Introduction
Extranodal NK/T-cell lymphoma, nasal type
(EN-NK/T-NT) is a highly aggressive non-Hodgkin lymphoma,
characterized by prominent tumor necrosis and angiocentric
invasion. Neoplastic cells are cytoplasmic CD3ε(+), CD56(+),
cytotoxic-molecule(+) and Epstein-Barr virus(+) (1). This type of lymphoma usually occurs in
the upper airway tract, mostly in the nasal and paranasal area, and
is more prevalent among Asian and South American populations
(2). In recent years, disease
outcome has improved with chemotherapy and radiotherapy, but the
prognosis remains poor, with a 5-year overall survival (OS) rate of
42–70% (3,4). The underlying pathogenesis of the
disease remains unclear, and no effective targeted therapy is
currently available.
Array comparative genomic hybridization and gene-
expression profiling in EN-NK/T-NT revealed that the 6q21 regions
most frequently deleted involve the downregulation of several
important tumor-suppressor genes, such as POPDC3, PREP, positive
regulatory domain containing I (PRDM1), ATG5, AIM1, HACE1 and FOXO3
(5,6). Recent studies identified PRDM1, a
tumor suppressive transcriptor, as being frequently inactivated in
EN-NK/T-NT (7). Its inactivation
may be associated with the combination of 6q21/PRDM1 gene deletion,
PRDM1 mutation, aberrant upregulation of miRNA and/or an epigenetic
mechanism (8–11). Our team previously reported that the
hypermethylation of PRDM1 played a predominant role in the
downregulation of PRDM1 expression, significantly affecting the
biological behavior of tumor cells in EN-NK/T-NT. Notably, the
PRDM1 expression exerted an effect on the outcome of patients
(10,11). Thus, PRDM1 expression has an
important clinical prognostic value, and its inactivation may be an
important pathogenetic mechanism for EN-NK/T-NT.
Gene-expression profiling also detected the
activation of several oncogenic pathways in EN-NK/T-NT, including
those of NF-κB, mitogen-activated protein kinase (MAPK), and Janus
kinase 3/activator of transcription 3 (JAK3/STAT3) (8,12,13).
The JAK3/STAT3 pathway is known to play a crucial role in
regulating cell growth, survival, motility and cell differentiation
through the regulation of several downstream genes (14). The STAT3 protein is a key factor in
JAK3/STAT3 pathway activation, and phospho-STAT3 (Tyr705) (p-STAT3)
the activated status of STAT3. The phosphorylated form of STAT3 was
detected in cell lines and the majority of the NK/T cell lymphoma
primary tumors, and it may be associated with STAT3 gene mutation
(15–17).
Although PRDM1 gene inactivation and the JAK3/STAT3
pathway play significant roles in the pathogenesis of EN-NK/T-NT,
the association between them and their roles in EN-NK/T-NT are
elusive. In the present study, oncogenic pathways were clarified in
EN-NK/T-NT. The expression of PRDM1, activation of STAT3, and their
genomic statuses were detected. In addition, potential clinical
roles of PRDM1 and p-STAT3 were evaluated.
Materials and methods
Patients and samples
Archived formalin-fixed paraffin- embedded (FFPE)
tumor blocks were collected from 58 Chinese patients (35 males and
23 females; median age, 47 years old) diagnosed with EN-NK/T-NT
from the Department of Pathology, Peking University First Hospital
(Beijing, China). Thirty of the primary EN-NK/T-NT cases included
in this study have been described in previous studies. The
diagnosis of EN-NK/T-NT was confirmed according to the WHO
classification (18). Follow-up
data were available for 51 patients. The follow-up period was
defined as the period from the date of initial diagnosis to the
death of the patient (from any cause) or last follow-up.
Cell culture, treatment and
viability
Three nasal NK/T cell lymphoma cell lines were used
in the present study: NKL (19),
NK92 (20) and YT (21). YT and NKL cells were obtained from
Beijing Hong Bo Kang Biological Technology (Beijing, China). NK92
cells were purchased from the Chinese Academy of Medical Sciences
(Beijing, China). The cell culture methods used have been described
in our previous study (9). Cells
were seeded at 2×105 cells/ml/well in 24-well plates and
treated with tofacitinib (50 and 100 nM) (cat. no. 14703; Cell
Signaling Technology, Inc., Danvers, MA, USA) and Stattic (1, 2, 5
and 10 µM) (cat. no. HY-13818; MedChemExpress, Inc., Shanghai,
China) at indicated concentrations for 24 and 48 h, with dimethyl
sulfoxide (DMSO) used as the control, before being subjected to
cell counting and MTS assays.
Immunohistochemistry (IHC)
IHC was performed using the DAKO EnVision Detection
Kit (Agilent Technologies, Inc., Santa Clara, CA, USA). The tissue
sections were subjected to heat-induced antigen retrieval in EDTA
buffer (pH 9.0), and a primary antibody against PRDM1 (dilution
1:100; cat no. 9115; Cell Signaling Technology; clone no. C14A4) or
p-STAT3 (dilution 1:100; cat no. 9145; Cell Signaling Technology;
clone no. D3A7) was used. A positive nuclear staining pattern was
interpreted as PRDM1 or p-STAT3 immunoreactivity. Based on the
studies by Garcia et al (22) and Nie et al (23,24),
the positive expression of PRDM1 nuclear staining was
semi-quantitatively graded as follows: Negative (0 to <10%
positive cells) and positive (>10 to 100% positive cells). A
high expression of p-STAT3 was defined as moderate/strong nuclear
staining in ≥50% of the tumor cell population and a low expression
of p-STAT3 as <50% nuclear staining (16,17).
Samples from the plasma cell myelomas and squamous epithelium of
the nasal mucosa were used as positive controls for PRDM1 staining,
and lung adenocarcinoma tissue and squamous epithelium of the nasal
mucosa were used as a positive control for p-STAT3. For the
negative control reactions, phosphate-buffered saline (PBS) was
used instead of the primary antibody.
Western blot analysis
Cell lysis buffer (Nanjing Keygen Biotech, Co.,
Ltd., Nanjing, China) was used to lyse YT, NK92 and NKL cells and
collect protein. BCA assay (Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) was used to quantify protein concentration. A
total of 40 µg of protein from each sample was separated by
electrophoresis in 10% sodium dodecyl sulphate polyacrylamide gels.
After electroblotting the gels were transferred to polyvinylidene
difluoride membranes (EMD Millipore, Billerica, MA, USA). The
membranes were blocked with 5% milk for 1 h at room temperature,
followed by incubation with a rabbit or mouse monoclonal antibody
against PRDM1 (dilution 1:1,000; cat. no. 9115; Cell Signaling
Technology; clone no. C14A4), p-STAT3 (Tyr705) (dilution 1:1,000;
cat. no. 9145; Cell Signaling Technology; clone no. D3A7), STAT3
(dilution 1:1,000; cat. no. 4914; Cell Signaling Technology; clone
no. 79D7), or β-actin (dilution 1:5,000; cat. no. TA346894;
ZSGB-BIO, Inc., Beijing, China) overnight at 4°C. Anti-rabbit and
anti-mouse horseradish peroxidase-conjugated secondary antibodies
(both dilution 1:5,000; cat. nos. ZB-2305 and ZB-2306; ZSGB-BIO,
Inc.) were used to incubate for 60 min at room temperature.
Enhanced chemiluminescence (EMD Millipore, Billerica, MA, USA) was
used to develop protein signals. The band intensity of western
blotting was measured by densitometry using the G:BOX Chemi XT4
(Syngene, Cambridge, UK). Protein expression was quantified by
densitometry and normalized to β-actin.
PanCancer pathways analysis
According to our IHC grading criteria, 8 PRDM1(+)
and 8 PRDM1(−) FFPE samples and 2 samples of normal nasal mucosa
were selected from the 58 NK/T lymphoma cases. Total RNA was
extracted using RNeasy Mini Kit (Qiagen GmbH, Hilden, Germany)
according to the manufacturer's instructions. After determining the
RNA quality using the Agilent 2100 Bioanalyzer (Agilent
Technologies, Inc.), 5 PRDM1(+) and 5 PRDM1 (−) specimens (P1, P2,
P3, P4, P5 and N1, N2, N3, N4, N5, respectively) that met the
criterion of NanoString analysis were identified. The 2 normal
nasal mucosa samples were used as blank controls (B1, B2). The
NanoString nCounter PanCancer Pathways Panel (NanoString
Technologies, Inc., Seattle, WA, USA) includes 770 essential genes
representing 13 Canonical Pathways: Notch, Wnt, Hedgehog, TGFβ,
MAPK, STAT, P13K, RAS, chromatin modification, transcriptional
regulation, DNA damage control, cell cycle (CC), and apoptosis. The
NanoString nCounter assay was performed according to the standard
protocol of NanoString with analysis and normalization of the raw
NanoString data conducted using nSolver Analysis Software v3.0
(NanoString Technologies, Inc.). All procedures associated with
mRNA quantification, including sample preparation, hybridization,
detection and scanning, were carried out as recommended by
NanoString Technologies, Inc.
