Introduction
Src family kinases (SFKs) comprise the largest
family of non-receptor tyrosine kinases (1). Activated SFKs initiate numerous
signaling pathways; under the control of extracellular stimuli,
these pathways serve important roles in various cellular functions,
including proliferation, migration and survival (2). SFK activation results in the
phosphorylation of Tyr416 in the kinase domain (3), and SFK activity is regulated by a
phosphotyrosine ligand with a higher affinity for the SH2 domain
(1). Studies have demonstrated
increased SFK activity in a number of human cancers (4), thus the analysis of SFK activation is
important to understand tumor progression.
CUB domain-containing protein 1 (CDCP1) is a type I
transmembrane protein, also known as SIMA135 (5), gp140 (6)
and Trask (7). Initially, CDCP1 was
reported as being predominantly expressed in colorectal cancers
(8). However, in an investigation of
the phosphoprotein associated with anchorage independence of lung
adenocarcinoma cells, it was observed that CDCP1 saved a principal
role as the SH2 domain-binding phosphoprotein of SFK in anoikis
inhibition of metastatic tumor cells (9). SFK activation results in the
phosphorylation of tyrosine residues in CDCP1; when this occurs in
the intracellular domain, protein kinase C δ (PKCδ) is recruited to
the plasma membrane, where it subsequently undergoes specific
phosphorylation by SFK at Tyr311 (9–11). This
SFK-CDCP1-PKCδ signaling is involved in the regulation of anoikis
resistance (9), cell migration
(12), extracellular matrix
degradation (13) and invasion
(14).
CDCP1 possesses three complement C1r/C1s, urchin
embryonic growth factor bone morphogenetic protein 1 (CUB) domains
in its extracellular domain (ECD), which may be involved in
protein-protein interactions (15–17).
Therefore, CDCP1 may form a homophilic complex at the plasma
membrane through its ECD. CUB1, the domain located furthest from
the transmembrane domain, is cleaved and released, and has been
detected in the urine of patients with cancer (18,19). CDCP1
cleavage in the ECD induces cell migration in triple-negative
breast cancer cells (18). These
findings demonstrated that the extracellular CUB domains of CDCP1
potentially regulate tumor cell migration by promoting CDCP1
signaling; however, the functional mechanism of this activation is
not yet understood. The present study investigated how CUB domains
stimulate CDCP1 signaling. Utilizing recombinant proteins, it was
demonstrated for the first time that the extracellular CUB2 domain
of CDCP1 is the formation site of the CDCP1 homophilic complex.
This provides novel information on the role of the CUB2 domain in
regulating intracellular CDCP1 signaling.
Materials and methods
Antibodies
Anti-HA (Y-11; cat. no. sc-805; 1:300),
anti-HA-horseradish peroxidase (HRP)-conjugated (Y-11 HRP; cat. no.
sc-805 HRP; 1:500), anti-c-Src (SRC2; cat. no. sc-18; 1:4,000) and
anti-PKCδ (C-20; cat. no. sc-937; 1:4,000) antibodies were
purchased from Santa Cruz Biotechnology. Inc. Anti-FLAG M2
fluorescein isothiocyanate-conjugated (cat. no. F4049; 1:600),
anti-FLAG M2 peroxidase-conjugated specific (cat. no. A8592;
1:4,000) and anti-α tubulin (cat. no. T5168; 1:10,000) antibodies
were purchased from Sigma-Aldrich (Merck KGaA). An
anti-maltose-binding protein (MBP) antibody (cat. no. E8032
1:10,000) was purchased from New England BioLabs, Inc.
Anti-phospho-Src family (Tyr416, and 2101; 1:2,000) and
anti-phospho-PKCδ (Tyr311; cat. no. 2055; 1:2,000)
antibodies were purchased from Cell Signaling Technology, Inc.
HRP-conjugated anti-mouse IgG (cat. no. NA931V; 1:4,000) and
anti-rabbit IgG (cat. no. NA934V; 1:4,000) antibodies were
purchased from GE Healthcare. Alexa Fluor 488 goat anti-mouse (cat.
no. A11011; 1:800) and Alexa Fluor 546 goat anti-rabbit (cat. no.
A11010; 1:800) antibodies were purchased from Thermo Fisher
Scientific, Inc. Rabbit polyclonal anti-CDCP1 antibody was prepared
as described previously (9).
Expression plasmids
pcDNA3.1 expression plasmids (Thermo Fisher
Scientific, Inc.,) encoding human CDCP1 with a C-terminal FLAG or
HA tag, and the CDCP1 rescue sequence that introduces silent
mutations that are not suppressed by CDCP1 siRNA (CDCP1res-F and
CDCP1res-HA, respectively), have been described previously
(13). The ECD-deleted mutant plasmid
with C-terminal FLAG tag (∆ECD-F) was generated by inverse PCR
using the KOD-Plus-Mutagenesis kit (Toyobo Life science) per the
manufacturer's protocol.
