Introduction
Glioblastoma is the most common malignant brain
cancer diagnosed in humans, with an average patient survival rate
of less than one year after diagnosis (1). Surgical resection followed by a
combination of radiation and chemotherapy is the standard treatment
for glioblastoma. Although innovative therapeutic strategies have
continually been investigated in the past two decades, there are
still no effective methods to prevent tumor recurrence. Therefore,
it is urgent to explore novel chemotherapeutic targets for
glioblastoma.
Nicotinamide phosphoribosyltransferase (NAMPT) is a
rate-limiting enzyme in the salvage pathway of nicotinamide adenine
dinucleotide (NAD) biosynthesis. NAD is an essential coenzyme
involved in cellular redox reactions and is a substrate for
NAD-dependent enzymes (2).
NAD-dependent enzymes, including histone deacetylases (SirT1-T7),
CD38 and poly(ADP-ribose) polymerase (PARP), play an important role
in the maintenance of organismal metabolic homeostasis and genomic
stability (3–5). Thus, NAD displays a wide range of
effects on cell cycle progression, apoptosis, DNA repair, circadian
rhythms, regulation of chromatin dynamics, telomerase activity,
intracellular calcium mobilization, and transcriptional regulation
(6–12). All of these effects are associated
with the regulation of tumorigenesis and development. NAMPT is a
rate-limiting enzyme for NAD synthesis; therefore, it plays an
important role in tumor generation and progression (13–15).
Previous studies have shown that NAMPT expression is
significantly elevated in various human malignant tumors, including
lymphomas, breast cancer, prostate adenocarcinoma, esophageal
cancer, gastric cancer, colorectal adenocarcinoma, ovarian serous
adenocarcinoma and glioblastoma (6).
Higher NAMPT expression is correlated with deeper tumor invasion,
presence of lymph node metastases, advanced clinical TMN stage and
shorter patient survival times (16–19). To
determine the function of NAMPT in glioblastoma, we searched a
database and further performed experiments to explore its role in
glioblastoma.
Materials and methods
Oncomine database
The Oncomine database (http://www.oncomine.com) was queried to assess the
expression of NAMPT. Set filter conditions included: i. Cancer
Type: Glioblastoma; ii. Gene: NAMPT; iii. Data Type: mRNA; iv.
Sample Type: Clinical Specimen; v. Analysis Type: Cancer vs. Normal
Analysis; vi. Critical setting condition: P-value
<1×10−4, fold change >2, gene rank=top 10%.
R2 platform
The Kaplan-Meier survival analysis was performed
using the ‘Tumor glioblatoma-TCGA-540 MAS5.0-u133a’ dataset in the
Genomics Analysis and Visualization Platform (R2 platform)
(https://hgserver1.amc.nl/cgi-bin/r2/main.cgi). The
median was used as the threshold to distinguish the high and low
expression of NAMPT.
Immunohistochemical microarray
analysis
A glioblastoma tissue microarray (US Biomax GL805b)
was obtained from GeneChem (Shanghai, China) containing two spots
each of tissues from 35 cases of glioblastoma, paracarcinoma
tissues from 2 cases (1.5-cm distance from the paracarcinoma tissue
to the cancer tissue) and normal neural tissues from 3 cases. A
two-step immunohistochemical protocol was used. The primary
antibody was an anti-NAMPT rabbit monoclonal antibody (dilution
1:500, cat. no. ab58640; Abcam), and the secondary antibody was
from an UltraSensitive SP KIT-9720 (Fujian MXB). Three random
high-magnification fields were observed under an optical microscope
(Caikon XDS-100, original magnification, ×200).
The immunohistochemical microarray staining was
scored on a semiquantitative scale by assessing the intensity and
proportion of staining. The staining intensity was scored using the
following scale: 0 (negative), 1 (weak), 2 (moderate), and 3
(strong). According to the positive rate of staining, the
proportion of staining was scored as 0 (0–4%), 1 (5–25%), 2
(26–50%), 3 (51–75%), or 4 (76–100%). The total score was
calculated by multiplying the proportion score and the intensity
score. Specimens with a total score of ≤6 were assigned to the
low-expression group. Specimens with a total score of >6 were
assigned to the high-expression group.
Gene set enrichment analysis
(GSEA)
GBM gene expression profile was downloaded from The
Cancer Genome Atlas (TCGA) database (https://www.cancer.gov/tcga). The queue were divided
into NAMPT high and low expression groups according to the median
expression of NAMPT. Gene set enrichment analysis was performed
using the GSEA software, version 2.0.1, obtained from the Broad
Institute (http://www.broad.mit.edu/gsea). Gene set permutations
were performed 1,000 times for each analysis. Family error rate
(FWER) and false discovery rate (FDR) were used to screen the
enriched pathways in each group.
Cell lines and cell culture
The human glioblastoma cell lines, U87 and U251,
were obtained from the American Type Culture Collection (ATCC). STR
profiling indicated that the U87 was a probable glioblastoma of
unknown origin. These cell lines were cultured with
Corning® DMEM medium (Corning Inc.) containing 10% FBS.
Cell lines were cultured at 37°C in a humidified atmosphere of 5%
CO2.
NAMPT shRNA design and lentivirus
construction
A small hairpin RNA (shRNA) targeting NAMPT with
high specificity was designed and synthesized by GeneChem
(Shanghai, China) and cloned into a lentiviral vector (GeneChem),
which also encoded green fluorescent protein (GFP). Then, the
lentivirus expressing NAMPT shRNA (LV-NAMPT-RNAi) was prepared and
collected. The NAMPT shRNA sequence was as follows:
5′-AACTTAGATGGTCTGGAAT-3′. A scrambled shRNA sequence was used as
the negative control: 5′-TTCTCCGAACGTGTCACGT-3′.
Lentivirus infection
The human glioblastoma cell lines U87 and U251 were
used for lentivirus infection. Briefly, cells in the logarithmic
phase were digested by trypsinase. A 4×104/ml cell
suspension was prepared, and 1 ml of the cell suspension was
inoculated into a 12-well plate. When the cell confluence was 20%,
the appropriate lentiviral particle solution containing the
lentivirus expressing either NAMPT or the scrambled shRNA was added
to the target plate (MOI=5). The infection efficiency was
determined by the GFP expression status as observed with a
fluorescence microscope (Olympus Corp., Tokyo, Japan) 72 h after
infection.
Quantitative real-time PCR (qPCR)
Total RNA was extracted and reverse transcribed. The
resulting cDNA was used as the template for RT-PCR using a standard
SYBR Green PCR kit (Takara) in a LightCycler 480 Real-Time PCR
machine (Roche). Independent experiments were conducted in
triplicate. GAPDH served as the internal control. Gene expression
was calculated using the ΔΔCq method (20).
