Introduction
Colorectal cancer (CRC) is the third most commonly
diagnosed malignant tumor and is the fourth leading cause of
cancer-related death worldwide (1).
It is largely diagnosed in individuals >50 years of age
(2). CRC patients in the early
stage [tumor-node-metastasis (TNM) stage I and II] present with a
prolonged 5-year survival following surgical excision; the 5-year
survival is up to 95% and 60–80% for stage I and II, respectively.
However, existing therapies for CRC usually exhibit a limited
effect on patient prognosis, and the 5-year survival of patients in
stages III and IV can be as low as 35 and 10% (3). Research has shown that CRC incidence
and mortality can be decreased significantly through screening
programs, while CRC screening is only offered to very few
individuals worldwide based on the CRC incidence rate, national
economic level and medical security system (4). A previous study demonstrated that
germline mutations enable next generation hereditary susceptibility
to CRC accounting for 6–7%. In addition, mutations in DNA repair
genes and signal transduction genes also contribute to the
occurrence of CRC. In addition to inherited genetic mutations,
environmental factors, such as the heavy consumption of alcohol,
smoking habit, increased body fat and diets high in fat, salt and
red and processed meat, also play important roles (5). However, more and more studies have
shown that CRC displays accumulated defects in the activation of
oncogenes and the inactivation of tumor-suppressor genes (TSGs)
(6). Except for classical CRC risk
factors, such as: KRAS, TP53, APC and markers for
microsatellite instability (MSI) (7,8), an
impressive body of literature indicates that multiple factors such
as hsa-miR-19a (9), PROK2 (10), B7-H3 (11), DSCC1 (12) and microRNAs (13) are involved in the occurrence of CRC.
A recent study revealed that the dysbiosis of microbial communities
in the human body are also associated with gastrointestinal tract
cancer, such as gastric and colorectal cancer. Moreover, a
hypothesis called ‘Alpha bug’ considers that some bacteria could
alter the primary bacterial community and the remodeled bacterial
community could promote CRC by strengthening the mucosal immune
response (14). Although numerous
studies have shown that the pathogenesis of CRC is multifactorial,
the detailed mechanisms remain unclear at present.
C2ORF68, which belongs to the UPF0561 family,
contains 166 amino acids, and the monoisotopic molecular weight is
18.6033 kDa (15). The predicted
potential NLS and NES sequences of C2ORF68 and the domain of the EF
hand indicate that c2orf68 could function as a cell cycle
regulator in the nucleus, which may explain its possible role in
CRC pathogenesis. Our early research suggests that C2ORF68 may
regulate colon cancer cell apoptosis, proliferation, migration and
cell cycle distribution through the PI3K/Akt/mTOR pathway (16). We also suggested that C2ORF68 may
promote the carcinogenesis of CRC and play a vital role in the
pathogenesis of CRC through activation of the Wnt/β-catenin
signaling pathway (17), such as
the upregulation of β-catenin, survivin, cyclin D1 and
c-Myc, and the downregulation of GSK-3β in CRC cells.
Although it was demonstrated that C2ORF68 may play a role in the
occurrence of CRC and may be a potential oncogene in CRC, its
specific molecular mechanism remains dim.
Our previous study indicated that C2ORF68 may play
an important role in the occurrence and development of CRC through
the PI3K/AKT/mTOR and Wnt/β-catenin signaling pathways. However,
its exact mechanisms are still mysterious. To elucidate the role
and status of C2ORF68 in the carcinogenesis of colorectal
adenocarcinoma, we carried out bioinformatic analyses. It was found
that 12 proteins interact with c2orf68, including KLHL15, GMNN,
SMG6, GNE, DDHD2, PIR, ELAVL1 (HuR), NAGK, XPO1, HSPA9, JOSD2 and
GUK1. HuR is an RNA-binding protein whose expression level is
widely upregulated in many types of human cancer, including CRC. A
previous study showed that the HuR expression level is closely
related to AKT phosphorylation and increased cytoplasmic abundance
of HuR in human cancer may be associated with oncogenic activation
of AKT signaling (18). To
determine the interaction between C2ORF68 and HuR, and their roles
in the occurrence of CRC, immunohistochemistry (IHC),
immunofluorescence (IF), flow cytometry, Transwell migration and
CCK-8 assays, co-immunoprecipitation (co-IP), qRT-PCR and western
blot analysis were performed. The results revealed that C2ORF68 has
a synergistic effect with HuR, and their role in the onset and the
development of CRC may be through the upregulated expression of
Bcl-2, c-Myc, cyclin A and cyclin D1 and the
downregulation of Bax, coonsequently promoting cell
proliferation and inhibiting cell apoptosis.
Materials and methods
Bioinformatic analysis
BioGRID is an interaction database with data based
on comprehensive curation efforts. The interactive proteins for
C2ORF68 were predicted through the BioGrid repository (https://thebiogrid.org/).
Tissue microarray
A tissue microarray including 90 rectal cancer
tissues and adjacent normal tissues were purchased from Shanghai
Xinchao (Shanghai Xinchao Biological Co. Ltd., Shanghai, China).
The Medical Research Ethics Committee of Sichuan University
approved the sample acquisition (Chengdu, China), and a written
informed consent was also obtained from all patients.
Immunohistochemistry (IHC) of the
rectal cancer tissue microarray
IHC was performed as previously described (19). Clinicopathological parameters of the
CRC patients are shown in Table I.
The primary antibody (Ab) used for IHC was mouse monoclonal C2ORF68
Ab (1:200; BIO014915; Beacombio, Birmingham, UK). The expression
level of C2ORF68 protein in rectal cancer tissues were scored by
three independent examiners. The level of C2ORF68 staining pattern
was scored according to four subgroups: i) negative (−); ii) weak
(+); iii) moderate (++); and ⅳ) strong (+++).
 | Table I.Association of the expression of
C2ORF68 with the clinicopathological parameters of the CRC
samples. |
Table I.
Association of the expression of
C2ORF68 with the clinicopathological parameters of the CRC
samples.
|
| Expression
intensity (n) |
|
|
|---|
|
|
|
|
|
|---|
| Parameter | +++ | ++ | ++ | – | χ2 | P-value |
|---|
| Sex |
|
Female | 16 | 25 | 11 | 2 |
|
|
|
Male | 14 | 16 | 4 | 2 | 1.568 | 0.698 |
| Age, years |
|
≤60 | 13 | 11 | 8 | 2 |
|
|
|
>60 | 17 | 30 | 7 | 2 | 4.283 | 0.233 |
| TNM stage |
| I | 4 | 13 | 2 | 1 |
|
|
| II | 13 | 19 | 3 | 1 |
|
|
|
III | 12 | 9 | 10 | 2 |
|
|
| IV | 1 | 0 | 0 | 0 | 13.882 | 0.115 |
| Differentiation
degree |
|
Well | 1 | 7 | 2 | 1 |
|
|
|
Moderate | 19 | 32 | 10 | 3 |
|
|
|
Low | 10 | 2 | 3 | 0 | 13.047 | 0.043 |
| Lymph node
metastasis |
|
Yes | 13 | 9 | 10 | 2 |
|
|
| No | 17 | 32 | 5 | 2 | 10.343 | 0.013 |
| Survival
status |
|
Surviving | 15 | 25 | 10 | 3 |
|
|
|
Deceased | 15 | 16 | 5 | 1 | 1.310 | 0.727 |
Cell lines and cell culture
Human colon cancer cell lines, SW480 and LoVo
[American Type Culture Collection (ATCC) Manassas, VA, USA], were
cultured in Dulbecco's modified Eagle's medium (DMEM; HyClone; GE
Healthcare Life Sciences, Logan, UT, USA) which contained 10% fetal
bovine serum (FBS), penicillin (100 IU/ml) and streptomycin (100
IU/ml). The cells were grown at 37°C in a humidified atmosphere
with 5% CO2. The cells were harvested when they were in
the exponential growth phase and then the experiments stated above
were performed.
Immunofluorescence (IF)
For immunostaining, the SW480 and LoVo cells were
fixed with 4% paraformaldehyde for 20 min, permeabilized with 0.5%
Triton X-100 for 15 min and blocked with 3% bovine serum albumin
(BSA) for 30 min at room temperature. The treated cells were then
incubated with mouse monoclonal anti-C2ORF68 (1:100) and rabbit
polyclonal anti-HuR (dilution 1:100; cat. no. ab200342; Abcam,
Cambridge, UK) overnight at 4°C, and finally incubated with
FITC-labeled and TRITC-labeled secondary Ab for 30 min under the
conditions of protection from light at room temperature. Each step
was followed by two 5-min washes in PBS. The nuclei were
counterstained using DAPI, and observed using an Olympus BX53
fluorescence microscope (Olympus, Hamburg, Germany).
siRNA selection and transient
transfection
siRNAs for c2orf68 (siRNA sequences,
5′-CUAUGAAGAGUCCGGUGAAdTdT-3′ and 3′-dTdTCUUCUCCAUAACUUACGAU-5′)
and HuR (siRNA sequences, 5′-GGUUGCGUUUAUCCGGUUUdTdT-3′ and
3′-dTdTCCAACGCAAAUAGGCCAAA-5′) were designed and synthesized by
RiboBio (Guangzhou, China). Using the blank and negative control
groups, the transfection was performed with 100 nM of siRNA and
Invitrogen™ Lipofectamine 2000™ (Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) to induce the knockdown of
c2orf68/HuR expression. After transfection for 6 h,
the cells were respectively incubated in fresh DMEM for 48 h to
detect the mRNA expression level and for 72 h to detect the protein
expression level.
