Introduction
Liver cancer is a serious global health problem, and
it accounts for 4% of all new cancer cases and 6% of
cancer-associated mortalities in men (1–3). Over
600,000 individuals succumb to liver cancer annually worldwide
despite advances in treatment approaches like hepatic resection,
image-guided tumor ablation and liver transplantation (4–6). The
poor prognosis of liver cancer is due to its complex molecular
mechanisms and limited molecular treatment targets (7). To date, few treatments with proven
survival benefits exist for patients advanced liver cancer
(8–10). The first drug approved for systemic
therapy was a multiple kinase inhibitor termed sorafenib (11). Sorafenib has been plagued with toxic
side-effects and limited efficacy (12,13),
therefore an urgent need for alternative liver cancer treatments
persists (14–16). Doctors and medical scientists have
gradually begun to recognize the anticancer therapeutic potential
of natural compounds with their relatively few side-effects and
curative effects for other diseases, including other types of
cancer (17).
Nitidine chloride (NC), an ingredient well-known in
traditional Chinese medicine, is present in the root of
Zanthoxylum nitidum (Roxb.) DC (18). Previous studies revealed that NC
could mediate a variety of physiological and pathological
processes, including inflammation modulation (19), analgesia and tumor suppression
(20,21). Previous studies on NC have focused
on its antitumor effects in liver cancer. Liao et al
(22) reported that NC inhibited
the cell growth of liver cancer cells via the Janus-activated
kinase 1/signal transducer and activator of transcription 3
(JAK1/STAT3) signaling pathway. NC also induced the apoptosis of
liver cancer cells via pathways associated with p53, p21,
Bax and Bcl-2 oncogenes (23). Research on the precise mechanisms by
which NC functions remains in its early stages, with studies
emphasizing the function of NC in tumor growth suppression
(24–26). While NC is predicted to be of
pivotal therapeutic significance, it may be challenging to uncover
its mechanisms at the molecular level as well as from a systemic
viewpoint.
To address these challenges, the cell growth
inhibitory effect of NC on HepG2 cells was assessed in
vitro; the tumor suppressive capacity of NC was also analysed
in vivo in a murine model. A series of bioinformatic
analyses were performed to uncover potential NC regulatory
mechanisms based on the change in expression profiles following NC
treatment on xenografted liver tumors. The clinical significance of
several key genes associated with the function of NC in liver
cancer were also analyzed. In doing so, the authors of the present
study attempted to identify potential mechanisms of NC for the
treatment of liver cancer.
Materials and methods
Preparation of NC and cell
culture
NC with a purity >95% was provided by Guangxi
Traditional Chinese Medicine Institute (Nanning, China). NC was
dissolved in dimethyl sulfoxide at a concentration of 10 mM for
subsequent testing. Human HepG2 cells were obtained from the
Shanghai Cell Bank of the Chinese Academy of Sciences (Shanghai,
China). HepG2 cells were cultured in Dulbecco's modified Eagle's
medium (DMEM; Gibco; Thermo Fisher Scientific, Inc., Waltham, MA,
USA) in a stable humidified atmosphere of 5% CO2 at
37°C. Authentication of the HepG2 cell line performed using short
tandem repeat profiling.
Inhibitory effect of NC on HepG2 cells
based on in vitro and in vivo models
The MTT method was performed to examine the
inhibitory effect of NC on HepG2. Dimethyl sulfoxide was used to
dissolve the purple formazan, the supernatant collected and then
the absorbance of formazan was measured at a wavelength of 570 nm.
The inhibition rate was detected at 24, 48 and 72 h. A total of 32
six-week-old nude specific-pathogen-free BALB/c mice (16 males and
16 females; weighing 18–20 g) were purchased from the Shanghai
Laboratory Animal Co., Ltd. (Shanghai, China) and housed in the
Experimental Animal Center of Guangxi Medical University (Nanning,
China). The mice were housed in specific-pathogen-free conditions
in a controlled temperature of 24°C, food and water were provided
ad libitum. Mice were fed a sterile diet with a 12/12 h
light/dark cycle. All procedures associated with the animal
experiments were approved by the Animal Ethics Committee of Guangxi
Medical University. The immunodeficient nude BALB/c mice were
randomly assigned into four treatment groups, including a low-NC
group (2.5 mg/kg/day), a medium-NC group (5 mg/kg/day), a high-NC
group (10 mg/kg/day) and a saline-only control group (0.2 ml
saline/day). Then, 0.2 ml HepG2 cells was subcutaneously injected
into the right axilla of these mice at a density of
1×1010 cells/ml. A total of 7 days after the cells were
injected, mice were intraperitoneally administered with NC (2.5, 5
or 10 mg/kg/day) or 0.2 ml saline/day for 14 days. When the
xenografted tumors reached 150–300 mm3, mice were
transferred for further experimentation. On the day after tumor
implantation, NC or saline was administered via intraperitoneal
injection once per day for 14 days. The length and width of the
xenografted tumors were measured at 7-day intervals using a
caliper, and xenografted tumor size was calculated. Following the
sacrifice of the animals, tumors were resected and weighed. Samples
were fixed with 10% methanol at room temperature for 24 h. Samples
were cut into 0.5-µm-thick sections, prepared on 1 mm3
slides, and stained with hematoxylin and eosin at 37°C for 4 min.
The sections were then observed under a light microscope
(magnification, ×100 and ×200) to examine morphology changes.
Gene profiling detected by
microarray
The high-NC group and the saline group were used to
microarray analysis. Human OneArray was provided by Aksomics, Inc.
(Shanghai, China). The sample preparation, array hybridization and
data analysis procedures were performed as previously reported
(27,28). Genes with a fold change (FC) of ≥1.5
were considered differentially expressed genes (DEGs).
Gene enrichment analysis and network
construction of DEGs following NC treatment
DEGs determined by the microarray analysis were
further submitted for Gene Ontology (GO) and Kyoto Encyclopedia of
Genes and Genomes (KEGG) analyses using the Database for
Annotation, Visualization, and Integrated Discovery tool (version
6.8; http://david.ncifcrf.gov/). Functional
enrichment terms at P<0.05 were selected for further evaluation.
To provide an intuitionistic and full-scale comprehension of the
interaction network, the Search Tool for the Retrieval of
Interacting Genes/Proteins database (https://string-db.org/; version 10.0) was utilized to
construct a protein-protein interaction (PPI) network. Interactions
among genes were calculated by setting a confidence threshold of
>0.9.
Connectivity Map (cMap)
To further elucidate potential regulatory mechanisms
of the tumor suppressive function of NC, a list of DEGs was
submitted to the cMap database (https://portals.broadinstitute.org/cmap/; version,
build 02) (29). Each imported
query was compared with the predefined signatures of therapeutic
compounds and ranked by the connectivity score (from −1 to 1),
indicating the relative similarity of DEG profiles produced
following NC treatment and the compound's treatment. A high
positive connectivity score indicated compounds with functions
similar to that of NC. Compounds with negative connectivity scores
were dissimilar.
DEGs in liver cancer based on The
Cancer Genome Atlas (TCGA)
A list of DEGs in hepatocellular carcinoma, a type
of liver cancer, and non-tumor samples was obtained from the online
database, Gene Expression Profiling Interactive Analysis (GEPIA),
to reveal potential mechanisms by which NC suppresses liver cancer
(30). The set of DEGs met the
following criteria: |Log2FC| >1 and q<0.05.
Considering the significant anti-proliferation effects of NC in
liver cancer cells, targets of NC are likely to be involved in the
initiation and progression of liver cancer (23). Genes that were simultaneously
upregulated in liver cancer and downregulated in the NC-treated
groups were selected as key genes, as such genes that may be
effective inhibitory targets of NC treatment for liver cancer. NC
could exert its significant effects via these targets. Among the
DEGs identified in the in-house microarray, the authors of the
present study proposed that the key genes involved in liver cancer
initiation and progression may be the effective targets of NC.
The Cancer Cell Line Encyclopedia
(CCLE)
The CCLE (version 2018; http://portals.broadinstitute.org/ccle), a database
with detailed and comprehensive genetic characterizations, provides
28 publicly accessible liver cancer cell lines (namely SNU449, C3A,
JHH7, HUH6, SNU761, LI7, HLF, JHH4, JHH1, JHH5, HUH7, SNU182,
SNU398, SNU423, SNU387, SNU475, JHH6, SKHEP1, SNU886, SNU878,
NCI-H684, PLC/PRF/5, Hep 3B2.1-7, HLE, PLCPRF5, HEPG2, HUH1 and
JHH2 cell lines) with mRNA expression data. The expression levels
of the aforementioned key genes were also validated using the data
for liver cancer cell lines downloaded from the CCLE.
Development of diagnostic and
prognostic signatures
Key genes were subjected to logistic regression
analysis to develop a diagnostic index, which was more powerful
than a single gene for distinguishing liver cancer from non-tumor
samples. A prognostic risk score was constructed using the online
database SurvExpress (version 2.0) (31) to validate the performance of
multi-gene biomarkers for clinical outcomes in human cancers.
Determination of alteration status of
key genes
To explore the underlying mechanisms relevant to
genetic alterations in key genes, their alteration status was
analyzed using the cBioPortal database (http://www.cbioportal.org/index.do) (32). The proportions of liver cancer
patients with alterations to each gene and their survival status
were obtained.
Statistical analysis
All statistical analyses were conducted using SPSS
24.0 (IBM Corp., Armonk, NY, USA). Associations between gene
expression level and clinicopathological parameters of liver cancer
patients were calculated using independent sample t-tests. One-way
analysis of variance (ANOVA) followed by Bonferroni post hoc
analysis was conducted to compare the difference of tumor weight
when the control group was compared with low-NC, medium-NC and
high-NC groups. The RNA-seq data obtained from TCGA and then
log2-transformed for further analysis. A differentially expressed
gene list was downloaded from GEPIA database. To explore the
clinical significance of key genes, an independent t-test was used
to compare the difference between HCC and non-tumor tissues.