Western blot analysis
Cell lysates were prepared in ice-cold RIPA lysis
buffer. Total protein concentration was determined using an
enhanced BCA protein assay kit. An amount of 30 µg of total protein
per lane was separated by 10% SDS-PAGE, transblotted onto PVDF
membranes, which were then blocked with 5% milk dissolved in TBST
for 1 h at room temperature and incubated overnight with primary
antibodies at 4°C. The primary antibodies used were as follows:
Rabbit anti-NAMPT (cat. no. ab58640; dilution 1:500; Abcam), mouse
anti-GAPDH (cat. no. ab8245; dilution 1:3,000; Abcam). Then the
blots were incubated with anti-rabbit (cat. no. ab205718; dilution
1:5,000; Abcam) or anti-mouse (cat. no. ab205719; dilution 1:5,000;
Abcam) HRP-labeled secondary antibody at room temperature for 1.5
h. The immunoactivity was detected using an ECL-Plus kit (Thermo
Fisher Scientific. Inc.). The immunoreactive bands were quantified
by densitometry with ImageJ software (National Institutes of
Health, Bethesda, MD, USA).
CCK-8 assay
Cells at a density of 2,000 cells/well in
logarithmic phase were cultured in 96-well culture plates. A 10-µl
volume of CCK-8 reagent (Sigma-Aldrich; Merck KGaA) was added at
various time points (24, 48, 72, 96 and 120 h). After cells were
cultured for 4 h, the absorbance of the plates was read at a
wavelength of 450 nm using a microplate reader (M2009PR; Tecan
Infinite). The experiment was repeated five times.
Colony formation assay
Three days after infection with the lentivirus,
cells were seeded at a density of 800 cells per well in 6-well
plates with triplicate wells for each group. Cell colonies were
grown in a humidified 5% CO2 incubator at 37°C for 16
days. Images were acquired with an inverted microscope at ×100
magnification. Then, cells were fixed, stained with Giemsa solution
and imaged. Colonies measuring 0.3 mm or more were counted.
Wound healing assay
Cells at a density of 50,000 cells/well were seeded
in 96-well wounding replicator and grown to over 90% confluence.
Then the culture medium was replaced with 0.5% FBS. Wounds were
generated using the tip with 200 µm width of a wounding replicator,
VP scientific, VP408FH. Cells were incubated for up to 24 h. Wound
closure was observed at 0, 8 and 24 h post scratching. These assays
were performed with three independent replicates. Cell images were
acquired with an inverted microscope (Caikon XDS-100, original
magnification, ×10). The migration rate was calculated as follows:
Migration rate=[(width of wound at 0 h-width of wound at the time
point tested)/width of wound at 0 h] ×100%.
Transwell chamber assay
A Corning invasion kit was used to perform this
assay according to the manufacturer's instructions. Cell
suspensions (105 cells per well in a 24-well culture
plate) in FBS-free DMEM were added to the top chamber, and DMEM
containing 30% FBS was added to the bottom chamber. Cells that
migrated through the filter were stained with Giemsa. Images of the
migrated cells were captured with a digital camera connected to an
inverted microscope (Caikon XDS-100). Five random fields of vision
at ×100 magnification and 9 random fields of vision at ×200
magnification were observed per well. The migrated cells were
counted in the ×200 images. The invasion assay was carried out with
three independent replicates.
In vivo tumorigenesis in nude
mice
The first step was to prepare tumor-forming cells
according to the method described above. U87 cells infected with a
lentiviral vector expressing NAMPT-shRNA and GFP sequences were
generated as the knockdown (KD) group; cells infected with
lentiviral vector expressing scrambled shRNA and GFP sequences were
generated as the negative control (NC) group. Mice were randomized
into the two groups. BALB/c nude mice (male, 4 weeks old, ~20 g)
were purchased from the HUST Animal Center, Wuhan, China. They were
maintained in a constant temperature, humidity, sterile environment
and routinely fed with food and water according to the national
regulations. A total of 1×107 logarithmically growing
U87 cells in 0.1 ml of PBS were subcutaneously injected into the
right flank of the mice, 10 mice in each group. According to the
Institutional Animal Care and Use Committees (IUCUC) protocol
(AN-5803), the maximal allowable size of the tumours could not
exceed 10% of mouse body weight. Tumor volumes were measured every
four days after injection with a caliper and calculated using the
formula V=π/6 (lxwxw), where l is the length and w is the width.
Forty days after injection, animals were anesthetized using an
intraperitoneal injection of 150 mg/kg pentobarbital sodium. Tumor
tissues were excised and weighed. Immunohistochemical staining of
Ki-67 and CD31 was performed on the transplanted tumors, and the
Ki-67 index and microvascular density (MVD) were calculated. All
animal experiments were approved by the Institutional Animal Care
and Use Committee of Huazhong University of Science and
Technology.
Statistical analysis
Data are expressed as the means ± standard errors of
the mean. Student's t-test was used to compare the data between two
groups. The data between three groups were compared by ANOVA, and
then Tukey test was used to compare the data between the two groups
if P<0.05. P<0.05 was considered to indicate a statistically
significant difference.