Strain, plasmid, plasmid extraction
and overexpression
The c2orf68 gene was synthesized and inserted
into the XhoI/EcoRI site of the pcDNA3.1 eukaryotic
expression vector (Pharma Co., Shanghai, China), which was verified
by restriction digestion followed by sequencing (Beijing Genomic
Institute, Beijing, China) in our previous study (17). The microbial strain and plasmids
used in the present study were Escherichia coli (E.
coli), DH5α and the pcDNA3.1-c2orf68 eukaryotic
expression vector. The plasmid extraction process was carried out
by TIANprep Plasmid Mini kit introduction (cat. no. DP103-02;
Tiangen Biotech Co., Ltd., Beijing, China). The LoVo cells at a
density of 3×105 cells/well in a 6-well plate
transfected with 5 µl interference fragment or negative control
(NC) vector using Lipofectamine 2000 which was then replaced with
fresh growth medium after 6 h. Following culture for 48 h, the
transfected LoVo cells were treated with 600 lg/ml of G418
(Sigma-Aldrich; Merck KGaA, Darmstadt, Germany). After 14 days, the
monoclonal cells were cultured in the presence of 300 lg/ml of
G418.
Co-immunoprecipitation (co-IP)
For co-IP, pcDNA3.1-c2orf68 eukaryotic expression
vector was transfected for 60 h in SW620 cells. Then the cells with
overexpression of c2orf68 were lysed with mild lysis buffer
containing several protease inhibitors for 30 min, and the cell
lysates were separated by centrifugation at 15,000 × g for 20 min.
Next, a modicum of cell lysates was reserved and utilized for
western blot analysis. Next, the mouse anti-C2ORF68 Ab and rabbit
anti-HuR Ab were added into the cell lysates for one night at 4°C.
On the following day, 100 µl protein A+G were added into the
compound and incubated for 4 h. Subsequently, the sediments were
gathered and the beads were washed twice using mild lysis buffer.
After that, the same volume of 2X SDS-PAGE loading buffer was used
to elute the protein which was absorbed on sepharose beads.
Finally, the protein was used for SDS-PAGE.
Cell proliferation assay
The cell proliferation assay was performed as
previously described (19). Plates
were read at an absorbance wavelength of 450 nm with the help of a
microplate reader (Model 680; Bio-Rad Laboratories, Hercules, CA,
USA). Each transfection group had six replicates, and the
experiment was repeated three times.
Flow cytometry
Flow cytometry was performed as previously described
(19). Then, the results were
analyzed by flow cytometry (FACSAria II Cell Sorter; BD
Biosciences, Franklin Lakes, NJ, USA). Each transfection group had
three replicates, and the experiment was repeated three times.
Cell migration assay
Cell migration assay was performed as previously
described (19). Migrated cells
were quantified by counting the stained cells under a microscope
(Model 680; Bio-Rad Laboratories) at ×200 magnification. For each
well, five random fields were selected to determine the total
number of migrated cells. The assay was performed in triplicate and
repeated three times.
qRT-PCR analysis
qRT-PCR analysis was performed as previously
described (19). The sequences of
forward and reverse primers are shown in Table II. The amplification was performed
on a Bio-Rad C1000 Touch Thermal Cycler (Bio-Rad Laboratories). The
GAPDH gene was used as an endogenous control, and the ΔΔCq
(20) method was used to quantify
the data, and the experiment was repeated three times.
 | Table II.Primer sequences for reverse
transcription-quantitative polymerase chain reaction. |
Table II.
Primer sequences for reverse
transcription-quantitative polymerase chain reaction.
| Gene | Primer sequence
(5′→3′) | Location | Product length,
bp |
|---|
| c2orf68 | F:
GAAGAGTCCGGTGAAAGCAG | 315–336 | 169 |
|
| R:
TACGCAACTTGAGGGCTTCT | 483–462 |
|
| HuR | F:
ACCCAGGATGAGTTACGA | 245–264 | 125 |
|
| R:
GCCCAAACCGAGAGAACAT | 369–349 |
|
| Bax | F:
AAGCTGAGCGAGTGTCTCAAG | 172–192 | 178 |
|
| R:
CAAAGTAGAAAAGGGCGACAAC | 349–328 |
|
| Bcl2 | F:
GTTTGATTTCTCCTGGCTGTCTC | 1–23 | 133 |
|
| R:
GAACCTTTTGCATATTTGTTTGG | 649–627 |
|
| c-Myc | F:
TCAAGAGGCGAACACACAAC | 1,631-1,550 | 110 |
|
| R:
GGCCTTTTCATTGTTTTCCA | 1,740-1,721 |
|
| Cyclin D1 | F:
GTGGCCTCTAAGATGAAGGAGA | 534–555 | 169 |
|
| R:
GGAAGTGTTCAATGAAATCGTG | 702–681 |
|
| Cyclin A | F:
TGTCTCATGGACCTTCACCA | 1,541-1,560 | 117 |
|
| R:
CTCTGGTGGGTTGAGGAGAG | 1,657-1,638 |
|
| GAPDH | F:
GGAAGGTGAAGGTCGGAGT | 179–197 | 117 |
|
| R:
TGAGGTCAATGAAGGGGTC | 295–277 |
|
Western blot analysis
Western blotting was performed as previously
described (19). The primary
antibodies used for western blotting were as follows: C2ORF68 (cat.
no. ab81363; Abcam, Cambridge, UK), Bcl-2 (cat. no. sc-492; Santa
Cruz Biotechnology, Inc., Dallas, TX, USA), Bax (cat. no. sc-623;
Santa Cruz Biotechnology, Inc.), c-Myc (cat. no. ab32072; Abcam),
(cat. no. ab200342; Abcam), cyclin D (cat. no. ab134175; Abcam),
cyclin A (cat. no. ab181591; Abcam) and β-actin (cat. no. sc-8432;
Santa Cruz Biotechnology, Inc.). All the primary antibodies used in
this step at 1:1,000 dilution. Signals detection were performed
through a Gel Imaging system (ProteinSimple, Santa Clara, CA, USA).
Subsequent densitometry analysis were conducted using ImageJ
(version 1.52g; National Institutes of Health, Bethesda, MD,
USA).
Statistical analysis
Statistical analysis was performed by SPSS software
(version 19.0; IBM Corp., Armonk, NY, USA). All experiments were
performed three times, and all data are expressed as mean ± SD.
Student's t-test and one-way ANOVA were used for statistical
analysis. Multiple comparison between the groups was performed
using the S-N-K method. A P-value <0.05 was considered to
indicate a statistically significant result.
Results
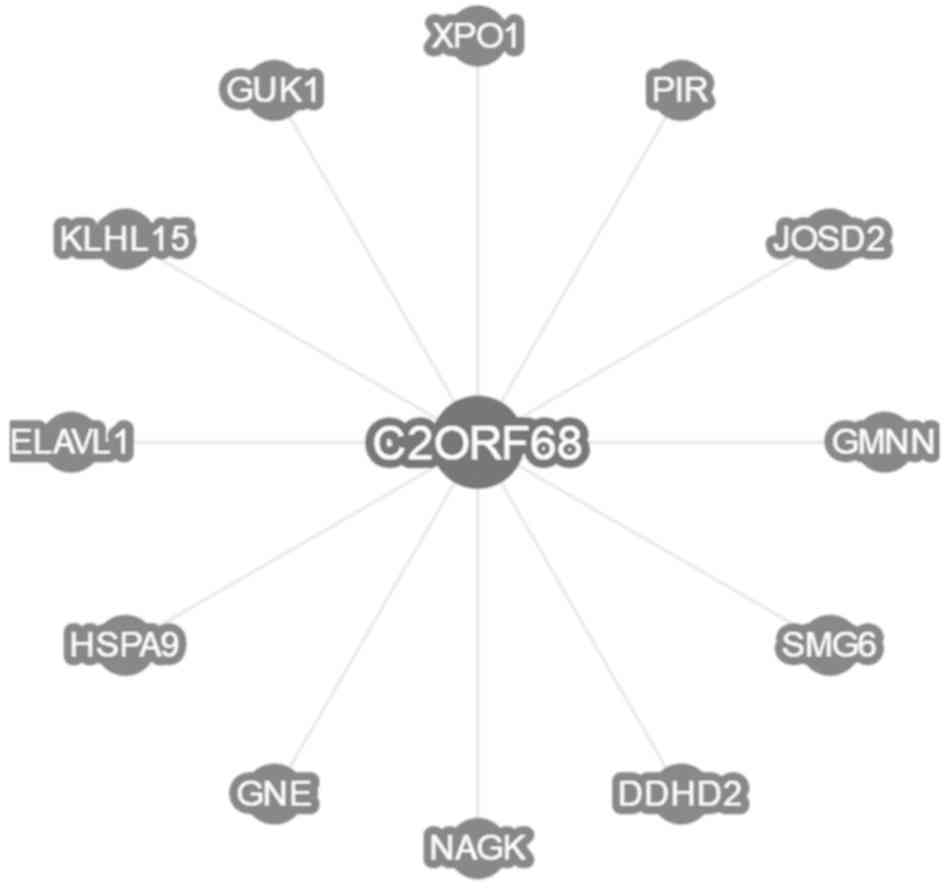
From the BioGrid database, it is found that there
are 12 proteins which may interact with C2ORF68, including KLHL15,
GMNN, SMG6, GNE, DDHD2, PIR, ELAVL1(HuR), NAGK, XPO1, HSPA9, JOSD2
and GUK1 (Fig. 1).
