Introduction
Ovarian cancer, one of the most common malignant
tumors of the female reproductive tract, has the third highest
incidence rate and the highest mortality rate (3%) among other
female malignant reproductive cancers (1). The 5-year survival rate of advanced
stage disease (FIGOII–IV) accounts for 30–44% (2). However, although the 5-year survival
rate is ~93% among patients who have early stage ovarian cancer
(FIGO I), these patients account for <15% of all ovarian cancer
patients (3). Therefore, an early
diagnosis of ovarian cancer is critical in improving the prognosis
of patients and reducing mortality.
Epigenetic modifications are closely related to
oncogene activation, tumor suppressor gene inactivation, DNA damage
repair defects and cancer stem cell differentiation, among which
DNA methylation and histone acetylation are the two most widely
used methods in diagnosing cancers (4). The study of epigenetic mechanisms of
tumors is helpful during clinical diagnosis and preventive
treatment of tumors (5). DNA
methylation which is the transfer of methyl groups to the 5th
carbon atom of cytosine producing 5-methylcytosine under the
catalysis of DNA methyltransferases, is an important epigenetic
mechanism. Abnormal hypermethylation of the DNA promoter region is
a mechanism underlying the inactivation of tumor suppressor genes
and it plays an important role in the development of a cancer
(6,7). 5-Aza-2′-deoxycitydine (5-aza-dc) is
presently recognized as a common demethylation drug and is commonly
used in clinical treatment of hematological diseases and lung
cancer. However, its high price and great side effects have
generated research focusing on finding a demethylation substitute
to 5-aza-dc. In addition to methylation, acetylation is another
important post-transcriptional regulation of histones. In general,
histone acetylation is related to the opening of chromatin and
helps promote gene transcription (8). Histone acetylation has been revealed
to be closely related to the proliferation, differentiation and
progression of tumor cells (9). The
regulation of histone acetylation is a reversible dynamic process
that relies on both histone acetyltransferases (HATs) and histone
deacetylases (HDACs) that regulate the conformation of the
chromatin structure and promote or suppress gene transcription
(10).
Ginsenosides, a type of steroid compound, mainly
distributed in medical materials of the Panax species, are
considered as active ingredients in ginseng and have long been used
as a traditional Chinese medicine (11,12).
As one of the most important ginsenoside monomers, ginsenoside Rg3
induces G2 phase cell cycle arrest, therefore inhibiting the
synthesis of proteins and ATPs in the pre-mitotic phase, slowing
down the proliferation and growth of cancer cells (13), and promoting tumor cell apoptosis
(14), as well as inhibiting cancer
cell infiltration and metastasis (15,16).
The aforementioned functions have all been confirmed in ovarian
cancer cells (17,18). However, the effect of ginsenoside
Rg3 on epigenetic modification in ovarian cancer still remains
unclear.
Therefore, the present study was conducted to
investigate the role of ginsenoside Rg3 on epigenetic modification
of ovarian cancer cells, providing a molecular basis for novel
diagnosis and treatment strategies of ovarian cancer.
Materials and methods
Drugs
Ginsenoside Rg3 (CAS no. 14197-60-5) with a
molecular weight of 785.02 kDa
(C42H72O13) was purchased from
Chengdu Mansite Biotechnology Co., Ltd. (Cdmust; Chengdu, China) in
July 2016. The HPLC purity of was ≥98%. It was diluted by culture
media and prepared to be used in experiments. Cisplatin (BP809) was
purchased from Sigma-Aldrich; Merck KGaA (Darmstadt, Germany).
Cell culture and treatment
Human normal ovarian epithelial cells HOSEpiC (cat.
no. BNCC340096; Bena Culture Collection, Beijing, China) and human
ovarian cancer SKOV3 cells (HTB-77) were purchased from the
American Type Culture Collection, (ATCC; Manassas, VA, USA). Cells
were cultured in Dulbecco's modified Eagle's medium (DMEM; high
glucose) (HyClone; GE Healthcare Life Sciences, Logan, UT, USA)
containing 10% (v/v) fetal bovine serum (FBS) (Gemini Bio-Products,
West Sacramento, CA, USA), 100 U/ml streptomycin and 100 µg/ml
penicillin) (Biological Industries, Beit Haemek, Israel) in an
incubator with 5% CO2 at 37°C. HOSEpiC cells were
treated with different concentrations of cisplatin (0, 5, 10, 20,
40 and 80 µg/ml) and ginsenoside Rg3 (0, 100, 200, 400, 800 and
1,600 µg/ml). SKOV3 cells were treated with different
concentrations of cisplatin (0, 2, 4, 8, 16 and 32 µg/ml) and
ginsenoside Rg3 (0, 25, 50, 100, 200 and 500 µg/ml).
Cell viability assay
A Cell Counting Kit-8 (CCK-8; Beyotime Institute of
Biotechnology, Haimen, China) assay was carried out to determine
cell viabilities. Briefly, cells (5×103/well) treated
with different concentrations of cisplatin and ginsenoside Rg3 were
inoculated in 96-well plates. After having been incubated for 12,
24 and 48 h, the cells were stained with 20 µl staining reagent for
1 h. The optical density (OD) values at 450 nm were read using a
MultiSkan 1500 microplate reader (Thermo Fisher Scientific, Inc.,
Waltham, MA, USA).
Cell apoptosis assay
Annexin V/propidium iodide (PI) double-staining
assay (Roche Diagnostics, Basel, Switzerland) was performed to
assess cell apoptosis rates. Briefly, SKOV3 cells were first
treated with different concentrations of ginsenoside Rg3 (0, 50,
100 and 200 µg/ml) for 48 h and were then stained with 5 µl Annexin
V and 5 µl PI for 5 min in the dark at 37°C. The analysis was
performed using BD CellQuest™ Pro Software (BD Biosciences,
Franklin Lakes, NJ, USA) without delay.
Cell metastasis ability assay
Wound healing assay was applied to determine cell
metastatic abilities. To be more specific, 1×105 SKOV3
cells were treated with different concentrations of ginsenoside Rg3
(0, 50, 100 and 200 µg/ml). Next, the cells were inoculated in each
well of 12-well plates and scratched gently to form a cell-free
area and then cultured in an incubator for 12 and 24 h at 37°C.
Finally, the diameters of cell-free areas were assessed under an
Olympus DSX100 optical microscope (Olympus Corp., Tokyo,
Japan).
