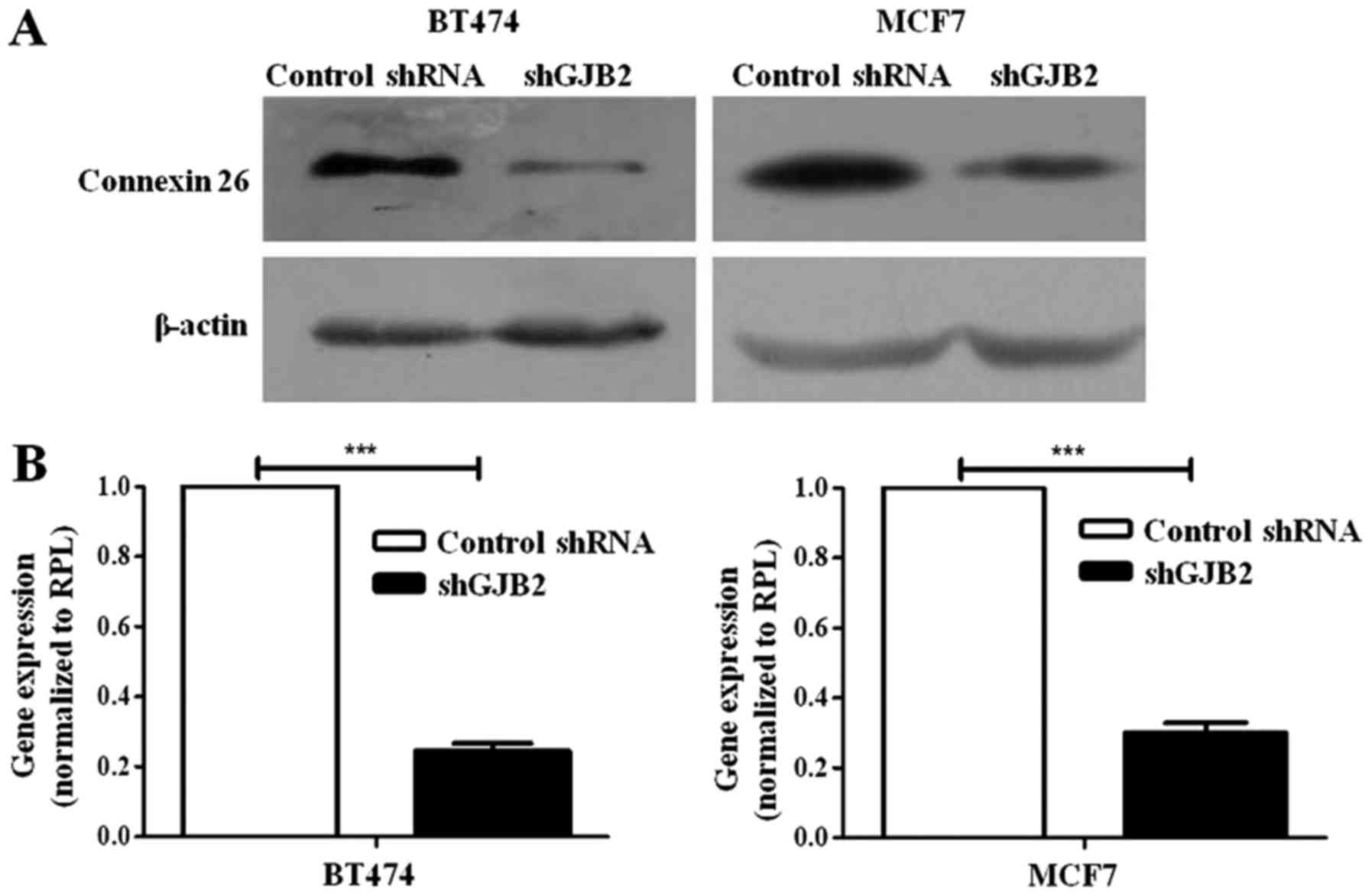
Generation of BT-474 and MCF7 cells in
which GJB2 was knocked down using shRNA
To generate cells in which GJB2 was knocked down,
the BT-474 and MCF7 cells were transfected with a shRNA construct
targeting GJB2 (shGJB2) or non-targeting shRNA vectors (control
shRNA; Origene, Rockville, Maryland, USA) using Lipofectamine 2000
(Invitrogen). Furthermore, the stable transfected cells were
generated by selection with puromycin (0.5 µg/ml; Sigma-Aldrich)
followed by flow cytometry-based sorting (MoFlo™ XDP; Beckman
Coulter, Inc., Brea, CA, USA) for GFP expression encoded by the
vector, and then were expanded and frozen for future use. Knockdown
was confirmed by western blot analysis and RT-qPCR analysis.
Western blot analysis
Cell lysates were prepared using lysis buffer 50 mM
Tris-HCl (pH 7.4) with 1% NP40 detergent, 0.5% sodium deoxycholate,
0.1% SDS, 1 mM sodium fluoride, 1 mM sodium orthovanadate, 150 mM
sodium chloride, 1 mM EDTA (all from Sigma-Aldrich) and protease
inhibitors (Calbiochem; Merck KGaA, Darmstadt, Germany). Protein
estimation was performed using Bradford reagent (Sigma-Aldrich) and
equal amount of protein was resolved by SDS-PAGE (12% gel) using
Bio-Rad apparatus (Bio-Rad, Hercules, CA, USA), transferred to PVDF
membranes (Millipore, Billerica, MA, USA). The blot was blocked
using 5% skimmed milk (Sigma-Aldrich) as blocking reagent for 1 h
at room temperature and probed with goat polyclonal anti-GJB2
antibody (1:1,000 dilution; ab59020; Abcam), followed by incubation
with HRP-coupled rabbit anti-goat secondary antibody (1:2,000
dilution; A5420; Sigma-Aldrich). Immunoblots were visualized using
femtoLUCENT PLUS-HRP kit (G-Biosciences, St. Louis, MO, USA). The
expression of β-actin (using mouse monoclonal antibody from
Sigma-Aldrich; A544; 1:2,000 dilution) was used as the loading
control in all western blot analyses.