Expression of C2ORF68 in the rectal
cancer tissue microarray
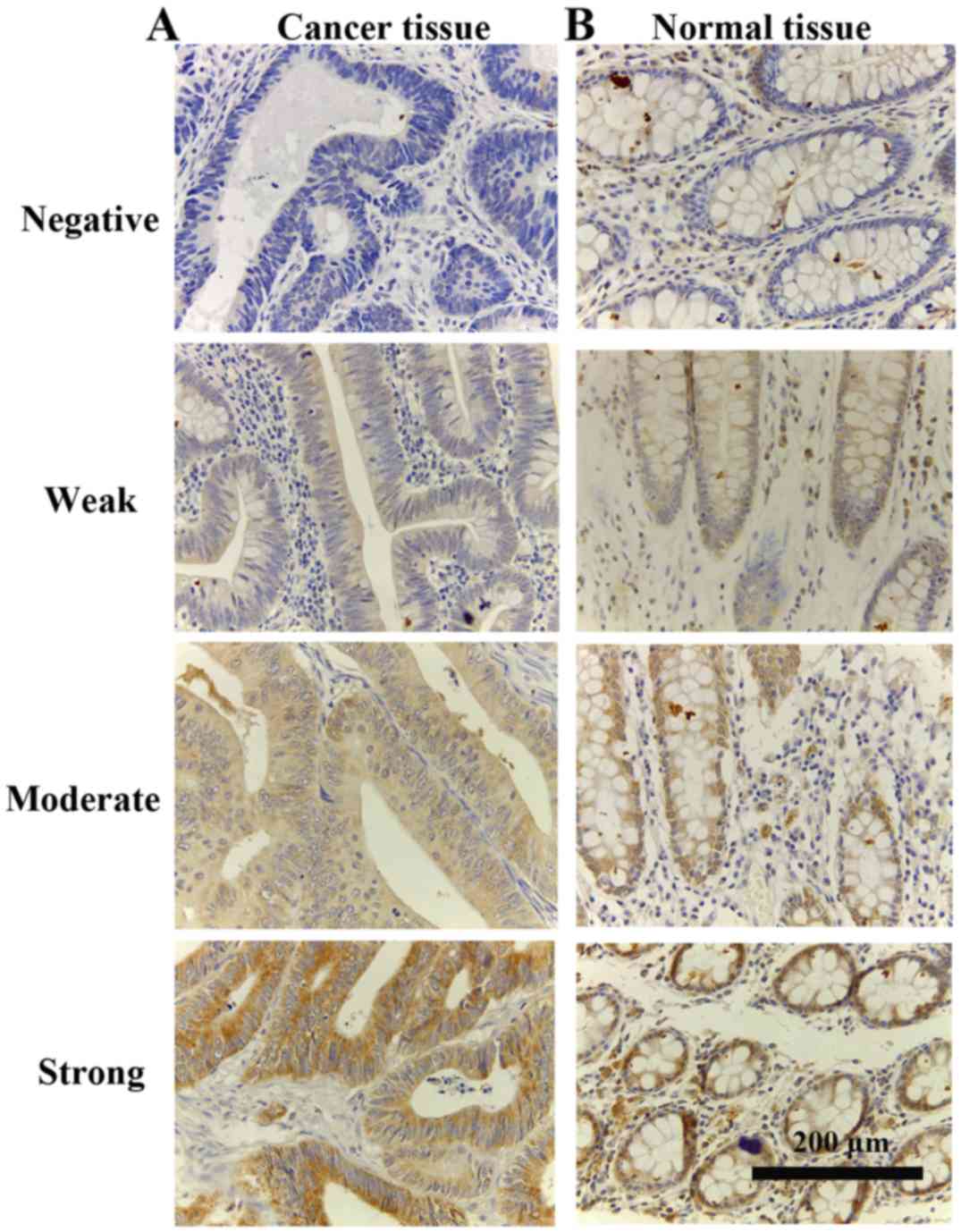
The representative cytoplasmic staining of C2ORF68
in the rectal cancer tissue microarray is shown in Fig. 2A-C (magnification, ×400). Our
previous study (15) demonstrated
that C2ORF68 presents two different staining patterns, including
nuclear staining and cytoplasmic staining. In line with the
observation in IF, C2ORF68 is a predominantly nuclear protein, but
cytoplasmic C2ORF68 localization may play a vital role in the
occurrence of CRC. In the rectal cancer tissue microarray (Fig. 2A), C2ORF68 protein expression was
detected in 95.56% (86/90) of the cancer samples. Among these,
4.44% (4/90), 16.67% (15/90), 45.56% (41/90) and 33.33% (30/90) of
these cases exhibited negative (−), weak (+), moderate (++) and
strong (+++) C2ORF68 protein staining, respectively. In contrast,
5.56% (5/90), 25.56% (23/90), 58.89% (53/90) and 10% (9/90) of
normal rectal specimens exhibited negative (−), weak (+), moderate
(++) and strong (+++) C2ORF68 protein staining, respectively
(Fig. 2C). We selected five
nonoverlapping views randomly from each image and calculated the
mean optical density. Then, the difference in mean optical density
between rectal cancer and adjacent normal tissue was analyzed by
statistical analysis. It was showed that compared with the adjacent
normal rectal tissues, the expression of C2ORF68 was significantly
increased in the rectal cancer tissues (P<0.05). In addition, we
also researched the relationship between the expression of C2ORF68
and clinical parameters. Significant associations were noted
between C2ORF68 expression and pathological grade (P<0.05) and
lymph node metastasis (P<0.05). However, the association between
C2ORF68 expression and age, sex and TNM stage was not statistically
significant.
Cellular localization of C2ORF68 and
HuR in SW480 and LoVo cells
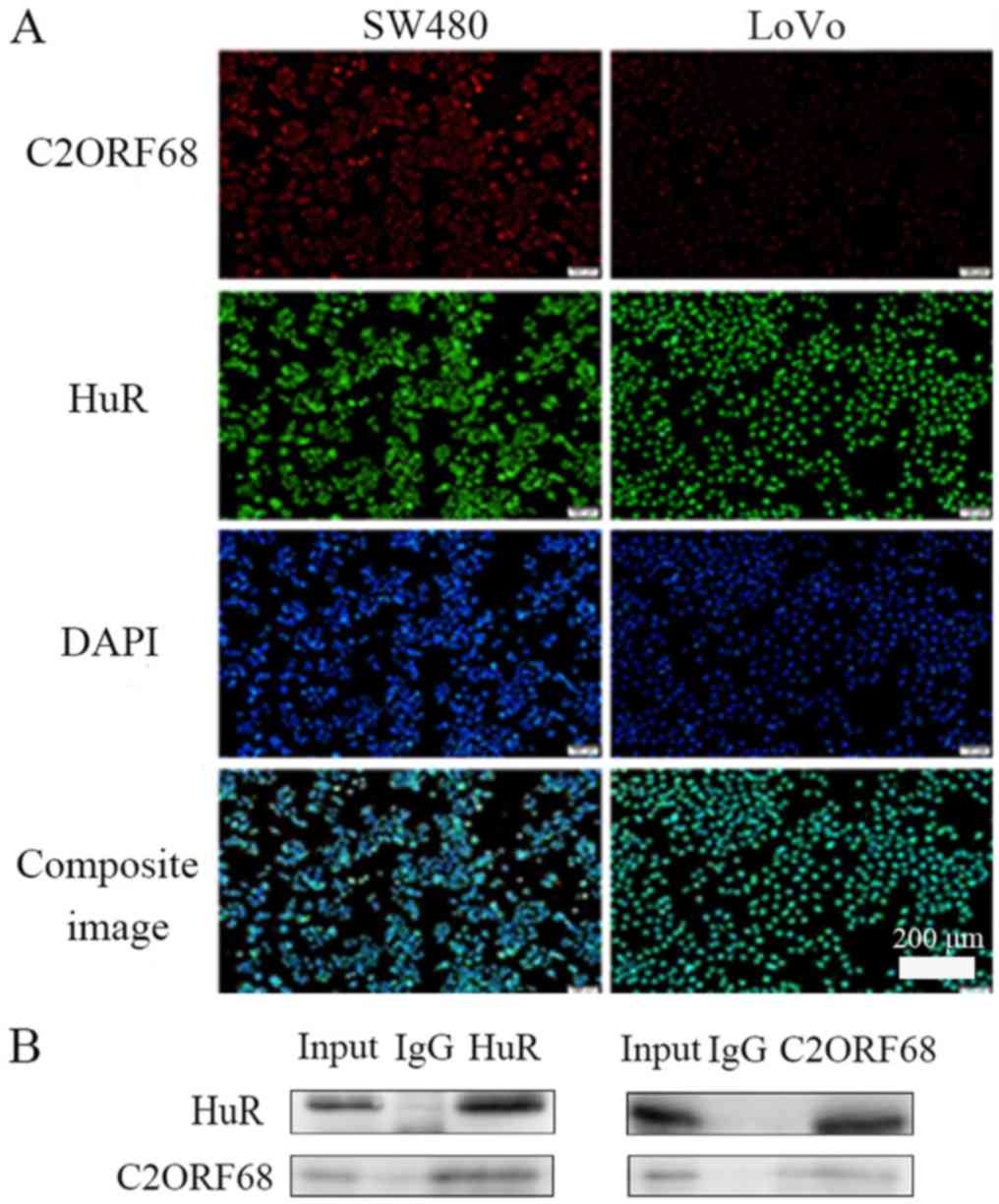
The results revealed that C2ORF68 and HuR were
localized mainly in the nucleus in both SW480 and LoVo cells
(Fig. 3A) (magnification, ×100).