Sanger sequencing
We were able to extract genomic DNA from 37 of the
58 FFPE specimens using DNeasy Blood & Tissue kit (Qiagen GmbH)
according to the manufacturer's instructions. Sanger sequencing was
used to confirm STAT3 SRC homology 2 (SH2)-domain mutations. STAT3
SH2-domain primer sequences were as follows: Exon 19-1 forward,
5′-TAGACGGGCCTGAGGATTT-3′ and reverse, 5′-CAAAGGGCCAGGATGTACTTT-3′;
Exon 19-2 forward, 5′-AGACTTGGCTTTCCCATTACTC-3′ and reverse,
5′-TTGCTAACAGGGCATCCATC-3′; Exon 20-1 forward,
5′-GCTCTCAGCAAGCCAGT-3′ and reverse, 5′-TCACTGAATCTTAGCAGGAAGG-3′;
Exon 20-2 forward, 5′-GGAGCGGGCCATCTTGA-3′ and reverse,
5′-CTCAGCAGCCACCAGCA-3′; Exon 21-1 forward,
5′-TCTCTGAGATGACCTAGCTGTAG-3′ and reverse,
5′-CCATGATCTTATAGCCCATGATGA-3′; Exon 21-2 forward,
5′-GTCCGTGGAACCATACACAA-3′ and reverse, 5′-GCTCTCTGGCCGACAATAC-3′;
Exon 21-3 forward, 5′-CTCTATCCTGACATTCCCAAGG-3′ and reverse,
5′-GCATTTGCCTATCTATCCTCCA-3′; Exon 22 forward,
5′-TGCGAAGTCACAGTCAGTAAG-3′ and reverse,
5′-GATTAACTCTCACCCAGTGTCC-3′. Polymerase chain reaction (PCR) was
performed using Platinum Taq Polymerase (cat. no. 10966-083; Thermo
Fisher Scientific, Inc., Waltham, MA, USA) cycled at 95°C for 10
min, 45 cycles at 95°C for 30 sec, 56°C for 30 sec, 72°C for 1 min,
and a final extension of 72°C for 10 min. PCR sequencing was
carried out using ABI BigDye Terminator v3.1 (cat. no. 4337457;
Thermo Fisher Scientific, Inc.) and cycled at 96°C for 1 min, 29
cycles at 96°C for 10 sec, 50°C for 5 sec and 60°C for 4 min. The
resulting products were run on an ABI 3730×l DNA analyzer (Applied
Biosystems; Thermo Fisher Scientific, Inc.).
Flow cytometry
The extent of drug-induced apoptosis and CC arrest
was determined by staining with an Annexin V Apoptosis Detection
kit I (BD Biosciences, San Jose, CA, USA) and Propidium Iodide Flow
Cytometry kit (Abcam, Cambridge, MA, USA) following the
manufacturer's instructions. Flow cytometry was performed using
FACSAria II instruments and a FACSCalibur flow cytometer (both from
BD Biosciences).
Statistical analysis
For the cell proliferation experiment, the data were
obtained from at least 3 independent experiments, each with
triplicate measurements (n≥3). Statistical analysis of the data was
performed using one-way ANOVA followed by Dunnett's post hoc test.
P-values of <0.05 were considered to indicate a statistically
significant difference. Fisher's exact test was used to analyze the
association between STAT3 mutation and the clinical parameters of
the subjects. The Spearman rank correlation coefficient test was
used to determine the correlation between PRDM1 and p-STAT3
expression. OS was calculated using the Kaplan-Meier method and
compared to log-rank tests. A two-sided P<0.05 was considered to
indicate a statistically significant difference. All analyses were
performed using SPSS 19.0 software (IBM Corp., Armonk, NY,
USA).
Results
p-STAT3 and PRDM1 expression in
EN-NK/T-NT
To determine the activation status of STAT3 and the
expression of PRDM1 in EN-NK/T-NT, IHC analysis of PRDM1 and
p-STAT3 was performed in 58 cases of EN-NK/T-NT. The results
revealed that 32.8% (19/58) of the cases exhibited weak, moderate
or strong nuclear staining of PRDM1 (≥10% positive cells) (Fig. 1A and C). p-STAT3 was highly
expressed in 72.4% (42/58) of the EN-NK/T-NT cases. The staining
intensity of p-STAT3-positive cells was strong in the majority of
cases (Fig. 1B and C). The
expression of PRDM1 and p-STAT3 in malignant NK cell lines was also
examined by western blot analysis. As revealed in Fig. 1D, the expression level of p-STAT3
was higher in YT and NKL than in NK92 cells, whereas PRDM1 was
highly expressed in the YT, but not in the other 2 cell lines.
These results demonstrated that STAT3 was frequently activated and
PRDM1 commonly downregulated in EN-NK/T-NT.
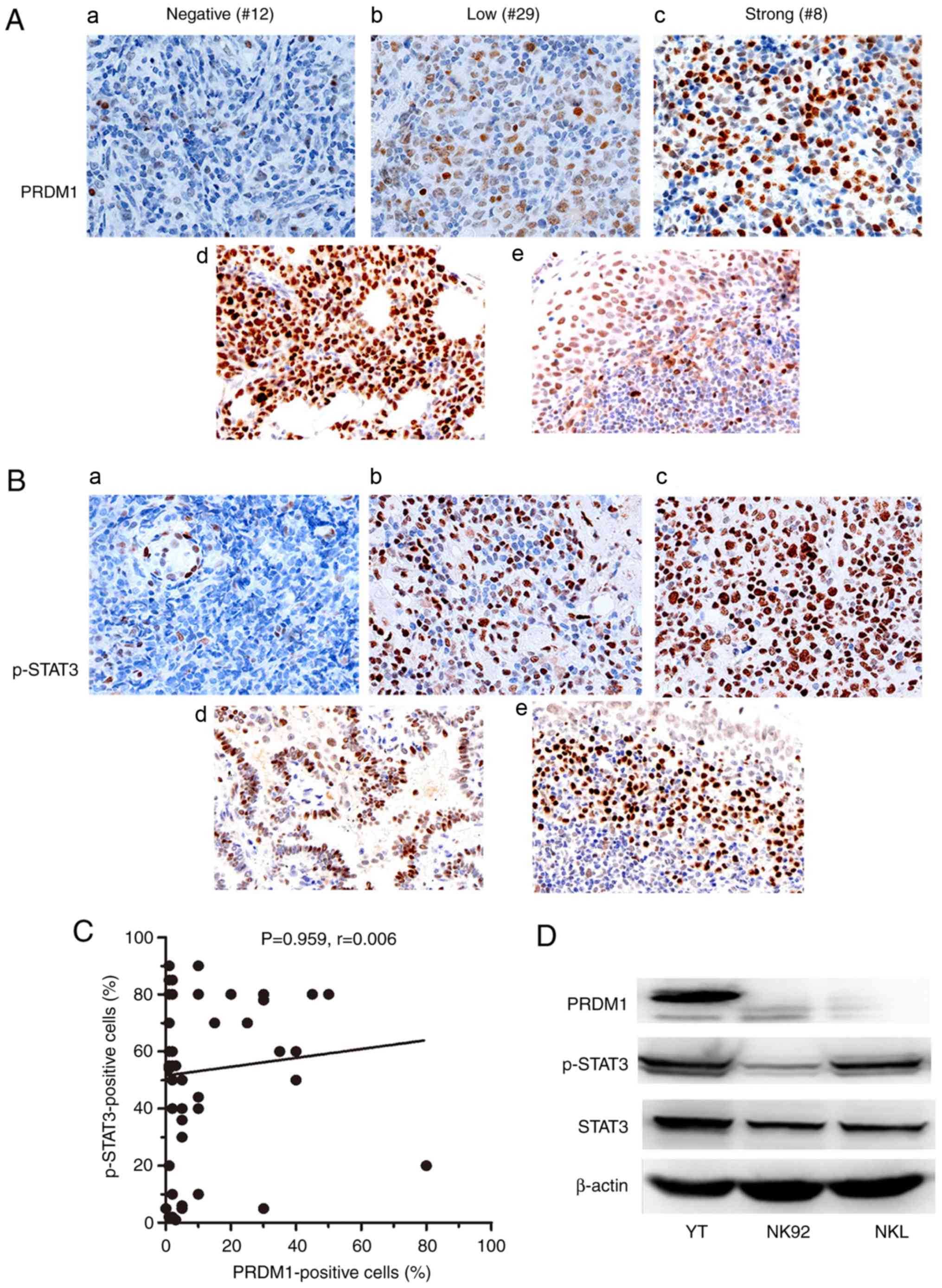 | Figure 1.p-STAT3 and PRDM1 expression in
EN-NK/T-NT. (A) Examples of IHC analysis of PRDM1 in EN-NK/T-NT
specimens and control samples. In EN-NK/T-NT specimens, PRDM1 was
commonly inactivated. Representative images of PRDM1 (a) negative,
(b) low, and (c) high expression. (d) Samples from plasma cell
myelomas, and (e) the squamous epithelium of nasal mucosa, were
used as positive controls for PRDM1 staining. (B) Examples of IHC
analysis of p-STAT3 in EN-NK/T-NT specimens and control samples. In
EN-NK/T-NT specimens, p-STAT3 was commonly activated.
