Introduction
Approximately 25% of newly diagnosed urothelial
carcinoma (UC) presents as biologically aggressive muscle-invasive
disease (1). Clinical studies have
shown a survival benefit of neoadjuvant or adjuvant cisplatin
(CDDP)-based chemotherapy for patients with muscle-invasive UC
before or after radical cystectomy (2). CDDP-based chemotherapy has generally
produced a complete or partial response in ~50–70% of patients with
UC (3). However, tumors treated
with CDDP ultimately acquire CDDP resistance. Although considerable
efforts have been devoted to solving CDDP resistance over the past
three decades (4,5), the mechanisms of CDDP resistance have
yet to be fully elucidated. Therefore, clarifying new molecular
mechanisms underlying CDDP resistance hold great importance in
improving chemotherapy outcomes for muscle-invasive UC.
Long non-coding RNAs (lncRNAs), which are >200
bp, are a subtype of non-coding RNAs. Although they lack
protein-coding capacity, lncRNAs play an essential role in the
development and progression of UC (6–8). The
transcribed ultraconserved regions (T-UCRs) are a relatively new
class of lncRNAs, which are highly conserved among vertebrates
(9). Considering that T-UCRs are
conserved across species, they allegedly play critical roles in
human development and disease, including cancer. T-UCRs are
differentially expressed and act in an oncogenic or
tumor-suppressive role according to the context of the cancer
(10,11). Based on this evidence, T-UCRs may
provide useful diagnostic markers for some specific types of cancer
and could represent potential therapeutic targets for cancer
treatment. Uc.63+ (278 bp) is located on the chromosome 2p15, in
the third intron of the XPO1 gene (9). A recent study revealed that Uc.63+ was
induced in a hypoxia-dependent manner (12). Furthermore, the expression of Uc.63+
was revealed to be upregulated in breast cancer and was correlated
with a poor prognosis (13). In
addition, we previously revealed that Uc.63+ promoted docetaxel
resistance through regulation of androgen receptor (AR) in prostate
cancer (14). These findings
indicated that Uc.63+ may contribute to progression in several
types of cancers. However, there is no evidence of the role of
Uc.63+ in UC. In the present study, we evaluated the expression and
functional role of Uc.63+ in UC and we analyzed the effect of
Uc.63+ on the expression of AR and CDDP resistance.
Materials and methods
Cell lines
Prostate cancer cell line, LNCaP, was purchased from
the American Type Culture Collection (ATCC; Manassas, VA, USA). The
three human UC cell lines (RT112, T24 and UMUC3) were provided by
the Vancouver Prostate Centre (Vancouver, BC, Canada). Cell lines
were authenticated by DNA fingerprinting using Amp FISTR
Amplification or Amp FISTR Profiler PCR Amplification protocols
(Life Technologies; Thermo Fisher Scientific, Inc., Waltham, MA,
USA). RT112 and T24 cell lines were maintained in RPMI-1640 medium
(Nissui Pharmaceutical Co. Ltd., Tokyo, Japan) containing 10% fetal
bovine serum (FBS; BioWhittaker, Walkersville, MD, USA), 2 mM
L-glutamine, 50 U/ml penicillin, and 50 g/ml streptomycin in a
humidified atmosphere of 5% CO2 and 95% air at 37°C.
LNCaP cells were withdrawn from hormone effects in the medium by
culture with medium containing charcoal/dextran-stripped FBS for 2
days before treatment. Dehydrotestosteron (DHT) (10 nM) or vehicle
control (0.5% ethanol) was added to the cells. CDDP-resistant UMUC3
cells were also provided by the Vancouver Prostate Centre. The
method used to establish CDDP-resistant UMUC3 cells was previously
described (15).
Tissue samples
We used 8 non-neoplastic bladder tissues and 16 UC
tissue samples for quantitative reverse transcription-polymerase
chain reaction (qRT-PCR) (Table I).
The samples were collected from patients at Hiroshima University
Hospital (Hiroshima, Japan) from April 2010 to October 2018. The
Institutional Review Board of Hiroshima University Hospital
approved the present study (IRB# E912). Appropriate written
informed consent was obtained from each patient. The present study
was conducted in accordance with the Ethical Guidance for Human
Genome/Gene Research of the Japanese Government.
 | Table I.Clinicopathological characteristics
of 16 BCa tissues. |
Table I.
Clinicopathological characteristics
of 16 BCa tissues.
| Number of
cases | 16 |
|---|
| Sex |
|
|
Male | 16 |
| Median
age (years) | 74 (63–92) |
| Race |
|
|
Asian | 16 |
| Histology |
|
|
Urothelial carcinoma | 16 |
| Tumor grade |
|
| G1 | 3 |
| G2 | 4 |
| G3 | 9 |
| Pathological T
stage |
|
|
pT1 | 7 |
|
pT2 | 4 |
|
pT3 | 5 |
| Pathological N
stage |
|
| 0 | 15 |
| 1 | 1 |
| Metastasis at time
of diagnosis |
|
|
Absence | 16 |
|
Presence | 0 |
Quantitative RT-PCR analysis
Extraction of total RNA, synthesis of cDNA and
qRT-PCR were performed as previously described (14). ACTB-specific PCR products,
which were amplified from the same RNA samples, served as internal
controls. The primer sequences and IDs are summarized in Table II.
 | Table II.Primer sequences for qRT-PCR. |
Table II.
Primer sequences for qRT-PCR.
|
| Forward primer | Reverse primer |
|---|
| Uc.63+ |
TTGCATAAAAGCCAAATGTCA |
CTGTTTGCTTGCCTGGTAAA |
| AR |
GACGCTTCTACCAGCTCACC |
GAAAGGATCTTGGGCACTTG |
| ACTB |
TCACCGAGCGCGGCT |
TAATGTCACGCACGATTTCCC |
RNA interference and expression
vector
Silencer® Select (Ambion, Austin, TX,
USA) against Uc.63+ was used for RNA interference as previously
described (14). Transfection was
performed using Lipofectamine RNAiMAX (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's instructions.
Cells were used 48 h after transfection in each of the experiments
and assays.
For constitutive expression of Uc.63+, cDNA was PCR
amplified and subcloned into pcDNA 3.1 (Invitrogen; Thermo Fisher
Scientific, Inc.) as previously described (14). The pcDNA-Uc.63+ expression vector
was transfected into RT112 and UMUC3 cells with FuGENE6 (Roche
Diagnostics, Basel, Switzerland) according to the manufacturer's
instructions.
