Introduction
As the most common primary malignant bone tumor,
osteosarcoma (OS) is a leading cause of cancer death among children
and adolescents, and accounts for ~60% of all malignant bone tumors
(1). The many advances made in
treating OS (for example, the use of surgery combined with
neoadjuvant and adjuvant chemotherapy) have significantly improved
patient outcomes, and 60–70% of OS patients now survive their
disease. However, several patients, who are resistant to available
chemotherapeutics or with metastatic disease at diagnosis, have far
worse clinical outcomes, and their survival rate is <30%
(2–4).
PSD95/Discs-large/ZO-1 (PDZ) domain containing 2
(PDZD2), also referred to as KIAA0300, PIN-1, PAPIN, activated in
prostate cancer (AIPC) and PDZ domain-containing protein 3 (PDZK3),
is a six-PDZ domain protein (5).
PDZD2 is ubiquitously expressed in multiple tissues, including
heart, brain, liver, spleen, lung, and kidney, but is expressed at
exceptionally high levels in the pancreas and certain cancer
tissues, such as prostate cancer (6–8). An
early study reported that the PDZD2 transcript is upregulated in
human prostate tumor cells and the AIPC protein is abundantly
expressed in high-grade premalignant lesions of the prostate and in
human prostate tumors, suggesting that activation of PDZD2
expression may be an early event in human prostate tumorigenesis
(7). During a recent evaluation of
single nucleotide polymorphisms (SNPs) associated with the risk for
renal cell carcinoma (RCC), Zhang et al (9) demonstrated that Rs10054504 (5p13.3),
which is located in intron 4 of PDZD2, was significantly associated
with the risk for RCC in a Chinese population. However, the role of
PDZD2 in osteosarcoma remains unclear.
The vast majority of RNA transcripts in mammalian
cells originate from genes that do not code for proteins, and are
processed to generate different classes of RNAs with different
sizes (10). The most investigated
type of such RNAs are microRNAs (miRNAs), which are small
non-coding RNA molecules of 18–22 nucleotides in length that
regulate gene expression at the post-transcriptional level by
interacting with complementary sequences in the 3′-UTRs of their
target mRNAs to inhibit their expression (11). Aberrant miRNA expression has been
recognized as a critical event during carcinogenesis, and depending
on the tumor type, may serve either to inhibit or enhance tumor
growth. For example, miR-7, miR-15/16, miR-124, and miR-363 have
been demonstrated to suppress tumor growth, while miR-155, miR-9,
miR-708, and miR-224 can function as oncogenes (12–14).
Tian et al (15) reported
that miR-15a expression is downregulated in osteosarcoma tissues.
miR-15a serves to inhibit cell proliferation, migration, and
invasion by targeting the TNFα-induced protein 1 gene. Decreased
levels of miR-382, which targets Kruppel-like factor 12 and
homeodomain interacting protein kinase 3, were reported in tumor
specimens from OS patients with poor response to chemotherapy,
compared with specimens obtained from patients with good response
to chemotherapy (16). miR-363 has
exhibited tumor suppressive effects in numerous types of cancer,
including colorectal cancer (17),
hepatocellular carcinoma (18),
gallbladder cancer (19) and breast
cancer (20). However, the tumor
suppressive function of miR-363 in OS requires further
investigation.
In the present study, a bioinformatics analysis was
performed and the results identified the PDZD2 gene as a direct
target of miR-363 in OS. Restoration of miR-363 expression and
knockdown of PDZD2 impaired the typical characteristics of OS tumor
cells, including their proliferation, evasion of apoptosis, and
metastasis.
Materials and methods
Cell lines and reagents
Three OS cell lines (MG-63, HOS, and Saos2) and one
normal human osteoblastic cell line (hFOB1.19) were used in the
present study. These cell lines were purchased from the cell bank
of the Type Culture Collection of the Chinese Academy of Sciences
(Shanghai, China). The OS cell lines were cultured in Dulbecco's
modified Eagle's medium (DMEM) supplemented with 10% fetal bovine
serum (FBS; Thermo Fisher Scientific, Inc., Waltham, MA, USA),
ampicillin, and streptomycin at 37°C with 5% CO2. The
hFOB 1.19 cells were routinely maintained in DMEM/Ham's F12 medium
(DMEM/F12; 1:1 w/w mix) containing 10% FBS and 300 μg/ml neomycin
(G418) at 34oC with 5% CO2. Antibodies targeting GAPDH, E-cadherin,
PDZD2, proliferating cell nuclear antigen (PCNA), cleaved caspase-3
and vimentin were obtained from Cell Signaling Technology, Inc.
(Danvers, MA, USA) and Abcam (Cambridge, MA, USA). The miR-363
mimics (5′-AAUUGCACGGUAUCCAUCUGUA-3′) and negative control
(5′-UUCUCCGAACGUGUCACGUTT-3′) oligonucleotides were purchased from
GenePharma Co., Ltd. (Shanghai, China). Small interfering RNA
(siRNA) targeting PDZD2 (siRNA-PDZD2) (139,
5′-GCUGAACUUUGCUGUGGAUUU-3′; 580, 5′ -CUCUGAACCAGGAGAAACAUU-3′; and
1027, 5′-GCUGGGAAUUCAGGUUAGUUU-3′), pcDNA 3.1-NEAT1, and the
negative controls were prepared by RiboBio Co., Ltd. (Guangzhou,
China). The psiCHECK2-UTR (wild-type and mutant) of PDZD2 was
prepared by GenePharma Co., Ltd.
RNA isolation and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
RT-qPCR was performed to determine the expression of
miR-363 and PDZD2 in tumor cells. Total RNA was extracted from
tissues or cell lines using TRIzol reagent (Thermo Fisher
Scientific, Inc.) according to a standard RNA isolation protocol.
Briefly, for the detection of mature miR-363, poly-A polymerase was
used to attach poly-A tails onto the 3′ end of miR-363 molecules,
and High-capacity cDNA Reverse Transcriptase kit (Applied
Biosystems; Thermo Fisher Scientific, Inc.) was used to reverse
transcribe the poly-A-tailed miRNAs by use of a unique oligo (dT)
adaptor primer, according to the manufacturer's protocol (Takara
Biotechnology Co., Ltd., Dalian, China). The reaction conditions
used for qPCR were as follows: incubation at 95°C for 60 sec,
followed by 40 cycles of 95°C for 5 sec, and 60°C for 34 sec.
miR-363 and PDZD2 levels were quantified by using SYBR Premix Ex
Taq (Takara Biotechnology Co., Ltd.) on an ABI 7500 Fast Sequence
Detection System (Applied Biosystems; Thermo Fisher Scientific,
Inc.). The levels of miR-363 and PDZD2 were normalized to those of
U6 and GAPDH, respectively. The primers were: miR-363, forward
5′-ACACTCCAGCTGGGAATTGCACGGTATCCATC-3′ and reverse
5′-CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGTACAGATG-3′; U6, forward
5′-CTCGCTTCGGCAGCACA-3′ and reverse 5′-AACGCTTCACGAATTTGCGT-3′;
PDZD2, forward 5′-TCTGTACTGTGTACCTCACCAA-3′ and reverse
5′-CCCTGCGCTTTTCACCATAG-3′; and GAPDH, forward
5′-TGTTCGTCATGGGTGTGAAC-3′ and reverse 5′-ATGGCATGGACTGTGGTCAT-3′.
