Introduction
Metallothioneins are a family of low molecular
weight (6–7 kDa), highly conserved, cysteine-rich non-enzymatic
cytosolic proteins which play a vital role in metal ion
homeostasis. In humans, the majority of metallothionein genes (16
of 19) are located in a cluster on chromosome 16q13, of which 12
are protein-coding (e.g. MT1A, MT1E and MT1F) and the
rest are pseudogenes (e.g. MT1CP, MT1L and MT1P1).
The main biological roles of metallothioneins in cells are directly
related to their ability to bind metal ions. Changes in
metallothionein expression alter their major cellular functions,
i.e. buffering and delivering zinc and copper, which are critical
for proliferating, differentiating and apoptotic cells (1). Due to their affinity for cadmium,
lead, or mercury, metallothioneins protect cells from heavy metal
toxicity (2,3). They also function as antioxidants
against DNA damage caused by free radicals (4).
Metallothioneins have a significant role in cancer
development. Gene expression changes have been associated with
tumor growth, metastasis and angiogenesis (5). Earlier research has indicated that
metallothioneins contribute to the development of resistance to
drugs or radiotherapy (6–8); however, recent research has shown that
they can also suppress cardiotoxicity induced by anticancer agents
(9). Various studies also revealed
that metallothionein expression is not universal for different
cancer types. Specifically, metallothionein genes are frequently
downregulated in liver cancer, squamous cell lung carcinoma, or
prostate tumors, which could be associated with promoter DNA
methylation (10–12). In contrast, increased
metallothionein expression has been reported in melanoma, ovarian
or breast tumors (13–15), while investigations of some other
tumors produced contradictory results.
Renal cell carcinoma (RCC) comprises ~2–3% of all
non-cutaneous cancers and is the most fatal type of urologic
malignancies with high mortality rates in Europe. Lithuania has the
third highest RCC incidence rate and is in the first place
according to mortality worldwide (16). A variety of histologically distinct
tumors falls under the RCC definition, with clear cell RCC (ccRCC)
being the most common subtype. Other commonly detected tumors are
papillary (pRCC) and chromophobe RCC (chRCC). As clinical outcomes
are closely related to tumor stage, grade and other parameters,
diagnostic methods for accurate cancer characterization, as well as
early detection, are critically important. At present, ~50% of
sporadic RCC cases are incidentally detected in asymptomatic
patients during examination for other diseases (17). Despite the use of highly sensitive
methods, such as computed tomography scan and magnetic resonance
imaging, RCC is often detected at already advanced stages when
treatment options become limited (18). The lack of diagnostic clinical tests
draws attention to genetic and epigenetic features as potential
biomarkers of RCC. Novel molecular biomarkers could potentially
improve early RCC diagnostics and advise the most beneficial
treatment.
In the present study, we investigated aberrant DNA
methylation of several protein-coding metallothionein genes aiming
to elucidate their importance in renal carcinogenesis. DNA
methylation status was compared with clinical-pathological patient
characteristics, as well as with gene expression levels. The Cancer
Genome Atlas (TCGA) dataset of kidney renal cell carcinoma (KIRC)
was used for the screening step. This study led to the
identification of the potential importance of MT1E and
MT1M genes in RCC development.
Materials and methods
Patients and samples
In total, 54 patients [22 males and 32 females with
the mean age of 63 (41–85) and 67 (27–85), respectively] diagnosed
with RCC, who underwent full or partial nephrectomy at the National
Cancer Institute (Vilnius, Lithuania) between July 2013 and January
2016, were involved in the study. The study cohort mainly consisted
of ccRCC and various cases of other histological subtypes (Table I). Tissues were sampled and grades
were assigned to tumors by an expert pathologist. The Fuhrman grade
describes adverse morphological characteristics of cell nuclei,
whereas the differentiation grade, defined according to the WHO
recommendations, is based on tissue histology in general. In total,
54 tumors, 10 paired pericancerous renal tissues (PRT; at a
distance of 1–2 cm from the tumor margin), and 33 paired
non-cancerous renal tissues (NRT; morphologically normal tissue at
>2 cm from the tumor margin and ≤1 cm from the surgical margin
if partial nephrectomy was performed) were included in the study
(Table I). For 10 patients, two
tumor foci were available for the molecular analysis. Approval to
conduct biomedical research (no. 158200-13-620-192) was obtained
from the Lithuanian Bioethics Committee (Vilnius, Lithuania) before
initiating the study, and all patients gave informed consent for
participation.
 | Table I.Clinicopathological characteristics
of the RCC study cohorts. |
Table I.
Clinicopathological characteristics
of the RCC study cohorts.
| Parameter |
| All cases
(n=54) |
| Methylation
analysis (n=30) |
| Gene expression
analysis (n=51) |
|---|
| Tissue samples,
n |
|
RCC |
| 54 |
| 30 |
| 51 |
|
PRT |
| 10 |
| 10 |
| 0 |
|
NRT |
| 33 |
| 30 |
| 9 |
| Histological tumor
subtype, n |
|
ccRCC | 41 | 22 | 41 |
|
|
|
|
chRCC |
| 3 |
| 1 |
| 3 |
|
pRCC |
| 1 |
| 1 |
| 1 |
|
OCT |
| 5 |
| 3 |
| 3 |
| Other
types | 4 | 3 | 3 |
|
|
|
| Tumor size, mean
(range), mm |
| 50 (14–130) |
| 54 (14–130) |
| 56 (20–130) |
| Pathological tumor
stage, n |
|
≤pT2 | 31 | 16 | 29 |
|
|
|
|
≥pT3 |
| 23 |
| 14 |
| 22 |
| Fuhrmann grade,
n |
| F2 | 19 | 8 | 19 |
|
|
|
| F3 |
| 23 |
| 16 |
| 23 |
|
Unknown |
| 12 |
| 6 |
| 9 |
| Differentiation
gradea, n |
|
≤G2 |
| 28 |
| 11 |
| 28 |
| G3 |
| 19 |
| 15 |
| 18 |
|
Unknown |
| 7 |
| 4 |
| 5 |
| Necrotic zones in
tumor, n |
|
Yes |
| 15 |
| 10 |
| 14 |
| No |
| 39 |
| 20 |
| 37 |
|
Sex | Female, n=32 | Male, n=22 | Female, n=16 | Male, n=14 | Female, n=30 | Male, n=21 |
| Age at diagnosis,
mean (range), years | 67 (27–85) | 63 (41–85) | 66 (27–85) | 62 (41–85) | 67 (27–85) | 63 (41–85) |
| Waist
circumferenceb, mean
(range), cm | 102 (79–135) | 100 (78–126) | 107 (93–135) | 98 (78–122) | 101 (79–135) | 100 (78–126) |
| Unknown, n | 2 | 2 | 1 | 2 | 2 | 2 |
| Raised fasting
plasma glucose levelc,
n |
|
Yes |
| 44 |
| 6 |
| 8 |
| No |
| 10 |
| 24 |
| 43 |
Nucleic acid extraction
Renal tissue samples (~60 mg) were mechanically
homogenized with cryoPREP™ CP02 Impactor using tissueTUBEs TT1
(Covaris, Inc., Woburn, MA, USA). For the isolation of genomic DNA,
up to 30 mg of tissue powder was treated with proteinase K (Thermo
Scientific™; Thermo Fisher Scientific, Vilnius, Lithuania) in 0.5
ml of lysis buffer (50 mM Tris-HCl pH 8.5, 1 mM EDTA, 0.5%
Tween-20; all from Carl Roth GmbH, Co., KG, Karlsruhe, Germany) for
up to 18 h at 55°C and DNA was extracted according to the standard
phenol-chloroform purification and ethanol precipitation.