For constitutive expression of human AR, cDNA was
PCR amplified from normal prostate tissues and subcloned into
pDON-5 Neo (Takara Biotechnology Co., Ltd., Dalian, China) using a
retrovirus vector with psPAX2 envelope and pMD2.G packaging
plasmids, according to the manufacturer's instructions.
Cell proliferation assay
To examine cell proliferation, an MTT assay was
performed as previously described (16). Cell proliferation was monitored
after 1, 2 and 4 days.
Cell death ELISA
Cells were seeded in 12-well plates
(1×105 cells). Mononucleosomes and oligonucleosomes in
the cytoplasmic fraction were assessed by a Cell Death Detection
ELISA kit (Roche Diagnostics) according to the manufacturer's
instructions. Absorbance was determined at 405 nm.
Western blot analysis
For western blot analysis, cells were lysed as
previously described (17).
Proteins in UC cell lines were extracted using RIPA buffer and
subjected to concentration measurements using the BCΑ kit. The
lysates (40 µg) were solubilized in Laemmli sample buffer by
boiling and then subjected to 10% SDS-polyacrylamide gel
electrophoresis followed by electrotransfer onto a nitrocellulose
membrane. Cleaved PARP (c-PARP) (cat. no. 9542; Cell Signaling
Technology, Inc., Danvers, MA, USA) and AR antibody (cat. no.
MA5-13423; Thermo Fisher Scientific, Inc.) were used as the primary
antibodies with a 1:1,000 dilution. β-actin (Sigma-Aldrich; Merck
KGaA, Darmstadt, Germany) was detected as a loading control.
Peroxidase-conjugated anti-mouse IgG (cat. no. 330; MBL, Nagoya,
Japan) or anti-rabbit IgG (cat. no. 458; MBL) were used as the
secondary antibodies with a 1:500 dilution. Immunocomplexes were
visualized with an ECL Western Blot Detection system (Amersham
Biosciences; GE Healthcare, Chicago, IL, USA).
Drug treatment
CDDP (Nippon Kayaku Co., Ltd., Tokyo, Japan) was
obtained and handled according to the manufacturer's
recommendations. Cell lines treated with vehicle (0.5% ethanol) or
escalating doses of CDDP were assessed for cell viability. An MTT
assay was performed at 48 h after CDDP chemotherapy (15). Drug sensitivity curves and
IC50 values were calculated using GraphPad Prism 4.0
software (GraphPad Software Inc., San Diego, CA, USA).
Statistical analysis
All experiments were repeated at least three times
with each sample in triplicate. The results are expressed as the
mean ± standard deviation (SD) of triplicate measurements. Sample
sizes for relevant experiments were determined by power analysis.
Statistical differences were evaluated using the Mann-Whitney U
test. A P-value of <0.05 was considered to indicate a
statistically significant result. One-way analysis of variance
(ANOVA) followed by Tukey's test was used for the comparison of
multiple groups. Spearman's correlation coefficient was used to
examine the degree of relations between two groups. Statistical
analyses were conducted primarily using GraphPad Prism software
(version 4.0; GraphPad Software Inc., La Jolla, CA, USA).
Results
Expression of Uc.63+ is upregulated in
UC tissues and UC cell lines
We analyzed the expression of Uc.63+ in 16 UC
tissues and 8 non-neoplastic urinary bladder tissues using qRT-PCR.
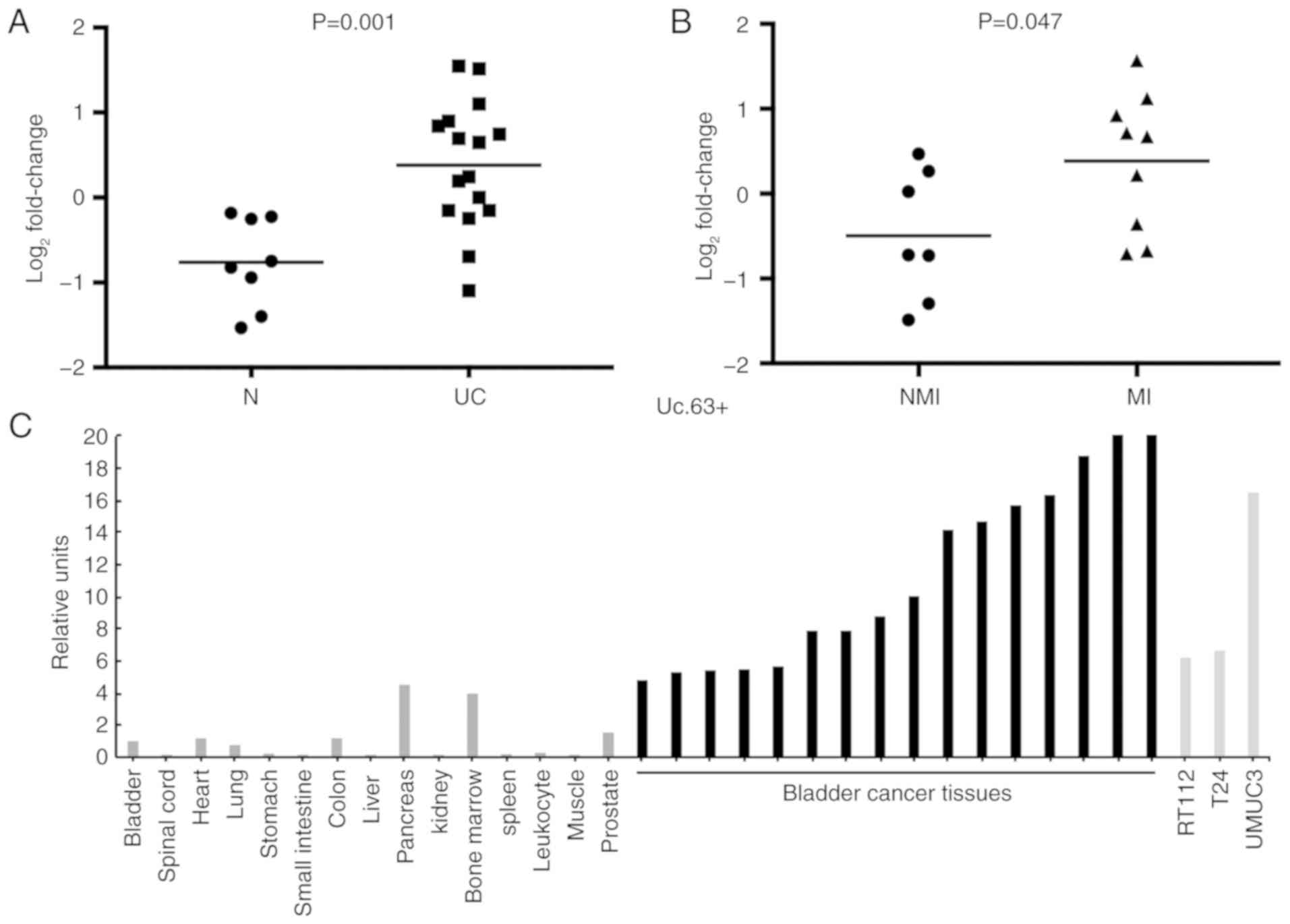
The expression of Uc.63+ was upregulated in 69% (11/16) of UC
tissues compared with that in non-neoplastic urinary bladder
tissues (P=0.001; Fig. 1A). Despite
the small sample sets, the expression of Uc.63+ was significantly
higher in muscle invasive UC than that in non-muscle invasive UC
(P=0.047) (Fig. 1B). As revealed in
Fig. 1C, the expression of Uc.63+
was higher in UC tissues and UC cell lines than that in the 15
types of normal tissue samples including a normal urinary bladder
tissue.