Relative fold changes in mRNA expression were calculated using the
formula 2−∆∆Cq (21).
Western blot analysis
Total proteins in tissues or cell lines were
extracted with RIPA cell lysis buffer (Santa Cruz Biotechnology,
Inc., Dallas, TX, USA), and the concentrations of these proteins
were determined with a BCA assay kit (Pierce; Thermo Fisher
Scientific, Inc.). Then each protein sample (30 µg) was separated
by 4–15% SDS-PAGE, followed by transfer onto nitrocellulose
membranes (Amersham; GE Healthcare, Chicago, IL, USA). The
membranes were blocked with 5% non-fat milk in Tris buffer saline
containing 0.1% Tween-20 (TBST) for 1 h at room temperature, and
subsequently incubated overnight at 4°C with primary antibodies
against GAPDH (cat. no. 5174; 1:1,000; Cell Signaling Technology,
Inc.), E-cadherin (cat. no. 14472; 1:1,000; Cell Signaling
Technology, Inc.), PDZD2 (cat. no. ab196631; 1:400; Abcam), PCNA
(cat. no. 13110; 1:1,000; Cell Signaling Technology, Inc.), cleaved
caspase-3 (cat. no. 9661; 1:1,000; Cell Signaling Technology,
Inc.), and vimentin (cat. no. 5741; 1:1,000; Cell Signaling
Technology, Inc.). After washing with TBST buffer, the protein
bands were incubated with horseradish peroxidase (HRP)-conjugated
secondary antibodies (cat. nos. ab205718 and ab205719; 1:20,000;
Abcam) for 1 h at room temperature. Finally, the blots were
visualized with ECL-Plus reagent (Millipore, Billerica, MA, USA).
GAPDH was used as a loading control in the western blot
analyses.
Immunofluorescence assays
Immunofluorescence assays were performed to estimate
the levels of E-cadherin, vimentin, and PDZD2 in tumor cells. The
cells were fixed with 4% formaldehyde in PBS for 15 min and then
washed three times with PBS. Next, the cells were permeabilized
with 100% methanol for 10 min at −20°C, blocked with 3% bovine
serum albumin (BSA) in PBS for 60 min, and then incubated overnight
with primary antibodies anti-PDZD2 (cat. no. ab196631; 1:200;
Abcam), anti-E-Cadherin (cat. no. 14472; 1:50; Cell Signaling
Technology, Inc.) and anti-vimentin (cat. no. 5741; 1:100; Cell
Signaling Technology, Inc.) at 4°C. Following incubation, the cells
were rinsed three times in PBS and placed onto coverslips that were
incubated with a fluorochrome-conjugated secondary antibody for 1 h
at room temperature in the dark. After staining the cell nuclei
with DAPI, the coverslips were mounted onto glass slides with
neutral gum and the cells were observed under an FV10i confocal
microscope (Olympus Corporation, Tokyo, Japan).
Immunohistochemistry
Immunohistochemistry assays were performed to
determine the expression of PDZD2 in tumor tissues as previously
described (22). Formalin-fixed,
paraffin-embedded tissue sections (2-µm thickness) were prepared
and subsequently incubated in xylene for 5 min, 100% ethanol for 10
min, and then in 95% ethanol for 10 min. Antigen unmasking was
performed, and the slides were then blocked with 3% hydrogen
peroxide for 30 min at room temperature. Next, the sections were
incubated overnight at 4°C with a primary antibody against PDZD2,
and then subsequently incubated with the secondary antibody. An
EnVision Detection System kit (Dako, Glostrup, Denmark) was used to
assess the immunostaining results.
Cell transfection
MG-63 cells were seeded into a 12-well plate and
cultured to ~60% confluence. The cells were then washed with PBS
and transfected with miR-363 mimics (50 nM), siRNA-PDZD2 (1 µg/ml)
or the corresponding negative control for 24, 48 and 72 h, using
100 nM Lipofectamine 2000 (Thermo Fisher Scientific, Inc.),
according to the manufacturer's instructions. Following
transfection for the indicated time period, the cells were
harvested for use in further experiments.
Cell Counting Kit-8 (CCK-8) assay
MG-63 cells, transfected with the indicated agents,
were harvested, washed with PBS, and then counted with a Cell
Counting Kit-8 (Dojindo Molecular Technologies, Inc., Kumamoto,
Japan). Other transfected cells were mixed with DMEM for use in
cell viability assays. Absorbance was measured at 450 nm with a
microplate reader.
EdU assay
Cell proliferation was assessed by use of an EdU
assay kit (Ruibo Biotechnology Co., Ltd., Guangzhou, China). Tumor
cells (1×105 cells/well) were incubated with 100 µl of
50 µM EdU per well for 2 h at 37°C in 24-well plates, and then were
fixed with 100 µl of fixing buffer (4% polyformaldehyde in PBS) for
30 min at room temperature. Subsequently, the cells were incubated
for 5 min with 50 µl of 2 mg/ml glycine, and washed with 100 µl
PBS. Next, the cells were treated with 0.5% Triton X and reacted
with 1X Apollo solution for 30 min. The cells were then incubated
with 100 µl Hoechst 33342 solution for 30 min in the dark, and
washed with 100 µl PBS. Finally, the cells were analyzed by
fluorescence microscopy (Lionheart; BioTek Instruments, Inc.,
Winooski, VT, USA).
Colony formation assay
MG-63 cells transfected with the indicated agents
were harvested and then re-suspended in complete medium containing
10% FBS. The re-suspended cells were then seeded into 12-well
plates (1×103 cells/well) and treated as indicated for
10 days. Following fixation in methanol for 15 min, the cells were
stained with 0.1% crystal violet, and visualized under a dissection
microscope (Olympus Corporation). Colonies consisting of ≥50 cells
were counted.
Apoptosis and cell cycle distribution
analysis
Cell apoptosis was analyzed using the Hoechst 33258
assay, flow cytometry or the terminal
deoxynucleotidyl-transferase-mediated dUTP nick end labelling
(TUNEL) assay. For the Hoechst assay, MG-63 cells were harvested,
washed with PBS, and then fixed with paraformaldehyde. The fixed
cells were then stained with 0.1 µg/ml Hoechst 33258 solution
(Sigma-Aldrich; Merck KGaA, Darmstadt, Germany), and changes in
nuclear morphology were observed under a fluorescence microscope
(Olympus Corporation). For flow cytometry, 2 µl of Annexin V mixed
with 2 µl of propidium iodide (PI; eBioscience; Thermo Fisher
Scientific, Inc.) were added to each tube of cells
(1×105 cells per tube) for 30 min, according to the
manufacturer's instructions. The cell staining was then quantified
by flow cytometry on a FACS Calibur instrument with CellQuest Pro
v5.2 software (BD Biosciences, San Jose, CA, USA). For the TUNEL
assay, tumor tissues were fixed in formalin, embedded in paraffin,
and cut into 4-µm sections. Then, tumor sections were stained using
an apoptosis in situ detection kit (Wako Pure Chemical
Industries, Ltd., Osaka, Japan), and according to the
manufacturer's instructions. Five non-overlapping fields from each
section were randomly selected and observed under a fluorescence
microscope. To determine the cell cycle phase distribution, cells
were stained with PI staining solution (10 µg/ml RNase A; 50 µg/ml
PI) at 37°C for 30 min in the dark, and then analyzed using a flow
cytometer equipped with Cell-Quest Pro v5.2 software (BD
Biosciences).