Representative images of (a) p-STAT3 negative, (b) low, and (c)
high expression. (d) Samples from adenocarcinoma tissue, and (e)
the inflammatory nasal mucosa, were used as positive controls for
p-STAT3 staining. (C) A scatter plot revealed no direct correlation
between PRDM1 immunostaining and p-STAT3 expression. (P=0.959,
r=0.005) (D) Western blot analysis of the expression level of
STAT3, p-STAT3 and PRDM1 in YT, NKL and NK92 cells. p-STAT3,
phospho-STAT3 (Tyr705); PRDM1, positive regulatory domain
containing I; EN-NK/T-NT, extranodal NK/T-cell lymphoma, nasal
type; IHC, immunohistochemistry. |
Activation of JAK3/STAT3 pathway and
its correlation with the status of PRDM1 in EN-NK/T-NT
To identify whether the JAK3/STAT3 pathway plays a
predominant role in EN-NK/T-NT tumors, a panel of 13 classical
oncogenic pathways was selected to perform gene expression analysis
using the NanoString nCounter® system. A total of 10 [5
PRDM1(+) and 5 PRDM1(−)] EN-NK/T-NT and 2 normal nasal mucosa
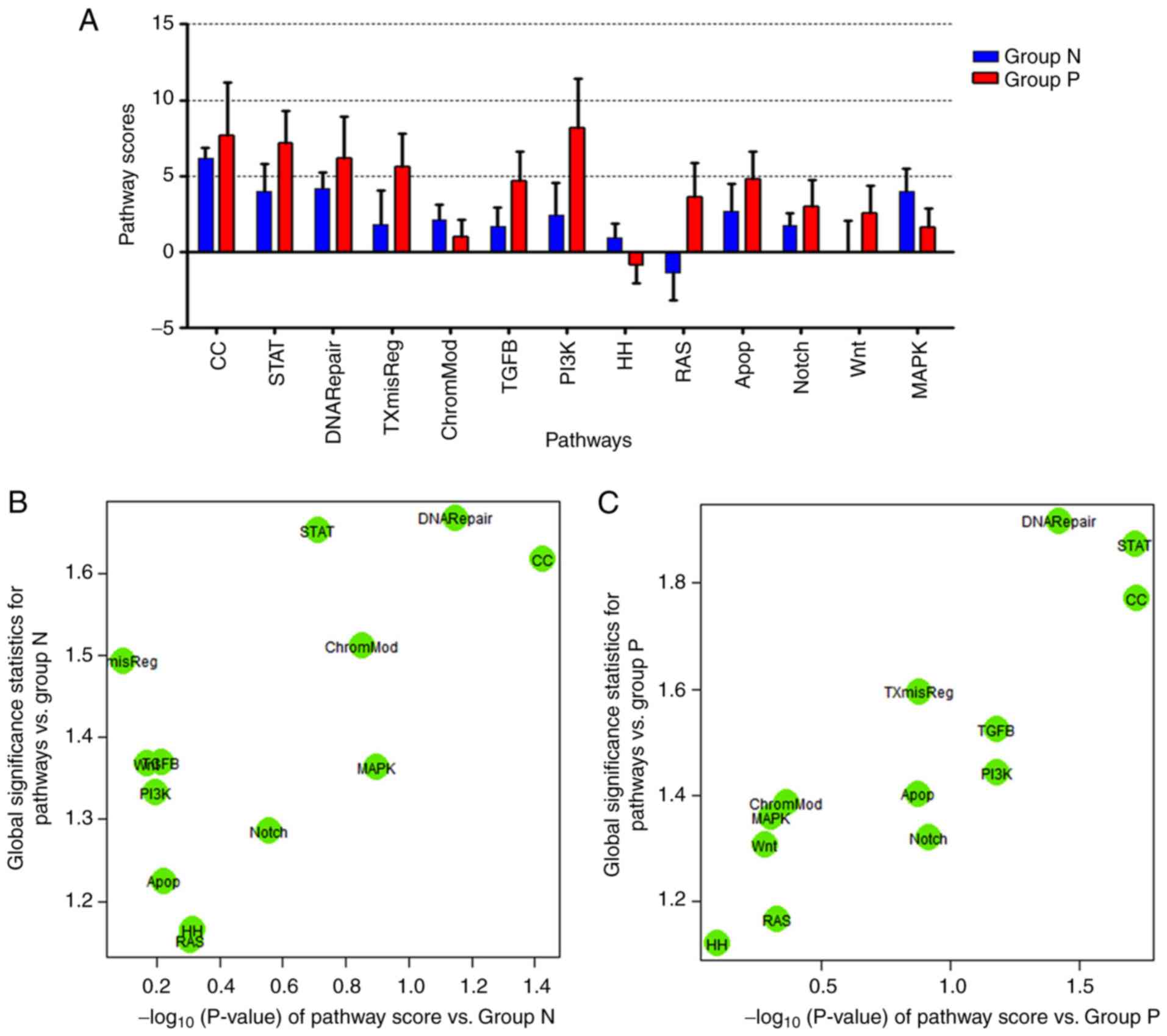
samples were assessed. First, the scoring of 13 pathways in each
case was analyzed using unsupervised hierarchical clustering. As
revealed in the heat map (Fig. 2A),
5 EN-NK/T-NT tumor samples (i.e., N4, N3, P1, P5 and P2) scored
highly in the Wnt, RAS, apoptosis, STAT, TXmisReg, TGFβ and PI3K
signaling pathways, suggesting the strong activation of these
pathways in EN-NK/T-NT. P3 and P4 also scored highly in the STAT
pathway (Fig. 2A), indicating the
aberrant activation of the STAT pathway in the detected EN-NK/T-NT
samples. The expression patterns of major genes in STAT pathways in
EN-NK/T-NT were further analyzed. Fig.
2B reveals that the STAT pathway scores were much higher in the
EN-NK/T-NT tumor samples than in the control (fold change >7).
The heat maps in Fig. 2C further
illustrated the expression levels of individual genes involved in
the JAK/STAT pathway in each case. As depicted in Fig. 2C, STAT pathway-activating genes
(including JAK3, STAT3, STAT1, STAT4, MYC, PIM1 and
PCNA) were increased, while STAT pathway-suppressing genes
(including IL17, PIK3CA, PIK3R and SOCS2) were
decreased in the majority of EN-NK/T-NT samples, supporting an
overall effect of JAK3/STAT3 activation on EN-NK/T-NT tissues.
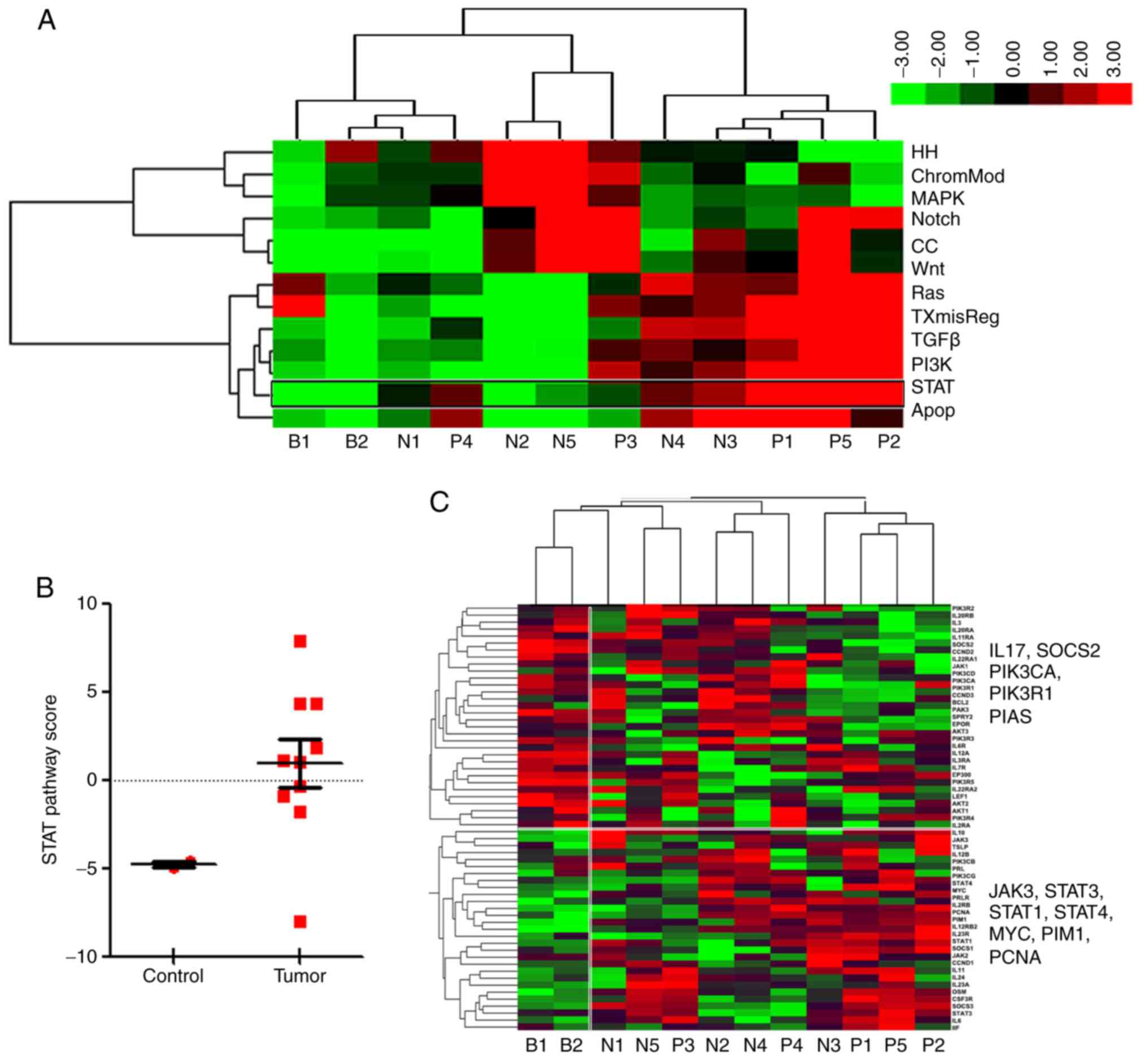 | Figure 2.Activation of JAK3/STAT3 pathway in
EN-NK/T-NT. NanoString nCounter analyzed 13 distinct oncogenic
pathways and scored them using unsupervised hierarchical clustering
between the tumor (group N+P) and blank groups (normal nasal mucosa
specimens, group B). (A) Heat map scoring of 13 pathways for each
case, demonstrated clear differences between normal and tumor
specimens. Red indicates high expression and green low. Expression
values were scaled within pathways. The 13 signaling pathways were
divided into 3 clusters. Five EN-NK/T-NT cases (right) exhibited a
cluster of high scores in the Wnt, RAS, apoptosis, STAT, TXmisReg,
TGFβ and PI3K signaling pathways. (B) Box plots of pathway scores
revealing clear differences in the STAT pathway scores between the
tumor and normal nasal mucosa groups (control). Tumor cases had
higher scores than the normal nasal mucosa cases (fold change
>7). (C) Heat map indicating the expression level of genes
associated with the STAT pathway. Red indicates high expression and
green low. Expression values were scaled for genes. The expression
of genes associated with STAT3 pathway activation was increased,
whereas that of genes involved in the suppression of its activation
was low in tumor samples. STAT3, signal transducer and activator of
transcription 3; EN-NK/T-NT, extranodal NK/T-cell lymphoma, nasal
type. |
The 10 cases were then divided into either the
PRDM1(+) or the PRDM1(−) group, according to the PRDM1 IHC staining
results. As depicted in the bar graph (Fig. 3A), STAT, DNA Repair and CC pathways
were highly upregulated in the two groups, although it was more
marked in the PRDM1(+) group. Consistently, linear associations of
pathway score analysis revealed that STAT, DNA repair and CC
pathways were strongly activated in both the PRDM1(−) and PRDM1(+)
groups (Fig. 3B and C). These
results indicated that the activation of the STAT pathway may be
independent from the PRDM1 expression in EN-NK/T-NT.
