Introduction
The DNA damage response causes numerous cascades of
post-translational modifications including ubiquitination,
SUMOylation, acetylation and phosphorylation that coordinate the
cell cycle, apoptosis and the DNA damage repair pathway (1,2).
Histone modification is one of the important events in the DNA
damage response pathway and regulates recruitment of other proteins
to DNA damage sites, chromatin remodeling and the enzymatic
signaling pathway during DNA damage (3,4).
Histone H2B can be post-translationally modified at
its N-terminal and C-terminal tail like other histone proteins.
Monoubiquitination at the Lys-120 (K120) residue of H2B is
associated with gene transcription, improper development,
tumorigenesis and defects in the differentiation of stem cells
(5,6). In humans, K120-monoubiquitination of
H2B is known to be catalyzed by Ring finger protein (RNF)20 and
RNF40 E3 ligases in conjunction with ubiquitin-conjugating enzyme
UbcH6 (7,8).
K120-monoubiquitination of H2B also regulates
chromatin compaction and RNF8/RNF168-dependent transcriptional
silencing triggered near double-strand break sites in mammals
(9–11). Accordingly, H2B monoubiquitination
is linked to positive regulation of transcription. UbcH6 and
RNF20/40, E2 conjugating enzyme and E3 ligase of H2B respectively,
are recruited to transcriptionally active sites. Monoubiquitination
of H2B also facilitates histone chaperone FACT and promotes
transcription elongation (12). In
addition, downregulation of H2B ubiquitination is also related with
cancer development. USP22 which is a deubiquitinase of
monoubiquityl H2B is related with aggressive growth, metastasis,
and therapy resistance, whereas RNF20, ubiquitinase of H2B was
reported to be a putative tumor-suppressor (13,14).
RNF20 and RNF40 are recruited to DNA double-strand break sites and
induce K120-monoubiquitination of H2B to regulates the DNA damage
response pathway in an ATM-dependent manner (15).
TRAIP (TRAF-interacting protein) has been identified
as a TNF receptor-associated factor 1 (TRAF1) and TNF
receptor-associated factor 2 (TRAF2)-binding partner which contains
N-terminal really interesting new gene (RING) domain motif
(16). Like other TNF
receptor-associated factor proteins, TRAF1 and TRAF2 contribute to
tumor necrosis factor (TNF) receptor-induced signal transduction
pathways including the NF-κB signaling pathway and TRAIP inhibits
NF-κB activation through interaction with TRAF proteins (16,17).
TRAIP is known to inhibit NF-κB activation by interacting with CYLD
or Syk independently of its E3 ubiquitin ligase activity (18–20).
Most RING containing proteins act as ubiquitin ligase or mediate
ubiquitin transfer reaction and TRAIP also has an E3 ubiquitin
ligase function on self-ubiquitination (18,21).
Recent studies have also noted that TRAIP is a regulator of mitotic
progression and DNA damage response (22,23).
In particularly, we and other authors have demonstrated that TRAIP
promotes homologous recombination and protects genomic stability
after replication stress, and deficiency of normal TRAIP is related
to genomic instability-related diseases such as primordial dwarfism
and lung cancer (23–26). One study demonstrated that the E3
ubiquitin ligase activity of TRAIP is essential for its role in the
DNA damage response by showing the promotion of RPA loading and
phosphorylation after replication stress (26). We also reported that RNF20 and RNF40
interact with TRAIP and recruit TRAIP to DNA damage sites to
regulate the DNA damage response pathway (25).
Since the RNF20/40 complex controls the DNA damage
response pathway by inducing H2B monoubiquitination and recruiting
TRAIP to the DNA damage sites, we aimed to ascertain whether TRAIP
participates in the regulation of H2B monoubiquitination in
response to DNA damage.
In the present study, we confirmed that TRAIP
controls H2B monoubiquitination through the RING domain and
C-terminus. In addition, we further confirmed that the nuclear
expression of TRAIP is correlated with H2B monoubiquitination.