Cell migration and invasion
analysis
The scratch wound assay was used to determine the
migration of MG-63 cells. Cells transfected with the indicated
agents were allowed to form separate monolayers, which were then
scratched by dragging a plastic tip across the monolayer's surface.
Five fields were randomly selected, and the distances traveled by
the migrated cells were measured under a light microscope. For the
invasion assay, transwells coated with matrigel were used. A total
of 2×104 MG-63 cells in serum-free media were added into
the upper chambers of the matrigel-coated transwell inserts. DMEM
with 10% FBS (1 ml) was added into the bottom chambers. After 24 h
incubation, cells that did not migrate or invade were removed using
a cotton swab, stained with crystal violet, and then counted under
an inverted microscope. The number of cells in each of five
randomly selected microscopic fields was counted. Each experiment
was repeated three times.
Xenograft animal model
Thirty male mice (4–5 weeks; 20–22 g; purchased from
Tianjin Purcell Biotechnology Co., Ltd., Tianjin, China) were
housed in a sterile room at 25°C, with a 12-h light/dark cycle and
free access to food and water. Human MG-63 cells (2×106)
transfected with the lentivirus vector containing miR-363 mimics or
the negative control were subcutaneously injected into the rear
flank of each nude mouse (10 mice per group). The tumor sizes were
measured at three-day intervals, and the tumor volumes were
calculated as follows: V (cm3) = width2
(cm2) × length (cm)/2. The protocols for all animal
studies were approved by the Institutional Animal Care and Use
Committee of Southern Medical University (Guangzhou, China).
Statistical analysis
Results are presented as the mean ± standard
deviation resulting from at least three independent repeats.
Student's t-test or one-way (followed by Tukey's multiple
comparisons test) or two-way (followed by Sidak's multiple
comparisons test) analysis of variance was used to compare
differences between two or more groups. All data were processed
with SPSS 19.0 software (IBM Corp., Armonk, NY, USA). P<0.05 was
considered to indicate a statistically significant difference.
Results
Restoration of miR-363 in OS cells
inhibits cell growth in vitro
Firstly, the role of miR-363 in OS was investigated
in vitro. The levels of miR-363 expression were evaluated in
three OS cell lines (MG-63, HOS and Saos2) and one human
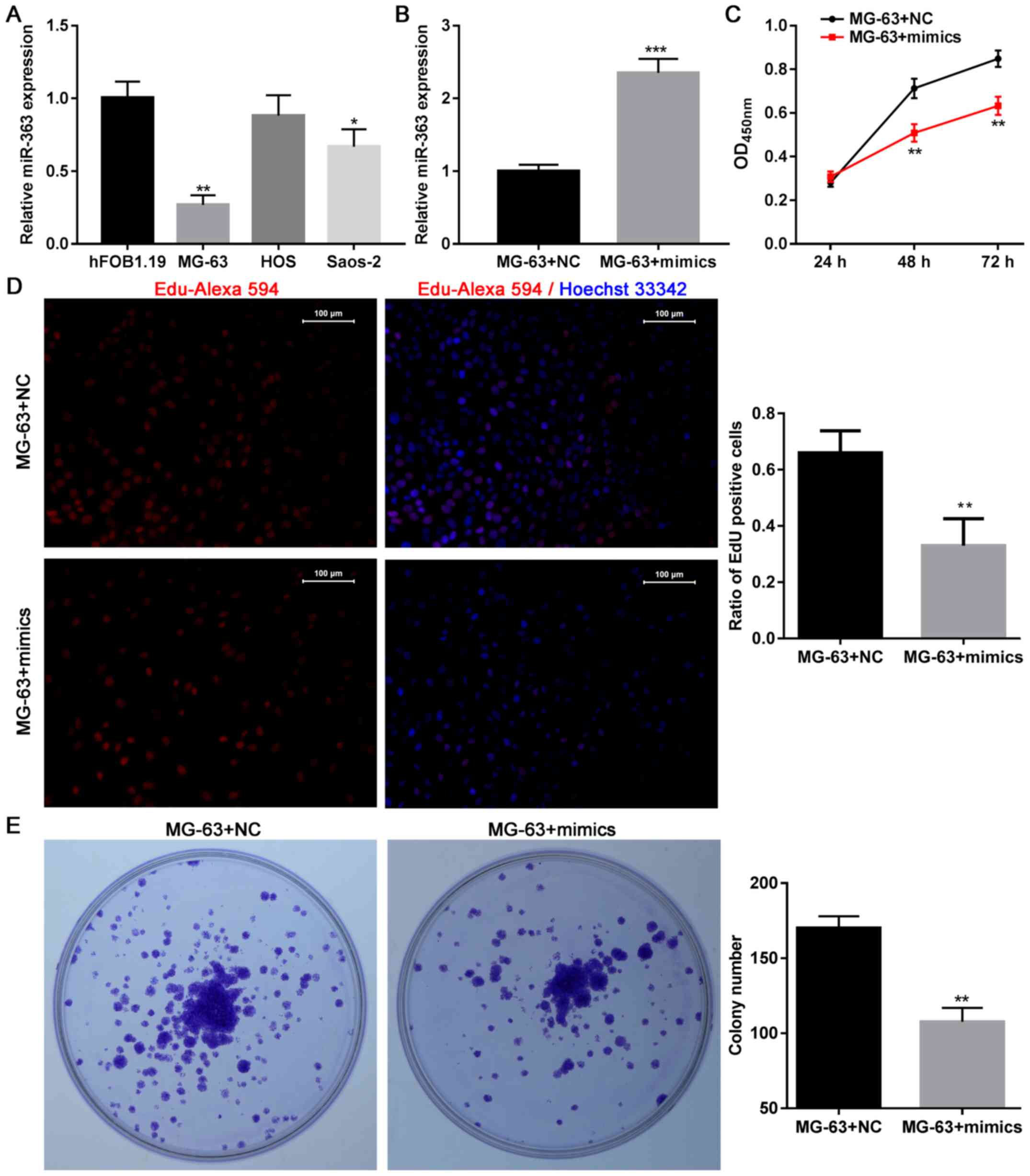
osteoblastic cell line (hFOB1.19). The present data demonstrated
that the levels of miR-363 were lower in two out of three tumor
cells lines tested, compared with the hFOB1.19 cells, and
particularly in the MG-63 cells (Fig.
1A). Thus, overexpression of miR-363 was forced in MG-63 cells
(Fig. 1B) and the subsequent
effects were investigated on OS cell growth. The results
demonstrated that overexpression of miR-363 significantly impaired
the viability of MG-63 cells at 48 and 72 h (Fig. 1C). Overexpression of miR-363
significant impaired cell viability at 48 and 72 h in SaOS-2 cells
as well (data not shown). In addition, the EdU assay indicated that
the proliferation of MG-63 cells was impaired following
transfection with miR-363 mimics (Fig.
