Introduction
Gastric carcinoma (GC) is one of the most common
types of malignancy and the second leading cause of
cancer-associated mortality worldwide (1,2).
Despite the advances in diagnosis and cancer therapy in the past
decade, the survival rate remains low (3). This may be attributed to a limited
understanding of the exact causes and mechanisms underlying GC.
Thus, it is an urgent to determine the potential molecular
mechanisms of GC and develop novel therapeutic strategies.
MicroRNAs (miRNAs/miRs) are endogenous noncoding
RNAs that are vital for cancer development by acting as tumor
suppressors or oncogenes (4–6). In
GC, numerous miRNAs exhibit aberrant expression and contribute to
the progression of GC and chemoresistance (7–10). For
example, miR-26a/b inhibits GC growth and angiogenesis by targeting
the hepatocyte growth factor-vascular endothelial growth factor
axis (11). In addition, miRNA-106a
induces multidrug resistance in GC by targeting runt related
transcription factor 3 (12).
Circulating miR-18a contributes to the detection and monitoring of
GC (13); however, the roles of
numerous miRNAs in GC remain unknown.
In the present study, miR-92a was analyzed. It has
been demonstrated that miR-92a is upregulated in GC (14); however, the role of miR-92a and its
associated mechanisms are unknown. The present study reported that
miR-92a is upregulated in GC tissues compared with in adjacent
tissues. Functional analyses revealed that suppression of miR-92a
significantly inhibited GC cell proliferation and induced
apoptosis. Further investigation suggested that inhibitor of growth
protein 2 (ING2) is a direct target of miR-92a. Additionally,
suppression of miR-92a sensitized GC cells to doxorubicin
treatment. Collectively, the results of the present study highlight
the oncogenic role of miR-92a in GC and may provide a potential
therapeutic basis for the treatment of GC.
Materials and methods
Clinical samples
A total of 10 GC tumor samples and their
corresponding adjacent tissues were collected from patients (45–60
years old) at The Third Affiliated Hospital of Harbin Medical
University form February 2015 to August 2017 (Harbin, China).
Informed consent was obtained from all patients and the present
study was approved by the Ethics Committee of Harbin Medical
University. All tissue samples were immediately frozen and
preserved in liquid nitrogen until further use.
Cell culture
GC cell lines, including AGS, SGC7901, MGC803 and
BGC823, and human gastric epithelial GES-1 cells were purchased
from the Cell Bank of Chinese Academy of Sciences (Shanghai,
China). Cells were cultured in Dulbecco's Modified Eagle's medium
(DMEM; Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA,
USA) with 10% fetal bovine serum (Invitrogen; Thermo Fisher
Scientific, Inc.), 50 U/ml penicillin and 50 µg/ml streptomycin
(Invitrogen; Thermo Fisher Scientific, Inc.). All cells were
maintained at 37°C in a humidified incubator at 5%
CO2.
Cell transfection
The sequences of the miR-92a mimics (cat. no.
miR10000092-1-5), mimics-negative control (NC; cat. no.
miR01101-1-5), miR-92a inhibitor (cat. no. miR20000092-1-5) and
inhibitor-NC (miR02201-1-5) were obtained from Guangzhou Ribobio
Co., Ltd. (Guangzhou, China). miR-92a mimics or inhibitor (20 nM)
was used for transfection with by Lipofectamine® 2000
(Invitrogen; Thermo Fisher Scientific, Inc.) with serum-free medium
according to experiments request. The sequences were as follows:
mimics-NC, 5′-UUUGUACUACACAAAAGUACUG-3′ and
3′-AAACAUGAUGUGUUUUCAUGAC-5′; miR-92a mimics,
5′-UAUUGCACUUGUCCCGGCCUGU-3′ and 3′-AUAACGUGAACAGGGCCGGACA-5′;
inhibitor-NC, 5′-CAGUACUUUUGUGUAGUACAAA-3′ and miR-92a inhibitor,
5′-ACAGGCCGGGACAAGUGCAAUA-3′. SGC7901 cells (8×103) were
seeded into 6 cm dishes 24 h prior to transfection. miR-92a,
inhibitor or inhibitor-NC were transfected using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) with serum-free medium according to the
conditions of the subsequent experiments. After 5 h, the cell
medium was replaced with complete medium. The cell lysates were
harvested 48 h post-transfection.
RNA extraction and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
Cells and tissues were lysed and total RNA was
extracted using TRIzol reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocols and
transcribed into cDNA using Superscriptase II (Invitrogen; Thermo
Fisher Scientific Inc.). qPCR was performed using Power SYBR Green
PCR master Mix (Thermo Fisher Scientific, Inc.). The following
primer sets were used for ING2 detection. ING2,
forward 5′-GCAGCAACTGTACTCGTCG-3′, reverse,
5′-GACTCCACGCACTCAAGGTA-3′; and β-actin, forward
5′-CATGTACGTTGCTATCCAGGC-3′, reverse,
5′-CTCCTTAATGTCACGCACGAT-3′.
miR-92a expression was measured using the TaqMan
MicroRNA assay kit (Thermo Fisher Scientific, Inc.). In brief, 1 µg
of RNA was reverse transcribed into cDNA using an miR-92a specific
stem-loop primer at 25°C for 10 min, 37°C for 120 min, 85°C for 5
min, and 4°C for 5 min. qPCR with miR-92a specific primers and
Taqman probes was performed on an ABI7500 real-time PCR system
(Applied Biosystems; Thermo Fisher Scientific, Inc.), over 35
cycles of 95°C, 30 sec at 60°C and 72°C for 35 sec. The data were
analyzed using 2−ΔΔCq method (15). The sequence of miR-92a and U6 were
as follows: miR-92a, forward, 5′-GCTGAGTATTGCACTTGTCCCG-3′,
reverse, 5′-GTGTCGTGGAGTCGGCAA-3′ and U6, forward
5′-CTCGCTTCGGCAGCACA-3′ and reverse,
5′-AACGCTTCACGAATTTGCGT-3′.