BioGrid predicted that there is an interaction between C2ORF68 and
HuR. It was shown that C2ORF68 interacts with HuR by Co-IP
(Fig. 3B).
Cell apoptosis, cell proliferation and
cell migration in LoVo+c2orf68,-HuR and LoVo+c2orf68 cells
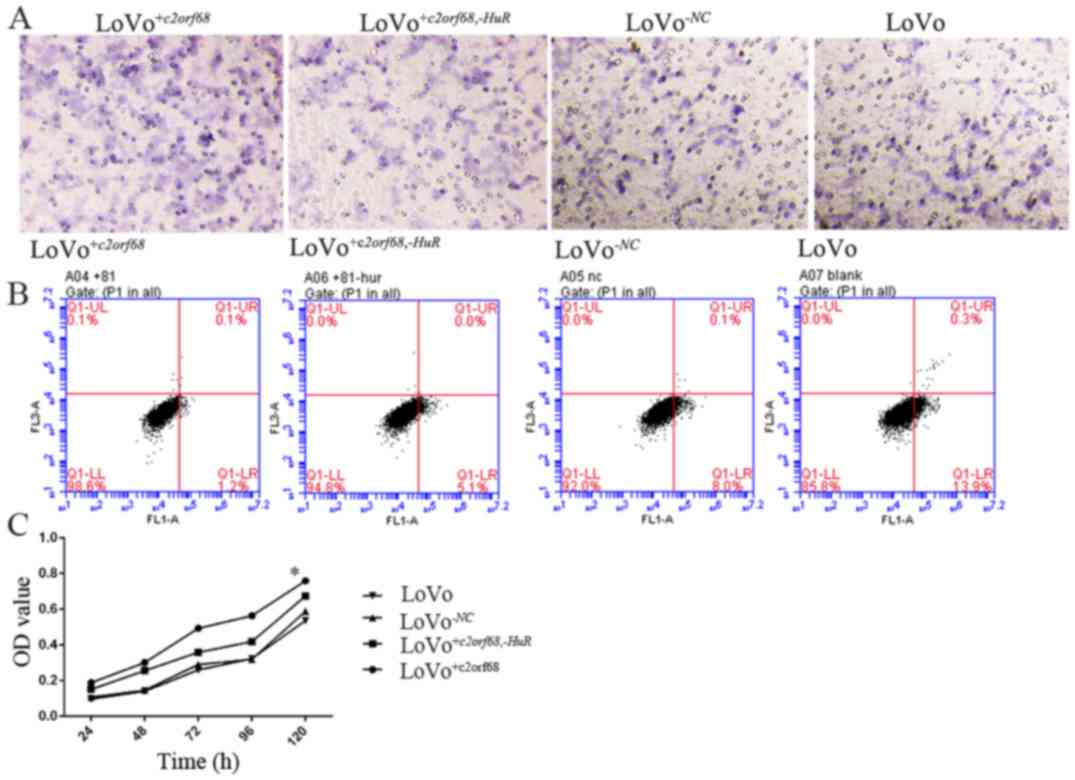
Following transfection for 24 h, flow cytometry was
performed to analyze c2orf68 and HuR-induced cell
cycle arrest and apoptosis in colon cancer cells. The cell
apoptosis rate of LoVo+c2orf68,-HuR and
LoVo+c2orf68 cells was significantly decreased compared to
that of the LoVo-NC and LoVo cells (Fig. 4B, P<0.05). There was no
statistical significance noted between
LoVo+c2orf68,-HuR and LoVo+c2orf68 cells,
LoVo-NC and LoVo cells. Compared with the control group,
LoVo+c2orf68 and LoVo+c2orf68,-HuR cell
proliferation was significantly increased (P<0.05); and the cell
proliferation of LoVo+c2orf68 cells was statistically
significantly different when compared with that of
LoVo+c2orf68,-HuR cells (Fig. 4C, P<0.05). Following transfection
for 24 h, the number of migrated LoVo+c2orf68,-HuR,
LoVo+c2orf68, LoVo-NC and LoVo cells was 223±24,
379±33, 239±31 and 244±26, respectively. The number of migrated
LoVo+c2orf68 cells was significantly higher than that of
LoVo+c2orf68,-HuR, LoVo-NC and LoVo cells
(Fig. 4A, P<0.05).
Cell apoptosis, cell proliferation and
cell migration of SW480-c2orf68,-HuR, SW480-c2orf68 and SW480-HuR
cells
The apoptosis rate of
SW480-c2orf68,-HuR cells was significantly higher
than that of SW480-c2orf68, SW480-HuR,
SW480-NC and SW480 cells (Fig.
5B, P<0.05). In addition, the apoptosis rate of
SW480-c2orf68 and SW480-HuR cells was significantly
higher than that of SW480-NC and SW480 cells, respectively
(Fig. 5B, P<0.05). Compared with
the control group, cell proliferation was significantly decreased
in the SW480-c2orf68,-HuR, SW480-c2orf68 and
SW480-HuR cells (Fig. 5C,
P<0.05); cell proliferation in the SW480-c2orf68,-HuR
cells was significantly less than that of the SW480-c2orf68
and SW480-HuR cells (Fig.
5C, P<0.05). The number of migrated cells of
SW480-c2orf68,-HuR, SW480-c2orf68, SW480-HuR.,
SW480-NC and SW480 were 122±16, 234±51, 233±48, 381±42 and
401±24, respectively. Compared with the control group, the number
of migrated cells were significantly inhibited in the
SW480-c2orf68,-HuR, SW480-c2orf68 and
SW480-HuR cells (Fig. 5A,
P<0.05) cells. However, the number of migrated
SW480-c2orf68,-HuR cells was significantly lower than that
of the SW480-c2orf68 and SW480-HuR cells (Fig. 5A, P<0.05).