1D), and colony formation assays revealed that cells
transfected with the miR-363 mimics had a decreased ability to form
colonies (Fig. 1E). Furthermore,
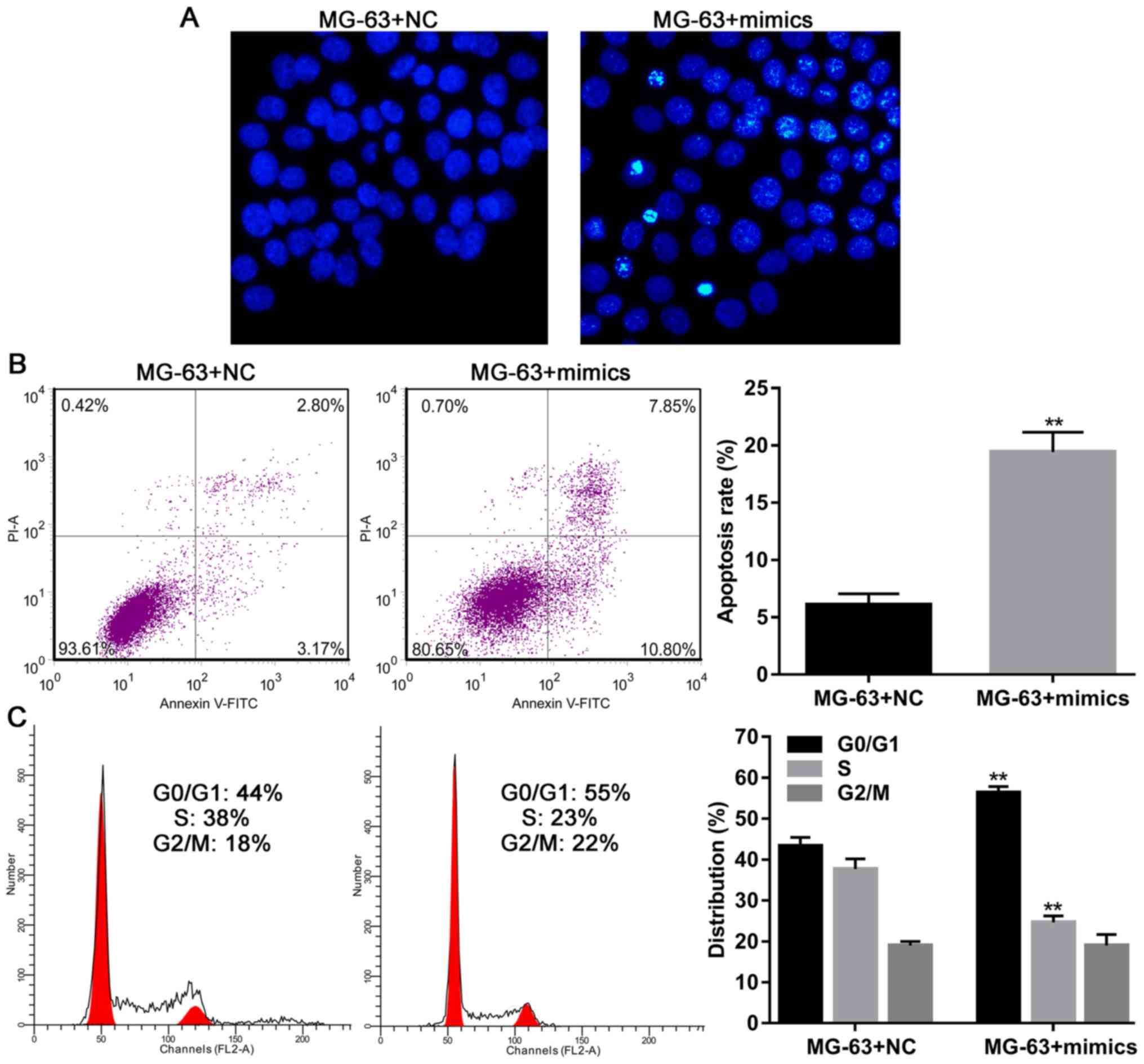
Hoechst staining and flow cytometry analysis demonstrated that
MG-63 cells overexpressing miR-363 had higher rates of apoptosis
(Fig. 2A and B), and flow cytometry
results demonstrated that miR-363 overexpression could induce G1/S
arrest in MG-63 cells (Fig. 2C). In
total, these findings indicated that miR-363 expression inhibited
the growth and proliferation of OS cells by inducing apoptosis and
G1/S arrest.
miR-363 overexpression impairs cell
migration and invasion by regulating the EMT phenotype
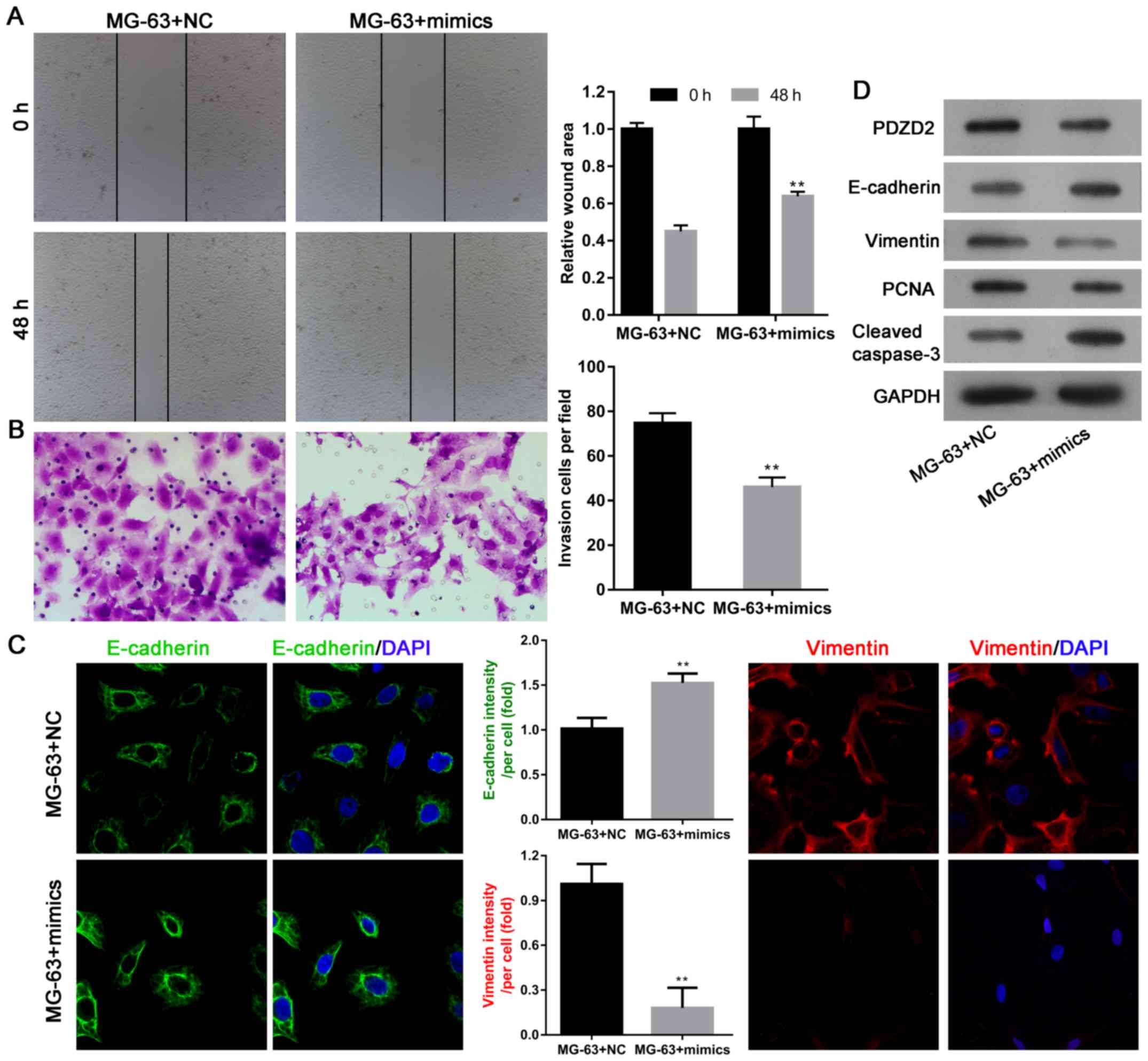
Next, the effects of miR-363 overexpression on the
migration and invasion abilities of MG-63 cells, and the potential
underlying mechanisms, were investigated. Scratch wound assays
demonstrated that MG-63 cell migration was significantly attenuated
following miR-363 mimics transfection, compared with transfection
with negative controls (Fig. 3A).