Results
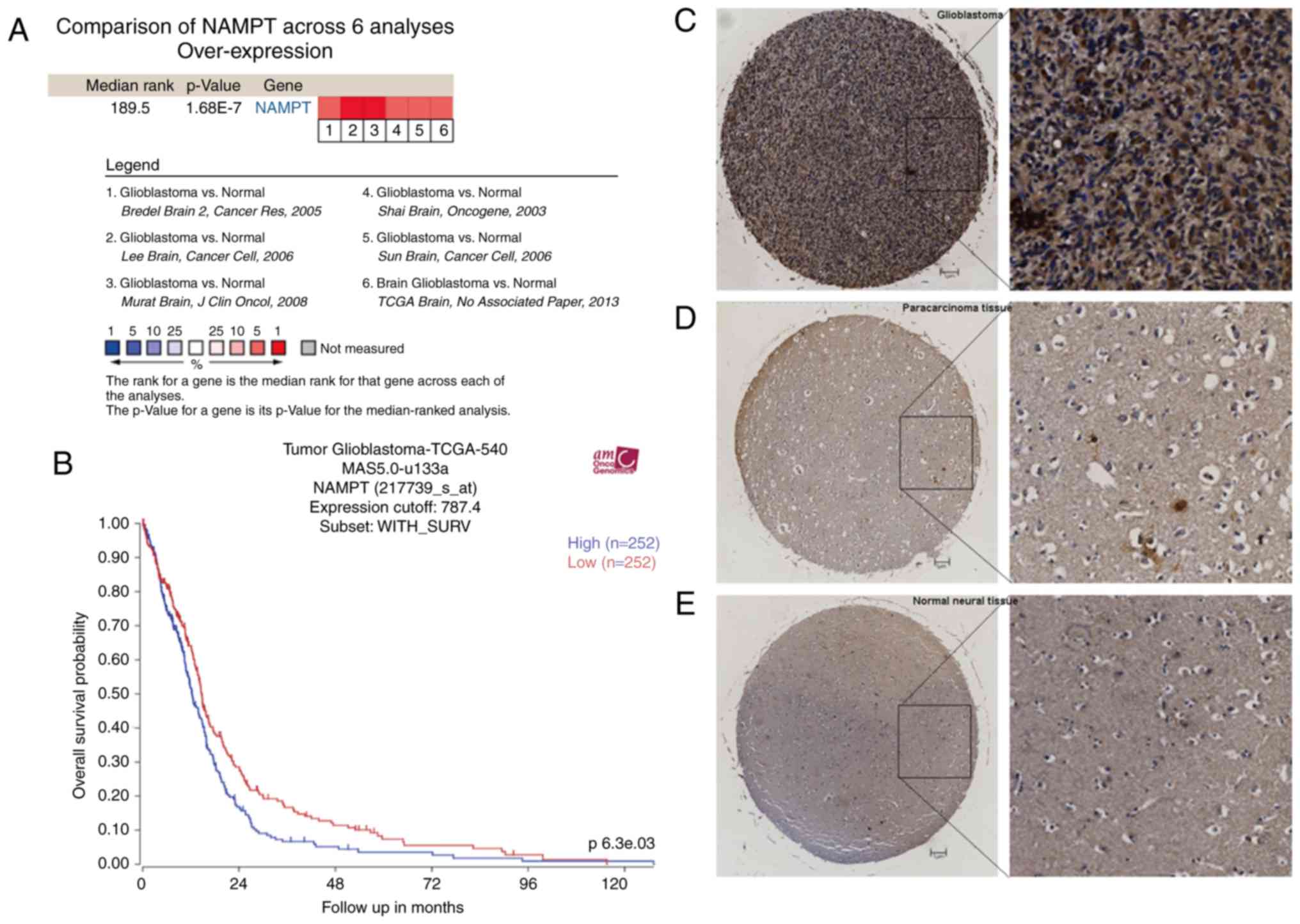
NAMPT is overexpressed in glioblastoma
and correlated with poor overall survival
Six studies in the Oncomine database, comprising a
total of 942 samples, met the criteria (21–25).
Meta-analysis of the 6 studies showed that NAMPT was statistically
overexpressed in glioblastoma relative to its expression in normal
neural tissue (P=1.68E-7) (Fig. 1A).
Immunohistochemical microarray analysis showed that NAMPT was
mainly localized in the cytoplasm (Fig.
1C-E). The average total staining scores of the glioblastoma
and nontumor tissues (paracarcinoma and normal neural tissues) were
7.54 and 4.600, respectively (P=0.000637). These results suggest
that the expression of NAMPT in human glioblastoma is higher than
that in normal tissues.
Data from the R2 platform (540 glioblastoma samples)
showed that patients with higher NAMPT expression had a lower
overall survival rate than patients with lower NAMPT expression.
The 2-year survival rates were 4 and 1% for the high- and
low-expression groups, respectively (P=0.0063) (Fig. 1B).
Biological pathways enriched in the
NAMPT high-expression phenotype as obtained by gene set enrichment
analysis (GSEA)
Gene set enrichment analysis (GSEA) was performed
using data from the TCGA GBM cohort comprising 169 samples. The
expression level of NAMPT was used as the phenotype label. A
pathway was classified as enriched by setting the FWER P-value to
<0.01 and the FDR to <0.05. The ‘GO acute inflammatory
response’, ‘positive regulation of I-κB kinase NF-κB signaling’,
‘I-κB kinase NF-κB signaling’, and ‘acute phase response’ pathways
were enriched in the NAMPT high-expression phenotype (Fig. 1F). Therefore, NAMPT may play a role
via these pathways.
A lentiviral-based shRNA strategy is
successfully used to inhibit NAMPT expression in GBM cells
A lentiviral vector expressing both shRNA targeting
human NAMPT and the GFP sequence was used to establish knockdown
