Introduction
Cervical cancer (CC) is the fourth most common
cancer in women worldwide, ~85% of which occur in low-income and
middle-income countries (1,2). There are various histologic types of
CC, with the majority classified as squamous cell cervical
carcinomas. Squamous cell CC is considered to be the result of a
multi-step process, involving a transition from precancerous
low-grade squamous intraepithelial lesions (LSIL) to high-grade
squamous intraepithelial lesion (HSIL) to invasive CC (3,4).
Multiple factors are associated with CC development, including
persistent infection with certain major subtypes of oncogenic human
papillomavirus (HPV) (5,6), smoking (7,8) and
immunosuppression (9). Persistent
infection with certain types of oncogenic HPV is central to the
etiology of CC (5), and vaccines
against HPV have been approved for use and progressively introduced
since 2006 (10). Based on
cytologic screening for high-risk types of HPV, tests have been
widely used for screening precancerous lesions and CC, and also
direct the subsequent follow-up investigations, which has
significantly decreased the incidence and mortality of CC (2,10).
However, the specificity of CC screening methods is not high
enough, and implementation is still limited and unsuiTable in many
poor regions (1,6,11).
Furthermore, the application of cervical biopsy under a colposcope,
which is the gold standard for diagnosis of CC and depends on the
results of the screening, is costly and has sometimes produced
unnecessary tests in previous years (4,12).
Accordingly, increased understanding of cervical carcinogenesis,
and early and easy-access diagnostic methods for CC remain
important. In past decades, research into cancer epigenetic
modifications, particularly DNA methylation, and their contribution
to tumor carcinogenesis has been continuously increasing (13–15);
however, the effects of epigenetic factors on CC remain largely
unknown and provide new research directions for investigating
cervical carcinogenesis and CC development.
Studies have established that proteins encoded by
the stem cell-related SOX family of genes are important
transcription factors (TF) containing high-mobility-group (HMG)
domain (16,17) that have important regulatory roles
in the development, differentiation and metabolism (18,19).
Abnormalities in these TFs may result in abnormal cell protein
expression. SRY-related HMG-box gene 11 (SOX11), which is involved
in embryonic development, cell proliferation, differentiation and
apoptosis, has been reported to influence the survival, growth and
transformation of tumor cells in certain solid malignancies
(20–22). In recent years, several studies have
reported that SOX11 acts as a tumor suppressor gene (TSG) (23–26).
For instance, SOX11 was downregulated (23) and improved disease-free survival
(24) in ovarian cancer.
Additionally, hypermethylation-induced silencing of SOX11
was detected in ovarian epithelial cell carcinoma, B cell lymphoma
(25) and nasopharyngeal carcinoma
(26), suggesting that DNA
methylation of SOX11 may have a significant role in the
development of malignant tumors. The aforementioned studies
demonstrated that dysfunctional SOX11 is associated with
tumorigenesis in several types of cancer. However, the expression
and function of the SOX11 gene in CC has, to the best of our
knowledge, not been investigated previously.
In the present study, it was demonstrated that
SOX11 was downregulated at the transcriptional and
translational levels, while the promoter region of the SOX11
gene was hypermethylated in CC tissues and cell lines compared with
normal tissue or Tara-1 cells, respectively. The association
between the methylation status of the SOX11 promoter and the
development of CC suggested that SOX11 may, at least in part,
function as a tumor suppressor in CC and contribute to the
carcinogenesis of CC.
Materials and methods
Study subjects and ethics
statement
Samples from patients with CC (n=54) admitted to the
Department of Gynecology at the Second Affiliated Hospital of Xi'an
Jiaotong University (Xi'an) from December 2016-November 2017 were
used in the present study. Biopsy samples were obtained by
colposcopy prior to surgery, chemotherapy or radiotherapy. Normal
cervix (NC) samples were collected from 30 hospitalized age-matched
patients with uterine fibroids during hysterectomy. LSIL and HSIL
samples were collected from 20 and 24 patients, respectively,
undergoing colposcopy and cervical biopsies or after cervical loop
electrosurgical excision procedure during the same period. All
hematoxylin and eosin (H&E) sections of the specimens were
reviewed and confirmed by two pathologists.
The design and implementation of the study were
approved by the Ethics Committee of Medical School of Xi'an
Jiaotong University (Xi'an, China; no. 2017-266). During the study,
a written informed consent was obtained from all participants.
Cell culture
The human CC cell lines HeLa, SiHa, C33A and CaSki
were purchased from American Type Culture Collection (Manassas, VA,
USA) and Tera-1, which served as a positive control, was a gift
from Dr Yue Li (Xi'an Jiaotong University Health Science Center,
Xi'an, China). As SOX11 is a stem cell-associated gene, Tera-1 is a
teratoma cell line that abundantly expresses stem cell-associated
genes. Tera-1 was selected as the positive control. Cells were
cultured in Dulbecco's modified Eagle's medium (DMEM;
Sigma-Aldrich; Merck KGaA, Darmstadt, Germany), except for the
CaSki cells which were maintained in McCoy's 5A medium
(Sigma-Aldrich; Merck KGaA) supplemented with 10% fetal bovine
serum (FBS; Invitrogen; Thermo Fisher Scientific, Inc., Waltham,
MA, USA) in a cell culture incubator at 37°C with 5%
CO2.
5-Aza-deoxycytidine (5-Aza-dC)
treatment
Cells were trypsinized, counted and seeded in
triplicate in 6-wells plate. After 24 h, the medium was replaced
with fresh medium containing 0 (PBS), 5, or 10 µM 5-Aza-dC
(Sigma-Aldrich; Merck KGaA). The medium was replaced every 24 h.
After 72 h, the total cellular RNA and protein were extracted for
subsequent experiments.
Bisulfite sequencing (BSQ) and
methylation-specific polymerase chain reaction (PCR)
Genomic DNA was extracted using the Universal
Genomic DNA Extraction kit ver. 3.0 (cat. no. DV811A; Takara
Biotechnology Co., Ltd., Dalian, China). Genomic DNA (2 µg/sample)
was bisulfite-modified, and the bisulfite-modified DNA was purified
according to the manufacturer's protocol (EpiTect BisuLfite kit;
Qiagen GmbH, Hilden, Germany). Then, the purified
bisulfite-modified DNA was amplified using the following primers:
SOX11 forward, 5′-AGAGAGATTTTAATTTTTTGTAGAAGGA-3′ and
reverse, 5′-CCCCCTTCCAAACTACACAC-3′. The modified DNA was amplified
by PCR using the following conditions: 95°C for 3 min, and then 35
cycles of 95°C for 30 sec, 54°C for 30 sec and 72°C for 30 sec,
followed by a 10-min incubation at 72°C. The PCR products were
analyzed by gel electrophoresis in 2.5% agarose to confirm that a
single band had been produced. Then, TA clones were established
according to the instructions of the protocol for the pEASY-T1
Cloning kit (Beijing Transgen Biotech Co., Ltd., Beijing, China).
The 10–15 positive monoclonal bacteria solution were sequenced by
Xi'an Qing Biological Co., Ltd. (Xi'an, China). The sequencing
primers were M13 forward, 5′-GTTTTCCCAGTCACGAC-3′ and reverse,
5′-CAGGAAACAGCTATGAC-3′. The sequencing results were analyzed using
the BiQ Analyzer 2.0 (Max Planck Institute for Informatics,
Saarbrücken, Germany) and were output as circle graphs.
