Introduction
Endometrial cancer (EC) comprises a biologically and
clinically heterogeneous group of tumors, generally classified as
Type I (endometrioid) or Type II (non-endometrioid) on the basis of
morphological characteristics. It was recently demonstrated that
these two groups are characterized by different genetic
alterations; consequently, the function of these variations has
been widely investigated, in order to identify diagnostic and
prognostic markers for improving the delineation of EC (1).
Recently, a novel classification system was
published by The Cancer Genome Atlas (TCGA) on the basis of genetic
and epigenetic modifications (2).
In addition, a previous TCGA-based analysis has confirmed that
>90% of endometrioid EC (EEC) harbor genetic alterations in the
phosphoinositide 3-kinase (PI3K) signaling pathway (3), primarily concerning PI3K catalytic
subunit α (PIK3CA), and phosphatase and tensin homolog
(PTEN) (3–12).
Furthermore, it has been identified that grade 3
(G3) EEC exhibits clinical and pathological behavior between type I
and type II, suggesting that EEC may evolve by mechanisms different
from those for other EEC grades (13–16).
In EEC, the frequency of PIK3CA mutations is
50% (17) making it one of the most
frequently mutated genes (18–21),
independently of histological types (17,22).
In total, >70% of PIK3CA mutations cluster at three
‘hotspot’ codons, exon 1 (p.R88Q), exon 9 (p.E542K, p.E545K/G/A,
p.Q546R) and exon 21 (p.H1047R/L/Y, p.M1043I/V) (17,23),
each of which induces its constitutive activation (3,24–26).
However, the association between genetic variations and endometrial
carcinogenesis remains poorly understood. Several studies have
identified different correlations between PIK3CA variations
and tumor grade, stage and type (5,6,26–29).
PTEN somatic mutations have been reported in
several types of gynecological tumor; in addition, PTEN
inactivation is present in endometriosis and endometrial
hyperplasia, and may be considered an early event in cancer
development (30–33). In EECs, PTEN mutations have
been identified (16), and PTEN
protein loss was revealed to be correlated with overall survival
(OS) (34).
As suggested by TCGA, the PI3K signaling pathway may
represent a potential target for targeted therapy for the treatment
of EC (3,23). Indeed, several PI3K pathway
inhibitors have been developed and are currently under
investigation in preclinical studies, and in early clinical trials
(35), with various results
depending on the therapeutic agent used and treatment set-up
(8,18,20,22,24,26,29,36–44).
Nevertheless, it is currently unclear how PIK3CA and
PTEN mutations affect the sensitivity to inhibitors in EC
(45–47). Consequently, efforts should be
focused on improving the characterization of the PIK3CA and
PTEN mutational profiles, to allow the establishment of
personalized targeted therapy.
Several approaches may be used for the
characterization of mutations [e.g. next-generation sequencing
(NGS), mass spectrometry (MS), Sanger sequencing, polymerase chain
reaction (PCR)-high-resolution melting analysis (HRMA) and
quantitative PCR (qPCR)]. Certain methods (i.e. NGS and MS) provide
a comprehensive overall evaluation with high specificity and
sensitivity, but require expensive instrumentation and procedures,
and trained personnel; other methods (i.e. Sanger sequencing,
PCR-HRMA and qPCR) are simpler and more cost-effective, but allow
the analysis of only a limited number of selected exons, and the
specificity and sensitivity differ (Sanger sequencing had high
specificity and low sensitivity, and vice versa for PCR-HRMA,
whereas qPCR is sensitive and specific, but allows the analysis of
only single known hotspots) (48–51).
The present pilot study had two main aims: i) To
develop an economical and efficient combined procedure on the basis
of PCR-HRMA and Sanger sequencing for evaluating the presence of
variants in PIK3CA (exons 1, 9 and 21) and PTEN
(exons 5, 6, 7 and 8) genes with the intention to overcome the
limitations of each technique; and ii) to assess the presence of
the primary mutations in PIK3CA and PTEN in a
selected homogeneous subgroup of G3 EECs.
Materials and methods
Tissue collection and DNA
extraction
A total of 18 available and suitable tissues were
selected from patients with G3 EEC and with at least 2 years of
follow up (up to 1 February 2018). All patients underwent surgical
treatment between May 2013 and November 2015, at the Obstetrics and
Gynecology Unit, Careggi University Hospital (Florence, Italy).
Patients were treated depending on International Federation of
Gynecology and Obstetrics (FIGO staging), grading and European
Society of Gynecological Oncology (ESGO) risk (52). Tumor samples were collected at the
time of surgery, following acquisition written informed consent
from the patients. The study was approved by the Ethics Committee
of Careggi University Hospital. Patients' demographic, clinical and
pathological features are presented in Table I.
 | Table I.Demographic and clinicopathological
features of patients with grade 3 endometrioid endometrial
cancer. |
Table I.
Demographic and clinicopathological
features of patients with grade 3 endometrioid endometrial
cancer.
| Sample | BMI | Age at surgery,
years | Menopause,
status | Menopause duration
years | Myometrial,
infiltration % | FIGO stage | Time disease-free,
months | Total follow-up,
months | LVSI | Lymph node
involvement (n) | Risk |
|---|
| S1 | Ow | 57 | Yes | 4 | >50 | IB | 22 | 58 | No | No | High |
| S2 | Nw | 74 | Yes | 29 | >50 | IB | – | 56 | No | No | High |
| S3 | Ob II | 65 | Yes | 15 | >50 | IIIC | – | 42 | No | 3 | Advanced |
| S4 | Ob II | 78 | Yes | 23 | >50 | IB | 6 | 38 | No | No | High |
| S5 | Ow | 60 | Yes | 9 | >50 | IB | – | 37 | Yes | No | High |
| S6 | Ob II | 68 | Yes | 16 | >50 | IIIA | – | 36 | Yes | No | High |
| S7 | Nw | 54 | Yes | 1 | >50 | IB | – | 36 | Yes | No | High |
| S8 | Ob I | 72 | Yes | 20 | >50 | II | 20 | 37 | No | No | High |
| S9 | Nw | 80 | Yes | 31 | >50 | IB | – | 36 | Yes | No | High |
| S10 | Ob III | 80 | Yes | 30 | >50 | IB | – | 34 | Yes | No | High |
| S11 | Ob I | 79 | Yes | 30 | >50 | II | – | 34 | No | No | High |
| S12 | Nw | 77 | Yes | 24 | >50 | IB | 25 | 32 | Yes | No | High |
| S13 | Nw | 43 | No | – | <50 | IA | – | 31 | No | No |
High-intermediate |
| S14 | Ob II | 74 | Yes | 15 | <50 | IA | – | 30 | Yes | No |
High-intermediate |
| S15 | Ow | 69 | Yes | 13 | <50 | IA | – | 28 | No | No |
High-intermediate |
Tumor samples were immediately fixed in formalin and
processed for paraffin embedding. The resulting formalin-fixed
paraffin-embedded (FFPE) tissues were examined histologically by
pathologists at Careggi University Hospital for the selection of
areas containing >70% of tumor cells.
DNA was extracted from slices using a QIAamp DNA
FFPE tissue kit (Qiagen GmbH, Hilden, Germany), according to the
manufacturer's protocol. DNA quality (ratio R) and quantity (DNA
concentration) were determined using a spectrophotometer
(NanoDrop® 1000 UV spectrophotometer; Thermo Fisher
Scientific, Inc., Waltham, MA, USA).
Primers and PCR conditions
Primers for selected exons of PIK3CA and
PTEN were custom-made by Sigma-Aldrich; Merck KGaA
(Darmstadt, Germany), and were designed to be suitable for Sanger
sequencing and PCR-HRMA using primer3web software (version 4.1.0;
primer3.ut.ee), checked for specificity and mismatch using
Primer-BLAST (www.ncbi.nlm.nih.gov/tools/primer-blast) and using SNP
Check (version 3; secure.ngrl.org.uk/SNPCheck/snpcheck.htm). Primer
sequences are presented in Table
II.
 | Table II.Primer sequences. |
Table II.
Primer sequences.
| Gene | Exon | Forward primer | Reverse primer | Product size,
bp |
|---|
| PIK3CA | 1 |
5′-TGTTACTCAAGAAGCAGAAAGGG-3′ |
5′-ACGAAGGTATTGGTTTAGACAGA-3′ | 231 |
|
| 9 |
5′-AGGGAAAATGACAAAGAACA-3′ |
5′-ACCTGTGACTCCATAGAAA-3′ | 124 |
|
| 21 |
5′-TGCTCCAAACTGACCAAACTG-3′ |
5′-TGCATGCTGTTTAATTGTGTGG-3′ | 299 |
| PTEN | 5 |
5′-TGTGAAGATCTTGACCAATGGC-3′ |
5′-AAATTCTCAGATCCAGGAAGAGG-3′ | 231 |
|
| 6 |
5′-ACGACCCAGTTACCATAGCA-3′ |
5′-TGTGAAACAACAGTGCCACT-3′ | 185 |
|
| 7 |
5′-CCTCAGTTTGTGGTCTGCC-3′ |
5′-GCCAGAGTAAGCAAAACACCT-3′ | 296 |
|
| 8 |
5′-TACCAGGACCAGAGGAAACC-3′ |
5′-AGCAAGTTCTTCATCAGCTGT-3′ | 280 |
PCR was performed using the custom primers, Taq PCR
core kit (Qiagen GmbH) and the third-generation DNA-intercalating
dye Syto 9 (Thermo Fisher Scientific, Inc.). Briefly, 10 ng DNA was
amplified using 2 µl master mix (10X), 0.5 µl each primer (10 µM),
0.4 µl dNTPs (10 mM each), 0.4 µl Syto9 (50 µM), 0.1 µl Taq DNA
polymerase (5 U/µl) and water in a final volume of 20 µl. Samples
were subjected to incubation at 95°C for 5 min, then 40
amplification cycles of 95°C for 30 sec, 56/60°C (56°C for
PTEN and 60°C for PIK3CA) for 45 sec and 72°C for 45
sec, and a final incubation at 72°C for 20 min in a GeneAmp PCR
system 9700 (Thermo Fisher Scientific, Inc.).