A system for the stable, siRNA-induced suppression
of CDCP1 expression was constructed using the Block IT Pol II miRNA
Expression Vector kit (Invitrogen; Thermo Fisher Scientific, Inc.,)
according to the manufacturer's instructions. The following primers
were used to generate the CDCP1 miR RNAi plasmid (miCDCP1):
Forward,
5′-TGCTGAATGTTGCTTTCTCGTGGCAGGTTTTGGCCACTGACTGACCTGCCACGAAAGCAAGATT-3′,
and reverse
5′-CCTGAATGTTGCTTTCGTGGCAGGTCAGTCAGTGGCCAAAACCTGCCACGAGAAAGCAACATTC-3′.
The LacZ miR RNAi plasmid was selected as the negative control, and
was constructed using the following primers: Forward,
5′-TGCTGAAATCGCTGATTTGTGTAGTCGTTTTGGCCACTGACTGACGACTACACATCAGCGATTT-3′,
and reverse,
5′-CCTGAAATCGCTGATGTGTAGTCGTCAGTCAGTGGCCAAAACGACTACACAAATCAGCGATTTC-3′.
Cell culture and transfection
Human lung adenocarcinoma A549 and human pancreatic
cancer BxPC3 cell lines were cultured in RPMI 1640 (Sigma-Aldrich;
Merck KGaA) supplemented with 10% fetal bovine serum (FBS; Gibco;
Thermo Fisher Scientific, Inc.) at 37°C, 5% CO2. A549
miCDCP1 cells with suppressed CDCP1 expression, and A549 miLacZ
cells (the control cells from the Block IT Pol II miRNA Expression
Vector kit) were used as described previously (9).
For transfection, cells were seeded at
5×106 cells/10-cm dish or 1×105 cells/well of
a 24-well plate, prior to a 24-h incubation period. The cells were
transfected with the expression plasmids (CDCP1res-F, 5 mg;
CDCP1res-HA, 5 µg; and ∆ECD-F, 1 µg) using Lipofectamine™ 2000
(Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's instructions. The transfected cells were selected in
the presence of 800 µg/ml G418 sulfate (EMD/Merck KGaA) for one
month prior to experimentation.
Immunofluorescence staining
For immunostaining, cells cultured on cover glasses
were washed with phosphate-buffered saline (PBS) at 37°C and fixed
with 4% paraformaldehyde in PBS for 10 min at room temperature. The
cells were washed again with PBS and permeabilized with 0.25%
Triton X-100 in PBS. The cells were the blocked with Blocking One
(Nacalai Tesque, Inc.,) for 40 min, and co-localization was
analyzed using a confocal laser-scanning microscope (FLUOVIEW
FV10i) and the FV10-ASW software ver. 4.1 ‘Co-Localization’
(Olympus Corporation). At a certain threshold, the FV10-ASW
software converts the pixels of the area co-stained with two
fluorescent substances into a white dot; the ratio of white dot
pixels to total pixels is then determined, and the pixel rate
corresponds to the co-localization area value.
Western blotting and
immunoprecipitation
Cell lysates were prepared in PLC buffer [10 mM
Tris-HCl (pH 7.5), 5 mM EGTA, 150 mM NaCl, 1% Triton X-100, 10%
glycerol and 1 mM sodium orthovanadate]. The lysates were
centrifuged at 20,630 × g for 20 min at 4°C, and the supernatants
were collected. Protein concentrations were measured using a BCA
Protein Assay kit (Pierce; Thermo Fisher Scientific, Inc.).
For immunoprecipitation, the lysates (1 µg/µl; 350
µl) were incubated with anti-FLAG M2 affinity gel (20 µl) or
anti-HA antibody conjugated with Protein G Sepharose 4 Fast Flow
gel (30 µl) on ice for 1 h. The anti-FLAG M2-protein or
anti-HA-protein complexes were then harvested and washed three
times with PLC lysis buffer. The total amount of each
immunoprecipitated sample was separated by 8% SDS-PAGE gel and
visualized using western blotting.
For western blotting, the samples were transferred
to polyvinylidene difluoride (PVDF) membranes (Immobilon-P; EMD
Millipore). After blocking with blocking buffer (Blocking One;
Nacalai Tesque) the membranes were probed with primary antibodies
(anti-CDCP1, anti-α tubulin, anti-FLAG M2 peroxidase-conjugated,
anti-HA, anti-HA-HRP-conjugated, anti-MBP, anti-c-Src,
Anti-phospho-Src family, anti-PKCδ and anti-phospho-PKCδ) at room
temperature for 1 h to detect the indicated proteins. After washing
three times with TBS-T (0.05% Tween-20 in TBS), the membranes were
further probed with the corresponding secondary antibodies at room
temperature for 30 min. The blots were visualized using the Western
Lightning Plus-ECL kit (PerkinElmer, Inc.) and images were captured
with the myECL imager (Thermo Fisher Scientific, Inc.). The
proteins were subsequently quantified using ImageJ software,
version 1.50i (National Institutes of Health).