Reverse transcription-quantitative PCR
(RT-PCR)
Total RNA was extracted using the RLT reagent with
1% β-mercaptoethanol in the RNeasy Mini Kit (cat. no. 74106; Qiagen
GmbH) according to the manufacturer's protocol. Total RNA (500 ng)
was reverse transcribed using the High-Capacity cDNA Reverse
Transcription Kit (cat. no. 4368814; Applied Biosystems; Thermo
Fisher Scientific, Inc.) with incubation for 10 min at 25°C, 120
min at 37°C and 5 min at 85°C. qPCR was performed on an Applied
Biosystems 7700 Prism RT-PCR machine (Applied Biosystems; Thermo
Fisher Scientific, Inc.) and conditions were as follows: Enzyme
activation for 10 min at 95°C, PCR cycle denaturation for 15 sec at
95°C and annealing/elongation 1 min at 60°C. The sequences of the
SOX11 primers were as follows: Forward,
5′-GGTGGATAAGGATTTGGATTCG-3′ and reverse, 5′-GCTCCGGCGTGCAGTAGT-3′.
Expression was normalized to the expression of 18S and transformed
using the relative standard curve method and comparative
quantification cycle (Ct) method (ΔΔCq) as described previously
(27).
Immunoblotting
Cells and CC cell line samples were washed twice
with cold PBS, followed by lysis buffer (cat. no. P0013D; Beyotime
Institute of Biotechnology, Haimen, China). Protein concentration
was quantified using a bicinchoninic acid kit (cat. no. P0009;
Beyotime Institute of Biotechnology) according to the
manufacturer's instructions. Protein lysates (25 µg) were separated
at 80 V on 10% acrylamide gel for ~2 h. Transfer to Immobilon-FL
(EMD Millipore, Billerica, MA, USA) membrane was performed at 20 V
for 1.5 h. Following blocking in the Odyssey blocking buffer (cat.
no. 927-40000; LI-COR Biosciences, Lincoln, NE, USA) for 50 min at
room temperature, a primary antibody rabbit polyclonal anti-human
SOX11 (1:500 dilution; cat. no. sc-20096; Santa Cruz Biotechnology,
Inc., Dallas, TX, USA), GAPDH (1:10,000 dilution; cat. no. G8795;
Sigma-Aldrich; Merck KGaA) and β-tubulin (1:5,000 dilution; cat.
no. sc-80011; Santa Cruz Biotechnology) were incubated at 4°C
overnight. Secondary antibody conjugated to Alexa Fluor®
680 dye (cat. no. A32734; Invitrogen; Thermo Fisher Scientific,
Inc.) or IRdye800 (cat. no. 610-731-002; Rockland Immunochemicals
Inc., Limerick, Pennsylvania, USA) was subsequently incubated with
the membrane for 1 h at room temperature to visualize the proteins
at 700 or 800 nm using a LI-COR Odyssey imaging system (LI-COR
Biosciences). The results were quantitatively analyzed using the
ImageJ software (National Institutes of Health, Bethesda, MD,
USA).
Immunohistochemistry
Human specimens were fixed with 4% paraformaldehyde
overnight at room temperature, then embedded in paraffin and
sectioned into 4 µm slices. The slides were baked at 65°C overnight
prior to deparaffinization in xylene twice for 20 min and hydrated
in a series of graded ethanol (100, 95, 90 and 80% ethanol; 5 min
each). The sections were boiled in citrate buffer (pH 6.0) by
heating in a pressure cooker for 1–2 Alexa Fluor® 680
min for antigen retrieval. Endogenous peroxidases were blocked for
10 min with freshly prepared 3% H2O2 at room
temperature. Following blocking with goat serum for 30 min at room
temperature, the SOX11 primary antibody (1:200 diluted in 1% bovine
serum albumin PBS solution; cat. no. sc-20096; Santa Cruz
Biotechnology, Inc.) was incubated with the slides at 4°C
overnight. The following day, the slides were incubated with a
secondary antibody labeled with horseradish peroxidase (1:50
dilution, cat. no. GAR-HRP; Pierce; Thermo Fisher Scientific, Inc.)
for 40 min at room temperature. Diaminobenzidine was used to color
the slides for ~20 min at room temperature. The sections were
observed under the microscope to control the reaction time when the
sections were incubated with the DAB reagent for signal
amplification. Then the sections were stained with H&E.
Histological analysis was performed by two blinded pathologists. At
least 10 high-power fields at ×1,000 magnification were examined
for each sample. The staining score was classified into four grades
based on the staining intensity as follows: 1, no cell staining; 2,
weak yellow; 3, moderate yellow; and 4, strong brown yellow. The
proportion of positively stained cells was classified into four
levels as follows: 1 (0–25%), 2 (26–50%), 3 (51–75%) and 4
(76–100%). The immunoreactivity score (IRS) was obtained by
multiplying the intensity and proportion values, and samples were
grouped into three levels as follows: Negative (−, score 1–4),
positive (+, score 5–9), strongly positive (++, score 10–16). The
mean score was used for comparison between groups.
Proliferation assay
Cell viability was measured using the PrestoBlue kit
(Invitrogen; Thermo Fisher Scientific, Inc.). A total of 2,000
cells in the logarithmic growth phase were seeded into a 96-well
plate in triplicate and allowed to adhere overnight. Throughout the
6-day period, the medium was replaced with fresh medium every day,
and the same concentration of 5-Aza-dC was added. PrestoBlue
reagent was used to assess the proliferative ability according to
the standard manufacturer's protocol. The fluorescence was read at
a wavelength of 570 nm using a FLUOstar Optima microplate
spectrophotometer (BMG Labtech GmbH, Ortenberg, Germany).
Statistical analysis
Statistical analyses were performed using the Prism
7 software (GraphPad Software Inc., La Jolla, CA, USA) and SPSS
version 22.0 for Windows (IBM Corp., Armonk, NY, USA). The unpaired
t-test with Welch's correction was used to analyze the difference
between two groups. The two-tailed Mann Whitney U test with
Bonferroni's correction for post hoc comparisons was performed
following the Kruskal-Wallis test and Tukey's post hoc test was
used following the one-way analysis of variance test to evaluate
the statistical comparison of multiple groups. The Pearson
χ2 analysis was used to analyze the association between
SOX11 expression and the clinical features, when the number was
<5, Fisher's exact test was used. Pearson's correlation test was
used to analyze the correlation between the SOX11 mRNA expression
level and its promoter methylation level. Error bars represent ±
standard error. P<0.05 was considered to indicate a
statistically significant difference.