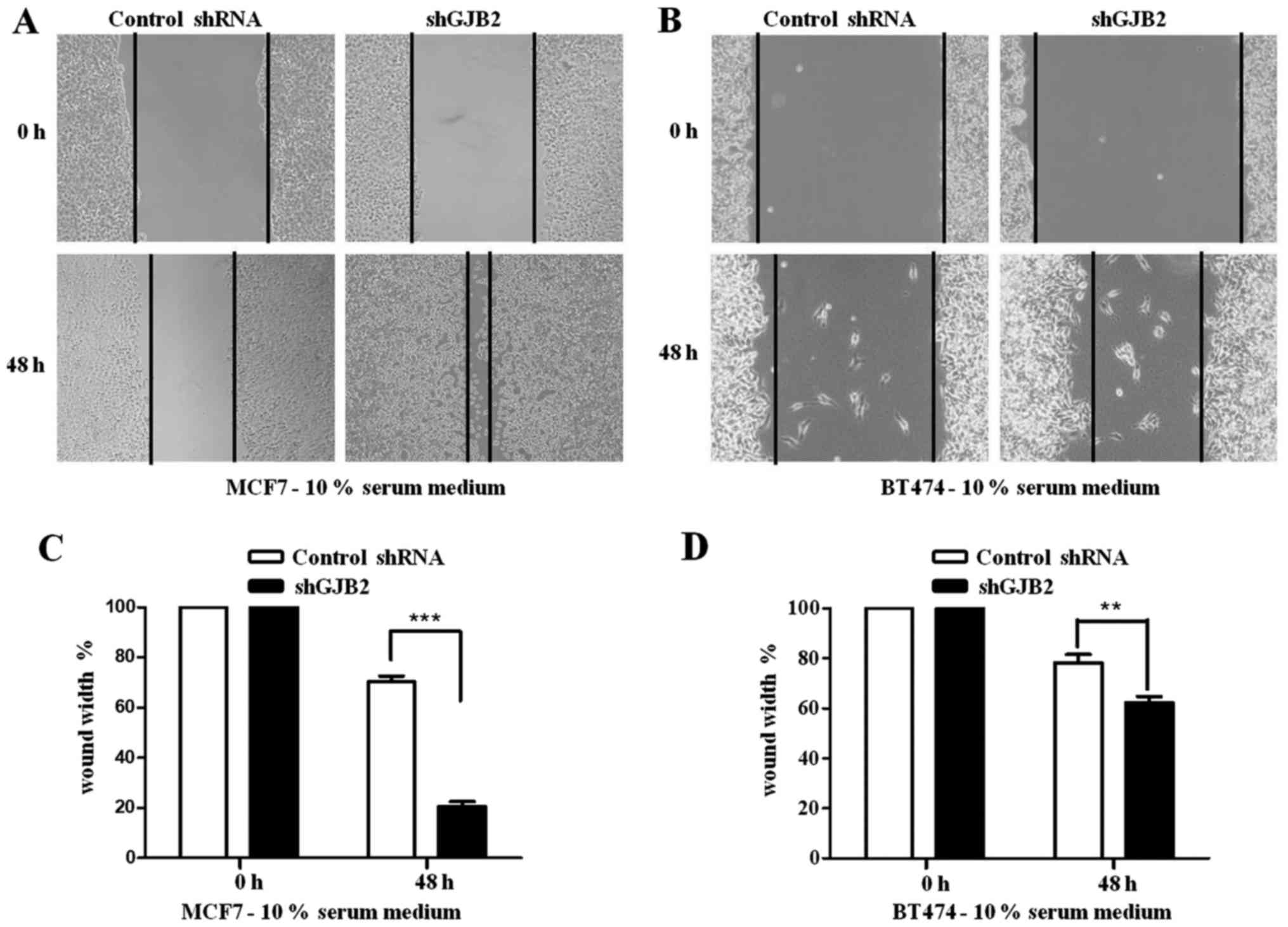
Migration assay
BT474 and MCF7 cells transfected with shGJB2 or
control shRNA were seeded in 6-well Petri plates. Following 12 h of
seeding, the cells were treated with 10 µg/ml of mitomycin C
(Calbiochem, Merck KGaA) in serum-free medium for 2 h to arrest
proliferation and then, two wounds were made using a P200 pipette
tip. Thereafter, the cells were cultured for 48 h in culture medium
with or without 10% FBS. Photomicrographs were captured using a
microscope (Carl Zeiss, Jena, Germany) at 0 and 48 h after the
wound generation. The distance migrated was quantified using ImageJ
capture software (National Institutes of Health, Bethesda, MA, USA)
and plotted as a difference of wound width between 0 and 48 h.