HRMA
Amplified samples were submitted to a
high-resolution melting (HRM) protocol (95°C for 5 min, 40°C for 1
min, ramping temperature from 72 to 84°C) in a Rotor-gene Q PCR
cycler (Qiagen GmbH). The melting curve of each sample was analyzed
using melting and HRM software (Rotor-Gene® Q-Pure
Detection series software, version 2.1.0, build 9; Qiagen
GmbH).
Sanger sequencing
The amplified and melted PCR products were purified
using a QIAquick PCR purification kit (Qiagen GmbH) and sequenced
on an ABI 310 genetic analyzer (Thermo Fisher Scientific, Inc.).
Briefly, 3 µl purified PCR was added to 1 µl BigDye Terminator, 3.2
µl forward/reverse primer (1 µM), 3.5 µl BigDye Terminator buffer
and water in a final volume of 20 µl, and amplified for 25 cycles
of 96°C for 10 sec, 59°C for 5 sec and 60°C for 2 min in the
GeneAmp PCR system 9700, purified using a DyeEx 2.0 Spin column
(Qiagen GmbH), according to the manufacturer's protocol, and 7 µl
was Sanger sequenced.
To confirm identified mutations, the samples were
also sequenced using reverse primers. The electropherogram of each
sequenced amplicon was independently evaluated by two individuals.
For validation of the combined procedure, in a subgroup of three
samples, NGS was performed using Ion Torrent S5 with HotSpot cancer
panel version 1 on Chip 520 (Thermo Fisher Scientific, Inc.) with a
coverage (mean depth) average of 1,000X, mean reads of 400,000 and
mean read length of 115 bp, according to the manufacturer's
protocol.
For validation of variations identified by PCR-HRMA,
but not by Sanger sequencing (sample S2 and S4), their
corresponding PCR products associated with PTEN exon 5 were
cloned using TOPO-TA cloning (Thermo Fisher Scientific, Inc.),
according to the manufacturer's protocol. A total of 20 colonies
was isolated and the plasmids were extracted using a QIAprep Spin
Miniprep kit (Qiagen GmbH). The extracted plasmids were sequenced
as aforementioned.
Mutation identification
The nucleotide changes were evaluated using the
Catalogue of Somatic Mutations in Cancer (COSMIC) database (version
86; cancer.sanger.ac.uk/cosmic), ensembl
(release 93; www.ensembl.org/index.html), PolyPhen-2 (version 2;
genetics.bwh.harvard.edu/pph2) and
Mutation Taster (MT) (version 2; current build, NCBI 37/Ensembl 69;
www.mutationtaster.org) software. Single
nucleotide polymorphisms (SNPs) were confirmed using UCSC Genome
Browser (genome.ucsc.edu/). The clinical impact
was evaluated using COSMIC by Functional Analysis Through Hidden
Markov Models score (pathogenic score, range: 0, not pathogenic, to
1, pathogenic), for single amino acid substitutions by PolyPhen-2
score (range: 0, not pathogenic, to 1, pathogenic) and for
synonymous single amino acid substitutions, complex variations
(insertions/deletions/frameshifts) and modifications in intronic
sequence by MT [evaluation: Disease-causing (DC) or SNP].
Results
Quality of DNA
The extracted DNAs had sufficient amount and quality
for PCR amplification and Sanger sequencing; in particular, the
concentration mean was 164.15±2.00 ng/µl; the median was
138.00±2.00 ng/µl; the range was between 18.00 and 299.10 ng/µl;
the value of R was 2.15 (mean) and 2.15 (median), and the range was
between 2.00 and 2.47.
Establishment of the combined PCR-HRMA
and Sanger sequencing procedure
The combined procedure is based on the sequential
execution of PCR-HRMA and Sanger sequencing, without other steps,
and avoids double PCR if the two procedures are performed
separately with a corresponding saving of DNA, time of execution
and limiting the number of mistakes due to performing the analysis
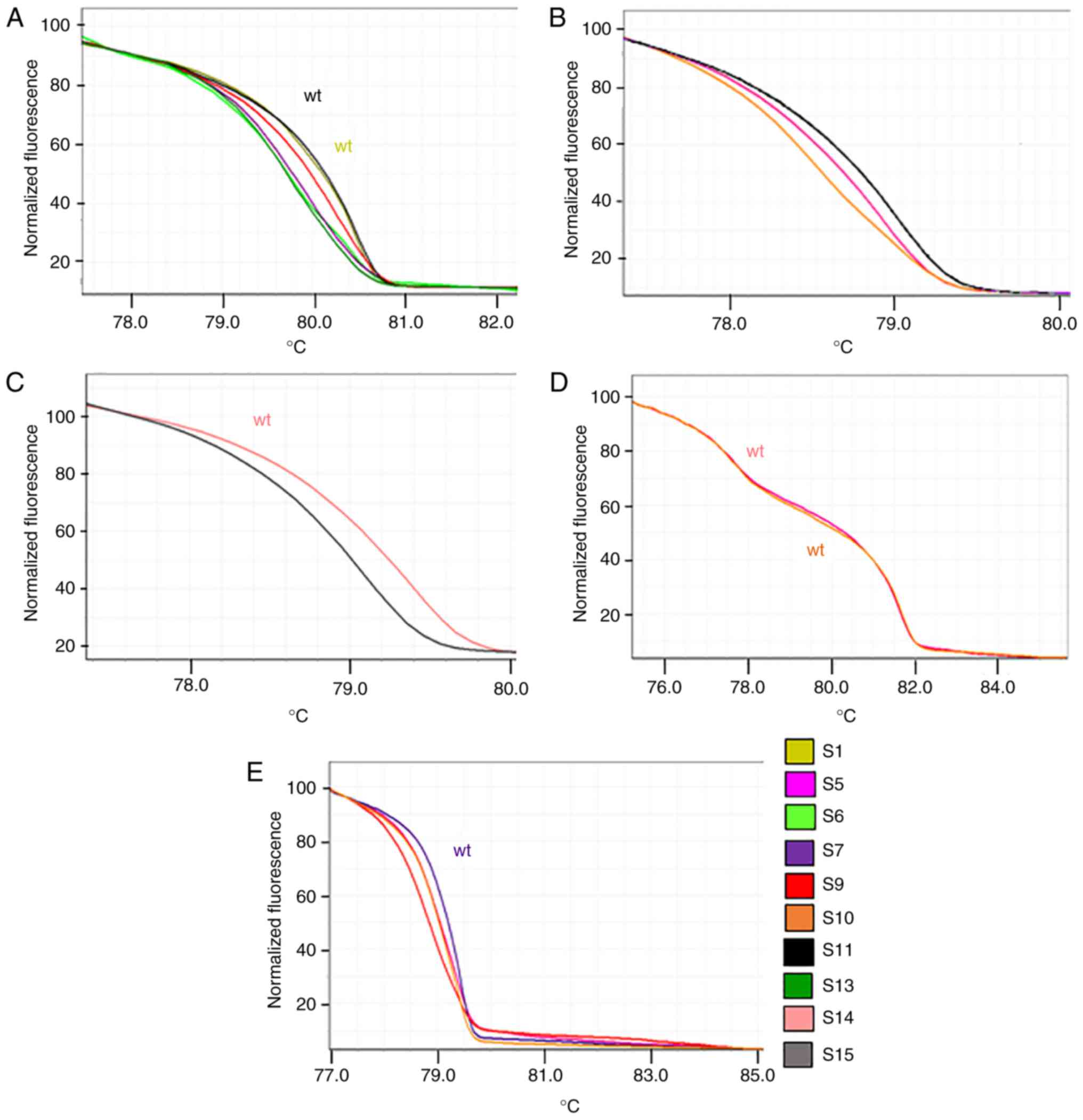
twice. All samples were amplified to verify the success of
amplification and to perform the HRMA. An appropriate curve was
obtained for all the samples and for each analyzed exon. The HRMA
suggested the presence of nucleotide alteration(s) without
providing direct information on the specific nucleotide change(s),
which induced changes in the shape of the HRM and melting curves.
In particular, the presence of nucleotide change(s) induced or a
shift in the peak or the appearance of additional peak(s) in the
melting curve and a change in the shape of the HRM curve. In order
to identify associations of the curve shape with a specific
nucleotide sequence, the amplicons were sequenced using the Sanger
procedure.