Results
SOX11 expression is downregulated
during the development of CC
SOX11 has been demonstrated to have different
properties in different human cancers. It is upregulated in gastric
(28) and prostate cancer (29), and other types of cancers, whereas
it is downregulated in medulloblastoma (30) and malignant gliomas (20). To determine the expression of SOX11
during the development of CC, SOX11 protein expression level was
analyzed by immunohistochemistry and western blot analysis in NC,
LSIL, HSIL and CC tissues. SOX11 was localized in the nuclei of all
positive cells with different levels in different specimens. SOX11
was highly expressed in NC and LSIL epithelial basal cells, and
weakly expressed or virtually absent in tumor parenchymal cells of
HSIL and CC. The IRS of SOX11 was as follows: NC, 9.933±0.948;
LSIL, 7.200±0.766; HSIL, 4.917±0.492; and CC, 3.074±0.301 (Fig. 1B). With the progression of cervical
lesions, the expression level of SOX11 gradually decreased, and
there were statistical differences in IRS levels of SOX11 in the
four groups (NC vs. LSIL, P=1.000; NC vs. HSIL, P=0.022; NC vs. CC,
P<0.0001; LSIL vs. HSIL, P=0.625; LSIL vs. CC, P<0.0001; HSIL
vs. CC, P=0.057; P-value are adjusted). In addition, the expression
level of SOX11 was determined by western blot analysis in randomly
selected tissues including 8 NC and 20 CC (Fig. 1C). The relative expression of the
SOX11protein is presented as mean SOX11 expression of 0.95±0.07 in
NC, and 0.17±0.04 in CC (Fig. 1D).
Thus, the expression of SOX11 protein in NC was 5.59 fold higher
than that in CC specimens (P<0.001). The association between
SOX11 expression based on the immunohistochemistry staining results
and the clinicopathological characteristics in the patients with CC
are summarized in Table I. The
Pearson χ2 analysis revealed a significant association
between SOX11 expression and the tumor grade (P=0.005) and HPV
status (P<0.001); however, there was no significant association
between SOX11 expression and other clinical features, including
age, International Federation of Gynecology and Obstetrics stage,
lymph node, myometrial invasion depth and histologic type. The
association between SOX11 expression and the HPV status in the
patients with LSIL and HSIL in Tables
II and III reveal a
significant association between SOX11 expression and HPV status
(P=0.032 and 0.035, respectively). These findings suggest that the
expression of SOX11 decreases with the development of CC
malignancy, which strongly suggested that the loss of SOX11
function, which may act as a tumor suppressor, promotes the
progression of CC.
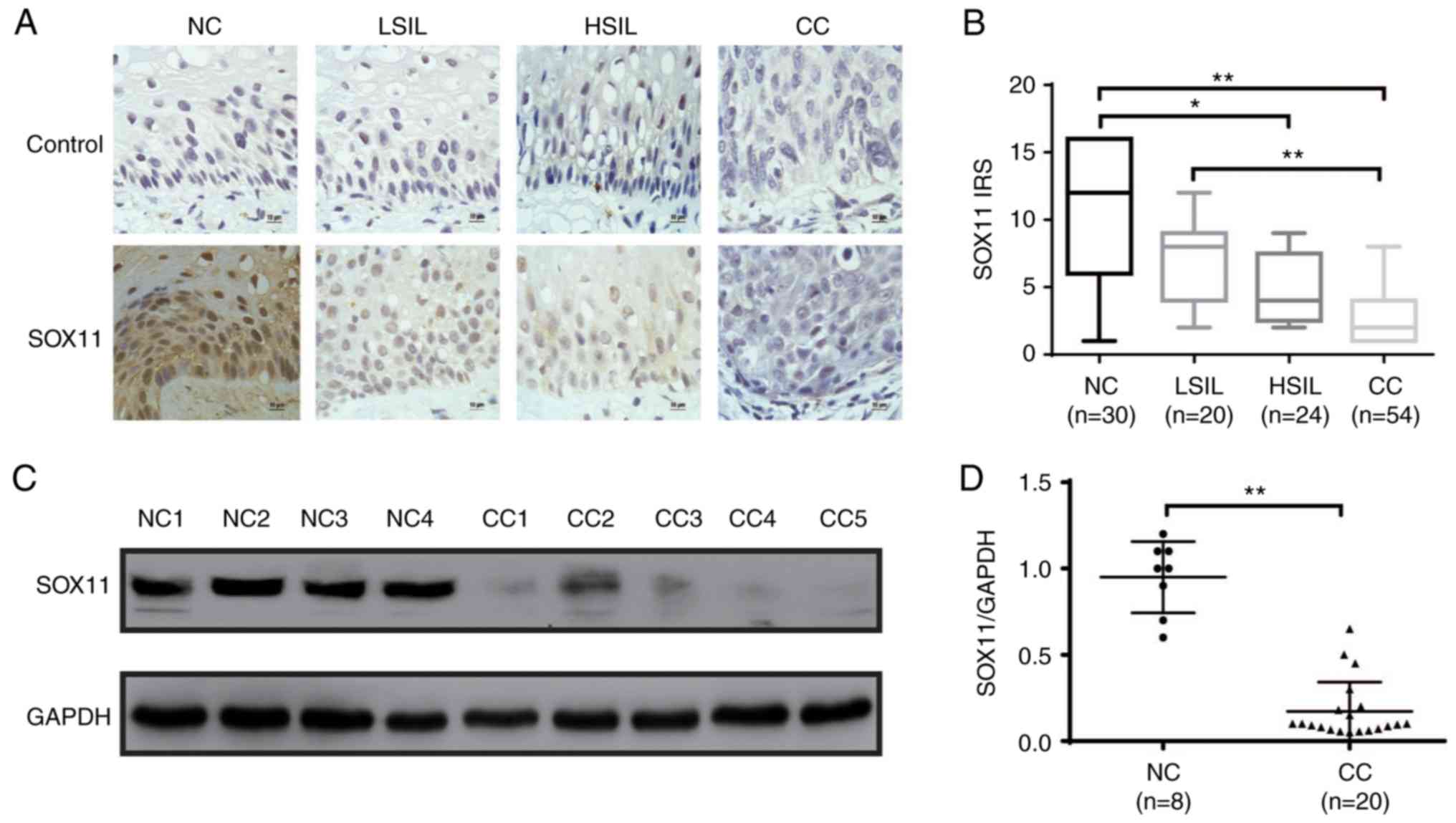 | Figure 1.SOX11 expression during the
development of CC. (A) Expression of SOX11 in normal cervix, LSIL,
HSIL and CC was evaluated by immunohistochemistry. PBS was used
instead of SOX11 antibody as control group. (B) IRS presented as
box plots and significance calculated by the Mann Whitney U test
with Bonferroni's correction (new P<0.0083 was required for
significance, the adjusted P-values are shown as *P≤0.05,
**P≤0.01). (C) SOX11 protein expression level was determined by
western blot analysis in randomly selected tissues (8 NC and 20
CC). (D) Relative expression of the SOX11/GAPDH was estimated by
densitometry. Bars represent standard error and data was analyzed
by unpaired t tests. *P≤0.05, **P≤0.01. NC, normal cervix; LSIL,
low-grade squamous intraepithelial lesion; HSIL, high-grade
squamous intraepithelial lesion; CC, cervical cancer; SOX11,
SRY-related HMG-box gene 11. |
 | Table I.Association between SOX11 expression
and clinicopathological characteristics in patients with cervical
cancer. |
Table I.