Cell invasion ability assay
Cell invasion abilities of ovarian cancer cells
treated with ginsenoside Rg3 were assessed using 24-well Transwell
chambers that contained 8-µm pore filters (Corning Inc., Corning,
NY, USA). In brief, 5×104 SKOV3 cells treated with
different concentrations of ginsenoside Rg3 (0, 50, 100 and 200
µg/ml) were cultured in DMEM culture media in Matrigel GFR-coated
(BD Biosciences) upper chambers of the Transwell. In addition, DMEM
culture media containing 10% FBS was filled into the lower
chambers. After having been incubated for 48 h, the bottom membrane
was stained with 0.1% crystal violet for 30 min at 37°C. The number
of invasive cells was calculated using Olympus DSX100 optical
microscope (Olympus Corp.) with a magnification of ×200.
Methylation assay
The methylation degrees of tumor inhibitors p53, p16
and hMLH1 in SKOV3 ovarian cancer cells, which were treated with
different concentrations of ginsenoside Rg3 (0, 50, 100 and 200
µg/ml), were assessed using methylation specific PCR (MSP). SKOV3
(1.5×104) cells were treated with ginsenoside Rg3 (0,
50, 100 and 200 µg/ml) for 48 h. The methylation was detected by EZ
DNA Methylation-Startup kit (Zymo Research Corp., Irvine, CA, USA)
for bisulfite conversion. Next, the converted DNA (30 ng) was
subjected for MSP amplification. The promoter regions of p53, p16
and human mutL homolog-1 (hMLH1) were respectively identified using
specific methylated (M) and unmethylated (U) allele-specific
primers and observed with MethPrimer 2.0 (Chinese Academy of
Medical Sciences, Beijing, China). The amplification process was
conducted as follows: Pre-denaturation at 95°C for 5 min; 35 (M) or
40 (U) cycles of denaturation at 95°C for 40 sec, and annealing at
65°C (M) or 56°C (U) for 45 sec; and a final extension at 72°C for
5 min.
Enzyme activity assays
The activity of DNMT was determined by EpiQuik DNMT
Activity/Inhibition Assay Ultra Kit (Colorimetric) (Epigentec Group
Inc., Farmingdale, NY, USA) in ovarian cancer cells treated with
ginsenoside Rg3 (0, 50, 100 and 200 µg/ml) or 50 µg/ml 5-aza-dc (as
a negative control). The samples were first co-incubated with
cytimidine substrate and then with 5′-methyl-cytimidine antibody.
The activity of HDAC was determined using Epigenase HDAC
Activity/Inhibition Direct Assay Kit (Colorimetric) (Epigentec
Group Inc.) in ovarian cancer cells treated with ginsenoside Rg3
(0, 50, 100 and 200 µg/ml) or 500 ng/ml HDAC inhibitor trichostatin
A (TSA) (Selleck Chemicals, Houston, TX, USA) (as a negative
control). The samples were co-incubated with acetylated substrate
and then with photographic developer. The OD values at 450 nm were
measured using a MultiSkan 1500 microplate reader (Thermo Fisher
Scientific, Inc.).
Real-time quantitative polymerase
chain reaction (RT-qPCR)
The mRNA expression levels of methylation related
factors were detected by RT-qPCR in SKOV3 ovarian cancer cells
treated with different concentrations of ginsenoside Rg3 (0, 50,
100 and 200 µg/ml) or 50 µg/ml 5-aza-dc (as a negative control).
Total RNA was extracted from different cells using TRIzol reagent
(Invitrogen; Thermo Fisher Scientific, Inc.), and
reversely-transcribed to cDNA using Transcriptase (Takara Bio,
Inc., Otsu, Japan). Next, the cDNA was amplified by
LightCycler® Multiplex Masters on a
LightCycler® 480 II System (both from Roche Diagnostics,
Indianapolis, IN, USA). The thermocycling conditions were: Initial
denaturation at 95°C for 5 min, 35 cycles (a denaturation at 95°C
for 25 sec, annealing at 56°C for 25 sec, an extension at 72°C for
35 sec) and a final extension at 72°C for 5 min. The primer
sequences of p53, p16, hMLH1, DNMT1, DNMT3a and DNMT3b are listed
in Table I.
 | Table I.The primer sequences used in the
present study. |
Table I.
The primer sequences used in the
present study.
| Name | Type | Sequence (5′-3′) |
|---|
| β-actin | F |
GTGGACATCCGCAAAGAC |
|
| R |
GAAAGGGTGTAACGCAACT |
| p53 | F |
GCCCCTCCTCAGCATCTTAT |
|
| R |
AAAGCTGTTCCGTCCCAGTA |
| p16 | F |
CAGGTCATGATGATGGGCAG |
|
| R |
GATGGCCCAGCTCCTCAG |
| hMLH1 | F |
CTACTTCCAGCAACCCCAGA |
|
| R |
AGAACCTCATGTCCCTGCTC |
| DNMT1 | F |
CCGACTACATCAAAGGCAGC |
|
| R |
AGGTTGATGTCTGCGTGGTA |
| DNMT3a | F |
GGGACCCCTACTACATCAGC |
|
| R |
CATTCTTGTCCCCAGCATCG |
| DNMT3b | F |
GGCCACCTTCAATAAGCTCG |
|
| R |
GTTGCGTGTTGTTGGGTTTG |
Western blotting
The protein expression levels of methylation and
acetylation-related factors were detected by performing western
blotting in SKOV3 ovarian cancer cells treated with different
concentrations of ginsenoside Rg3 (0, 50, 100 and 200 µg/ml), 50
µg/ml 5-aza-dc (as a negative control) or 500 ng/ml HDAC inhibitor
TSA (as a positive control). Cells were lysed by RIPA lysis buffer
(Wuhan Boster Biological Technology, Ltd., Wuhan, China) for 20 min
and centrifuged on ice at 12,000 × g for 10 min. The supernatant
with proteins was first quantified using a BCA protein assay kit
(Pierce; Thermo Fisher Scientific, Inc.) and then subjected to 15%
sodium dodecyl sulfate-polyacrylamide gel electrophoresis
(SDS-PAGE), for protein separation. Next, the separated proteins
were transferred to polyvinylidene fluoride (PVDF) membranes
(Thermo Fisher Scientific, Inc.). The membranes were blocked using
5% non-fat dry milk at 37°C for 1 h, and were first probed with
specific primary antibodies overnight at 4°C and then with
secondary antibody for 1 h at 37°C. The immunoblots were visualized
using enhanced chemiluminescense (ECL) detection reagents (Pierce;
Thermo Fisher Scientific, Inc.) and analyzed by Bio-Rad ChemiDoc
XRS densitometry with Image Lab™ Software version 6.0.1 (Bio-Rad
Laboratories, Inc., Hercules, CA, USA). The primary antibodies were
purchased from Cell Signaling Technology, Inc. (Danvers, MA, USA):
Rabbit anti-p53 (dilution 1:1,000; cat. no. 2527), p16 (dilution
1:1,000; cat. no. 80772), hMLH1 (dilution 1:1,000; cat. no. 4256),
DNMT1 (dilution 1:1,000; cat. no. 5032), DNMT3a (dilution 1:1,000;
cat. no. 32578), DNMT3b (dilution 1:1,000; cat. no. 57868),
acetyl-H3 K14 (dilution 1:1,000; cat. no. 7627), acetyl-H3 K9
(dilution 1:1,000; cat. no. 9649), acetyl-H4 K12 (dilution 1:1,000;
cat. no. 13944), acetyl-H4 K5 (dilution 1:1,000; cat. no. 8647),
acetyl-H4 K16 (dilution 1:1,000; cat. no. 13534) and β-actin
(dilution 1:1,000; cat. no. 4970). The secondary antibodies were
anti-rabbit IgG and HRP-linked antibody (dilution 1:5,000; cat. no.