The total RNA was extracted using mirVana™ miRNA
Isolation kit (Ambion; Thermo Fisher Scientific, Inc., Waltham, MA,
USA) as previously described (12).
Briefly, ~10 mg of homogenized tissue were treated with 0.5 ml of
lysis/binding buffer and 50 µl of miRNA homogenate additive for 10
min in an ice-water bath. The total RNA was extracted with 0.5 ml
of acid-phenol:chloroform and purified using the supplied filter
cartridges.
Concentration and purity of extracted nucleic acids
were evaluated spectrophotometrically with NanoDrop 2000 (NanoDrop
Technologies; Thermo Fisher Scientific, Inc.). The integrity of
randomly selected DNA samples and all RNA samples was analyzed in
1–1.5% agarose gels prepared with 1X TAE buffer (Thermo Fisher
Scientific, Inc.) and only intact samples were used for the
molecular analysis.
DNA methylation analysis
Bisulfite treatment was applied to 400 ng of
extracted DNA using EZ DNA Methylation kit (Zymo Research Corp.,
Irvine, CA, USA) according to the manufacturer's protocol, except
that the initial incubation of samples was performed at 42°C for 15
min. Methylation-specific PCR (MSP) was used for the qualitative
promoter methylation analysis of genes MT1E, MT1G, MT1F and
MT1M. Primers specific for methylated or unmethylated DNA
(Table II) were designed with
Methyl Primer Express Software v1.0 (Applied Biosystems; Thermo
Fisher Scientific, Inc.) or selected from the previous publication
(12). One microliter of
bisulfite-modified DNA was added to 24 µl of MSP mix containing
1.25 U AmpliTaq Gold DNA Polymerase, 1X Gold PCR buffer, 1 µl of
360 GC Enhancer, 2.5 mM MgCl2 (all from Applied
Biosystems; Thermo Fisher Scientific, Inc.), 0.4 mM of each dNTP
(Thermo Fisher Scientific, Inc.) and 0.5 µM of each primer
(Metabion, Martinsried, Germany). Thermocycling conditions were
optimized prior to the study and included 35–38 cycles with the
primer annealing step at 56–58°C (Table II). Methylation-positive,
methylation-negative, and non-template controls were routinely
included. Amplification products were analyzed in 3% agarose gels
with 1X TAE buffer and visualized under UV light after ethidium
bromide staining (Carl Roth GmbH, Co., KG).
 | Table II.Primers used for methylation-specific
PCR (MSP) and amplification conditions. |
Table II.
Primers used for methylation-specific
PCR (MSP) and amplification conditions.
| Gene symbol | Primer ID | Primer sequence
(5′-3′) | Product size
(bp) | Amplicon location
from TSS (bp) | PCR cycles | Primer annealing T,
(°C) | (Refs.) |
|---|
| MT1E | M-F |
GGATTTCGGGAATATCGC | 217 | −113/+104 | 38 | 56 | (12) |
|
| M-R |
ACGAAAATCGAACCGAAC |
|
|
|
|
|
|
| U-F |
TTTGGATTTTGGGAATATTGT | 220 | −116/+104 |
|
|
|
|
| U-R |
ACAAAAATCAAACCAAACACA |
|
|
|
|
|
| MT1F | M-F |
GTATTCGGAATTTTAAGGGGC | 134 | −262/-129 | 35 | 57 | This study |
|
| M-R |
CGAACCGTCCCTTTAAAATC |
|
|
|
|
|
|
| U-F |
TAGGTATTTGGAATTTTAAGGGGT | 139 | −265/-127 |
|
|
|
|
| U-R |
CACAAACCATCCCTTTAAAATC |
|
|
|
|
|
| MT1G | M-F |
TCGTATACGGGGGGTATAGC | 131 | −232/-102 | 37 | 58 | This study |
|
| M-R |
GCGATCCCGACCTAAACT |
|
|
|
|
|
|
| U-F |
AAGTTGTATATGGGGGGTATAGT | 137 | −235/-99 |
|
|
|
|
| U-R |
CCCACAATCCCAACCTAAACT |
|
|
|
|
|
| MT1M | M-F |
GGATATTGCGTATTATTCGGC | 112 | −240/-129 | 38 | 56 | This study |
|
| M-R |
ATAAATACCGAACGCACCATC |
|
|
|
|
|
|
| U-F |
TTGGGGATATTGTGTATTATTTGGT | 116 | −244/-129 |
|
|
|
|
| U-R |
ATAAATACCAAACACACCATCCC |
|
|
|
|
|
Gene expression analysis
For cDNA synthesis, 250 ng of extracted RNA was
reverse transcribed (RT) using High-Capacity cDNA Reverse
Transcription kit with RNase Inhibitor following the manufacturer's
protocol (Applied Biosystems; Thermo Fisher Scientific, Inc.).
Expression levels of genes MT1E, MT1G, MT1M and endogenous
control HPRT1 were evaluated by means of quantitative PCR
(RT-qPCR) using TaqMan Gene Expression assays (Hs01582977_gH,
Hs01584215_g1, Hs00828387_g1, and Hs02800695_m1, respectively;
Applied Biosystems; Thermo Fisher Scientific, Inc.). The reaction
mix (20 µl) consisted of 1X TaqMan Universal Master Mix II no UNG
(Applied Biosystems; Thermo Fisher Scientific, Inc.), 0.4X TaqMan
Gene Expression assay and 2 µl of RT product. Amplification was
performed with Mx3005P™ qPCR System (Agilent Technologies, Inc.,
Santa Clara, CA, USA) in triplicates per gene. Thermocycling
consisted of 95°C for 10 min, followed by 45 cycles of 95°C for 15
sec and 60°C for 1 min. Non-template controls were included in each
run. Samples with HPRT1 amplification at cycle ≥35 were
considered of low quality and were excluded from the analysis. Data
preprocessing was performed using GenEx 6.0.1 software (Multid
Analyses AB, Göteborg, Sweden). Relative gene expression values,
transformed to a linear scale, were used for statistical
analysis.
The Cancer Genome Atlas dataset of
renal clear cell carcinoma
For the overview analysis of the metallothionein
gene family, the TCGA KIRC dataset was used (19). Global DNA methylation profiling data
using Illumina Infinium HumanMethylation450K (HM450) platform and
RNA expression data obtained by RNA-seq were utilized in the study.
Gene-specific Level 3 datasets were acquired from the cBioPortal
(http://www.cbiopotal.org) and MethHC (http://methhc.mbc.nctu.edu.tw) data analyses portals
in September 2018 (20,21).