Association between SOX11 expression
and clinicopathological characteristics in patients with cervical
cancer.
|
|
| SOX11
expression |
|
|---|
|
|
|
|
|
|---|
| Factor | n | Negative | Positive | P-value |
|---|
| Age, years |
|
|
| 0.483 |
|
<45 | 8 | 7 | 1 |
|
|
≥45 | 46 | 35 | 11 |
|
| Grade |
|
|
| 0.005 |
| I | 20 | 11 | 9 |
|
|
II–III | 34 | 31 | 3 |
|
| International
federation of gynecology and obstetrics stage |
|
|
| 0.591 |
| I | 6 | 4 | 2 |
|
| II | 42 | 34 | 8 |
|
|
III–IV | 6 | 4 | 2 |
|
| Lymph node |
|
|
| 0.855 |
| N0 | 10 | 8 | 2 |
|
| N1 | 44 | 34 | 10 |
|
| Myometrial invasion
depth |
|
|
| 0.562 |
|
<1/2 | 22 | 18 | 4 |
|
|
≥1/2 | 32 | 24 | 8 |
|
| Histologic
type |
|
|
| 0.595 |
|
Squamous carcinoma | 40 | 37 | 3 |
|
|
Adenocarcinoma | 14 | 12 | 2 |
|
| HPV infection |
|
|
| <0.001 |
|
Positive | 43 | 40 | 3 |
|
|
Negative | 11 | 4 | 7 |
|
| Total | 54 |
|
|
|
 | Table II.Association between SOX11 expression
and clinicopathological characteristics in patients with low-grade
squamous intraepithelial lesion. |
Table II.
Association between SOX11 expression
and clinicopathological characteristics in patients with low-grade
squamous intraepithelial lesion.
|
|
| Sox11
expression |
|
|---|
|
|
|
|
|
|---|
| HPV infection | n | Negative | Positive | P-value |
|---|
| Positive | 16 | 14 | 2 | 0.032 |
| Negative | 4 | 1 | 3 |
|
 | Table III.Association between SOX11 expression
and clinicopathological characteristics in patients with high-grade
squamous intraepithelial lesion. |
Table III.
Association between SOX11 expression
and clinicopathological characteristics in patients with high-grade
squamous intraepithelial lesion.
|
|
| Sox11
expression |
|
|---|
|
|
|
|
|
|---|
| HPV infection | n | Negative | Positive | P-value |
|---|
| Positive | 20 | 17 | 3 | 0.035 |
| Negative | 4 | 1 | 3 |
|
Promoter region of SOX11 is
hypermethylated in CC
The hypermethylation of promoter CpG islands is one
of the essential mechanisms of transcriptional silencing of TSGs,
and also the most important early, common event in the process of
cervical disease progression to cancer (13,31–34).
To further investigate the mechanism of the downregulation of SOX11
during the development of CC, 10 NC and 10 CC specimens were
randomly selected to determine the methylation status of the SOX11
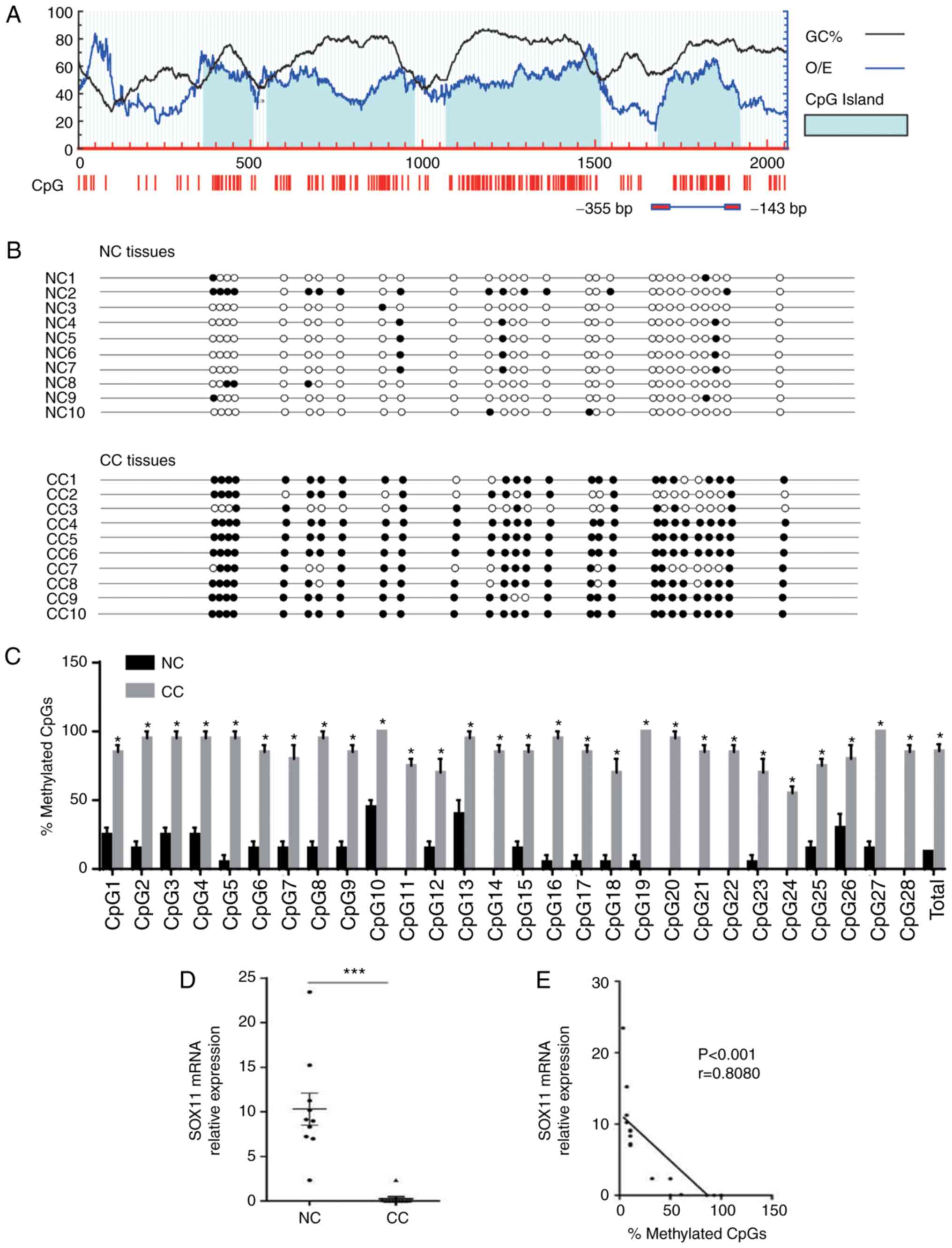
promoter. Four CpG islands of SOX11 were identified with a GC
content >50% and observed/expected CpG ratio >0.6 when to
promoter region (2,000 bp upstream of the transcription start site)
of SOX11 was analyzed using MethPrimer 2.0 (urogene.org/methprimer2/; Fig. 2A). Subsequent sequencing experiments
were performed on the fourth CpG island, which had previously been
reported to be determinative for SOX11 expression in multiple cell
lines (25) and includes 28 CpG
sites (Fig. 2A). The results are
presented in Fig. 2B, with the CpG
sites are indicated with circles (solid circle indicate methylation
and unshaded circle indicates no methylation). The SOX11 promoter
was hypermethylated in CC (total mean level was 85.71%), which was
significantly higher than that in the NC samples (total mean level
was 12.68%) at each CpG site (Fig.