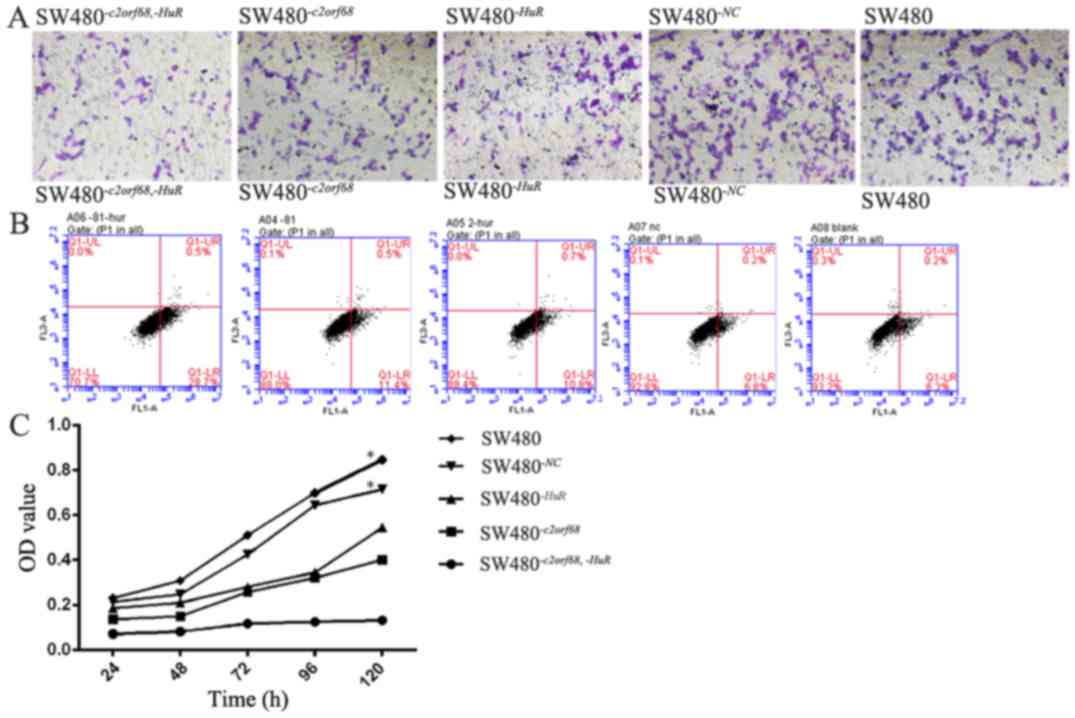 | Figure 5.(A) Compared with the blank group,
the number of migrated cells was observably inhibited in the
SW480−c2orf68,−HuR, SW480−c2orf68 and
SW480−HuR cells. However, the number of
SW480−c2orf68,-HuR cells that migrated was observably
less than that of SW480−c2orf68 and SW480−HuR
cells (P<0.05). (B) The apoptosis rates of
SW480−c2orf68,−HuR, SW480−c2orf68 and
SW480−HuR cells were significantly increased
(P<0.05). (C) Compared with the blank group, cell proliferation
was significantly decreased in the SW480−c2orf68,−HuR,
SW480−c2orf68 and SW480−HuR cells
(P<0.05). Furthermore, the proliferation of
SW480−c2orf68,-HuR cells was significantly lesser than
that of SW480−c2orf68 and SW480−HuR cells
(P<0.05). *P<0.05. |
mRNA and protein expression of
C2orf68, HuR, Bcl-2, Bax, c-Myc, cyclin D and cyclin A in
LoVo+c2orf68,-HuR and LoVo+c2orf68 cells
As shown in Fig. 6C,
following transfection for 48 h, c2orf68 gene expression was
significantly overexpressed in the LoVo+c2orf68 (P<0.01)
and LoVo+c2orf68,-HuR cells (P<0.05) when compared with
the control group. Similarly, HuR, Bcl-2 and c-Myc
were significantly overexpressed in the LoVo+c2orf68
(P<0.01, P<0.01 and P<0.05, respectively) and
LoVo+c2orf68,-HuR cells (all P<0.05). Cyclin D and
Cyclin A were significantly overexpressed in the
LoVo+c2orf68 cells (P<0.05), while the upregulation of
Cyclin D and Cyclin A mRNA expression levels in
LoVo+c2orf68,-HuR cells exhibited no significance. The mRNA
expression of c2orf68, HuR, Bcl-2, c-Myc and Cyclin A
in LoVo+c2orf68 cells was significantly increased, while
cyclin D in LoVo+c2orf68,-HuR cells exhibited no
statistical difference with LoVo+c2orf68 cells. In contrast,
the mRNA expression level of Bax was decreased in the
LoVo+c2orf68 (P<0.05) and LoVo+c2orf68,-HuR
(P<0.01) cells, when compared to the control. Compared with the
LoVo+c2orf68,-HuR cells, the mRNA expression of Bax
in the LOVO+c2orf68 cells was significantly decreased
(P<0.05).
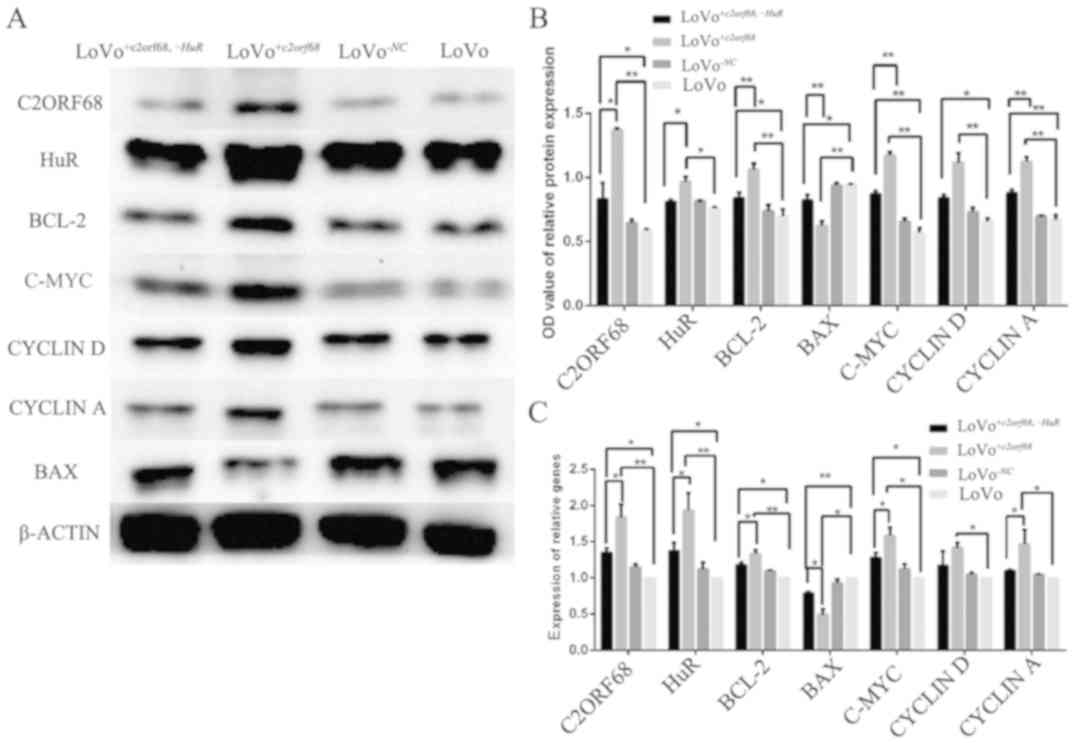 | Figure 6.(A and B) The protein expression
levels of C2ORF68, Bcl-2, c-Myc, cyclin D and cyclin A were
overexpressed in LoVo+c2orf68,−HuR and
LoVo+c2orf68 cells; while HuR was overexpressed in
LoVo+c2orf68 cells when compared to the LoVo cells.
Compared with the LoVo+c2orf68,−HuR cells, the protein
expression of C2ORF68, HuR, Bcl-2, c-Myc, cyclin D and cyclin A in
LoVo+c2orf68 cells was significantly increased. The
protein expression level of Bax was decreased in
LoVo+c2orf68,−HuR and LoVo+c2orf68 cells when
compared to the LoVo cells. Compared with
LoVo+c2orf68,−HuR cells, the protein expression of Bax
in LoVo+c2orf68 cells was significantly decreased. (C)
In LoVo+c2orf68,−HuR and LoVo+c2orf68 cells,
the mRNA levels of c2orf68, HuR, Bcl-2, c-Myc were significantly
overexpressed in both LoVo+c2orf68 and
LoVo+c2orf68,−HuR cells. Cyclin D and cyclin A were
significantly overexpressed in LoVo+c2orf68 cells, while
the upregulation of cyclin D and cyclin A mRNA expression levels in
LoVo+c2orf68,−HuR cells exhibited no significant
difference. *P<0.05, **P<0.01. |
As shown in Fig. 6A and
B, following transfection for 48–72 h, compared to the blank
group, the protein expression level of C2ORF68 was significantly
increased in the LoVo+c2orf68,-HuR (P<0.05) and
LoVo+c2orf68 (P<0.01) cells. In addition, HuR, BCL-2,
C-MYC and Cyclin A protein expression levels were overexpressed in
the LoVo+c2orf68,-HuR (NS, P<0.05, P<0.01 and
P<0.01, respectively) and LoVo+c2orf68 cells (P<0.05,
P<0.01, P<0.01 and P<0.01, respectively) when compared to
the blank group; compared with the LoVo+c2orf68,-HuR cells,
the protein expression of C2ORF68, HuR, BCL-2, C-MYC and Cyclin A
in LoVo+c2orf68 cells was increased (P<0.05, P<0.05,
P<0.01, P<0.01 and P<0.01, respectively). In contrast, the
protein expression level of BAX was decreased in the
LoVo+c2orf68,-HuR (P<0.05) and LoVo+c2orf68 cells
(P<0.01). Compared with LoVo+c2orf68,-HuR cells, the
protein expression of BAX (P<0.01) in LoVo+c2orf68 cells
was significantly decreased.
mRNA and protein expression of
c2orf68, HuR, Bcl-2, Bax, c-Myc, cyclin D and cyclin A in
SW480-c2orf68,-HuR, SW480-c2orf68 and SW480-HuR cells
As shown in Fig. 7C,
in SW480-c2orf68,-HuR, SW480-c2orf68 and
SW480-HuR cells, the mRNA expression level of c2orf68,
HuR, Bcl-2, c-Myc, cyclin D and cyclin A significantly
decreased (P<0.05). Furthermore, the inhibition rate of
c2orf68, HuR, Bcl-2, c-Myc and cyclin A in
SW480-c2orf68,-HuR cells was significantly higher than that
in SW480-c2orf68 and SW480-HuR cells (P<0.05). The
inhibition rate of cyclin D in SW480-c2orf68,-HuR
cells compared with SW480-HuR cells, was statistically
significant (P<0.05). However, compared with
SW480-c2orf68 cells, the inhibition of cyclin D was
not statistically significant. Meanwhile, the increase in
Bax mRNA expression was significantly higher in
SW480-c2orf68,-HuR cells than in SW480-c2orf68 and
SW480-HuR cells (P<0.05). Furthermore, the mRNA
expression level of Bax was increased in
SW480-c2orf68,-HuR, SW480-c2orf68 and
SW480-HuR cells.