Furthermore, the expression levels of these two proteins are
related to patient survival. Taken together, these results suggest
that TRAIP is a novel regulator of H2B monoubiquitination in DNA
damage response and cancer development in lung adenocarcinoma.
Materials and methods
Plasmids and antibodies
A Myc-tagged TRAIP expression plasmid was previously
described (25). To remove the
off-targeting effects of RNAi-mediated knockdown of TRAIP, we
constructed an siRNA-resistant TRAIP wild-type expression vector
(TRAIP-R) by site-directed mutagenesis using the Myc-tagged TRAIP
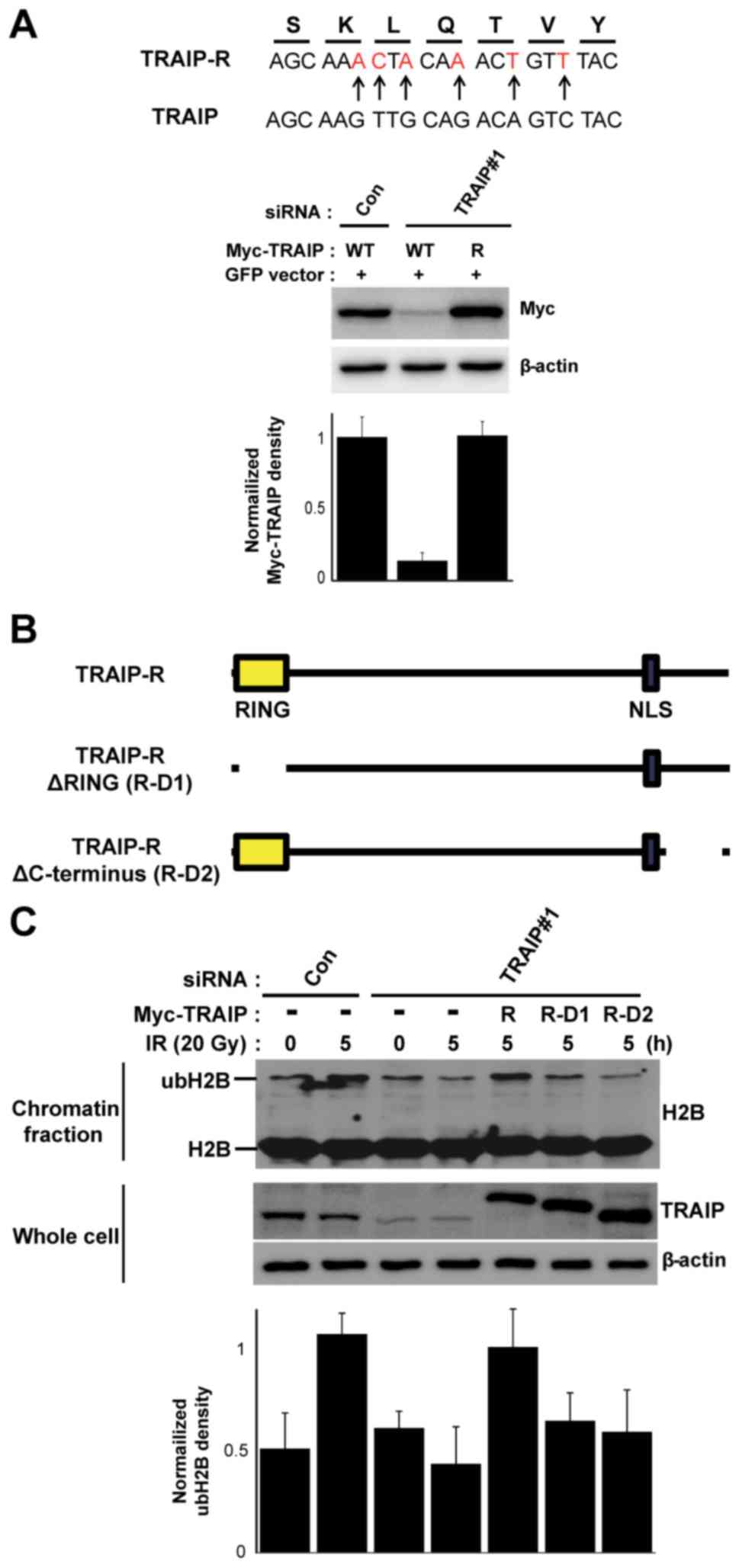
expression plasmid as shown in Fig.
3A. TRAIP-R-D1 and TRAIP-R-D2 expression plasmids (Fig. 3B) were generated by mutagenesis
using the TRAIP-R vector. Rabbit anti-human TRAIP polyclonal
antibody was raised by immunizing rabbits with the GST-TRAIP fusion
protein as previously described (25). Antibodies against Myc (cat. no.
11814150001; Sigma-Aldrich; Merck KGaA, Darmstadt, Germany),
histone H3 (cat. no. sc-10809; Santa Cruz Biotechnology, Dallas,
TX, USA) and β-actin (cat. no. A5316; Sigma-Aldrich; Merck KGaA)
were used. Anti-Histone H2B (cat. no. 07-371) and anti-ubiquityl
Histone H2B (cat. no. 05-1312) antibodies were purchased from
Sigma-Aldrich; Merck KGaA). The dilution ratio of each antibody for
western blotting was as follows: Anti-TRAIP, 1:400; anti-Myc,
1:2,000; anti-H3, 1:1,000: Anti-Histone H2B, 1:2,000;
anti-ubiquityl Histone H2B 1:2,000; and anti-β-actin, 1:5,000.
HRP-conjugated secondary antibodies specific to rabbit (cat. no.
A0545; Sigma-Aldrich; Merck KGaA) or mouse (cat. no. A9917;
Sigma-Aldrich; Merck KGaA) IgG were used at a dilution of
1:2,000.
Cell culture, siRNAs, transfection and
irradiation
HeLa cells were cultured in Dulbecco's modified
Eagle's medium (DMEM; WelGENE Inc., Gyeongsangbuk-do, Korea),
supplemented with 10% fetal bovine serum (FBS; Invitrogen; Thermo
Fisher Scientific, Inc., Waltham, MA, USA), 100 U/ml penicillin and
100 µg/ml streptomycin (Invitrogen; Thermo Fisher Scientific,
Inc.). The cells were maintained at 37°C in a humidified atmosphere
containing 5% CO2. The control siRNA was previously
described (27,28). The TRAIP siRNA sequences were as
follows: TRAIP#1, 5′-GCAGCAGGAUGAGACCAAAUU-3′ and TRAIP#2,
5′-GCAAGUUGCAGACAGUCUAUU-3′. All siRNAs were purchased from Noble
Bio (Hwaseong, Korea). Cells were transfected with 50 nM siRNAs
using DharmaFECT 1 (Dharmacon, Inc., Lafayette, CO, USA) and DNA
transfection was performed using Lipofectamine 2000 (Invitrogen;
Thermo Fisher Scientific, Inc.) as per the instructions of the
manufacturer. Twenty-four hours after plasmid transfection or 48 h
after siRNA transfection, transfected cells were irradiated at the
indicated doses by a gamma-irradiator (IBL437C; CIS Bio Inc.,
Saclay, France) and 137Cs was used for ionized radiation
source.
Mutagenesis and reconstruction
experiments
Myc-tagged siRNA-resistant TRAIP was generated by
silent mutagenesis of siRNA-targeted sequence using a Myc-tagged
TRAIP expression plasmid. Myc-tagged siRNA resistant D1 and D2 of
TRAIP were generated by further mutagenesis using a Myc-tagged
siRNA-resistant TRAIP plasmid. HeLa cells were transfected with
siRNAs using DharmaFECT 1 (Dharmacon, Inc.) as per the instructions
of the manufacturer. Twenty-four hours after siRNA transfection,
siRNA-resistant plasmid DNA transfection was performed using
Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) as
per the instructions of the manufacturer. Twenty-four hours after
DNA transfection, cells were irradiated. Irradiated cells were
harvested and analyzed by chromatin fraction and western blotting
after indicated times since cells were irradiated.
Western blotting
HeLa cells were transfected with siRNA and
expression plasmids as indicated in each figure legend. Forty-eight
hours after siRNA or 24 h after DNA transfection, cells were lysed
in NETN buffer [0.5% Nonidet P-40, 20 mM Tris (pH 8.0), 50 mM NaCl,
50 mM NaF, 100 µM Na3VO4, 1 mM dithiothreitol
and 50 µg/ml phenylmethylsulfonyl fluoride] for 20 min on ice.