7074; Cell Signaling Technology, Inc.).
Statistical analysis
The statistical analysis was carried out using SPSS
19.0 (IBM Corp., Armonk, NY, USA). The data were obtained from at
least three repeated experiments. The significance of difference
was analysed by Dunnett's post hoc test. P<0.05 or P<0.01 was
considered to indicate a statistically significant difference.
Results
Ginsenoside Rg3 inhibits cell
proliferation and promotes apoptosis of ovarian cancer cells
The cell proliferation abilities using CCK-8 assay
were detected in order to investigate the effect of ginsenoside Rg3
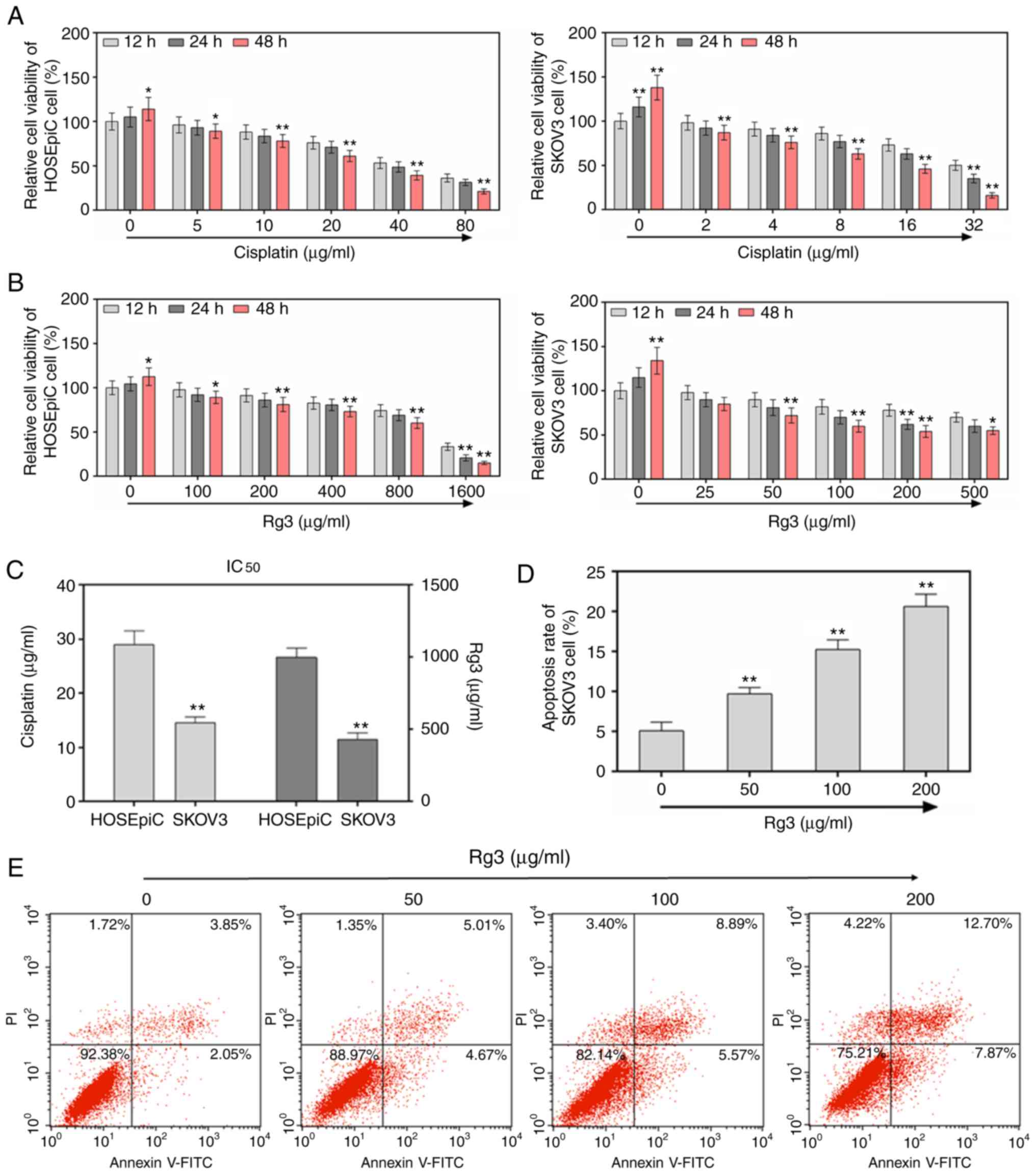
on SKOV3 ovarian cancer cell proliferation. First, the cytotoxic
effects of classical anticancer drugs cisplatin (Fig. 1A) and Rg3 (Fig. 1B) were investigated in both normal
HOSEpiC and ovarian cancer cells SKOV3. It was revealed that Rg3
had almost no cytotoxicity to HoSEpiC and SKOV3 cells with an
IC50 of 1,000 and 400 µg/ml, respectively, compared to
cisplatin (IC50 of HOSEpiC, 29 µg/ml; IC50 of
SKOV3, 14 µg/ml) (Fig. 1C).
Therefore, the concentrations of Rg3 used in subsequent experiments
were significant to the study. The results of the present
experiment revealed that cell proliferation abilities were
inhibited by ginsenoside Rg3 treatment in dose-dependent (0, 25,
50, 100 and 200 µg/ml) and time-dependent (12, 24 and 48 h)
manners. It was revealed that cell viability was significantly
decreased when the cells were incubated with different
concentrations of ginsenoside Rg3 for 48 h, compared to cells being
incubated for 12 h (P<0.01, Fig.