2C). The mRNA expression level of SOX11 in CC was significantly
lower than that in NC tissues (P<0.001; Fig. 2D). Pearson's correlation analysis of
the SOX11 mRNA expression level and its promoter methylation level
revealed that the SOX11 mRNA expression level in CC was negatively
correlated with hypermethylation in the promoter region (r=−0.8080;
P<0.001; Fig. 2E). The
expression of SOX11 protein was downregulated, with its promoter
region hypermethylated in CC. These results suggest that
hypermethylation of the SOX11 promoter may be involved in the
mechanism of downregulation of SOX11 in CC.
Methylation status of SOX11 promoter
in CC cell lines
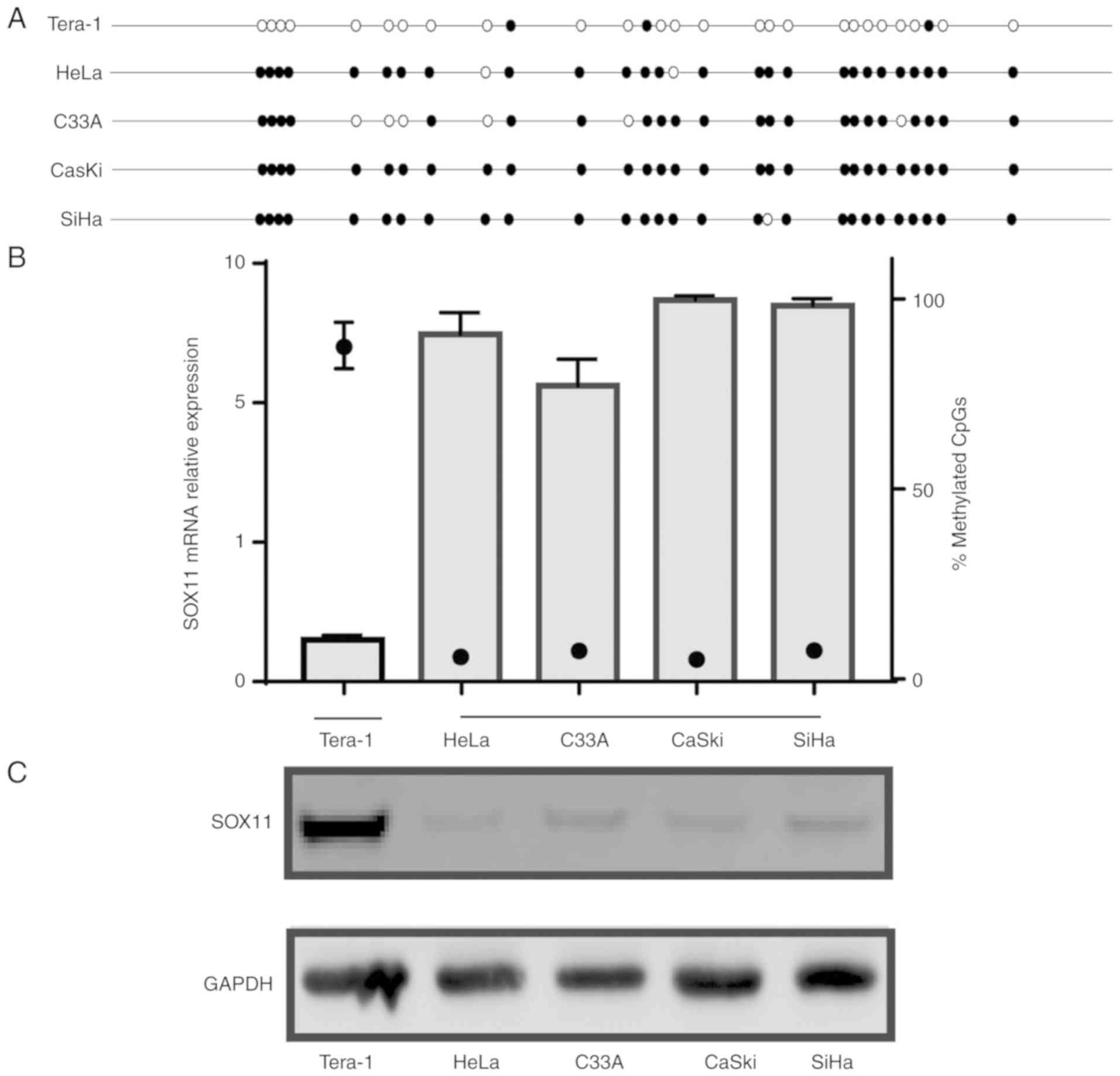
The transcriptional and translational level of SOX11
and its promoter methylation status was examined in CC cell lines.
A marked difference was observed between the SOX11 expression and
promoter methylation level. The Tera-1 cell line as a positive
control. The results revealed high levels of SOX11 promoter
methylation in HeLa, C33A, CaSki and SiHa (90.71, 77.14, 99.64 and
98.21%, respectively) cell lines, consistent with a lack of
SOX11 mRNA and protein expression. By contrast, SOX11
promoter methylation was low in the Tera-1 (10.36%), and its mRNA
and protein expression was significantly higher than those in the
four CC cell lines (Fig. 3). These
findings suggest that SOX11 expression in the CC cell lines is
negatively associated with the methylation level of the promoter,
which is consistent with the results in human tissue specimens.