Crude lysates were cleared by centrifugation at 12,300 × g for 15
min at 4°C. A total of 15 µg samples of supernatants or chromatin
fraction were subjected to each lane of 12% SDS-PAGE gels. After
electrophoresis, the separated proteins were transferred to a
polyvinylidene difluoride (PVDF) membrane and blocked using 1X
Tris-buffered saline with 0.5% Tween-20 (TBST) with 1% w/v non-fat
dry milk for 30 min at room temperature. The membranes were
incubated with primary antibodies in blocking buffer for 16 h at
4°C. After washing with 1X TBST, secondary antibodies were applied
with blocking buffer and further incubated for 1 h at room
temperature. Visualization reagent (AbSignal, ABC-3001) was
purchased from AbClon Inc. (Seoul, Korea). Band densities were
quantified using Image Lab (version 5.2; Bio-Rad Laboratories,
Hercules, CA, USA).
Chromatin fraction
Cells were collected and washed in
phosphate-buffered saline (PBS). The collected cells were lysed in
NETN buffer [0.5% Nonidet P-40, 20 mM Tris (pH 8.0), 50 mM NaCl, 50
mM NaF, 100 µM Na3VO4, 1 mM dithiothreitol
and 50 µg/ml phenylmethylsulfonyl fluoride] at 4°C for 30 min.
Crude lysates were cleared by centrifugation at 16,000 × g at 4°C
for 10 min and the pellet was suspended in HCl (0.2 M) for 1 h. The
resuspended mixture was centrifuged at 16,000 × g at 4°C for 10
min, and then the lysate which is the chromatin fraction was
neutralized with Tris-HCl (1 M at pH 8.0) for western blotting. We
used histone 3 (H3) as a control for the chromatin fraction.
Tissue microarrays and
immunohistochemistry
A tissue microarray containing 75 cases of human
lung adenocarcinoma tissues and each matched normal adjacent tissue
(HLug-Ade150Sur-01) with no information regarding the treatment and
therapy received by patients was obtained from US Biomax, Inc.
(Rockville, MD, USA). Immunohistochemical staining was performed as
previously described (21) with a
rabbit polyclonal anti-TRAIP antibody (PA5-27699; Thermo Fisher
Scientific, Inc.) and a rabbit monoclonal anti-ubiquityl-H2B
antibody (cat. no. 5546S; Cell Signaling Technology, Inc., Danvers,
MA, USA) at a concentration of 1:200 and 1:500, respectively. The
staining intensity was assigned an arbitrary value, on a 4-scale