Scratch wound-healing assay
To investigate the wound-healing effect, A549
miCDCP1 cells transfected with CDCP1res-F, ∆ECD-F or mock
(pcDNA3.1) plasmids were seeded in RPMI (10% FBS) at
1×106 cells/well in 6-well, and incubated until
confluent. Each monolayer was scratched to create a wound of ~600
µm. After washing three times with PBS to remove cell debris, 5 ml
RPMI (10% FBS) was added to each well. Images were captured by
phase-contrast microscopy (BZ-X 710; Keyence Corporation) at 0 and
12 h for BxPC3 cells, and after 24 h for A549 cells. Wound recovery
was measured with the BZ-X analyzer. Cell migration was evaluated
as the mean of the migration length in three independent
experiments. BxPC3 cell migration was analyzed in the same manner,
with the only difference being the timing of image capture (at 0
and 12 h).
MBP protein expression and far-western
blotting
To create MBP constructs, the amplified CUB2 and
CUB3 domain sequences of CDCP1 were inserted into the
pMAL-c5× expression vector (New England BioLabs, Inc.). PCR
was performed using the following primers: rMBP-CUB2 forward,
5′-CGCATATGTGCACAGACCACCGGTACTGC-3′, and reverse,
5′-GCGAATTCAACGCCTTCCTCTTTGAAATAAG-3′; and rMBP-CUB3 forward,
5′-GCCATATGGAGGAAGGCGTTTTCACGGTGAC-3′, and reverse,
5′-CGGAATTCTGGGGTAAGTGTCACCGAGAAGAG-3′. Protein expression and
affinity purification with amylose resin were performed in
accordance with the manufacturer's protocol.
For far-western blotting, each rMBP protein (3 µg)
was blotted onto a PVDF membrane. After blocking with blocking
buffer for 1 h, each cell lysate expressing CDCP1res-F or ΔECD-F
(10 µg/ml with TBS-Tween20) was incubated with the membrane for 1
h. After washing three times with TBS-Tween 20, the membrane was
used for western blotting.
Statistical analysis
All data are presented as the mean ± standard
deviation; ANOVA followed by Tukey's test was used for multiple
comparisons among sample groups. P<0.05 was considered to
indicate a statistically significant difference.
Results
CDCP1 forms a homophilic complex via
the ECD
A previous study indicated that CDCP1 was capable of
forming dimers within the cell (20).
To verify this phenomenon, FLAG-tagged CDCP1 (CDCP1res-F) and
HA-tagged CDCP1 (CDCP1res-HA) (Fig.
1A) were co-expressed in A549 miCDCP1 cells, where endogenous
CDCP1 expression was stably suppressed (26.1±4.0%; Fig. 1B). CDCP1res-F was immunoprecipitated
with a anti-FLAG M2 antibody. As shown in Fig. 1C, CDCP1res-HA was
co-immunoprecipitated with CDCP1res-F (lane 5), and the same result
was observed by immunoprecipitation with the anti-HA antibody (lane
8). This indicated that differentially tagged CDCP1 molecules
formed a complex with each other. To determine whether the ECD of
CDCP1 was important for complex formation, ∆ECD-F (which still
contained the N-terminal signal sequence and is expressed at the
cell membrane) was co-expressed with CDCP1res-HA (Fig. 1A) and subjected to immunoprecipitation
and western blotting with anti-FLAG M2 and anti-HA antibodies.
CDCP1res-HA was associated with CDCP1res-F; however, by comparison,
less association was detected with CDCP1res-HA and ∆ECD-F (Fig. 1C, lanes 6 and 9). In addition, two
control IgG antibodies did not immunoprecipitate with CDCP1res-F
and CDCP1res-HA (Fig. 1C, lanes 11
and 14).