Statistical analysis
Statistical analysis was performed using STATISTICA
v8.0 (StatSoft Inc., Tulsa, OK, USA). The two-sided Fisher's exact
test was used for two-group comparisons of categorical data, while
the Mann-Whitney U test was used for continuous data. Heterogeneity
index (HI) was calculated to estimate the discordance rate of
methylation status of paired tumor foci. Correlations of gene
expression levels with quantitative clinicopathological or
molecular parameters were evaluated by calculating Spearman's
RS and/or Pearson's RP correlation
coefficients. Receiver Operating Characteristic (ROC) curve
analysis was performed and the area under the curve (AUC) was
calculated in order to evaluate the clinical utility of the test.
Logistic regression analysis was applied for the putative biomarker
combination. P-level of <0.0500 was considered significant. Data
visualization was developed using GraphPad Prism v5.03 software
(GraphPad Software, Inc., La Jolla, CA, USA).
Results
Metallothionein gene analysis in the
TCGA dataset
For the screening step, DNA methylation and gene
expression data were extracted for 8 protein-coding metallothionein
genes from the TCGA KIRC dataset. In total, data of 520 ccRCC and
209 healthy tissue samples were available for the analysis. Due to
the ubiquitous expression of some of the metallothioneins in
various tissues and induction by a variety of stimuli (including
hormones and growth factors), we focused on MT1 group genes
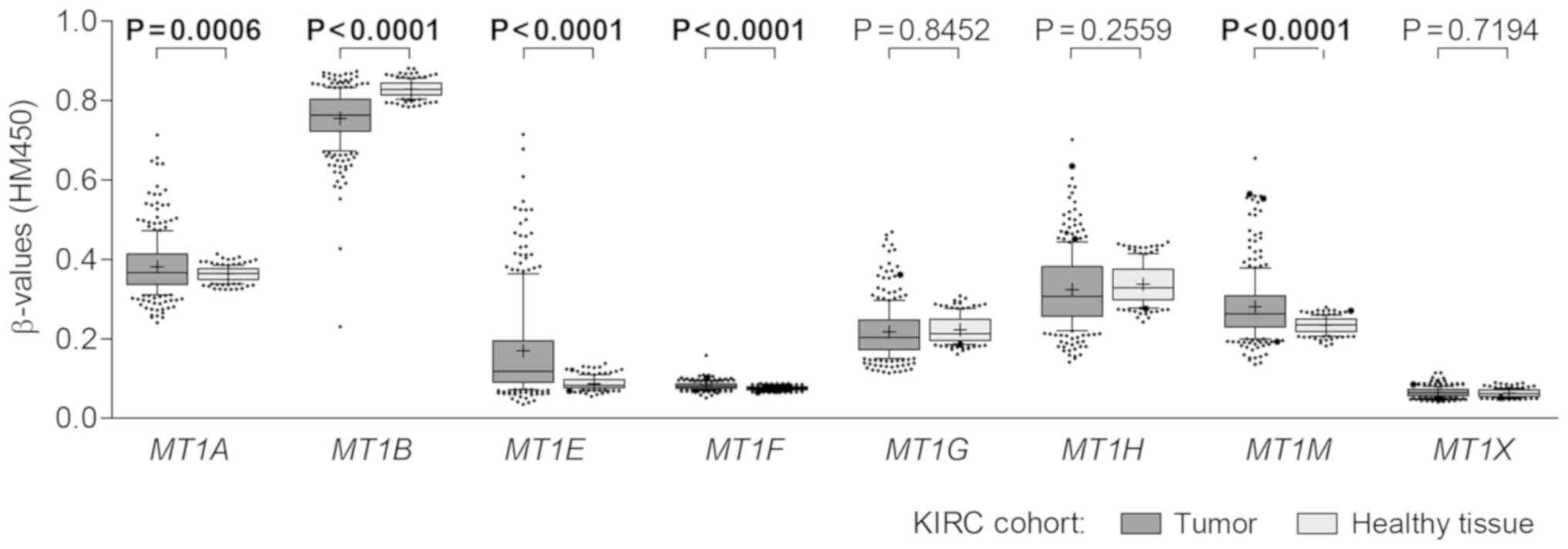
only. Significant differences in methylation levels between ccRCC
and healthy tissues were identified for 5 genes, of which MT1A,
MT1E, MT1F and MT1M had higher methylation levels in
tumors, while MT1B was highly, but still differentially
methylated in both tissue types (all P<0.0500; Fig. 1). Furthermore, despite the variable
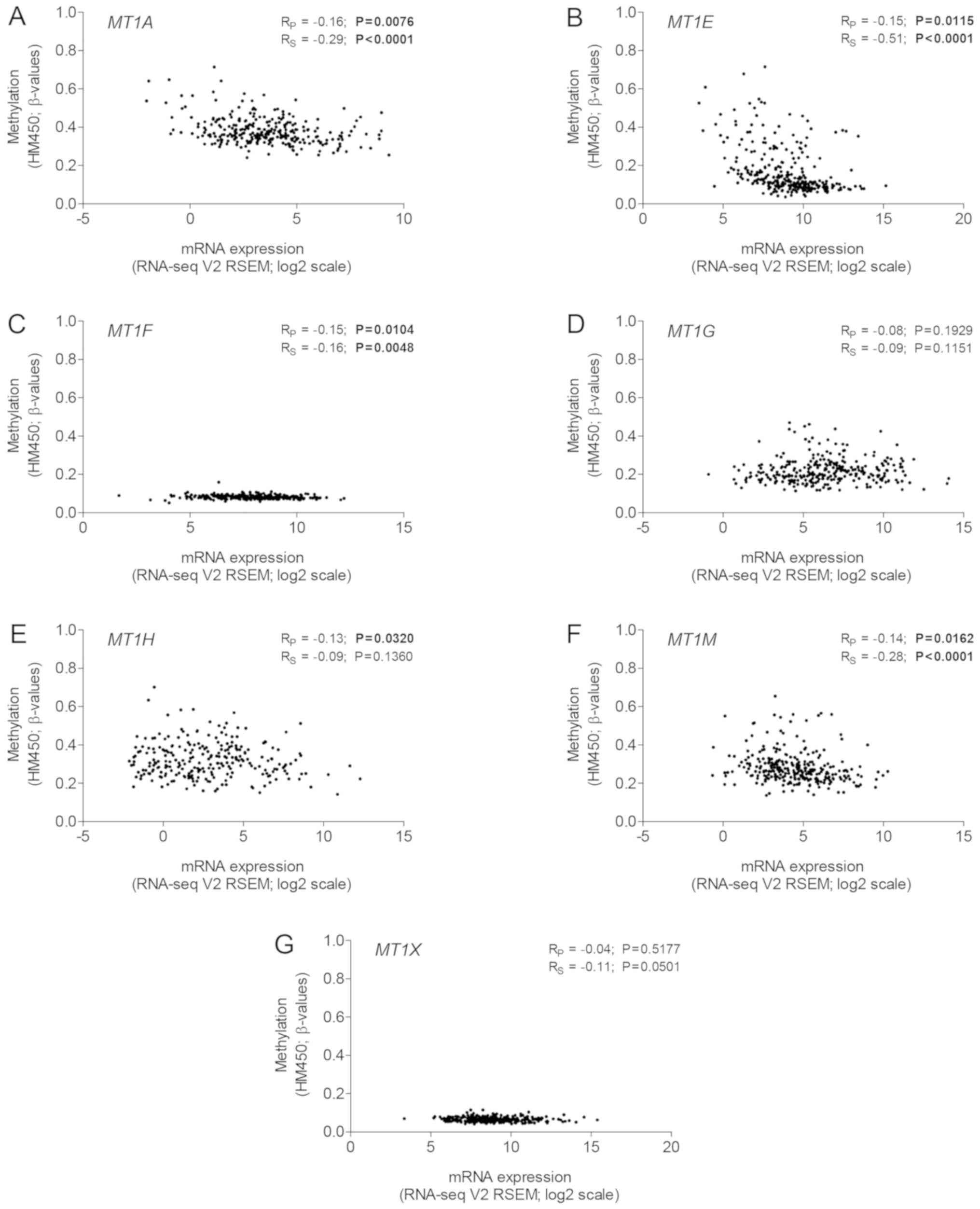
range, the methylation intensity of MT1A, MT1E, MT1F and
MT1M was correlated with downregulated gene expression (all
P<0.0500; Fig. 2). Expression of
MT1B was absent in almost all tumor samples, therefore, it
could not be compared with the promoter methylation (data not
shown).