(KD) cells, while a lentiviral vector expressing both scrambled
shRNA and the GFP sequence was used to establish negative control
(NC) cells in the U87 and U251 cell lines. Cells without lentivirus
infection were considered blank control (CON) cells. GFP expression
was observed in more than 90% of cells infected with the lentivirus
72 h after infection (Fig. 2A and
B).
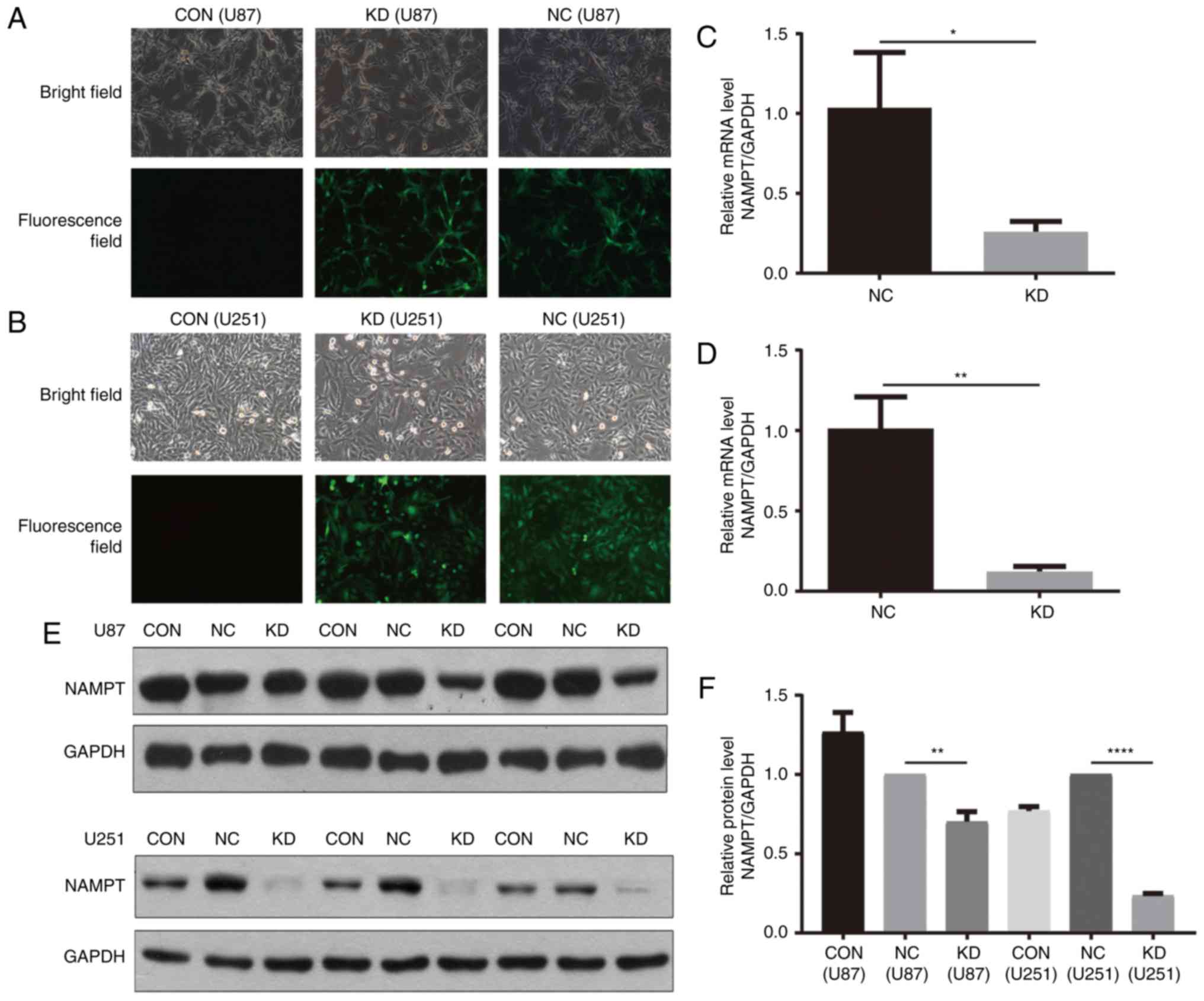 | Figure 2.A lentivirus-based shRNA strategy was
effectively used to knock down NAMPT in U87 and U251 GBM cell
lines. (A and B) Observation of cells via optical and fluorescence
microscopy after lentiviral infection (magnification, ×200). (C and
D) NAMPT-shRNA was effective for NAMPT knockdown in U87 (C) and
U251 (D) GBM cells, as verified by RT-PCR. (E and F) NAMPT-shRNA
effectively knocked down NAMPT expression in U87 and U251 GBM
cells, as verified by western blotting. Western blot band (E) and
quantification by Compass software (F) (*P<0.05, **P<0.01,
****P<0.0001). NAMPT, nicotinamide phosphoribosyltransferase;
GBM, glioblastoma. Groups: KD, cells transfected with shRNA
targeting human NAMPT; NC, cells transfects with scrambled shRNA;
CON, cells without lentivirus infection considered as blank control
cells. |
RT-PCR and western blot analysis were performed to
assess the knockdown efficiency. RT-PCR results showed that
NAMPT-shRNA was effective in both U251 and U87 GBM cells, with a
knockdown efficiency of approximately 74.9% in U87 GBM cells
(Fig. 2C) and 87.7% in U251 GBM cells
(Fig. 2D). The location (Fig. 2E) and quantification (Fig. 2F) of the western blot band further
confirmed the above results.
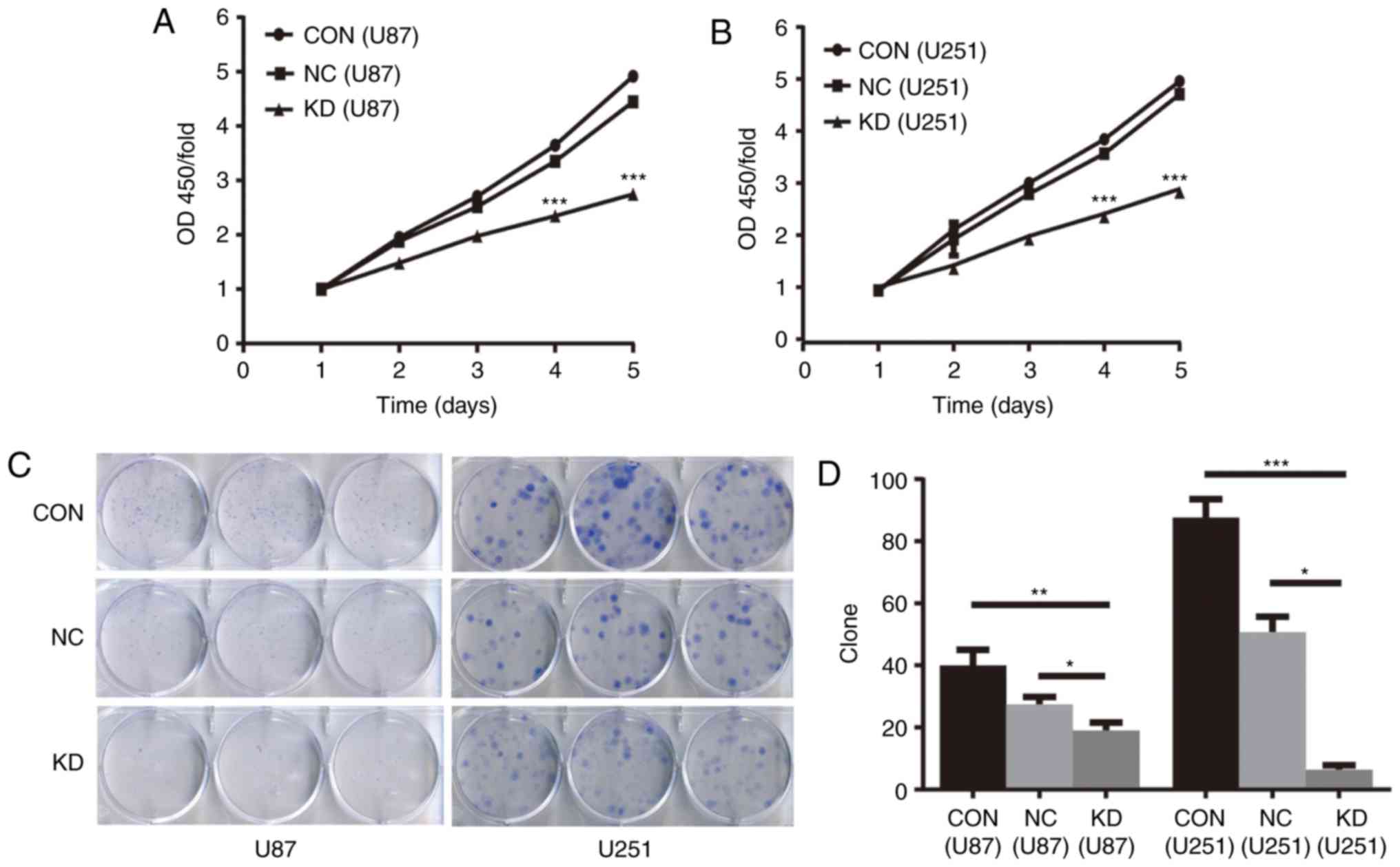
NAMPT knockdown inhibits cell
proliferation and colony formation in GBM cells
The CCK-8 assay showed that the proliferation of
cells in the KD group was significantly inhibited at the 4 and 5
days in U87 and U251 GBM cells (P<0.001 KD vs. NC for both cell
lines) (Fig. 3A and B).