Genomic alterations of STAT3 and PRDM1
in EN-NK/T-NT
The strong p-STAT3 expression in the majority of
EN-NK/T-NT cases prompted us to investigate the underlying
mechanism of STAT activation. Recent studies have revealed that
STAT3 mutations are highly associated with STAT3 activation in
lymphoma cells. Since STAT3 mutations occur almost exclusively in
its SH2 coding region, the SH2 domain-coding region of STAT3 was
sequenced in 3 NK/T lymphoma cell lines (YT, NKL and NK92) and 37
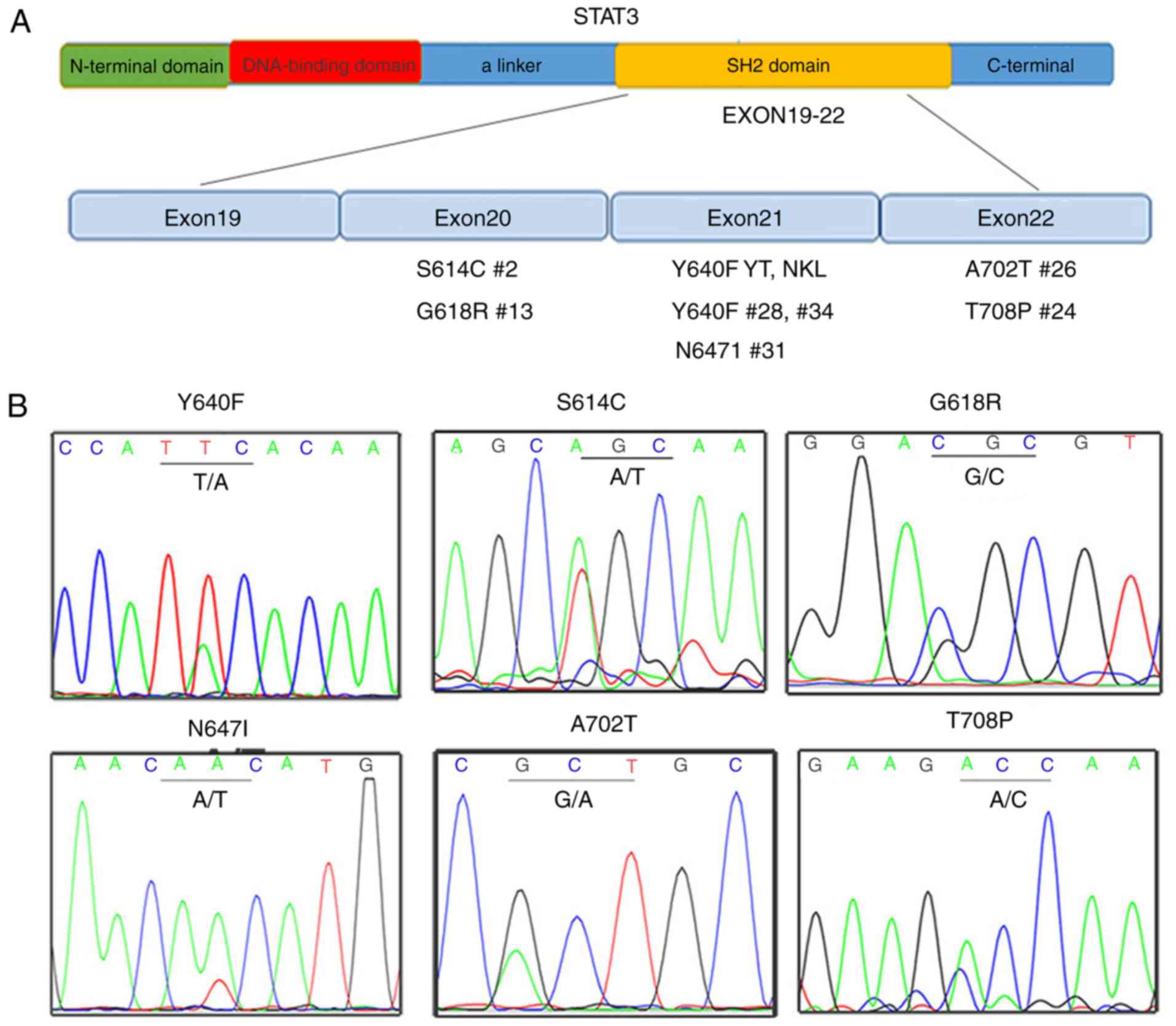
EN-NK/T-NT cases. Fig. 4A
summarized all 6 missense mutation types detected in 2 lymphoma
cell lines (Y640F in YT and NKL, but not NK92) and 7 EN-NK/T-NT
cases (e.g., S614C in case no. 2). Fig.
2B presents the sequencing results of these missense mutations.
The STAT3 missense mutation rate was 18.92% (7/37) (Table I). In addition, a silent mutation
(G618G, GGG>GGT) was detected in 2 EN-NK/T-NT cases (case nos. 8
and 15). Notably, all 7 EN-NK/T-NT cases with STAT3 missense
mutations exhibited a marked p-STAT3 expression (Table I), strongly indicating that STAT3
gene mutations in the SH2 domain is a driving force for the
activation of the JAK3/STAT3 pathway in EN-NK/T-NT.
 | Table I.Clinicopathological features of 37
extranodal NK/T-cell lymphoma, nasal type. |
Table I.
Clinicopathological features of 37
extranodal NK/T-cell lymphoma, nasal type.
| Sample no. | Sex | Age (years) | Biopsy site | Ann Arbor
stage | PRDM1
expression | p-STAT3
expression | STAT3 mutation | Outcome |
|---|
| Case 1 | M | 6 | Nasal | IV | High | High | No | Alive |
| Case 2 | M | 49 | Nasal | I | High | High | S614C | Alive |
| Case 3 | M | 63 | Nasal | II | Negative | High | No | Alive |
| Case 4 | F | 22 | Nasal | III | Negative | Negative | No | Alive |
| Case 5 | M | 35 | Nasal | III | Negative | High | No | Died |
| Case 6 | M | 66 | Skin | IV | Negative | Negative | No | Died |
| Case 7 | M | 51 | Nasal | III | Negative | Negative | No | Alive |
| Case 8 | M | 53 | Nasal | III | High | High | G618G | Alive |
| Case 9 | F | 41 | Inguinal lymph
nodes | IV | Negative | High | No | Alive |
| Case 10 | F | 43 | Nasal | IV | High | High | No | Died |
| Case 11 | M | 52 | Nasal | IV | High | High | No | Died |
| Case 12 | M | 36 | Nasal | II | Negative | Negative | No | Alive |
| Case 13 | M | 29 | Skin | III | Low | High | G618R | Alive |
| Case 14 | M | 55 | Nasal | IV | Negative | Low | No | Died |
| Case 15 | M | 48 | Nasal | II | High | Low | G618G | Alive |
| Case 16 | F | 62 | Nasal | III | High | low | No | Died |
| Case 17 | M | 77 | Nasal | II | High | High | No | Alive |
| Case 18 | M | 47 | Nasal | IV | Negative | High | No | Died |
| Case 19 | M | 29 | Nasal | III | High | High | No | Died |
| Case 20 | F | 25 | Nasal | IV | High | High | No | Died |
| Case 21 | M | 66 | Tonsil | I | Low | High | No | Alive |
| Case 22 | M | 80 | Nasal | IV | Low | Low | No | Died |
| Case 23 | F | 62 | Axillary lymph
node | IV | Negative | High | No | Died |
| Case 24 | F | 38 | Skin | NA | Negative | High | T708p | Died |
| Case 25 | F | 81 | Nasal | III | High | High | No | Died |
| Case 26 | M | 49 | Intestine | IV | Negative | High | A702t | Died |
| Case 27 | F | 55 | Bone marrow | II | Negative | Negative | No | Alive |
| Case 28 | M | 28 | Skin | IV | Negative | High | Y640f | Died |
| Case 29 | M | 45 | Nasal | I | Low | High | No | Alive |
| Case 30 | M | 73 | Skin | IV | Negative | High | No | Died |
| Case 31 | F | 41 | Nasal | Iv | Negative | High | N647i | Died |
| Case 32 | F | 23 | Nasal | III | Negative | Negative | No | Alive |
| Case 33 | F | 54 | Nasal | IV | Negative | Negative | No | Died |
| Case 34 | M | 12 | Nasal | IV | Negative | High | Y640f | Died |
| Case 35 | M | 63 | Tonsil | I | Low | High | No | Alive |
| Case 36 | M | 71 | Nasal | I | Negative | High | No | Alive |
| Case 37 | F | 29 | Inguinal lymph
nodes | II | Negative | High | No | NA |
With respect to the PRDM1 genomic status, no
correlation between the STAT3 gene mutation and 6q21/PRDM1 gene
deletion was identified. Despite that, STAT3 activation, PRDM1
loss, and 6q21 and/or PRDM1 heterozygous deletion could
simultaneously be detected in the NKL cell line and in 9 EN-NK/T-NT
cases (case nos. 22, 28, 34, 36, 41, 42, 48, 52 and 55), suggesting
that genetic alterations of STAT3 and PRDM1 could be independent
events in the development of EN-NK/T-NT (Table II).
 | Table II.Summary of PRDM1 expression,
combination of gene deletion, p-STAT3 expression and STAT3 mutation
in EN-NK/T-NT. |
Table II.