Acridine orange/ethidium bromide
(AO/EB) fluorescence staining
SGC7901 cells were transfected with miR-92a
inhibitor or inhibitor-NC. Cells (5×103) were incubated
with AO/EB mixing solution for 5 min (Beijing Solarbio Science and
Technology Co., Ltd., Beijing, China) at room temperature. A total
of eight fields were randomly analyzed to examine the cellular
morphology alterations under a fluorescence microscope
(magnification, ×200). The percentage of apoptotic cells was
calculated by the following formula: Apoptotic rate (%) = number of
apoptotic cells/number of all cells counted.
Dual-luciferase reporter assay
Wild-type ING2 (ING2-WT) and mutant
ING2 (ING2-Mut) 3′-untranslated region (UTR) sequences were
separately inserted into the SpeI and HindIII sites
of pMIR-REPORT Luciferase vectors (Ambion; Thermo Fisher
Scientific, Inc.). SGC7901 cells were seeded in 6-well plates and
transfected with the specific vectors using Lipofectamine 2000 for
48 h. Luciferase activity was assessed using the Dual
Luciferase-reporter 1000 assay system immediately after
transfection (Promega Corporation, Madison, WI, USA).
Renilla activity was used for normalization.
Antibodies and western blotting
Cells were lysed with radioimmunoprecipitation assay
lysis buffer containing a protease inhibitor cocktail (Roche
Diagnostics, Basel, Switzerland). Protein concentration was
measured using a BCA Protein assay kit (Beyotime Institute of
Biotechnology, Shanghai, China). Equal amounts of protein (40 µg)
were separated by 12.5% SDS-PAGE and transferred to a
polyvinylidene difluoride transfer membrane (Thermo Fisher
Scientific, Inc.). Following blocking with 5% skim milk for 2 h at
room temperature, the blots were probed with primary antibodies
against β-actin (cat. no. 3700, dilution: 1:20,00), B-cell lymphoma
2(Bcl-2)-associated X (Bax; cat. no. 2772, dilution: 1:1,000),
Bcl-2-associated death promoter (Bad; cat. no. 9292, dilution:
1:500), p53 (cat. no. 2524, dilution: 1:500) and cleaved caspase-3
(cat. no. 9953, dilution: 1:1,000) (Cell Signaling Technology,
Inc., Dallas, TX, USA), proliferating cell nuclear antigen (PCNA)
(cat. no. sc-25280, dilution: 1:1,000), cyclin dependent kinase
(CDK)4 (Cat. sc-70832, dilution: 1:1,000) and CDK6 (cat. no.
sc-7961, dilution: 1:1,000) (Santa Cruz Biotechnology, Inc.,
Dallas, TX, USA) and ING2 (cat. no. ab109504, dilution: 1:1,000)
(Abcam, Cambridge, UK). Following washing with PBST and incubating
with rabbit or mouse secondary antibodies (Cell Signaling
Technology, Inc.), the blots were visualized by an enhanced
chemiluminescence reagent (GE Healthcare, Chicago, IL, USA). Three
individual experiments were performed.
Cell Counting Kit-8 (CCK-8) viability
assay
SGC7901 cells transfected with miR-92a inhibitor or
inhibitor-NC were seeded into 96-well plates with a density of
5×103 per well and cultured for 4 days. Cell viability
was assessed with a CCK-8 kit (Dojindo Molecular Technologies,
Inc., Kumamoto, Japan) at days 0, 2 and 4 at 450 nm with a
microplate reader (Thermo Fisher Scientific, Inc.). To assess the
cell viability upon doxorubicin treatment, the cells transfected
with miR-92a inhibitor or inhibitor-NC were seeded into 96-well
plates with a density of 5×103 per well and treated with
various concentrations 5, 10, 20, 50, 100, 200, 300 and 500 nM) of
doxorubicin for 24 h. Cell viability was measured at 450 nm with a
microplate reader.
Colony formation assay
A total of 8×102 SGC7901 cells
transfected with miR-92a inhibitor or inhibitor-NC were counted and
seeded into 6 cm dishes. After 10 days of culturing, the colonies
were stained with 0.1% crystal violet in 20% methanol for 15 min at
room temperature. The samples were imaged and the numbers of
visible colonies were counted.
Ki-67and γH2AX immunofluorescence
staining
SGC7901 cells were seeded on cover slips and
transfected with miR-92a inhibitor or inhibitor-NC. After 48 h, the
cells were fixed with 4% PFA at room temperature for 10 min and
then incubated with Ki-67 antibody (cat. no. 9129, Cell Signaling
Technology, Inc.) or γH2AX (cat. no. 7631, Cell Signaling
Technology, Inc.) for 1 h and then incubated with Alexa-488
conjugated anti-rabbit IgG (cat. no. 4412, Cell Signaling
Technology, Inc.) at room temperature for 20 min. H2AX expression
was analyzed following treatment with 80 nM doxorubicin for 24 h at
37°C. The cells were then counterstained with 1 µg/ml DAPI at room
temperature for 10 min to stain the cell nuclei. All cover slips
were mounted using Prolong Diamond Antifade Mountant (Applied
Biosystems; Thermo Fisher Scientific, Inc.). Nine random fields per
slips were captured for analysis using a fluorescence microscope
(magnification, ×200).
Target gene prediction
Potential miRNA-target gene interactions were
predicted using www.Targetscan.org; release 7.2) and microrna.org.
Establishment of doxorubicin-resistant
SGC7901 cell lines
The SGC7901 cell line was continuously exposed to
increasing doses of doxorubicin (5, 10 and 15 µM) at 37°C in a
humidified incubator at 5% CO2 (Aladdin Biochemical
Technology Co. Shanghai, China) for ~9 weeks. The starting dose was
5 µM and this was increased to 10 µM after 4 weeks, to 15 µM after
another 4 weeks and continued at 15 µM for the last 4 weeks. The
established resistant SGC7901 cell line was then maintained in DMEM
medium with 10% (v/v) FBS and 10 µM doxorubicin.
Statistical analysis
Data were obtained from at least 3 independent
experiments and are presented as the mean ± standard deviation. For
the clinical tissue test, the data was evaluated by a paired
Student's t test, and data from the other experiments were
evaluated by unpaired Student's t-test or analysis of variance
followed by a Tukey's post-hoc test to compare multiple groups.