Receiver operating characteristic (ROC) curves and corresponding
area under the ROC curves (AUC) were calculated for each gene to
reveal the utility of genes for distinguishing liver cancer from
non-tumor samples. Kaplan-Meier (K-M) plots of key genes were
obtained from the GEPIA database. For all tests, P<0.05
indicated that the difference between groups was statistically
significant.
Results
Inhibitory effect of NC on HepG2 cells
based on in vitro and in vivo models
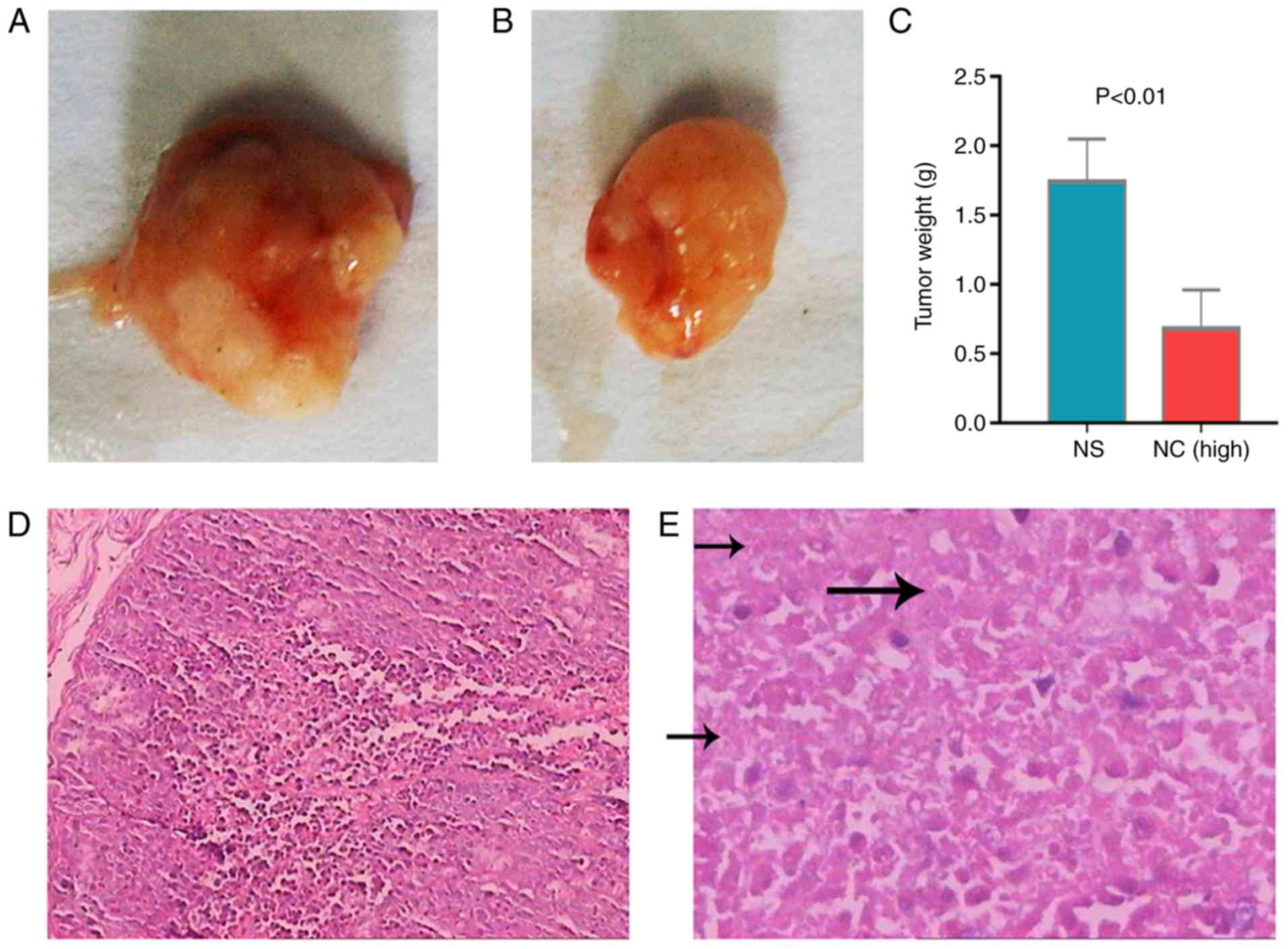
The tumor growth inhibitory effect of NC indicated
that the xenografts of untreated mice were significant heavier that
of NC-treated mice (Fig. 1A and B).
Tumor weight in the high-NC group was significantly lower compared
with the saline group (Fig. 1C).
Furthermore, there were many necrotic tumor cells in NC-treated
group compared with control group (Fig.
1D and E). Microarray analysis revealed that 280 genes were
altered post-NC treatment, including 138 upregulated and 142
downregulated genes (Fig. 1F).
These genes were sent for further pathway and cMap inquiries. The
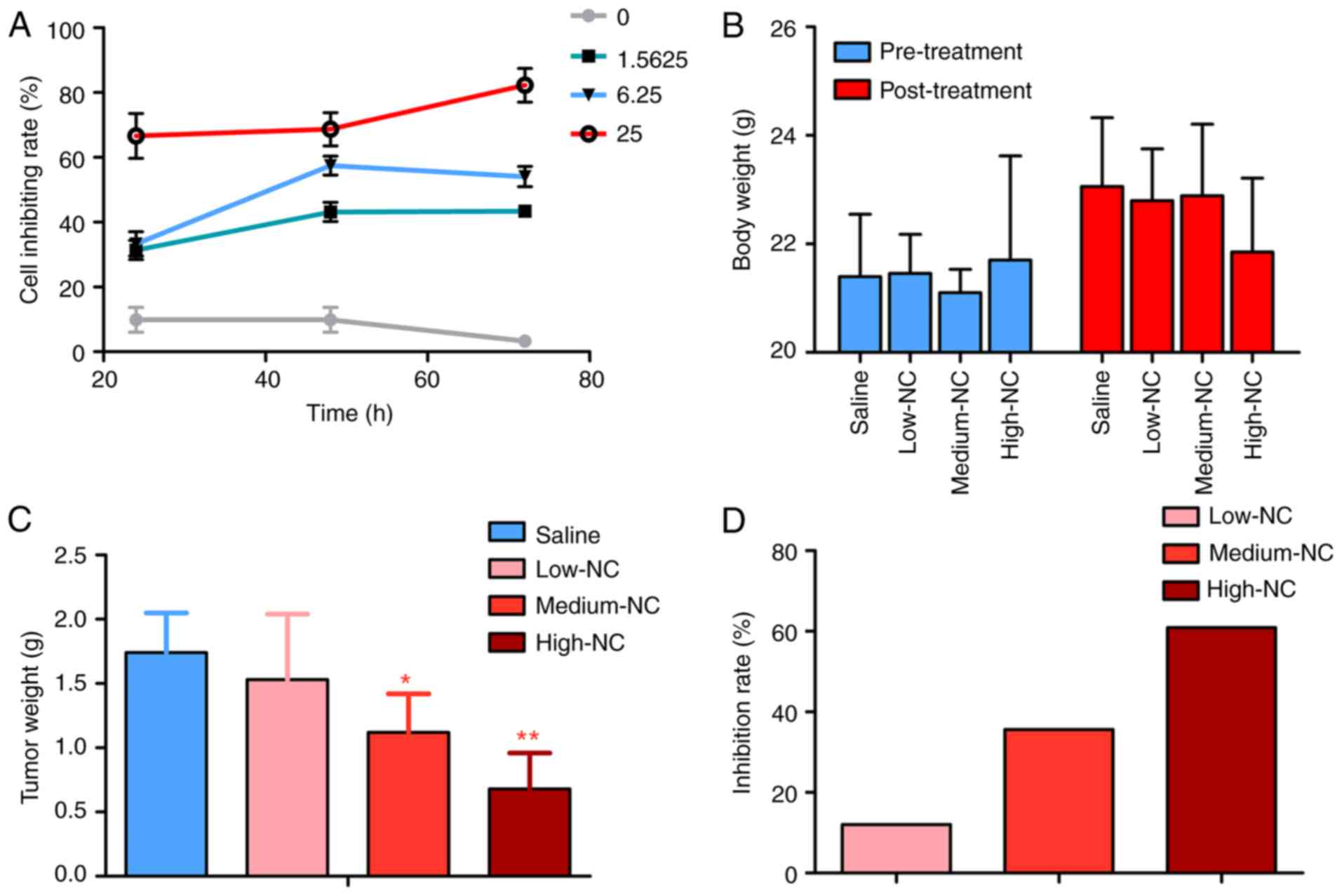
cell growth inhibitory effect of NC on HepG2 cells in vitro
was also confirmed. The higher NC concentrations markedly increased
the inhibition rate of HepG2 cells (Fig. 2A). The body weight of nude mice was
markedly changed following NC treatments (Fig. 2B). Tumor weights were significantly
higher in the saline group compared with the medium-NC (P=0.012)
and high-NC groups (P<0.001; Fig.
2C). The inhibition rate of NC increased in what appeared to be
a dose dependent manner (Fig.
2D).