(intensity score) as follows: Non-stained (0), weak (1), moderate (2), and strong (3). The H-score was calculated by
multiplying the intensity score and the fraction score (percentage
of counted samples at each scale), producing the range 0–300, and
repeated on three different areas.
Statistical analysis
Statistical analysis was performed using GraphPad
Prism 5 program (GraphPad Software, Inc., San Diego, CA, USA).
Values were generally represented as mean of H-scores ± standard
error of the mean (SEM). A Box-and-Whisker plot was also used to
compare the distribution of each sample. Comparison of TRAIP
expression in the nucleus of tumor and adjacent non-tumor groups
was analyzed using t-test. Pearson correlation coefficient (r)
analysis was used to compare the expression of TRAIP and H2B in the
nucleus of tumor cells. The method of Kaplan-Meier and the log-rank
test were used for comparing overall survival between groups. A
two-sided P-value of <0.05 was considered to indicate a
statistically significant result.
Results
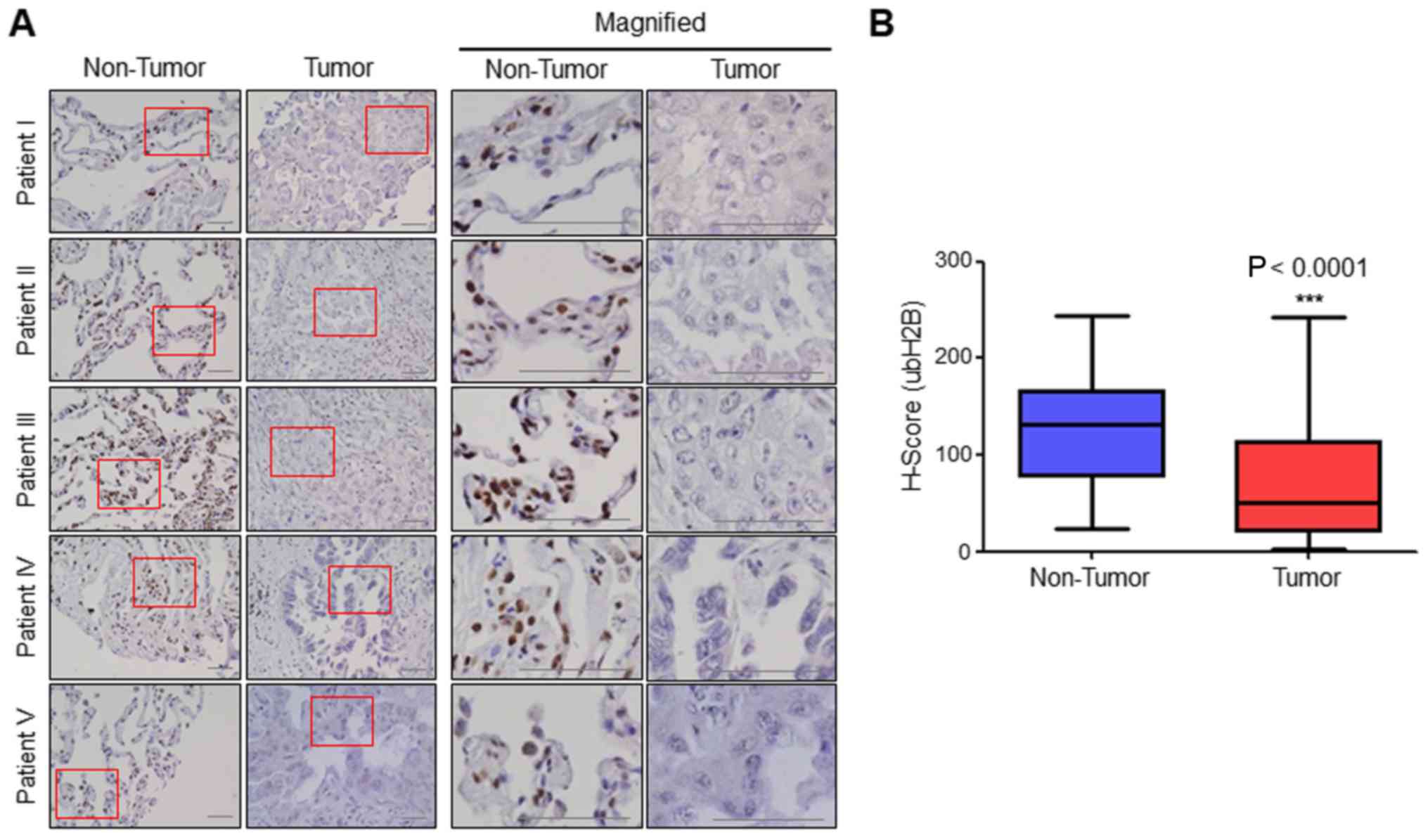
H2B monoubiquitination is
downregulated in human lung cancer patient samples
To determine whether H2B monoubiquitination is
related with lung cancer, we determined the expression levels of
H2B monoubiquitination in human lung cancer patient tissues and
normal adjacent tissues (≥2 cm away from every cancer tissue) using
a tissue microarray containing 75 cases of each. We found that the
expression levels of H2B monoubiquitination in tumors were much
lower than that noted in the non-tumor tissues (Fig. 1A). H-scoring in every tissue
confirmed that H2B monoubiquitination was greatly decreased in
human lung cancer patients (median H-score, 72.3) compared with
that in each matched normal adjacent tissue (median H-score, 123.3)
(Fig. 1B). The comparison of the
results indicated that H2B monoubiquitination in 60.3% of the
cancer tissues was at least 1.5-fold lower than that in each
corresponding matched normal tissue.
TRAIP depletion decreases ionizing
radiation (IR)-induced H2B monoubiquitination
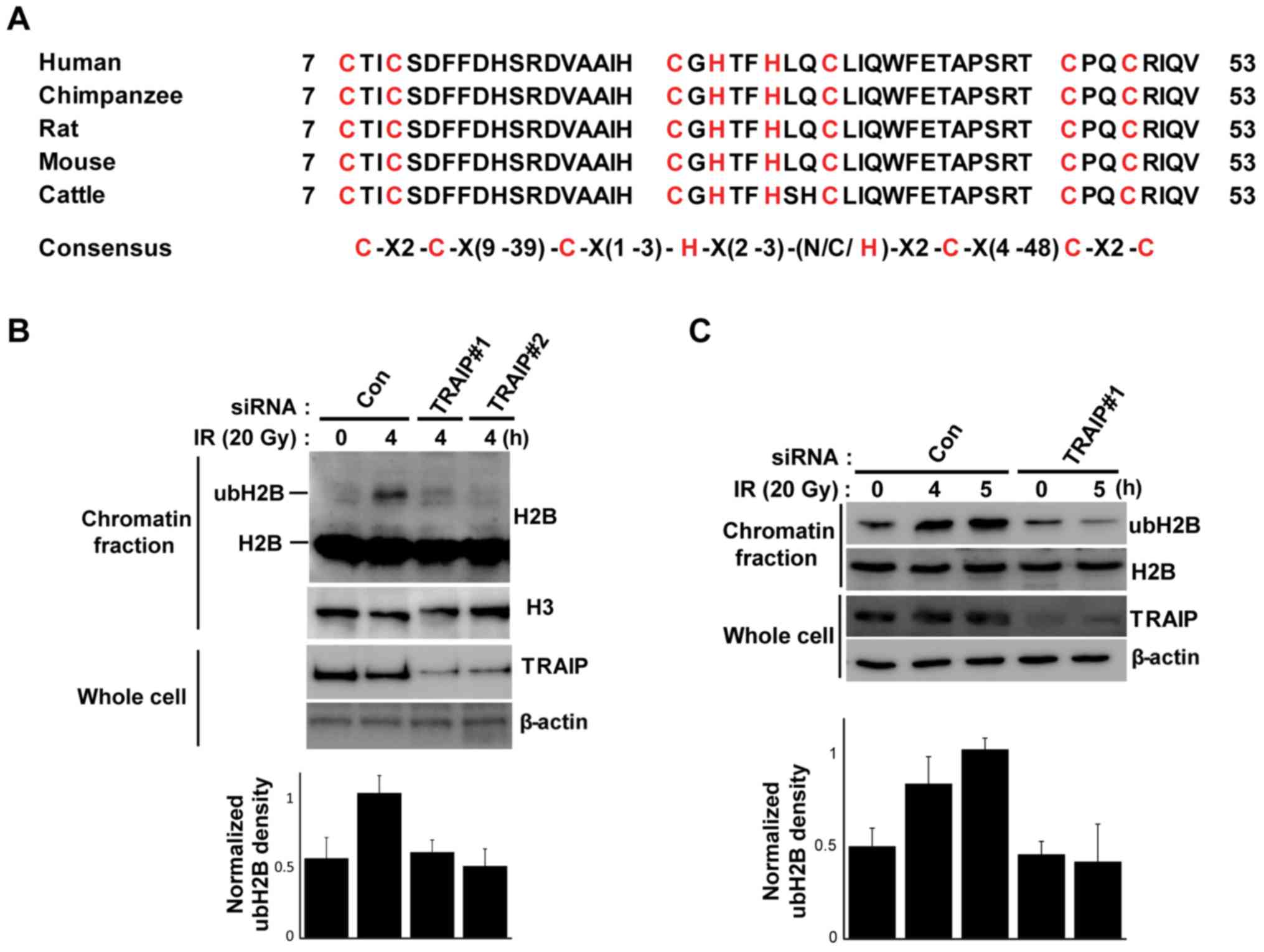
The structure of the RING finger domain of TRAIP is
conserved widely across mammals (Fig.