The soft agar colony formation assay showed that in
the KD, NC and CON groups of U87 and U251 GBM cells the average
colony numbers were 19, 27 and 40 and 6, 51 and 88, respectively
(P<0.05 KD vs. NC for both cell lines) (Fig. 3C and D).
NAMPT knockdown inhibits cell
migration and invasion and induces apoptosis in the GBM cell
lines
As shown by the wound healing assay in U87 cells,
the migration rates of KD cells at 8 and 24 h were 0.14±0.08 and
0.4±0.06, respectively, lower than those of the CON group
(0.21±0.08 and 0.64±0.05) and NC (0.19±0.07 and 0.54±0.07) cells
(Fig. 4A and B).
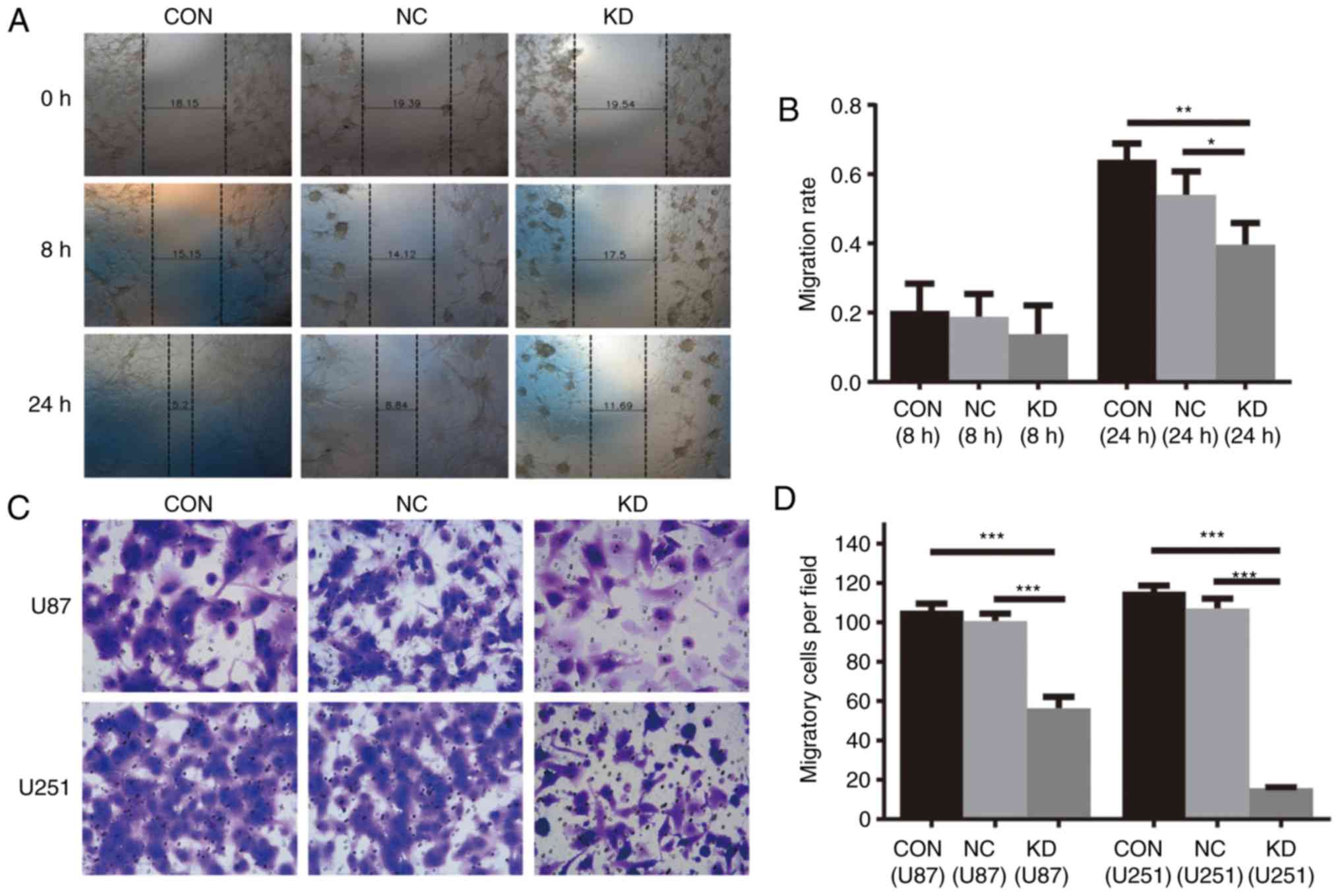 | Figure 4.(A and B) Cell migration was
decreased by NAMPT knockdown in U87 GBM cells as analyzed by wound
healing assay. Representative images (magnification, ×100). (A) and
histogram showing the means ± SDs of cell migration rate from three
separate experiments (*P<0.05, **P<0.01). (C and D) Cell
invasion was decreased by NAMPT knockdown in U87 and U251 GBM
cells, as determined by a Transwell assay. Representative images
(magnification, ×200). (C) and histogram showing the means ± SDs of
the number of migrated cells in three separate experiments (D)
(***P<0.001). (E-G) Apoptosis was induced by NAMPT knockdown in
U87 and U251 GBM cells. Representative images (E) and histograms
showing the means ± SDs of the percentage of apoptotic cells in
three separate experiments in U87 (F) and U251 (G) GBM cells
(***P<0.001, ****P<0.0001). NAMPT, nicotinamide
phosphoribosyltransferase; GBM, glioblastoma. Groups: KD, cells
transfected with shRNA targeting human NAMPT; NC, cells transfects
with scrambled shRNA; CON, cells without lentivirus infection
considered as blank control cells. |
The Transwell assay showed that the average numbers
of migrated cells in the KD, NC and CON groups were 56±5.55,
101±3.88 and 106±3.74 in the U87 cells and 16±0.44, 107±5.00 and
116±2.92 in the U251 cells, respectively (P<0.001, KD vs. NC and
CON) (Fig. 4C and D).
The apoptosis assay showed that the percentage of
apoptotic U87 GBM cells in the KD group was 7.68±0.3386%, which was
higher than that in the NC group (3.74±0.3103%) (P<0.001).
Similarly, the percentage of apoptotic U251 GBM cells in the KD
group was 7.43±0.4921%, which was also higher than that in the NC
group (3.88±0.3553%, P<0.001) (Fig.
4E-G).