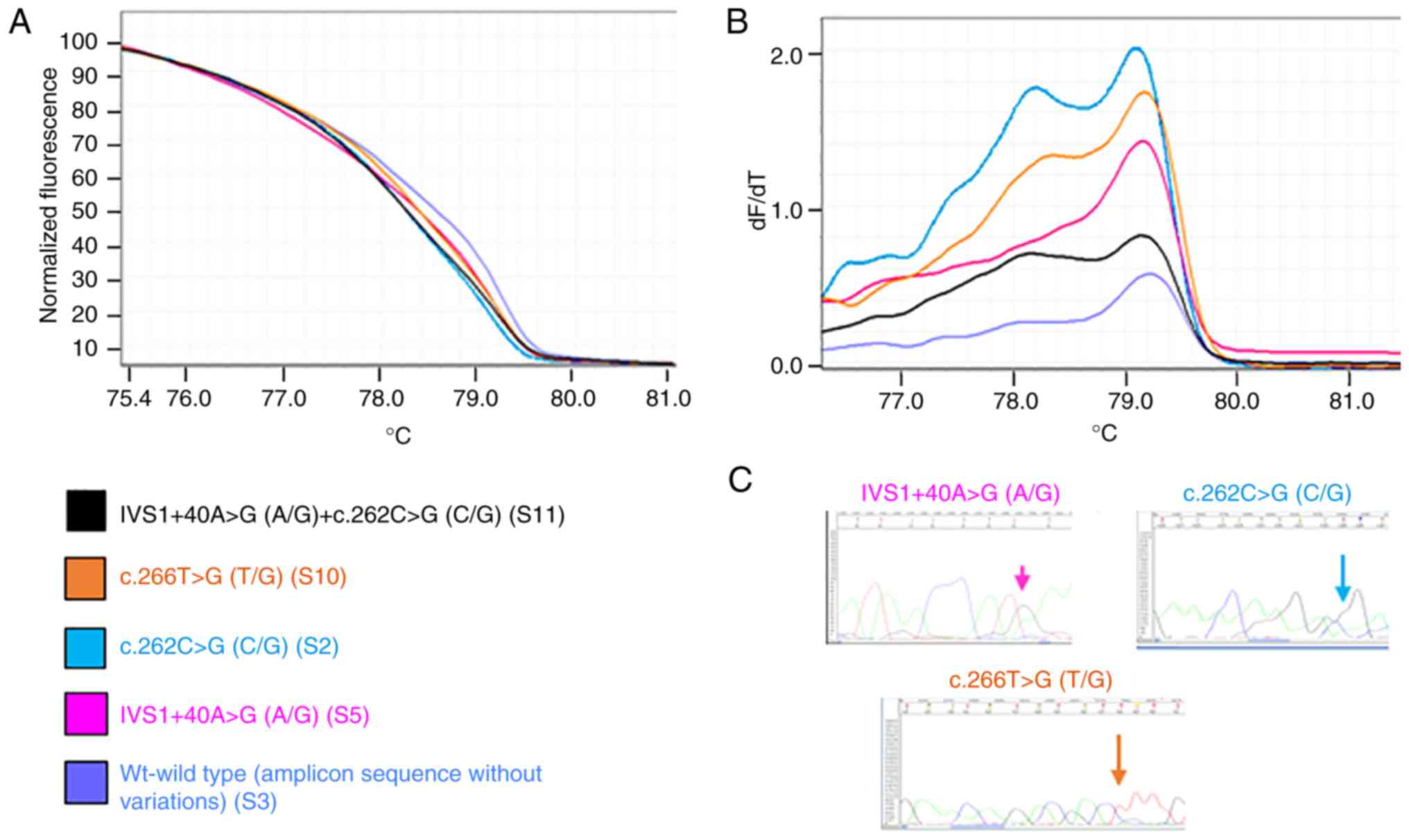
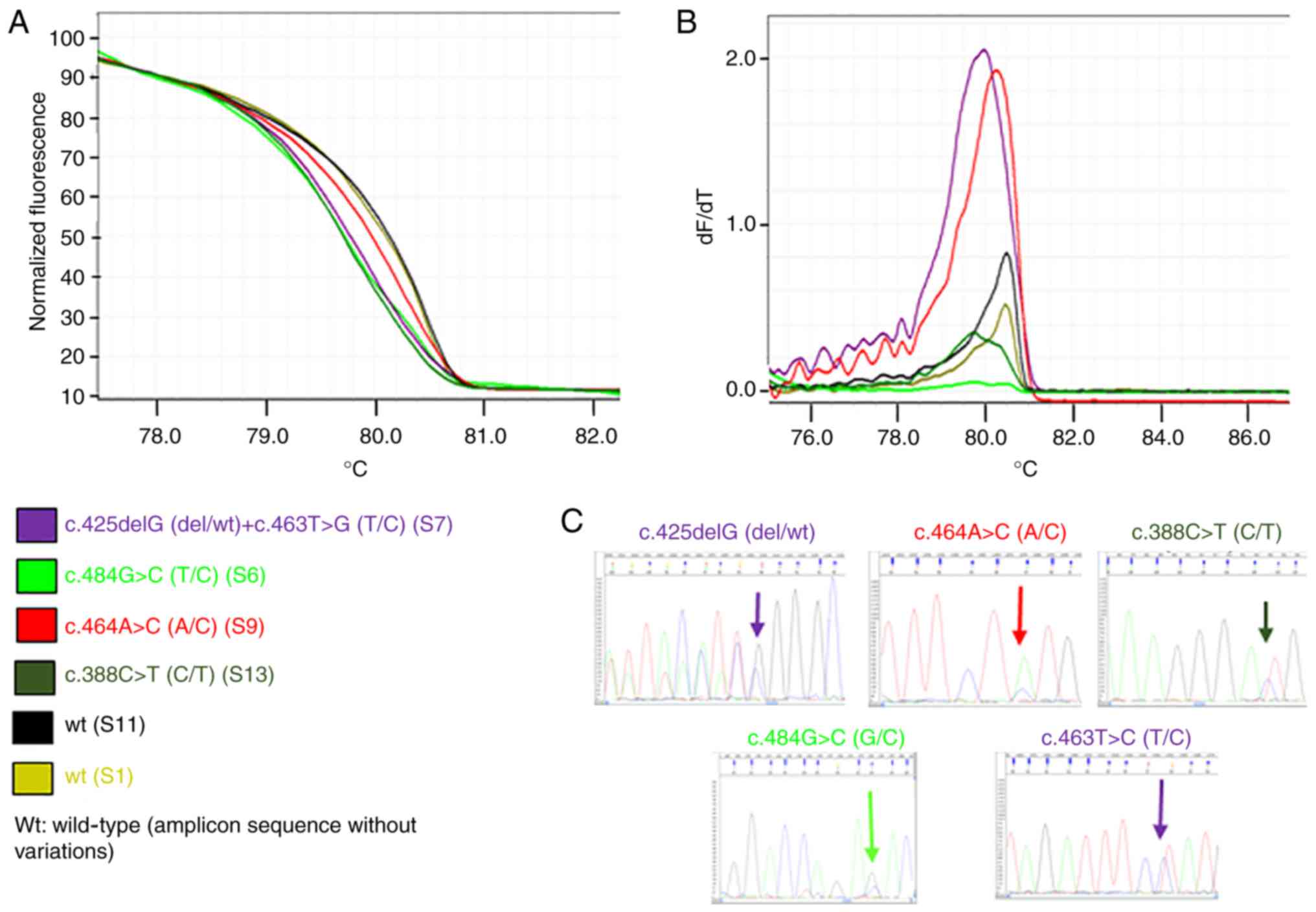
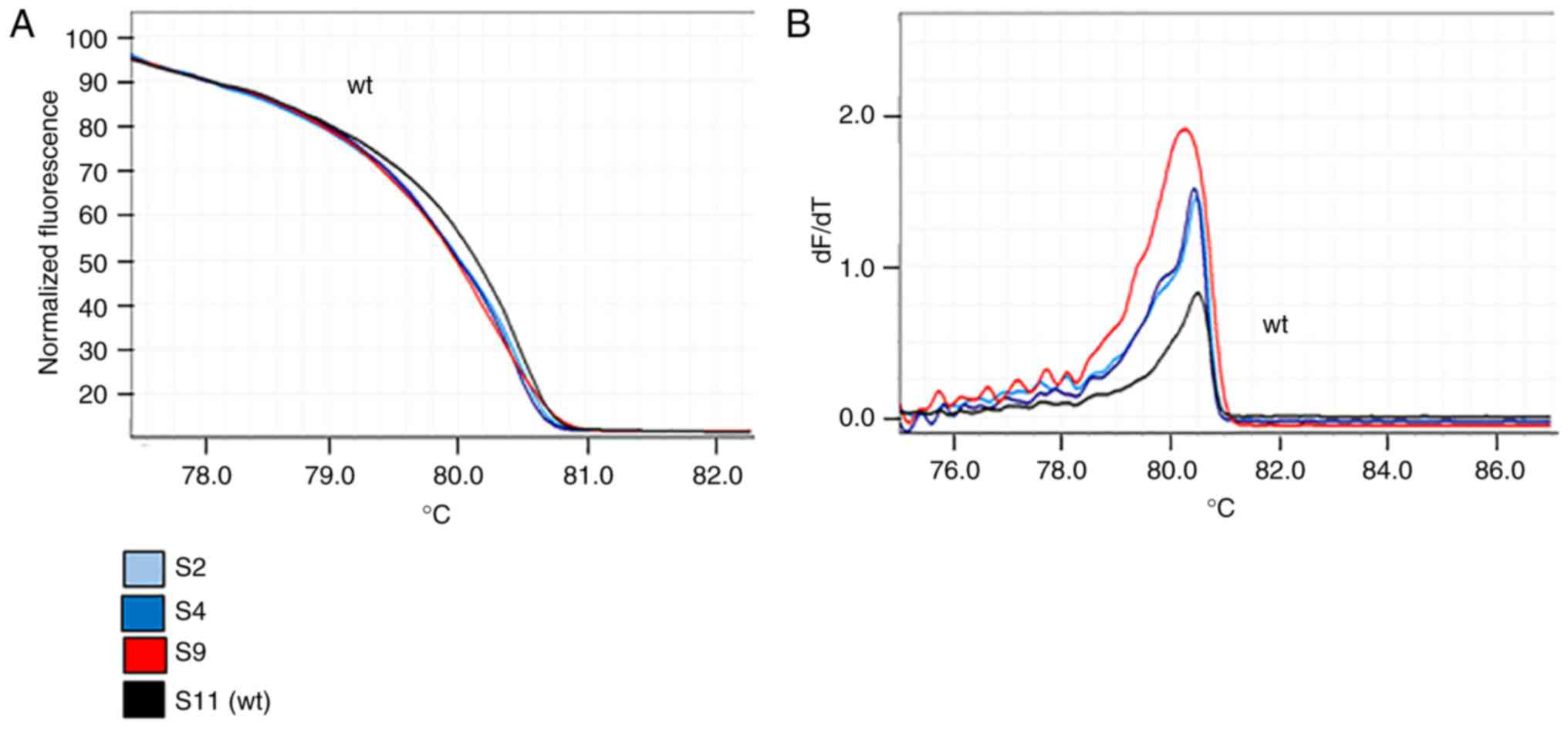
Figs. 1 and 2 present representative results of the HRM
and melting curves, and the corresponding specific base
modification identified by Sanger sequencing, for PIK3CA
exon 1 and for PTEN exon 5, respectively.