GO term enrichment and KEGG
pathways
Results of gene functional enrichment analysis are
summarized in Table I. Results of
GO term enrichment analysis demonstrated that DEGs were most
significantly associated with the biological process terms
‘leukocyte migration’, ‘embryonic foregut morphogenesis’ and
‘response to antibiotic’. Unsurprisingly, an
inflammation-associated term ranked first on the list. DEGs were
also most significantly associated with the cellular component
terms ‘nucleus’, ‘nucleoplasm’ and ‘cytosol’ (Fig. 3B). The top three most significant
terms associated with molecular function were ‘protein kinase
binding’, ‘DNA binding’ and ‘protein binding’. The results of the
KEGG enrichment analysis indicated that the transforming growth
factor (TGF)-β signaling pathway, the phosphatidylinositol
4,5-bisphosphate 3-kinase/RAC-α serine/threonine-protein kinase
(PI3K-Akt) signaling pathway, and several tumor-associated
signalling pathways were associated with the DEGs. These results
depict the functional terms of DEGs. Using gene functional
enrichment analysis, the potential molecular functions of NC were
elucidated. NC may exert antitumor function via these pathways.
‘TGF-beta signaling pathway’, ‘PI3K-Akt signaling pathway’ and
‘Arrhythmogenic right ventricular cardiomyopathy (ARVC)’ were the
three most significant KEGG pathways. The TGF-β signaling pathway,
which is involved in regulating biological processes, including
cell growth, proliferation, differentiation, migration and
apoptosis (33), was the top result
of the enrichment analysis. Another important signaling pathway,
PI3K-Akt, which has been demonstrated to significantly stimulate
cell proliferation while inhibiting apoptosis (34), was suppressed by NC treatment.
 | Table I.Gene functional enrichment analysis
of differentially expressed genes. |
Table I.
Gene functional enrichment analysis
of differentially expressed genes.
| Category | Term | Count | Gene ratio (%) | P-value |
|---|
| Biological
process |
GO:0050900-leukocyte migration | 7 | 2.702703 | 0.004638 |
| Biological
process |
GO:0048617-embryonic foregut
morphogenesis | 3 | 1.158301 | 0.005437 |
| Biological
process | GO:0046677-response
to antibiotic | 4 | 1.544402 | 0.007613 |
| Biological
process | GO:0016337-single
organismal cell-cell adhesion | 6 | 2.316602 | 0.009284 |
| Biological
process |
GO:0006355-regulation of transcription,
DNA-templated | 30 | 11.58301 | 0.015146 |
| Biological
process | GO:0060021-palate
development | 5 | 1.930502 | 0.015870 |
| Biological
process |
GO:1903959-regulation of anion
transmembrane transport | 3 | 1.158301 | 0.019216 |
| Biological
process | GO:0031016-pancreas
development | 3 | 1.158301 | 0.028706 |
| Biological
process | GO:0050821-protein
stabilization | 6 | 2.316602 | 0.029689 |
| Biological
process | GO:0030335-positive
regulation of cell migration | 7 | 2.702703 | 0.030093 |
| Cellular
component |
GO:0005634-nucleus | 90 | 34.749030 | 0.001786 |
| Cellular
component |
GO:0005654-nucleoplasm | 53 | 20.463320 | 0.001803 |
| Cellular
component |
GO:0005829-cytosol | 56 | 21.621620 | 0.015614 |
| Cellular
component |
GO:0005801-cis-Golgi network | 4 | 1.544402 | 0.015682 |
| Cellular
component |
GO:0005737-cytoplasm | 81 | 31.27413 | 0.022047 |
| Cellular
component |
GO:0030425-dendrite | 10 | 3.861004 | 0.026234 |
| Cellular
component |
GO:0031012-extracellular matrix | 9 | 3.474903 | 0.034230 |
| Cellular
component |
GO:0005913-cell-cell adherens
junction | 9 | 3.474903 | 0.051794 |
| Cellular
component | GO:0005925-focal
adhesion | 10 | 3.861004 | 0.059138 |
| Cellular
component |
GO:0005667-transcription factor
complex | 6 | 2.316602 | 0.096043 |
| Molecular
function | GO:0019901-protein
kinase binding | 14 | 5.405405 | 0.001355 |
| Molecular
function | GO:0003677-DNA
binding | 35 | 13.513510 | 0.006048 |
| Molecular
function | GO:0005515-protein
binding | 134 | 51.737450 | 0.007031 |
| Molecular
function | GO:0042826-histone
deacetylase binding | 6 | 2.316602 | 0.010778 |
| Molecular
function |
GO:0044822-polyadenylated RNA binding | 24 | 9.266409 | 0.022243 |
| Molecular
function | GO:0046872-metal
ion binding | 37 | 14.285710 | 0.042665 |
| Molecular
function | GO:0050660-flavin
adenine dinucleotide binding | 4 | 1.544402 | 0.050953 |
| Molecular
function |
GO:0061631-ubiquitin conjugating enzyme
activity | 3 | 1.158301 | 0.054528 |
| Molecular
function |
GO:0003682-chromatin binding | 10 | 3.861004 | 0.070868 |
| Molecular
function | GO:0042393-histone
binding | 5 | 1.930502 | 0.075467 |
| KEGG pathway |
hsa04350-transforming growth factor-β
signaling pathway | 5 | 1.930502 | 0.030985 |
| KEGG pathway | hsa04151-PI3K-Akt
signaling pathway | 10 | 3.861004 | 0.055178 |
| KEGG pathway |
hsa05412-arrhythmogenic right ventricular
cardiomyopathy | 4 | 1.544402 | 0.079302 |
| KEGG pathway |
hsa04022-cGMP-protein kinase G signaling
pathway | 6 | 2.316602 | 0.086341 |
| KEGG pathway | hsa04141-protein
processing in endoplasmic reticulum | 6 | 2.316602 | 0.091529 |
| KEGG pathway | hsa03013-RNA
transport | 6 | 2.316602 | 0.096875 |
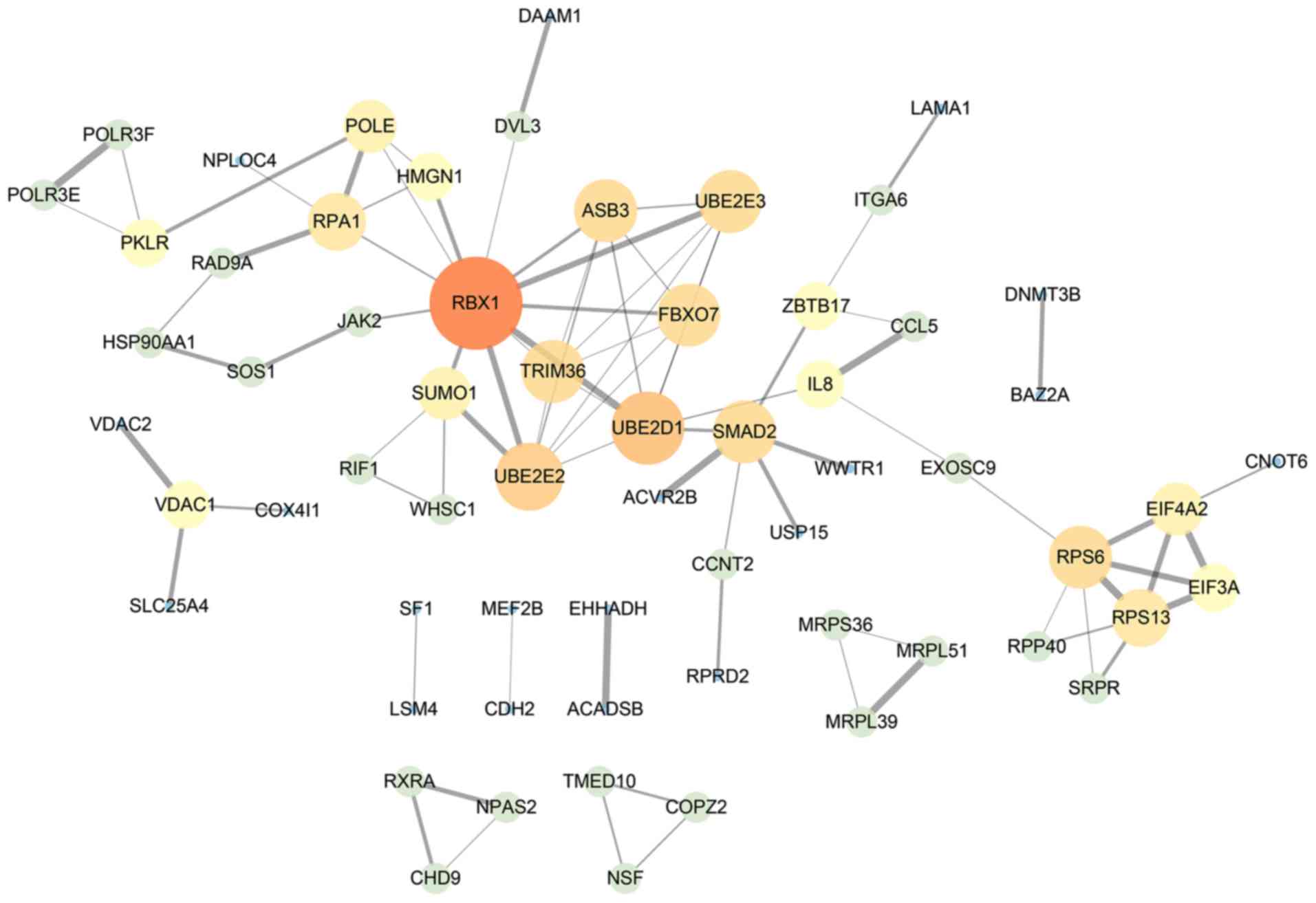
Network analysis
The PPI network was constructed according to gene
interactions with a confidence threshold >0.9. In total, 84 PPI
associations in 61 nodes were identified. RBX1 was the core
gene in the network, suggesting it may be central to the action of
NC on HepG2 cells (Fig. 3).
Analysis of NC by cMap
The results of the cMap analysis revealed a series
of compounds with either similar or opposite effects to that of NC.