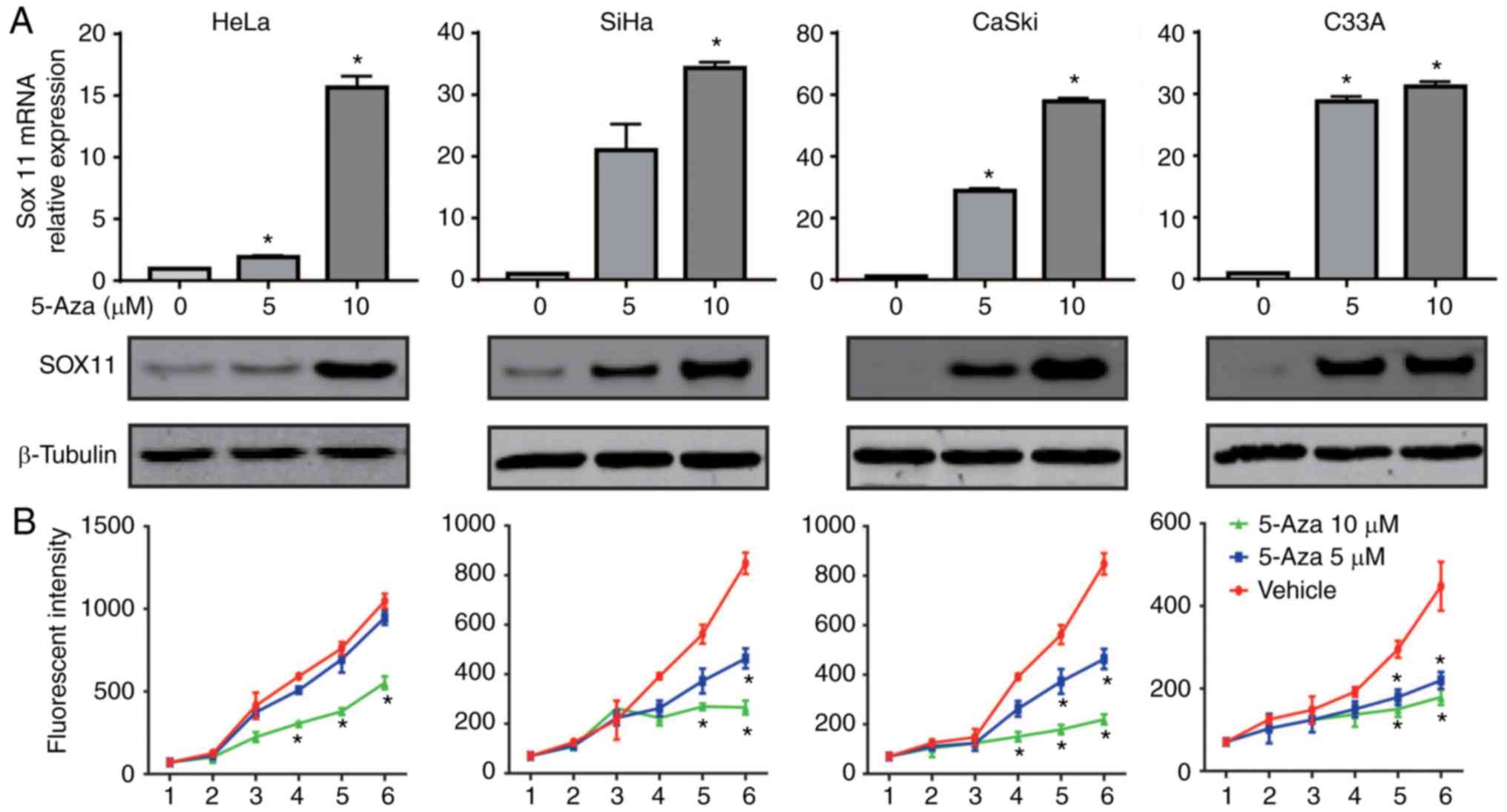
SOX11 expression is enhanced by
5-Aza-dC and increased SOX11 expression inhibits the proliferation
of CC cells
As the silencing of TSGs by aberrant methylation of
the gene promoter is reversible (35), to further investigate the role of
hypermethylated promoter level in the expression regulation of
SOX11, HeLa, C33A, CaSki and SiHa cell lines were treated with
different doses of the demethylating agent 5-Aza-dC. The mRNA and
protein expression levels of SOX11 under different
conditions were measured by RT-qPCR and western blot analysis. As
shown in Fig. 4A, when DNA of
cervical cells was demethylated with different doses of 5-Aza-dC,
the SOX11 mRNA level was increased from 0.95±0.05 to
15.65±0.65 in HeLa, 0.95±0.07 to 34.30±0.70 in C33A, 1.12±0.11 to
57.85±1.05 in CaSki and 1.00±0.12 to 31.2±0.80 in SiHa cells
treated with 10 µM compared with no 5-Aza-dC treatment. Similarly,
the SOX11 protein expression level was gradually increased from
0.78 to 1.42 in HeLa, 0.52 to 1.28 in C33A, 0.38 to 1.36 in CaSki
and 0.70 to 1.38 in SiHa cells comparing no 5-Aza-dC treatment to
10 µM. Cell viability was also to determine the contribution of the
SOX11 expression level to CC cell growth. The proliferation ability
of cervical cells was significantly suppressed by 5-Aza-dC
(Fig. 4B). These results suggest
that the hypermethylation of the promoter reduces SOX11
expression in the four CC cell lines and activation of SOX11
by demethylation of the promoter significantly inhibited the
proliferation of cervical cells.
Discussion
The development of CC is a multi-gene, multi-step
carcinogenesis process that involves complex genetic and epigenetic
mechanisms. DNA methylation is a common form of epigenetic
modification and silencing of TSGs via hypermethylation of their
promoter, particularly CpG islands, is associated with the genesis
of multiple tumors (23,36). Paz et al (37) reported that at least one
hypermethylated gene exists in each of 70 tumor cell lines
analyzed. In addition, studies have demonstrated that cancer
stemness genes in the tumor are downregulated due to methylation,
such as SOX9 in gastric cancer (38), and Krüppel-like factor 4 in multiple
tumors including bladder (39) and
colorectal cancer (40), and CC
(41). Accordingly, changes in gene
methylation status are one of the key factors involved in
carcinogenesis.
Currently, the role of SOX11 in tumor development is
controversial, as it has been associated with both improved and
worsened survival (23–25,42,43).
In recent years, researchers have focused on the relevance of the
SOX11 expression in carcinogenesis (26,28).
Xu et al (28) reported that
the silencing of SOX11 as TSG in gastric cancer cell lines
and primary tissues was due to the hypermethylation of the
SOX11 promoter region, which could be a novel target for the
treatment of GC. Sernbo et al (23) reported that the re-expression of
SOX11 using the demethylating drug 5-Aza-dC in epithelial
ovarian cancer inhibited the growth of ovarian cancer cell lines.
Additionally, hypermethylation of SOX11 contributed to the
downregulation of SOX11 and promoted cell growth and
invasion in nasopharyngeal carcinoma (26). These findings suggest that aberrant
DNA methylation of SOX11 has a major role in the development
of certain malignant tumors.
However, the regulatory mechanism of SOX11
expression is not precisely the same, or even be opposite, in
different malignancies. Histone acetylation can affect the
corresponding chromosome structure, change the level of gene
transcription, and subsequently, affect the cell cycle,
differentiation and apoptosis regulation, which can ultimately lead
to tumor formation. A highly acetylated state usually leads to
transcriptional activation and a deacetylated state often leads to
transcriptional silencing. The levels of histone H3-acetylation at
the SOX11 locus is also associated with transcriptional activity
(44). Vegliante et al
(45) treated mantle cell lymphomas
(MCL) cells with the histone acetylation inhibitor (SAHA) and/or
methyltransferase inhibitor (Aza). The treatment with SAHA can
reversed the expression of the SOX11 gene despite the
methylation state in SOX11 low-expressing cell lines, while Aza did
not increase the expression of SOX11, suggesting that the
repression of SOX11 through promoter methylation is not the
only mechanism involved, as the expression of SOX11 is associated
with histone acetylation. Wasik et al (46) demonstrated high SOX11 mRNA
expression in the majority of the MCL examined. The mRNA and
protein expression of SOX11 in the MCL cell lines, Granta 519 and
Rec-1, was decreased following the administration of the
demethylating 5-Aza-CdR agent, which further indicated that the
regulation of SOX11 gene expression in MCL was not caused by
high methylation of the promoter region. Additionally, there may be
other regulatory mechanisms, such as the common regulatory
mechanisms of the SOXC group (SOX11, SOX4 and SOX12) (46). In the present study, it was
demonstrated SOX11 expression was regulated by its promoter
methylation status in CC. More clinical samples and experimental
data are required to be studied to confirm the involvement of such
a mechanism.