The comparison of HRMA and sequencing data confirmed
that samples with the same HRM curve shared the same nucleotide
sequence. Differences in HRM curves for the same amplicon suggested
the presence of different nucleotide sequences. Figs. 3 and 4 present the principal variations detected
by HRM curves along with the corresponding nucleotide change(s) for
each exon of PIK3CA and PTEN analyzed. Notably, for
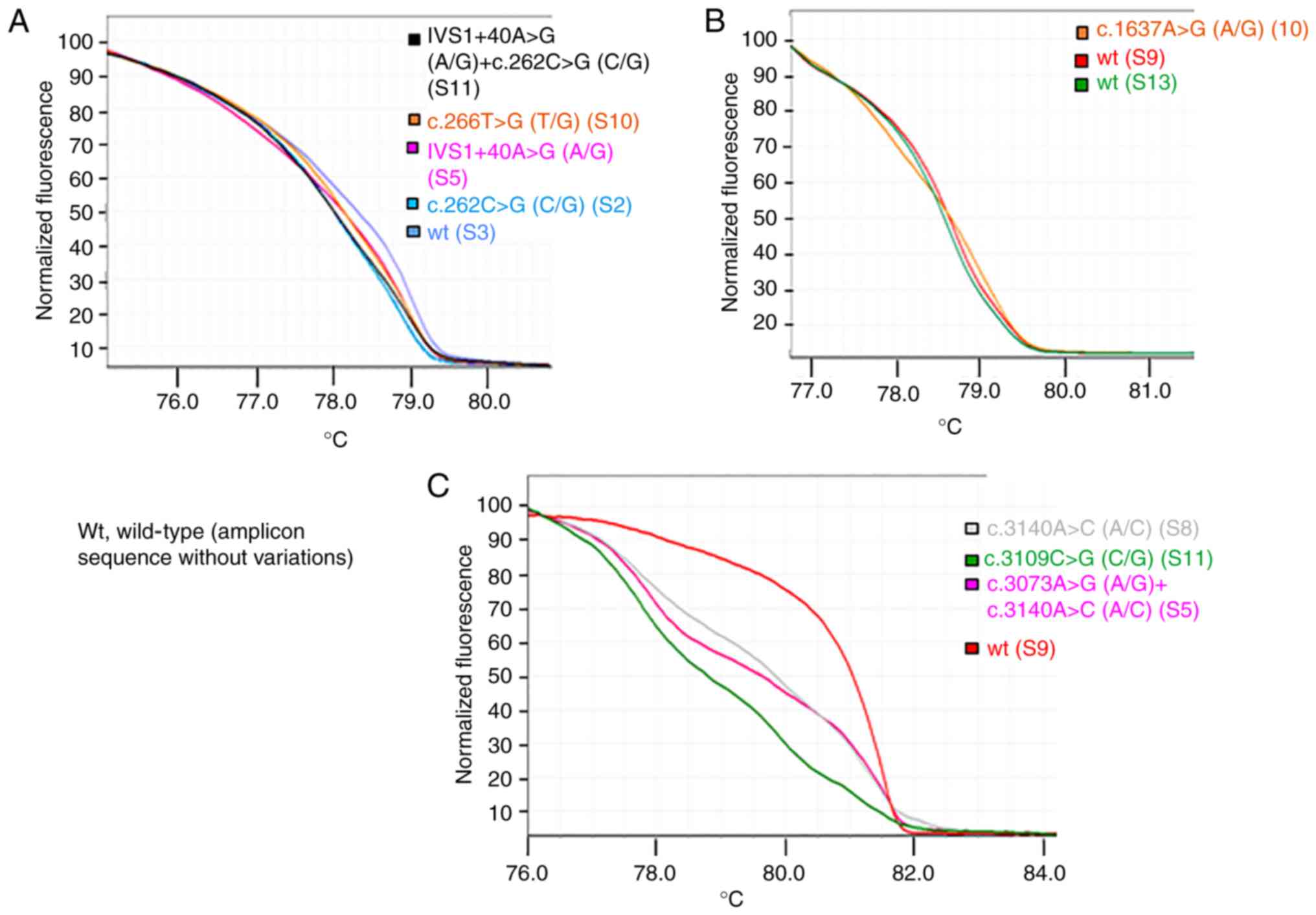
PTEN exon 5, it was observed that three samples shared the
same HRM and melting curves (Fig.
5), but the corresponding Sanger sequences revealed the
presence of a specific variant only for one sample (sample S9;
p.Y155S), suggesting that the two samples (samples S2 and S4) may
have the same variation as sample S9, but at lower levels. To
verify this hypothesis, the PCR products of samples S2 and S4 were
cloned into a plasmid and sequenced, which confirmed the presence
of the suggested variant at low level (two and three positive
colonies, respectively, in samples S2 and S4).
This combined approach allowed the avoidance of a
double PCR (if the two methodologies were applied separately),
retaining DNA for use in further analysis. Furthermore, the use of
HRMA alone suggested the presence of homoduplex or heteroduplex
variations in the analyzed exons [PIK3CA exon 1 (61%), exon
9 (6%) and exon 21 (33%), and PTEN exon 5 (44%), exon 6
(78%), exon 8 (50%) and none in exon 7), but provided information
on the type of variation. Sanger sequencing provided this specific
information on the type of alteration, allowing the
characterization of the nucleotide variation, but with low
sensitivity. The merged information provided the sensitivity of
HRMA and the specificity of Sanger sequencing (for example, for
PTEN exon 5, the mutation detection frequency increased from 33 to
44%).
To further evaluate this approach, three samples
were validated using this combined procedure and by NGS technology
in which all the PTEN and PIK3CA exons were analyzed,
as well as 48 other cancer-associated genes. The two technologies
yielded the same results: For sample S16 only, a single-base
mutation in PTEN exon 5 was detected.
Evaluation of variants on PIK3CA and
PTEN
PIK3CA
A total of seven different variations in 11 samples
(61%) were identified, all of which were heterozygous, except for
IVS1+40A>G (intronic) which in three samples was homozygous and
7/11 were pathogenic (Table III).
In particular, in exon 1, two mutations were identified, namely
p.L89R (6%) and p.R88G (18%), as well as the SNP IVS1+40A>G
(rs3729674; 44%); in exon 9, the mutation p.Q546R (cosm12459; 6%),
was identified; and in exon 21, the mutation p.H1047P (cosm249874;
22%) and the variant p.E1037Q (12%) were detected. The highest
number of mutations (three mutations and one variant) was observed
in exon 21 compared with exon 1 (two mutations) and exon 9 (one
mutation). All five samples at FIGO stage III (IIIA and IIIC) did
not exhibit any variation in PIK3CA.
 | Table III.PIK3CA variants. |
Table III.
PIK3CA variants.
|
|
| PIK3CA
variants | Pathogenic
effect | Reported by
COSMIC |
|---|
|
|
|
|
|
|
|---|
| Sample | FIGO stage | Location | Nucleotide
change | Predicted protein
change | Yes/no | Predicted by | Other types of
cancer | In EC |
|---|
| S1 | IB | Exon 1 | IVS1+40A>G |
| No (SNP) |
UCSC/dbSNP/ClinVar | – | – |
| S2 | IB | Exon 1 | c.262C>G | p.R88G | Yes | PolyPhen-2 (score
0.994) | – | – |
| S3 | IIIC | – | – | – | – | – | – | – |
| S4 | IB | Exon 1 | IVS1+40A>G |
| No (SNP) |
UCSC/dbSNP/ClinVar | – | – |
| S5 | IB | Exon 1 | IVS1+40A>G |
| No (SNP) |
UCSC/dbSNP/ClinVar | – | – |
|
|
| Exon 21 | c.3073A>G | p.T1025A | Yes | COSMIC (FATHMM
score 1.00) | Yes | Yes
(first)a |
|
|
| Exon 21 | c.3140A>C | p.H1047P | No (SNP) | Cosmic (FATHMM
score 0.96) | Yes | No |
| S6 | IIIA | – | – | – | – | – | – | – |
| S7 | IB | Exon 1 | IVS1+40A>G |
| No (SNP) |
UCSC/dbSNP/ClinVar | – | – |
|
|
| Exon 21 | c.3140A>C | p.H1047P | Yes | Cosmic (FATHMM
score 0.96) | Yes | No |
| S8 | II | Exon 1 | IVS1+40A>G
homo |
| No (SNP) |
UCSC/dbSNP/ClinVar | – | – |
|
|
| Exon 21 | c.3140A>C | p.H1047P | Yes | Cosmic (FATHMM
score 0.96) | Yes | No |
| S9 | IB | Exon 1 | IVS1+40A>G
homo |
| No (SNP) |
UCSC/dbSNP/ClinVar | – | – |
| S10 | IB | Exon 1 | c.266T>G | p.L89R | Yes | PolyPhen-2 (score
1.000) | – | – |
|
|
| Exon 9 | c.1637A>G | p.Q546R | Yes | Cosmic (FATHMM
score 0.97) | Yes | Yes
(third)a |
|
|
| Exon 21 | c.3140A>C | p.H1047P | Yes | Cosmic (FATHMM
score 0.96) | Yes | No |
| S11 | II | Exon 1 | c.262C>G | p.R88G | Yes | PolyPhen-2 (score
0.994) | – | – |
|
|
| Exon 1 | IVS1+40A>G |
| No (SNP) |
UCSC/dbSNP/ClinVar | – | – |
|
|
| Exon 21 | c.3109G>C | p.E1037Q | Yes | PolyPhen-2 (score
0.011) | – | – |
|
|
|
|
|
|
| Mutation Taster
(disease-causing) | – | – |
| S12 | IB | Exon 1 | IVS1+40A>G
homo |
| No (SNP) |
UCSC/dbSNP/ClinVar | – | – |
| S13 | IA | Exon 1 | c.262C>G | p.R88G | Yes | PolyPhen-2 (score
0.994) | – | – |
|
|
| Exon 21 | c.3109G>C | p.E1037Q | No | PolyPhen-2 (score
0.011) | – | – |
|
|
|
|
|
|
| Mutation Taster
(disease-causing) | – | – |
| S14 | IA | – | – | – | – | – | – | – |
| S15 | IA | – | – | – | – | – | – | – |
| S16 | IIIA | – | – | – | – | – | – | – |
| S17 | IIIC | – | – | – | – | – | – | – |
| S18 | IIIC | – | – | – | – | – | – | – |
PTEN
A total of 12 different variations were identified
and 16/18 (89%) samples exhibited at least one variation, among
which 12 (67%) presented a pathological variation (Table IV). In particular, exon 6 exhibited
the highest number of variations. The most represented alteration
was p.Q171Q (78%), followed by p.R189K (cosm1745951; 44%). Exon 5
exhibited pathogenic mutations in six samples (33%): p.Y155S (18%),
p.D162H (cosm5274; 12%), p.Y155H (cosm5038; 6%), p.G143fs*4
(cosm30623; 6%), R130* (cosm5152; 6%) and G129Stop (cosm18663; 6%).