The top ten results were parthenolide, trichostatin A,
thapsigargin, disulfiram, vorinostat, ciclopirox, antimycin A,
scriptaid, hycanthone and phenoxybenzamine, as presented in
Table II. Among them, antimycin A
may exhibit opposite effects to NC. The remaining compounds may
have similar functions to that of NC. These findings suggest that
NC's efficacy may be based on activities similar or opposite to
those of known compounds.
 | Table II.cMap analysis of the top 10 molecules
similar to nitidine chloride. |
Table II.
cMap analysis of the top 10 molecules
similar to nitidine chloride.
| Rank | cMap name | Mean | n | Enrichment | P-value | Specificity |
|---|
| 1 | Parthenolide | 0.684 | 4 | 0.944 | <0.00001 | 0.0359 |
| 2 | Trichostatin A | 0.376 | 182 | 0.526 | <0.00001 | 0.2417 |
| 3 | Thapsigargin | 0.632 | 3 | 0.940 | 0.00030 | 0.1019 |
| 4 | Disulfiram | 0.556 | 5 | 0.817 | 0.00046 | 0.0116 |
| 5 | Vorinostat | 0.319 | 12 | 0.536 | 0.00084 | 0.3970 |
| 6 | Ciclopirox | 0.539 | 4 | 0.828 | 0.00133 | 0.0386 |
| 7 | Antimycin A | −0.379 | 5 | −0.763 | 0.00142 | 0.0000 |
| 8 | Scriptaid | 0.587 | 3 | 0.906 | 0.00174 | 0.0833 |
| 9 | Hycanthone | 0.618 | 4 | 0.793 | 0.00362 | 0.0622 |
| 10 |
Phenoxybenzamine | 0.559 | 4 | 0.792 | 0.00364 | 0.2822 |
Key genes influenced by NC in liver
cancer
By screening TCGA data from the GEPIA database,
2,218 DEGs were identified. Among them, 1,485 genes were
upregulated and 733 downregulated. In the list of upregulated
genes, it was determined that eight genes were significantly
suppressed by NC treatment, including LSM4, CDKN2AIPNL, MFSD5,
ITGA6, HMGN1, ZCRB1, RTN2 and C1ORF54 (Fig. 4A). However, no genes that are
differentially downregulated in liver cancer were overexpressed in
the NC-treated groups (Fig. 4B). A
heat map demonstrated the expression profiles of the eight genes;
it revealed that the eight genes were upregulated in HCC tissues
(Fig. 4C).
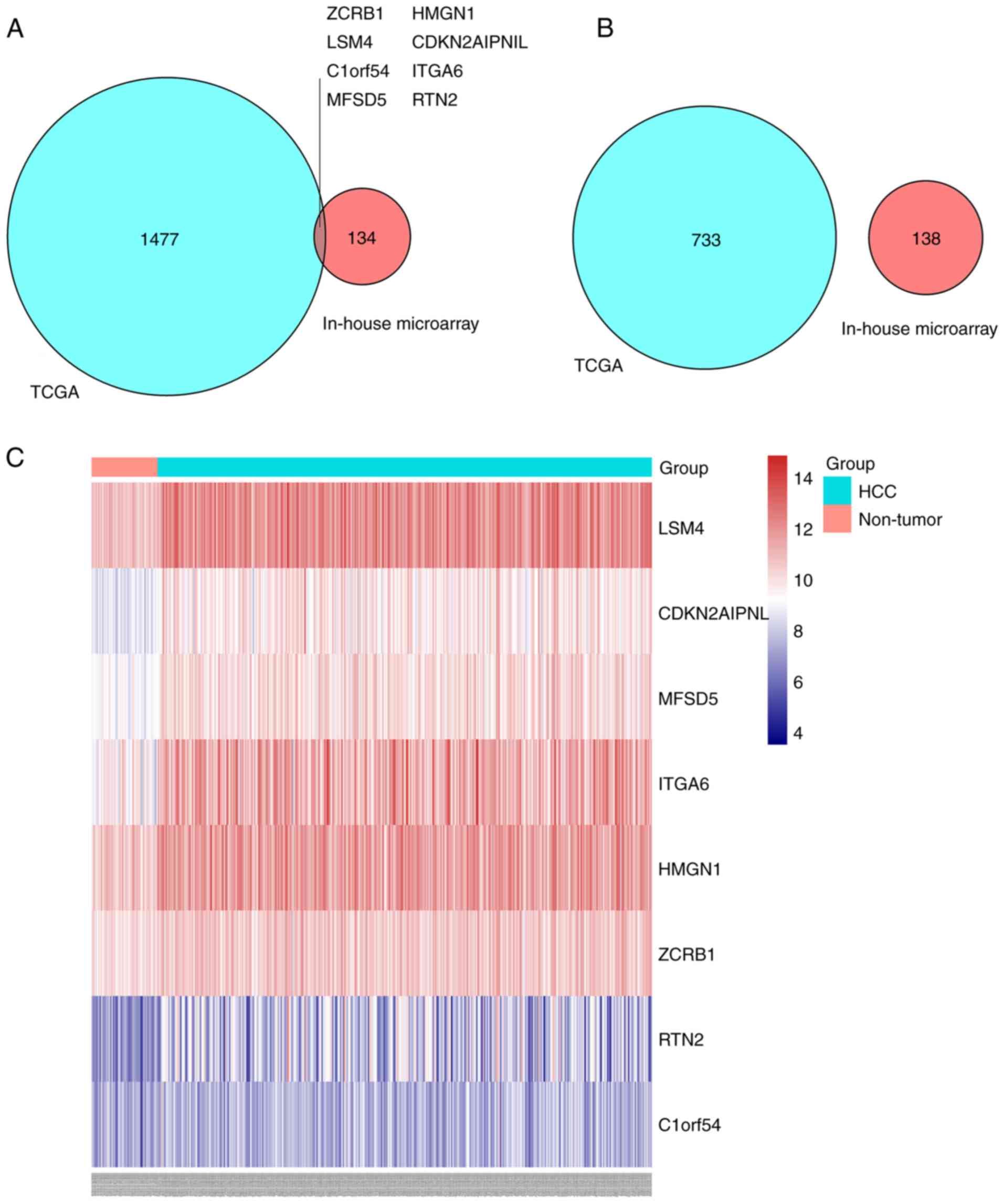 | Figure 4.Intersection of DEGs of TCGA and DEGs
of the in-house microarray. Venn diagrams of (A) genes that were
differentially upregulated in liver cancer and inhibited in the
NC-treated groups, and (B) genes that were differentially
downregulated in liver cancer and overexpressed in the NC-treated
groups. (C) A heat map presenting the expression profiles of the
eight key genes. TCGA, The Cancer Genome Atlas; DEG, differentially
expressed gene. LSM4, U6 snRNA-associated Sm-like protein LSm4;
MFSD5, molybdate-anion transporter; HMGN1, non-histone chromosomal
protein HMG-14; RTN2, reticulon-2; CDKN2AIPNL, CDKN2AIP
N-terminal-like protein; ITGA6, integrin α-6; ZCRB1, zinc finger
CCHC-type and RNA-binding motif-containing protein 1; C1orf54,
uncharacterized protein C1orf54. |
Clinical significance of the eight key
genes
Revealing the clinical significance of these eight
key genes could illuminate how NC suppresses tumors in HepG2 cells.
The AUC values of these genes in the diagnosis of liver cancer, as
well as the prediction of progress and prognosis of liver cancer
were comprehensively analyzed. As presented in Fig. 5, these genes were overall
significantly elevated in liver cancer compared with non-tumor
tissues, although C1ORF54 expression levels only revealed a
trend toward statistical significance. AUC values for the
expression levels of the other seven genes were >0.7,
illustrating a moderate potential for distinguishing liver cancer
from non-tumor liver samples.