Uc.63+ acts as an oncogene and is
associated with cell proliferation and apoptosis
To clarify the biological roles of Uc.63+ in UC, we
investigated the effects on the knockdown of Uc.63+ using small
interfering RNA (siRNA) that was previously designed to
specifically target Uc.63+ (14).
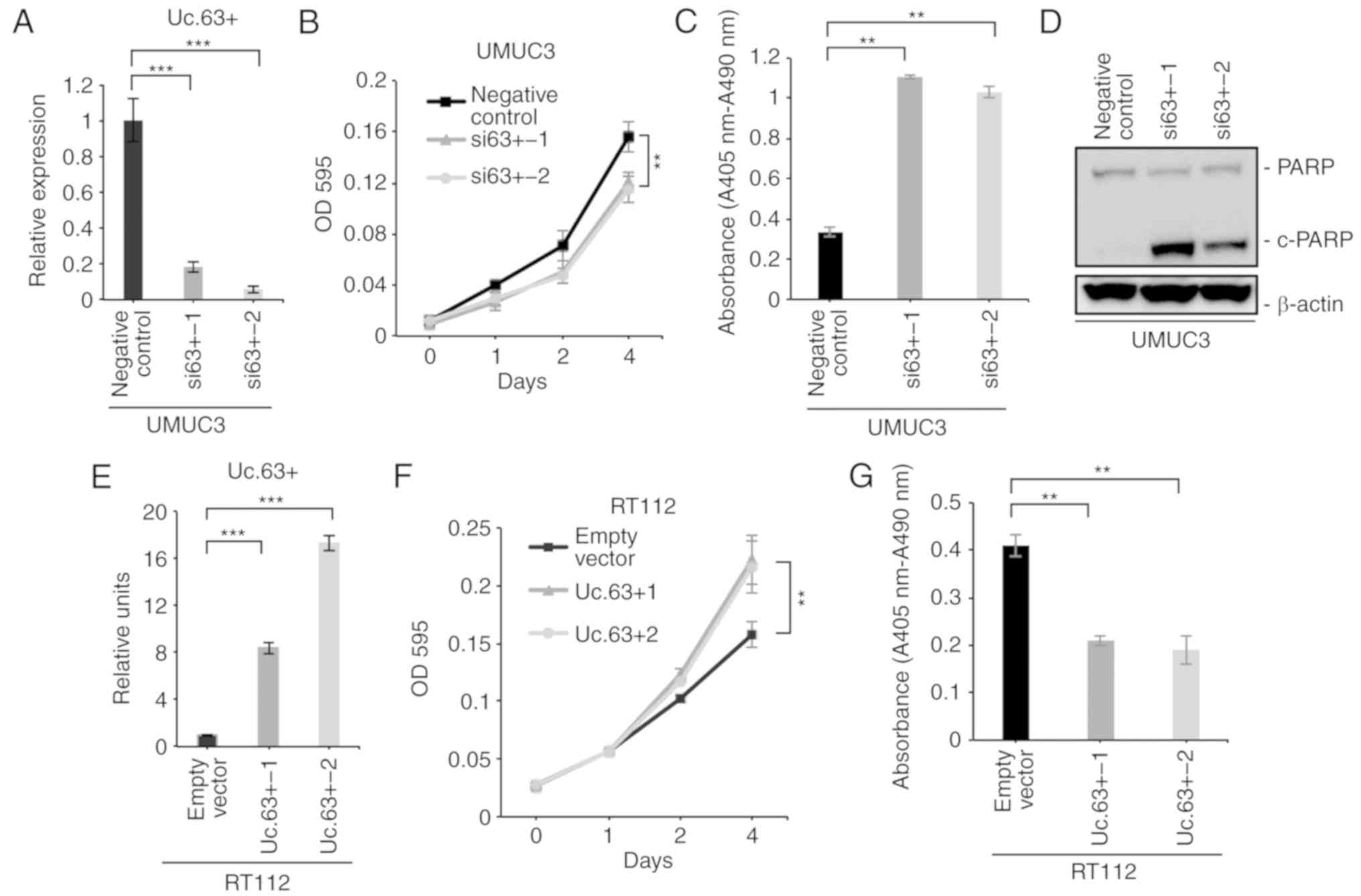
We confirmed a significantly lower expression of Uc.63+ with two
different siRNAs than with the negative control in UMUC3 cells
(Fig. 2A). Next, we performed a
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT)
assay and observed that the downregulation of Uc.63+ significantly
suppressed cell proliferation of UMUC3 (Fig. 2B). A recent study revealed that
Uc.63+ induced apoptosis in breast cancer (13). Therefore, we analyzed the effect of
knockdown of Uc.63+ on apoptosis. Cell d eath ELISA revealed that
knockdown of Uc.63+ significantly induced apoptosis compared to the
negative control (Fig. 2C). Western
blotting revealed that knockdown of Uc.63+ enhanced the expression
of cleaved PARP which was used as the activation status of
apoptosis (Fig. 2D). To further
verify whether Uc.63+ plays an important role in cell proliferation
and apoptosis, we transfected the Uc.63+ expression vector into
RT112, which had low expression of Uc.63+. As anticipated, the MTT
assay revealed that the overexpression of Uc.63+ significantly
increased cell proliferation (Fig. 2E
and F). Cell death ELISA revealed that the overexpression of
Uc.63+ significantly suppressed apoptosis compared to the empty
vector (Fig. 2G). These results
indicated that Uc.63+ was involved in cell proliferation and
apoptosis in UC.