2A). This conservation implies the importance of RING domain
for the function of TRAIP, and RING finger domain usually interacts
with ubiquitination enzyme and has E3 ligase activity (29). In addition, we confirmed that the
RNF20-RNF40 complex, which is an E3 ubiquitin ligase for H2B
monoubiquitination at the sites of DNA damage, regulates
translocation of TRAIP to DNA damage site leading to the regulation
of the DNA damage response pathway and homologous repair which is
induced by H2B monoubiquitination (25). Based on these results, we
hypothesized that TRAIP may participate in monoubiquitination of
H2B during the DNA damage response. To verify this hypothesis, we
depleted TRAIP with specific siRNAs and the cells were exposed to
IR. IR increased H2B monoubiquitination without any effect on the
expression of H2B in the chromatin fraction, but the IR-mediated
H2B monoubiquitination was decreased in the TRAIP-knockdown cells
(Fig. 2B). We also confirmed that
IR induced H2B monoubiquitination at the K120 residue using the
anti-ubiquityl H2B (Lys-120) antibody and that knockdown of TRAIP
diminished the K120-H2B monoubiquitination (Fig. 2C). These results indicated that
TRAIP is necessary for the DNA damage-mediated
K120-monoubiquitination of H2B.
RING domain and C-terminal of TRAIP is
crucial for IR-mediated H2B monoubiquitination
Since previous reports noted that the RING domain of
TRAIP has the E3 ubiquitin ligase activity and C-terminal region of
TRAIP is involved in translocating its DNA damage sites (18,25),
we next examined which domain of TRAIP is involved in the
IR-mediated H2B monoubiquitination. First, to remove the
off-targeting effects of RNAi-mediated knockdown of TRAIP, we
constructed the siRNA-resistant TRAIP wild-type expression vector
(TRAIP-R) with silent mutation and checked the siRNA-resistant
feature. HeLa cells were transfected with non-targeting (control)
or TRAIP-targeting siRNA and 24 after siRNA transfection, the cells
were secondly transfected with a plasmid carrying wild-type TRAIP
(WT) or siRNA-resistant wild-type TRAIP (TRAIP-R) as indicated
(Fig. 3A). The reconstitution using
the TRAIP-R in TRAIP-depleted cells successfully rescued the
IR-mediated H2B monoubiquitination (Fig. 3C). These results confirmed again
that TRAIP is a key regulator for H2B monoubiquitination during the
DNA damage response. We then generated TRAIP-deletion mutants
without the RING domain (R-D1) or C-terminus (R-D2) using the
TRAIP-R as the template (Fig. 3B).
As shown in Fig. 3C, both deletion
mutants, R-D1 and R-D2 failed to rescue the IR-mediated H2B
monoubiquitination, suggesting that both RING domain (E3 ubiquitin
ligase activity) and C-terminus (TRAIP recruitment to DNA lesion)
are essential for inducing H2B monoubiquitination.
Expression of nuclear TRAIP positively
correlates with H2B monoubiquitination and lower expression of both
is associated with poor prognosis in patients with lung cancer
We then investigated a possible correlation between
the expression of TRAIP and H2B monoubiquitination in human lung
cancer patient tissues to evaluate the clinical importance of the
relationship. For this purpose, we examined the immunostaining
patterns of nuclear TRAIP and H2B monoubiquitination in the same
region of every tissue using consecutive sections. As we previously
reported (25), nuclear TRAIP
expression in lung cancer patient tissues was lower than that in
each matched normal adjacent tissue. Intriguingly, we also observed
a positive correlation between the nuclear expression of TRAIP and
H2B monoubiquitination in the tumor tissues (Fig. 4A). Decreased H2B monoubiquitination
was found in the patient tissues with a lower nuclear expression of
TRAIP while increased H2B monoubiquitination in the tumors with a
higher nuclear expression of TRAIP. A scatter plot and Pearson
correlation coefficient analysis revealed a significant positive
correlation (r=0.4052) between the nuclear expression of TRAIP and
H2B monoubiquitination (Fig. 4B).
In addition, comparison among three patient groups divided by the
sum of H-scores for nuclear TRAIP and H2B monoubiquitination
indicated that patient survival of the high expression group was
considerably longer than the other two groups, middle (P=0.0059)
and low group (P=0.026) (Fig. 4C)
even though neither nuclear TRAIP nor H2B monoubiquitination alone
showed any meaningful correlation with patient survival (Fig. 4D and E), suggesting that decreased
expression of both nuclear TRAIP and H2B monoubiquitination is
significantly associated with shorter survival of lung cancer
patients. Together these results indicated that the expression of
nuclear TRAIP positively correlates with H2B monoubiquitination and
a decrease in both nuclear TRAIP and H2B monoubiquitination is
significantly associated with a poor prognosis in patients with
lung cancer.