Furthermore, a significant association between SOX11
expression and the presence of HPV was revealed in patients with
CC, LSIL and HSIL. Persistent infection with HPV is a key factor
during the carcinogenesis of CC. The E6 gene of HPV can induce the
degradation of tumor protein 53 (TP53) via ubiquitin-proteasome
pathway, which sequentially inhibits the function of TP53 as a
tumor suppressor gene. Chang et al (47) used a luciferase assay and
glutathione S-transferase pull-down experiments to demonstrate that
SOX11 could interact with TP53 in vitro and promote the
transcriptional activity of TP53. These findings suggested that the
expression of SOX11 during the initiation and development of CC may
be associated with the E6 gene of HPV and the tumor suppressor gene
TP53; however the specific mechanism requires further
investigation.
There are also certain limitations of the present
study. Immunohistochemistry was performed to detect SOX11 and the
expression was quantified. The sections were obtained from the
adjacent sections of which the pathological types had been
confirmed by H&E staining. However, LSIL and HSIL sample
lysates could not be obtained to determine the protein level of
SOX11 by western blotting and to detect the methylation state of
SOX11. The aberrant expression of SOX11 was reversed by the
wide-spectrum demethylating drug, 5-Aza-dC, which suggested the
effects 5-Aza-dC are associated with SOX11, at least in part. A
knock-in SOX11 on CC cells using the inducible Crispr-Cas9 system
to investigate the specific effects of SOX11 demethylation on cell
proliferation and also investigating the role of SOX11 in the
invasive potential CC cells are planned for future studies.
In conclusion, SOX11 expression was significantly
downregulated in CC compared with normal tissue, suggesting that
SOX11 may have a role as a tumor suppressor gene. In addition,
hypermethylation of the SOX11 promoter in CC samples and
cell lines was observed compared with NC and Tara-1 cells.
Additionally, the downregulation of SOX11 were reversed by a
demethylating drug at the mRNA and protein levels, and cell
viability was also inhibited. The SOX11 promoter methylation
is anticipated to be a new molecular marker for the diagnosis of CC
and a treatment target. More experiments are required to confirm
the function of SOX11 in CC. Studies with larger sample sizes and a
long-term follow-up period are required to further investigate the
clinical significance of SOX11 in CC.
Acknowledgements
The authors would like to thank all the participants
who provided tissues for the study.
Funding
The present study was supported by grants from the
Sci-tech Program Foundation of Shaanxi Province (grant no.
2017S-013) and the National Natural Science Foundation of China
(grant no. 81702578).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
YL, YC, XL, NS, MD and YY collected the samples. YL
and YC performed the experiments; XL and RT analyzed and
interpreted the data; YG, XL, RT and NS drafted the article; XL, YG
and XW designed the work; NS polished the language; XL, MD, RT, NS
and YY revised it critically. YG ORCID no. 0000-0002-7894-7312. All
authors read and approved the manuscript and agree to be
accountable for all aspects of the research in ensuring that the
accuracy or integrity of any part of the work are appropriately
investigated and resolved.
Ethics approval and consent to
participate
The design and implementation of the study were
approved by the Ethics Committee of Medical School of Xi'an
Jiaotong University (Shannxi, China; no. 2017-266).
Patient consent for publication
During the study, a written informed consent for the
publication was obtained from all participants.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Ginsburg O, Bray F, Coleman MP, Vanderpuye
V, Eniu A, Kotha SR, Sarker M, Huong TT, Allemani C, Dvaladze A, et
al: The global burden of women's cancers: A grand challenge in
global health. Lancet. 389:847–860. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: Sources, methods and major
patterns in GLOBOCAN 2012. Int J Cancer. 136:E359–E386. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Snijders PJ, Steenbergen RD, Heideman DA
and Meijer CJ: HPV-mediated cervical carcinogenesis: Concepts and
clinical implications. J Pathol. 208:152–164. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Voltaggio L, Cimino-Mathews A, Bishop JA,
Argani P, Cuda JD, Epstein JI, Hruban RH, Netto GJ, Stoler MH,
Taube JM, et al: Current concepts in the diagnosis and pathobiology
of intraepithelial neoplasia: A review by organ system. CA Cancer J
Clin. 66:408–436. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bosch FX and de Sanjosé S: Chapter 1:
Human papillomavirus and cervical cancer-burden and assessment of
causality. J Natl Cancer Inst Monogr. 3–13. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Denny L, de Sanjose S, Mutebi M, Anderson
BO, Kim J, Jeronimo J, Herrero R, Yeates K, Ginsburg O and
Sankaranarayanan R: Interventions to close the divide for women
with breast and cervical cancer between low-income and
middle-income countries and high-income countries. Lancet.
389:861–870. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Castellsagué X and Muñoz N: Chapter 3:
Cofactors in human papillomavirus carcinogenesis-role of parity,
oral contraceptives, and tobacco smoking. J Natl Cancer Inst
Monogr. 20–28. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Louie KS, Castellsague X, de Sanjose S,
Herrero R, Meijer CJ, Shah K, Munoz N and Bosch FX; International
Agency for Research on Cancer Multicenter Cervical Cancer Study
Group, : Smoking and passive smoking in cervical cancer risk:
Pooled analysis of couples from the IARC multicentric case-control
studies. Cancer Epidemiol Biomarkers Prev. 20:1379–1390. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Denslow SA, Rositch AF, Firnhaber C, Ting
J and Smith JS: Incidence and progression of cervical lesions in
women with HIV: A systematic global review. Int J STD AIDS.
25:163–177. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Markowitz LE, Tsu V, Deeks SL, Cubie H,
Wang SA, Vicari AS and Brotherton JM: Human papillomavirus vaccine
introduction-the first five years. Vaccine. 30 (Suppl 5):F139–F148.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bruni L, Diaz M, Barrionuevo-Rosas L,
Herrero R, Bray F, Bosch FX, de Sanjosé S and Castellsagué X:
Global estimates of human papillomavirus vaccination coverage by
region and income level: A pooled analysis. Lancet Glob Health.
4:e453–e463. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Solomon D, Breen N and McNeel T: Cervical
cancer screening rates in the United States and the potential
impact of implementation of screening guidelines. CA Cancer J Clin.
57:105–111. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Saavedra KP, Brebi PM and Roa JC:
Epigenetic alterations in preneoplastic and neoplastic lesions of
the cervix. Clin Epigenetics. 4:132012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Siegel EM, Riggs BM, Delmas AL, Koch A,
Hakam A and Brown KD: Quantitative DNA methylation analysis of
candidate genes in cervical cancer. PLoS One. 10:e01224952015.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Laird PW: The power and the promise of DNA
methylation markers. Nat Rev Cancer. 3:253–266. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Gubbay J, Collignon J, Koopman P, Capel B,
Economou A, Münsterberg A, Vivian N, Goodfellow P and Lovell-Badge
R: A gene mapping to the sex-determining region of the mouse Y
chromosome is a member of a novel family of embryonically expressed
genes. Nature. 346:245–250. 1990. View
Article : Google Scholar : PubMed/NCBI
|
|
17
|
Sinclair AH, Berta P, Palmer MS, Hawkins
JR, Griffiths BL, Smith MJ, Foster JW, Frischauf AM, Lovell-Badge R
and Goodfellow PN: A gene from the human sex-determining region
encodes a protein with homology to a conserved DNA-binding motif.