Uc.63+ modulates the expression of
AR
Several studies have reported that AR is involved in
the development and progression of UC (18,19).
Previously, we revealed that overexpression of Uc.63+ enhanced the
expression of AR (14). Given these
findings, we analyzed the association between Uc.63+ and AR in UC.
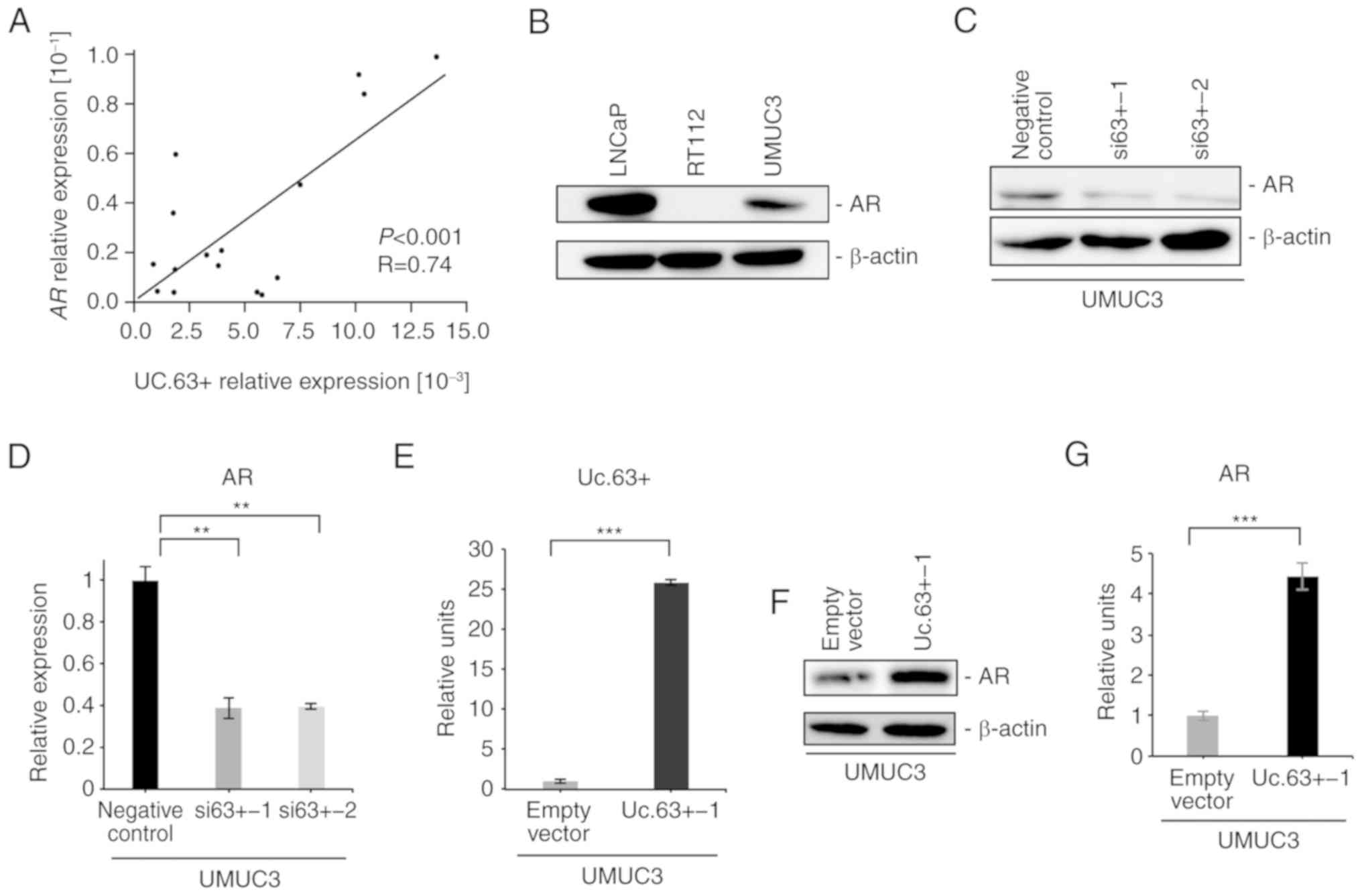
We investigated the expression of AR in 16 UC tissues by qRT-PCR
and found that the expression of AR was significantly correlated
with the expression of Uc.63+ (P<0.001, R=0.74) (Fig. 3A). Western blotting revealed that
the expression of AR was detected in UMUC3 cells, which was
consistent with previous studies (19,20)
(Fig. 3B). To further investigate
the interaction between Uc.63+ and AR, we examined the effect of
Uc.63+ deregulation on the expression of AR. Knockdown of Uc.63+
suppressed the expression of AR in UMUC3 cells at the mRNA and
protein levels (Fig. 3C and D). We
confirmed a significantly higher expression of Uc.63+ with Uc.63+
expression vector than with the empty vector in UMUC3 cells
(Fig. 3E). Overexpression of Uc.63+
induced the expression of AR in UMUC3 cells at the mRNA and protein
levels in UMUC3 cell lines (Fig. 3F and
G). In addition, upregulation of Uc.63+ did not affect the
expression of AR in RT112 cells (data not shown). These results
indicated that Uc.63+ modulated the expression of AR in UMUC3
cells.