Transwell invasion assays demonstrated that the invasion ability of
MG-63 cells was also impaired following miR-363 overexpression
(Fig. 3B). With regard to the
underlying mechanism, results from immunofluorescence and western
blot analyses revealed that expression of the epithelial marker
E-cadherin was upregulated following miR-363 overexpression, while
the mesenchymal marker vimentin was downregulated (Fig. 3C and D). Furthermore, expression of
the cell proliferation marker PCNA was decreased, while cleavage of
caspase-3 was increased following miR-363 overexpression (Fig. 3D), suggesting that the tumor
suppressive effect of miR-363 was dependent on PCNA, caspase-3, and
the EMT phenotype.
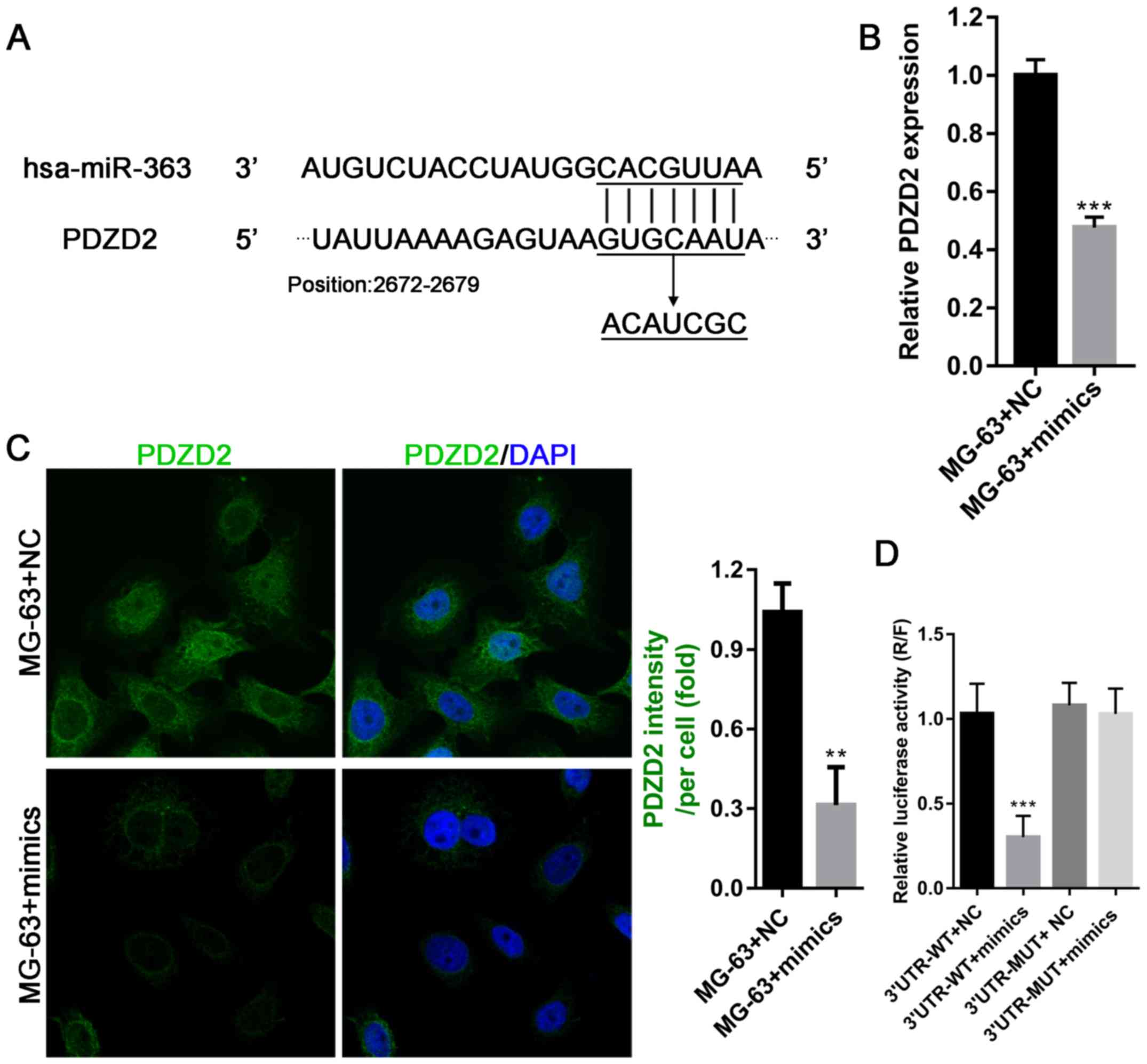
PDZD2 is a direct target of miR-363
and promotes cell proliferation in vitro
The online prediction software miRWalk (http://www.umm.uni-heidelberg.de/apps/zmf/mirwalk/)
was used to discover potential targets of miR-363, and the gene
PDZD2 was identified. Further analysis revealed that the 3′
untranslated region (UTR) of PDZD2 mRNA harbored a binding site for
miR-363 (Fig. 4A); therefore, the
interaction between PDZD2 and miR-363 was investigated in OS cells.
The results demonstrated that overexpression of miR-363 in MG-63
cells significantly inhibited their ability to produce PDZD2 mRNA
and protein (Fig. 4B and C). To
confirm PDZD2 as a direct target of miR363, a luciferase reporter
vector with the full-length 3′UTR (wild-type or mutant) of PDZD2
was transfected into MG-63 cells for use in luciferase reporter
assays. The results revealed that miR-363 mimics significantly
impaired the luciferase activity of the wild-type PDZD2 3′UTR, but
not the mutant control, indicating that miR-363 directly targeted
PDZD2 in OS cells to decrease PDZD2 protein levels (Fig. 4D).
Next, the effects of decreased PDZD2 protein levels
in OS cells were investigated. After efficiently knocking down
PDZD2 expression in MG-63 cells by siRNA (Fig. 5A), the apoptosis (Fig. 5B), colony formation (Fig. 5C), and invasion capabilities of the
cells were examined (Fig. 5D). The
results demonstrated that knockdown of PDZD2 induced apoptosis and
attenuated the colony formation and invasion capabilities of the
cells. These effects were associated with a decrease in PCNA
expression, increased cleavage of caspase-3, and reduced expression
of the EMT phenotype (Fig. 5E).
These findings suggested that PDZD2 could induce tumor suppression
via miR363 in OS cells.