1B). When the concentration of ginsenoside was 500 µg/ml
(>200 µg/ml), the inhibition rate was revealed to decrease
again. Thus, 50, 100 and 200 µg/ml ginsenoside Rg3 were selected to
treat cells in subsequent experiments. The effect of ginsenoside
Rg3 on cell apoptosis of SKOV3 ovarian cancer cells was then
detected by carrying out Annexin V/PI assay. The cell apoptosis
rates were revealed to be significantly promoted by ginsenoside Rg3
treatment (50, 100 and 200 µg/ml) (P<0.01, Fig. 1D and E).
Ginsenoside Rg3 inhibits cell invasion
and metastatic abilities of ovarian cancer cells
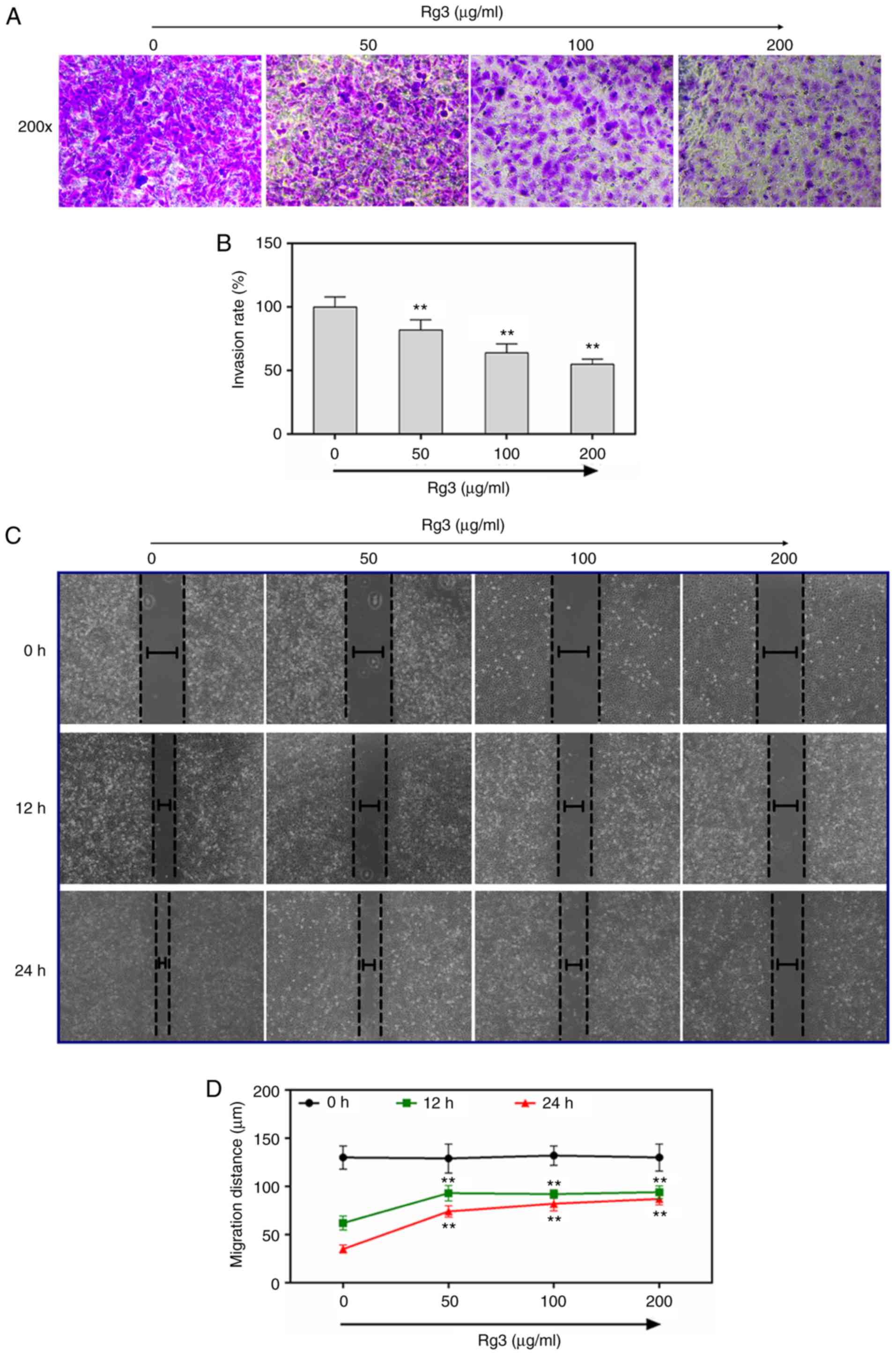
The effect of ginsenoside Rg3 on cell invasion and
metastatic abilities of SKOV3 ovarian cancer cells was determined
respectively by performing Transwell and wound healing assays. The
Transwell images with 200-fold amplification demonstrated that the
invasion rates of SKOV3 ovarian cancer cells significantly
decreased as concentration of ginsenoside Rg3 increased from 50, to
100 and to 200 µg/ml (P<0.01, Fig.
2A and B). The wound healing images revealed that the
metastatic rates of SKOV3 ovarian cancer cells also significantly
decreased as concentration of ginsenoside Rg3 increased from 50, to
100 and to 200 µg/ml, both at 12 and 24 h. (P<0.01, Fig. 2C and D).