Uc.63+ promotes CDDP resistance
through AR
A recent study revealed that AR activity modulated
CDDP sensitivity (21). We
transfected the AR expression vector into RT112 cells, which do not
express AR (Fig. 3B). The
expression of AR was upregulated with AR expression vector compared
to that with the empty vector in RT112 cells as determined by
western blotting (Fig. 4A). We
measured cell viability in RT112 cells with empty vector or AR
expression vector under 0, 5, 10, 20 and 40 µM of CDDP using an MTT
assay. Overexpression of AR decreased CDDP sensitivity in RT112
cells (Fig. 4B), which was
consistent with a previous study (21). To clarify the effect of Uc.63+ on
CDDP resistance, we measured the cell viability in UMUC3 cells with
deregulation of Uc.63+ under 0, 5, 10, 20 and 40 µM of CDDP using
an MTT assay. The IC50 value of UMUC3 cells transfected
with the negative control was higher than that of UMUC3 cells
transfected with siRNAs for Uc.63+ (Fig. 4C). As anticipated, overexpression of
Uc.63+ promoted CDDP resistance in UMUC3 cells (Fig. 4D). Conversely, overexpression of
Uc.63+ did not affect CDDP sensitivity in RT112 cells (Fig. 4E). Collectively, these results
indicated that Uc.63+ may in some way be involved in the
acquisition of CDDP resistance through AR regulation.
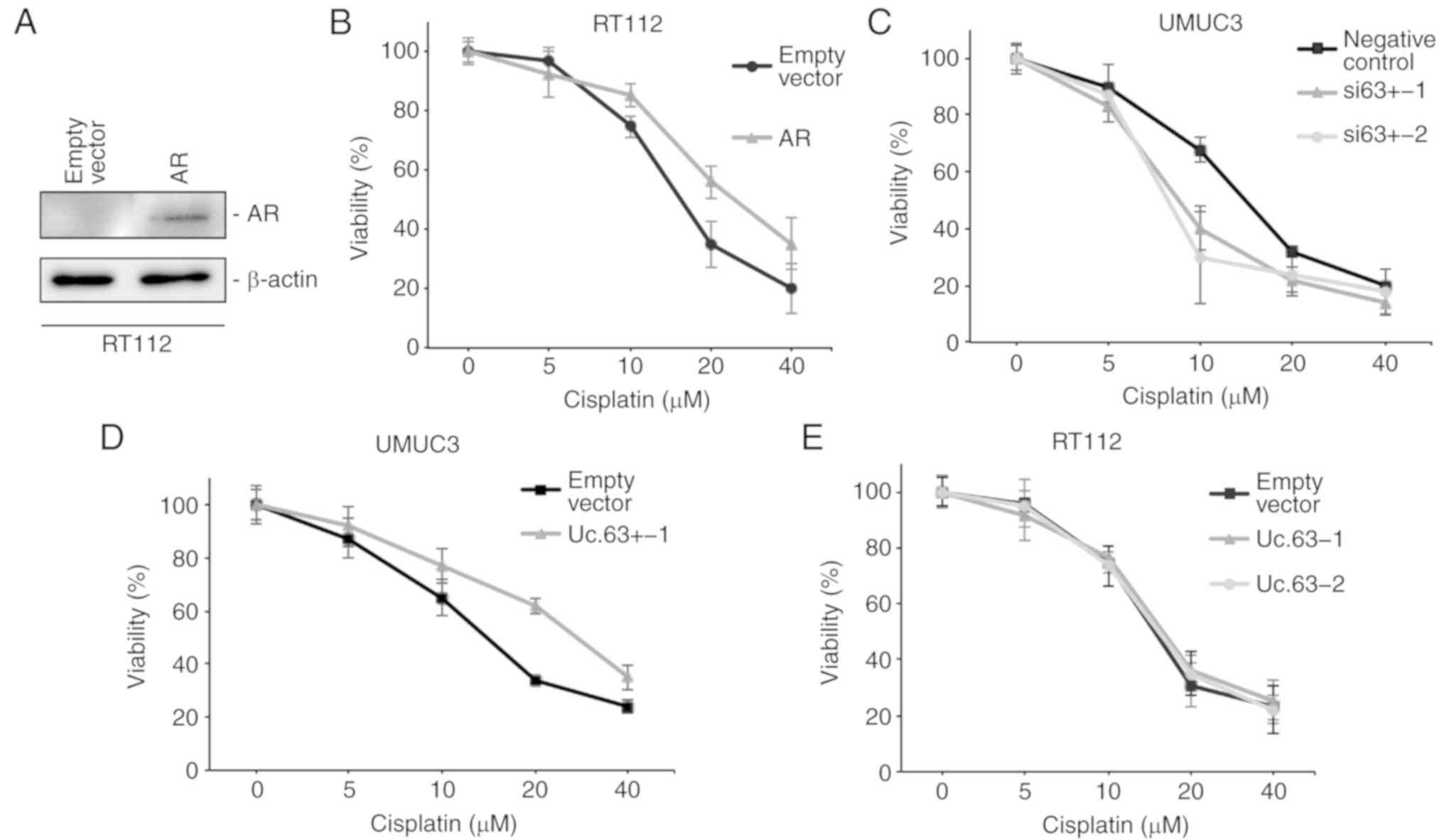 | Figure 4.Effect of Uc.63+ on cisplatin
sensitivity in bladder cancer cell lines. (A) Western blotting of
AR in RT112 cells transfected with empty vector or AR expression
vector. β-actin was used as a loading control. (B) Dose-dependent
effect of cisplatin on the viability of RT112 cells transfected
with empty vector or AR expression vector. The IC50
value of the empty vector, 15.3 µM; the IC50 value of
AR, 25.9 µM. (C) Dose-dependent effect of cisplatin on the
viability of UMUC3 cells transfected with negative control or two
different siRNAs. The IC50 of the negative control, 14.1
µM; the IC50 value of si63+-1, 8.5 µM; the
IC50 value of si63+-2, 7.7 µM. (D) Dose-dependent effect
of cisplatin on the viability of UMUC3 cells transfected with empty
vector or Uc.63+ expression vector. The IC50 value of
the empty vector, 13.9 µM; the IC50 value of Uc.63+-1,
27.1 µM. (E) Dose-dependent effect of CDDP on the viability of
RT112 cells transfected with empty vector or Uc.63+ expression
vector. The IC50 value of the empty vector, 14.7 µM, the
IC50 value of Uc.63+-1, 15.6 µM; the IC50
value of Uc.63+-2, 15.2 µM. AR, androgen receptor; siRNA, small
interfering RNA. |
Inhibition of Uc.63+ reverses CDDP
resistance
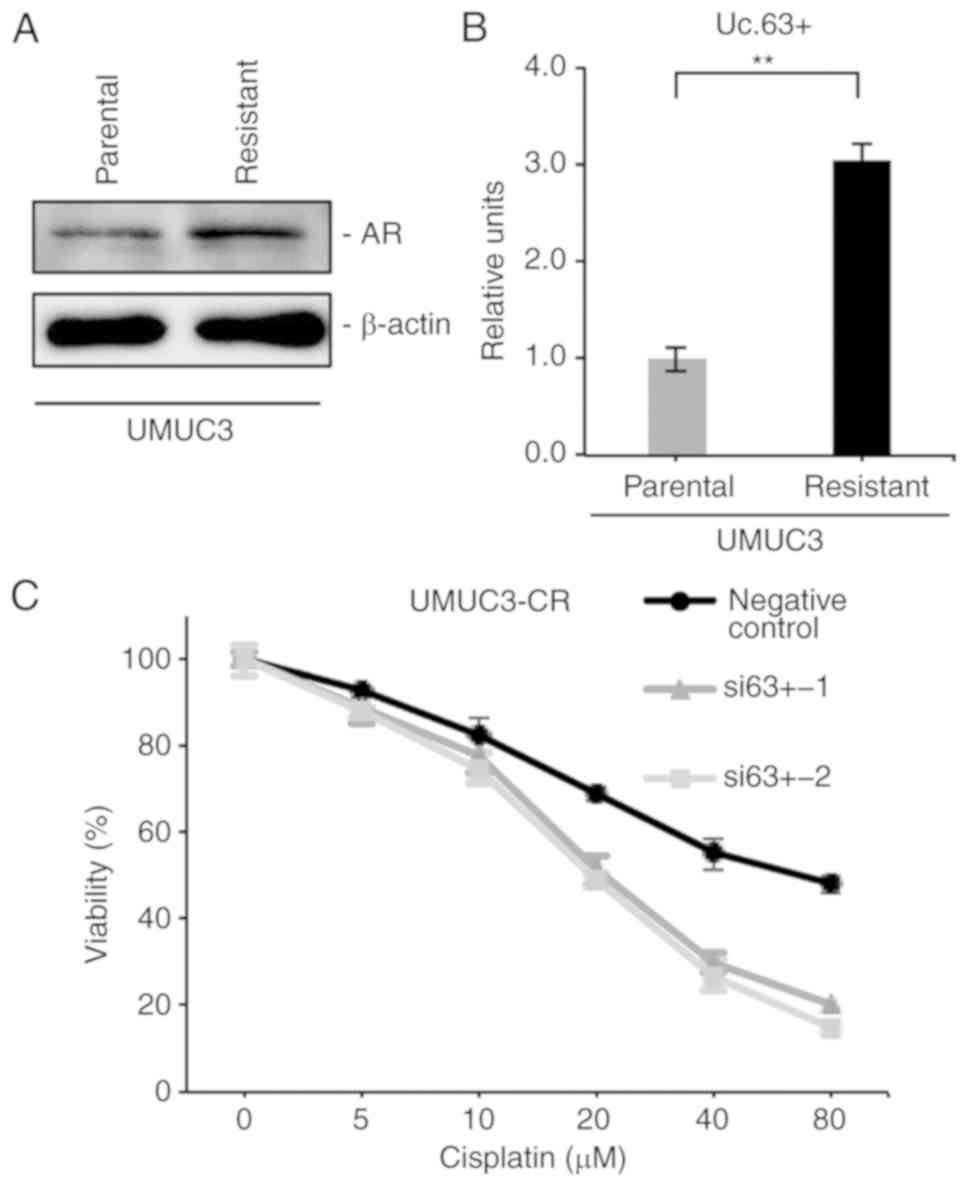
We previously established CDDP-resistant UMUC3 cells
(UMUC3-CR) by long-term culture with low to increasing doses of
CDDP (15) and confirmed that
UMUC3-CR cells were more resistant to CDDP than were parental UMUC3
cells (data not shown). Recent evidence demonstrated that the
expression of AR was enhanced in CDDP-resistant UC cell lines
(21). Western blotting revealed
that the expression of AR was upregulated in UMUC3-CR cells
compared with that in parental UMUC3 cells (Fig. 5A). Furthermore, we investigated the
involvement of Uc.63+ in CDDP resistance. qRT-PCR revealed that the