DNA methylation analysis of selected
metallothionein genes
Based on the TCGA KIRC dataset analysis and with
regard to the literature review, four metallothionein genes,
MT1E, MT1F, MT1G, and MT1M, were selected for the
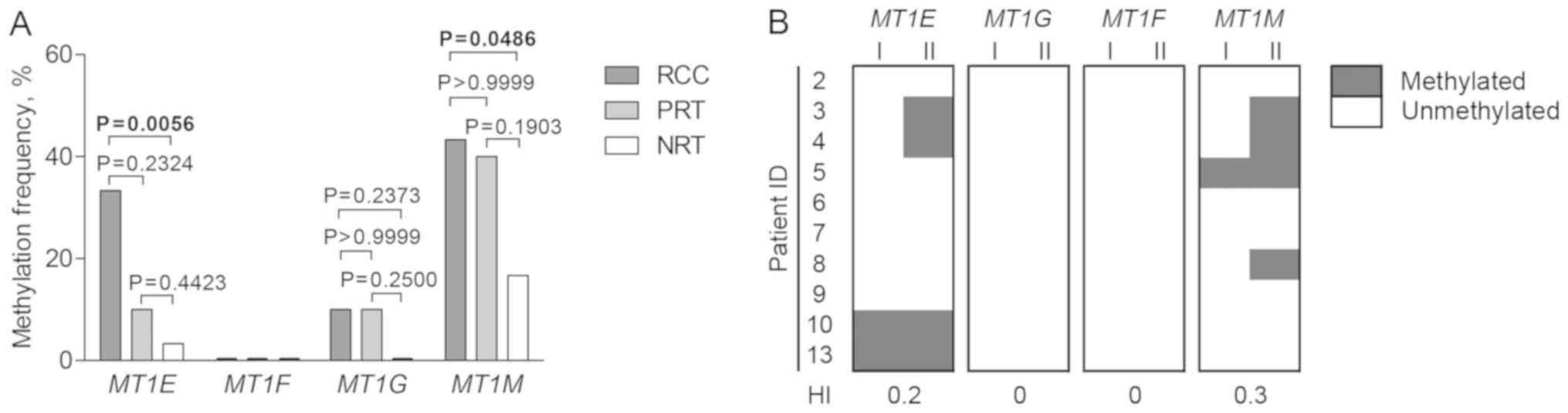
qualitative analysis of promoter DNA methylation. MT1E,
MT1G, and MT1M were methylated in ≤43.3% of tumors and
less frequently in NRT, but only MT1E and MT1M showed
significant differences (P=0.0056 and P=0.0486, respectively). The
three genes were also methylated in PRT indicating the field
cancerization phenomenon of the tumor-adjacent area (Fig. 3A). Interfocal variation of
methylation status was present in MT1E and MT1M gene
promoters, but no heterogeneity of MT1G and MT1F was
detected (Fig. 3B). Only
unmethylated promoter status of MT1F was observed in all
analyzed tissues.
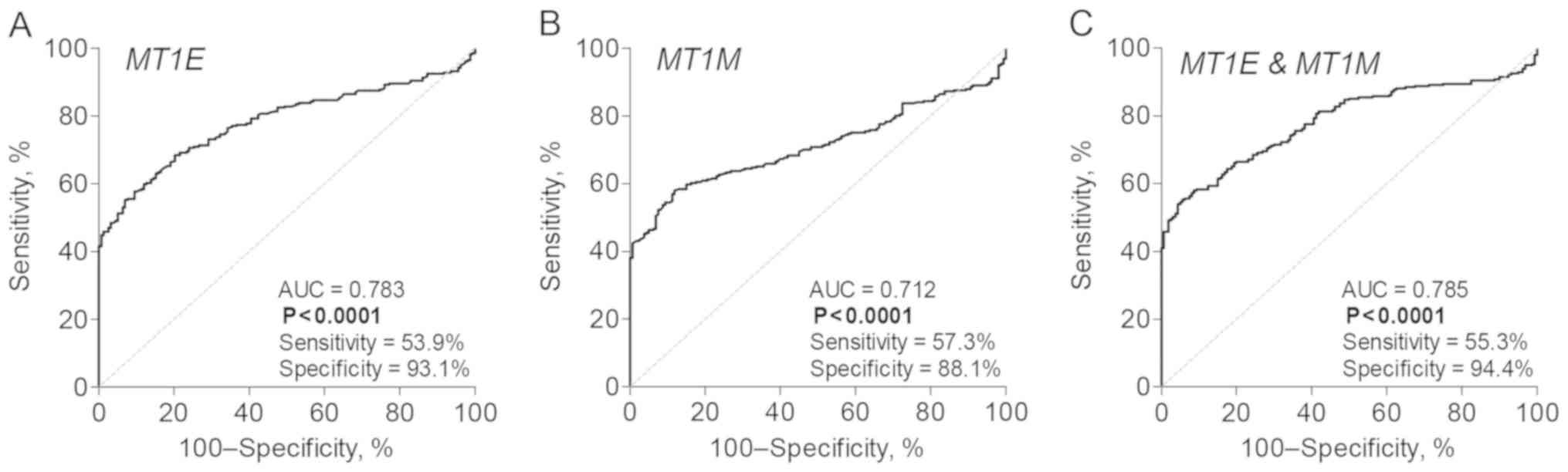
In our cohort, the sensitivity and specificity of
the MT1E and MT1M gene combination were 53.3 and
83.3%, respectively. The ROC curve analysis of the TCGA KIRC
dataset revealed comparable diagnostic values of the two genes
individually or in combination, with the specificity reaching up to
94.4% (Fig. 4).