Ginsenoside Rg3 inhibits methylation
levels in ovarian cancer cells
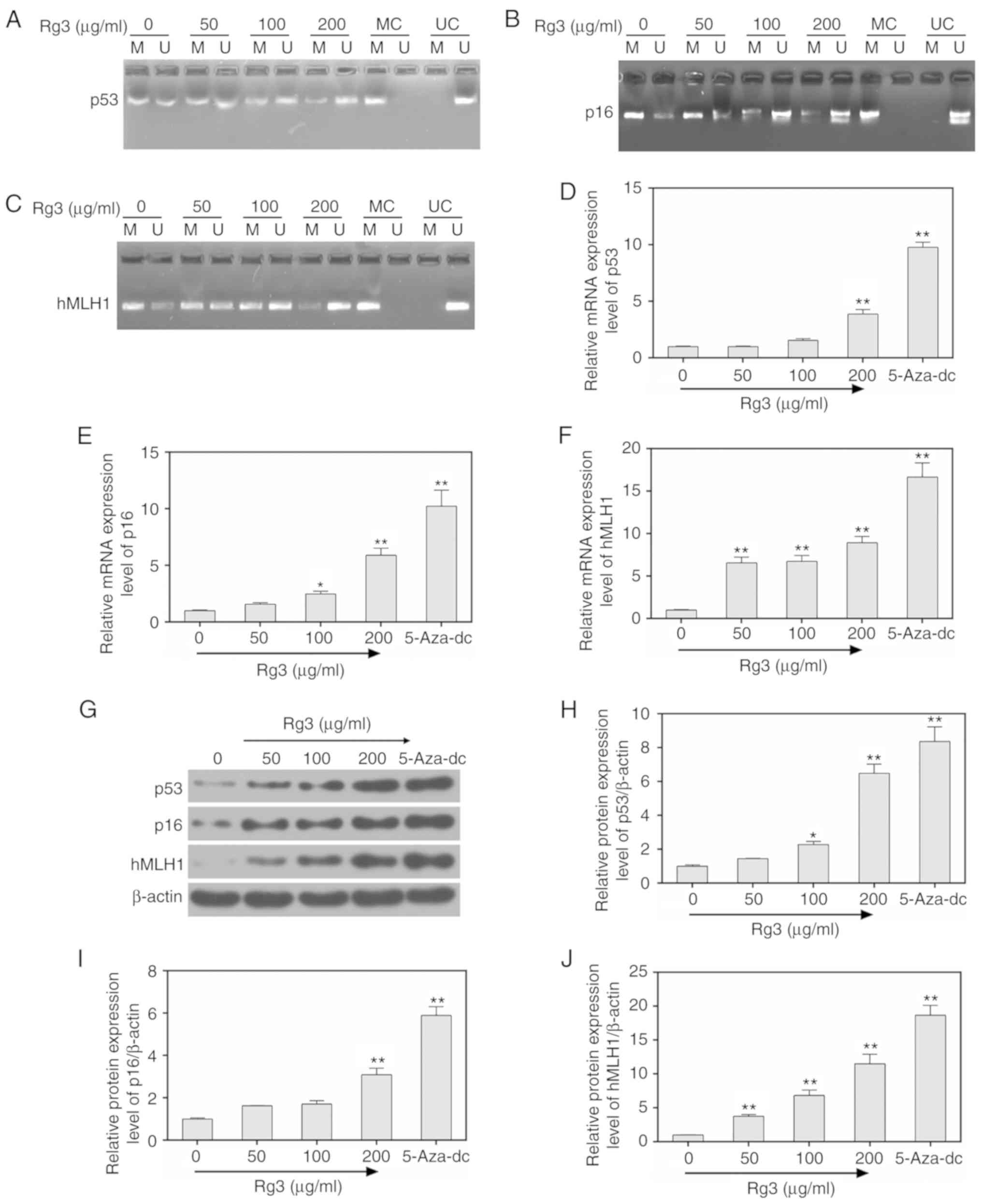
By performing MSP detection, the methylation levels
of p53, p16 and hMLH1 were revealed to be significantly decreased
by ginsenoside Rg3 treatment (0, 50, 100 and 200 µg/ml), while the
un-methylation levels were significantly increased (Fig. 3A-C). In addition, the mRNA and
protein levels of p53, p16 and hMLH1 were respectively assessed by
carrying out RT-qPCR and western blot assays. The data demonstrated
that the expression of p53, p16 and hMLH1 were significantly
increased by ginsenoside Rg3 treatment, compared to those in the
control group (cells not treated with ginsenoside Rg3) (P<0.05,
Fig. 3D-J). In addition, treating
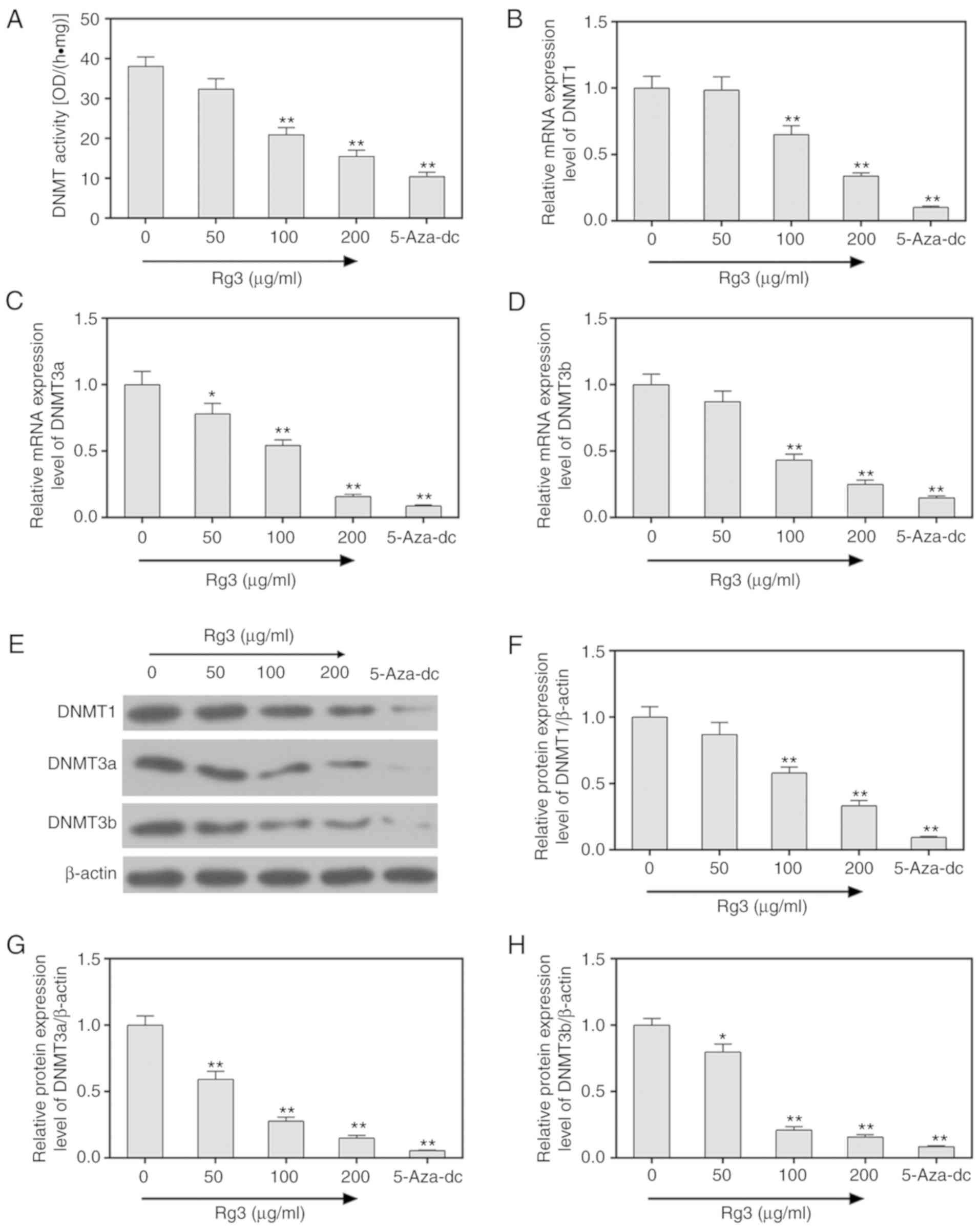
5-aza-dc as a positive control, the activity of DNA
methyltransferases (DNMTs) was assessed using EpiQuik DNMT Activity
Assay Kit. The results revealed that ginsenoside Rg3 significantly
decreased the DNMT activities in SKOV3 ovarian cancer cells
(P<0.05, Fig. 4A). The mRNA and
protein levels of DNMT1, DNMT3a and DNMT3b were also determined by
performing RT-qPCR and western blot assays, and it was revealed
that the expression of DNMT1, DNMT3a and DNMT3b were significantly
decreased by ginsenoside Rg3 (P<0.05, Fig. 4B-H).