Exon 8 had an intronic variant (IVS8+32T>G; 44%) and in one
sample it was associated with p.I300fs*2. Exon 7 did not present
any mutation according to Kafshooz et al (53). No particular trend was observed for
the PTEN mutation, except for the non-menopausal patient
that had PTEN R130* and the presence of the PTEN T167fs* in
the obese class III patient.
 | Table IV.PTEN variants. |
Table IV.
PTEN variants.
|
|
| PTEN variants | Pathogenic
effect | Reported by
COSMIC |
|---|
|
|
|
|
|
|
|---|
| Sample | FIGO stage | Location | Nucleotide
change | Predicted protein
change | Yes/no | Predicted by | Other types of
cancer | In EC |
|---|
| S1 | IB | Exon 6 | c.513G>A | p.Q171Q | (Yes) | Mutation Taster
(disease-causing) | – | – |
|
|
| Exon 8 | IVS8+32T>G |
| No (SNP) |
UCSC/dbSNP/ClinVar | – | – |
| S2 | IB | Exon 5 | c.464A>C | p.Y155S | Yes | PolyPhen-2 (score
1.000) | – | – |
|
|
| Exon 6 | c.566G>A | p.R189K | Yes | COSMIC (FATHMM
score 0.88) | Yes | No |
|
|
| Exon 6 | c.513G>A | p.Q171Q | (Yes) | Mutation Taster
(disease-causing) | – | – |
|
|
| Exon 8 | IVS8+32T>G
homo |
| No (SNP) |
UCSC/dbSNP/ClinVar | – | – |
| S3 | IIIC | Exon 5 | c.484G>C | p.D162H | Yes | COSMIC (FATHMM
score 0.99) | Yes | No |
|
|
| Exon 6 | c.513G>A | p.Q171Q | (Yes) | Mutation Taster
(disease-causing) | – | – |
| S4 | IB | Exon 5 | c.464A>C | p.Y155S | Yes | PolyPhen-2 (score
1.000) | – | – |
|
|
| Exon 6 | c.513G>A | p.Q171Q | (Yes) | Mutation Taster
(disease-causing) | – | – |
|
|
| Exon 6 | c.566G>A | p.R189K | Yes | COSMIC (FATHMM
score 0.88) | Yes | No |
| S5 | IB | Exon 6 | c.513G>A | p.Q171Q | (Yes) | Mutation Taster
(disease-causing) | – | – |
|
|
| Exon 6 | c.566G>A | p.R189K | Yes | COSMIC (FATHMM
score 0.88) | Yes | No |
|
|
| Exon 8 | IVS8+32T>G
homo |
| No (SNP) |
UCSC/dbSNP/ClinVar |
|
|
| S6 | IIIA | Exon 5 | c.484G>C | p.D162H | Yes | COSMIC (FATHMM
score 0.99) | Yes | No |
|
|
| Exon 6 | c.513G>A | p.Q171Q | (Yes) | Mutation Taster
(disease-causing) | – | – |
|
|
| Exon 8 | IVS8+32T>G |
| No (SNP) |
UCSC/dbSNP/ClinVar | – | – |
| S7 | IB | Exon 5 | c.425delG | p.G143Afs*4 | No | COSMIC (FATHMM
score no.) | (ovary) | No |
|
|
| Exon 5 | c.463T>C | p.Y155H | Yes | COSMIC (FATHMM
score 0.97) | Yes | Yes
(first)a |
|
|
| Exon 6 | c.513G>A | p.Q171Q | (Yes) | Mutation Taster
(disease-causing) | – | – |
|
|
| Exon 6 | c.566G>A | p.R189K | Yes | COSMIC (FATHMM
score 0.88) | Yes | No |
| S8 | II | Exon 6 | c.513G>A | p.Q171Q | (Yes) | Mutation Taster
(disease-causing) | – | – |
|
|
| Exon 6 | c.566G>A | p.R189K | Yes | COSMIC (FATHMM
score 0.88) | Yes | No |
| S9 | IB | Exon 5 | c.464A>C | p.Y155S | Yes | PolyPhen-2 (score
1.000) | – | – |
|
|
| Exon 6 | c.513G>A | p.Q171Q | (Yes) | Mutation Taster
(disease-causing) | – | – |
|
|
| Exon 6 | c.566G>A | p.R189K | Yes | COSMIC (FATHMM
score 0.88) | Yes | No |
|
|
| Exon 8 | c.899_900delCT | p.I300Nfs*2 | Yes | PolyPhen-2 (score
0.059), associated with an amino acid alteration | – | – |
|
|
|
|
|
|
| Mutation Taster
(disease-causing) | – | – |
|
|
| Exon 8 | IVS8+32T>G |
| No (SNP) |
UCSC/dbSNP/ClinVar | – | – |
| S10 | IB | Exon 6 | c.497_498insT | p.T167Nfs*13 | Yes | PolyPhen-2 (score
0.999), associated with an amino acid alteration | – | – |
|
|
|
|
|
|
| Mutation Taster
(disease-causing) | – | – |
|
|
| Exon 6 | c.513G>A | p.Q171Q | (Yes) | Mutation Taster
(disease-causing) | – | – |
|
|
| Exon 8 | IVS8+32T>G
homo |
| No (SNP) |
UCSC/dbSNP/ClinVar |
|
|
| S11 | II | Exon 6 | c.513G>A | p.Q171Q | (Yes) | Mutation Taster
(disease-causing) | – | – |
| S12 | IB | Exon 6 | c.513G>A | p.Q171Q | (Yes) | Mutation Taster
(disease-causing) | – | – |
| S13 | IA | Exon 5 | c.388C>T | p.R130* |
| COSMIC (FATHMM
score 0.95) | Yes | Yes
(first)a |
|
|
| Exon 6 | c.513G>A | p.Q171Q | (Yes) | Mutation Taster
(disease-causing) |
|
|
|
|
| Exon 6 | c.566G>A | p.R189K | Yes | COSMIC (FATHMM
score 0.88) | Yes | No |
| S14 | IA | Exon 8 | IVS8+32T>G |
| No (SNP) |
UCSC/dbSNP/ClinVar | – | – |
| S15 | IA | Exon 6 | c.513G>A | p.Q171Q | (Yes) | Mutation Taster
(disease-causing) | – | – |
|
|
| Exon 6 | c. 549G>A | p.K183K | (Yes) | Mutation Taster
(disease-causing) | – | – |
|
|
| Exon 6 | c.566G>A | p.R189K | Yes | COSMIC (FATHMM
score 0.88) | Yes | No |
|
|
| Exon 8 | IVS8+32T>G |
|
|
UCSC/dbSNP/ClinVar | – | – |
| S16 | IIIA | – | – | – | – | – | – | – |
| S17 | IIIC | Exon 5 | c.385G>T | p.G129Stop | Yes | COSMIC (FATHMM
score 0.99) | Yes | Yes
(third)a |
| S18 | IIIC | – | – | – | – | – | – | – |
Variant/mutational load
Regarding the overall variant evaluation, the
majority of the samples (89%) exhibited at least one variation
(Table V). Regarding SNPs, 50% of
the samples exhibited one SNP and 18% presented with two SNPs.
 | Table V.Overall evaluation of PIK3CA
and PTEN variants in the samples analyzed. |
Table V.
Overall evaluation of PIK3CA
and PTEN variants in the samples analyzed.
| Sample | PIK3CA
variants | PTEN
variants |
|---|
| S1 |
IVS1+40A>G | p.Q171Q,
IVS8+32T>G |
| S2 | p.R88G | p.R189K,
p.Y155S§, p.Q171Q, IVS8+32T>G |
| S3 |
| p.D162H,
p.Q171Q |
| S4 |
IVS1+40A>G | p.R189K,
p.Y155S§, p.Q171Q |
| S5 | p.T1025A,
p.H1047P, IVS1+40A>G | p.R189K,
p.Q171Q, IVS8+32T>G |
| S6 |
| p.D162H,
p.Q171Q, IVS8+32T>G |
| S7 | p.H1047P,
IVS1+40A>G | p.Y155H,
p.R189K, p.G143Afs*4, p.Q171Q |
| S8 | p.H1047P,
IVS1+40A>G | p.R189K,
p.Q171Q |
| S9 |
IVS1+40A>G | p.R189K,
p.Q171Q, p.Y155S, p.I300Nfs*2, IVS8+32T>G |
| S10 | p.Q546R,
p.H1047P, p.L89R |
p.T167Nfs*13, p.Q171Q,
IVS8+32T>G |
| S11 | p.R88G,
p.E1037Q, IVS1+40A>G | p.Q171Q |
| S12 |
IVS1+40A>G | p.Q171Q |
| S13 | p.R88G,
p.E1037Q | p.R130*,
p.R189K, p.Q171Q |
| S14 |
|
IVS8+32T>G |
| S15 |
| p.R189K,
p.Q171Q, p.K183K, IVS8+32T>G |
| S16 |
|
|
| S17 |
|
p.G129Stop |
| S18 |
|
|
Regarding the mutations, 67% of the samples had
confirmed mutations at least in one of the two genes, whereas 18%
had mutations in the two genes, and these values increased to 72
and 33%, respectively, if the mutations predicted with PolyPhen-2
were considered, and to 82 and 39%, respectively, if the MT
prediction was considered.
It was observed that 28% of the samples exhibited a
single confirmed mutation, 16% exhibited two mutations, 22%
exhibited three mutations and 6% exhibited four mutations.
Considering the overall variant load (excluding SNPs), 18% of the
samples exhibited none, one or four mutations, 10% exhibited two or
five mutations, and 26% exhibited three mutations.
No specific mutation/variant load distribution was
observed according to clinicopathological characteristics.