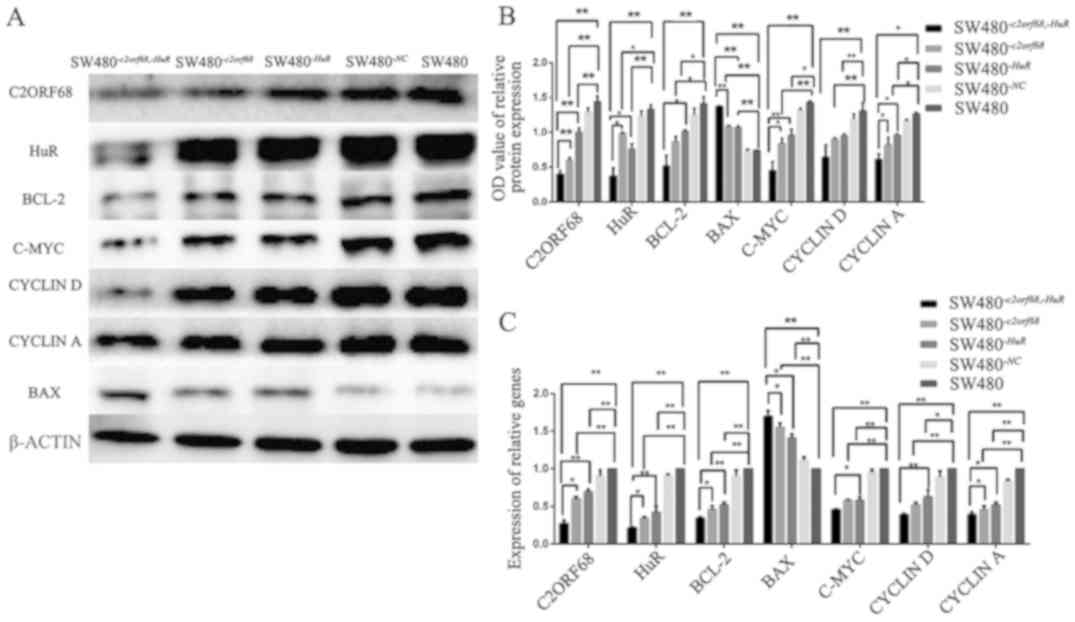 | Figure 7.(A and B) The protein expression
levels of C2ORF68, HuR, Bcl-2, c-Myc, cyclin D and cyclin D were
significantly decreased. The inhibition rate of C2ORF68, HuR,
Bcl-2, c-Myc, cyclin D and cyclin A was significantly higher in the
SW480−c2orf68,−HuR cells than in
SW480−c2orf68 and SW480−HuR cells. However,
the inhibition rate of CYCLIND was not statistically significant in
SW480−c2orf68,−HuR cells, compared with
SW480−c2orf68 and SW480−HuR cells. Meanwhile,
the protein expression level of BAX significantly increased in
SW480−c2orf68,−HuR, SW480−c2orf68 and
SW480−HuR cells. (C) In the
SW480−c2orf68,−HuR, SW480−c2orf68 and
SW480−HuR cells, the mRNA expression levels of c2orf68,
HuR, Bcl-2, c-Myc, cyclin D and cyclin A were significantly
decreased. Furthermore, the inhibition rate of c2orf68, HuR, Bcl 2
and cyclin A in SW480−c2orf68 −HuR cells was
significantly higher than that in the SW480−c2orf68 and
SW480−HuR cells. The inhibition rate of cyclin D in
SW480−c2orf68,−HuR cells compared with
SW480−HuR cells, was statistically significant. However,
compared with SW480−c2orf68 cells, the inhibition of
cyclin D was not statistically significant. Furthermore, the mRNA
expression level of Bax was significantly increased in the
SW480−c2orf68,−HuR, SW480−c2orf68 and
SW480−HuR cells (P<0.01, P<0.01, P<0.01), and
the expression of BAX was also increased in the
SW480−c2orf68,−HuR cells when compared with the
SW480−c2orf68 (P<0.05) and SW480−HuR
(P<0.05) cells. *P<0.05, **P<0.01. |
As shown in Fig. 7A and
B, following transfection for 48–72 h, compared to the blank
group, the protein expression level of C2ORF68 was significantly
decreased in the SW480-c2orf68,-HuR (P<0.01),
SW480-c2orf68 (P<0.01) and SW480-HuR cells
(P<0.01). Similarly, the protein expression levels of HuR,
BCL-2, C-MYC, Cyclin D and Cyclin A were also decreased in the
SW480-c2orf68,-HuR, SW480-c2orf68 and
SW480-HuR cells. Furthermore, the inhibition rate of C2ORF68
(P<0.01, P<0.01), HuR (P<0.05, P<0.05), C-MYC
(P<0.05, P<0.01) and Cyclin A (P<0.05, P<0.05)was
significantly higher in the SW480-c2orf68,-HuR cells than
that in the SW480-c2orf68 and SW480-HuR cells.
However, the inhibition rate of Cyclin D in
SW480-c2orf68,-HuR cells, compared with SW480-c2orf68
and SW480-HuR cells, was not statistically significant.
Meanwhile, the protein expression level of BAX was significantly
increased in the SW480-c2orf68,-HuR, SW480-c2orf68
and SW480-HuR cells when compared with the blank group.
Compared with the SW480-c2orf68 and SW480-HuR cells,
the protein expression level of BAX was significantly higher in the
SW480-c2orf68,-HuR cells (P<0.01, P<0.01).
Discussion
In the present study, it was shown that the
expression level of C2ORF68 was significantly upregulated in rectal
cancer tissues compared with its adjacent normal tissues by IHC.
This indicates that c2orf68 may be involved the occurrence
of rectal cancer. In addition, upregulated expression of C2ORF68
was significantly correlated with a variety of important
clinicopathological parameters, including pathological grade and
lymph node metastasis. It was suggested that C2ORF68 may play a
role in the development and metastasis of CRC. As a result, a
statistical significance was found between the expression of
C2ORF68 and pathological grade. This indicates that the expression
level of C2ORF68 may be associated with the malignant potential of
cancer. That is, the higher the expression level of C2ORF68, the
higher is the degree of malignancy of rectal carcinoma. These
present results suggest that the c2orf68 gene is associated
with the occurrence and development of rectal cancer and may be a
potential carcinogenic factor in rectal cancer. However, the
detailed mechanism for c2orf68 upregulation in rectal cancer
remains to be clarified.
Through bioinformatic analyses, we discovered that
there are 12 proteins which interact with C2ORF68, including
KLHL15, GMNN, SMG6, GNE, DDHD2, PIR, ELAVL1 (HuR), NAGK, XPO1,
HSPA9, JOSD2 and GUK1(BioGrid database). At the same time, by our
experiments, including IF and Co-IP, we verified that C2ORF68
co-localized with HuR in SW480 and LoVo cell lines, and C2ORF68
could interact with HuR. HuR, a member of the Hu/ELAV
family, is predominantly located in the nucleus and translocates to
the cytoplasm when cells are stimulated by endogenous factors or
external stimuli (21,22). HuR (ELAV1) is an RNA binding
protein, which has been shown to regulate the expression of
multiple genes by different post-transcriptional mechanisms, such
as mRNA decay and protein translation (23). HuR modulates posttranscriptional
processing of target premRNAs or mRNA stabilization and translation
through interaction with AU-rich elements (ARE) within
3′-untranslated regions (UTRs) of the target mRNAs to
transformation (24). Furthermore,
it has been shown that HuR stabilizes mRNAs that encode p53
and WEE1 (25), activates
ATF2 (26), Jun D
(27) and XIAP (28) and enhances the translation of mRNAs
that encode c-Myc (29),
ICH-1 (30) and IL-1β
(31). Many of these transcripts
are reported to participate in certain key cellular processes
including cell proliferation, cell apoptosis, angiogenesis, immune
response and metastasis. HuR is also increased in malignant cells
when compared with corresponding normal cells, and it has been
found to be associated with adverse clinicopathological factors in
several different cancer types, such as gastric, gallbladder
breast, urothelial and non-small cell lung cancer (32).