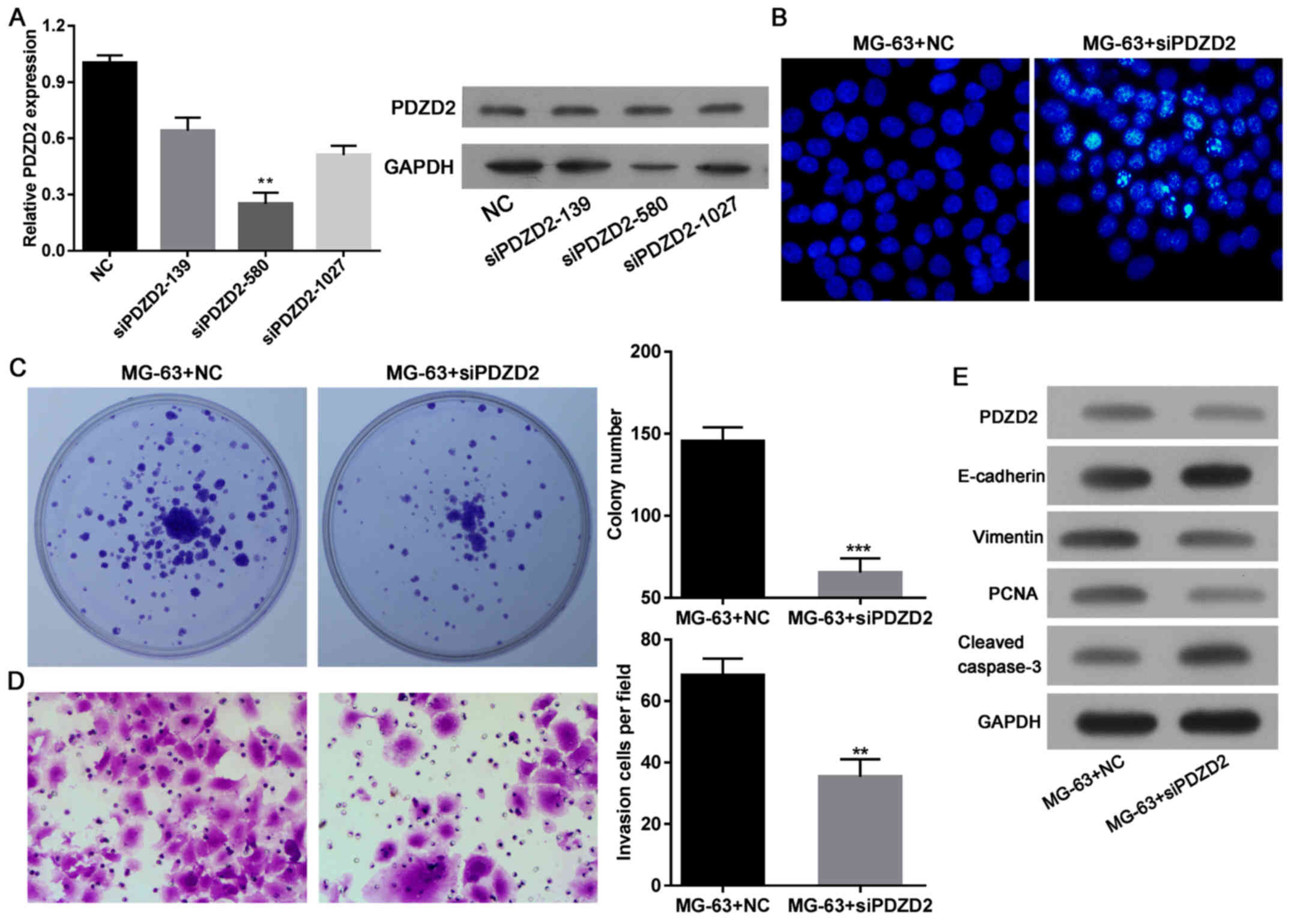 | Figure 5.PDZD2 knockdown inhibits osteosarcoma
cell growth and EMT. (A) Successful knockdown of PDZD2 was
established by evaluating three separate siRNAs, and selecting the
most efficient construct. (B) Cell apoptosis (magnification, ×200),
(C) colony formation and (D) invasion (magnification, ×200) were
evaluated in the MG-63 cells following PDZD2 knockdown. (E) Protein
levels of PDZD2, EMT markers E-cadherin and vimentin, PCNA, and
cleaved caspase-3 were determined by western blot analysis. Data
are presented as mean ± standard deviation. **P<0.01 and
***P<0.001 compared with NC group. PZDZ2, PDZ domain containing
2; EMT, epithelial-mesenchymal transition; siRNA, small interfering
RNA; PCNA, proliferating cell nuclear antigen; NC negative
control. |
miR-363 expression restricts tumor
growth with reduced PDZD2 levels in vivo
Because miR-363 and PDZD2 were demonstrated to
regulate OS cells in vitro, we sought to determine the
function of miR-363 in OS in vivo. MG-63 cells were
transfected with miR-363 mimics or the negative control, and then
injected into nude mice to produce a xenograft model of human MG-63
tumors. The results demonstrated that the mice injected with MG-63
cells overexpressing miR-363 developed significantly smaller tumors
that displayed delayed growth (Fig. 6A
and B). The successful upregulation of miR-363 (Fig. 6C) and downregulation of PDZD2
(Fig. 6D and E) was confirmed at
the mRNA and protein levels in the tumor tissues. The tumor cell
apoptosis rates were also estimated in the tumor tissues, and the
results demonstrated that miR-363 markedly induced apoptosis of OS
cells in vivo (Fig. 6F).
These effects were associated with the decreased expression of
PCNA, increased cleavage of caspase-3, and impairment of the EMT
phenotype in the miR-363-overexpressing tumors compared with
controls (Fig. 6G).