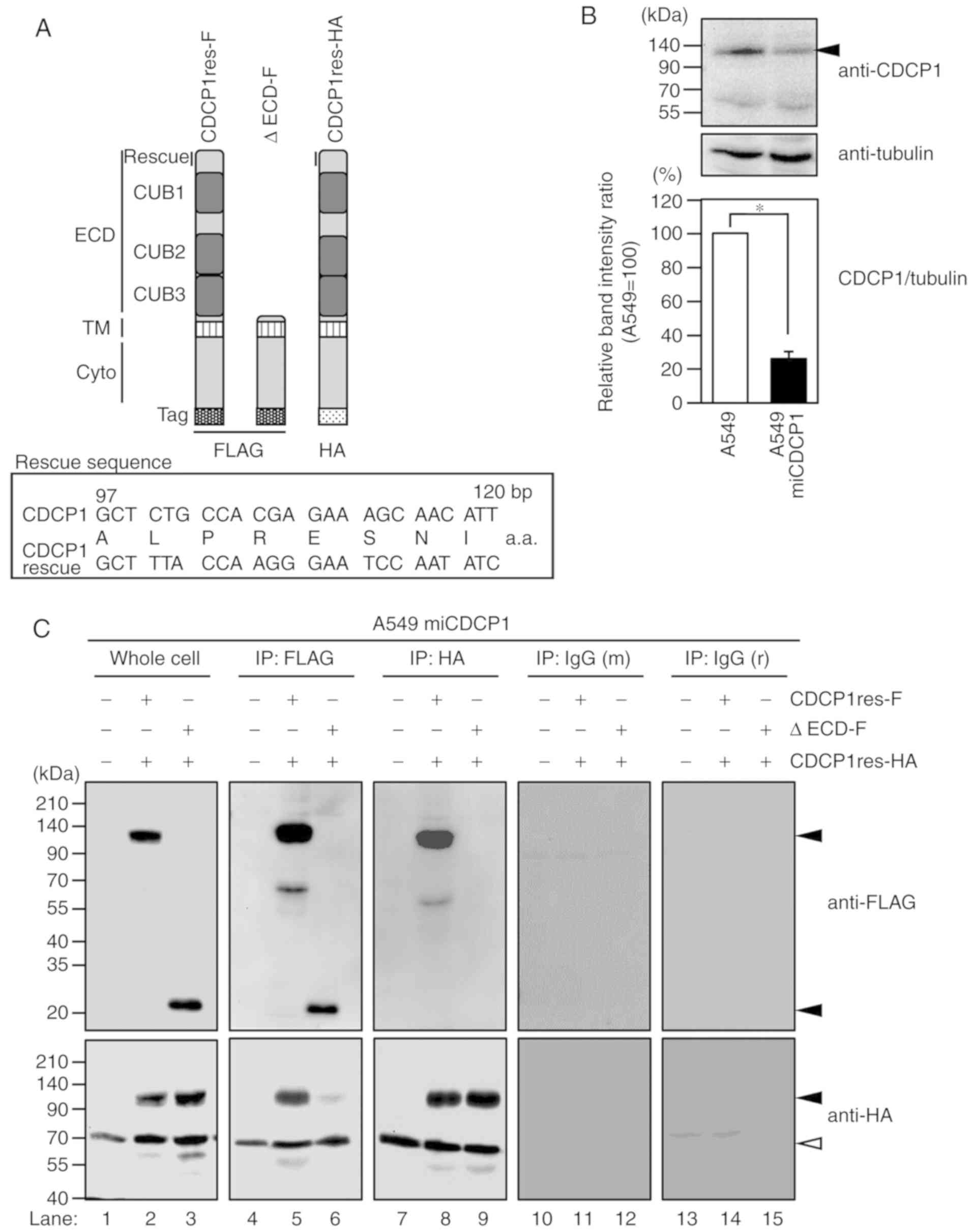 | Figure 1.CDCP1 forms a homophilic complex via
its ECD. (A) Schematic of the CDCP1 variants. CDCP1res-F and
CDCP1res-HA (135 kDa) include the CUB domain in the ECD. ΔECD-F (20
kDa) lacks the ECD, but contains the signal sequence. The FLAG or
HA tag is attached to the C-terminus. Rescue, target CDCP1 DNA
sequence and the rescue mutant sequence that is not suppressed by
CDCP1 small interfering RNA (13).
(B) Stable suppression of CDCP1 in A549 miCDCP1 cells. Cells were
cultured in RPMI 1640 with 800 µg/ml G418 sulfate. Cell lysates
were subjected to western blotting using an anti-CDCP1 antibody.
Tubulin served as an internal control. The suppression ratio is
shown in the bar graph. Mean ± standard deviation; n=3 for each
bar; *P<0.05, tukey's test. (C) A549 miCDCP1 cell lysates
co-expressing either CDCP1res-F and CDCP1res-HA or ΔECD-F and
CDCP1res-HA were immunoprecipitated with anti-FLAG M2, anti-HA,
control IgG (m) or IgG (r) antibodies. Whole cell lysates and
precipitates were subjected to immunoblotting with anti-FLAG and
anti-HA antibodies. Black arrowheads indicate CDCP1 variants. White
arrowheads indicate nonspecific bands. CDCP1, CUB domain-containing
protein; ECD, extracellular domain; CUB, complement C1r/C1s, urchin
embryonic growth factor, bone morphogenetic protein 1; TM,
transmembrane domain; Cyto, cytoplasmic domain; bp, base pair;
a.a., amino acids; IgG (m), control mouse IgG; IgG (r), control
rabbit IgG; -F, FLAG-tagged; -HA, HA-tagged. |
CDCP1 homophilic complex formation on cells was also
investigated. CDCP1res-HA and either CDCP1res-F or ΔECD-F was
co-expressed in A549 miCDCP1 cells, and localization was examined
by immunostaining. CDCP1res-F and CDCP1res-HA were detected on the
plasma membrane and ruffling edge (Fig.