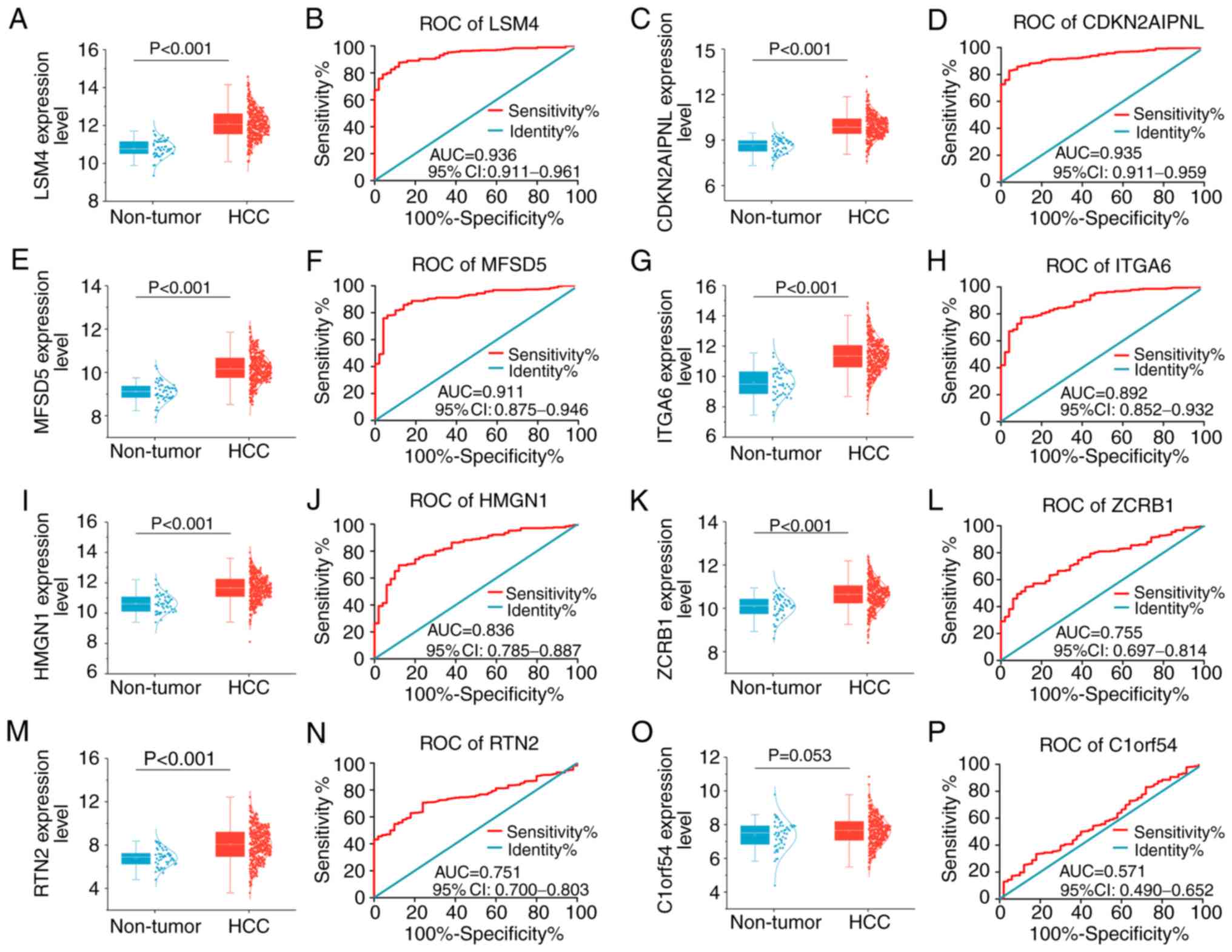 | Figure 5.Expression levels and receiver
operating characteristic curves of eight key genes between liver
cancer and non-tumor samples. Gene expression was assessed in liver
cancer and non-tumor samples. (A) Scatter plot and (B) ROC curve of
LSM4 expression. (C) Scatter plot and (D) ROC curve of CDKN2AIPNL
expression. (E) Scatter plot and (F) ROC curve of MFSD5 expression.
(G) Scatter plot and (H) ROC curve of ITGA6 expression. (I) Scatter
plot and (J) ROC curve of HMGN1 expression. (K) Scatter plot and
(L) ROC curve of ZCRB1 expression. (M) Scatter plot and (N) ROC
curve of RTN2 expression. (O) Scatter plot and (P) ROC curve of
C1ORF54 expression. ROC, receiver operating characteristic; HCC,
hepatocellular carcinoma; AUC, area under the ROC curves; CI,
confidence interval; LSM4, U6 snRNA-associated Sm-like protein
LSm4; MFSD5, molybdate-anion transporter; HMGN1, non-histone
chromosomal protein HMG-14; RTN2, reticulon-2; CDKN2AIPNL, CDKN2AIP
N-terminal-like protein; ITGA6, integrin α-6; ZCRB1, zinc finger
CCHC-type and RNA-binding motif-containing protein 1; C1orf54,
uncharacterized protein C1orf54. |
The patients with liver cancer were divided into
different groups based on clinical parameters to analyze
differences in gene expression. Clinicopathological relevance
analyses were performed for several variables, including age (≥60
years old vs. <60 years old; Fig.
6A), sex (male vs. female; Fig.
6B), pathologic stage (stage III/IV vs. stage I/II; Fig. 6C) and histologic grade (grade III/IV
vs. grade I/II; Fig. 6D).
HMGN1 was significantly overexpressed in younger patients.
No noteworthy differences were observed between female and male
patients with respect to the eight key genes. With respect to
pathological stage, LSM4, MFSD5, ITGA6, HMGN1 and
ZCRB1 were significantly upregulated in patients at advanced
stages. As for histological grade, LSM4, CDKN2AIPNL, MFSD5
and HMGN1 were significantly overexpressed in poorly
differentiated tumor samples.
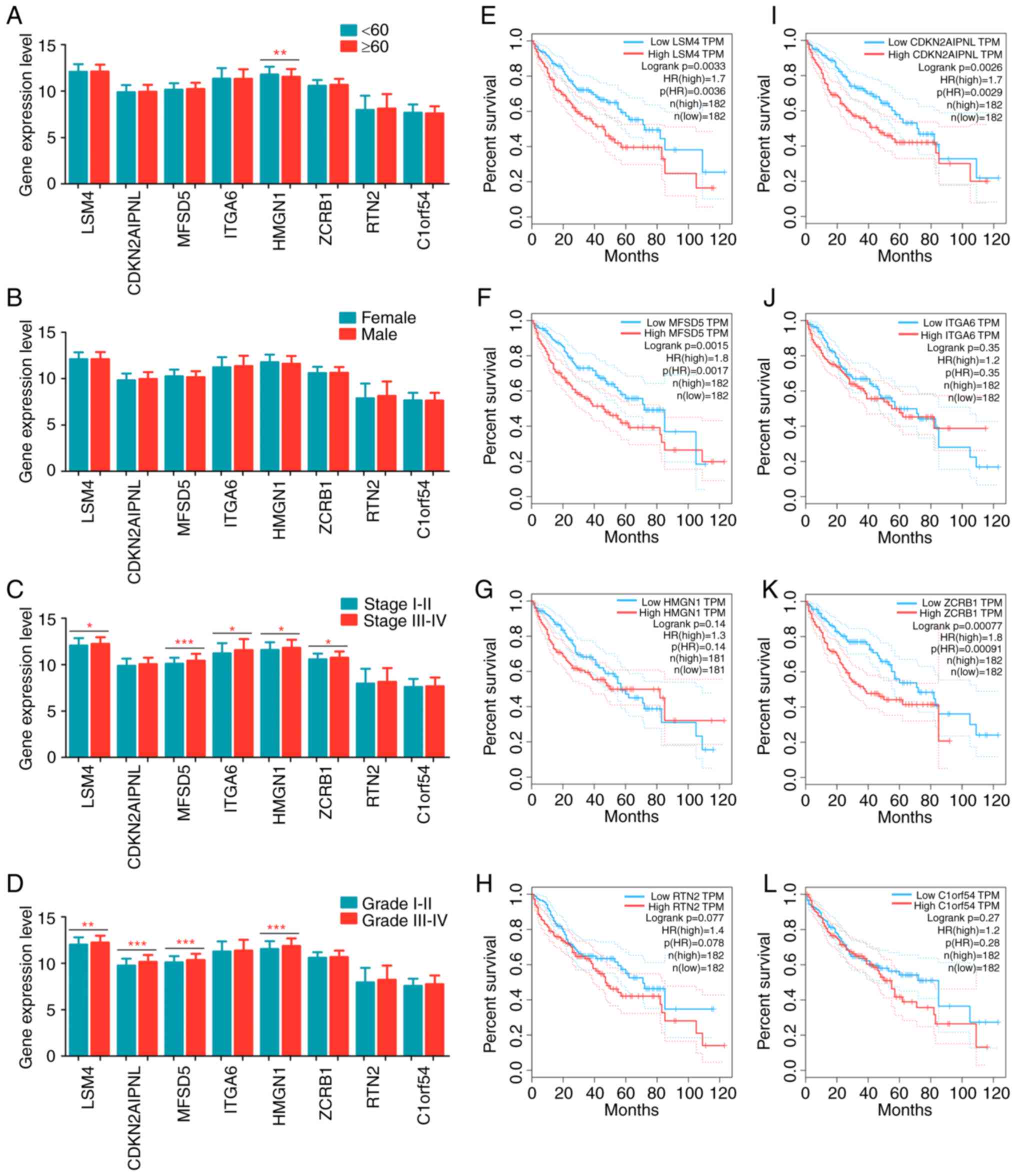 | Figure 6.The clinical significance of eight
key genes in liver cancer. Gene expression levels between (A)
patients <60 and ≤60 years old, (B) female and male, (C) tumor
stages I–II and III–IV, and (D) tumor grades I–II and III–IV.
Kaplan-Meier plots of (E) LSM4, (F) MFSD5, (G)
HMGN1, (H) RTN2, (I) CDKN2AIPNL, (J)
ITGA6, (K) ZCRB1 and (L) C1ORF54. HR, hazard
ratio; LSM4, U6 snRNA-associated Sm-like protein LSm4; MFSD5,
molybdate-anion transporter; HMGN1, non-histone chromosomal protein
HMG-14; RTN2, reticulon-2; CDKN2AIPNL, CDKN2AIP N-terminal-like
protein; ITGA6, integrin α-6; ZCRB1, zinc finger CCHC-type and
RNA-binding motif-containing protein 1; C1orf54, uncharacterized
protein C1orf54; TPM, transcripts per million kilobases.
*P<0.05, **P<0.01 and ***P<0.001, indicating significant
difference. |
Survival curves were drawn using K-M analysis and a
log-rank test was used to evaluate the prognostic values of these
genes. Inferior overall survival was associated with high
expression levels of LSM4, CDKN2AIPNL, MFSD5 or ZCRB;
however, no difference in overall survival was observed when the
expression levels of ITGA6, HMGN1, RTN2 and C1ORF54
were assessed (Fig. 6E-L). These
findings indicated that these genes exerted an indispensable
function in the initiation and progression of liver cancer. More
importantly, these genes were significantly reduced in the
NC-treated groups (Fig. 4A). These
results suggest that NC may act as a tumor suppressor in liver
cancer progression by inhibiting the activities of these genes.
Expression profiles in liver cancer
cell lines
The authors of the present study analyzed gene
expression values in the cell lines and found that all key genes
had high expression levels with the exception of C1ORF54 (Fig. 7A).