expression of Uc.63+ was higher in UMUC3-CR cells than in parental
UMUC3 cells (Fig. 5B). We measured
the cell viability in UMUC3-CR cells with knockdown of Uc.63+ under
0, 5, 10, 20, 40 and 80 µM of CDDP. The IC50 value of
UMUC3-CR cells transfected with the negative control was higher
than that of UMUC3 cells transfected with siRNAs (Fig. 5C). Collectively, these results
indicated that inhibition of Uc.63+ re-sensitized the UMUC3-CR
cells to CDDP.
Discussion
Transcribed ultraconserved regions (T-UCRs) are
highly conserved among most of the vertebrates, implying that
T-UCRs could play an important role in biological processes
compared with other non-coding RNAs (9). Several recent studies have shown that
cancer-specific T-UCRs are involved in cancer progression and
tumorigenesis in some types of cancer (11,22).
To date, it has been reported that upregulation of Uc.8+ increased
the expression of MMP9 and promoted cancer progression in
urothelial carcinoma (UC) (23).
However, the role of T-UCRs in UC remains unclear. The present
study, which is the first, to the best of our knowledge, to
investigate the expression and biological role of Uc.63+ in UC,
revealed that the expression of Uc.63+ was upregulated in UC
compared with 15 types of normal tissue samples including normal
bladder tissue. In addition, we revealed that Uc.63+ was involved
in cell proliferation and apoptosis in vitro. These results
indicated that Uc.63+ was more likely to contribute to cancer
progression than tumorigenesis in UC.
Some lines of evidence have recently indicated that
androgen receptor (AR) signaling plays a pivotal role in
carcinogenesis and cancer progression in UC (20,24).
Retrospective clinical studies have revealed that androgen
deprivation therapy prevents the recurrence of UC in male patients
(25,26). One recent study revealed that lncRNA
XIST interacted with miR-124 to promote cell proliferation,
invasion and migration through AR in UC (27). Although the essential role of AR in
the development and progression of UC has been reported, little
evidence is available with regard to the regulation of AR in UC. In
the present study, the expression of AR was significantly disrupted
by the overexpression or knockdown of Uc.63+. Furthermore, qRT-PCR
revealed that there was a positive association between Uc.63+ and
AR in UC tissues. Previously, we revealed that Uc.63+ modulated AR
expression in prostate cancer (14). Collectively, these data indicated
that Uc.63+ may play an important role in the AR pathway in both UC
and prostate cancer. However, the mechanism of how Uc.63+ regulates
the expression of AR remains unclear. To date, there is well known
evidence indicating that microRNA-T-UCR interactions contribute to
tumorigenesis and cancer progression in some types of cancer
(22,28). There may be numerous miRNAs that are
potentially regulated by Uc.63+, which may explain how Uc.63+
regulates the expression of AR. Although further studies are
required to elucidate the mechanism of regulation of AR by Uc.63+,
the interaction between Uc.63+ and AR may play a decisive role in
cancer progression in UC.