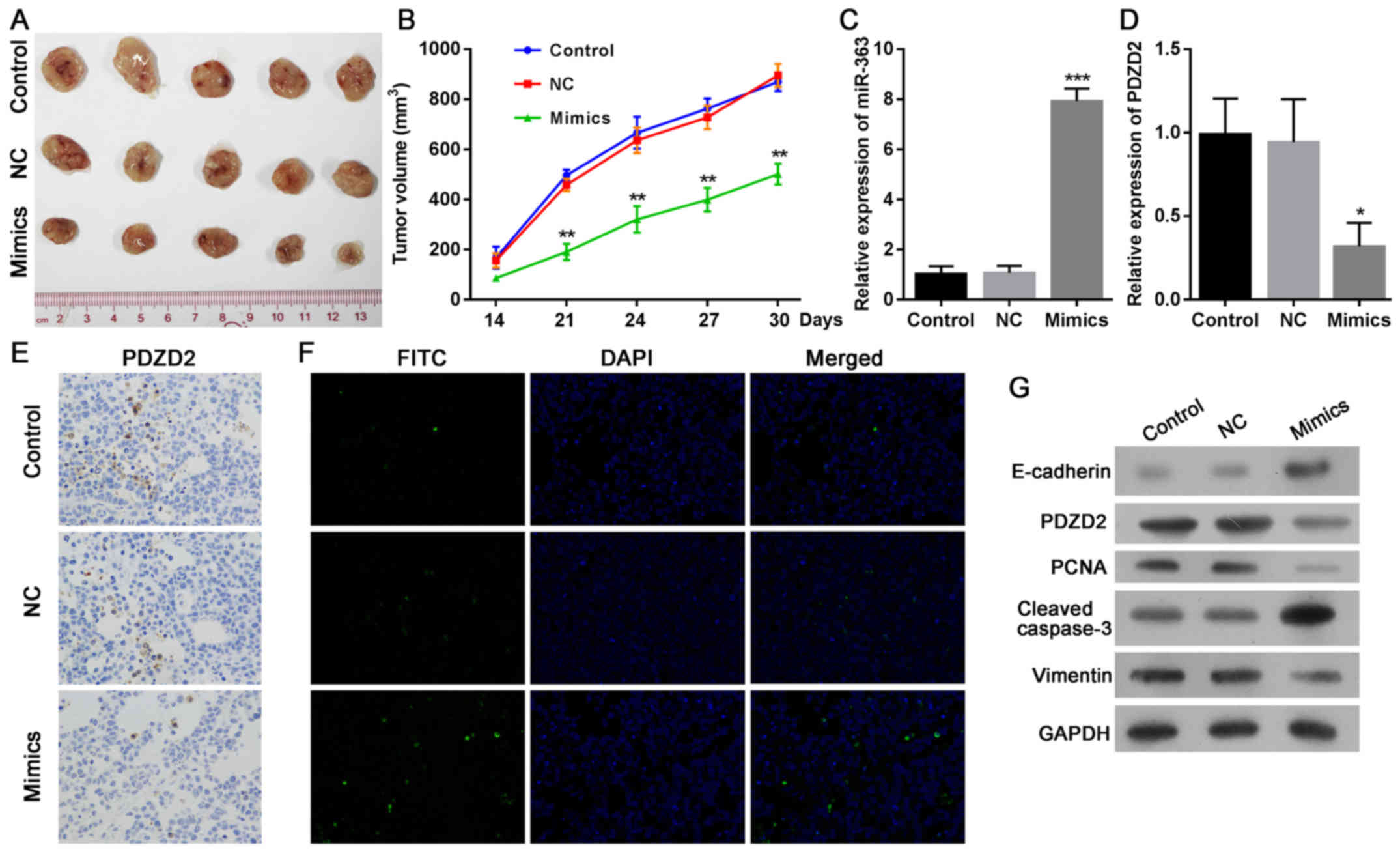 | Figure 6.mir-363 induces the regression of
osteosarcoma tumors in vivo. MG-63 cells
(~2×106), transfected with miR-363 mimics or negative
control, were subcutaneously injected into the rear flanks of nude
mice (5 mice per group). (A) Images of the tumors at the end of the
experiment. (B) Quantification of the tumor volumes
(mm3) over time. (C) Expression levels of miR-363 and
(D) PDZD2 mRNA in tumor tissues were analyzed by reverse
transcription-quantitative polymerase chain reaction. (E) PDZD2
protein expression in tumor tissues was determined by
immunohistochemistry (magnification, ×200). (F) The apoptosis rates
in the tumor sections were estimated by TUNEL staining
(magnification, ×200). (G) Protein expression levels of PDZD2,
E-cadherin, vimentin, PCNA and cleaved caspase-3 expression were
determined by western blot analysis. Data are presented as mean ±
standard deviation. *P<0.05, **P<0.01 and ***P<0.001
compared with NC group. PZDZ2, PDZ domain containing 2; PCNA,
proliferating cell nuclear antigen; NC negative control. |
Discussion
Osteosarcoma is an aggressive malignant neoplasm
that arises from primitive transformed cells of mesenchymal origin.
The survival rate of patients with metastatic disease is <30%,
and further efforts, including novel treatment protocols, are
needed to improve patient outcomes (23). Although recent studies have provided
additional insights into both the oncogenic and tumor suppressive
functions of miRNAs during the development of osteosarcoma, the
exact molecular mechanisms underlying the miRNA/target interaction
in OS remain to be fully identified (24). The in vitro and in
vivo analyses included in the present study demonstrated that
the tumor suppressor miR-363 was downregulated in OS cells and
promoted tumor growth by directly targeting PDZD2. Although
preliminary results from our group have indicated that the miR-363
levels are reduced in OS tissues when compared to normal tissues
(n=8, data not shown), further confirmation of these results is
underway in our group with larger samples.
PDZD2 was originally thought to be an oncogene whose
expression is upregulated in prostate tumor cell lines and human
primary prostate tumors. The activation of PDZD2 expression is an
early event in human prostate tumorigenesis (7). The present study demonstrated that
PDZD2 expression was associated with OS tumor progression. A recent
study of single nucleotide polymorphisms associated with renal cell
carcinoma indicated that rs10054504 at PDZD2 was significantly
associated with the risk for renal cell carcinoma in a Chinese
population (22). However, the
efficiency of therapies that target PDZD2 remains unclear. Human
secreted PDZD2 (sPDZD2) was demonstrated to have tumor suppressive
effects. sPDZD2 induced the senescence of prostate cancer cells via
transcriptional activation of mutant or wild-type p53, and
sensitized cancer cells to apoptosis induction via genotoxic stress
(25). In addition, the
antiproliferative effect of sPDZD2 in human cancer cells was
demonstrated to be mediated by induction of S phase cell cycle
arrest (26). The present study
reported that inhibition of PDZD2 promoted the apoptosis of OS
cells and abrogated the migration and invasion capabilities of OS
cell lines. Furthermore, knockdown of PDZD2 significant reduced
expression of the cell proliferation marker PCNA, induced the
cleavage of caspase-3, and impaired the EMT phenotype in OS cells.
Further studies will be required to fully elucidate that
miR-363/PDZD2-related downstream signals and their functions in OS
cells.
Dysregulation of miRNA during the development of
cancer is a hallmark of malignant transformation. The tumor
suppressor miR-363 belongs to the miR-92a family, which is a group
of highly conserved miRNAs that include miR-25, miR-92a-1,
miR-92a-2, and miR-363, and are associated with the formation of
vascular endothelial cells (27).
The aberrant expression of miR-92a family members has been reported
in multiple types of cancer, and is related to carcinogenesis and
tumor development (28). Zhou et
al (29) found that the
expression levels of miR-92a were significantly higher in
colorectal cancer (CRC) tissues, and were correlated with an
advanced clinical stage, lymph node metastasis and distant
metastasis. miR-363-3p was demonstrated to be downregulated in CRC
tissue specimens with lymph node metastasis, to promote CRC cell
migration/invasion and induce EMT by increasing SRY-box 4 (Sox4)
expression in vitro and in vivo (30). In prostate cancer, a high level of
miR-363 in PC-3 cells promoted cell proliferation, induced cell
transformation, and promoted EMT by targeting or inhibiting MYC
proto-oncogene (31). The current
study demonstrated that miR-363 levels in osteosarcoma cells were
downregulated, which in turn resulted in upregulated expression of
its target gene, PDZD2. Restoration of miR-363 expression in OS
cells impaired cell viability, colony formation, migration and
invasion, while it induced apoptosis and cell cycle arrest via
PDZD2. These effects might be associated with decreases in PCNA,
caspase-3, and the EMT phenotype. Similarly, Wang et al
(32) demonstrated that miR-363
inhibited osteosarcoma cell proliferation and invasion by targeting
SOX4. These findings indicate that miR-363 may function as a
universal tumor suppressor in multiple types of cancer.
In conclusion, the present study reported PDZD2 as a
novel target of the tumor suppressor miR-363 in OS cells.