2A, inserts a, b, and f). CDCP1res-F and CDCP1res-HA were also
primarily co-localized at the plasma membrane and the periphery of
cells (Fig. 2A, insert d). Although
ΔECD-F was expressed around the plasma membrane (Fig. 2A, insert e), it was localized mainly
at a region of the cell surface that was different from the region
at which CDCP1res-HA was localized (Fig.
2A, insert h). To monitor the level of co-localization of CDCP1
variants at the cell surface, the co-localization areas were
quantified. The co-localization of each CDCP1 variant on the cell
membrane is shown as the area of white dots detected by FV10-ASW
software (Fig. 2B, inserts a and b).
The co-localization area of CDCP1res-F and CDCP1res-HA (6.89+1.95%)
was greater than that of ΔECD-F and CDCP1res-HA (1.63+0.76%)
(Fig. 2B). Taken together, these
results suggested that the ECD is required for CDCP1 homophilic
complex formation at the cell surface.
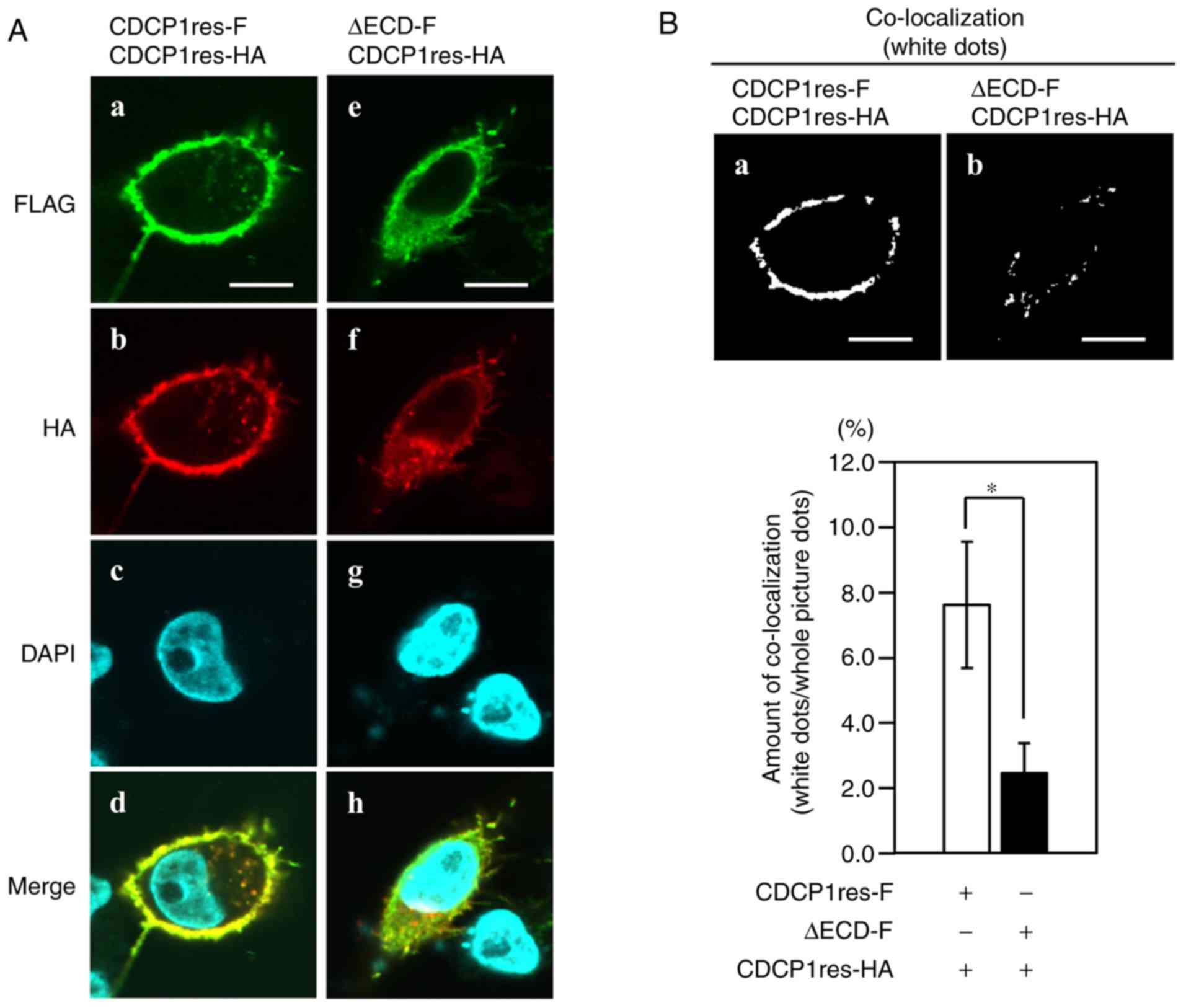 | Figure 2.Localization of CDCP1 variants in
A549 miCDCP1 cells. (A) CDCP1 variants were either FLAG or
HA-tagged. Nuclei were stained with DAPI and the indicated
antibodies: A and e, anti-FLAG (green); b and f, anti-HA (red); c
and g, DAPI (blue); d and h, merge. Images were captured using a
confocal microscope (magnification, ×60). Scale bar=10 µm. (B)
Co-localization is indicated by white dots (a, b, co-localization)
and the co-localization ratio was evaluated using FV10-ASW
software. Scale bar=10 µm. Data are presented as the mean ±
standard deviation; n=3 for each bar; *P<0.05, tukey's test.