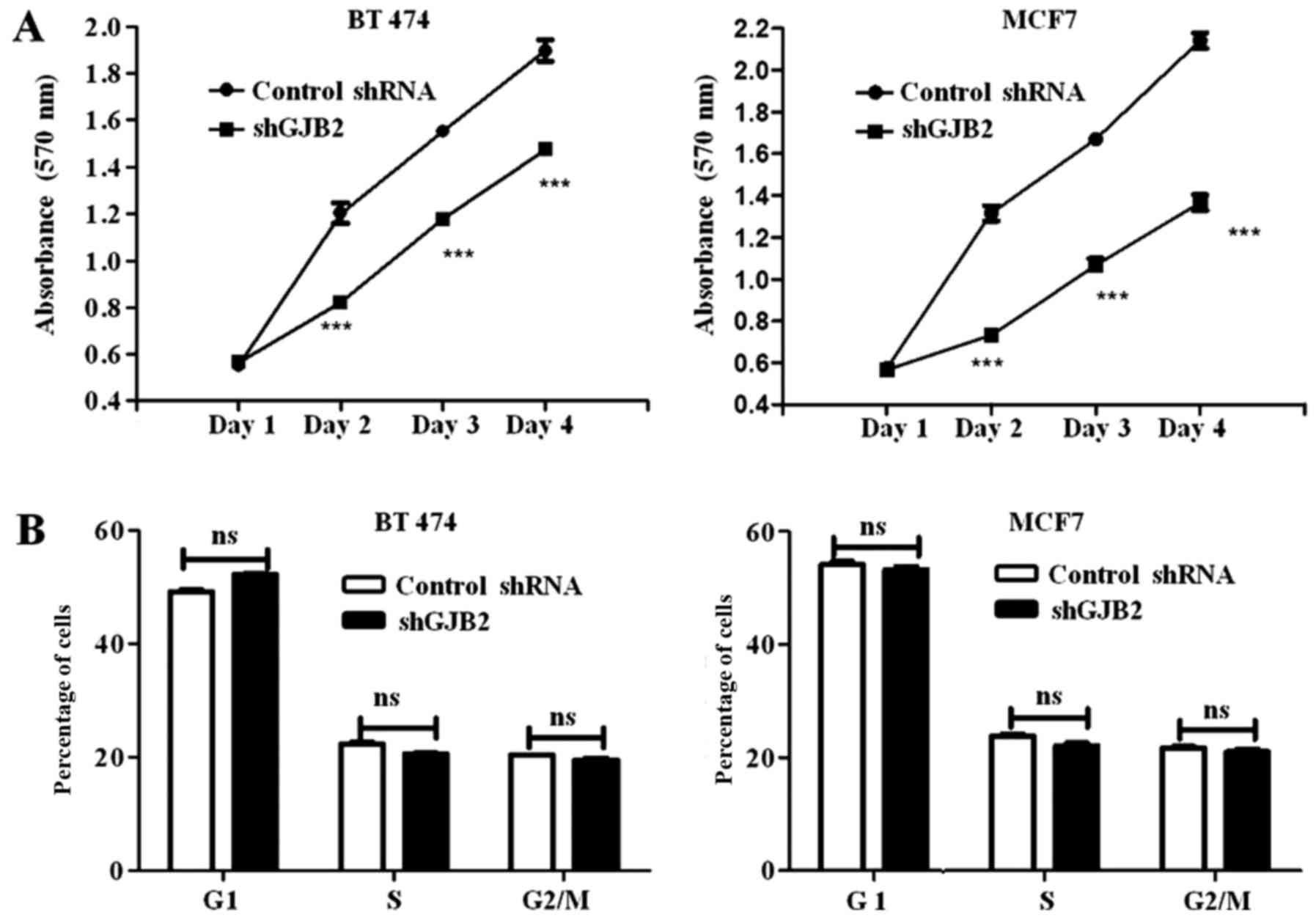
Proliferation assay and cell cycle
analysis
Cell proliferation was estimated by MTT assay which
was performed in triplicate in 96-well plates (Nunc, Roskilde,
Denmark) using BT474 and MCF7 cells stably transfected with shGJB2
or control shRNA. MTT assay was performed every 24 h up to 4 days.
Briefly, MTT (5 mg/ml) reagent (Sigma-Aldrich) was added to each
well and the plate was incubated for 4 h until the formazan
crystals were formed. Crystals were dissolved in DMSO and the plate
was read in ELISA reader at 575 nm. Cell proliferation was
expressed by plotting the absorbance of both shGJB2 transfected
cells, and control shRNA transfected cells.
For cell cycle analysis, thje BT-474 and MCF7 cells
transfected with control shRNA or shGJB2 (5×105) were
seeded into 6-well plates and following 48 h of culturing, the
cells were trypsinized and 5×105 cells were fixed by the
addition of 70% ethanol overnight at −20°C. The cells were then
washed twice with PBS, and treated with RNAse A (100 µg/ml;
Sigma-Aldrich) in PBS and incubated at 37°C for 4 h. Furthermore,
the cells were washed twice with PBS, re-suspended in 300 µl of PBS
containing propidium iodide (20 µg/ml; Sigma-Aldrich) and analyzed
in a flow cytometer (BD FACSAria; Becton-Dickinson, Oakville, ON,
Canada).
In vivo tumor formation assay
Animal experiments were performed following the
approval from the Institutional Animal Ethics Committee, Indian
Institute of Science (Bangalore, India). Four- to five-week-old
female athymic nude mice were used for the in vivo animal
experiments (n=14; weight prior to experimentation, 15–18 g; weight
upon sacrifice, 28–30 g). The animals were housed under specific
pathogen-free conditions (temperature of 22±2°C; humidity of
30–70%; 12-h light/dark cycle and fed standard sterile mouse chow
and had access to sterile water ad libitum). Mice were
divided into two groups (n=7 per group). The mice were injected
with shGJB2-transfected BT474 cells or control shRNA-transfected
BT474 cells (107 cells) into the 5th mammary fat pad.
Once the tumor attained a volume of 100 mm3, tumor size
was measured regularly (each week) with digital vernier callipers.
After 8 weeks, the tumors were dissected out and the animals were
re-sutured and the experiment was continued for 5 more weeks to
examine metastasis to other organs. The maximum tumor diameter
observed in a single tumor in this study was 1.8 cm. The dissected
tumors were subjected to RNA isolation and RT-qPCR analysis for
GJB2 to confirm the knockdown. Part of the tumor was processed for
immunohistochemical analysis for MIB-1 (1:100 dilution; cat. no.
M7240; Dako, Glostrup, Denmark) as described above to estimate the
proliferation index.
Statistical analysis
Statistical analysis was performed using a paired
Student's t-test for data displayed in Fig. 1A; ANOVA followed by Tukey's post hoc
test for data in Fig. 1D and an
unpaired Student's t-test for data analysis of all other figures.