P<0.05 was considered to indicate a statistically significant
difference. Statistical values were calculated using SPSS 19.0
software (IBM Corp., Armonk, NY, USA) and illustrated using the
GraphPad Prism 5.0 (GraphPad Software, Inc., La Jolla, CA,
USA).
Results
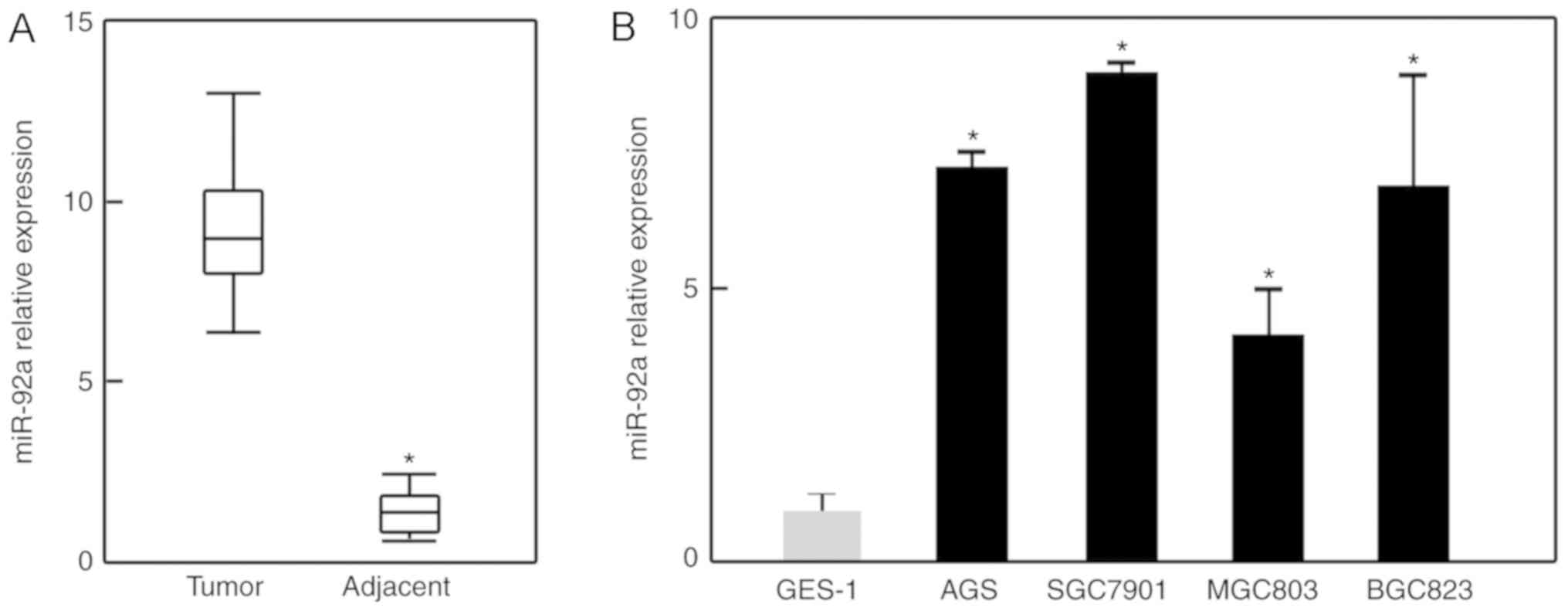
miR-92a is overexpressed in GC
To determine the expression of miR-92a in GC, 10 GC
tissues and their corresponding adjacent tissues were collected.
Via RT-qPCR, miR-92a was observed to be upregulated by ~6.1-fold in
GC tissues than in adjacent tissues (Fig. 1A). In addition, the expression of
miR-92a was analyzed in several GC cell lines. As presented in
Fig. 1B, the expression levels of
miR-92a were significantly increased in all GC cell lines compared
with in human gastric epithelial GES-1 cells. These observations
suggest a potential role of miR-92a in GC. As SGC7901 cells
expressed the highest levels of miR-92a, further experiments were
conducted using this cell line.
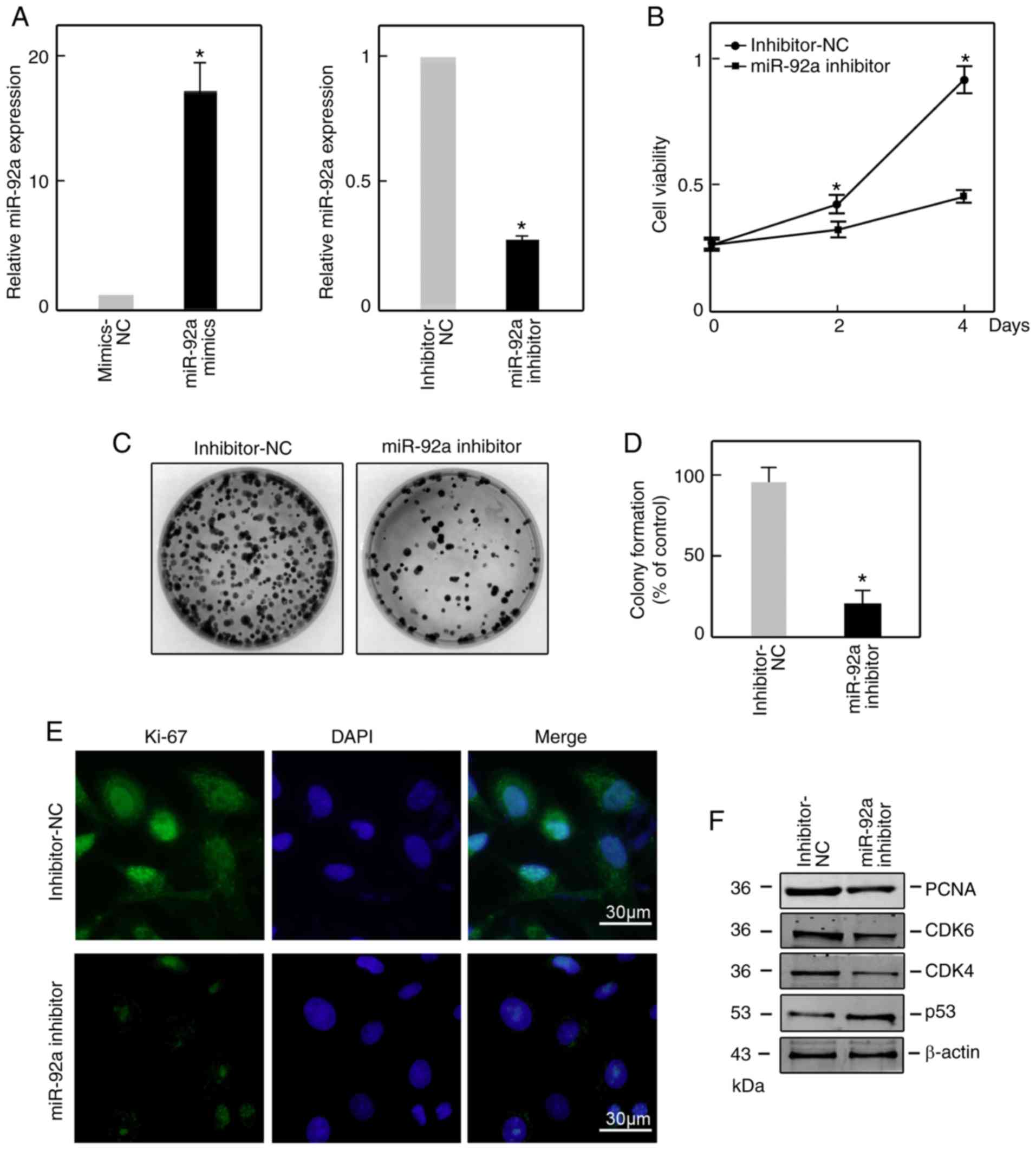
miR-92a contributes to the
proliferative ability of SGC7901 cells
To investigate the function of miR-92a in GC,
SGC7901 cells were transiently transfected with an miR-92a
inhibitor or mimics to suppress or upregulate the expression of
miR-92a, respectively. The suppression and overexpression
efficiencies were confirmed by RT-qPCR at 48 h post-transfection
(Fig. 2A). A CCK-8 assay was