Summary of PRDM1 expression,
combination of gene deletion, p-STAT3 expression and STAT3 mutation
in EN-NK/T-NT.
| Sample | PRDM1 protein | 6q21 and/or PRDM1
HD | p-STAT3 | STAT3 mutation |
|---|
| YT | + | − | + | Y640F |
| NKL | − | + | + | Y640F |
| NK92 | − | + | − | − |
| Case 22 | − | + | + | − |
| Case 26 | − | + | + | A702T |
| Case 28 | − | + | + | Y640F |
| Case 29 | + | − | + | − |
| Case 33 | − | − | − | − |
| Case 34 | − | + | + | Y640F |
| Case 35 | + | − | + | − |
| Case 36 | − | + | + | − |
| Case 37 | − | + | − | ND |
| Case 38 | − | + | − | ND |
| Case 39 | − | + | − | ND |
| Case 40 | − | − | + | ND |
| Case 41 | − | + | + | ND |
| Case 42 | − | + | + | ND |
| Case 43 | − | − | + | ND |
| Case 44 | − | + | − | ND |
| Case 45 | − | + | − | ND |
| Case 46 | + | + | + | ND |
| Case 47 | − | − | + | ND |
| Case 48 | − | + | + | ND |
| Case 49 | − | − | + | ND |
| Case 50 | − | − | − | ND |
| Case 51 | + | − | + | ND |
| Case 52 | − | + | + | ND |
| Case 53 | + | + | + | ND |
| Case 54 | − | − | − | ND |
| Case 55 | − | + | + | ND |
Stratified clinicopathological
significance of STAT3 activation and PRDM1 expression in
EN-NK/T-NT
To explore the clinicopathological significance of
STAT3 mutations and PRDM1 activation and expression in EN-NK/T-NT,
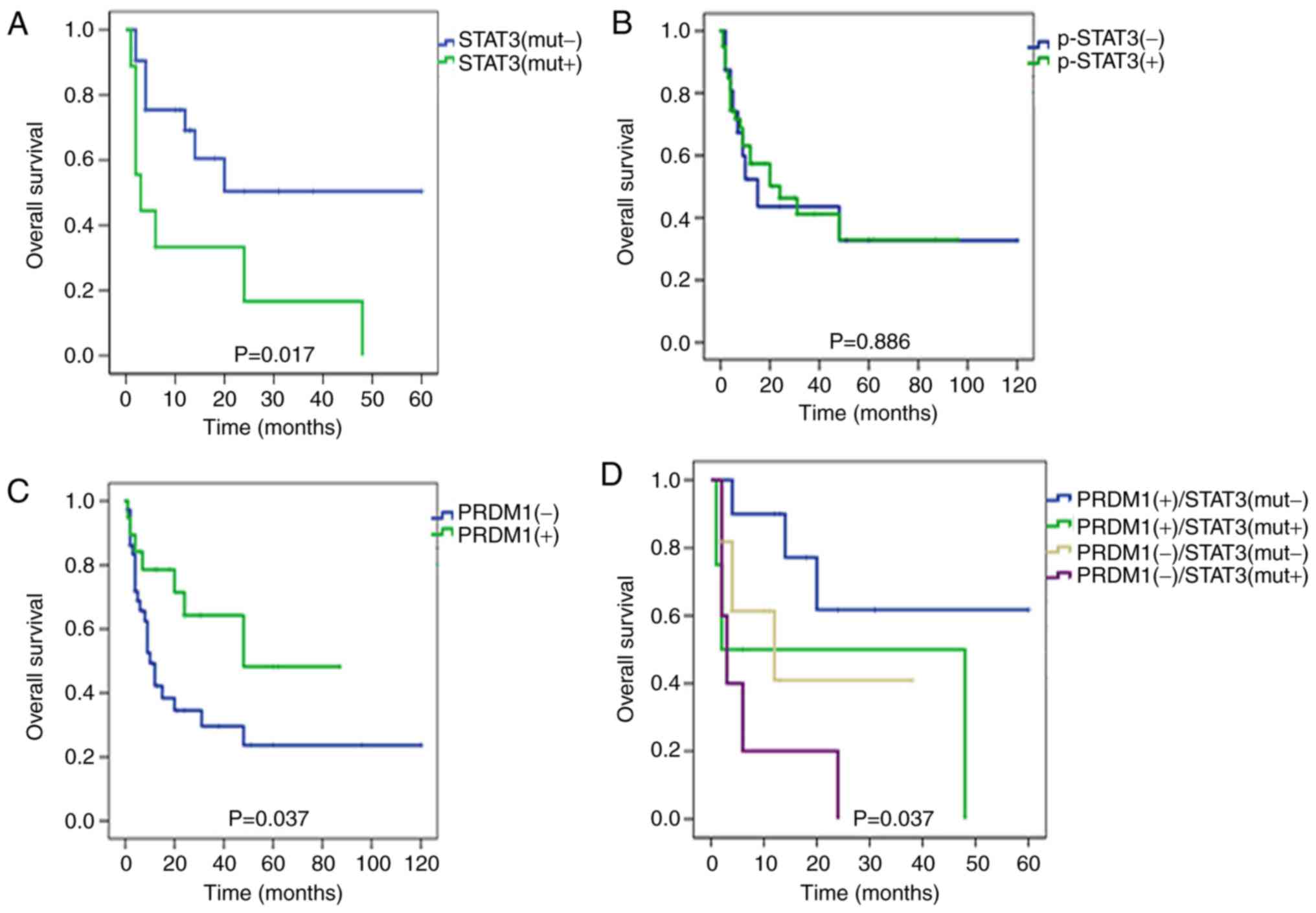
the survival effect of STAT3 expression and mutations was analyzed.
Kaplan-Meier single-factor analysis and the log-rank test revealed
that patients in the mutated STAT3 group [STAT3(mut+)] had a
considerably shorter survival time than those in the non-mutation
group [STAT3(mut-)] (Fig. 5A;
P=0.017) while the p-STAT3 level did not predict OS time in
EN-NK/T-NT (Fig. 5B). In addition,
STAT3(mut+) and p-STAT3(+) were revealed to be correlated with
angiocentric infiltration but no other pathological factor (i.e.,
site, Ann Arbor Stage, outcome or necrosis) in EN-NK/T-NT (Table III). These findings indicated that
STAT3(mut+) is a predictor for poor prognosis in patients with
EN-NK/T-NT. The survival effect of PRDM1 expression was also
analyzed. PRDM1(+) staining predicted a favorable effect on OS time
(Fig. 5C; P=0.037). Furthermore,
EN-NK/T-NT cases in this cohort were divided into 4 groups:
PRDM1(+)/STAT3(mut-), PRDM1(+)/STAT3(mut+), PRDM1(−)/STAT3(mut-)
and PRDM1(−)/STAT3(mut+). Notably, the majority of cases with
PRDM1(−)/STAT3(mut+) resulted in a poorer prognosis (Fig. 5D; P=0.037), as compared to the other
groups, suggesting that detecting STAT3 mutations and PRDM1
expression may be useful for routine pathological diagnosis and
patient prognosis.
 | Table III.Correlation of p-STAT3 expression and
STAT3 mutation with clinical factors and prognostic value. |
Table III.
Correlation of p-STAT3 expression and
STAT3 mutation with clinical factors and prognostic value.
|
| p-STAT3
expression | STAT3 mutation |
|---|
|
|
|
|
|---|
|
| n | % | Low | High | P-value | n | % | No | Yes | P-value |
|---|
| STAT3 mutation | 37 |
|
|
|
|
|
|
|
|
|
|
Yes | 7 | 18.9 | 0 | 7 | 0.31 |
|
|
|
|
|
| No | 30 | 81.1 | 8 | 22 |
|
|
|
|
|
|
| PRDM1 | 58 |
|
|
|
| 37 |
|
|
|
|
|
PRDM1(+) | 19 | 32.8 | 2 | 17 | NA | 15 | 40.5 | 13 | 2 | 0.39 |
|
PRDM1(−) | 39 | 67.2 | 14 | 25 |
| 22 | 59.5 | 17 | 5 |
|
| Sex | 58 |
|
|
|
| 37 |
|
|
|
|
|
Male | 35 | 60.3 | 7 | 28 | 0.11 | 24 | 64.9 | 19 | 5 | 1.00 |
|
Female | 23 | 39.7 | 9 | 14 |
| 13 | 35.1 | 11 | 2 |
|
| Age (years) | 58 |
|
|
|
| 37 |
|
|
|
|
|
≤60 | 44 | 75.9 | 15 | 29 | 0.11 | 26 | 70.3 | 19 | 7 | 0.08 |
|
>60 | 14 | 24.1 | 1 | 13 |
| 11 | 29.7 | 11 | 0 |
|
| Site | 58 |
|
|
|
| 37 |
|
|
|
|
|
Nasal | 44 | 75.9 | 4 | 10 | 1.00 | 27 | 73 | 24 | 3 | 0.06 |
|
Extranasal | 14 | 24.1 | 12 | 32 |
| 10 | 27 | 6 | 4 |
|
| Ann Arbor
Stage | 36 |
|
|
|
| 17 |
|
|
|
|
|
I/II | 10 | 27 | 4 | 6 | 0.69 | 5 | 29.4 | 5 | 0 | 0.26 |
|
III/IV | 27 | 73 | 8 | 19 |