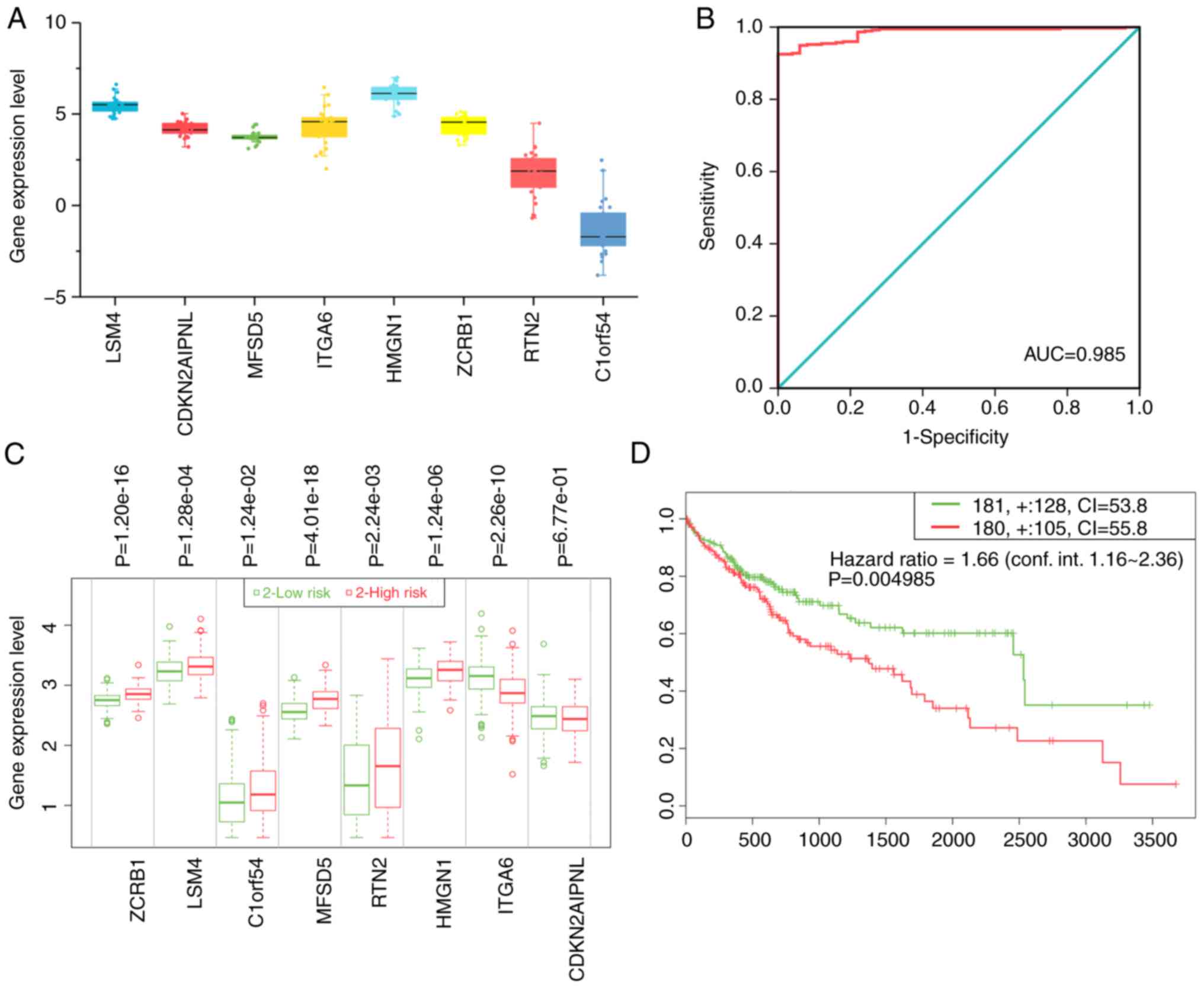 | Figure 7.The overall clinical value of key
genes. (A) Relative expression levels of key genes in liver cancer
cells. (B) ROC of diagnostic index. (C) The expression levels of
the eight key genes and (D) the Kaplan-Meier plot in the high- and
low-risk groups. ROC, receiver operating characteristic; AUC, area
under the ROC curves; CI, confidence interval; LSM4, U6
snRNA-associated Sm-like protein LSm4; MFSD5, molybdate-anion
transporter; HMGN1, non-histone chromosomal protein HMG-14; RTN2,
reticulon-2; CDKN2AIPNL, CDKN2AIP N-terminal-like protein; ITGA6,
integrin α-6; ZCRB1, zinc finger CCHC-type and RNA-binding
motif-containing protein 1; C1orf54, uncharacterized protein
C1orf54. |
Development of diagnostic index and
prognostic index
A diagnostic index according to the following
formula was constructed: Expression value of LSM4x2.305 +
expression value of CDKN2AIPNLx2.738 + expression value of
ITGA6x2.336-expression value of
ZCRB1x2.433-expression value of C1ORF54x1.336. The
resulting diagnostic index significantly improved the ability of
the authors to distinguish liver cancer from non-tumor tissues
[AUC=0.985; 95% confidence interval (CI)=0.975–0.994; P<0.001;
Fig. 7B]. These eight genes were
also used to construct a prognostic signature using SurvExpress.
The prognostic signature based on these genes was able to separate
patients with liver cancer into two groups with significantly
different prognostic statuses (hazard ratio=1.66; 95% CI=1.16–2.36;
P=0.005; Fig. 7C and D). Patients
in the high-risk group exhibited a worse prognosis.
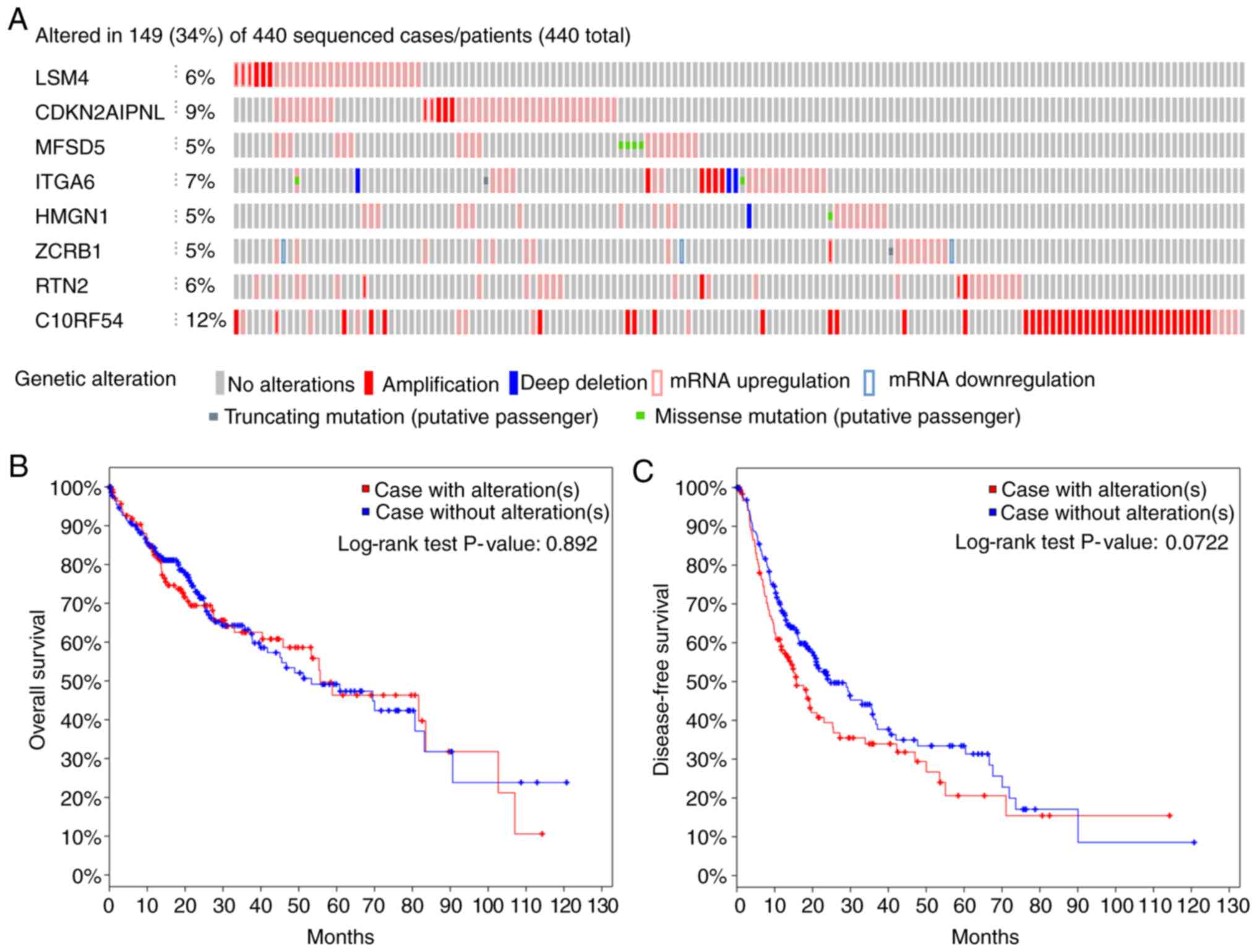
Genetic mutation status
Out of 440 patients with liver cancer, genetic
alterations in the key genes was detected in 149 (34%) of them
using the online database cBioPortal (Fig. 8A). Among them, C1ORF54 was
the gene with the highest alteration rate. However, K-M analysis
yielded no differences between cases with and without alterations
(Fig. 8B and 8C).