performed to analyze viable cells at day 0, 2 and 4
post-transfection. The data revealed that suppression of miR-92a
significantly suppressed cell viability compared with in the
inhibitor-NC-transfected group by a CCK-8 assay (Fig. 2B). The colony formation assay
revealed significantly fewer visible colonies upon miR-92a
knockdown (Fig. 2C and D).
Furthermore, Ki-67 immunofluorescence staining exhibited notably
lower signals in miR-92a inhibitor-transfected SGC7901 cells
compared with in inhibitor-NC-transfected cells (Fig. 2E). Consistently, western blotting
demonstrated that the protein expression levels of PCNA, CDK4 and
CDK6 were notably reduced, while p53 was upregulated (Fig. 2F). All these results indicate that
miR-92a contributes to the maintenance of proliferation in SGC7901
cells.
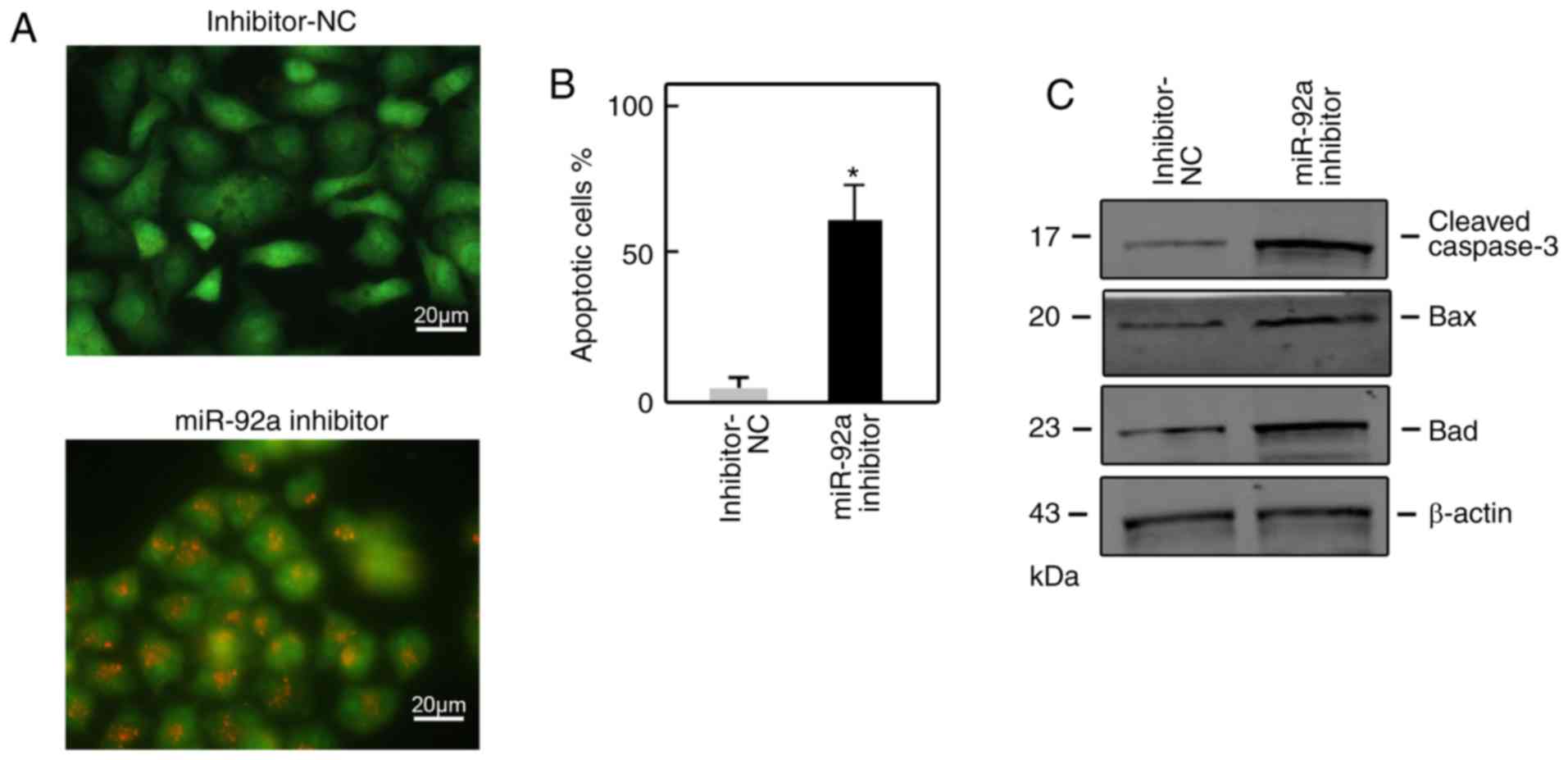
Suppression of miR-92a induces the
apoptosis of SGC7901 cells
To investigate the role of miR-92a in cell
apoptosis, we subjected miR-92a inhibitor-transfected SGC7901 cells
to AO/EB staining. The results revealed that >50% of apoptotic
cells were counted upon miR-92a inhibitor transfection; however,
<5% of apoptotic cells were counted upon inhibitor-NC
transfection (Fig. 3A and B). In
addition, proapoptotic proteins, including Bax, Bad and cleaved
caspase-3, were determined to be increased via the suppression of
miR-92a compared with in control cells (Fig. 3C).