CDCP1, CUB domain-containing protein 1; ECD, extracellular domain;
-F, FLAG-tagged; -HA, HA-tagged. |
CDCP1 ECD regulates lung cancer cell
migration
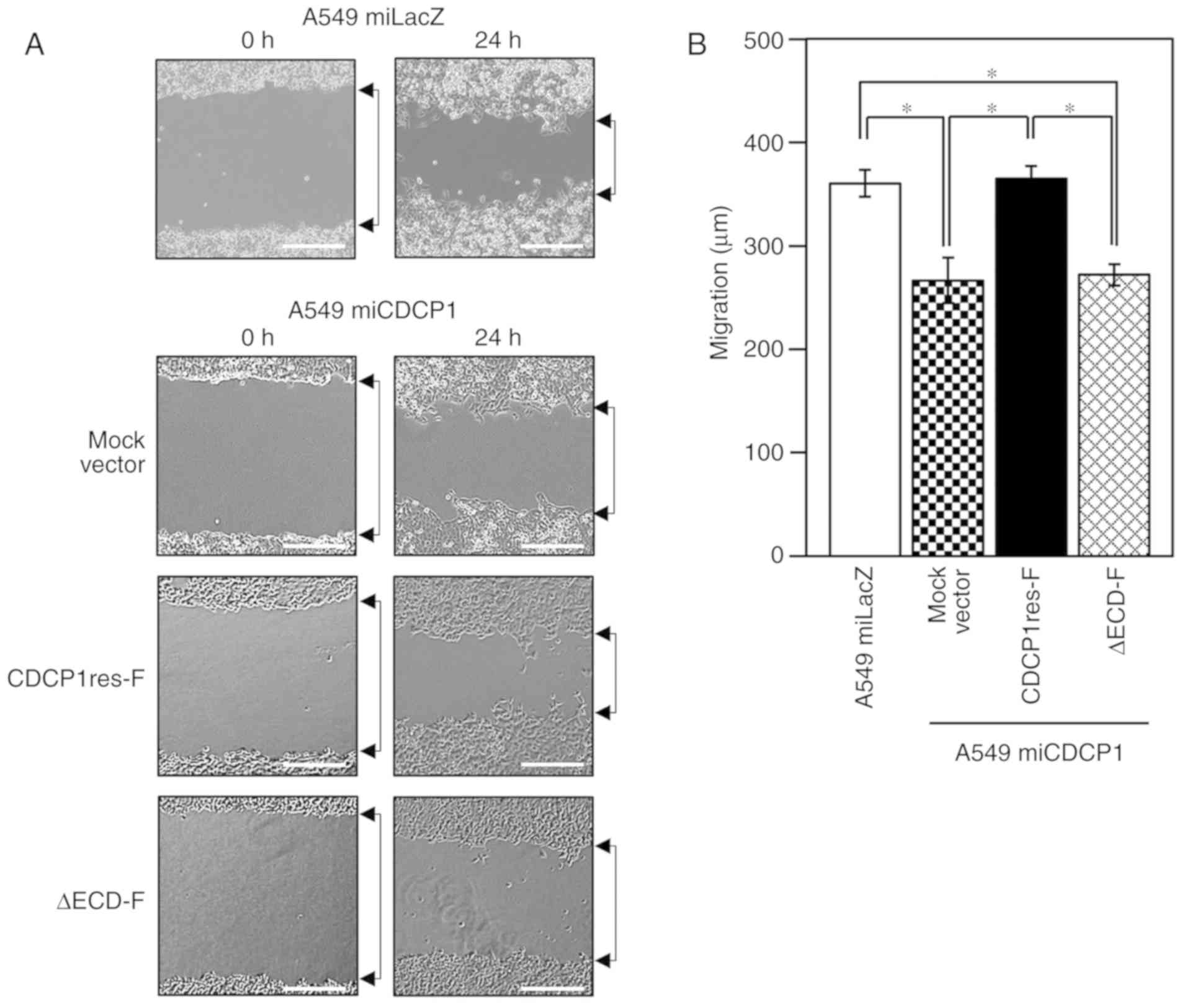
Previous reports have suggested that CDCP1 is
important for the regulation of cancer cell migration (13). It was thus hypothesized that CDCP1
formed a homophilic complex via the ECD on the cell surface to
regulate cell migration. A scratch wound-healing assay was used to
assess the significance of the CDCP1 ECD in cancer cell migration
(Fig. 3A). Cells with CDCP1
suppression exhibited significantly reduced migratory properties,
compared with CDCP1-expressing cells (Fig. 3B, miLacZ vs. miCDCP1 Mock vector), as
supported by a previous report (13).