DNA methylation and
clinicopathological parameters
Aberrant promoter methylation of MT1E, MT1G
and MT1M was further analyzed according to
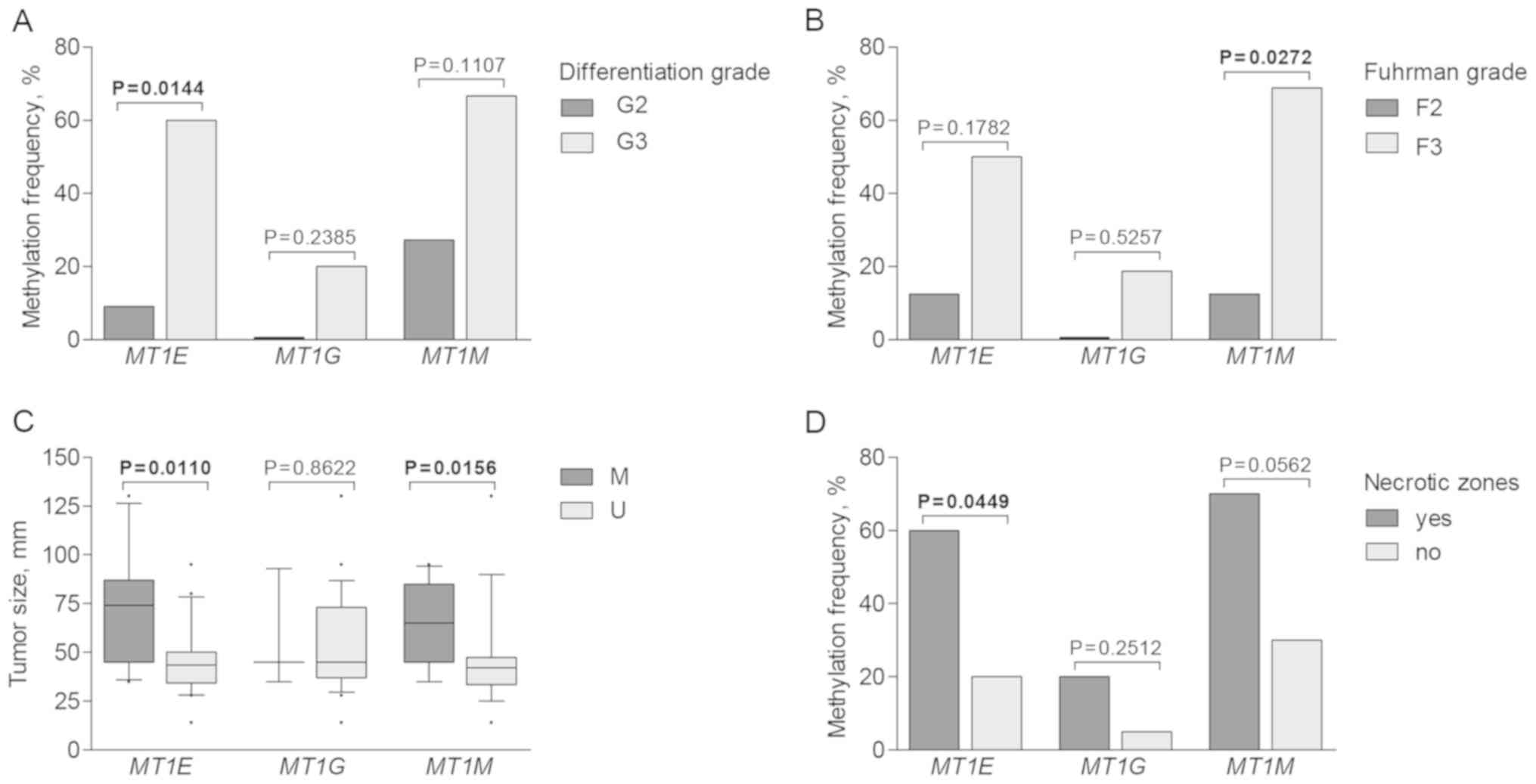
clinicopathological patient characteristics. Methylation was more
commonly detected in cases with advanced disease parameters. Higher
MT1E methylation frequency was observed in tumors with
higher differentiation grade (P=0.0144), while MT1M was more
commonly methylated in RCC cases characterized with higher Fuhrman
grade (P=0.0272; Fig. 5A and B).
Tumors with methylated MT1E and MT1M promoters were
significantly larger than those with unmethylated promoter status
(P=0.0110 and P=0.0156, respectively; Fig. 5C). Furthermore, methylation of
metallothionein genes was recurrently observed in tumors having
necrotic zones; however, only MT1E showed significant
difference (P=0.0449; Fig. 5D). No
associations were detected between metallothionein gene methylation
and patient age, sex, or pathological tumor stage (data not
shown).
Gene expression analysis and
association with DNA methylation
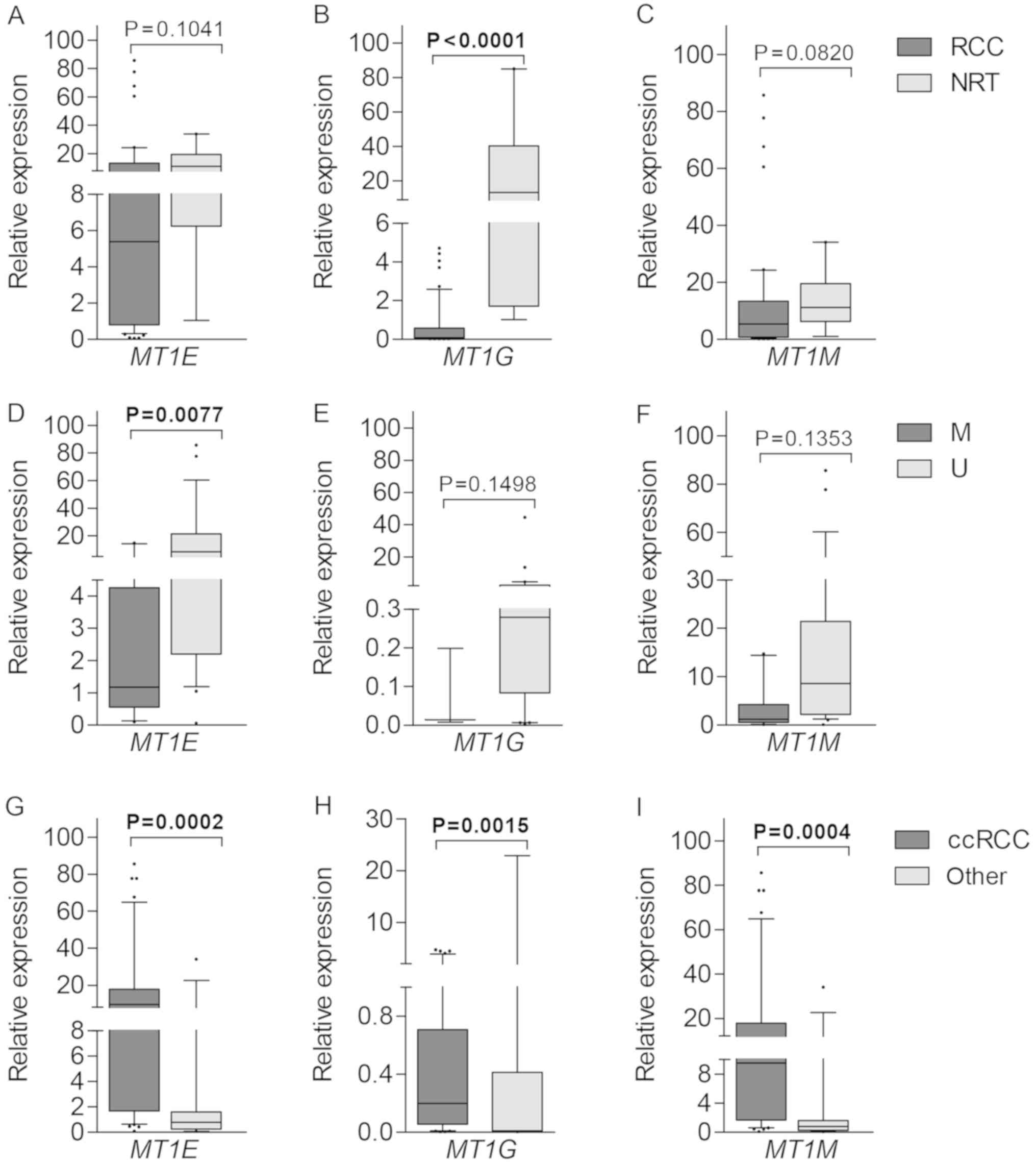
Gene expression at the transcriptional level was
quantified by means of RT-qPCR. Lower expression levels of MT1E,
MT1G and MT1M were detected in tumors as compared to NRT
samples, however, only MT1G showed significant difference
(P<0.0001; Fig. 6A-C).