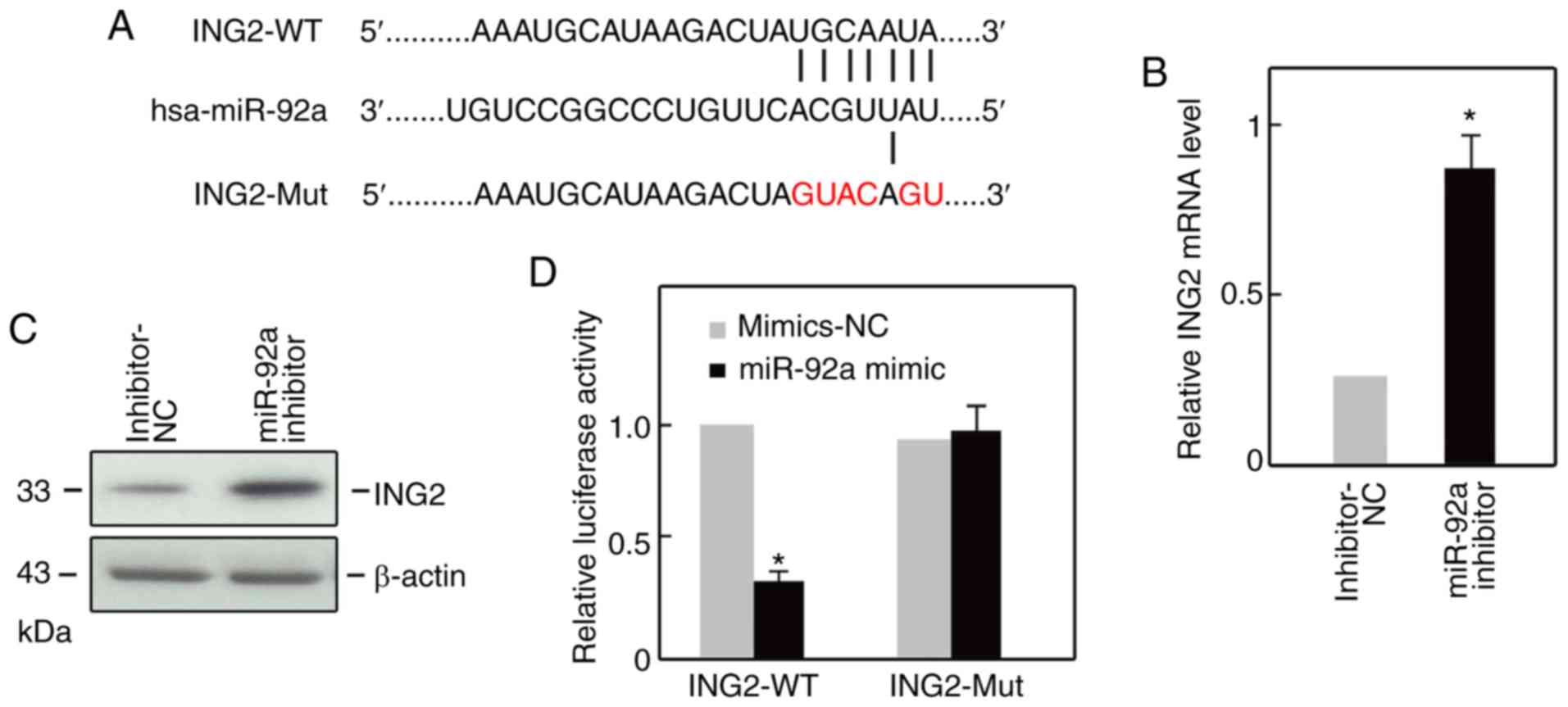
ING2 is a direct target of
miR-92a
Conventionally, miRNAs suppress gene expression by
binding to the 3′-UTR sequence of target mRNA (16). By employing bioinformatics databases
(TargetScan, microrna.org), we observed that
ING2 was a potential target of miR-92a (Fig. 4A). Additionally, ING2 expression at
the mRNA and protein levels was notably increased following miR-92
inhibitor transfection compared with the control (Fig. 4B and C). To investigate whether ING2
was a direct target of miR-92a, a luciferase reporter construct
containing the ING2-WT or ING2-Mut of ING2. Co-transfection
of the miR-92a mimic and ING2-WT resulted in increased luciferase
activity (Fig. 4D); however,
miR-92a inhibitor did not markedly alter the luciferase activities
in the ING2-Mut group (Fig. 4D).
Thus, these findings support the result that ING2 is a direct
target of miR-92a.