GraphPad prism version 5 software (GraphPad Software, Inc., La
Jolla, CA, USA) was used for all statistical tests and plotting of
the graphs. The results are presented as the means ± SEM.
Results
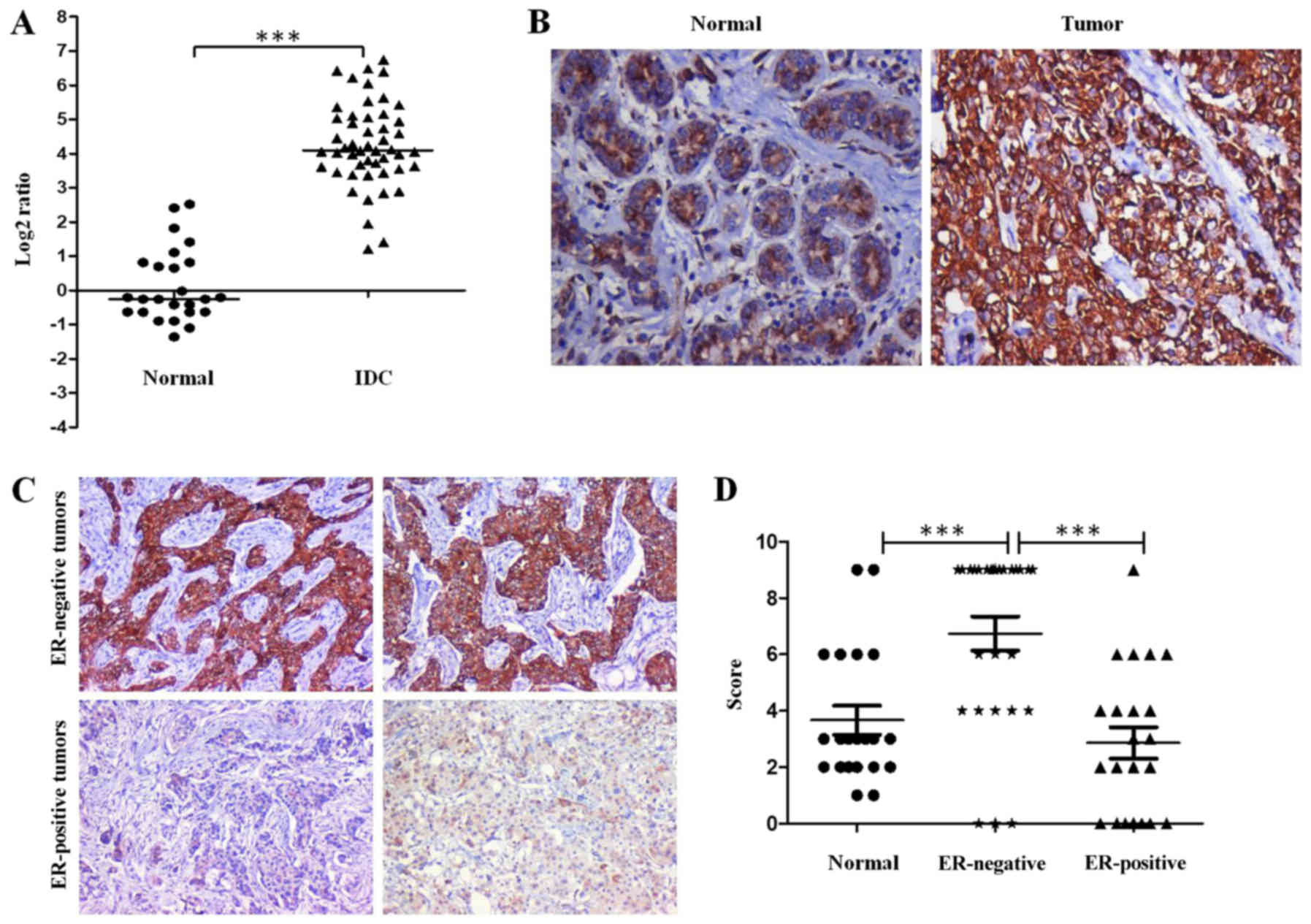
GJB2 gene is upregulated in breast
cancer tissues
Previous microarray data from our laboratory
(30) on human breast cancer
tissues and normal breast tissues (GEO Accession no. GSE40206)
revealed the upregulation of the GJB2 gene in various categories of
breast cancer. In this study, we validated these data using RT-qPCR
analysis from the RNA isolated from breast cancer tissues and
adjacent normal breast tissues. The results of RT-qPCR confirmed
the upregulation of the GJB2 gene in the breast cancer tissues.
GJB2 was significantly upregulated in the invasive ductal carcinoma
samples (n=50) when compared with the normal tissue samples (n=25;
median log2 fold change of 4.1; P<0.001) (Fig. 1A).
GJB2 protein expression is negatively
associated with the estrogen receptor status in breast cancer
tissues
Since we observed an increased expression of GJB2
gene in breast cancer tissues in the above-mentioned result, we
examined the expression of GJB2 protein in a total of 49 grade III
invasive ductal carcinoma tissue samples (27 ER-negative and 22
ER-positive) and 21 normal breast tissue samples using
immunohistochemistry. A representative staining pattern of GJB2 in
normal and breast cancer tissues is depicted in Fig. 1B. The analysis revealed intense
cytoplasmic and membrane staining. The staining for GJB2 was more
intense in the majority of the breast cancer tissues compared with
the normal breast tissue samples. Notably, we observed that the
staining for GJB2 was more intense (6–9 score) in the majority of
the ER-negative breast cancer tissues (19 of 27 samples, 70%)
whereas, the majority of the ER-positive breast cancer samples (17
of 22 samples, 77%) exhibited weak to moderate staining (score
<6) (Fig. 1C and D). In
addition, the majority of the normal breast tissue samples
exhibited weak to moderate staining. No significant association
between the expression of GJB2 and HER2 or the triple-negative
status was observed. The scoring of different samples used for the
analysis is shown in Table I.