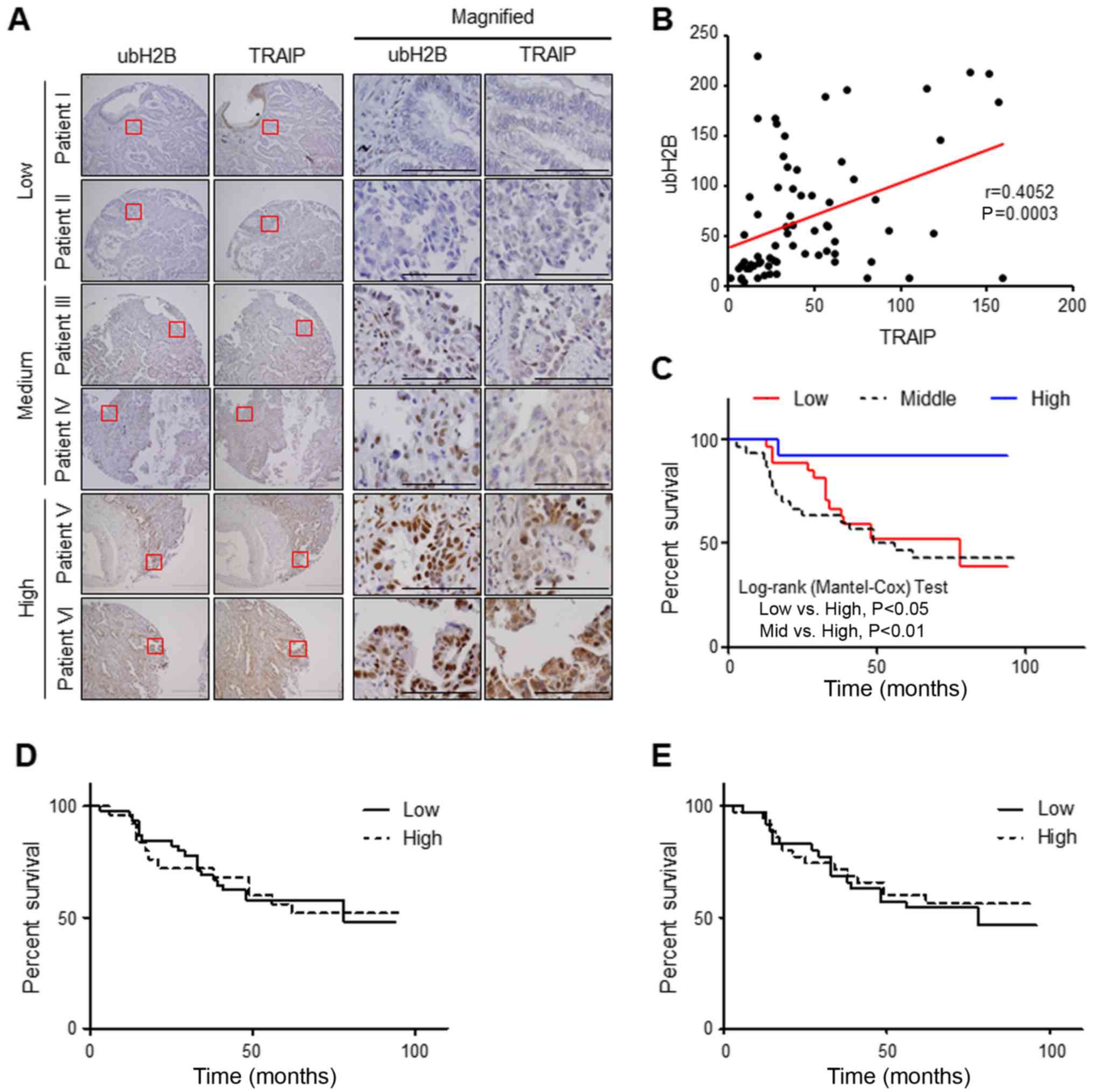 | Figure 4.Correlation between nuclear TRAIP
expression and ubH2B in lung cancer patient samples. (A)
Immunohistochemical staining of TRAIP and ubH2B in human lung
cancer patient tissues. Red squares indicate the magnified region
shown in the right panel. Scale bar, 200 µm and scale bar in the
magnified figure, 50 µm. (B) Pearson correlation coefficient (r)
between the nuclear expression of TRAIP and ubH2B. The graph
presents the scatter plot of the H-score of nuclear TRAIP (x-axis)
and ubH2B (y-axis). P=0.0003 (n=70). (C) Patients in the high
expression group of both analytical factors show longer overall
survival than in the middle (P=0.0059) or low group (P=0.0226). No
significant difference between low and middle groups (P=0.53) was
noted. High expression group (blue solid line, n=13) with sum
H-score (sum of H-scores for TRAIP and ubH2B) >180, middle group
(black dot line, n=30) with 80< sum-score <180, and low group
(red solid line, n=27) with sumH-scoring <80. (D) No significant
difference between low and high groups (P=0.9974). High group
(dotted line, n=35) with H-score for TRAIP expression >50, low
group (solid line, n=35) with <50. (E) No significant difference
between low and high groups (P=0.8806). High group (dotted line,
n=25) with H-score for ubH2B expression >50, low group (solid
line, n=45) with <50. TRAIP, TRAF-interacting protein; ubH2B,
H2B monoubiquitination. |
Discussion
Lung cancer is the most common cancer in regards to
incidence accounting for 13% of all newly diagnosed cancer cases,
and lung cancer-related mortality accounts for 18% of the total
cancer-related deaths globally (30). Lung cancer is caused by diverse
factors, such as smoking, air pollution, diet and genetic mutations
(31). Non-small cell lung cancer
(NSCLC) is the main lung cancer subtype with 85% of patients and
has two major subtypes, lung adenocarcinoma (LUAD) and lung
squamous cell carcinoma (LUSC) (32). Although many different types of
interventions including targeted therapy have been developed to
deal with cancer to date, most patients with lung cancer are
treated with cytotoxic chemotherapeutic drugs such as cisplatin,
carboplatin, docetaxel, and gemcitabine, which mainly induce DNA
damage (33).
A primary hallmark of cancer is genomic instability
that is induced by the accumulation of DNA damage. Cellular DNA
damage caused by various carcinogens including cytotoxic drugs
stimulates many types of enzymes and signaling molecules leading to
cell cycle arrest, regulation of DNA replication and the repair of
DNA damage, which is called DNA damage response (DDR) (34). Many molecules participating at DDR
have been classified by their roles as DNA damage sensors,
recruitment mediators, transducers, and effectors to initiate
signaling pathways to impact various cellular responses (35). The combination of radiotherapy and
DNA damage-inducing chemotherapy is the classic and effective
strategy for overall tumor treatment, even though serious side
effects occur. To diminish unwanted adverse reactions, novel drugs
targeting the molecules involved in DDR has been recently
developed; for example, olaparib (Lynparza®), the
poly(ADP-ribose) polymerase (PARP) inhibitor (36).
Histone modifications including methylation,
ubiquitination, phosphorylation and acetylation have been
considered the essential events for DDR by forming recruitment
platforms for downstream effectors and guiding the activity of
chromatin remodelers (4). In
particularly, H2B monoubiquitination is an important histone
modification in transcriptional regulation and chromatin
organization even in the absence of DNA damage (37), and also has a pivotal role in the
recognition of DNA damage sites as a regulator of damage checkpoint
activation and timely initiation of repair (38,39).