| 12 | 70.6 | 8 | 4 |
|
| Outcome | 50 |
|
|
|
| 35 |
|
|
|
|
|
Alive | 22 | 39.3 | 7 | 15 | 0.49 | 15 | 42.9 | 13 | 2 | 0.67 |
|
Dead | 34 | 60.7 | 8 | 26 |
| 20 | 57.1 | 15 | 5 |
|
| Angiocentric
infiltration | 58 |
|
|
|
| 37 |
|
|
|
|
| No | 34 | 58.6 | 13 | 21 | 0.04 | 20 | 51.4 | 18 | 1 | 0.04 |
|
Yes | 24 | 41.4 | 3 | 21 |
| 17 | 48.6 | 12 | 6 |
|
| Necrosis | 58 |
|
|
|
| 37 |
|
|
|
|
| No | 20 | 34.5 | 7 | 13 | 0.36 | 16 | 43.2 | 14 | 2 | 0.67 |
|
Yes | 38 | 65.5 | 9 | 29 |
| 21 | 56.8 | 16 | 5 |
|
| OS |
| Mean ± SD |
|
| 23.56±31.9 | 20.05±22.7 | 0.69 |
|
| 16.41±15.02 | 7.71±8.05 | 0.05 |
Effects of inhibition of JAK3/STAT3
pathway on EN-NK/T-NT cells
The potential therapeutic application of suppressing
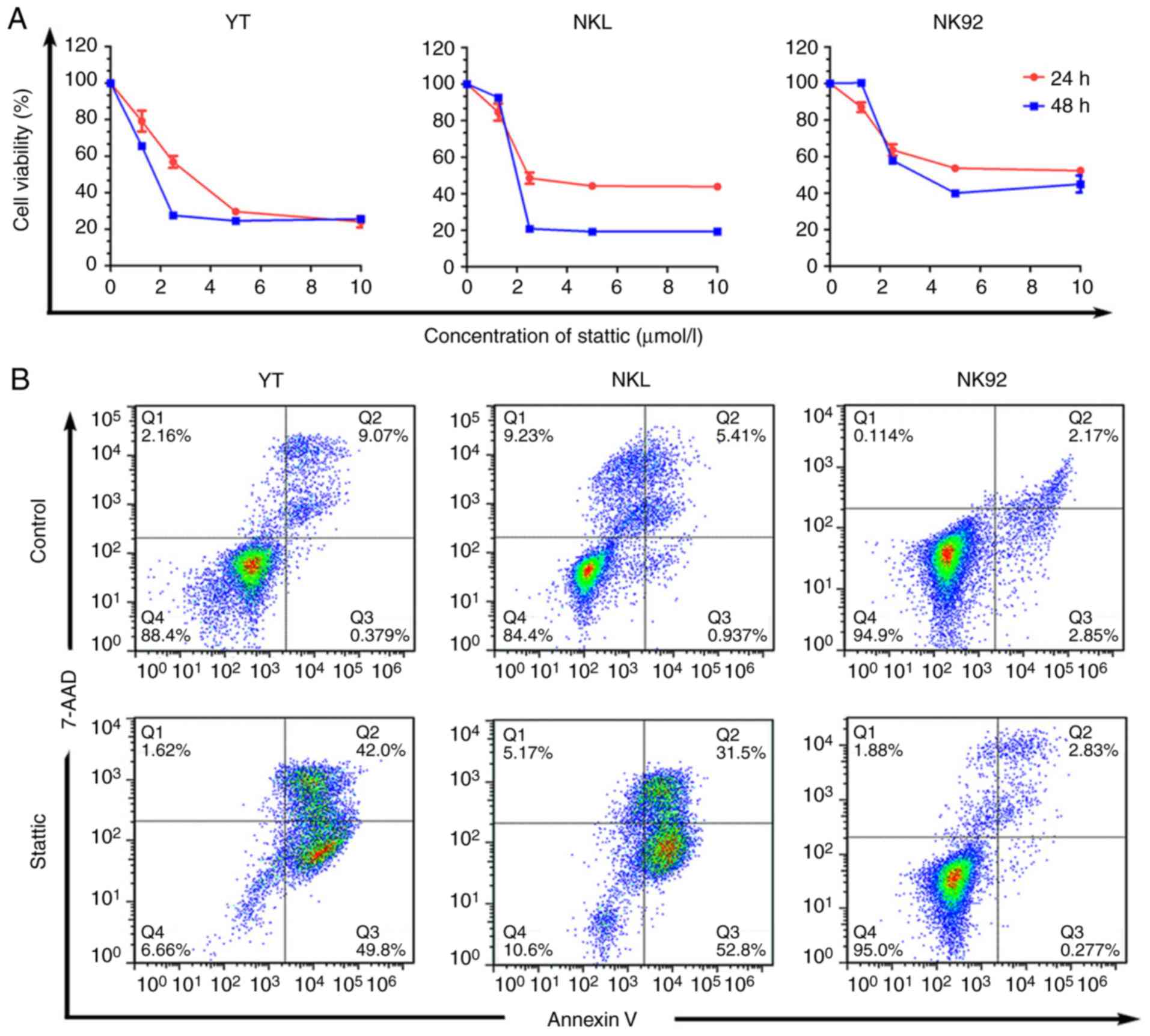
the JAK3/STAT3 pathway in EN-NK/T-NT was explored. The cell
viability of YT and NKL cells harboring STAT3 Y640F was inhibited
by the STAT3 inhibitor Stattic (Fig.
6A). However, Stattic had a low toxicity in NK92 cells with
wild-type STAT3, which usually exhibit relatively lower p-STAT3
levels than YT and NKL cells (Fig.
6A). To evaluate whether Stattic induces apoptosis, the
apoptosis of various Stattic-treated cells was analyzed by flow
cytometry. The treatment of YT and NKL cells with Stattic
significantly increased apoptotic cells, when compared with
DMSO-treated cells. However, no increase in the number of apoptotic
cells was observed in NK92 cells (Fig.
6B). In addition, YT, NKL and NK92 cells were treated with
tofacitinib, a JAK inhibitor, to further identify the biological
effect of the JAK3/STAT3 pathway on NK/T cells. As revealed in
Fig. 7A, not only did the number of
NK92 cells significantly decrease following treatment with 50 and
100 nM tofacitinib, but also the number of YT and NKL slightly
decreased. However, tofacitinib did not alter the number of
apoptotic cells in the 3 cell lines (not shown). Next, the effect
of tofacitinib on the CC was investigated. Flow cytometric analysis
demonstrated that 100 nM tofacitinib significantly increased the
percentage of YT, NKL and NK92 cells at the G0/G1 phase with a
concomitant reduction in the percentage of cells in the S (YT, NKL
and NK92) and G2 (NK92) phases in the treated NK/T lymphoma cells,
as compared with the control group (Fig. 7B). These findings indicated that
suppressing the JAK3/STAT3 pathway may have a therapeutic effect in
EN-NK/T-NT.
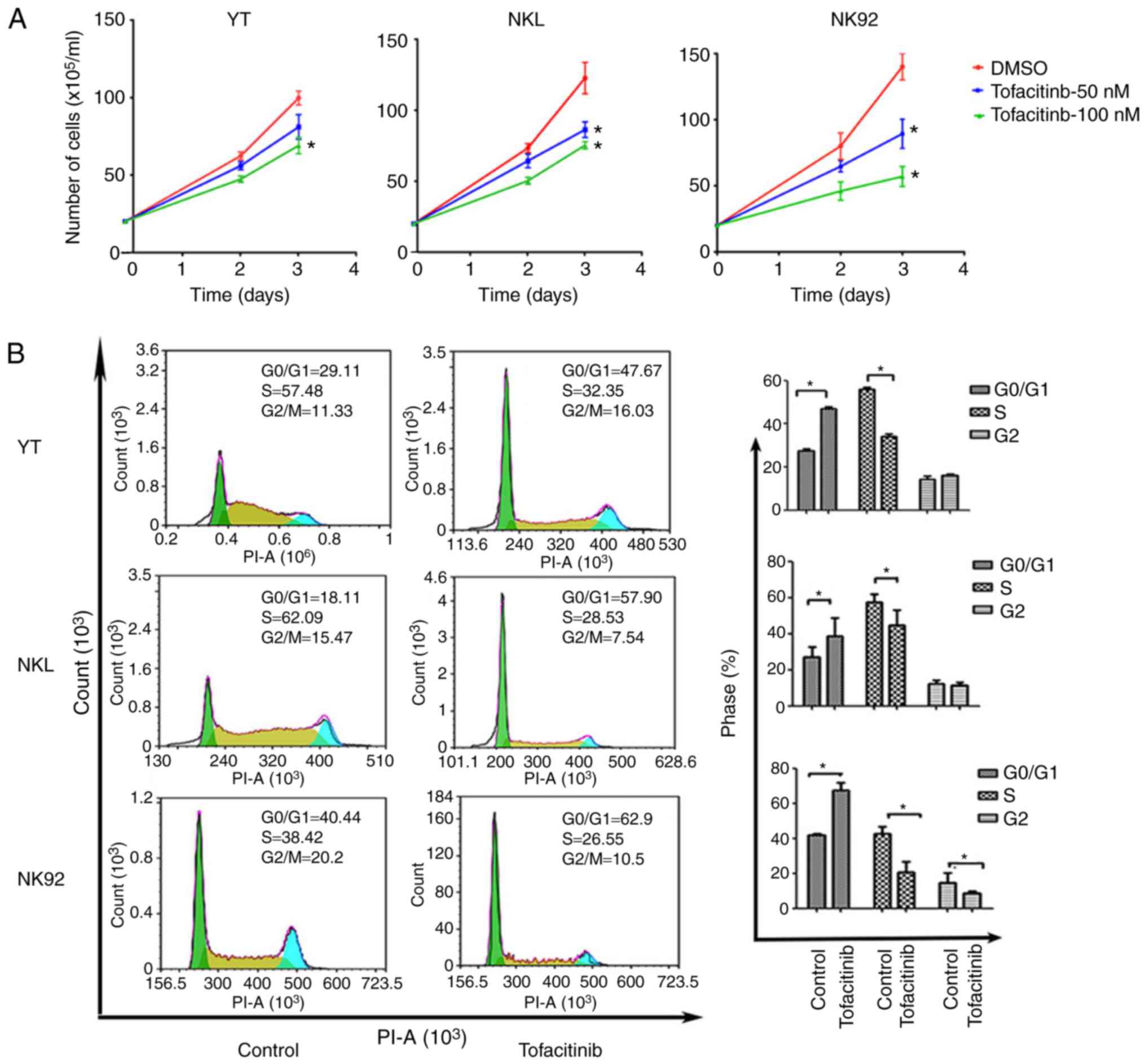 | Figure 7.Tofacitinib, a JAK3 inhibitor,
affects EN-NK/T-NT cell growth and CC. (A) YT, NKL and NK92 cells
were treated with the indicated concentrations of tofacitinib, and
viable cells were counted at the indicated time-points, using the
trypan blue exclusion test. Growth of YT, NKL, and NK92 cells were
significantly inhibited following treatment with tofacitinib at 50
and 100 nM. Values are expressed as the means ± SE of the results
from at least 3 independent experiments, each with triplicate
measurements (n≥3); *P<0.05 as compared with the DMSO-treated
cells. (B) The 3 cell lines were treated with DMSO or 100 nM
tofacitinib for 48 h, in order to assess CC profiles using flow
cytometry. Following treatment, a significant increase was observed
in the percentage of cells at the G1 phase and a concomitant
reduction in the percentage of cells at the S and G2 phases in the
3 cell types. Values are presented as the mean ± SE from triplicate
experiments (P<0.05 as compared with DMSO-treated cells).
EN-NK/T-NT, extranodal NK/T-cell lymphoma, nasal type; SE, standard
error; DMSO, dimethyl sulfoxide; CC, cell cycle. |
Discussion
NK cell malignancy is a multistep process, often
involving the cooperation of tumor suppressor gene inactivation and
oncogene activation. Previous studies have revealed that PRDM1 is a
tumor suppressor gene in NK/T cell lymphomas, and PRDM gene
deletion is a common event (6,7).