Cisplatin (CDDP) has been widely used for the
treatment of muscle-invasive UC. However, the elevated incidence of
CDDP resistance is the main obstacle in clinical practice.
Molecular mechanisms of CDDP resistance are believed to be caused
by multiple factors including drug transport, detoxification, DNA
repair and apoptosis (4,5). A recent study revealed that AR
activation resulted in induction of CDDP resistance and modulated
CDDP sensitivity (21). Another
study reported that the expression of AR was increased in
CDDP-resistant cell lines in endometrial carcinoma (29). In the presents study, we revealed
that upregulation of AR decreased CDDP sensitivity in RT112 cells,
which is an AR-negative cell line, and that the disrupted
expression of Uc.63+ modulated CDDP sensitivity in UMUC3 cells,
which is an AR-positive cell line. Overexpression of Uc.63+ had no
effect on CDDP sensitivity in RT112 cells. Collectively, these
results indicated that Uc.63+ may be responsible for CDDP
resistance through AR in UC. To the best of our knowledge, these
findings represent the first evidence that CDDP resistance can be
regulated by a specific T-UCR. In current cancer treatments,
different types of chemotherapeutic agents are often combined to
improve efficacy and to minimize toxicity. In the present study,
inhibition of Uc.63+ re-sensitized UMUC3-CR cells to CDDP
treatment. Furthermore, we observed that the expression of Uc.63+
was higher in UC tissues than in various normal tissue samples,
indicating that the combination therapy of a Uc.63 inhibitor and
CDDP may be a promising strategy to overcome CDDP resistance with
fewer adverse effects.
There are some limitations in the present study. One
of the biggest issues of the present study was that the sample size
for Uc.63+ expression by qRT -PCR was relatively small, which
signifies that our results may not apply to most patients with UC.
Therefore, a study with a larger number of patients with UC will be
necessary to further verify this current data. The other issue is
that we could not show solid evidence that underpins the
involvement of Uc.63+ in apoptosis. In order to confirm this
novelty, validation of the effect of modulation of Uc.63+ on cell
cycle distribution and morphological change in addition to cell
death ELISA and analysis of the expression of c-PARP should be
performed.
In summary, our results revealed that the expression
of Uc.63+ was upregulated in UC tissues and UC cell lines. Uc.63+
acted as an oncogene and was associated with cell proliferation. We
also revealed that Uc.63+ modulated the expression of AR and
promoted CDDP resistance through AR. Furthermore, inhibition of
Uc.63+ reversed CDDP resistance in vitro. The data presented
here highlight the potential of Uc.63+ as a therapeutic target in
patients with CDDP treatment.
Acknowledgements
We would like to thank Mr. Shinichi Norimura for his
excellent technical assistance. The present study was carried out
with the kind cooperation of the Research Center for Molecular
Medicine of the Faculty of Medicine of Hiroshima University. We
also thank the Analysis Center of Life Science of Hiroshima
University for the use of their facilities.
Funding
The present study was supported by Grants-in-Aid for
Scientific Research (nos. JP15H04713 and JP16K08691) and by the
Challenging Exploratory Research (nos. 26670175 and JP16K15247)
from the Japan Society for the Promotion of Science.
Availability of data and materials
All data generated or analyzed during the present
study are included in this article.
Authors' contributions
YSe, NS, KS, NO and WY designed the study. YSh, AI,
TH and AM provided the patients' clinical information. YSe, RH and
JT performed the experiments and acquired the data. YSh, NS, AI,
KS, NO and WY interpreted the results. YSe, TH, NS KS, NO, JT, AM
and WY drafted and edited the manuscript. All authors read and
approved the manuscript and agree to be accountable for all aspects
of the research in ensuring that the accuracy or integrity of any
part of the work are appropriately investigated and resolved.
Ethics approval and consent to
participate
The Institutional Review Board of Hiroshima
University Hospital approved the present study (IRB# E912).
Appropriate written informed consent was obtained from each
patient. The present study was conducted in accordance with the
Ethical Guidance for Human Genome/Gene Research of the Japanese
Government.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
AR
|
androgen receptor
|
|
UC
|
urothelial carcinoma
|
|
CDDP
|
cisplatin
|
|
lncRNAs
|
long non-coding RNAs
|
|
MTT
|
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
|
|
siRNA
|
small interfering RNA
|
|
T-UCR
|
transcribed-ultraconserved region
|
|
UMUC3-CR
|
CDDP-resistant UMUC3 cells
|
References
|
1
|
Lobo N, Mount C, Omar K, Nair R,
Thurairaja R and Khan MS: Landmarks in the treatment of
muscle-invasive bladder cancer. Nat Rev Urol. 14:565–574. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Keegan KA, Zaid HB, Patel SG and Chang SS:
Increasing utilization of neoadjuvant chemotherapy for
muscle-invasive bladder cancer in the United States. Curr Urol Rep.
15:3942014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
von der Maase H, Sengelov L, Roberts JT,
Ricci S, Dogliotti L, Oliver T, Moore MJ, Zimmermann A and Arning
M: Long-term survival results of a randomized trial comparing
gemcitabine plus cisplatin, with methotrexate, vinblastine,
doxorubicin, plus cisplatin in patients with bladder cancer. J Clin
Oncol. 23:4602–4608. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Amable L: Cisplatin resistance and
opportunities for precision medicine. Pharmacol Res. 106:27–36.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Galluzzi L, Vitale I, Michels J, Brenner
C, Szabadkai G, Harel- Bellan A, Castedo M and Kroemer G: Systems
biology of cisplatin resistance: Past, present and future. Cell
Death Dis. 5:e12572014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Martens-Uzunova ES, Bottcher R, Croce CM,
Jenster G, Visakorpi T and Calin GA: Long noncoding RNA in
prostate, bladder, and kidney cancer. Eur Urol. 65:1140–1151. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Chandra Gupta S and Nandan Tripathi Y:
Potential of long non-coding RNAs in cancer patients: From
biomarkers to therapeutic targets. Int J Cancer. 140:1955–1967.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Taheri M, Omrani MD and Ghafouri-Fard S:
Long non-coding RNA expression in bladder cancer. Biophys Rev.