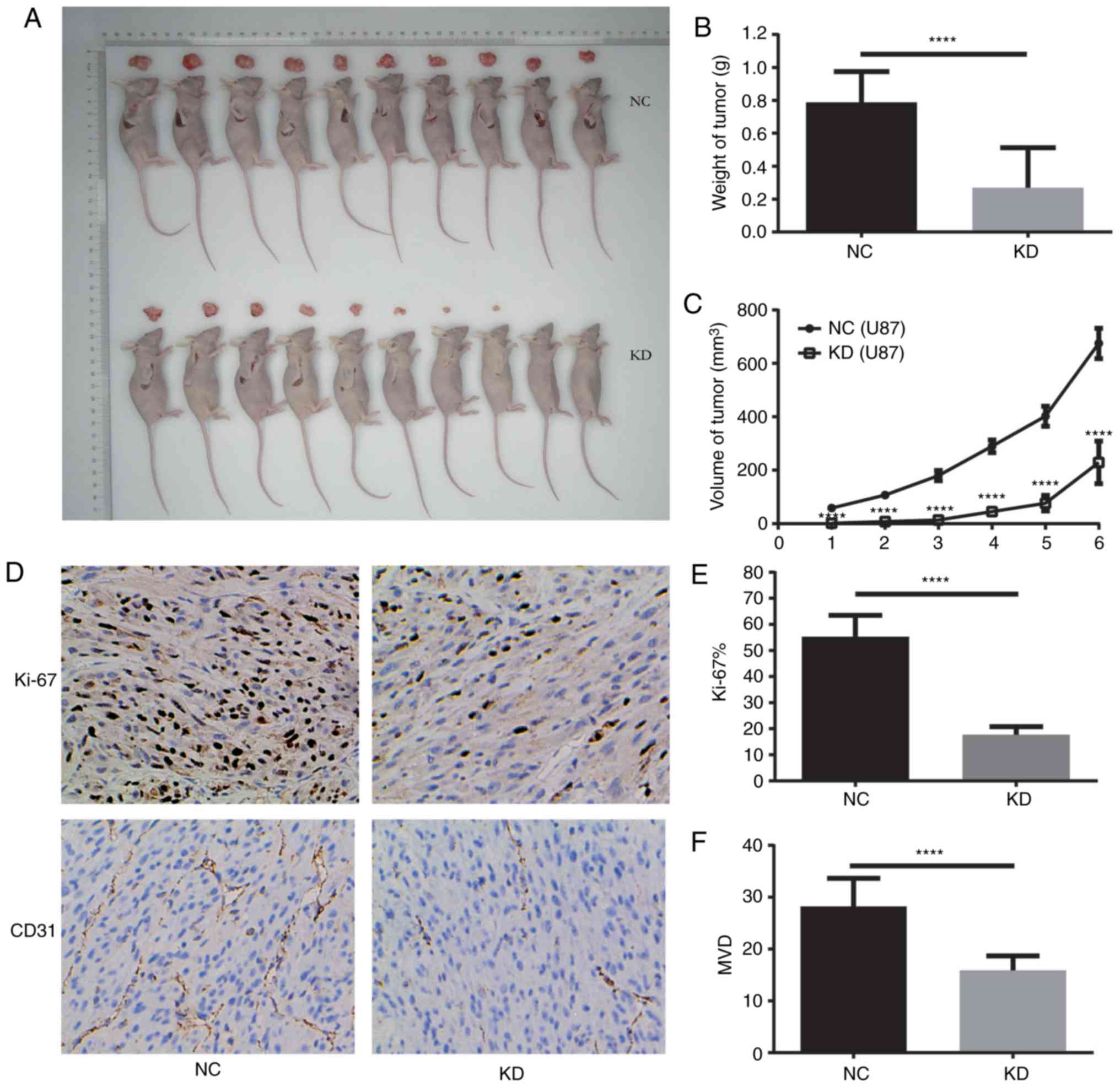
NAMPT knockdown inhibits the growth of
transplanted U87 GBM cells in nude mice
The growth curve of subcutaneously transplanted
tumors in nude mice showed that tumors derived from the KD cells
grew much more slowly than those derived from the NC tumors. Forty
days after inoculation, the maximum diameter of the KD and NC
transplanted tumors were 11.40 and 18.50 mm, the maximum weights of
the KD and NC tumors were 0.643 and 1.049 g, respectively. The
average weights of the KD and NC transplanted tumors were
0.269±0.244 and 0.788±0.186 g, respectively (P<0.0001) (Fig. 5A-C).
The Ki-67 indices of the KD and NC tumors was
17.7±3.1 and 55.3±8.2%, and the MVD in the KD and NC tumors was
15.9±2.8 and 28.2±5.4, respectively. These results suggest that
NAMPT knockdown inhibits the growth and microvessel formation of
U87 glioblastoma xenografts in nude mice (Fig. 5D-F).
Discussion
Manyprevious studies have reported that NAMPT
expression is increased in human malignant tumors and that high
expression of NAMPT is associated with advanced tumor stage and
reduced patient survival time (6,16–19). Hufton et al (26) reported that NAMPT expression was
6-fold higher than that in benign tissues. Olesen et al
(18) found that NAMPT was
overexpressed in blood diseases and was associated with more
aggressive phenotypes of malignant lymphoma. Nakajima et al
(27,28) reported that NAMPT may be a useful
biomarker for predicting the progression of gastric and colorectal
cancer. Maldi et al (16)
reported that NAMPT promoted the growth, metastasis and
dedifferentiation of melanoma cells. Gujar et al
demonstrated that NAMPT is highly expressed in glioblastoma tumors
and patient-derived glioblastoma stem-like cells (GSCs), and NAMPT
knockdown inhibited the in vivo tumorigenicity of GSCs
(29). To further study the effect of
NAMPT on glioblastoma, we performed a series of studies. First, we
searched the Oncomine database and found that NAMPT was
statistically overexpressed in glioblastoma tissues relative to its
expression in normal neural tissues. Then, using the R2 platform,
we found that high expression of NAMPT was associated with poor
prognosis in glioblastoma. Next, we carefully selected a
glioblastoma tissue microarray containing tissues from 35 cases
(each with two spots). The microarray analysis results indicated
that the expression of NAMPT in glioblastoma was higher than that
in normal neutral tissue. In conclusion, we believe that NAMPT is
overexpressed in glioblastoma and that high NAMPT expression is
associated with poor prognosis in glioblastoma.