Our study revealed that cell apoptosis increased,
cell proliferation and cell migration decreased when c2orf68
was inhibited in SW480 cells. These results are consistent with our
previous study (16). In addition,
we also demonstrated that cell apoptosis increased while cell
proliferation and cell migration decreased in SW480-HuR and
SW480-c2orf68,-HuR cells. This indicates that both
c2orf68 and HuR can regulate cell apoptosis and
proliferation in CRC cells. The cell apoptosis rate, cell
proliferation and cell migration in SW480-c2orf68,-HuR cells
which has more significant results than that in SW480-HuR
cells revealed that c2orf68 and HuR may have a
synergistic effect in regulating cell apoptosis, cell proliferation
and cell migration. This study differed from our previous study
(16), which focused on the
PI3K/Akt/mTOR signaling pathway and its downstream molecules, such
as Akt, PI3K, Bcl-2, c-Myc, cyclin D1 and bax when
c2orf68 was inhibited. A recent study showed that the HuR
expression level is closely related to AKT phosphorylation and
PI3K/AKT/NF-κB signaling can notably elevate HuR gene
transcription (18). This study
focused on the relationship between C2ORF68 and HuR and the
downstream molecules of HuR, such as Bcl-2, Bax, c-Myc,
cyclin D and cyclin A in SW480-c2orf68,-HuR,
SW480-c2orf68 and SW480-HuR cells. According to our
results, Bcl-2, c-Myc, cyclin D and Cyclin A
decreased, and Bax increased in the
SW480-c2orf68,-HuR, SW480-c2orf68 and
SW480-HuR cells. In addition, Bcl-2, c-Myc, cyclin D
and cyclin A was significantly decreased and Bax was
significantly increased in SW480-c2orf68,-HuR cells. This
shows that c2orf68 and HuR may co-regulate Bcl-2,
c-Myc, cyclin D, cyclin A and Bax, resulting in the cell
apoptosis and cell proliferation CRC cells.
In the present study, the mRNA and protein
expression of HuR was downregulated when c2orf68 was
inhibited in SW480 cells, and the mRNA and protein expression of
HuR was upregulated when c2orf68 was overexpressed in LoVo
cells. Furthermore, when HuR was inhibited, the mRNA and
protein expression levels of c2orf68 were also decreased.
That is, HuR and c2orf68 had a synergistic
effect.
The present study revealed that cell apoptosis
decreased while cell proliferation and cell migration were
increased in LoVo+c2orf68 cells. These results are
consistent with our previous study (17), and it was confirmed that
c2orf68 can regulate cell apoptosis and proliferation. In
addition, it was also revealed that cell apoptosis was decreased
when cell proliferation and cell migration were increased in
LoVo+c2orf68,-HuR cells. However, cell apoptosis was
significantly lower and cell proliferation and cell migration were
significantly higher in LoVo+c2orf68 cells, compared with
LoVo+c2orf68,-HuR cells. All these results suggest again
that c2orf68 and HuR may have a synergistic effect in
promoting cell proliferation and migration, and in inhibiting cell
apoptosis in the role of CRC. Unlike our previous study, which
focused on the Wnt signaling pathway and its molecules such as
β-catenin, survivin, cyclin D1, c-Myc and GSK-3β
(17), the present study focused on
the relationship between C2ORF68 and HuR and the downstream
molecules of HuR such as Bcl-2, Bax, c-Myc, cyclin D
and cyclin A. In LoVo+c2orf68 and
LoVo+c2orf68,-HuR cells, Bcl-2, c-Myc, cyclin D and
cyclin A increased, while Bax decreased. In addition,
Bcl-2, c-Myc, cyclin D and cyclin A were
significantly upregulated and Bax was significantly
decreased in LoVo+c2orf68 cells, compared with
LoVo+c2orf68,-HuR cells. Previously in this manuscript, we
described that Bcl-2, c-Myc, cyclin D and cyclin A
were decreased, while Bax was increased in the
SW480-c2orf68,-HuR, SW480-c2orf68 and
SW480-HuR cells. In addition, Bcl-2, c-Myc, cyclin D
and cyclin A were significantly decreased and Bax was
significantly increased in the SW480-c2orf68,-HuR cells,
compared with the SW480-c2orf68 and SW480-HuR cells.
It is known that all these genes are involved in key cellular
processes such as cell proliferation and cell cycle. The expression
of cyclin D1 is increased in many tumors and it promotes cell
proliferation by regulating cell cycle progression through the G1/S
restriction point (33). The
transcription factor c-Myc, which is a leucine zipper protein
regulating the expression of 10–15% of human genes, plays an
important role in cell proliferation, differentiation, growth and
survival. Its overexpression is associated with cancer occurrence
and development (34). BCL-2, an
integral outer mitochondrial membrane protein, serving as an
anti-apoptosis protein, belongs to the Bcl-2 family, and was
firstly discovered in B cell malignancies and regulates the
intrinsic mitochondrial apoptosis pathway (35). Activated Bax then oligomerizes at
the mitochondria to induce outer mitochondrial membrane (OMM)
permeabilization and releases into the cytosol apoptotic factors
which promote caspase activation and subsequent apoptosis execution
(36). Cyclins are fundamental
regulators of the cell cycle, playing an important role in
tumorigenesis, Cyclin A is required for cells to progress through
the S phase (37). As a result, it
is believed that c2orf68 and HuR may have a
synergistic effect and may co-regulate cell proliferation and
apoptosis by regulating the downstream molecules of HuR,
such as upregulating Bcl-2, c-Myc, cyclin D and cyclin
A gene expression and downregulating Bax gene
expression, resulting in the development of cancer.
In conclusion, the c2orf68 gene may be a
potential oncogene and may play a potential carcinogenic effect in
the pathogenesis of colorectal cancer. Furthermore, it may be
associated with lymph node metastasis of colorectal cancer. The
carcinogenesis of c2orf68 may be related to the promotion of
cell proliferation and inhibition of cell apoptosis. C2orf68
may have a synergistic effect with HuR, and the possible
mechanism of c2orf68 and HuR involves the
co-promotion of cell proliferation and migration, and the
co-inhibition of cell apoptosis. However, the exact mechanism
remains mysterious. C2orf68 and HuR may co-regulate cell
proliferation and apoptosis by upregulating Bcl-2, c-Myc, cyclin
D and cyclin A gene expression and downregulating
Bax gene expression, resulting in the development of
colorectal cancer. Our previous study indicated that C2ORF68 can
regulate cancer cell proliferation and apoptosis through
PI3K/AKT/mTOR signaling (16). At
the same time, a recent study (18)
showed that the HuR expression level is closely related to AKT
phosphorylation and cytoplasmic abundance of HuR in human cancer
may be associate with oncogenic activation of AKT signaling.
According to the above conclusions, we may hypothesize that
C2ORF68, HuR and AKT signaling could make up a mutually reinforcing
loop, regulating cancer cell proliferation and apoptosis. However,
the specific mechanism warrants further research.
Acknowledgements
I need to express my sincere gratitude to all
authors for the assistance with this article, especially to
Professor Yao Chen, whose constant encouragement directed me
through all stages of the writing of this manuscript. Without her
help, I could not have gotten to this point. Furthermore, I also
extend my heartfelt gratitude to the other authors, and thanks for
their efforts.
Funding
The present study was supported by Chengdu
Department of Science and Technology (2018-YF05-00-038-SN), Sichuan
Province, China.
Availability of data and materials
The datasets used during the present study are
available from the corresponding author upon reasonable
request.
Authors' contributions
LS, KS, TJ and YC conceived and designed the study.
ZL and KH performed the experiments. ZL and KH wrote the paper. LS,
KS, TJ and YC reviewed and edited the manuscript. All authors read
and approved the manuscript and agree to be accountable for all
aspects of the research in ensuring that the accuracy or integrity
of any part of the work are appropriately investigated and
resolved.
Ethics approval and consent to
participate
The Medical Research Ethics Committee of Sichuan
University approved the sample acquisition (Chengdu, China), and a
written informed consent was also obtained from all patients.
Patient consent for publication
Not applicable.
Competing interests
The authors state that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2017. CA Cancer J Clin. 67:7–30. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lin J, Chuang CC and Zuo L: Potential
roles of microRNAs and ROS in colorectal cancer: Diagnostic
biomarkers and therapeutic targets. Oncotarget. 8:17328–17346.