Overexpression of miR-363 or inhibition of PDZD2 promoted
caspase-3-related apoptosis of tumor cells, inhibited EMT, and
induced the regression of osteosarcoma tumors.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Author's contributions
FH, LF and QY conceived and supervised the study,
designed experiments, performed experiments, analyzed data, wrote
the manuscript and made manuscript revisions. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
All protocols involving the use of animals were
approved by the Institutional Animal Care and Use Committee of
Southern Medical University (Guangzhou, China).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Geller DS and Gorlick R: Osteosarcoma: A
review of diagnosis, management, and treatment strategies. Clin Adv
Hematol Oncol. 8:705–718. 2010.PubMed/NCBI
|
|
2
|
Lamoureux F, Trichet V, Chipoy C,
Blanchard F, Gouin F and Redini F: Recent advances in the
management of osteosarcoma and forthcoming therapeutic strategies.
Expert Rev Anticancer Ther. 7:169–181. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Gaspar N, Hawkins DS, Dirksen U, Lewis IJ,
Ferrari S, Le Deley MC, Kovar H, Grimer R, Whelan J, Claude L, et
al: Ewing sarcoma: Current management and future approaches through
collaboration. J Clin Oncol. 33:3036–3046. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Misaghi A, Goldin A, Awad M and Kulidjian
AA: Osteosarcoma: A comprehensive review. SICOT J. 4:122018.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Thomas MK, Tsang SW, Yeung ML, Leung PS
and Yao KM: The roles of the PDZ-containing proteins bridge-1 and
PDZD2 in the regulation of insulin production and pancreatic
beta-cell mass. Curr Protein Pept Sci. 10:30–36. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ma RY, Tam TS, Suen AP, Yeung PM, Tsang
SW, Chung SK, Thomas MK, Leung PS and Yao KM: Secreted PDZD2 exerts
concentration-dependent effects on the proliferation of INS-1E
cells. Int J Biochem Cell Biol. 38:1015–1022. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Chaib H, Rubin MA, Mucci NR, Li L, Day ML,
Rhim JS and Macoska JA; Taylor JMG, : Activated in prostate cancer:
A PDZ domain-containing protein highly expressed in human primary
prostate tumors. Cancer Res. 61:2390–2394. 2001.PubMed/NCBI
|
|
8
|
Yeung ML, Tam TS, Tsang AC and Yao KM:
Proteolytic cleavage of PDZD2 generates a secreted peptide
containing two PDZ domains. EMBO Rep. 4:412–418. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhang N, Wu Y, Gong J, Li K, Lin X, Chen
H, Yu Y, Gou Y, Hou J, Jiang D, et al: Germline genetic variations
in PDZD2 and ITPR2 genes are associated with clear cell renal cell
carcinoma in Chinese population. Oncotarget. 8:24196–24201.
2017.PubMed/NCBI
|
|
10
|
Panwar B, Arora A and Raghava GP:
Prediction and classification of ncRNAs using structural
information. BMC Genomics. 15:1272014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Shukla GC, Singh J and Barik S: MicroRNAs:
Processing, maturation, target recognition and regulatory
functions. Mol Cell Pharmacol. 3:83–92. 2011.PubMed/NCBI
|
|
13
|
Hayes J, Peruzzi PP and Lawler S:
MicroRNAs in cancer: Biomarkers, functions and therapy. Trends Mol
Med. 20:460–469. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Di Leva G, Garofalo M and Croce CM:
MicroRNAs in cancer. Annu Rev Pathol. 9:287–314. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Tian X, Zhang J, Yan L, Dong JM and Guo Q:
MiRNA-15a inhibits proliferation, migration and invasion by
targeting TNFAIP1 in human osteosarcoma cells. Int J Clin Exp
Pathol. 8:6442–6449. 2015.PubMed/NCBI
|
|
16
|
Xu M, Jin H, Xu CX, Sun B, Mao Z, Bi WZ
and Wang Y: miR-382 inhibits tumor growth and enhance
chemosensitivity in osteosarcoma. Oncotarget. 5:9472–9483. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Tsuji S, Kawasaki Y, Furukawa S, Taniue K,
Hayashi T, Okuno M, Hiyoshi M, Kitayama J and Akiyama T: The
miR-363-GATA6-Lgr5 pathway is critical for colorectal
tumourigenesis. Nat Commun. 5:31502014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhou P, Huang G, Zhao Y, Zhong D, Xu Z,
Zeng Y, Zhang Y, Li S and He F: MicroRNA-363-mediated
downregulation of S1PR1 suppresses the proliferation of
hepatocellular carcinoma cells. Cell Signal. 26:1347–1354. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wang SH, Zhang WJ, Wu XC, Weng MZ, Zhang
MD, Cai Q, Zhou D, Wang JD and Quan ZW: The lncRNA MALAT1 functions
as a competing endogenous RNA to regulate MCL-1 expression by
sponging miR-363-3p in gallbladder cancer. J Cell Mol Med.
20:2299–2308. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhang R, Li Y, Dong X, Peng L and Nie X:
MiR-363 sensitizes cisplatin-induced apoptosis targeting in Mcl-1
in breast cancer. Med Oncol. 31:3472014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ding L, Ren J, Zhang D, Li Y, Huang X, Hu
Q, Wang H, Song Y, Ni Y and Hou Y: A novel stromal lncRNA signature
reprograms fibroblasts to promote the growth of oral squamous cell
carcinoma via LncRNA-CAF/interleukin-33. Carcinogenesis.
39:397–406. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Luetke A, Meyers PA, Lewis I and Juergens
H: Osteosarcoma treatment - where do we stand? A state of the art
review. Cancer Treat Rev. 40:523–532. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Liang W, Gao B, Fu P, Xu S, Qian Y and Fu
Q: The miRNAs in the pathgenesis of osteosarcoma. Front Biosci.
18:788–794. 2013. View
Article : Google Scholar
|
|
25
|
Tam CW, Cheng AS, Ma RY, Yao KM and Shiu
SY: Inhibition of prostate cancer cell growth by human secreted PDZ
domain-containing protein 2, a potential autocrine prostate tumor
suppressor. Endocrinology. 147:5023–5033. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Tam CW, Liu VW, Leung WY, Yao KM and Shiu
SY: The autocrine human secreted PDZ domain-containing protein 2
(sPDZD2) induces senescence or quiescence of prostate, breast and
liver cancer cells via transcriptional activation of p53. Cancer
Lett. 271:64–80. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Li M, Guan X, Sun Y, Mi J, Shu X, Liu F
and Li C: miR-92a family and their target genes in tumorigenesis
and metastasis. Exp Cell Res. 323:1–6. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Olive V, Jiang I and He L: mir-17-92, a
cluster of miRNAs in the midst of the cancer network. Int J Biochem
Cell Biol. 42:1348–1354. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhou T, Zhang G, Liu Z, Xia S and Tian H:
Overexpression of miR-92a correlates with tumor metastasis and poor
prognosis in patients with colorectal cancer. Int J Colorectal Dis.
28:19–24. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Hu F, Min J, Cao X, Liu L, Ge Z, Hu J and
Li X: MiR-363-3p inhibits the epithelial-to-mesenchymal transition
and suppresses metastasis in colorectal cancer by targeting Sox4.
Biochem Biophys Res Commun. 474:35–42. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Chen Y, Lu X, Wu B, Su Y, Li J and Wang H:
MicroRNA 363 mediated positive regulation of c-myc translation
affect prostate cancer development and progress. Neoplasma.
62:191–198. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wang K, Yan L and Lu F: miR-363-3p
Inhibits Osteosarcoma Cell Proliferation and Invasion via Targeting
SOX4. Oncol Res. 27:157–163. 2019. View Article : Google Scholar : PubMed/NCBI
|