H2B monoubiquitination has been known to be generated by
RNF20/RNF40, RING finger proteins, with E3 ubiquitin ligase
activity (40–43). TRAIP also exists in the nucleus and
the cytoplasm, but DNA damage increases its nuclear translocation,
especially to damaged sites (23,25,44).
We also reported that TRAIP induces DDR and repair and that
RNF20/RNF40 regulates translocation of TRAIP to DNA double-strand
breaks (25). In the present study,
we demonstrated that TRAIP is a novel regulator of H2B
monoubiquitination, further supporting that TRAIP is an essential
component in DDR. Furthermore, in the present study, a positive
correlation was confirmed between nuclear expression of TRAIP and
H2B monoubiquitination in lung cancer patient samples and that
decreased expression of both nuclear TRAIP and H2B
monoubiquitination was closely related with shorter survival of
lung cancer patients (Fig. 4). As
discussed above, genomic instability is one of primary hallmarks of
cancer development and progression, and is also increased by
therapeutic treatment of cancer patients by chemotherapy or
radiotherapy. These results strongly suggest that dysregulation of
TRAIP and H2B monoubiquitination related with DNA repair are
involved in lung cancer pathogenesis, which may influence patient
survival. We also conjecture the relationship among RNF20, RNF40
and TRAIP during the DDR pathway. Therefore, future studies to
investigate the role of TRAIP in DDR and its expression patterns
related with other DDR molecules such as RNF20 and RNF40 in cancer
patient samples are warranted to elucidate the detailed mechanisms
of DDR and its dysregulation in cancer and then to pave the way for
developing novel therapeutic interventions.
Acknowledgements
The authors would like to thank the laboratory staff
at Kyung Hee University and UNIST.
Funding
The present study was supported by the National
Research Foundation of Korea (NRF) grants funded by the Korea
government (MEST) (NRF-2018R1A2B2003129 and NRF-2015R1A4A1042399)
and by Global Ph.D. Fellowship Program through the NRF funded by
the Ministry of Education (2016H1A2A1909739).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
YGH, MY and MC performed the experiments, analyzed
the data and drafted the manuscript. SGL and HK designed and
interpreted the study, and wrote and edited the manuscript. All
authors read and approved the manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Durocher D and Jackson SP: DNA-PK, ATM and
ATR as sensors of DNA damage: Variations on a theme? Curr Opin Cell
Biol. 13:225–231. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kerzendorfer C and O'Driscoll M: Human DNA
damage response and repair deficiency syndromes: Linking genomic
instability and cell cycle checkpoint proficiency. DNA Rep.
8:1139–1152. 2009. View Article : Google Scholar
|
|
3
|
Uckelmann M and Sixma TK: Histone
ubiquitination in the DNA damage response. DNA Repair. 56:92–101.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Dantuma NP and van Attikum H:
Spatiotemporal regulation of posttranslational modifications in the
DNA damage response. EMBO J. 35:6–23. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Fuchs G, Shema E, Vesterman R, Kotler E,
Wolchinsky Z, Wilder S, Golomb L, Pribluda A, Zhang F, Haj-Yahya M,
et al: RNF20 and USP44 regulate stem cell differentiation by
modulating H2B monoubiquitylation. Mol Cell. 46:662–673. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Minsky N, Shema E, Field Y, Schuster M,
Segal E and Oren M: Monoubiquitinated H2B is associated with the
transcribed region of highly expressed genes in human cells. Nat
Cell Biol. 10:483–488. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhu B, Zheng Y, Pham AD, Mandal SS,
Erdjument-Bromage H, Tempst P and Reinberg D: Monoubiquitination of
human Histone H2B: The factors involved and their roles in
HOX gene regulation. Mol Cell. 20:601–611. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Thorne AW, Sautiere P, Briand G and
Crane-Robinson C: The structure of ubiquitinated Histone H2B. EMBO
J. 6:1005–1010. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Weake VM and Workman JL: Histone
ubiquitination: Triggering gene activity. Mol Cell. 29:653–663.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Shanbhag NM, Rafalska-Metcalf IU,
Balane-Bolivar C, Janicki SM and Greenberg RA: ATM-dependent
chromatin changes silence transcription in cis to DNA double-strand
breaks. Cell. 141:970–981. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Xu Z, Song Z, Li G, Tu H, Liu W, Liu Y,
Wang P, Wang Y, Cui X, Liu C, et al: H2B ubiquitination regulates
meiotic recombination by promoting chromatin relaxation. Nucleic
Acids Res. 44:9681–9697. 2016.PubMed/NCBI
|
|
12
|
Pavri R, Zhu B, Li G, Trojer P, Mandal S,
Shilatifard A and Reinberg D: Histone H2B monoubiquitination
functions cooperatively with FACT to regulate elongation by RNA
polymerase II. Cell. 125:703–717. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang XY, Varthi M, Sykes SM, Phillips C,
Warzecha C, Zhu W, Wyce A, Thorne AW, Berger SL and McMahon SB: The
putative cancer stem cell marker USP22 is a subunit of the human
SAGA complex required for activated transcription and cell-cycle
progression. Mol Cell. 29:102–111. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Shema E, Tirosh I, Aylon Y, Huang J, Ye C,
Moskovits N, Raver-Shapira N, Minsky N, Pirngruber J, Tarcic G, et
al: The Histone H2B-specific ubiquitin ligase RNF20/hBRE1 acts as a
putative tumor suppressor through selective regulation of gene
expression. Genes Dev. 22:2664–2676. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Moyal L, Lerenthal Y, Gana-Weisz M, Mass
G, So S, Wang SY, Eppink B, Chung YM, Shalev G, Shema E, et al:
Requirement of ATM-dependent monoubiquitylation of Histone H2B for
timely repair of DNA double-strand breaks. Mol Cell. 41:529–542.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lee SY and Choi Y: TRAF-interacting
protein (TRIP): A novel component of the tumor necrosis factor
receptor (TNFR)-and CD30-TRAF signaling complexes that inhibits
TRAF2-mediated NF-kappaB activation. J Exp Med. 185:1275–1286.