Downregulation of MT1E correlated with methylated promoter
status (P=0.0077), while no such associations were detected for
MT1G and MT1M (both P>0.0500; Fig. 6D-F). All three genes were expressed
at significantly higher levels in ccRCC in comparison to the mixed
group of other histological tumor subtypes (P=0.0002, P=0.0015 and
P=0.0004 for MT1E, MT1G and MT1M, respectively;
Fig. 6G-I). No other associations
were observed between gene expression and clinical-pathological
parameters.
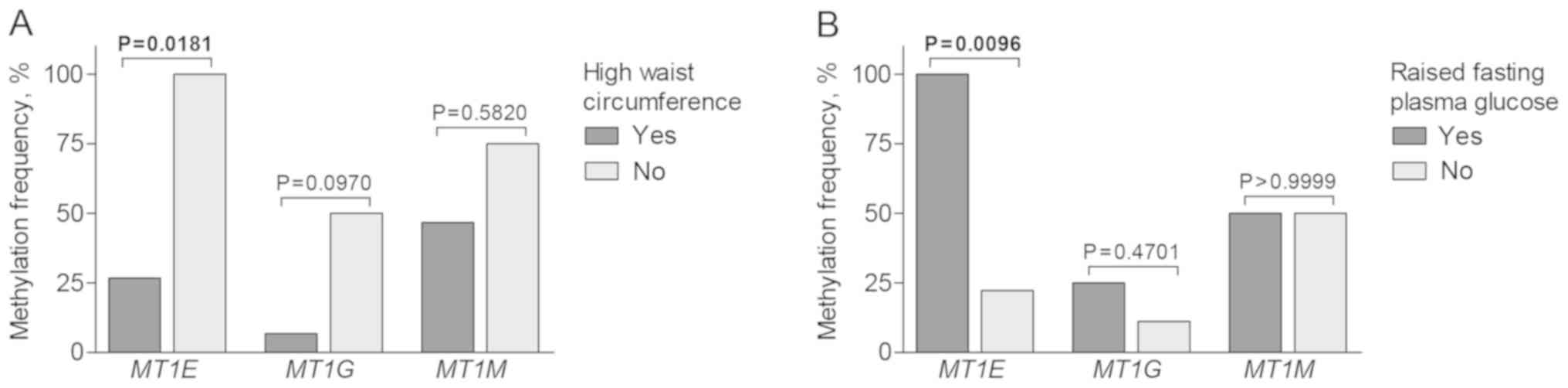
Associations with metabolic
syndrome-related parameters
Metabolic syndrome is concomitant with particular
tumor types and is considered to be a risk factor of kidney cancer.
In the present study, molecular alterations of metallothionein
genes were associated with particular parameters used to diagnose
metabolic syndrome. Methylated MT1E status in ccRCC subtype
was more common in cases with raised fasting plasma glucose level
(P=0.0096), but less frequent in patients with high waist
circumference (P=0.0181; Fig. 7).
Notably, expression of MT1G and MT1M was positively
correlated with women's waist circumference (RS=0.38,
P=0.0490 and RS=0.47, P=0.0119, respectively), whereas
MT1M was upregulated in men having raised fasting glucose
level (P=0.0348; data not shown). Due to the missing data for the
majority of the cases, associations with other metabolic
syndrome-related parameters (such as hypertriglyceridemia or
high-density lipoprotein level) were not analyzed.
Discussion
In recent years, metallothioneins have emerged as
important players in human carcinogenesis. Due to their unique
function of metal ion buffering and delivery in a cell, these small
cysteine-rich proteins have been shown to be pivotal regulators of
various cellular processes, such as proliferation, differentiation,
or apoptosis. Besides metal ion homeostasis and detoxification,
metallothioneins protect cells against oxidative stress and DNA
damage by scavenging free radicals. Numerous studies have reported
deregulation of metallothionein expression in human tumors and,
thus, their important role in various aspects of carcinogenesis has
been proposed (10–15). However, the mechanisms responsible
for the deregulated expression have been sparsely investigated.
In the present study, we investigated several
protein-coding metallothionein genes in renal tumors aiming to
evaluate their promoter DNA methylation for potential clinical
utility. Four metallothionein genes, namely MT1E, MT1G, MT1F
and MT1M, were selected for epigenetic analysis in renal
cell carcinoma (RCC) and paired non-cancerous renal tissues. To the
best of our knowledge, this is the first study to report aberrant
methylation of MT1E and MT1M genes in RCC. We showed
that methylation of MT1E and MT1M genes was
tumor-specific, which indicates the potential clinical value of the
two biomarkers in RCC diagnostics. Moreover, these results were
supported by our preliminary observations made by analyzing the
TCGA KIRC cohort (19). Until now,
the two genes have been investigated in several other cancer types
mostly at transcriptional and/or translational levels; however, the
data concerning their promoter methylation are limited. In the
present study, MT1E methylation associated with
downregulated gene expression was observed in RCC, which is in
accordance with our previous results obtained in prostate tumors
(12). Other studies have also
reported epigenetic silencing of MT1E in endometrial tumors,
melanoma, and several other cancer localizations (22,23).