For the rescue experiments, CDCP1res-F and ∆ECD-F vectors were
transfected into A549 miCDCP1 cells. Cell migration was recovered
by the CDCP1res-F vector, but not by the ∆ECD-F mutant vector
(Fig. 3B). This result demonstrated
that the CDCP1 ECD is required for cancer cell migration.
CDCP1 forms a homophilic complex via
the CUB2 domain
Given the reported importance of CUB1 domain release
in CDCP1 dimerization (18,21), it was hypothesized that the
extracellular CUB2 and/or CUB3 domains of CDCP1 served a role in
the regulation of homophilic complex formation. To test this
hypothesis, recombinant CUB2 or CUB3 domains were fused with the
MBP (rMBP; rMBP-CUB2 and rMBP-CUB3, respectively) (Fig. 4A). The rMBP fusion proteins (3 µg
each) were immobilized onto a PVDF membrane (Fig. 4B, rMBP proteins) and blotted with A549
miCDCP1 cell lysates expressing CDCP1res-F or ΔECD-F. Each protein
was visualized using an anti-FLAG M2 antibody. The results showed
that CDCP1res-F was able to bind rMBP-CUB2; however, it was
minimally bound to rMBP-CUB3 (Fig.
4B, CDCP1res-F). By contrast, ΔECD-F did not bind to either
rMBP fusion protein (Fig. 4B,
ΔECD-F), and neither CDCP1 variant bound to rMBP (Fig. 4B, CDCP1res-F and ΔECD-F). To confirm
the interaction between CDCP1 and purified recombinant CUB
proteins, each CUB protein was added to the culture medium of A549
miCDCP1 cells expressing CDCP1res-HA, and the binding capacity of
the CUB products was determined using immunostaining. CDCP1res-HA
localization was detected at the plasma membrane and the ruffling
edge, with minimal differences in CDCP1-HA expression levels
between samples (Fig. 4C, inserts a,
e and i). rMBP-CUB2 was co-localized with CDCP1res-HA at the plasma
membrane (Fig. 4C, insert g), but
rMBP and rMBP-CUB3 rarely co-localized (Fig. 4C, inserts c and k). The
co-localization of each rMBP fusion protein with CDCP1res-HA was
determined using FV10-ASW software, and is indicated by the white
dotted region (Fig. 4C, inserts d, h
and l). The co-localization area of rMBP-CUB2 with CDCP1res-HA
(3.32±0.46%) was greater than that of rMBP or rMBP-CUB3 with
CDCP1res-HA (0.17±0.08 and 0.78±0.39%, respectively) (Fig. 4D). These results indicated that
rMBP-CUB2 was the primary binding partner of CDCP1.
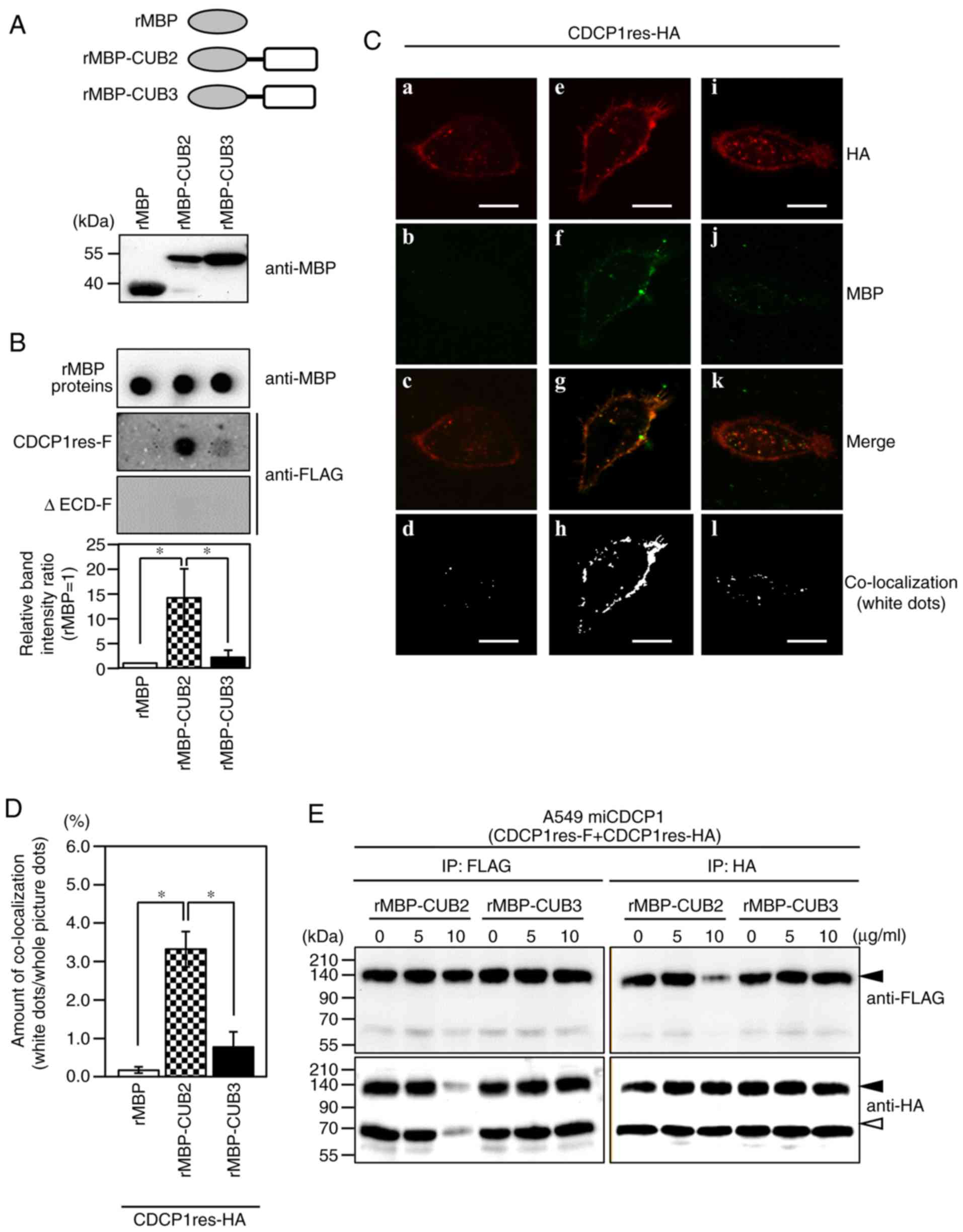 | Figure 4.The CUB2 domain is a binding site for