Discussion
Healthy cells carefully control cell proliferation,
thereby ensuring homeostasis of the cell cycle, and uncontrolled
proliferation is considered a fundamental trait of cancerous cells
(35). It is crucial to understand
the underlying mechanisms of proliferation and apoptosis to seek
out and develop effective agents for combating the proliferation of
cancer cells. In the present study, the authors identified DEGs
between the high-NC and saline groups using microarray technology
on HepG2 cell xenografts in nude mice. Next, functional annotation
was conducted and functional connections between drugs to reveal
the molecular characteristics of NC function were identified. TCGA
database was mined and eight key genes with potential significant
clinical value in modulating proliferation were uncovered. These
eight genes may be the effectors upon which NC applies its
antitumor function. These findings could help identify novel
potential targets for the treatment of liver cancer by NC.
Recent studies have demonstrated that NC is an
important antitumor compound (24,36).
Of all the functions in which NC has been implicated, the authors
of the present study consider inhibiting proliferation and inducing
apoptosis are the most important. The process of liver cancer
treatment by NC may involve multiple genes. Thus, DEGs (identified
by analyzing gene expression prior to and after NC treatment) were
studied to provide a theoretical basis for delineating the
mechanism by which NC inhibits proliferation and tumor cell cycles,
and induces apoptosis. GO and KEGG pathway analyses predicted that
the downregulated and upregulated genes were associated with
inflammation- or cancer-associated terms. The first biological
process term, ‘leukocyte migration,’ indicated that the immunologic
microenvironment was significantly influenced by NC. Indeed, NC is
a well-known inhibitor of inflammation (18,19).
The PI3K-Akt signaling pathway, which regulates cell
growth, survival, metabolism and apoptosis, was also implicated
(37,38). A growing body of research has
indicated that inhibiting PI3K/Akt signaling could contribute to
the suppression of liver cancer development (39–41).
Of interest, inflammation- and tumor-progression-associated
pathways were prominent in the results. The leading enriched KEGG
pathway, TGF-β, has also been considered to act as a
pro-tumorigenic factor in liver cancer by remodeling chronic
inflammation-mediated hepatocarcinogenesis, and promoting liver
cancer growth, metastasis and angiogenesis (42–44).
The findings of the present study suggest that NC acts against
liver cancer molecular targets and the tumor growth
environment.
In addition to interfering with signaling pathways,
NC may also have molecular functions similar to those of
established compounds. Based on the results of validation
experiments using cMap, NC was considered to exert a similar
function to parthenolide, which could explain the molecular
function of NC. Parthenolide has been revealed to possess
anti-inflammatory and anticancer activities (45–47).
Carlisi et al (48) reported
that parthenolide sensitizes liver cancer cells to tumor necrosis
factor-associated, apoptosis-inducing ligands through the
inhibition of STAT3 activation. Of note, NC also inhibited the
activation of the JAK1/STAT3 signaling pathway in liver cancer
(22). This comparison further
supports an anti-inflammatory and antitumor capacity of NC on liver
cancer cells. The results of the present study also provide clues
to other biological NC mechanisms against liver cancer.
Among the DEGs identified in the in-house
microarray, key liver cancer initiation and progression genes
suspected of being targeted by NC were processed. NC was observed
to suppress a total of eight overexpressed genes in liver cancer.
Consistent with the hypothesis of the present study, high
expression levels of certain genes were significantly associated
with advanced stage and grade of liver cancer, and could serve as
potential biomarkers for predicting the overall survival of
patients with liver cancer. NC may enact its anti-proliferative
function through these genes. A study by our group also provided
certain clues as to NC functional mechanisms (49). Ou et al (23) demonstrated that NC induced apoptosis
in ovarian cancer cells by activating the Fas signaling pathway.
Kim et al (46) reported
that NC suppressed the growth of acute myeloid leukemia cells by
inhibiting the phosphorylation of AKT and ERK. A study has also
proposed that the upregulation of p53, p21 and Bax,
and the downregulation of Bcl-2 mediate NC apoptotic
function in liver cancer (23).
Many studies have addressed different targets for NC antitumor
function (25,26,50–52).
The present study integrated the classical functional experiments
and big data mining, which could provide novel insights into the
molecular characteristics of NC. Generally, cancer can be regarded
as a disorder in the equilibrium between cell growth and death, and
these genes are frequently involved in cancer disorders (49,3). In
recent years, the research and development of new drugs have been
based on the pharmacological action of drugs on multiple targets
and through multiple pathways to achieve long-term low toxicity and
to reduce drug resistance (53,54).
Clinical significance was also explored by treating the eight key
genes as a set. Additional survival analysis revealed that the gene
set was highly correlated with a reduced overall survival. This
result provides a useful conceptual framework for understanding the
complex biology of NC effects on liver cancer.
However, there are several limitations of the
present study. First, only two cell plates used for the microarray.
Hence, the preliminary study is required a confirmation in the
future. Second, several bioinformatics analyses were performed
based on prediction algorithms, future in vitro or in
vivo experiments are needed to explore the exact regulatory
mechanisms.
In conclusion, the present study determined that NC
exhibited tumor growth inhibitory effects in vitro and in
vivo. It was also demonstrated that different key genes are
potentially involved in NC-mediated antitumor function, implicating
several prospective proliferative signaling pathways. The present
study demonstrates the antitumor function of NC and provides
insights into the potential complex molecular functions of NC
against liver cancer. In brief, NC has the potential for future
clinical application when its in-depth molecular mechanisms have
been fully explored.
Acknowledgments
Not applicable.
Funding
The present study was financially supported by funds
from the National Natural Science Foundation of China (grant no.
NSFC81560489), the Natural Science Foundation of Guangxi, China
(grant no. 2017GXNSFAA198017), the Innovation Project of Guangxi
Graduate Education (grant no. YCSW2018104), the Guangxi First-class
Discipline Project for Pharmaceutical Sciences (grant no.
GXFCDP-PS-2018), the Guangxi Medical University Training Program
for Distinguished Young Scholars, and the Medical Excellence Award
Funded by the Creative Research Development Grant from the First
Affiliated Hospital of Guangxi Medical University.
Availability of data and materials
The datasets used during the present study are
available from the corresponding author upon reasonable
request.
Authors' contributions
YWD and GC conceived and designed the experiments.
PL and LML performed the experiments. PL and HY analyzed the data.
PL, HY, LML and GC wrote the manuscript. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
All procedures associated with the animal
experiments were approved by the Animal Ethics Committee of Guangxi
Medical University (Nanning, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2018. CA Cancer J Clin. 68:7–30. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Laursen L: A preventable cancer. Nature.
516:S2–3. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
3
|
Fujiwara N, Friedman SL, Goossens N and
Hoshida Y: Risk factors and prevention of hepatocellular carcinoma
in the era of precision medicine. J Hepatol. 68:526–549. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
El-Serag HB: Epidemiology of viral
hepatitis and hepatocellular carcinoma. Gastroenterology.
142:1264–1273 e1. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Liu Z, Wang C, Jiao X, Zhao S, Liu X, Wang
Y and Zhang J: miR-221 promotes growth and invasion of
hepatocellular carcinoma cells by constitutive activation of NFκB.
Am J Transl Res. 8:4764–4777. 2016.PubMed/NCBI
|
|
7
|
Wong CH, Wong CS and Chan SL: Targeting
angiogenic genes as a therapeutic approach for hepatocellular
carcinoma. Curr Gene Ther. 15:97–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Forner A, Reig M and Bruix J:
Hepatocellular carcinoma. Lancet. 391:1301–1314. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wu CH, Lan CH, Wu KL, Wu YM, Jane WN,
Hsiao M and Wu HC: Hepatocellular carcinoma-targeted nanoparticles
for cancer therapy. Int J Oncol. 52:389–401. 2018.PubMed/NCBI
|
|
10
|
Shi JY, Ma LJ, Zhang JW, Duan M, Ding ZB,
Yang LX, Cao Y, Zhou J, Fan J, Zhang X, et al: FOXP3 is a HCC
suppressor gene and Acts through regulating the TGF-β/Smad2/3
signaling pathway. BMC Cancer. 17:6482017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Raoul JL, Kudo M, Finn RS, Edeline J, Reig
M and Galle PR: Systemic therapy for intermediate and advanced
hepatocellular carcinoma: Sorafenib and beyond. Cancer Treat Rev.
68:16–24. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Nada Y, Rashad N, Eissa M, Ghonaim A,
Farag K, Saadawi I, Sheha A, El Gewaity M and Abdel-Rahman O:
Outcomes of treatment with sorafenib in Egyptian patients with
hepatocellular carcinoma: A retrospective cohort study. Expert Rev
Gastroenterol Hepatol. 12:99–107. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Bruix J, Takayama T, Mazzaferro V, Chau
GY, Yang J, Kudo M, Cai J, Poon RT, Han KH, Tak WY, et al: Adjuvant
sorafenib for hepatocellular carcinoma after resection or ablation
(STORM): A phase 3, randomised, double-blind, placebo-controlled
trial. Lancet Oncol. 16:1344–1354. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kudo M: Systemic therapy for
hepatocellular carcinoma: 2017 update. Oncology. 93 (Suppl
1):S135–S146. 2017. View Article : Google Scholar
|
|
15
|
Yuan W, Sun Y, Liu L, Zhou B, Wang S and
Gu D: Circulating lncRNAs serve as diagnostic markers for
hepatocellular carcinoma. Cell Physiol Biochem. 44:125–132. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wu J, Li A, Yang J, Lu Y and Li J:
Efficacy and safety of TACE in combination with sorafenib for the
treatment of TACE-refractory advanced hepatocellular carcinoma in
Chinese patients: A retrospective study. Onco Targets Ther.
10:2761–2768. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Mondal S, Bandyopadhyay S, Ghosh MK,
Mukhopadhyay S, Roy S and Mandal C: Natural products: Promising
resources for cancer drug discovery. Anticancer Agents Med Chem.