2017.PubMed/NCBI
|
|
3
|
Fang Y, Liang X, Jiang W, Li J, Xu J and
Cai X: Cyclin B1 suppresses colorectal cancer invasion and
metastasis by regulating e-cadherin. PLoS One. 10:e01268752015.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Schreuders EH, Ruco A, Rabeneck L, Schoen
RE, Sung JJ, Young GP and Kuipers EJ: Colorectal cancer screening:
A global overview of existing programmes. Gut. 64:1637–1649. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Jeyakumar A, Dissabandara L and Gopalan V:
A critical overview on the biological and molecular features of red
and processed meat in colorectal carcinogenesis. J Gastroenterol.
52:407–418. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Sakamoto N, Feng Y, Stolfi C, Kurosu Y,
Green M, Lin J, Green ME, Sentani K, Yasui W, McMahon M, et al:
BRAFV600E cooperates with CDX2 inactivation to promote
serrated colorectal tumorigenesis. Elife. 6:e203312017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Raskov H, Pommergaard HC, Burcharth J and
Rosenberg J: Colorectal carcinogenesis-update and perspectives.
World J Gastroenterol. 20:18151–18164. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lech G, Słotwiński R, Słodkowski M and
Krasnodębski IW: Colorectal cancer tumour markers and biomarkers:
Recent therapeutic advances. World J Gastroenterol. 22:1745–1755.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Huang L, Wang X, Wen C, Yang X, Song M,
Chen J, Wang C, Zhang B, Wang L, Iwamoto A, et al: Hsa-miR-19a is
associated with lymph metastasis and mediates the TNF-α induced
epithelial-to-mesenchymal transition in colorectal cancer. Sci Rep.
5:133502015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kurebayashi H, Goi T, Shimada M, Tagai N,
Naruse T, Nakazawa T, Kimura Y, Hirono Y and Yamaguchi A:
Prokineticin 2 (PROK2) is an important factor for angiogenesis in
colorectal cancer. Oncotarget. 6:26242–26251. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ingebrigtsen VA, Boye K, Nesland JM,
Nesbakken A, Flatmark K and Fodstad Ø: B7-H3 expression in
colorectal cancer: Associations with clinicopathological parameters
and patient outcome. BMC Cancer. 14:6022014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yamaguchi K, Yamaguchi R, Takahashi N,
Ikenoue T, Fujii T, Shinozaki M, Tsurita G, Hata K, Niida A, Imoto
S, et al: Overexpression of cohesion establishment factor DSCC1
through E2F in colorectal cancer. PLoS One. 9:e857502014.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Thomas J, Ohtsuka M, Pichler M and Ling H:
MicroRNAs: Clinical relevance in colorectal cancer. Int J Mol Sci.
16:28063–28076. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lam SY, Yu J, Wong SH, Peppelenbosch MP
and Fuhler GM: The gastrointestinal microbiota and its role in
oncogenesis. Best Pract Res Clin Gastroenterol. 31:607–618. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wang K and Chen Y: Analysis of a novel
protein in human colorectal adenocarcinoma. Mol Med Rep. 8:529–534.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wen X, Zhu J, Dong L and Chen Y: The role
of c2orf68 and PI3K/Akt/mTOR pathway in human colorectal cancer.
Med Oncol. 31:922014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Chen Y, Jiang T, Shi L and He K: hcrcn81
promotes cell proliferation through Wnt signaling pathway in
colorectal cancer. Med Oncol. 33:32016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kang MJ, Ryu BK, Lee MG, Han J, Lee JH, Ha
TK, Byun DS, Chae KS, Lee BH, Chun HS, et al: NF-kappaB activates
transcription of the RNA-binding factor HuR, via PI3K-AKT
signaling, to promote gastric tumorigenesis. Gastroenterology.
135:2030–2042.e1-e3. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
He K, Shi L, Jiang T, Li Q, Chen Y and
Meng C: Association between SET expression and glioblastoma cell
apoptosis and proliferation. Oncol Lett. 12:2435–2444. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2-ΔΔCT method. Methods. 25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Woodhoo A, Iruarrizaga-Lejarreta M, Beraza
N, García-Rodríguez JL, Embade N, Fernández-Ramos D, Martínez-López
N, Gutiérrez-De Juan V, Arteta B, Caballeria J, et al: Human
antigen R contributes to hepatic stellate cell activation and liver
fibrosis. Hepatology. 56:1870–1882. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhang LF, Lou JT, Lu MH, Gao C, Zhao S, Li
B, Liang S, Li Y, Li D and Liu MF: Suppression of miR-199a
maturation by HuR is crucial for hypoxia-induced glycolytic switch
in hepatocellular carcinoma. EMBO J. 34:2671–2685. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Jakstaite A, Maziukiene A, Silkuniene G,
Kmieliute K, Gulbinas A and Dambrauskas Z: HuR mediated
post-transcriptional regulation as a new potential adjuvant
therapeutic target in chemotherapy for pancreatic cancer. World J
Gastroenterol. 21:13004–13019. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhu H, Berkova Z, Mathur R, Sehgal L,
Khashab T, Tao RH, Ao X, Feng L, Sabichi AL, Blechacz B, et al: HuR
suppresses fas expression and correlates with patient outcome in
liver cancer. Mol Cancer Res. 13:809–819. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lal S, Burkhart RA, Beeharry N,
Bhattacharjee V, Londin ER, Cozzitorto JA, Romeo C, Jimbo M, Norris
ZA, Yeo CJ, et al: HuR posttranscriptionally regulates WEE1:
Implications for the DNA damage response in pancreatic cancer
cells. Cancer Res. 74:1128–1140. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Xiao L, Rao JN, Zou T, Liu L, Marasa BS,
Chen J, Turner DJ, Zhou H, Gorospe M and Wang JY: Polyamines
regulate the stability of activating transcription factor-2 mRNA
through RNA-binding Protein HuR in intestinal epithelial cells. Mol
Biol Cell. 18:4579–4590. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zou T, Rao JN, Liu L, Xiao L, Yu TX, Jiang
P, Gorospe M and Wang JY: Polyamines regulate the stability of JunD
mRNA by modulating the competitive binding of its 3′ untranslated
region to HuR and AUF1. Mol Cell Biol. 30:5021–5033. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhang X, Zou T, Rao JN, Liu L, Xiao L,
Wang PY, Cui YH, Gorospe M and Wang JY: Stabilization of XIAP mRNA
through the RNA binding protein HuR regulated by cellular
polyamines. Nucleic Acids Res. 42:41432014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Liu L, Rao JN, Zou T, Xiao L, Wang PY,
Turner DJ, Gorospe M and Wang JY: Polyamines regulate c-Myc
translation through Chk2-dependent HuR phosphorylation. Mol Biol
Cell. 20:4885–4898. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Winkler C, Doller A, Imre G, Badawi A,
Schmid T, Schulz S, Steinmeyer N, Pfeilschifter J, Rajalingam K and
Eberhardt W: Attenuation of the ELAV1-like protein HuR sensitizes
adenocarcinoma cells to the intrinsic apoptotic pathway by
increasing the translation of caspase-2L. Cell Death Dis.
5:e13212014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Aguado A, Rodríguez C, Martínez-Revelles
S, Avendaño MS, Zhenyukh O, Orriols M, Martínez-González J, Alonso
MJ, Briones AM, Dixon DA and Salaices M: HuR mediates the
synergistic effects of angiotensin II and IL-1β on vascular COX-2
expression and cell migration. Br J Pharmacol. 172:3028–3042. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Elebro J, Ben Dror L, Heby M, Nodin B,
Jirström K and Eberhard J: Prognostic effect of hENT1, dCK and HuR
expression by morphological type in periampullary adenocarcinoma,
including pancreatic cancer. Acta Oncol. 55:286–296. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Li X, Zhang Q, Fan K, Li B, Li H, Qi H,
Guo J, Cao Y and Sun H: Overexpression of TRPV3 correlates with
tumor progression in non-small cell lung cancer. Int J Mol Sci.
17:4372016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Sharma T, Bansal R, Haokip DT, Goel I and
Muthuswami R: SMARCAL1 negatively regulates C-Myc transcription by
altering the conformation of the promoter region. Sci Rep.
9:179102015.
|
|
35
|
Choi JE, Kang SH, Lee SJ and Bae YK:
Prognostic significance of Bcl-2 expression in non-basal
triple-negative breast cancer patients treated with
anthracycline-based chemotherapy. Tumor Biol. 35:12255–12263. 2014.
View Article : Google Scholar
|
|
36
|
Ouyang YB and Giffard RG: microRNAs affect
BCL-2 family proteins in the setting of cerebral ischemia.
Neurochem Int. 77:2–8. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Kokontis JM, Lin HP, Jiang SS, Lin CY,
Fukuchi J, Hiipakka RA, Chung CJ, Chan TM, Liao S, Chang CH, et al:
Androgen suppresses the proliferation of androgen receptor-positive
castration-resistant prostate cancer cells via inhibition of Cdk2,
CyclinA, and Skp2. PLoS One. 9:e1091702014. View Article : Google Scholar : PubMed/NCBI
|