Karube et al using semi-quantitative RT-PCR detected PRDM1
expression in one representative donor and neoplastic samples and
revealed that average expression of PRDM1 levels in neoplastic
samples were significantly lower than those in normal NK cells
(6). PRDM1 expression has been
revealed to be associated with the stage of the disease and has a
positive effect on prognosis (10).
The present study also revealed PRDM1 low expression in the
majority of EN-NK/T-NT cases, and confirmed the effect of PRDM1
expression on the prognosis of EN-NK/T-NT.
Previous studies have revealed that the JAK/STAT
signaling pathway is crucial for disease development. Gene-
expression profiling also revealed that the JAK3/STAT3 signaling
pathway was one of the discriminating pathways in NK/T lymphoma
(13). The JAK family consists of 4
types: JAK1, JAK2, JAK3 and TyK2. Somatic mutations in JAK1, JAK2
and JAK3 result in constitutively active kinases in
myeloproliferative diseases and leukemia/lymphomas. Somatic
activating mutations in JAK1 and JAK2 have been mainly detected in
acute lymphoblastic leukemia and myeloproliferative neoplasms
(25). However, mutations in JAK3
have been mainly found in T- and NK cell lymphoma. In ENKTL, JAK3
phosphorylation has been detected in most cases. JAK3 activating
mutations were identified in approximately one-third of the cases.
In addition, a high expression of p-STAT3 has been detected in a
variety of tumors, including colorectal, gastric and breast cancer,
and non-Hodgkin lymphoma (25–27).
Bouchekioua et al (28) and
Nairismägi et al (29) using
western blot analysis on the expression level of p-STAT3 expression
in NK/T cell lines determined that p-STAT3 have a low/no expression
in normal NK cells. However, p-STAT3 has been revealed to be
overexpressed in tumor cell lines, as well as in most EN-NK/T-NT
primary tumors (16,17). In the present study, NanoString
results revealed that the STAT pathway was activated in 70% of
EN-NK/T-NT cases, IHC results revealed that p-STAT3 was highly
expressed in 72.4% of EN-NK/T-NT samples, and western blot analysis
results revealed that p-STAT3 was activated in NK/T cell lines.
Therefore, JAK3/STAT3 was activated in the majority of the
EN-NK/T-NT cases, demonstrating its important role.
It is well known that the JAK3/STAT3 pathway can be
activated by two main mechanisms in EN-NK/T-NT, either by
Epstein-Barr virus infection or by driver gene mutation (12,17,30,31).
Recently, next generation sequencing revealed that the frequencies
of JAK3 (A572V, A573V) mutations were variable and relatively low
(17,30,32–34).
Sanger sequencing of the cases in the present study did not reveal
these single nucleotide variants (data not shown), which was
consistent with a recent study (35). By contrast, the STAT3 mutation rate
was high and stable. In 4 published sequencing data, the mutation
frequencies of the STAT3 gene was 12% (3/25), 26% (9/34), 10.5%
(11/105) and 8% (2/25) in patients and/or cell lines, respectively
(17,30,32,33).
Notably, all STAT3 mutations occurred at the SH2 domain. Therefore,
frequent mutations in this domain can be presumed to be
gain-of-oncogenic-function type mutations. In the present study,
the SH2 domains of STAT3 in cell lines and primary tumors were
sequenced. The STAT3 mutation was detected in 18.92% of EN-NK/T-NT
cases, with an increased p-STAT3 expression also identified in
those cases, demonstrating that the activation of the JAK3/STAT3
pathway may be partly caused by STAT3 mutations. Among the 6 types
of missense mutations, T708P in STAT3 had not been previously
reported. Notably, EN-NK/T-NT patients with STAT mutations had a
considerably poorer OS. Therefore, STAT3 mutation is a prognostic
marker for EN-NK/T-NT patients, but p-STAT3 is not.
Certain studies have indicated that STAT3 could
mediate the upregulation of PRDM1, which in return inhibits STAT3
phosphorylation in B cells (36).
ChIP-seq data have also confirmed that STAT3 has the potential to
bind to multiple areas within PRDM1 (37). However, in the present study, the
JAK3/STAT3 pathway was activated in both the PRDM1(+) and PRDM1(−)
groups, although it was more marked in the PRDM1(+) group. The NKL
cell line with PRDM1 deletion exhibited a high p-STAT3 expression,
with the NK92 cell line with PRDM1 deletion exhibiting no p-STAT3
expression. In the present cohort, a high expression of p-STAT3 was
detected in the majority of the EN-NK/T-NT cases, but there were
only a few cases with a high expression of PRDM1. These results
indicated that PRDM1 inactivation and JAK3/STAT3 pathway activation
could be independent events in the process of EN-NK/T-NT
pathogenesis and progression, despite their possible influence on
each other at the regulation level.
Stattic (38), a
STAT3 inhibitor, inhibited cell viability and induced apoptosis in
STAT3-mutant YT and NKL cells. However, Stattic had a low toxicity
in NK92 cells with wild-type STAT3 and did not affect apoptosis.
Similarly, Stattic was effective against STAT3-mutant NTCL cells
but less effective in STAT3 wild-type NTCL with a high p-STAT3
expression (30). At present, JAK
inhibitors are expected to be effective in interrupting this
signaling pathway in cells with a STAT3 expression (39). However, it is unclear whether the
inhibitor may effectively inhibit this pathway in STAT3-mutant
cells. Therefore, STAT3 mutant YT, NKL cells and WT NK92 cells were
treated with tofacitinib, a pan-JAK inhibitor that has been
revealed to have a functional specificity for JAK1/3 over JAK2 in
cell assays (40). In consistency
with the results of a previous study by Ando et al (41), tofacitinib was found to induce G1 CC
arrest and inhibit cell growth in NK92 and STAT3-mutant cells.
These data indicated that JAK1/3 inhibitors may have a therapeutic
effect on EN-NK/T-NT patients with STAT3 mutations. Furthermore,
tofacitinib has been approved by the US Food and Drug
Administration for myeloproliferative disorders, and is therefore
available for trials on patients with EN-NK/T-NT, while Stattic has
still not been made clinically available (42). However, to facitinib blocks JAK3
activity in EN-NK/T-NT, both in vitro and in vivo,
and its clinical usage in cancer therapy has been limited to
pan-JAK inhibition activity (29).
Further development of small-molecule inhibitors targeting STAT3
may synergize with highly selective JAK3 inhibitors to improve the
outcome of these malignant diseases.
In conclusion, the activation of the JAK3/STAT3
pathway and inactivation of PRDM1 were two common events in
EN-NK/T-NT observed in the samples of the present study. Notably,
these two factors could stratify the clinicopathologic features of
EN-NK/T-NT. In practice, PRDM1 immunostaining and STAT3 gene
mutation could serve as potential biomarkers for prognosis in
patients with EN-NK/T-NT. In addition, the JAK3/STAT3 pathway
inhibitor could be a new measure to deal with this highly
aggressive malignancy.
Acknowledgements
Not applicable.
Funding
The present study was supported by a research grant
from the National Nature Sciences Foundation of China (grant no.
81470359; Beijing, China).
Availability of data and materials
The datasets used during the present study are
available from the corresponding author upon reasonable
request.
Authors' contributions
TL and BZ conceived and designed the experiments.
JL, LL and DL performed the experiments. LL and JL performed the
data acquisition. LN, YZ and SH analyzed and interpreted the data.
TL and JL wrote the paper. LN and LL were responsible for
manuscript revision/review. All authors read and approved the
manuscript and agree to be accountable for all aspects of the
research in ensuring that the accuracy or integrity of any part of
the work are appropriately investigated and resolved.
Ethics approval and consent to
participate
This study was approved by the Institute Research
Medical Ethics Committee of the ethical standards of the Medical
Ethics Committee of Peking University First Hospital (approval no.
2013[571]). A written informed consent was obtained from the
patients at the time of admission, with which the tissue, blood and
other samples might be used for scientific research but did not
relate to patient's privacy.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
NKTCL
|
NK/T-cell lymphoma
|
|
EN-NK/T-NT
|
extranodal NK/T-cell lymphoma, nasal
type
|
|
STAT3
|
signal transducer and activator of
transcription 3
|
|
p-STAT3
|
phospho-STAT3 (Tyr705)
|
|
PRDM1
|
positive regulatory domain containing
I
|
|
FFPE
|
formalin-fixed paraffin-embedded
|
|
CC
|
cell cycle
|
|
OS
|
overall survival
|
|
WT
|
wild-type
|
|
mut
|
mutated
|
References
|
1
|
Lee J, Kim WS, Park YH, Park SH, Park KW,
Kang JH, Lee SS, Lee SI, Lee SH, Kim K, et al: Nasal-type NK/T cell
lymphoma: Clinical features and treatment outcome. Br J Cancer.
92:1226–1230. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Tse E and Kwong YL: The diagnosis and
management of NK/T-cell lymphomas. J Hematol Oncol. 10:852017.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Tse E and Kwong YL: How I treat NK/T-cell
lymphomas. Blood. 121:4997–5005. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Makita S and Tobinai K: Clinical features
and current optimal management of natural killer/T-cell lymphoma.
Hematol Oncol Clin North Am. 31:239–253. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Iqbal J, Kucuk C, Deleeuw RJ, Srivastava
G, Tam W, Geng H, Klinkebiel D, Christman JK, Patel K, Cao K, et
al: Genomic analyses reveal global functional alterations that
promote tumor growth and novel tumor suppressor genes in natural
killer-cell malignancies. Leukemia. 23:1139–1151. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Karube K, Nakagawa M, Tsuzuki S, Takeuchi
I, Honma K, Nakashima Y, Shimizu N, Ko YH, Morishima Y, Ohshima K,
et al: Identification of FOXO3 and PRDM1 as tumor-suppressor gene
candidates in NK-cell neoplasms by genomic and functional analyses.