Discussion
Currently, there is increasing interest in
investigating the genome alterations in tumors, with such research
made possible by the development of specific and sensitive
technologies (e.g. NGS or MassArray). These promising technologies
are useful for understanding the mechanisms underlying
carcinogenesis and cancer evolution, but are costly and require
computational support for data analysis. For clinical application,
the identification of genetic variations must be performed rapidly
and cost-effectively, as usually performed by Sanger
sequencing/pyrosequencing or PCR-HRMA procedures. Typically, qPCR
is performed for confirmation analysis.
It is established that PCR-HRMA has a lower cost and
is an accurate tool for mutation scanning, allowing the
identification of variations through comparison of HRM curves;
consequently, it is used as screening method or for the detection
of known hotspot mutations. Its ability in discriminating known
mutations is due to the use of reference material, or if this
material is not available or novel variations are present in the
samples analyzed, the technique is not able to identify the
nucleotide change, but only to detect the presence (54–56).
The HRMA curve shape depends on the number and type of nucleotide
changes, with different nucleotide changes yielding different
curves. The procedure alone is highly sensitive (overall
sensitivity 0.1–5%) (50,51), but lacks specificity, since it does
not allow for the direct evaluation of the specific nucleotide
changes. Consequently, for the identification of nucleotide
variations, the procedure requires the use of reference materials
with the same alteration to be investigated along with the target
samples, or further investigations by other techniques (such as
sequencing or qPCR).
Sanger sequencing is the reference procedure for the
identification of nucleotide changes for clinical purposes; even if
this technology is unable to reveal variations at low levels
(overall sensitivity ~20%) (48,49),
sometimes pyrosequencing is used due its increased sensitivity.
However, this methodology is relatively expensive compared with
Sanger sequencing, even if more sensitive (57).
Currently, other high-throughput methodologies are
beginning to be used in the clinic for the identification of
genetic variation in clinical samples, such as NGS and MS that
allow the evaluation of multiple genes/exons and several samples in
the same run. Usually these procedures improve specificity and
sensitivity, but are expensive either for their required
instrumentation and methodological procedures, provide abundant
information that is not useful for clinical purposes, and require
trained personnel for data analysis. In particular, MS allows the
evaluation of known variations on specific exons/genes; instead NGS
allows the evaluation of genes or genome and the identifications of
new genetic variants. qPCR, which has high sensitivity and
specificity, allows the identification of well-known and
characterized variations, but is expensive and time-consuming if
several genes or variations require analysis, and is not suitable
for the detection of unknown variations. It is typically used for
confirmation when Sanger sensitivity fails.
The present study establishes a combined procedure
that overcomes the limitations of each technique alone (low
sensitivity of Sanger sequencing and low specificity of HRMA) and
combines the strengths (high specificity of Sanger sequencing and
high sensitivity of HRMA) of these techniques. It also allows a
double check for the presence of variants in the main exons of
PIK3CA and PTEN, in order to improve the reliability
of the results. The target of this approach is the identification
of variants in single samples and in amplicons of between 100 and
200 bp in length; consequently, it cannot be compared with
high-throughput procedures. Furthermore, this combined approach, in
comparison with the application of either method alone, may
decrease the amount of DNA used in the analysis (a single PCR is
performed for the two methodologies) and the number of samples
analyzed by Sanger sequencing. In particular, if a reference
wild-type (wt) sample is used in the PCR-HRMA step, only the
samples presenting HRM curves different from the wt should be
analyzed by Sanger sequencing. Furthermore, PCR-HRMA can also only
suggest the presence of novel variants, but they cannot be
identified; by contrast, the combined use of PCR-HRMA and Sanger
analysis can immediately confirm and identify the novel variations.
This combined procedure may even be useful for defining patient
eligibility for targeted therapy; it is known that targeted therapy
is dependent on the identification of specific mutations by Sanger
sequencing, and its efficacy is determined by the number of mutated
cells and by the development of any new mutations that can
originate from tumor heterogeneity or de novo. Owing to the
high sensitivity of HRMA, suspected cases with a low proportion of
mutations may still be identified and monitored (Table VI).
 | Table VI.Principal characteristics of the main
procedures used for identification of variants in DNA sequence. |
Table VI.
Principal characteristics of the main
procedures used for identification of variants in DNA sequence.
|
| Procedure |
|---|
|
|
|
|---|
|
|
High-throughput | Single
amplicon |
|---|
|
|
|
|
|---|
|
| MassArray | NGS | qPCR | PCR-HRMA | Sanger
sequencing | Pyrosequencing | PCR-HRMA+Sanger
sequencing |
|---|
| Cost per
sample | Medium | High | Medium | Low | Medium | Medium | Medium |
| Sensitivity | High | High | High | High | Low | Medium | High |
| Specificity | High | High | High | Low | High | High | High |
| Discovery of novel
variant | No | Yes | No | Yes | Yes | Yes/noa | Yes |
| Identification of
sequence change | Yes | Yes | Yes | No | Yes | Yes | Yes |
| Multiple hotspot
within amplicon length | No in the same
well/yes in a run | Yes | No/yesb | Yes | Yes | Yes | Yes |
| Multi-exon analysis
in the same preparation | Yes | Yes | No | No | No | No | No |
| Multi-sample
analysis in the same preparation | Yes | Yes | No | No | No | No | No |
| Trained
personnel | Yes | Yes | No | No | No | Medium | No |
| Cost of
instrumentation | High | High |
No/mediumc | No | No | Medium | No |
| Data analysis | Yes | Yes | No | No | No | No | No |
The present study focused on the analysis of
specific exons of PIK3CA and PTEN, since these are
established as the main genes involved in alterations in the PI3K
signaling pathway, and they have been widely investigated for the
characterization of EC as specific targets for personalized
therapies on the basis of PI3K pathway inhibitors (17,18,20,29,35,40).
Indeed, early clinical data on several tumor types suggested that
PIK3CA and PTEN mutations may affect the success of
PI3K/protein kinase B (AKT)/mammalian target of rapamycin
(mTOR)-targeted therapies (58–61).
Patients with gynecological tumors and PIK3CA mutations have
demonstrated a 30% response rate (RR) in early-phase clinical
trials with PI3K/AKT/mTOR inhibitors, compared with a 10% RR in
patients lacking PIK3CA mutations (42). It is conceivable that loss of PTEN
function can be similarly predictive for therapy efficacy, whereas
simultaneous mutations in the mitogen-activated protein kinase
signaling pathway may lead to resistance to treatment (58,60).
Identifying actionable molecular aberrations has been a critical
step for several major therapeutic advances in cancer medicine.
A combined technique was established in the present
study for the examination of G3 EEC in order to investigate the
involvement of major mutations of the PI3K signaling pathway in
this EC subtype, and to ultimately implement the clinical and
pathological knowledge of this grade of EEC regardless of differing
characteristics (e.g. FIGO stage and ESGO risk). The present pilot
study demonstrated that the approach was successful in analyzing G3
EEC, specifically for the genetic profile of selected PIK3CA
and PTEN exons.
It was observed, in the cohort of the present study,
that several samples were wt for specific exons (from 22% of
PTEN exon 6 to 100% of PTEN exon 7) and that the
overall number of variations was broad (in a single sample there
were up to three variations for PTEN exon 6 and up to seven
variations overall).
The focus was on the PIK3CA exons 1, 9, and
21, and on the PTEN exons 5, 6, 7 and 8 since these are
typically the more frequently mutated exons in EECs; indeed, in a
previous study, Rudd et al (17), when investigating all PIK3CA
and PTEN exons, identified that no other exons were involved
in G3 EEC. Furthermore, to verify this result and to partially
validate the combined methodological approach, three samples were
analyzed by NGS obtained the same results. Nevertheless, not
examining all exons of PTEN and PIK3CA is a
limitation of the present study, and perhaps something to perform
in the future.
Using the approach developed in the present study,
the overall evaluation revealed that 89% of samples exhibited at
least one variation in either of the two genes, and that 83% had
mutations. Regarding PIK3CA, 39% of samples exhibited
mutations, among which the most frequent were H1047P (27%),
p.T1025A (7%) and p.Q546R (7%) (7,17,21,22).
With regard to PTEN, 67% of the samples had mutations,
including p.R189K (53%), p.D132H (13%), p.Y155H (7/%) and p.R130*
(7%).
As aforementioned, the overall mutational load was
determined as at least one mutation in 83% of the samples and
coexistence of mutations in the two genes in 33% of the samples. An
increased mutation frequency was identified for each single gene
compared with the coexistence of mutations, in contrast with
results reported previously (27),
particularly those reported by Oda et al (6), which may be due to the specific
evaluation of the G3 subgroup of EEC in the present study.
It was observed that the FIGO stage III tumors did
not exhibit any mutation in PIK3CA, and that the patient
with class III obesity had PIK3CA p.Q546 and PTEN
p.T167fs* mutations, suggesting an association supported by a
previous study of the association of obesity and EC through the
involvement of the PI3K signaling pathway (62).
The preliminary data of the present study indicated
potential for the use of a combined technological approach for the
identification of variants in PIK3CA and PTEN.
Although they are preliminary results owing to the limited number
of samples analyzed, the data confirm the involvement of PI3K
pathway alterations in G3 EEC; however, further investigation is
required.
In conclusion, the present study has demonstrated
the suitability and reliability of a combined approach (PCR-HRMA
and Sanger sequencing) for the evaluation of variants in selected
exons of PIK3CA and PTEN in G3 EEC, suggesting that
this approach may be useful for improving the classification and
personalized treatment of patients with EC.
Acknowledgements
Not applicable.