Ginsenoside Rg3 promotes acetylation
levels in ovarian cancer cells
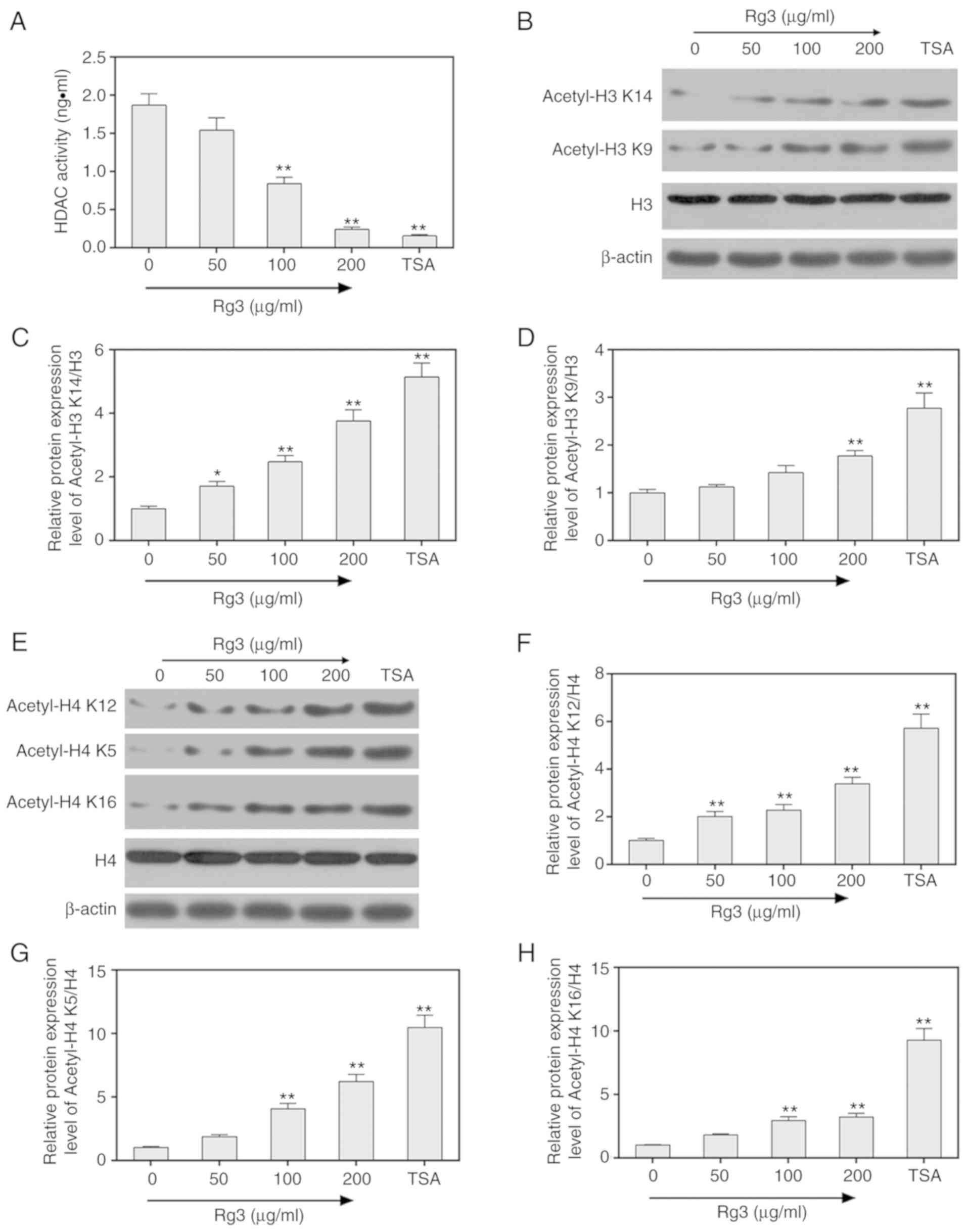
The activity of deacetylases HDAC was detected by
chemical colorimetry, and the results revealed that HDAC activity
was markedly decreased by ginsenoside Rg3 treatment (0, 50, 100 and
200 µg/ml) in SKOV3 ovarian cancer cells (P<0.05, Fig. 5A). TSA was treated as a positive
control. Subsequently, the protein levels of acetylated H3 K14 and
K9 were assessed by western blot assays. The results revealed that
acetylated H3 K14 and K9 were significantly increased when SKOV3
ovarian cells were treated with 0, 50, 100 and 200 µg/ml
ginsenoside Rg3 (P<0.05, Fig.
5B-D). The protein levels of acetylated H4 K12, K5 and K16 were
significantly increased when SKOV3 ovarian cells were treated with
0, 50, 100 and 200 µg/ml ginsenoside Rg3 (P<0.05, Fig. 5E-H).
Discussion
Despite surgery and chemotherapy that have been
applied in the treatment of ovarian cancer, the overall survival
rate among ovarian cancer patients has not significantly improved
(19) and the incidence and
mortality of cancer is still increasing (20). Thus, the diagnosis and treatment of
ovarian cancer is still a research hotspot.