12:49–75. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hu J, Zhang WD, Liu RH, Zhang C, Shen YH,
Li HL, Liang MJ and Xu XK: Benzophenanthridine alkaloids from
Zanthoxylum nitidum (Roxb.) DC, and their analgesic and
anti-inflammatory activities. Chem Biodivers. 3:990–995. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Khan H, Hadda TB and Touzani R: Diverse
therapeutic potential of nitidine, a comprehensive review. Curr
Drug Metab. 19:986–991. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Fang Z, Tang Y, Jiao W, Xing Z, Guo Z,
Wang W, Shi B, Xu Z and Liu Z: Nitidine chloride inhibits renal
cancer cell metastasis via suppressing AKT signaling pathway. Food
Chem Toxicol. 60:246–251. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kim LH, Khadka S, Shin JA, Jung JY, Ryu
MH, Yu HJ, Lee HN, Jang B, Yang IH, Won DH, et al: Nitidine
chloride acts as an apoptosis inducer in human oral cancer cells
and a nude mouse xenograft model via inhibition of STAT3.
Oncotarget. 8:91306–91315. 2017.PubMed/NCBI
|
|
22
|
Liao J, Xu T, Zheng JX, Lin JM, Cai QY, Yu
DB and Peng J: Nitidine chloride inhibits hepatocellular carcinoma
cell growth in vivo through the suppression of the
JAK1/STAT3 signaling pathway. Int J Mol Med. 32:79–84. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ou X, Lu Y, Liao L, Li D, Liu L, Liu H and
Xu H: Nitidine chloride induces apoptosis in human hepatocellular
carcinoma cells through a pathway involving p53, p21, Bax and
Bcl-2. Oncol Rep. 33:1264–1274. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Chen S, Yang L and Feng J: Nitidine
chloride inhibits proliferation and induces apoptosis in ovarian
cancer cells by activating the Fas signalling pathway. J Pharm
Pharmacol. 70:778–786. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Cheng Z, Guo Y, Yang Y, Kan J, Dai S,
Helian M, Li B, Xu J and Liu C: Nitidine chloride suppresses
epithelial-to-mesenchymal transition in osteosarcoma cell migration
and invasion through Akt/GSK-3β/Snail signaling pathway. Oncol Rep.
36:1023–1029. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lin J, Shen A, Chen H, Liao J, Xu T, Liu
L, Lin J and Peng J: Nitidine chloride inhibits hepatic cancer
growth via modulation of multiple signaling pathways. BMC Cancer.
14:7292014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhang Y, Dang YW, Wang X, Yang X, Zhang R,
Lv ZL and Chen G: Comprehensive analysis of long non-coding RNA
PVT1 gene interaction regulatory network in hepatocellular
carcinoma using gene microarray and bioinformatics. Am J Transl
Res. 9:3904–3917. 2017.PubMed/NCBI
|
|
28
|
Zhang Y, He RQ, Dang YW, Zhang XL, Wang X,
Huang SN, Huang WT, Jiang MT, Gan XN, Xie Y, et al: Comprehensive
analysis of the long noncoding RNA HOXA11-AS gene interaction
regulatory network in NSCLC cells. Cancer Cell Int. 16:892016.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lamb J, Crawford ED, Peck D, Modell JW,
Blat IC, Wrobel MJ, Lerner J, Brunet JP, Subramanian A, Ross KN, et
al: The Connectivity Map: Using gene-expression signatures to
connect small molecules, genes, and disease. Science.
313:1929–1935. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Tang Z, Li C, Kang B, Gao G, Li C and
Zhang Z: GEPIA: A web server for cancer and normal gene expression
profiling and interactive analyses. Nucleic Acids Res. 2017.
View Article : Google Scholar
|
|
31
|
Aguirre-Gamboa R, Gomez-Rueda H,
Martinez-Ledesma E, Martínez-Torteya A, Chacolla-Huaringa R,
Rodriguez-Barrientos A, Tamez-Peña JG and Treviño V: SurvExpress:
An online biomarker validation tool and database for cancer gene
expression data using survival analysis. PLoS One. 8:e742502013.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Gao J, Aksoy BA, Dogrusoz U, Dresdner G,
Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al:
Integrative analysis of complex cancer genomics and clinical
profiles using the cBioPortal. Sci Signal. 6:pl12013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Rao S and Mishra L: Targeting TGF-β
signaling in liver cancer. Hepatology. Dec 14–2018.(Epub ahead of
print). doi: 10.1002/hep.30426.
|
|
34
|
Liu M and Wang J, Qi Q, Huang B, Chen A,
Li X and Wang J: Nitidine chloride inhibits the malignant behavior
of human glioblastoma cells by targeting the PI3K/AKT/mTOR
signaling pathway. Oncol Rep. 36:2160–2168. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Kim LH, Khadka S, Shin JA, Jung JY, Ryu
MH, Yu HJ, Lee HN, Jang B, Yang IH, Won DH, et al: Nitidine
chloride acts as an apoptosis inducer in human oral cancer cells
and a nude mouse xenograft model via inhibition of STAT3.
Oncotarget. 8:91306–91315. 2017.PubMed/NCBI
|
|
37
|
Wang Z, Jiang W, Zhang Z, Qian M and Du B:
Nitidine chloride inhibits LPS-induced inflammatory cytokines
production via MAPK and NF-kappaB pathway in RAW 264.7 cells. J
Ethnopharmacol. 144:145–150. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Lamarca A, Mendiola M and Barriuso J:
Hepatocellular carcinoma: Exploring the impact of ethnicity on
molecular biology. Crit Rev Oncol Hematol. 105:65–72. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Bupathi M, Kaseb A, Meric-Bernstam F and
Naing A: Hepatocellular carcinoma: Where there is unmet need. Mol
Oncol. 9:1501–1509. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Zhu YJ, Zheng B, Wang HY and Chen L: New
knowledge of the mechanisms of sorafenib resistance in liver
cancer. Acta Pharmacol Sin. 38:614–622. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Liu L, Liao JZ, He XX and Li PY: The role
of autophagy in hepatocellular carcinoma: Friend or foe.
Oncotarget. 8:57707–57722. 2017.PubMed/NCBI
|
|
42
|
Mazzocca A, Antonaci S and Giannelli G:
The TGF-β signaling pathway as a pharmacological target in a
hepatocellular carcinoma. Curr Pharm Des. 18:4148–4154. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Neuzillet C, de Gramont A,
Tijeras-Raballand A, de Mestier L, Cros J, Faivre S and Raymond E:
Perspectives of TGF-β inhibition in pancreatic and hepatocellular
carcinomas. Oncotarget. 5:78–94. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Yang P, Li QJ, Feng Y, Zhang Y, Markowitz
GJ, Ning S, Deng Y, Zhao J, Jiang S, Yuan Y, et al:
TGF-β-miR-34a-CCL22 signaling-induced Treg cell recruitment
promotes venous metastases of HBV-positive hepatocellular
carcinoma. Cancer Cell. 22:291–303. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Li-Weber M, Palfi K, Giaisi M and Krammer
PH: Dual role of the anti-inflammatory sesquiterpene lactone:
Regulation of life and death by parthenolide. Cell Death Differ.
12:408–409. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Kim SL, Park YR, Lee ST and Kim SW:
Parthenolide suppresses hypoxia-inducible factor-1alpha signaling
and hypoxia induced epithelial-mesenchymal transition in colorectal
cancer. Int J Oncol. 51:1809–1820. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Zhang X, Chen Q, Liu J, Fan C, Wei Q, Chen
Z and Mao X: Parthenolide Promotes Differentiation of Osteoblasts
Through the Wnt/β-catenin signaling pathway in inflammatory
environments. J Interferon Cytokine Res. 37:406–414. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Carlisi D, D'Anneo A, Angileri L,
Lauricella M, Emanuele S, Santulli A, Vento R and Tesoriere G:
Parthenolide sensitizes hepatocellular carcinoma cells to TRAIL by
inducing the expression of death receptors through inhibition of
STAT3 activation. J Cell Physiol. 226:1632–1641. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Liu LM, Xiong DD, Lin P, Yang H, Dang YW
and Chen G: DNA topoisomerase 1 and 2A function as oncogenes in
liver cancer and may be direct targets of nitidine chloride. Int J
Oncol. 53:1897–1912. 2018.PubMed/NCBI
|
|
50
|
Li P, Yan S, Dong X, Li Z, Qiu Y, Ji C,
Zhang J, Ji M, Li W, Wang H, et al: Cell cycle arrest and apoptosis
induction activity of nitidine chloride on acute myeloid leukemia
cells. Med Chem. 14:60–66. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Liu N, Li P, Zang S, Liu Q, Ma D, Sun X
and Ji C: Novel agent nitidine chloride induces erythroid
differentiation and apoptosis in CML cells through c-Myc-miRNAs
axis. PLoS One. 10:e01168802015. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Zhai H, Hu S, Liu T, Wang F, Wang X, Wu G,
Zhang Y, Sui M, Liu H and Jiang L: Nitidine chloride inhibits
proliferation and induces apoptosis in colorectal cancer cells by
suppressing the ERK signaling pathway. Mol Med Rep. 13:2536–2542.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Mody K and Abou-Alfa GK: Systemic therapy
for advanced hepatocellular carcinoma in an evolving landscape.
Curr Treat Options Oncol. 20:32019. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Abou-Alfa GK, Meyer T, Cheng AL,
El-Khoueiry AB, Rimassa L, Ryoo BY, Cicin I, Merle P, Chen Y, Park
JW, et al: Cabozantinib in patients with advanced and progressing
hepatocellular carcinoma. N Engl J Med. 379:54–63. 2018. View Article : Google Scholar : PubMed/NCBI
|