Inhibition of NAMPT expression by small-molecule
inhibitors or gene knockdown has demonstrated antitumor effects
both in vitro and in vivo (30–33). Wang
et al discovered that inhibition of NAMPT expression in
prostate cancer cells suppressed growth and invasion in
vitro and the growth of tumor xenografts in vivo
(33). Yang et al (34) generated fibrosarcoma HT1080 and 293
cells with stable knockdown of NAMPT using siRNA. The stable cells
with knockdown of NAMPT expression were more sensitive to etoposide
and had increased levels of cleaved caspase 3 than the
corresponding parental cells. Jung et al reported that NAD
metabolism regulates glioblastoma stem cell maintenance (35). To explore whether NAMPT has an
influence on glioblastoma cells, we performed several cytological
and animal experiments. We used a lentivirus expressing NAMPT-shRNA
to knock down NAMPT expression in two human glioblastoma cell
lines, U87 and U251, and confirmed the knockdown effectiveness by
RT-PCR and western blotting. Then, we performed CCK-8 and soft agar
assays and observed that cell proliferation was inhibited by NAMPT
knockdown. In addition, apoptosis analysis showed that NAMPT
knockdown induced apoptosis. The wound healing and Transwell assays
showed that cell migration and invasion were decreased by NAMPT
knockdown. Our data confirmed that NAMPT knockdown markedly
inhibited cell proliferation, migration and invasion and induced
apoptosis in U87 and U251 GBM cell lines. To further confirm our
conclusion in vivo, we established a subcutaneous xenograft
model in nude mice. The growth of xenografts was significantly
slowed by NAMPT knockdown. Immunohistochemical staining of
xenografts showed that the Ki-67 index and microvessel density
(MVD) of NAMPT knockdown tumors were lower than those of negative
control tumors. These results showed that NAMPT knockdown exerts a
significant inhibitory effect on the tumorigenesis of GBM cells
in vitro and in vivo.
The exact molecular mechanism by which NAMPT
regulates glioblastoma generation and progression is complicated
and remains unclear. Studies have shown that NAMPT may perform its
function through NAD, which is an essential cofactor involved in
cellular redox reactions and a substrate for NAD-dependent enzymes
such as poly(ADP-ribose) polymerases (PARPs), sirtuins, and cyclic
ADP (cADP) ribose synthases. PARP-1 is an important regulator of
cell stress and responses to DNA damage. Venkateshaiah et al
found that NAMPT inhibition lowers cellular PARP-1 activity and
cell viability in myeloma cells (36). Sirtuin1 can attenuate p53, PTEN, and
retinoblastoma protein activity; stabilize N-Myc; promote
epithelial-to-mesenchymal transition; and increase cell migration
(17). Revollo et al found
that increased NAMPT expression increases cellular NAD levels and
enhanced Sirtuin1-mediated transcription in murine cells (37). Our GSEA results indicated that the GO
terms ‘acute inflammatory response’, ‘positive regulation of IκB
kinase NF-κB signaling’, ‘IκB kinase NF-κB signaling’, and ‘acute
phase response’ were enriched in the NAMPT high-expression
phenotype. Therefore, we deduced that NAMPT may play its role
through these pathways. However, we have not experimentally
verified the specific role and mechanism of NAMPT in these
pathways, which may be one of our next research directions.
In summary, our study demonstrated that NAMPT is
significantly overexpressed in glioblastoma tissues and that high
NAMPT expression indicates worse prognosis. In vitro and
in vivo experiments showed that NAMPT knockdown reduced cell
proliferation, migration, and invasion and induced apoptosis.
Therefore, NAMPT may be considered an oncogene in glioblastoma and
may be a potential prognostic and therapeutic target for
glioblastoma. In the future, we may further study the antitumor
mechanism of NAMPT through four pathways derived from the GSEA
analysis, and ascertain whether NAMPT knockdown or NAMPT inhibitors
combined with chemotherapy, radiotherapy or targeted drugs can play
an unexpected antitumor role in glioblastoma.
Acknowledgements
Not applicable.
Funding
This study was funded by the National Natural
Science Foundation (grant no. 1772680).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
QG performed the cell biology and animal experiments
and wrote the manuscript. MZ conducted the cell biology and animal
experiments, and managed the project. NH performed the
bioinformatic data analysis and contributed to the project
management. LS contributed to the management of the project and
performed the cell biology experiments. LY contributed to the
animal experiments and project management. XZ helped conduct the
cell biology experiments and modify the manuscript. YZ contributed
to the cell biology experiments and modified the manuscript. SY
contributed to the animal experiments and managed the project. All
authors read and approved the final manuscript and agreed to be
accountable for all aspects of the work in ensuring that questions
related to the accuracy or integrity of any part of the work are
appropriately investigated and resolved.
Ethics approval and consent to
participate
The animal experiments in the present study were
approved by the Ethics Committee for Animal Experimentation of
Genechem (no. GSZE0116844).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Iacob G and Dinca EB: Current data and
strategy in glioblastoma multiforme. J Med Life. 2:386–393.
2009.PubMed/NCBI
|
|
2
|
Garten A, Schuster S, Penke M, Gorski T,
de Giorgis T and Kiess W: Physiological and pathophysiological
roles of NAMPT and NAD metabolism. Nat Rev Endocrinol. 11:535–546.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Imai S, Armstrong CM, Kaeberlein M and
Guarente L: Transcriptional silencing and longevity protein Sir2 is
an NAD-dependent histone deacetylase. Nature. 403:795–800. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kim MY, Mauro S, Gevry N, Lis JT and Kraus
WL: NAD+-dependent modulation of chromatin structure and
transcription by nucleosome binding properties of PARP-1. Cell.
119:803–814. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Van Gool F, Galli M, Gueydan C, Kruys V,
Prevot PP, Bedalov A, Mostoslavsky R, Alt FW, De Smedt T and Leo O:
Intracellular NAD levels regulate tumor necrosis factor protein
synthesis in a sirtuin-dependent manner. Nat Med. 15:206–210. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Shackelford RE, Mayhall K, Maxwell NM,
Kandil E and Coppola D: Nicotinamide phosphoribosyltransferase in
malignancy: A review. Genes Cancer. 4:447–456. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lin SJ and Guarente L: Nicotinamide
adenine dinucleotide, a metabolic regulator of transcription,
longevity and disease. Curr Opin Cell Biol. 15:241–246. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Sebastian C, Satterstrom FK, Haigis MC and
Mostoslavsky R: From sirtuin biology to human diseases: An update.
J Biol Chem. 287:42444–42452. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Chiarugi A, Dolle C, Felici R and Ziegler
M: The NAD metabolome-a key determinant of cancer cell biology. Nat
Rev Cancer. 12:741–752. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chinnadurai G: The transcriptional
corepressor CtBP: A foe of multiple tumor suppressors. Cancer Res.