Blood. 118:3195–3204. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Küçük C, Iqbal J, Hu X, Gaulard P, De
Leval L, Srivastava G, Au WY, McKeithan TW and Chan WC: PRDM1 is a
tumor suppressor gene in natural killer cell malignancies. Proc
Natl Acad Sci USA. 108:20119–20124. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Huang Y, de Leval L and Gaulard P:
Molecular underpinning of extranodal NK/T-cell lymphoma. Best Pract
Res Clin Haematol. 26:57–74. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Liang L, Nong L, Zhang S, Zhao J, Ti H,
Dong Y, Zhang B and Li T: The downregulation of PRDM1/Blimp-1 is
associated with aberrant expression of miR-223 in extranodal
NK/T-cell lymphoma, nasal type. J Exp Clin Cancer Res. 33:72014.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Liang L, Zhang Z, Wang Y, Nong L, Zheng Y,
Qu L, Zhang B and Li T: The genetic deletion of 6q21 and PRDM1 and
clinical implications in extranodal NK/T cell lymphoma, nasal type.
Biomed Res Int. 2015:4354232015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhang Z, Liang L, Li D, Nong L, Liu J, Qu
L, Zheng Y, Zhang B and Li T: Hypermethylation of PRDM1/Blimp-1
promoter in extranodal NK/T-cell lymphoma, nasal type: An evidence
of predominant role in its downregulation. Hematol Oncol.
35:645–654. 2017. View
Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lee S, Park HY, Kang SY, Kim SJ, Hwang J,
Lee S, Kwak SH, Park KS, Yoo HY, Kim WS, et al: Genetic alterations
of JAK/STAT cascade and histone modification in extranodal
NK/T-cell lymphoma nasal type. Oncotarget. 6:17764–17776.
2015.PubMed/NCBI
|
|
13
|
Ng SB, Selvarajan V, Huang G, Zhou J,
Feldman AL, Law M, Kwong YL, Shimizu N, Kagami Y, Aozasa K, et al:
Activated oncogenic pathways and therapeutic targets in extranodal
nasal-type NK/T cell lymphoma revealed by gene expression
profiling. J Pathol. 223:496–510. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wang Y, Shen Y, Wang S, Shen Q and Zhou X:
The role of STAT3 in leading the crosstalk between human cancers
and the immune system. Cancer Lett. 415:117–128. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chen YW, Guo T, Shen L, Wong KY, Tao Q,
Choi WW, Au-Yeung RK, Chan YP, Wong ML, Tang JC, et al:
Receptor-type tyrosine-protein phosphatase κ directly targets STAT3
activation for tumor suppression in nasal NK/T-cell lymphoma.
Blood. 125:1589–1600. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Coppo P, Gouilleux-Gruart V, Huang Y,
Bouhlal H, Bouamar H, Bouchet S, Perrot C, Vieillard V, Dartigues
P, Gaulard P, et al: STAT3 transcription factor is constitutively
activated and is oncogenic in nasal-type NK/T-cell lymphoma.
Leukemia. 23:1667–1678. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Küçük C, Jiang B, Hu X, Zhang W, Chan JK,
Xiao W, Lack N, Alkan C, Williams JC, Avery KN, et al: Activating
mutations of STAT5B and STAT3 in lymphomas derived from γδ-T or NK
cells. Nat Commun. 6:60252015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Cazzola M: Introduction to a review
series: The 2016 revision of the WHO classification of tumors of
hematopoietic and lymphoid tissues. Blood. 127:2361–2364. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Robertson MJ, Cochran KJ, Cameron C, Le
JM, Tantravahi R and Ritz J: Characterization of a cell line, NKL,
derived from an aggressive human natural killer cell leukemia. Exp
Hematol. 24:406–415. 1996.PubMed/NCBI
|
|
20
|
Gong JH, Maki G and Klingemann HG:
Characterization of a human cell line (NK-92) with phenotypical and
functional characteristics of activated natural killer cells.
Leukemia. 8:652–658. 1994.PubMed/NCBI
|
|
21
|
Yodoi J, Teshigawara K, Nikaido T, Fukui
K, Noma T, Honjo T, Takigawa M, Sasaki M, Minato N, Tsudo M, et al:
TCGF (IL-2)-receptor inducing factor(s). I. Regulation of IL 2
receptor on a natural killer-like cell line (YT cells). J Immunol.
134:1623–1630. 1985.PubMed/NCBI
|
|
22
|
Garcia JF, Roncador G, Garcia JF, Sánz AI,
Maestre L, Lucas E, Montes-Moreno S, Fernandez Victoria R,
Martinez-Torrecuadrara JL, Marafioti T, et al: PRDM1/BLIMP-1
expression in multiple B and T-cell lymphoma. Haematologica.
91:467–474. 2006.PubMed/NCBI
|
|
23
|
Nie K, Gomez M, Landgraf P, Garcia JF, Liu
Y, Tan LH, Chadburn A, Tuschl T, Knowles DM and Tam W:
MicroRNA-mediated down-regulation of PRDM1/Blimp-1 in
Hodgkin/Reed-Sternberg cells: A potential pathogenetic lesion in
Hodgkin lymphomas. Am J Pathol. 173:242–252. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Nie K, Zhang T, Allawi H, Gomez M, Liu Y,
Chadburn A, Wang YL, Knowles DM and Tam W: Epigenetic
down-regulation of the tumor suppressor gene PRDM1/Blimp-1 in
diffuse large B cell lymphomas: A potential role of the microRNA
let-7. Am J Pathol. 177:1470–1479. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yuan J, Zhang F and Niu R: Multiple
regulation pathways and pivotal biological functions of STAT3 in
cancer. Sci Rep. 5:176632015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wu ZL, Song YQ, Shi YF and Zhu J: High
nuclear expression of STAT3 is associated with unfavorable
prognosis in diffuse large B-cell lymphoma. J Hematol Oncol.
4:312014.
|
|
27
|
Khanna P, Chua PJ, Bay BH and Baeg GH: The
JAK/STAT signaling cascade in gastric carcinoma (Review). Int J
Oncol. 47:1617–1626. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Bouchekioua A, Scourzic L, de Wever O,
Zhang Y, Cervera P, Aline-Fardin A, Mercher T, Gaulard P, Nyga R,
Jeziorowska D, et al: JAK3 deregulation by activating mutations
confers invasive growth advantage in extranodal nasal-type natural
killer cell lymphoma. Leukemia. 28:338–348. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Nairismägi M, Gerritsen ME, Li ZM, Wijaya
GC, Chia BKH, Laurensia Y, Lim JQ, Yeoh KW, Yao XS, Pang WL, et al:
Oncogenic activation of JAK3-STAT signaling confers clinical
sensitivity to PRN371, a novel selective and potent JAK3 inhibitor,
in natural killer/T-cell lymphoma. Leukemia. 32:1147–1156. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Sim SH, Kim S, Kim TM, Jeon YK, Nam SJ,
Ahn YO, Keam B, Park HH, Kim DW, Kim CW and Heo DS: Novel
JAK3-activating mutations in extranodal NK/T-cell lymphoma, nasal
type. Am J Pathol. 187:980–986. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Xu Y, Shi Y, Yuan Q, Liu X, Yan B, Chen L,
Tao Y and Cao Y: Epstein-Barr virus encoded LMP1 regulates cyclin
D1 promoter activity by nuclear EGFR and STAT3 in CNE1 cells. J Exp
Clin Cancer Res. 32:902013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Dobashi A, Tsuyama N, Asaka R, Togashi Y,
Ueda K, Sakata S, Baba S, Sakamoto K, Hatake K and Takeuchi K:
Frequent BCOR aberrations in extranodal NK/T-Cell lymphoma, nasal
type. Genes Chromosomes Cancer. 55:460–471. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Jiang L, Gu ZH, Yan ZX, Zhao X, Xie YY,
Zhang ZG, Pan CM, Hu Y, Cai CP, Dong Y, et al: Exome sequencing
identifies somatic mutations of DDX3X in natural killer/T-cell
lymphoma. Nat Genet. 47:1061–1066. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Koo GC, Tan SY, Tang T, Poon SL, Allen GE,
Tan L, Chong SC, Ong WS, Tay K, Tao M, et al: Janus kinase
3-activating mutations identified in natural killer/T-cell
lymphoma. Cancer Discov. 2:591–597. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Kimura H, Karube K, Ito Y, Hirano K,
Suzuki M, Iwata S and Seto M: Rare occurrence of JAK3 mutations in
natural killer cell neoplasms in Japan. Leuk Lymphoma. 55:962–963.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Ryu JG, Lee J, Kim EK, Seo HB, Park JS,
Lee SY, Moon YM, Yoo SH, Park YW, Park SH, et al: Treatment of
IL-21R-Fc control autoimmune arthritis via suppression of STAT3
signal pathway mediated regulation of the Th17/Treg balance and
plasma B cells. Immunol Lett. 163:143–150. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Barnes NA, Stephenson S, Cocco M, Tooze RM
and Doody GM: BLIMP-1 and STAT3 counterregulate microRNA-21 during
plasma cell differentiation. J Immunol. 189:253–260. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Heidelberger S, Zinzalla G, Antonow D,
Essex S, Basu BP, Palmer J, Husby J, Jackson PJ, Rahman KM,
Wilderspin AF, et al: Investigation of the protein alkylation sites
of the STAT3:STAT3 inhibitor Stattic by mass spectrometry. Bioorg
Med Chem Lett. 23:4719–4722. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Quintás-Cardama A and Verstovsek S:
Molecular pathways: Jak/STAT pathway: Mutations, inhibitors, and
resistance. Clin Cancer Res. 19:1933–1940. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Banerjee S, Biehl A, Gadina M, Hasni S and
Schwartz DM: JAK-STAT signaling as a target for inflammatory and
autoimmune diseases: Current and future prospects. Drugs.
77:521–546. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Ando S, Kawada JI, Watanabe T, Suzuki M,
Sato Y, Torii Y, Asai M, Goshima F, Murata T, Shimizu N, et al:
Tofacitinib induces G1 cell-cycle arrest and inhibits tumor growth
in Epstein-Barr virus-associated T and natural killer cell lymphoma
cells. Oncotarget. 7:76793–76805. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Vainchenker W, Leroy E, Gilles L, Marty C,
Plo I and Constantinescu SN: JAK inhibitors for the treatment of
myeloproliferative neoplasms and other disorders. F1000Res.
7:822018. View Article : Google Scholar : PubMed/NCBI
|