Funding
The present study was supported by grants from the
Tuscan Tumor Institute, Florence (Italy) and the University of
Florence (Italy).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
FM performed Sanger sequencing, evaluation of Sanger
electropherograms, analyses and interpretation of HRMA and Sanger
data, identification of nucleotide changes using databases (COSMIC,
PolyPhen-2 and mutation taster), and was a major contributor in
writing the manuscript. IT collected the clinical and
pathophysiological data for each patient, evaluated the criteria
for tissue collection and selection, and set up a database
containing patient information. FS was involved in patient
recruitment, collection of the clinical and pathophysiological data
for each patient, and evaluation of the criteria for tissue
collection and selection. EP performed the FFPE sample selection,
the histological examination of selected FFPE slices, and the
selection of appropriate slices for molecular analysis. FC
performed the FFPE sample collection, FFPE slices and the
histological examination of selected FFPE slices. MF performed the
patient recruitment, the surgery and treatment on enrolled women,
and collection of informed consent. FP co-ordinated the clinical
team for patient recruitment and supervised the data analysis. SP
performed DNA extraction and PCR, evaluation of Sanger
electropherograms and contributed in editing of the manuscript. IN
performed the patient recruitment, the surgery and treatment on
enrolled women, the collection of informed consents, follow-up of
patients and contributed in editing of the manuscript. All authors
read, revised and approved the final manuscript.
Ethics approval and consent to
participate
Research is performed on humans following
international and national regulations in accordance with The
Declaration of Helsinki, or any other relevant set of ethical
principles. This research involved human subjects or tissues, and
the authors state that informed consent for participation in the
study or use of their tissue was obtained from all
participants.
Patient consent for publication
The manuscript did not contain identifying
information, including names, initials, date of birth, hospital
numbers or images related to patients.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
EC
|
endometrial cancer
|
|
TCGA
|
The Genome Cancer Atlas
|
|
PI3K
|
phosphoinositide 3-kinase
|
|
EEC
|
endometrioid EC
|
|
AKT
|
protein kinase B
|
|
PTEN
|
phosphatase and tensin homolog
|
|
PIK3CA
|
PI3K catalytic subunit α
|
|
mTOR
|
mammalian target of rapamycin
|
|
G
|
grade
|
|
SNP
|
single nucleotide polymorphism
|
|
PCR
|
polymerase chain reaction
|
|
HRM
|
high-resolution melting
|
|
HRMA
|
high-resolution melting analysis
|
|
MT
|
Mutation Taster
|
References
|
1
|
Murali R, Soslow RA and Weigelt B:
Classification of endometrial carcinoma: More than two types.
Lancet Oncol. 15:e268–e278. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Cancer Genome Atlas Research Network, ;
Kandoth C, Schultz N, Cherniack AD, Akbani R, Liu Y, Shen H,
Robertson AG, Pashtan I, Shen R, et al: Integrated genomic
characterization of endometrial carcinoma. Nature. 497:67–73. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Marchiò C, De Filippo MR, Ng CK,
Piscuoglio S, Soslow RA, Reis-Filh JS and Weigelt B: PIKing the
type and pattern of PI3K pathway mutations in endometrioid
endometrial carcinomas. Gynecol Oncol. 137:321–328. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
McConechy MK, Ding J, Cheang MCU, Wiegand
K, Senz J, Tone A, Yang W, Prentice L, Tse K, Zeng T, et al: Use of
mutation profiles to refine the classification of endometrial
carcinomas. J Pathol. 228:20–30. 2012.PubMed/NCBI
|
|
5
|
Salvesen HB, Carter SL, Mannelqvist M,
Dutt A, Getz G, Stefansson IM, Raeder MB, Sosh ML, Engelsen IB,
Trovik J, et al: Integrated genomic profiling of endometrial
carcinoma associates aggressive tumors with indicators of PI3
kinase activation. Proc Natl Acad Sci USA. 106:4834–4839. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Oda K, Stokoe D, Taketani Y and McCormick
F: High frequency of coexistent mutations of PIK3CA, PTEN
genes in endometrial carcinoma. Cancer Res. 65:10669–10673. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Catasus L, D'Angelo E, Pons C, Espinosa I
and Prat J: Expression profiling of 22 genes involved in the
PI3K-AKT pathway identifies two subgroups of high-grade endometrial
carcinomas with different molecular alterations. Mod Path.
23:694–702. 2010. View Article : Google Scholar
|
|
8
|
Hayes MP, Wang H, Espinal-Witter R,
Douglas W, Solomon GJ, Baker SJ and Ellenson HL: PIK3CA,
PTEN mutations in uterine endometrioid carcinoma and complex
atypical hyperplasia. Clin Cancer Res. 12:5932–5935. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
van der Zee M, Jia Y, Wang Y,
Heijmans-Antonissen C, Ewing PC, Franken P, DeMayo FJ, Lydon JP,
Burger CW, Fodde R, et al: Alterations in Wnt-β-catenin and
Pten signalling play distinct roles in endometrial cancer
initiation and progression. J Pathol. 230:48–58. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Sal V, Demirkiran F, Erenel H, Tokgozoglu
N, Kahramanoglu I, Bese T, Turan H, Sofiyeva N, Calay Z, Arvas M
and Guralp O: Expression of PTEN and β-catenin and their
relationship with clinicopathological and prognostic factors in
endometrioid type endometrial cancer. Int J Gynecol Cancer.
26:512–520. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
McConechy MK, Ding J, Senz J, Yang W,
Melnyk N, Tone AA, Prentice LM, Wiegand KC, McAlpine JN, Shah SP,
et al: Ovarian and endometrial endometrioid carcinomas have
distinct CTNNB1 and PTEN mutation profiles. Mod Pathol. 27:128–134.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Machin P, Catasus L, Pons C, Muñoz J,
Matias-Guiu X and Prat J: CTNNB1 mutations and beta-catenin
expression in endometrial carcinomas. Hum Pathol. 33:206–212. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Tejerizo-García A, Jiménez-López JS,
Muñoz-González JL, Bartolomé-Sotillos S, Marqueta-Marqués L,
López-González G and Gómez JF: Overall survival and disease-free
survival in endometrial cancer: Prognostic factors in 276 patients.
Onco Targets Ther. 6:1305–1313. 2013.
|
|
14
|
Mang C, Birkenmaier A, Cathomas G and
Humburg J: Endometrioid endometrial adenocarcinoma: An increase of
G3 cancers? Arch Gynecol Obstet. 295:1435–1440. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Voss MA, Ganesan R, Ludeman L, McCarthy K,
Gornall R, Schaller G, Wei W and Sundar S: Should grade 3
endometrioid endometrial carcinoma be considered a type 2 cancer-a
clinical and pathological evaluation. Gynecol Oncol. 124:15–20.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yasuda M: Immunohistochemical
characterization of endometrial carcinomas: Endometrioid, serous
and clear cell adenocarcinomas in association with genetic
analysis. J Obstet Gyneacol Res. 40:2137–2176. 2014.
|
|
17
|
Rudd ML, Price JC, Fogoros S, Godwin AK,
Sgroi DC, Merino MJ and Bell DW: A unique spectrum of somatic
PIK3CA (p110Aα) mutations within primary endometrial
carcinomas. Clin Cancer Res. 17:1331–1340. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Mackay HJ, Eisenhauer EA, Kamel-Reid S,
Tsao M, Clarke B, Karakasis K, Werner HM, Trovik J, Akslen LA,
Salvesen HB, et al: Molecular determinants of outcome with
mammalian target of rapamycin inhibition in endometrial cancer.
Cancer. 15:603–610. 2014. View Article : Google Scholar
|
|
19
|
Mori N, Kyo S, Sakaguchi J, Mizumoto Y,
Ohno S, Maida Y, Hashimoto M, Takakura M and Inoue M: Concomitant
activation of AKT with extracellular-regulated kinase 1/2 occurs
independently of PTEN or PIK3CA mutations in
endometrial cancer and may be associated with favorable prognosis.
Cancer Sci. 98:1881–1888. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Shoji K, Oda K, Kashiyama T, Ikeda Y,
Nakagawa S, Sone K, Miyamoto Y, Hiraike H, Tanikawa M, Miyasaka A,
et al: Genotype-dependent efficacy of a dual PI3K/mTOR inhibitor,
NVP-BEZ235, and an mTOR inhibitor, RAD001, in endometrial
carcinomas. PLoS One. 7:e374312012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Millis SZ, Ikeda S, Reddy S, Gatalica Z
and Kurzrock R: Landscape of phosphatidylinositol-3-kinase pathway
alterations across 19784 diverse solid tumors. JAMA Oncol.
2:1565–1573. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Catasus L, Gallardo A, Cuatrecasas M and
Prat J: Concomitant PI3K-AKT and p53 alterations in endometrial
carcinomas are associated with poor prognosis. Mod Pathol.
22:522–529. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Konopka B, Janiec-Jankowska A, Kwiatkowska
E, Najmoła U, Bidziński M, Olszewski W and Goluda C: PIK3CA
mutations and amplification in endometrioid endometrial carcinomas:
Relation to other genetic defects and clinicopathologic status of
the tumors. Hum Pathol. 42:1710–1719. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
German S, Aslam HM, Saleem S, Raees A,
Anum T, Alvi AA and Haseeb A: Carcinogenesis of PIK3CA. Hered
Cancer Clin Pract. 11:52013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
English DP, Bellone S, Cocco E, Bortolomai
I, Pecorelli S, Lopez S, Silasi DA, Schwartz PE, Rutherford T and
Santin AD: Oncogenic PIK3CA gene mutations and HER2/neu gene
amplifications determine the sensitivity of uterine serous
carcinoma cell lines to GDC-0980, a selective inhibitor of Class I
PI3 kinase and mTOR kinase (TORC1/2). Am J Obstet Gynecol.