RT-qPCR analysis of the expression of GJB2 in normal, ER-negative
and ER-positive breast tumour samples revealed no association
between the expression of GJB2 and ER status (data not shown). We
also performed experiments in which breast cancer cells (MCF7 and
BT474) were treated with estradiol and assessed the expression of
GJB2. However, no regulation of GJB2 by estradiol was observed
(data not shown).
 | Table I.Immunohistochemistry of GJB2. |
Table I.
Immunohistochemistry of GJB2.
| ER-negative | ER-positive | Normal |
|---|
|
|
|
|---|
| Patient no. | Intensity | Cells +ve (%) | Score | Patient no. | Intensity | Cells +ve (%) | Score | Patient no. | Intensity | Cells +ve (%) | Score |
|---|
| 1 | 3 | 75 | 9 | 1 | −ve |
|
| 1 | 2 | 60 | 6 |
| 2 | 3 | 60 | 9 | 2 | −ve |
|
| 2 | 3 | 70 | 9 |
| 3 | 3 | 50 | 6 | 3 | 2 | 40 | 4 | 3 | 1 | 70 | 3 |
| 4 | 2 | 20 | 4 | 4 | −ve |
|
| 4 | 3 | 70 | 3 |
| 5 | 3 | 70 | 9 | 5 | 2 | 30 | 4 | 5 | 2 | 70 | 6 |
| 6 | −ve |
|
| 6 | −ve |
|
| 6 | 1 | 60 | 3 |
| 7 | 3 | 70 | 9 | 7 | −ve |
|
| 7 | 1 | 40 | 2 |
| 8 | −ve |
|
| 8 | 2 | 10 | 2 | 8 | 1 | 50 | 2 |
| 9 | 2 | 20 | 4 | 9 | 1 | 20 | 2 | 9 | 1 | 90 | 3 |
| 10 | 3 | 70 | 9 | 10 | 2 | 75 | 6 | 10 | 1 | 90 | 3 |
| 11 | 2 | 35 | 4 | 11 | 2 | 80 | 6 | 11 | 2 | 60 | 6 |
| 12 | 2 | 30 | 4 | 12 | 2 | 80 | 6 | 12 | 2 | 65 | 6 |
| 13 | 2 | 30 | 4 | 13 | 2 | 80 | 6 | 13 | 1 | 10 | 1 |
| 14 | 3 | 50 | 6 | 14 | 3 | 80 | 9 | 14 | 1 | 10 | 1 |
| 15 | 3 | 80 | 9 | 15 | 1 | 60 | 3 | 15 | 1 | 50 | 3 |
| 16 | 3 | 60 | 9 | 16 | 1 | 60 | 3 | 16 | 1 | 30 | 2 |
| 17 | 3 | 65 | 9 | 17 | 2 | 40 | 4 | 17 | 1 | 20 | 2 |
| 18 | 3 | 90 | 9 | 18 | −ve |
|
| 18 | 3 | 80 | 9 |
| 19 | 3 | 60 | 9 | 19 | 1 | 45 | 2 | 19 | 1 | 50 | 2 |
| 20 | 3 | 75 | 9 | 20 | 2 | 45 | 4 | 20 | 1 | 10 | 2 |
| 21 | 3 | 60 | 9 | 21 | 2 | 20 | 2 | 21 | 1 | 80 | 3 |
| 22 | 3 | 75 | 9 | 22 | −ve |
|
|
|
|
|
|
| 23 | −ve |
|
|
|
|
|
|
|
|
|
|
| 24 | 3 | 70 | 9 |
|
|
|
|
|
|
|
|
| 25 | 3 | 40 | 6 |
|
|
|
|
|
|
|
|
| 26 | 3 | 75 | 9 |
|
|
|
|
|
|
|
|
| 27 | 3 | 80 | 9 |
|
|
|
|
|
|
|
|
Knockdown of GJB2 promotes the
migration and inhibits the proliferation of breast cancer
cells
Since the above-mentioned results revealed high
levels of GJB2 in breast cancer tissues, we further investigated
the role of GJB2 in breast cancer cell migration and proliferation.
We generated BT474 and MCF7 breast cancer cells in which GJB2 was
knocked down using shRNA in (Fig.
2). We then performed a scratch assay with the BT474 and MCF7
cells stably transfected with shGJB2. The shGJB2-transfected BT474
and MCF7 cells demonstrated a significant increase in migration
(80% in MCF7 cells; Fig. 3A and C;
and 38% in BT474 cells; Fig. 3B and
D), when compared to the control shRNA-transfected cells (30%
in MCF7 cells and 28% in BT474 cells) after 48 h.
To further investigate the functional link between
the proliferation of cancer cells and GJB2, we performed a
proliferation assay and analyzed the doubling time for the control
shRNA-transfected cells the cells in which GJB2 was knocked down.
We observed that the knockdown of GJB2 significantly decreased the
proliferation of the cells at all time-points (P<0.001; Fig. 4A). We also observed that the
doubling time was increased (data not shown) in the
shGJB2-transfected cells (40.02±2.85 h in BT474 cells and
54.65±1.98 h in MCF7 cells) compared to the control
shRNA-transfected cells (29.79±1.9 h in BT474 cells and 35.43±2.6 h
in MCF7 cells). Furthermore, we also performed a cell cycle
analysis to determine the effects of the knockdown of GJB2 on the
cell cycle profile. Cell cycle analysis did not reveal any
significant difference in any of the cell cycle phases (G1, S and
G2/M) between the control shRNA-transfected and the cells in which
GJB2 was knocked down (Fig.
4B).