The presence of MT1E methylation in metastases of melanoma
patients, as well as in several invasive melanoma cell lines,
suggest its potential involvement in cancer progression (23). In this study, MT1E was more
frequently methylated in tumors of higher differentiation grade,
larger size, or having necrotic zones, all of which are indicative
of advanced disease. Furthermore, MT1E methylation status
was associated with patient waist circumference and raised fasting
plasma glucose, i.e. two of the parameters used for diagnosing
metabolic syndrome, which is considered as one of the RCC risk
factors. This hints that MT1E methylation may be an early
event in renal carcinogenesis and, thus, lays the grounds for
future investigations.
MT1M was the most commonly methylated gene in
our cohort (43%). Its aberrant methylation was also observed in
pericancerous renal tissues (PRT) (40%) indicating the field
cancerization phenomena in RCC, i.e. when histologically normal
tissue adjacent to cancer is primed to undergo transformation
(24). Promoter methylation of
MT1E and MT1G was also present in tumor-surrounding
tissues suggesting that such epigenetic alterations might precede
the development of RCC and predispose to multifocal tumors. In
clinical practice, detection of aberrant methylation in
normal-appearing biopsy specimens may be indicative of a missed
cancerous lesion nearby and could justify the need for a repeat
biopsy. Furthermore, together with MT1E, interfocal
heterogeneity of MT1M methylation status was observed in
several RCC cases, which is most likely attributable to discrepant
grades of different tumor foci. To date, epigenetic analysis of
MT1M in RCC has not been reported; however, it is one of the
most studied metallothionein genes in various other cancer types.
Downregulation of MT1M has been observed in hepatocellular,
esophagus squamous cell carcinomas, breast, and other tumors and
was associated with various clinicopathological parameters
describing cancer aggressiveness (25–27).
In this study, MT1M methylation was more frequently detected
in tumors of larger size and higher Fuhrman grade, which is in
accordance with previous observations in other tumors reporting its
putative role in cancer progression (25,27,28).
In the present study, methylation of MT1G and
MT1F was rare or absent. However, MT1G was the only
metallothionein with significantly decreased gene expression in RCC
as compared to healthy tissues. Aberrant MT1G methylation as
a novel biomarker of RCC was first reported by Dalgin et al
(29). In previous studies,
MT1G downregulation and epigenetic silencing have been
associated with poor clinical outcome and/or drug resistance in
various tumors (6,30,31).
In addition, another mechanism, loss of heterozygosity, has been
reported as being potentially responsible for the downregulated
expression (32); however, it was
not evaluated in the present study. We did not detect any
correlations between MT1G promoter methylation or
transcriptional expression and clinicopathological variables, which
may be related to the low MT1G methylation frequency
observed in our cohort. As no aberrant methylation of MT1F
was detected in any renal tissues, this gene was omitted from our
gene expression analysis. According to recent studies (5), MT1F overexpression rather than
downregulation seems to be more commonly observed in cancer, which
is in agreement with the lack of promoter methylation in our
data.
Previous studies have demonstrated that
metallothionein expression is quite specific to tumor localization
as summarized by Si et al (5). Even in the same type of cancer,
results can be contradictory due to the unique tumor
microenvironment and varying external stimuli. In this study,
despite promoter methylation status, MT1E, MT1G, and
MT1M were expressed at significantly higher levels in ccRCC
as compared to other histological subtypes of renal tumors.
Considering their previously reported role in drug resistance,
expression and/or methylation analysis of the three metallothionein
genes in renal tumors could potentially provide additional
information for treatment decision making.
In conclusion, this study has revealed the potential
clinical value of aberrant promoter methylation of metallothionein
genes in RCC. Methylated promoter status of MT1E and
MT1M, together with other clinicopathological disease
parameters, may serve for more accurate RCC characterization and
personalized treatment selection at the time of diagnosis. However,
further validation of these putative biomarkers are needed in
larger independent cohorts, including liquid-biopsy samples, such
as plasma or urine. DNA methylation analysis in body fluids not
only enables patient monitoring by acquiring serial samples but is
also considered to better reflect all tumor foci (including
metastases), unlike tissue biopsy, which poorly accounts for RCC
heterogeneity. As the diversity of metallothionein functions in the
cell has been coming to light of late, functional analysis may
provide significant insights on how the observed epigenetic
deregulation is translated to the protein level, and also may
indicate potential targets for drug development.
Acknowledgements
The authors would like to thank the patients who
participated in this study and the staff of National Center of
Pathology for tissue sampling and histopathologic evaluation. We
also thank Mr. Kerry S. Keys for language editing. The Cancer
Genome Atlas project (http://cancergenome.nih.gov) is acknowledged for data
availability.
Funding
The present study was supported by the grant no.
S-MIP-17-54 from the Research Council of Lithuania. RM and KD were
supported by the European Social Fund according to the activity
‘Development of students’ ability to carry out R&D activities'
under Measure No. 09.03.3-LMT-K-712 ‘Development of Scientific
Competences of Scientists, other Researchers and Students through
Practical Research Activities’ (grant no. 09.03.3-LMT-K-712-09-0156
to KD).
Availability of data and materials
The RCC datasets analyzed in the current study are
available from the corresponding author on reasonable request. The
KIRC dataset used for general epigenetic overview of the
metallothionein gene family is publicly available from the TCGA
Research Network (http://cancergenome.nih.gov/).
Authors' contributions
RM performed the experiments and the data analysis,
and drafted the manuscript. AZ and AB collected and analyzed the
clinical data. FJ coordinated the patient selection, supervised the
clinical data analysis and was involved in the conception of the
study. SJ contributed to study implementation, supplied the
samples, and participated in establishing research schemes. KD
conceived and designed the study, supervised the experimental part
of the study, and was a major contributor in writing the
manuscript. All authors read and approved the manuscript and agree
to be accountable for all aspects of the research in ensuring that
the accuracy or integrity of any part of the work are appropriately
investigated and resolved.
Ethics approval and consent to
participate
Approval to conduct biomedical research (no.
158200-13-620-192) was obtained from the Lithuanian Bioethics
Committee (Vilnius, Lithuania) before initiating the study, and all
patients provided informed consent for participation.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Krężel A and Maret W: The functions of
metamorphic metallothioneins in zinc and copper metabolism. Int J
Mol Sci. 18:E12372017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Klaassen CD, Liu J and Diwan BA:
Metallothionein protection of cadmium toxicity. Toxicol Appl
Pharmacol. 238:215–220. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Agrawal S, Flora G, Bhatnagar P and Flora
SJ: Comparative oxidative stress, metallothionein induction and
organ toxicity following chronic exposure to arsenic, lead and
mercury in rats. Cell Mol Biol. 60:13–21. 2014.PubMed/NCBI
|
|
4
|
Dutsch-Wicherek M, Sikora J and
Tomaszewska R: The possible biological role of metallothionein in
apoptosis. Front Biosci. 13:4029–4038. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Si M and Lang J: The roles of
metallothioneins in carcinogenesis. J Hematol Oncol. 11:1072018.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Sun X, Niu X, Chen R, He W, Chen D, Kang R
and Tang D: Metallothionein-1G facilitates sorafenib resistance
through inhibition of ferroptosis. Hepatology. 64:488–500. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Gansukh T, Donizy P, Halon A, Lage H and
Surowiak P: In vitro analysis of the relationships between
metallothionein expression and cisplatin sensitivity of non-small
cellular lung cancer cells. Anticancer Res. 33:5255–5260.