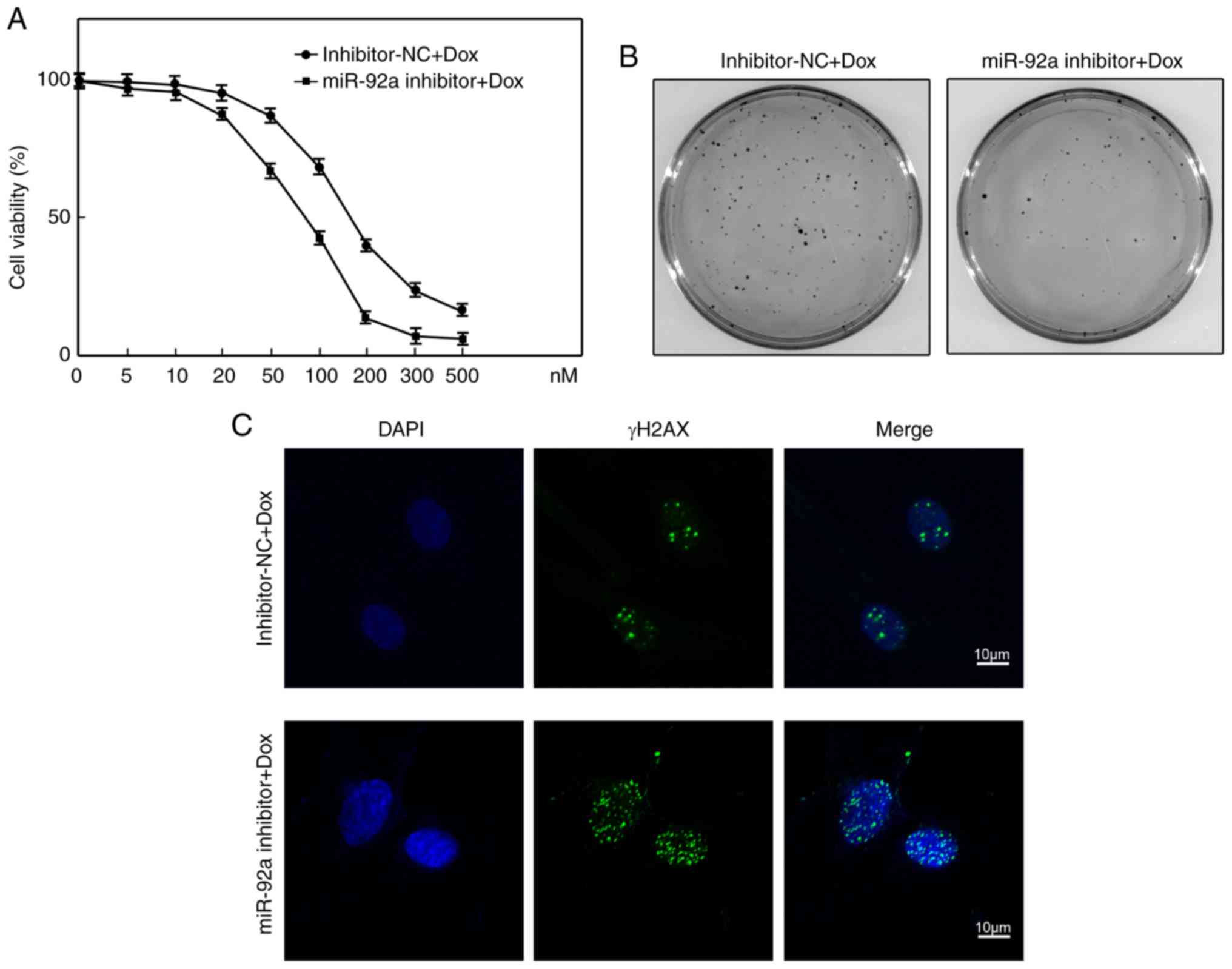
Suppression of miR-92a sensitizes
SGC7901 cells to doxorubicin treatment
It was proposed that suppression of miR-92a led to
reduced proliferation and the induction of apoptosis in SGC7901
cells via upregulation of ING2; thus, whether suppression of
miR-92a sensitizes SGC7901 cells to doxorubicin treatment was
investigated. Compared with in inhibitor-NC-transfected cells,
miR-92a inhibitor-transfected cells were more sensitive to
doxorubicin-induced growth inhibition as determined by the CCK-8
assay (Fig. 5A). According to
Fig. 5A, the half-maximal
inhibitory concentration (IC50) of inhibitor-NC
transfected SGC7901 cells was ~147.6 nM; however, a notably lower
IC50 (~82.1 nM) was calculated in SGC7901 cells
following knockdown of miR-92a. The colony formation assay also
demonstrated that suppression of miR-92a markedly reduced SGC7901
cell survival upon doxorubicin treatment, compared with control
cells (Fig. 5B). The cytotoxic
effect of doxorubicin is known to occur via the induction of DNA
damage (16). Bioinformatics
analysis revealed that ING2 was a potential target of
miR-92a (Fig. 4A). Additionally, we
reported that ING2 at the mRNA and protein levels was increased by
miR-92 inhibitor transfection (Fig. 4B
and C). To validate whether ING2 was a direct target of
miR-92a, a luciferase reporter construct containing ING2-WT or
ING2-Mut of the ING2 was generated. Co-transfection of the
miR-92a mimics and ING2-WT resulted in increased luciferase
activity (Fig. 4D). However, the
miR-92a inhibitor did not change the luciferase activities in the
ING2-Mut group (Fig. 4D). Thus,
these findings support ING2 as a direct target of miR-92a. Thus,
DNA damage foci formation was examined by γH2AX immunofluorescence
staining. We observed that miR-92a downregulation was associated
with numerous DNA damage foci than in control cells (Fig. 5C). These data suggest that
suppression of miR-92a sensitizes SGC7901 cells to doxorubicin
treatment.
Discussion
GC poses great challenge in clinical therapy and is
associated with poor prognosis (17); however, increasing evidence has
demonstrated that numerous genes are involved in GC (18,19).
Therefore, understanding the potential molecular mechanisms of GC
oncogenesis is required.
miRNAs have been predicted to regulate >60% of
human protein-coding genes (20).
Numerous studies have reported that miRNAs serve key roles in GC
via downregulation of a variety of genes (21,22).
Mir-92a has been identified to be involved in the regulation of the
cell cycle and cell signaling, which is critical for cancer
progression (23,24). miRNA-92a functions as an oncogene in
colorectal cancer by targeting phosphatase and tensin homolog
(25). miR-92a also promotes lymph
node metastasis of human esophageal squamous cell carcinoma via
E-cadherin (26). A high level of
circulating miR-92a expression correlates with poor prognosis in
patients with non-small cell lung cancer (27). A study using miRNA-locked nucleic
acid in situ hybridization reported that miR-92a is
upregulated expressed in GC (14).