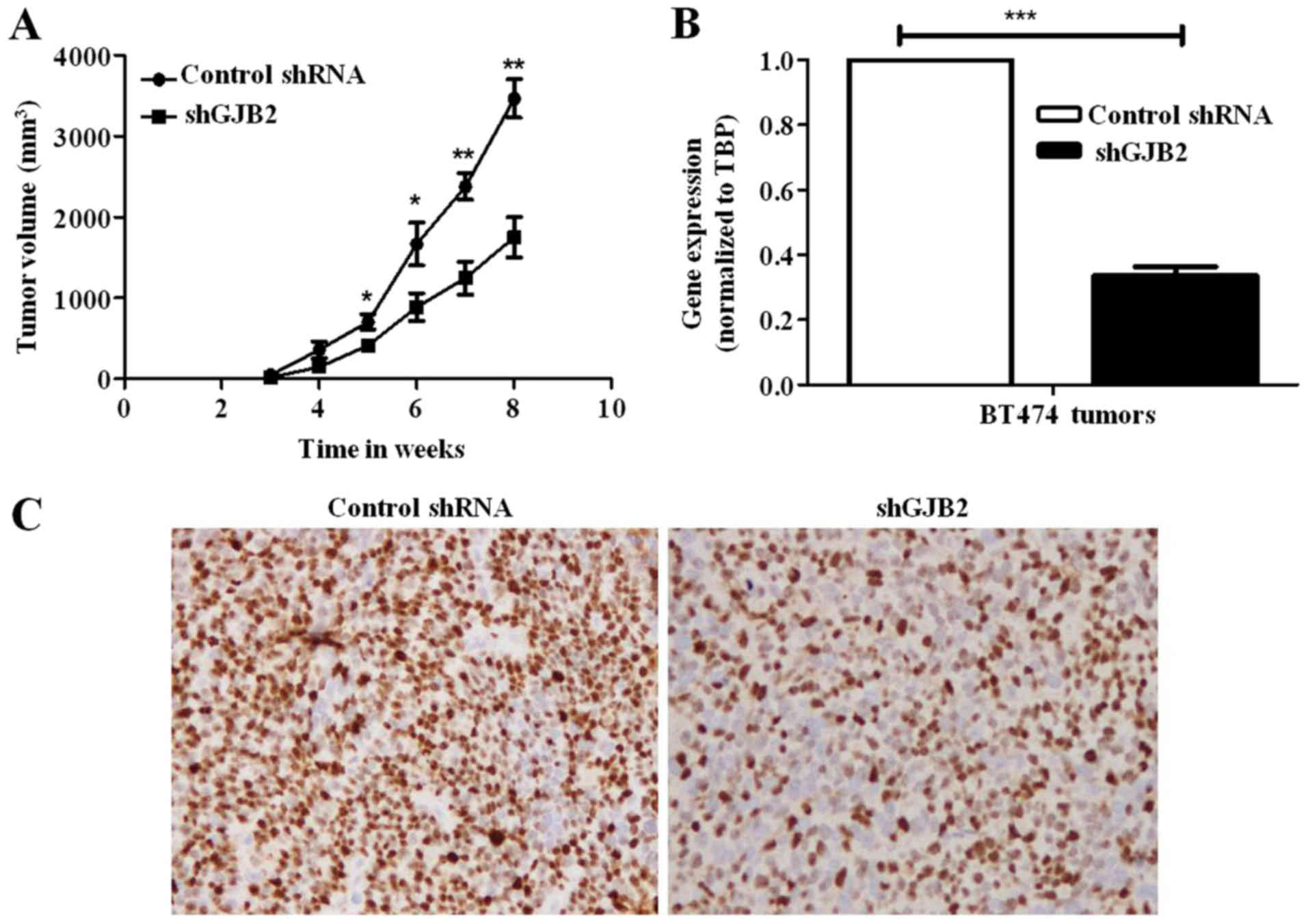
Knockdown of GJB2 suppresses tumor
formation and the proliferation of breast tumor cells in vivo
To further confirm the above-mentioned results in
vivo, we performed an orthotopic xenograft tumor formation
assay using immunocompromised mice. Consistent with the
above-mentioned in vitro data, we observed a significant
reduction in the growth of tumors derived from cells in which GJB2
was knocked down (49.5%; P<0.001) compared to the tumors derived
from control shRNA-transfected cells (Fig. 5A). We also confirmed the knockdown
of GJB2 in tumors by subjecting the RNA isolated from tumors to
RT-qPCR analysis (Fig. 5B).
Furthermore, we also performed immunohistochemistry for MIB-1 to
examine the proliferation rate in these tumors. There was a
significant decrease in the proliferation rate in the tumors
derived from cells in which GJB2 was knocked down (almost 50%)
compared to the tumors derived from control shRNA-transfected cells
(Fig. 5C). There was no observable
metastasis of both the tumors derived from shGJB2-transfected cells
and those derived from control shRNA-transfected cells to any other
organs (data not shown).
Discussion
Previous studies have revealed that in some types of
cancer, some of the connexins and other gap junction proteins are
downregulated (31–33). However, GJB2 (connexin 26)
overexpression has been reported to be expressed in various
carcinomas, including breast cancer (17,18,21,24–26).
Our data derived from both RT-qPCR and immunohistochemical analysis
of human breast cancer tissue samples confirmed these findings. In
addition, our analysis revealed a negative association of GJB2
protein expression with the ER status of breast cancer tissue
samples. Little is known about the regulation of connexins by
estrogen. However, Saito et al (34) demonstrated that the activation of
ER-α by estrogen resulted in the downregulation of connexins,
leading to the suppression of gap junctional intercellular
communication and the promotion of tumor progression in endometrial
carcinoma. In addition, a recent study indicated that other forms
of connexions, such as connexin 43 are regulated by estradiol in
glioma cell lines (35). Hence, we
hypothesized that GJB2 protein may also be regulated by estrogen
receptor in breast cancers. Notably, we did not observe the
regulation of GJB2 RNA by the estrogen treatment of breast cancer
cells (data not shown). In addition, the association between GJB2
and the ER status was observed only in tumor tissues by
immunohistochemistry. However, no differences in the RNA expression
of GJB2 in ER-positive and -nevative breast cancer tissues were
observed (data not shown). This could be attributed to the
post-transcriptional regulation of GJB2, which may be indirectly
associated with the ER status of the tumors. Furthermore, breast
cancer cells treated with estradiol did not exhibit evidence of
GJB2 regulation (data not shown). This indicated that other
variables, including the tumor microenvironment may be involved in
the regulation of GJB2, which requires further investigations.