10:1205–1213. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bejerano G, Pheasant M, Makunin I, Stephen
S, Kent WJ, Mattick JS and Haussler D: Ultraconserved elements in
the human genome. Science. 304:1321–1325. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lujambio A, Portela A, Liz J, Melo SA,
Rossi S, Spizzo R, Croce CM, Calin GA and Esteller M: CpG island
hypermethylation-associated silencing of non-coding RNAs
transcribed from ultraconserved regions in human cancer. Oncogene.
29:6390–6401. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Goto K, Ishikawa S, Honma R, Tanimoto K,
Sakamoto N, Sentani K, Oue N, Teishima J, Matsubara A and Yasui W:
The transcribed-ultraconserved regions in prostate and gastric
cancer: DNA hypermethylation and microRNA-associated regulation.
Oncogene. 35:3598–3606. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ferdin J, Nishida N, Wu X, Nicoloso MS,
Shah MY, Devlin C, Ling H, Shimizu M, Kumar K, Cortez MA, et al:
HINCUTs in cancer: Hypoxia-induced noncoding ultraconserved
transcripts. Cell Death Differ. 20:1675–1687. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Marini A, Lena AM, Panatta E, Ivan C, Han
L, Liang H, Annicchiarico-Petruzzelli M, Di Daniele N, Calin GA,
Candi E, et al: Ultraconserved long non-coding RNA uc.63 in breast
cancer. Oncotarget. 8:35669–35680. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Sekino Y, Sakamoto N, Goto K, Honma R,
Shigematsu Y, Sentani K, Oue N, Teishima J, Matsubara A and Yasui
W: Transcribed ultraconserved region Uc.63+ promotes resistance to
docetaxel through regulation of androgen receptor signaling in
prostate cancer. Oncotarget. 8:94259–94270. 2017.PubMed/NCBI
|
|
15
|
Hayashi T, Seiler R, Oo HZ, Jäger W,
Moskalev I, Awrey S, Dejima T, Todenhöfer T, Li N, Fazli L, et al:
Targeting HER2 with T-DM1, an antibody cytotoxic drug conjugate, is
effective in HER2 over expressing bladder cancer. J Urol.
194:1120–1131. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Oue N, Naito Y, Hayashi T, Takigahira M,
Kawano-Nagatsuma A, Sentani K, Sakamoto N, Zarni Oo H, Uraoka N,
Yanagihara K, et al: Signal peptidase complex 18, encoded by
SEC11A, contributes to progression via TGF-alpha secretion in
gastric cancer. Oncogene. 33:3918–3926. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Sekino Y, Oue N, Shigematsu Y, Ishikawa A,
Sakamoto N, Sentani K, Teishima J, Matsubara A and Yasui W: KIFC1
induces resistance to docetaxel and is associated with survival of
patients with prostate cancer. Urol Oncol. 35:31 e13–31 e20. 2017.
View Article : Google Scholar
|
|
18
|
Miyamoto H, Zheng Y and Izumi K: Nuclear
hormone receptor signals as new therapeutic targets for urothelial
carcinoma. Curr Cancer Drug Targets. 12:14–22. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Miyamoto H, Yang Z, Chen YT, Ishiguro H,
Uemura H, Kubota Y, Nagashima Y, Chang YJ, Hu YC, Tsai MY, et al:
Promotion of bladder cancer development and progression by androgen
receptor signals. J Natl Cancer Inst. 99:558–568. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Shiota M, Takeuchi A, Yokomizo A,
Kashiwagi E, Tatsugami K, Kuroiwa K and Naito S: Androgen receptor
signaling regulates cell growth and vulnerability to doxorubicin in
bladder cancer. J Urol. 188:276–286. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kashiwagi E, Ide H, Inoue S, Kawahara T,
Zheng Y, Reis LO, Baras AS and Miyamoto H: Androgen receptor
activity modulates responses to cisplatin treatment in bladder
cancer. Oncotarget. 7:49169–49179. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Fabris L and Calin GA: Understanding the
genomic ultraconservations: T-UCRs and cancer. Int Rev Cell Mol
Biol. 333:159–172. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Olivieri M, Ferro M, Terreri S, Durso M,
Romanelli A, Avitabile C, De Cobelli O, Messere A, Bruzzese D,
Vannini I, et al: Long non-coding RNA containing ultraconserved
genomic region 8 promotes bladder cancer tumorigenesis. Oncotarget.
7:20636–20654. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Li Y, Izumi K and Miyamoto H: The role of
the androgen receptor in the development and progression of bladder
cancer. Jpn J Clin Oncol. 42:569–577. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Shiota M, Kiyoshima K, Yokomizo A,
Takeuchi A, Kashiwagi E, Dejima T, Takahashi R, Inokuchi J,
Tatsugami K and Eto M: Suppressed recurrent bladder cancer after
androgen suppression with androgen deprivation therapy or
5alpha-reductase inhibitor. J Urol. 197:308–313. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Izumi K, Ito Y, Miyamoto H, Miyoshi Y, Ota
J, Moriyama M, Murai T, Hayashi H, Inayama Y, Ohashi K, et al:
Expression of androgen receptor in non-muscle-invasive bladder
cancer predicts the preventive effect of androgen deprivation
therapy on tumor recurrence. Oncotarget. 7:14153–14160. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Xiong Y, Wang L, Li Y, Chen M, He W and Qi
L: The long non-coding RNA XIST interacted with MiR-124 to modulate
bladder cancer growth, invasion and migration by targeting androgen
receptor (AR). Cell Physiol Biochem. 43:405–418. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Calin GA, Liu CG, Ferracin M, Hyslop T,
Spizzo R, Sevignani C, Fabbri M, Cimmino A, Lee EJ, Wojcik SE, et
al: Ultraconserved regions encoding ncRNAs are altered in human
leukemias and carcinomas. Cancer Cell. 12:215–229. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Chen L, Chang WC, Hung YC, Chang YY, Bao
BY, Huang HC, Chung WM, Shyr CR and Ma WL: Androgen receptor
increases CD133 expression and progenitor-like population that
associate with cisplatin resistance in endometrial cancer cell
line. Reprod Sci. 21:386–394. 2014. View Article : Google Scholar : PubMed/NCBI
|