2013.PubMed/NCBI
|
|
8
|
Smith DJ, Jaggi M, Zhang W, Galich A, Du
C, Sterrett SP, Smith LM and Balaji KC: Metallothioneins and
resistance to cisplatin and radiation in prostate cancer. Urology.
67:1341–1347. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Jing L, Yang M, Li Y, Yu Y, Liang B, Cao
L, Zhou X, Peng S and Sun Z: Metallothionein prevents doxorubicin
cardiac toxicity by indirectly regulating the uncoupling proteins
2. Food Chem Toxicol. 110:204–213. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Park Y and Yu E: Expression of
metallothionein-1 and metallothionein-2 as a prognostic marker in
hepatocellular carcinoma. J Gastroenterol Hepatol. 28:1565–1572.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Theocharis S, Karkantaris C, Philipides T,
Agapitos E, Gika A, Margeli A, Kittas C and Koutselinis A:
Expression of metallothionein in lung carcinoma: Correlation with
histological type and grade. Histopathology. 40:143–151. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Demidenko R, Daniunaite K, Bakavicius A,
Sabaliauskaite R, Skeberdyte A, Petroska D, Laurinavicius A,
Jankevicius F, Lazutka JR and Jarmalaite S: Decreased expression of
MT1E is a potential biomarker of prostate cancer
progression. Oncotarget. 8:61709–61718. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Weinlich G, Eisendle K, Hassler E, Baltaci
M, Fritsch PO and Zelger B: Metallothionein-overexpression as a
highly significant prognostic factor in melanoma: A prospective
study on 1,270 patients. Br J Cancer. 94:835–841. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hengstler JG, Pilch H, Schmidt M,
Dahlenburg H, Sagemüller J, Schiffer I, Oesch F, Knapstein PG,
Kaina B and Tanner B: Metallothionein expression in ovarian cancer
in relation to histopathological parameters and molecular markers
of prognosis. Int J Cancer. 95:121–127. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Tai SK, Tan OJ, Chow VT, Jin R, Jones JL,
Tan PH, Jayasurya A and Bay BH: Differential expression of
metallothionein 1 and 2 isoforms in breast cancer lines with
different invasive potential: Identification of a novel nonsilent
metallothionein-1H mutant variant. Am J Pathol. 163:2009–2019.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wong MCS, Goggins WB, Yip BHK, Fung FDH,
Leung C, Fang Y, Wong SYS and Ng CF: Incidence and mortality of
kidney cancer: Temporal patterns and global trends in 39 countries.
Sci Rep. 7:156982017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Rabjerg M, Mikkelsen MN, Walter S and
Marcussen N: Incidental renal neoplasms: Is there a need for
routine screening? A Danish single-center epidemiological study.
APMIS. 122:708–714. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Linehan WM, Bratslavsky G, Pinto PA,
Schmidt LS, Neckers L, Bottaro DP and Srinivasan R: Molecular
diagnosis and therapy of kidney cancer. Annu Rev Med. 61:329–343.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ricketts CJ, De Cubas AA, Fan H, Smith CC,
Lang M, Reznik E, Bowlby R, Gibb EA, Akbani R, Beroukhim R, et al:
The cancer genome atlas comprehensive molecular characterization of
renal cell carcinoma. Cell Rep. 23:313–326. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Cerami E, Gao J, Dogrusoz U, Gross BE,
Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et
al: The cBio cancer genomics portal: An open platform for exploring
multidimensional cancer genomics data. Cancer Discov. 2:401–404.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Huang WY, Hsu SD, Huang HY, Sun YM, Chou
CH, Weng SL and Huang HD: MethHC: A database of DNA methylation and
gene expression in human cancer. Nucleic Acids Res. 43:D856–D861.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Tse KY, Liu VW, Chan DW, Chiu PM, Tam KF,
Chan KK, Liao XY, Cheung AN and Ngan HY: Epigenetic alteration of
the metallothionein 1E gene in human endometrial carcinomas. Tumour
Biol. 30:93–99. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Faller WJ, Rafferty M, Hegarty S, Gremel
G, Ryan D, Fraga MF, Esteller M, Dervan PA and Gallagher WM:
Metallothionein 1E is methylated in malignant melanoma and
increases sensitivity to cisplatin-induced apoptosis. Melanoma Res.
20:392–400. 2010.PubMed/NCBI
|
|
24
|
Chai H and Brown RE: Field effect in
cancer-an update. Ann Clin Lab Sci. 39:331–337. 2009.PubMed/NCBI
|
|
25
|
Ding J and Lu SC: Low metallothionein 1M
expression association with poor hepatocellular carcinoma prognosis
after curative resection. Genet Mol Res. 15:2016. View Article : Google Scholar :
|
|
26
|
Oka D, Yamashita S, Tomioka T, Nakanishi
Y, Kato H, Kaminishi M and Ushijima T: The presence of aberrant DNA
methylation in noncancerous esophageal mucosae in association with
smoking history: A target for risk diagnosis and prevention of
esophageal cancers. Cancer. 115:3412–3426. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Jadhav RR, Ye Z, Huang RL, Liu J, Hsu PY,
Huang YW, Rangel LB, Lai HC, Roa JC, Kirma NB, et al: Genome-wide
DNA methylation analysis reveals estrogen-mediated epigenetic
repression of metallothionein-1 gene cluster in breast cancer. Clin
Epigenetics. 7:132015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Fu CL, Pan B, Pan JH and Gan MF:
Metallothionein 1M suppresses tumorigenesis in hepatocellular
carcinoma. Oncotarget. 8:33037–33046. 2017.PubMed/NCBI
|
|
29
|
Dalgin GS, Drever M, Williams T, King T,
DeLisi C and Liou LS: Identification of novel epigenetic markers
for clear cell renal cell carcinoma. J Urol. 180:1126–1130. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Guo R, Wu G, Li H, Qian P, Han J, Pan F,
Li W, Li J and Ji F: Promoter methylation profiles between human
lung adenocarcinoma multidrug resistant A549/cisplatin (A549/DDP)
cells and its progenitor A549 cells. Biol Pharm Bull. 36:1310–1316.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Nguyen A, Jing Z, Mahoney PS, Davis R,
Sikka SC, Agrawal KC and Abdel-Mageed AB: In vivo gene expression
profile analysis of metallothionein in renal cell carcinoma. Cancer
Lett. 160:133–140. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Chan KY, Lai PB, Squire JA, Beheshti B,
Wong NL, Sy SM and Wong N: Positional expression profiling
indicates candidate genes in deletion hotspots of hepatocellular
carcinoma. Mod Pathol. 19:1546–1554. 2006. View Article : Google Scholar : PubMed/NCBI
|