Nature. 346:240–244. 1990. View
Article : Google Scholar : PubMed/NCBI
|
|
18
|
Sarkar A and Hochedlinger K: The sox
family of transcription factors: Versatile regulators of stem and
progenitor cell fate. Cell Stem Cell. 12:15–30. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Castinetti F, Davis SW, Brue T and Camper
SA: Pituitary stem cell update and potential implications for
treating hypopituitarism. Endocr Rev. 32:453–471. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Weigle B, Ebner R, Temme A, Schwind S,
Schmitz M, Kiessling A, Rieger MA, Schackert G, Schackert HK and
Rieber EP: Highly specific overexpression of the transcription
factor SOX11 in human malignant gliomas. Oncol Rep. 13:139–144.
2005.PubMed/NCBI
|
|
21
|
Stuart JE, Lusis EA, Scheck AC, Coons SW,
Lal A, Perry A and Gutmann DH: Identification of gene markers
associated with aggressive meningioma by filtering across multiple
sets of gene expression arrays. J Neuropathol Exp Neurol. 70:1–12.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lopez FJ, Cuadros M, Cano C, Concha A and
Blanco A: Biomedical application of fuzzy association rules for
identifying breast cancer biomarkers. Med Biol Eng Comput.
50:981–990. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Sernbo S, Gustavsson E, Brennan DJ,
Gallagher WM, Rexhepaj E, Rydnert F, Jirström K, Borrebaeck CA and
Ek S: The tumour suppressor SOX11 is associated with improved
survival among high grade epithelial ovarian cancers and is
regulated by reversible promoter methylation. BMC Cancer.
11:4052011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Brennan DJ, Ek S, Doyle E, Drew T, Foley
M, Flannelly G, O'Connor DP, Gallagher WM, Kilpinen S, Kallioniemi
OP, et al: The transcription factor Sox11 is a prognostic factor
for improved recurrence-free survival in epithelial ovarian cancer.
Eur J Cancer. 45:1510–1517. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Gustavsson E, Sernbo S, Andersson E,
Brennan DJ, Dictor M, Jerkeman M, Borrebaeck CA and Ek S: SOX11
expression correlates to promoter methylation and regulates tumor
growth in hematopoietic malignancies. Mol Cancer. 9:1872010.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhang S, Li S and Gao JL: Promoter
methylation status of the tumor suppressor gene SOX11 is associated
with cell growth and invasion in nasopharyngeal carcinoma. Cancer
Cell Int. 13:1092013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2−ΔΔCT method. Methods. 25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Xu X, Chang X, Li Z, Wang J, Deng P, Zhu
X, Liu J, Zhang C, Chen S and Dai D: Aberrant SOX11 promoter
methylation is associated with poor prognosis in gastric cancer.
Cell Oncol. 38:183–194. 2015. View Article : Google Scholar
|
|
29
|
Yao Z, Sun B, Hong Q, Yan J, Mu D, Li J,
Sheng H and Guo H: The role of tumor suppressor gene SOX11 in
prostate cancer. Tumour Biol. 36:6133–6138. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Czapiewski P, Gorczynski A, Radecka K,
Wiewiora C, Haybaeck J, Adam P, Fend F, Zakrzewska M, Zakrzewski K,
Liberski PP, et al: Expression of SOX11, PAX5, TTF-1 and ISL-1 in
medulloblastoma. Pathol Res Pract. 212:965–971. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Esteller M: Epigenetics in cancer. N Engl
J Med. 358:1148–1159. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Jones PA and Baylin SB: The epigenomics of
cancer. Cell. 128:683–692. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Jones A, Lechner M, Fourkala EO,
Kristeleit R and Widschwendter M: Emerging promise of epigenetics
and DNA methylation for the diagnosis and management of women's
cancers. Epigenomics. 2:9–38. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Szalmás A and Kónya J: Epigenetic
alterations in cervical carcinogenesis. Semin Cancer Biol.
19:144–152. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Ronnekleiv-Kelly SM, Sharma A and Ahuja N:
Epigenetic therapy and chemosensitization in solid malignancy.
Cancer Treat Rev. 55:200–208. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Fang G, Liu J, Wang Q, Huang X, Yang R,
Pang Y and Yang M: MicroRNA-223-3p regulates ovarian cancer cell
proliferation and invasion by targeting SOX11 expression. Int J Mol
Sci. 18:E12082017. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Paz MF, Fraga MF, Avila S, Guo M, Pollan
M, Herman JG and Esteller M: A systematic profile of DNA
methylation in human cancer cell lines. Cancer Res. 63:1114–1121.
2003.PubMed/NCBI
|
|
38
|
Sun M, Uozaki H, Hino R, Kunita A,
Shinozaki A, Ushiku T, Hibiya T, Takeshita K, Isogai M, Takada K,
et al: SOX9 expression and its methylation status in gastric
cancer. Virchows Arch. 460:271–279. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Li H, Wang J, Xiao W, Xia D, Lang B, Wang
T, Guo X, Hu Z, Ye Z and Xu H: Epigenetic inactivation of KLF4 is
associated with urothelial cancer progression and early recurrence.
J Urol. 191:493–501. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Zhao W, Hisamuddin IM, Nandan MO, Babbin
BA, Lamb NE and Yang VW: Identification of Krüppel-like factor
4 as a potential tumor suppressor gene in colorectal cancer.
Oncogene. 23:395–402. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Yang WT and Zheng PS: Promoter
hypermethylation of KLF4 inactivates its tumor suppressor function
in cervical carcinogenesis. PLoS One. 9:e888272014. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Wang X, Asplund AC, Porwit A, Flygare J,
Smith CI, Christensson B and Sander B: The subcellular Sox11
distribution pattern identifies subsets of mantle cell lymphoma:
Correlation to overall survival. Br J Haematol. 143:248–252. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Fernàndez V, Salamero O, Espinet B, Solé
F, Royo C, Navarro A, Camacho F, Beà S, Hartmann E, Amador V, et
al: Genomic and gene expression profiling defines indolent forms of
mantle cell lymphoma. Cancer Res. 70:1408–1418. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Usui A, Iwagawa T, Mochizuki Y, Iida A,
Wegner M, Murakami A and Watanabe S: Expression of Sox4 and Sox11
is regulated by multiple mechanisms during retinal development.
FFBS Lett. 587:358–363. 2013. View Article : Google Scholar
|
|
45
|
Vegliante MC, Royo C, Palomero J,
Salaverria I, Balint B, Martín-Guerrero I, Agirre X, Lujambio A,
Richter J, Xargay-Torrent S, et al: Epigenetic activation of SOX11
in lymphoid neoplasms by histone modifications. PLoS One.
6:e213822011. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Wasik AM, Lord M, Wang X, Zong F,
Andersson P, Kimby E, Christensson B, Karimi M and Sander B: SOXC
transcription factors in mantle cell lymphoma: The role of promoter
methylation in SOX11 expression. Sci Rep. 3:14002013. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Chang Y, Zhang WN, Teng LI, et al: SOX11
interacts with p53 and increases its transcriptional activity.
http://en.cnki.com.cn/Journal_en/A-A006-SWTX-2010-05.htmLett
Biotechnol. 2010.
|