However, the negative association of GJB2 protein with the ER
status in our study indicated an alternate mechanism of GJB2
regulation by ER, which is not yet understood. Further studies are
warranted in order to elucidate the mechanisms of GJB2 regulation
in ER-negative breast tumors.
It is well known that gap junctional intercellular
communication (GJIC), mediated by connexins play an important role
in normal mammary gland development and homeostasis, and also in
the progression of breast cancer (15). In line with this, our study revealed
that the knockdown of GJB2 promoted the migration and inhibited the
proliferation of both breast cancer cell lines, which are
ER-positive. This result suggested a role for GJB2 in the
inhibition of the migration of ER-positive cells. This is of
importance, since ER-positive tumors exhibit the marginal
expression of GJB2, almost similar to normal tissues. Hence, it is
likely that ER-positive tumors have a greater migratory capacity
than ER-negative tumors. Further studies on a larger cohort of
patients are required in order to establish an association between
GJB2 and tumor cell migration. In addition, tumors in
immunocompromised mice formed from cells in which GJB2 was knocked
down were significantly smaller compared to those derived from
GJB2-expressing cells, suggesting a growth advantage when GJB2 is
overexpressed. The lack of GJIC due to the loss of connexins during
cancer progression may allow tumor cells to physically detach from
the microenvironment and migrate. Our data revealed the increased
expression of GJB2 in the majority of the IDC (grade 3) breast
tissues, which are highly proliferative. In agreement with this,
our data on the knockdown of GJB2 in breast cancer cells revealed
significant decrease in the proliferation of cells, as well as
tumour growth in immunocompromised mice. Hence, it is clear from
the available studies, as well as from our results that depending
on the stages of breast cancer, GJB2 can act as tumor suppressor or
promote tumor growth.
In conclusion, in the present study, we found that
GJB2 was overexpressed in invasive breast cancer tissue samples
compared to normal breast tissues and the expression of GJB2 was
associated with the ER status. In addition, the knockdown of GJB2
in breast cancer cells affected the migration properties of the
breast cancer cells. Furthermore, GJB2 knockdown reduced the
proliferation of the breast cancer cells in vitro and in
vivo. Hence, GJB2 appears to be an important regulator of
breast tumorigenesis. Further studies are warranted in order to
elucidate the exact role of GJB2 in breast tumorigenesis.
Acknowledgements
The authors are grateful to Dr Vani Santosh for
assisting with the immunohistochemistry of mouse tumors. In
addition, the authors are grateful for the infrastructure support
to the Department of Molecular Reproduction, Development and
Genetics (MRDG) and the Indian Institute of Science (IISc) by the
DST-FIST, the UGC and the DBT-IISc partnership. The authors also
acknowledge the flow cytometric and animal facility at the Indian
Institute of Science (IISc) for their support with the
experiments.
Funding
The present study received funding by the Department
of Biotechnology (DBT) in a program support and fellowship to SD.
AS received funding from UGC, Government of India for D.S Kothari
postdoctoral fellowship.
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
AS, design and execution of experiments, analysis of
data and manuscript writing; SD, tumor tissue data acquisition and
realtime PCR analysis; GM, immunohistochemistry and pathological
evaluation; PK, conceived, guidance, data analysis and manuscript
writing. All authors read and approved the manuscript, and agree to
be accountable for all aspects of the research in ensuring that the
accuracy or integrity of any part of the work are appropriately
investigated and resolved.
Ethics approval and consent to
participate
For the use of patient samples, all patients
provided informed consent following the approval of the KMIO
Medical Ethics Committee. Animal experiments were performed with
the approval from the Institutional Animal Ethics Committee, Indian
Institute of Science (Bangalore, India).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Charles AC, Naus CC, Zhu D, Kidder GM,
Dirksen ER and Sanderson MJ: Intercellular calcium signaling via
gap junctions in glioma cells. J Cell Biol. 118:195–201. 1992.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Monaghan P and Moss D: Connexin expression
and gap junctions in the mammary gland. Cell Biol. 20:121–125.
1996.
|
|
3
|
Goodenough DA, Goliger JA and Paul DL:
Connexins, connexons, and intercellular communication. Annu Rev
Biochem. 65:475–502. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhu D, Caveney S, Kidder GM and Naus CC:
Transfection of C6 glioma cells with connexin 43 cDNA: Analysis of
expression, intercellular coupling, and cell proliferation. Proc
Natl Acad Sci USA. 88:1883–1887. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Naus CC, Elisevich K, Zhu D, Belliveau DJ
and Del Maestro RF: In vivo growth of C6 glioma cells transfected
with connexin43 cDNA. Cancer Res. 52:4208–4213. 1992.PubMed/NCBI
|
|
6
|
Eghbali B, Kessler JA, Reid LM, Roy C and
Spray DC: Involvement of gap junctions in tumorigenesis:
Transfection of tumor cells with connexin 32 cDNA retards growth in
vivo. Proc Natl Acad Sci USA. 88:10701–10705. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Aasen T: Connexins: Junctional and
non-junctional modulators of proliferation. Cell Tissue Res.
360:685–699. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Carette D, Gilleron J, Chevallier D,
Segretain D and Pointis G: Connexin a check-point component of cell
apoptosis in normal and physiopathological conditions. Biochimie.
101:1–9. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
King TJ and Bertram JS: Connexins as
targets for cancer chemoprevention and chemotherapy. Biochim
Biophys Acta. 1719:146–160. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kotini M and Mayor R: Connexins in
migration during development and cancer. Dev Biol. 401:143–151.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Defamie N, Chepied A and Mesnil M:
Connexins, gap junctions and tissue invasion. FEBS Lett.