Our study aimed to investigate the potential role of miR-92a in GC.
Consistent with previous studies, we observed that miR-92a is
upregulated in GC tissues compared with adjacent tissues by
RT-qPCR. Suppression of miR-92a inhibited cell proliferation and
induced cell apoptosis in SGC7901 cells. High expression of miR-92a
is associated with poor prognosis and shorter overall survival of
patients with GC (14). The results
of the present study provides a key role for miR-92a in GC
tumorigenesis, and targeting miR-92a may have potential for
clinical application. Furthermore, previous studies revealed that
higher expression of miR-92a is closely associated with poorer
outcome of initial chemotherapy (28); upregulated tumor miR-92a expression
is associated with decreased survival in GC patients (28). Similarly, our results indicated that
knockdown of miR-92a promoted GC cells to doxorubicin-induced cell
death. The present study proposed a key role for miR-92a in GC
tumorigenesis, and targeting miR-92a may have potential for
clinical application.
ING2 is a member of the ING family and is
characterized as a type-II tumor suppressor gene (29). ING2 has important functions in cell
apoptosis, cell cycle arrest, cell senescence, chromatin
modification and the DNA damage response (30–34).
ING2 expression has been reported to be reduced in various
types of cancer (35). ING2 can be
sumoylated by small ubiquitin-related modifier 1 (Sin3a) on lysine
195; this sumoylation is required for the interaction between ING2
and SIN3 transcription regulator family member A to mediate the
expression of gens downstream p53 (36). Additionally, ING2 also binds to Ski
novel via its PHD domain to suppress cell proliferation (37). The present study reported ING2 as a
direct target of miR-92a; knockdown of miR-92a increased ING2
expression, as well as p53 expression. These finding suggests that
the oncogenic effects of miR-92a in GC occur at least partially via
the suppression of ING2 expression.
ING2 is also involved in the DNA damage response
(34). In response to doxorubicin
treatment, ING2 stabilizes the mSin3a-histone deacetylase 1 complex
at the promoter regions of proliferation-associated genes by its
PHD domain and further inhibits the expression of these genes
(33). As ING2 was determined to be
a direct target of miR-92a, the effects of miR-92a following
doxorubicin treatment were investigated. It was reported that
suppression of miR-92a induced more DNA damage foci and sensitized
SGC7901 cells to doxorubicin treatment.
Collectively, the present stud provided novel
insight into the biological function of miR-92a in GC and its
potential molecular mechanism. Our findings indicate that miR-92a
contributed to cell proliferation, apoptosis and doxorubicin
chemosensitivity in GC by inhibiting ING2 expression. Targeting
miR-92a may be a potential therapeutic strategy to enhance the
effects of doxorubicin in GC.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
XCT, XYZ, SBS and DQW made substantial contributions
to conception and design, and acquisition of data, or analysis and
interpretation of data. All authors were involved in drafting the
manuscript or revising it critically for important intellectual
content, and have given final approval of the version to be
published.
Ethics approval and consent to
participate
The present stud was approved by the Ethics
Committee of Harbin Medical University (Harbin, China) and informed
consent was obtained from all patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Yuasa Y: Control of gut differentiation
and intestinal-type gastric carcinogenesis. Nat Rev Cancer.
3:592–600. 2003. View
Article : Google Scholar : PubMed/NCBI
|
|
2
|
Panani AD: Cytogenetic and molecular
aspects of gastric cancer: Clinical implications. Cancer Lett.
266:99–115. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Coburn N, Cosby R, Klein L, Knight G,
Malthaner R, Mamazza J, Mercer CD and Ringash J: Staging and
surgical approaches in gastric cancer: A systematic review. Cancer
Treat Rev. 63:104–115. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Luo X, Yang B and Nattel S: MicroRNAs and
atrial fibrillation: Mechanisms and translational potential. Nat
Rev Cardiol. 12:80–90. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Frampton AE, Castellano L, Colombo T,
Giovannetti E, Krell J, Jacob J, Pellegrino L, Roca-Alonso L, Funel
N, Gall TM, et al: MicroRNAs cooperatively inhibit a network of
tumor suppressor genes to promote pancreatic tumor growth and
progression. Gastroenterology. 146:268–277. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Emmrich S, Katsman-Kuipers JE, Henke K,
Khatib ME, Jammal R, Engeland F, Dasci F, Zwaan CM, den Boer ML,
Verboon L, et al: miR-9 is a tumor suppressor in pediatric AML with
t(8;21). Leukemia. 28:1022–1032. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhang HH, Gu GL, Zhang XY, Li FZ, Ding L,
Fan Q, Wu R, Shi W, Wang XY, Chen L, et al: Primary analysis and
screening of microRNAs in gastric cancer side population cells.
World J Gastroenterol. 21:3519–3526. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Chen Z, Saad R, Jia P, Peng D, Zhu S,
Washington MK, Zhao Z, Xu Z and El-Rifai W: Gastric adenocarcinoma
has a unique microRNA signature not present in esophageal
adenocarcinoma. Cancer. 119:1985–1993. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Guo B, Liu L, Yao J, Ma R, Chang D, Li Z,
Song T and Huang C: miR-338-3p suppresses gastric cancer
progression through a PTEN-AKT axis by targeting P-REX2a. Mol
Cancer Res. 12:313–321. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wang F, Li T, Zhang B, Li H, Wu Q, Yang L,
Nie Y, Wu K, Shi Y and Fan D: MicroRNA-19a/b regulates multidrug
resistance in human gastric cancer cells by targeting PTEN. Biochem
Biophys Res Commun. 434:688–694. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Si Y, Zhang H, Ning T, Bai M, Wang Y, Yang
H, Wang X, Li J, Ying G and Ba Y: miR-26a/b inhibit tumor growth
and angiogenesis by targeting the HGF-VEGF axis in gastric
carcinoma. Cell Physiol Biochem. 42:1670–1683. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhang Y, Lu Q and Cai X: MicroRNA-106a
induces multidrug resistance in gastric cancer by targeting RUNX3.