69:731–734. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Malavasi F, Deaglio S, Funaro A, Ferrero
E, Horenstein AL, Ortolan E, Vaisitti T and Aydin S: Evolution and
function of the ADP ribosyl cyclase/CD38 gene family in physiology
and pathology. Physiol Rev. 88:841–886. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Burkle A and Virag L: Poly(ADP-ribose):
PARadigms and PARadoxes. Mol Aspects Med. 34:1046–1065. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Rodgers JT, Lerin C, Gerhart-Hines Z and
Puigserver P: Metabolic adaptations through the PGC-1 alpha and
SIRT1 pathways. FEBS Lett. 582:46–53. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
van der Horst A, Tertoolen LG, de
Vries-Smits LM, Frye RA, Medema RH and Burgering BM: FOXO4 is
acetylated upon peroxide stress and deacetylated by the longevity
protein hSir2(SIRT1). J Biol Chem. 279:28873–28879. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Bordone L, Motta MC, Picard F, Robinson A,
Jhala US, Apfeld J, McDonagh T, Lemieux M, McBurney M, Szilvasi A,
et al: Sirt1 regulates insulin secretion by repressing UCP2 in
pancreatic beta cells. PLoS Biol. 4:e312006. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Maldi E, Travelli C, Caldarelli A,
Agazzone N, Cintura S, Galli U, Scatolini M, Ostano P, Miglino B,
Chiorino G, et al: Nicotinamide phosphoribosyltransferase (NAMPT)
is over-expressed in melanoma lesions. Pigment Cell Melanoma Res.
26:144–146. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Huang WS, Chen CN, Sze CI and Teng CC:
Visfatin induces stromal cell-derived factor-1 expression by β1
integrin signaling in colorectal cancer cells. J Cell Physiol.
228:1017–1024. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Olesen UH, Hastrup N and Sehested M:
Expression patterns of nicotinamide phosphoribosyltransferase and
nicotinic acid phosphoribosyltransferase in human malignant
lymphomas. APMIS. 119:296–303. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Reddy PS, Umesh S, Thota B, Tandon A,
Pandey P, Hegde AS, Balasubramaniam A, Chandramouli BA, Santosh V,
Rao MR, et al: PBEF1/NAmPRTase/Visfatin: A potential malignant
astrocytoma/glioblastoma serum marker with prognostic value. Cancer
Biol Ther. 7:663–668. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Shai R, Shi T, Kremen TJ, Horvath S, Liau
LM, Cloughesy TF, Mischel PS and Nelson SF: Gene expression
profiling identifies molecular subtypes of gliomas. Oncogene.
22:4918–4923. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Bredel M, Bredel C, Juric D, Harsh GR,
Vogel H, Recht LD and Sikic BI: Functional network analysis reveals
extended gliomagenesis pathway maps and three novel MYC-interacting
genes in human gliomas. Cancer Res. 65:8679–8689. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lee J, Kotliarova S, Kotliarov Y, Li A, Su
Q, Donin NM, Pastorino S, Purow BW, Christopher N, Zhang W, et al:
Tumor stem cells derived from glioblastomas cultured in bFGF and
EGF more closely mirror the phenotype and genotype of primary
tumors than do serum-cultured cell lines. Cancer Cell. 9:391–403.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Sun L, Hui AM, Su Q, Vortmeyer A,
Kotliarov Y, Pastorino S, Passaniti A, Menon J, Walling J, Bailey
R, et al: Neuronal and glioma-derived stem cell factor induces
angiogenesis within the brain. Cancer Cell. 9:287–300. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Murat A, Migliavacca E, Gorlia T, Lambiv
WL, Shay T, Hamou MF, de Tribolet N, Regli L, Wick W, Kouwenhoven
MC, et al: Stem cell-related ‘self-renewal’ signature and high
epidermal growth factor receptor expression associated with
resistance to concomitant chemoradiotherapy in glioblastoma. J Clin
Oncol. 26:3015–3024. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Hufton SE, Moerkerk PT, Brandwijk R, de
Bruine AP, Arends JW and Hoogenboom HR: A profile of differentially
expressed genes in primary colorectal cancer using suppression
subtractive hybridization. Febs Lett. 463:77–82. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Nakajima TE, Yamada Y, Hamano T, Furuta K,
Gotoda T, Katai H, Kato K, Hamaguchi T and Shimada Y: Adipocytokine
levels in gastric cancer patients: Resistin and visfatin as
biomarkers of gastric cancer. J Gastroenterol. 44:685–690. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Nakajima TE, Yamada Y, Hamano T, Furuta K,
Matsuda T, Fujita S, Kato K, Hamaguchi T and Shimada Y:
Adipocytokines as new promising markers of colorectal tumors:
Adiponectin for colorectal adenoma, and resistin and visfatin for
colorectal cancer. Cancer Sci. 101:1286–1291. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Gujar AD, Le S, Mao DD, Dadey DY, Turski
A, Sasaki Y, Aum D, Luo J, Dahiya S, Yuan L, et al: An
NAD+-dependent transcriptional program governs
self-renewal and radiation resistance in glioblastoma. Proc Natl
Acad Sci USA. 113:E8247–E8256. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Lee YC, Yang YH, Su JH, Chang HL, Hou MF
and Yuan SS: High visfatin expression in breast cancer tissue is
associated with poor survival. Cancer Epidemiol Biomarkers Prev.
20:1892–1901. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Bi TQ, Che XM, Liao XH, Zhang DJ, Long HL,
Li HJ and Zhao W: Overexpression of Nampt in gastric cancer and
chemopotentiating effects of the Nampt inhibitor FK866 in
combination with fluorouracil. Oncol Rep. 26:1251–1257.
2011.PubMed/NCBI
|
|
32
|
Shackelford RE, Bui MM, Coppola D and
Hakam A: Over-expression of nicotinamide phosphoribosyltransferase
in ovarian cancers. Int J Clin Exp Pathol. 3:522–527.
2010.PubMed/NCBI
|
|
33
|
Wang B, Hasan MK, Alvarado E, Yuan H, Wu H
and Chen WY: NAMPT overexpression in prostate cancer and its
contribution to tumor cell survival and stress response. Oncogene.
30:907–921. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Yang H, Yang T, Baur JA, Perez E, Matsui
T, Carmona JJ, Lamming DW, Souza-Pinto NC, Bohr VA, Rosenzweig A,
et al: Nutrient-sensitive mitochondrial NAD+ levels
dictate cell survival. Cell. 130:1095–1107. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Jung J, Kim LJ, Wang X, Wu Q, Sanvoranart
T, Hubert CG, Prager BC, Wallace LC, Jin X, Mack SC and Rich JN:
Nicotinamide metabolism regulates glioblastoma stem cell
maintenance. JCI insight. 2(pii): 900192017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Venkateshaiah SU, Khan S, Ling W, Bam R,
Li X, van Rhee F, Usmani S, Barlogie B, Epstein J and Yaccoby S:
NAMPT/PBEF1 enzymatic activity is indispensable for myeloma cell
growth and osteoclast activity. Exp Hematol. 41:547–557.e2. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Revollo JR, Grimm AA and Imai S: The
regulation of nicotinamide adenine dinucleotide biosynthesis by
Nampt/PBEF/visfatin in mammals. Curr Opin Gastroenterol.
23:164–170. 2007. View Article : Google Scholar : PubMed/NCBI
|