209:465.e1–e9. 2013. View Article : Google Scholar
|
|
26
|
Dong Y, Yang X, Wong O, Zhang X, Liang Y,
Zhang Y, Wong W, Nong L, Liao Q and Li T: PIK3CA mutations
in endometrial carcinomas in Chinese women: Phosphatidylinositol
3′-kinase pathway alterations might t be associated with favorable
prognosis. Hum Pathol. 43:1197–1205. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Herrero-Gonzalez S and Di Cristofano A:
New routes to old places: PIK3R1, PIK3R2 join PIK3CA,
PTEN as endometrial cancer genes. Cancer Discov. 1:106–107.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Lin DI: Improved survival associated with
somatic PIK3CA mutations in copy-number low endometrioid
endometrial adenocarcinoma. Oncol Lett. 10:2743–2748. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wik E, Ræder MB, Krakstad C, Trovik J,
Birkeland E, Hoivik EA, Mjos S, Werner HM, Mannelqvist M,
Stefansson IM, et al: Lack of estrogen receptor-α is associated
with epithelial-mesenchymal transition and PI3K alterations in
endometrial carcinoma. Clin Cancer Res. 19:1094–1105. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Smith IN and Briggs JM: Structural
mutation analysis of PTEN and its genotype-phenotype correlations
in endometriosis and cancer. Proteins. 84:1625–1643. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Akiyama-Abe A, Minaguchi T, Nakamura Y,
Michikami H, Shikama A, Nakao S, Sakurai M, Ochi H, Onuki M,
Matsumoto K, et al: Loss of PTEN expression is an independent
predictor of favourable survival in endometrial carcinomas. Br J
Cancer. 109:1703–1710. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Djordjevic B, Hennessy BT, Li J, Barkoh
BA, Luthra R, Mills GB and Broaddus RR: Clinical assessment of PTEN
loss in endometrial carcinoma: Immunohistochemistry outperforms
gene sequencing. Mod Pathol. 25:699–708. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Catasus L, Gallardo A, Cuatrecasas M and
Prat J: PIK3CA mutations in the kinase domain (exon 20) of
uterine endometrial adenocarcinomas are associated with adverse
prognostic parameters. Mod Pathol. 21:131–139. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Li W, Wang Y, Fang X, Zhou M, Li Y, Dong Y
and Wang R: Differential expression and clinical significance of
DNA methyltransferase 3B (DNMT3B), phosphatase and tensin homolog
(PTEN) and human MutL homologs 1 (hMLH1) in endometrial carcinomas.
Med Sci Monit. 23:938–947. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Bradford LS, Rauh-Hain A, Clark RM,
Groeneweg JW, Zhang L, Borger D, Zukerberg LR, Growdon WB, Foster R
and Rueda BR: Assessing the efficacy of targeting the
phosphatidylinositol 3-kinase/AKT/mTOR signaling pathway in
endometrial cancer. Gynecol Oncol. 133:346–352. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Morice P, Leary A, Creutzberg C,
Abu-Rustum N and Darai E: Endometrial cancer. Lancet.
387:1094–1108. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Matias-Guiu X and Prat J: Molecular
pathology of endometrial carcinoma. Histopathology. 62:111–123.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
McIntyre JB, Nelson GS, Ghatage P, Morris
D, Duggan MA, Lee CH, Doll CM and Köbel M: PIK3CA missense mutation
is associated with unfavorable outcome in grade 3 endometrioid
carcinoma but not in serous endometrial carcinoma. Gynecolc Oncol.
132:188–193. 2014. View Article : Google Scholar
|
|
39
|
Miyake T, Yoshino K, Enomoto T, Takata T,
Ugaki H, Kim A, Fujiwara K, Miyatake T, Fujita M and Kimura T:
PIK3CA gene mutations and amplifications in uterine cancers,
identified by methods that avoid confounding by PIK3CA pseudogene
sequences. Cancer Lett. 261:120–126. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Garcia-Dios DA, Lambrechts D, Coenegrachts
L, Vandenput I, Capoen A, Webb PM, Ferguson K, ANECS, Akslen LA,
Claes B, et al: High-throughput interrogation of PIK3CA, PTEN,
KRAS, FBXW7 and TP53 mutations in primary endometrial carcinoma.
Gynecol Oncol. 128:327–334. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Tsikouras P, Bouchlariotou S, Vrachnis N,
Dafopoulos A, Galazios G, Csorba R and von Tempelhoff GF:
Endometrial cancer: Molecular and therapeutic aspects. Eu J Obstet
Gynecol Reprod Biol. 169:1–9. 2013. View Article : Google Scholar
|
|
42
|
Janku F, Wheler JJ, Westin SN, Moulder SL,
Naing A, Tsimberidou AM, Fu S, Falchook GS, Hong DS, Garrido-Laguna
I, et al: PI3K/AKT/mTOR inhibitors in patients with breast and
gynecologic malignancies harboring PIK3CA mutations. J Clin
Oncol. 30:777–782. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Bian X, Gao J, Luo F, Rui C, Zheng T, Wang
D, Wang Y, Roberts TM, Liu P, Zhao JJ, et al: PTEN deficiency
sensitizes endometrioid endometrial cancer to compound PARP-PI3K
inhibition but not PARP inhibition as monotherapy. Oncogene.
37:341–351. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Philip CA, Laskov I, Beauchamp MC, Marques
M, Amin O, Bitharas J, Kessous R, Kogan L, Baloch T, Gotlieb WH, et
al: Inhibition of PI3K-AKT-mTOR pathway sensitizes endometrial
cancer cell lines to PARP inhibitors. BMC Cancer. 17:6382017.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Hasegawa K, Kagabu M, Mizuno M, Oda K,
Aoki D, Mabuchi S, Kamiura S, Yamaguchi S, Aoki Y, Saito T, et al:
Phase II basket trial of perifosine monotherapy for recurrent
gynecologic cancer with or without PIK3CA mutations. Invest
New Drugs. 35:800–812. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Mirantes C, Dosil MA, Eritja N, Felip I,
Gatius S, Santacana M, Matias-Guiu X and Dolcet X: Effects of the
multikinase inhibitors Sorafenib and Regorafenib in PTEN deficient
neoplasias. Eur J Cancer. 63:74–87. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
McCormick A, Earp E, Leeson C, Dixon M,
O'Donnell R, Kaufmann A and Edmondson RJ: Phosphatase and Tensin
Homolog is a potential target for ovarian cancer sensitization to
cytotoxic agents. Int J Gynecol Cancer. 26:632–639. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Tsiatis AC, Norris-Kirby A, Rich RG, Hafez
MJ, Gocke CD, Eshleman JR and Murphy KM: Comparison of Sanger
sequencing, pyrosequencing, and melting curve analysis for the
detection of KRAS mutations: Diagnostic and clinical
implications. J Mol Diagn. 12:425–432. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Ihle MA, Fassunke J, König K, Grünewald I,
Schlaak M, Kreuzberg N, Tietze L, Schildhaus HU, Büttner R and
Merkelbach-Bruse S: Comparison of high resolution melting analysis,
pyrosequencing, next generation sequencing and immunohistochemistry
to conventional Sanger sequencing for the detection of p.V600E and
non-p V600E BRAF mutations. BMC Cancer. 14:132014.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Li BS, Wang XY, Ma FL, Jiang B, Song XX
and Xu AG: Is high resolution melting analysis (HRMA) accurate for
detection of human disease-associated mutations? A meta analysis.
PLoS One. 6:e280782011. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Vossen RH, Aten E, Roos A and den Dunnen
JT: High-resolution melting analysis (HRMA): More than just
sequence variant screening. Hum Mutat. 30:860–866. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Colombo N, Creutzberg C, Amant F, Bosse T,
González-Martín A, Ledermann J, Marth C, Nout R, Querleu D, Mirza
MR, et al: ESMO-ESGO-ESTRO consensus conference on endometrial
cancer diagnosis, treatment and follow-up. Int J Gynecol Cancer.
26:2–30. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Kafshooz L, Kafshdooz T, Tabrizi AD,
Mohaddes Ardabili SM, Ardabili M, Arkbazadeh A, Gharesouran J,
Ghojazadeh M and Farajnia S: Role of exon 7 PTEN gene in
endometrial carcinoma. Asian Pac J cancer Prev. 16:4521–4524. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Simi L, Pratesi N, Vignoli M, Sestini R,
Cianchi F, Valanzano R, Nobili S, Mini E, Pazzagli M and Orlando C:
High-resolution melting analysis for rapid detection of KRAS,
BRAF, and PIK3CA gene mutations in colorectal cancer. Am
J Clin Pathol. 130:247–253. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Ousati Ashtiani Z, Mehrsai AR, Pourmand MR
and Pourmand GR: High resolution melting analysis for rapid
detection of PIK3CA gene mutations in bladder cancer: A mutated
target for cancer therapy. Urol J. 15:26–31. 2018.PubMed/NCBI
|
|
56
|
Martínez-Carretero C, Pascual FI, Rus A
and Bernardo I: Detection of EGFR mutations in patients with
non-small cell lung cancer by high resolution melting. Comparison
with other methods. Clin Chem Lab Med. 55:1970–1978. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Tsiatis AC, Norris-Kirby A, Rich G, Hafez
MJ, Gocke CG, Eshleman JR and Murphy KM: Comparison of Sanger
sequencing, pyrosequencing, and melting curve analysis for the
detection of KRAS mutations: Diagnostic and clinical
implications. J Mol Diagn. 12:425–432. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Engelman A: Targeting PI3K signaling in
cancer: Opportunities, challenges and limitations. Nat Rev Cancer.
8:550–562. 2009. View Article : Google Scholar
|
|
59
|
Di Nicolantonio F, Arena S, Tabernero J,
Grosso S, Molinari F, Macarulla T, Russo M, Cancelliere C, Zecchin
D, Mazzucchelli L, et al: Deregulation of the PI3K and KRAS
signaling pathways in human cancer cells determines their response
to everolimus. J Clin Invest. 120:2558–2866. 2010. View Article : Google Scholar
|
|
60
|
Ni X, Zhang W and Huang RS:
Pharmacogenomics discovery and implementation in genome-wide
association studies era. Wiley Interdiscip Rev Syst Biol Med.
5:1–9. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Tsimberidou AM, Iskander NG, Hong DS,
Wheler JJ, Falchook GS, Fu S, Piha-Paul S, Naing A, Janku F, Luthra
R, et al: Personalized medicine in a phase I clinical trials
program: The MD Anderson Cancer Center initiative. Clin Cancer Res.
15:6373–6383. 2012. View Article : Google Scholar
|
|
62
|
Westin SN, Ju Z, Broaddus RR, Krakstad C,
Li J, Pal N, Lu KH, Coleman RL, Hennessy BT, Klempner SJ, et al:
PTEN loss is a context-dependent outcome determinant in obese and
non-obese endometrioid endometrial cancer patients. Mol Oncol.
9:1694–1703. 2015. View Article : Google Scholar : PubMed/NCBI
|