As a ginsenoside monomer, ginsenoside Rg3 has been
reported to be helpful for ovarian cancer treatment. In this
research, we studied whether the effect of ginsenoside Rg3 on
ovarian cancer cells was related to the molecular mechanism of
methylation and histone acetylation. It was first ascertained that
ginsenoside Rg3 had almost no obvious cytotoxicity effect, and then
it was revealed that Rg3 inhibited cell proliferation, invasion and
metastasis and promoted cell apoptosis in a dose-dependent manner
and the effects of Rg3 on migration/invasion were not completely
due to the effects on cell viability. 5-Aza-dc is a commonly used
demethylation drug in clinical treatment of many cancers. However,
5-aza-dc also produces severe side effects. In the present study
the de-methylation function of ginsenoside Rg3 with 5-aza-dc was
compared in ovarian cancer cells.
As two tumor suppressor genes, p53 and p16
(alternatively named multiple tumor suppressor 1 (MTS1) or
cyclin-dependent kinase inhibitor 2A (CDKN2A) are highly efficient
transcription factors that not only bind to 300 promoter elements
in the genome, but also extensively alter gene expression patterns
(21). Normal p53 and p16 proteins
act as cell cycle checkpoints and participate in the process of DNA
repair and apoptosis (22). The
activities of p53 and p16 are regulated by post-translational
modifications, for example, methylation, acetylation or
phosphorylation (23,24). The inactivation of p53 or p16 is
considered to be one of the main features of malignant tumors
(25). MLH1 is responsible for DNA
mismatch repair, and the methylated hMLH1 has been frequently
identified in many cancers, for instance, colorectal and esophageal
cancer (26,27). In this study, an MSP assay was used
to detect the methylation levels in the promoter region of p53, p16
and hMLH1. It was revealed that the methylation levels of CpG
islands in p53, p16 and hMLH1 decreased, and that the levels of
p53, p16 and hMLH1 significantly increased when ovarian cancer
cells were treated with ginsenoside Rg3 or 5-aza-dc. This indicated
that ginsenoside Rg3 promoted tumor suppressor function of p53, p16
and hMLH1 in ovarian cancer cells by inhibiting
promoter-methylation and expression levels.
The enzymes, which catalyze the DNA methylation
reaction, mainly include DNMT1, DNMT3a and DNMT3b. DNMT1 is the
main enzyme that maintains the methylation reaction. After DNA
replication is completed, DNMT1 catalyzes the transfer of methyl
groups to newly synthesized DNA duplexes. DNMT3a and DNMT3b are
responsible for catalyzing the methylation formation (de
novo methylation) reaction on DNA duplex (28). In the present study, it was revealed
that ginsenoside Rg3 was able to inhibit the expression levels of
DNMT1, DNMT3a and DNMT3b. These findings indicated that ginsenoside
Rg3 could be used as a potential de-methylation drug in cancer
treatment.
Apart from promoter-methylation,
histone-deacetylation modification was also frequently found in
some cancers such as gastric cancer, colorectal cancer. The balance
of histone acetylation and deaetylation is an indispensable
condition for tumor development. HDACs are a family of proteases
that catalyze the hydrolysis of acetyl groups at the ends of lysine
residues in various substrates such as nucleosomal histones
(29,30). HDACs are the histone deacetylases
which can inhibit gene transcription and promote tumor
proliferation (31). TSA is a
commonly used HDAC inhibitor, and it can inhibit tumor cell
proliferation (32). The effect of
ginsenoside Rg3 on deacetylatiion in ovarian cancer cells was
investigated by detecting HDAC activity and the acetylation levels
of histone H3 and H4, and the results revealed that ginsenoside Rg3
inhibited HDAC activity and increased the levels of H3 K14, H3 K9,
H4 K12, H4 K5 and H4 K16. This indicated that ginsenoside Rg3 not
only participated in methylation modification, but also in
acetylation modification. Collectively, ginsenoside Rg3 functioned
as tumor suppressor in ovarian cancer cells. Notably, the antitumor
effect of ginsenoside Rg3 on other ovarian cancer cell lines should
be investigated and it is also essential to undertake animal
experiments to demonstrate the role of ginsenoside Rg3 in
vivo.
In conclusion, it was revealed that ginsenoside Rg3
inhibited ovarian cancer cell proliferation, metastasis and
invasion and promoted cell apoptosis by regulating methylation and
acetylation. Thus, ginsenoside Rg3 could be applied as a methylase
or histone deacetylase inhibitor to inhibit cell proliferation in
ovarian cancer treatment. The methylation levels of tumor
suppressor genes, or acetylation levels of histones may be
considered as novel markers in diagnosing ovarian cancer.
Acknowledgements
Not applicable.
Funding
The present study was supported by the 2018 Zhejiang
Chinese Medicine Foundation for Outstanding Young Talents (grant
no. 2018ZQ009).
Availability of data and materials
The analyzed data sets generated during the study
are available from the corresponding author on reasonable
request.
Authors' contributions
LZ, HS, substantially contributed to the conception
and design of the manuscript and drafted the article or critically
revised it for important intellectual content. LC, WG, CF and PZ
acquired, analyzed and interpreted the data. All authors read and
approved the manuscript and agree to be accountable for all aspects
of the research in ensuring that the accuracy or integrity of any
part of the work are appropriately investigated and resolved.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Henderson JT, Webber EM and Sawaya GF:
U.S. preventive services task force evidence syntheses formerly
systematic evidence reviews Screening for ovarian cancer: An
updated evidence review for the U.S. preventive services task
force. Agency for Healthcare Research and Quality (US). Report No.:
17-05231-EF-1. 2018.
|
|
2
|
Woolas RP, Xu FJ, Jacobs IJ, Yu YH, Daly
L, Berchuck A, Soper JT, Clarke-Pearson DL, Oram DH and Bast RC Jr:
Elevation of multiple serum markers in patients with stage I
ovarian cancer. J Natl Cancer Inst. 85:1748–1751. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Smith RA, Manassaram-Baptiste D, Brooks D,
Doroshenk M, Fedewa S, Saslow D, Brawley OW and Wender R: Cancer
screening in the United States, 2015: A review of current American
cancer society guidelines and current issues in cancer screening.
CA Cancer J Clin. 65:30–54. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Rothbart SB and Strahl BD: Interpreting
the language of histone and DNA modifications. Biochim Biophys
Acta. 1839:627–643. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Vinci MC, Polvani G and Pesce M:
Epigenetic programming and risk: The birthplace of cardiovascular
disease? Stem Cell Rev. 9:241–253. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Perlin E and Moquin RB: Serum DNA levels
in patients with malignant disease. Am J Clin Pathol. 58:601–602.