588:1331–1338. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
McLachlan E, Shao Q, Wang HL, Langlois S
and Laird DW: Connexins act as tumor suppressors in
three-dimensional mammary cell organoids by regulating
differentiation and angiogenesis. Cancer Res. 66:9886–9894. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wang WK, Chen MC, Leong HF, Kuo YL, Kuo CY
and Lee CH: Connexin 43 suppresses tumor angiogenesis by
down-regulation of vascular endothelial growth factor via
hypoxic-induced factor-1α. Int J Mol Sci. 16:439–451. 2015.
View Article : Google Scholar
|
|
14
|
Aasen T, Mesnil M, Naus CC, Lampe PD and
Laird DW: Gap junctions and cancer: Communicating for 50 years. Nat
Rev Cancer. 16:775–788. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Banerjee D: Connexin's connection in
breast cancer growth and progression. Int J Cell Biol.
2016:90259052016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kelsell DP, Dunlop J, Stevens HP, Lench
NJ, Liang JN, Parry G, Mueller RF and Leigh IM: Connexin 26
mutations in hereditary non-syndromic sensorineural deafness.
Nature. 387:80–83. 1997. View
Article : Google Scholar : PubMed/NCBI
|
|
17
|
Pfeffer F, Koczan D, Adam U, Benz S, von
Dobschuetz E, Prall F, Nizze H, Thiesen HJ, Hopt UT and Löbler M:
Expression of connexin26 in islets of Langerhans is associated with
impaired glucose tolerance in patients with pancreatic
adenocarcinoma. Pancreas. 29:284–290. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Villaret DB, Wang T, Dillon D, Xu J, Sivam
D, Cheever MA and Reed SG: Identification of genes overexpressed in
head and neck squamous cell carcinoma using a combination of
complementary DNA subtraction and microarray analysis.
Laryngoscope. 110:374–81. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kanczuga-Koda L, Sulkowski S, Koda M and
Sulkowska M: Alterations in connexin26 expression during colorectal
carcinogenesis. Oncology. 68:217–222. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Tate AW, Lung T, Radhakrishnan A, Lim SD,
Lin X and Edlund M: Changes in gap junctional connexin isoforms
during prostate cancer progression. Prostate. 66:19–31. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Haass NK, Wladykowski E, Kief S, Moll I
and Brandner JM: Differential induction of connexins 26 and 30 in
skin tumors and their adjacent epidermis. J Histochem Cytochem.
54:171–182. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Tan LW, Bianco T and Dobrovic A: Variable
promoter region CpG island methylation of the putative tumor
suppressor gene connexin 26 in breast cancer. Carcinogenesis.
23:231–236. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Jamieson S, Going JJ, D'Arcy R and George
WD: Expression of gap junction proteins connexin 26 and connexin 43
in normal human breast and in breast tumours. J Pathol. 184:37–43.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Naoi Y, Miyoshi Y, Taguchi T, Kim SJ, Arai
T, Tamaki Y and Noguchi S: Connexin26 expression is associated with
lymphatic vessel invasion and poor prognosis in human breast
cancer. Breast Cancer Res Treat. 106:11–17. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ito A, Koma Y, Uchino K, Okada T,
Ohbayashi C, Tsubota N and Okada M: Increased expression of
connexin 26 in the invasive component of lung squamous cell
carcinoma: Significant correlation with poor prognosis. Cancer
Lett. 234:239–248. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Inose T, Kato H, Kimura H, Faried A,
Tanaka N, Sakai M, Sano A, Sohda M, Nakajima M, Fukai, et al:
Correlation between connexin 26 expression and poor prognosis of
esophageal squamous cell carcinoma. Ann Surg Oncol. 16:1704–1710.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Teleki I, Krenacs T, Szasz MA, Kulka J,
Wichmann B, Leo C, Papassotiropoulos B, Riemenschnitter C, Moch H
and Varga Z: The potential prognostic value of connexin 26 and 46
expression in neoadjuvant treated breast cancer. BMC Cancer.
13:502013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real time quantitative PCR and
the 2−ΔΔCT method. Methods. 25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Sehgal P, Kumar N, Kumar Praveen VR, Patil
S, Bhattacharya A, Kumar Vijaya M, Mukherjee G and Kondaiah P:
Regulation of protumorigenic pathways by insulin like growth factor
binding protein 2 and its association along with β-catenin in
breast cancer lymph node metastasis. Mol Cancer. 12:632013.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Bashir M, Damineni S, Mukhrejee G and
Kondaiah P: Actavin-A signalling promotes epithelial-mesenchymal
transition, invasion, and metastatic growth of breast cancer. NPJ
Breast Cancer. 1:150072015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Lee SW, Tomasetto C, Paul D, Keyomarsi K
and Sager R: Transcriptional downregulation of gap-junction
proteins blocks junctional communication in human mammary tumor
cell lines. J Cell Biol. 118:1213–1221. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Janssen-Timmen U, Traub O, Dermietzel R,
Rabes HM and Willecke K: Reduced number of gap junctions in rat
hepatocarcinomas detected by monoclonal antibody. Carcinogenesis.
7:1475–1482. 1986. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Mesnil M: Connexins and cancer. Biol Cell.
94:493–500. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Saito T, Tanaka R, Wataba K, Kudo R and
Yamasaki H: Overexpression of estrogen receptor-alpha gene
suppresses gap junctional intercellular communication in
endometrial carcinoma cells. Oncogene. 23:1109–1116. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Moinfar Z, Dambach H, Schoenebeck B,
Forster E, Prochnow N and Faustmann PM: Estradiol receptors
regulate differential connexin 43 expression in F98 and C6 glioma
cell lines. PLoS One. 11:e01500072016. View Article : Google Scholar : PubMed/NCBI
|