1997. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Rothe M, Pan MG, Henzel WJ, Ayres TM and
Goeddel DV: The TNFR2-TRAF signaling complex contains two novel
proteins related to baculoviral inhibitor of apoptosis proteins.
Cell. 83:1243–1252. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Besse A, Campos AD, Webster WK and Darnay
BG: TRAF-interacting protein (TRIP) is a RING-dependent ubiquitin
ligase. Biochem Biophys Res Commun. 359:660–664. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Regamey A, Hohl D, Liu JW, Roger T,
Kogerman P, Toftgard R and Huber M: The tumor suppressor CYLD
interacts with TRIP and regulates negatively nuclear factor kappaB
activation by tumor necrosis factor. J Exp Med. 198:1959–1964.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhou Q and Geahlen R: The protein-tyrosine
kinase Syk interacts with TRAF-interacting protein TRIP in breast
epithelial cells. Oncogene. 28:1348–1356. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Deshaies RJ and Joazeiro CA: RING domain
E3 ubiquitin ligases. Annu Rev Biochem. 78:399–434. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chapard C, Meraldi P, Gleich T, Bachmann
D, Hohl D and Huber M: TRAIP is a regulator of the spindle assembly
checkpoint. J Cell Sci. 127:5149–5156. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Harley ME, Murina O, Leitch A, Higgs MR,
Bicknell LS, Yigit G, Blackford AN, Zlatanou A, Mackenzie KJ, Reddy
K, et al: TRAIP promotes DNA damage response during genome
replication and is mutated in primordial dwarfism. Nat Genet.
48:36–43. 2016. View
Article : Google Scholar : PubMed/NCBI
|
|
24
|
Feng W, Guo Y, Huang J, Deng Y, Zang J and
Huen MS: TRAIP regulates replication fork recovery and progression
via PCNA. Cell Discov. 2:160162016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Soo Lee N, Jin Chung H, Kim HJ, Yun Lee S,
Ji JH, Seo Y, Hun Han S, Choi M, Yun M, Lee SG, et al: TRAIP/RNF206
is required for recruitment of RAP80 to sites of DNA damage. Nat
Commun. 7:104632016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Hoffmann S, Smedegaard S, Nakamura K,
Mortuza GB, Räschle M, Ibañez de Opakua A, Oka Y, Feng Y, Blanco
FJ, Mann M, et al: TRAIP is a PCNA-binding ubiquitin ligase that
protects genome stability after replication stress. J Cell Biol.
212:63–75. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kim H, Huang J and Chen J: CCDC98 is a
BRCA1-BRCT domain-binding protein involved in the DNA damage
response. Nat Struct Mol Biol. 14:710–715. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kim H, Chen J and Yu X: Ubiquitin-binding
protein RAP80 mediates BRCA1-dependent DNA damage response.
Science. 316:1202–1205. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lorick KL, Jensen JP, Fang S, Ong AM,
Hatakeyama S and Weissman AM: RING fingers mediate
ubiquitin-conjugating enzyme (E2)-dependent ubiquitination. Proc
Natl Acad Sci USA. 96:11364–11369. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Dela Cruz CS, Tanoue LT and Matthay RA:
Lung cancer: Epidemiology, etiology, and prevention. Clin Chest
Med. 32:605–644. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Molina JR, Yang P, Cassivi SD, Schild SE
and Adjei AA: Non-small cell lung cancer: Epidemiology, risk
factors, treatment, and survivorship. Mayo Clin Proc. 83:584–594.
2008. View
Article : Google Scholar : PubMed/NCBI
|
|
33
|
Jung Y and Lippard SJ: Direct cellular
responses to platinum-induced DNA damage. Chem Rev. 107:1387–1407.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Pearl LH, Schierz AC, Ward SE, Al-Lazikani
B and Pearl FM: Therapeutic opportunities within the DNA damage
response. Nat Rev Cancer. 15:166–180. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Jackson SP and Bartek J: The DNA-damage
response in human biology and disease. Nature. 461:1071–1078. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
European Medicines Agency: Lynparza
recommended for approval in ovarian cancer. https://www.ema.europa.eu/en/news/lynparza-recommended-approval-ovarian-cancer
|
|
37
|
Chen S, Jing Y, Kang X, Yang L, Wang DL,
Zhang W, Zhang L, Chen P, Chang JF, Yang XM, et al: Histone H2B
monoubiquitination is a critical epigenetic switch for the
regulation of autophagy. Nucleic Acids Res. 45:1144–1158.
2017.PubMed/NCBI
|
|
38
|
Jackson SP and Durocher D: Regulation of
DNA damage responses by ubiquitin and SUMO. Mol Cell. 49:795–807.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Schwertman P, Bekker-Jensen S and Mailand
N: Regulation of DNA double-strand break repair by ubiquitin and
ubiquitin-like modifiers. Nat Rev Mol Cell Biol. 17:379–394. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Mattiroli F, Vissers JH, van Dijk WJ, Ikpa
P, Citterio E, Vermeulen W, Marteijn JA and Sixma TK: RNF168
ubiquitinates K13-15 on H2A/H2AX to drive DNA damage signaling.
Cell. 150:1182–1195. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Kalb R, Mallery DL, Larkin C, Huang JT and
Hiom K: BRCA1 is a histone-H2A-specific ubiquitin ligase. Cell Rep.
8:999–1005. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Wood A, Krogan NJ, Dover J, Schneider J,
Heidt J, Boateng MA, Dean K, Golshani A, Zhang Y, Greenblatt JF, et
al: Bre1, an E3 ubiquitin ligase required for recruitment and
substrate selection of Rad6 at a promoter. Mol Cell. 11:267–274.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Wang H, Wang L, Erdjument-Bromage H, Vidal
M, Tempst P, Jones RS and Zhang Y: Role of histone H2A
ubiquitination in polycomb silencing. Nature. 431:873–878. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Wallace HA, Merkle JA, Yu MC, Berg TG, Lee
E, Bosco G and Lee LA: TRIP/NOPO E3 ubiquitin ligase promotes
ubiquitylation of DNA polymerase η. Development. 141:1332–1341.
2014. View Article : Google Scholar : PubMed/NCBI
|