FEBS Lett. 587:3069–3075. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Tsujiura M, Komatsu S, Ichikawa D,
Shiozaki A, Konishi H, Takeshita H, Moriumura R, Nagata H,
Kawaguchi T, Hirajima S, et al: Circulating miR-18a in plasma
contributes to cancer detection and monitoring in patients with
gastric cancer. Gastric Cancer. 18:271–279. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ren C, Wang W, Han C, Chen H, Fu D, Luo Y,
Yao H, Wang D, Ma L, Zhou L, et al: Expression and prognostic value
of miR-92a in patients with gastric cancer. Tumour Biol.
37:9483–9491. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Thomson DW, Bracken CP and Goodall GJ:
Experimental strategies for microRNA target identification. Nucleic
Acids Res. 39:6845–6853. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Pernot S, Voron T, Perkins G,
Lagorce-Pages C, Berger A and Taieb J: Signet-ring cell carcinoma
of the stomach: Impact on prognosis and specific therapeutic
challenge. World J Gastroenterol. 21:11428–11438. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Singh SS, Yap WN, Arfuso F, Kar S, Wang C,
Cai W, Dharmarajan AM, Sethi G and Kumar AP: Targeting the PI3K/Akt
signaling pathway in gastric carcinoma: A reality for personalized
medicine? World J Gastroenterol. 21:12261–12273. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Okugawa Y, Toiyama Y, Hur K, Toden S,
Saigusa S, Tanaka K, Inoue Y, Mohri Y, Kusunoki M, Boland CR and
Goel A: Metastasis-associated long non-coding RNA drives gastric
cancer development and promotes peritoneal metastasis.
Carcinogenesis. 35:2731–2739. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Harapan H, Fitra F, Ichsan I, Mulyadi M,
Miotto P, Hasan NA, Calado M and Cirillo DM: The roles of microRNAs
on tuberculosis infection: Meaning or myth? Tuberculosis (Edinb).
93:596–605. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wu W, Takanashi M, Borjigin N, Ohno SI,
Fujita K, Hoshino S, Osaka Y, Tsuchida A and Kuroda M: MicroRNA-18a
modulates STAT3 activity through negative regulation of PIAS3
during gastric adenocarcinogenesis. Br J Cancer. 108:653–661. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhu P, Zhang J, Zhu J, Shi J, Zhu Q and
Gao Y: miR-429 induces gastric carcinoma cell apoptosis through
Bcl-2. Cell Physiol Biochem. 37:1572–1580. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhou C, Shen L, Mao L, Wang B, Li Y and Yu
H: miR-92a is upregulated in cervical cancer and promotes cell
proliferation and invasion by targeting FBXW7. Biochem Biophys Res
Commun. 458:63–69. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ke TW, Wei PL, Yeh KT, Chen WT and Cheng
YW: miR-92a promotes cell metastasis of colorectal cancer through
PTEN-mediated PI3K/AKT pathway. Ann Surg Oncol. 22:2649–2655. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhang G, Zhou H, Xiao H, Liu Z, Tian H and
Zhou T: MicroRNA-92a functions as an oncogene in colorectal cancer
by targeting PTEN. Dig Dis Sci. 59:98–107. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Chen ZL, Zhao XH, Wang JW, Li BZ, Wang Z,
Sun J, Tan FW, Ding DP, Xu XH, Zhou F, et al: MicroRNA-92a promotes
lymph node metastasis of human esophageal squamous cell carcinoma
via E-cadherin. J Biol Chem. 286:10725–10734. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Xu X, Zhu S, Tao Z and Ye S: High
circulating miR-18a, miR-20a, and miR-92a expression correlates
with poor prognosis in patients with non-small cell lung cancer.
Cancer Med. 7:21–31. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ranade AR, Cherba D, Sridhar S, Richardson
P, Webb C, Paripati A, Bowles B and Weiss GJ: MicroRNA 92a-2*: A
biomarker predictive for chemoresistance and prognostic for
survival in patients with small cell lung cancer. J Thorac Oncol.
5:1273–1278. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ludwig S, Klitzsch A and Baniahmad A: The
ING tumor suppressors in cellular senescence and chromatin. Cell
Biosci. 1:252011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Unoki M, Kumamoto K, Robles AI, Shen JC,
Zheng ZM and Harris CC: A novel ING2 isoform, ING2b, synergizes
with ING2a to prevent cell cycle arrest and apoptosis. FEBS Lett.
582:3868–3874. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wang Y, Wang J and Li G: Leucine
zipper-like domain is required for tumor suppressor ING2-mediated
nucleotide excision repair and apoptosis. FEBS Lett. 580:3787–3793.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Pedeux R, Sengupta S, Shen JC, Demidov ON,
Saito S, Onogi H, Kumamoto K, Wincovitch S, Garfield SH, McMenamin
M, et al: ING2 regulates the onset of replicative senescence by
induction of p300-dependent p53 acetylation. Mol Cell Biol.
25:6639–6648. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Shi X, Hong T, Walter KL, Ewalt M,
Michishita E, Hung T, Carney D, Peña P, Lan F, Kaadige MR, et al:
ING2 PHD domain links histone H3 lysine 4 methylation to active
gene repression. Nature. 442:96–99. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Bua DJ, Martin GM, Binda O and Gozani O:
Nuclear phosphatidylinositol-5-phosphate regulates ING2 stability
at discrete chromatin targets in response to DNA damage. Sci Rep.
3:21372013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Han XR, Bai XZ, Sun Y and Yang Y: Nuclear
ING2 expression is reduced in osteosarcoma. Oncol Rep.
32:1967–1972. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Ythier D, Larrieu D, Binet R, Binda O,
Brambilla C, Gazzeri S and Pedeux R: Sumoylation of ING2 regulates
the transcription mediated by Sin3A. Oncogene. 29:5946–5956. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Sarker KP, Kataoka H, Chan A, Netherton
SJ, Pot I, Huynh MA, Feng X, Bonni A, Riabowol K and Bonni S: ING2
as a novel mediator of transforming growth factor-beta-dependent
responses in epithelial cells. J Biol Chem. 283:13269–13279. 2008.
View Article : Google Scholar : PubMed/NCBI
|