1972. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ibanez de Caceres I, Battagli C, Esteller
M, Herman JG, Dulaimi E, Edelson MI, Bergman C, Ehya H, Eisenberg
BL and Cairns P: Tumor cell-specific BRCA1 and
RASSF1A hypermethylation in serum, plasma, and peritoneal
fluid from ovarian cancer patients. Cancer Res. 64:6476–6481. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Gurard-Levin ZA and Almouzni G: Histone
modifications and a choice of variant: A language that helps the
genome express itself. F1000Prime Rep. 6:762014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hake SB, Xiao A and Allis CD: Linking the
epigenetic ‘language’ of covalent histone modifications to cancer.
Br J Cancer. 90:761–769. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Strahl BD and Allis CD: The language of
covalent histone modifications. Nature. 403:41–45. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Gillis CN: Panax ginseng pharmacology: A
nitric oxide link? Biochem Pharmacol. 54:1–8. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Liao B, Newmark H and Zhou R:
Neuroprotective effects of ginseng total saponin and ginsenosides
Rb1 and Rg1 on spinal cord neurons in vitro. Exp Neurol.
173:224–234. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Xu TM, Xin Y, Cui MH, Jiang X and Gu LP:
Inhibitory effect of ginsenoside Rg3 combined with cyclophosphamide
on growth and angiogenesis of ovarian cancer. Chin Med J.
120:584–588. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wang JH, Nao JF, Zhang M and He P:
20(s)-ginsenoside Rg3 promotes apoptosis in human ovarian cancer
HO-8910 cells through PI3K/Akt and XIAP pathways. Tumour Biol.
35:11985–11994. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Liu T, Zhao L, Zhang Y, Chen W, Liu D, Hou
H, Ding L and Li X: Ginsenoside 20(S)-Rg3 targets HIF-1alpha to
block hypoxia-induced epithelial-mesenchymal transition in ovarian
cancer cells. PLoS One. 9:e1038872014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Liu T, Zhao L, Hou H, Ding L, Chen W and
Li X: Ginsenoside 20(S)-Rg3 suppresses ovarian cancer migration via
hypoxia-inducible factor 1 alpha and nuclear factor-kappa B
signals. Tumour Biol. 39:10104283176922252017.PubMed/NCBI
|
|
17
|
Xu TM, Cui MH, Xin Y, Gu LP, Jiang X, Su
MM, Wang DD and Wang WJ: Inhibitory effect of ginsenoside Rg3 on
ovarian cancer metastasis. Chin Med J. 121:1394–1397. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zheng X, Chen W, Hou H Li J, Li H, Sun X,
Zhao L and Li X: Ginsenoside 20(S)-Rg3 induced autophagy to inhibit
migration and invasion of ovarian cancer. Biomed Pharmacother.
85:620–626. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Powell CB, Kenley E, Chen LM, Crawford B,
McLennan J, Zaloudek C, Komaromy M, Beattie M and Ziegler J:
Risk-reducing salpingo-oophorectomy in BRCA mutation
carriers: Role of serial sectioning in the detection of occult
malignancy. J Clin Oncol. 23:127–132. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lee Y, Miron A, Drapkin R, Nucci MR,
Medeiros F, Saleemuddin A, Garber J, Birch C, Mou H, Gordon RW, et
al: A candidate precursor to serous carcinoma that originates in
the distal fallopian tube. J Pathol. 211:26–35. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Sallum LF, Andrade L, Ramalho S, Ferracini
AC, de Andrade Natal R, Brito ABC, Sarian LO and Derchain S: WT1,
p53 and p16 expression in the diagnosis of low- and high-grade
serous ovarian carcinomas and their relation to prognosis.
Oncotarget. 9:15818–15827. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Heckl M, Schmoeckel E, Hertlein L,
Rottmann M, Jeschke U and Mayr D: The ARID1A, p53 and β-Catenin
statuses are strong prognosticators in clear cell and endometrioid
carcinoma of the ovary and the endometrium. PLoS One.
13:e01928812018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Marinas MC, Mogos DG, Simionescu CE,
Stepan A and Tanase F: The study of p53 and p16 immunoexpression in
serous borderline and malignant ovarian tumors. Rom J Morphol
Embryol. 53:1021–1025. 2012.PubMed/NCBI
|
|
24
|
Hu G, Li P, Li Y, Wang T, Gao X, Zhang W
and Jia G: Methylation levels of P16 and TP53 that are involved in
DNA strand breakage of 16HBE cells treated by hexavalent chromium.
Toxicol Lett. 249:15–21. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Geraldes C, Goncalves AC, Cortesao E,
Pereira MI, Roque A, Paiva A, Ribeiro L, Nascimento-Costa JM and
Sarmento-Ribeiro AB: Aberrant p15, p16, p53, and DAPK gene
methylation in myelomagenesis: Clinical and prognostic
implications. Clin Lymphoma, Myeloma Leuk. 16:713–720.e712. 2016.
View Article : Google Scholar
|
|
26
|
Morak M, Ibisler A, Keller G, Jessen E,
Laner A, Gonzales-Fassrainer D, Locher M, Massdorf T, Nissen AM,
Benet-Pagès A and Holinski-Feder E: Comprehensive analysis of the
MLH1 promoter region in 480 patients with colorectal cancer and
1150 controls reveals new variants including one with a heritable
constitutional MLH1 epimutation. J Med Genet. 55:240–248. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Li J, Ye D, Wang L, Peng Y, Li Q, Deng H
and Zhou C: Role of MLH1 methylation in esophageal cancer
carcinogenesis and its clinical significance. Onco Targets Ther.
11:651–663. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Nelissen EC, van Montfoort AP, Dumoulin JC
and Evers JL: Epigenetics and the placenta. Hum Reprod Update.
17:397–417. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Vancurova I, Gatla HR and Vancura A:
HDAC/IKK inhibition therapies in solid tumors. Oncotarget.
8:34030–34031. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Booth L, Roberts JL, Poklepovic A,
Kirkwood J and Dent P: HDAC inhibitors enhance the immunotherapy
response of melanoma cells. Oncotarget. 8:83155–83170. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Yano M, Yasuda M, Sakaki M, Nagata K,
Fujino T, Arai E, Hasebe T, Miyazawa M, Miyazawa M, Ogane N, et al:
Association of histone deacetylase expression with histology and
prognosis of ovarian cancer. Oncol Lett. 15:3524–3531.
2018.PubMed/NCBI
|
|
32
|
Gatla HR, Zou Y, Uddin MM, Singha B, Bu P,
Vancura A and Vancurova I: Histone deacetylase (HDAC) inhibition
induces ikappaB kinase (IKK)-dependent interleukin-8/CXCL8
expression in ovarian cancer cells. J Biol Chem. 292:5043–5054.
2017. View Article : Google Scholar : PubMed/NCBI
|