the CDCP1 homophilic complex. (A) Upper panel: Schematic structure
of the CUB domain fused with MBP. Lower panel: Expression of each
MBP construct was confirmed by western blotting using an anti-MBP
antibody. (B) Far-western blotting indicating the association
between MBP constructs and CDCP1 variant proteins. MBP constructs
were detected using an anti-MBP antibody (rMBP proteins), and each
CDCP1 variant was layered onto the dot-blotted rMBP proteins. The
association between rMBP proteins and each CDCP1 variant
(CDCP1res-F and ΔECD-F) was detected by western blotting with an
anti-FLAG antibody, and the association ratio is shown in the bar
graph. Mean ± standard deviation; n=3 for each bar; *P<0.05,
tukey's test. (C) A549 miCDCP1 cells were transfected with
CDCP1res-HA and incubated with each of the indicated rMBP proteins
(rMBP, rMBP-CUB2, and rMBP-CUB3). After 24 h, the cells were
immunostained with anti-HA (a, e, and i: Red) and anti-MBP (b, f,
and j: Green) antibodies. The merged image is shown in c, g, and k.
Co-localization is indicated by white dots, and was determined
using tFV10-ASW software (d, h, and l). Scale bar=10 µm. (D) The
co-localization ratio was also evaluated using FV10-ASW software;
mean ± standard deviation; n=3 for each bar; *P<0.05, tukey's
test. (E) A549 miCDCP1 cells-6 h post-transfection with CDCP1res-F
and CDCP1res-HA, the medium was changed and 0, 5, or 10 µg/ml
rMBP-CUB2 or rMBP-CUB3 was added; 24 h post-transfection, cell
lysates were harvested and immunoprecipitated with anti-FLAG M2 and
anti-HA antibodies. Precipitates were subjected to immunoblotting
with anti-FLAG and anti-HA antibodies. The black arrowhead
indicates CDCP1, and the white arrowhead indicates nonspecific
bands. CDCP1, CUB domain-containing protein 1; rMBP, recombinant
maltose binding protein; ECD, extracellular domain; -F,
FLAG-tagged; -HA, HA-tagged. |
The potential rMBP-CUB2-induced inhibition of CDCP1
homophilic complex formation was also investigated. A549 miCDCP1
cells were transfected with CDCP1res-F and CDCP1res-HA expression
vectors, and rMBP-CUB2 or rMBP-CUB3 proteins were added at the
indicated concentrations (0, 5 and 10 µg/ml). No significant
difference in the CDCP1 expression level was observed in these cell
lines using rMBP-CUB proteins (Fig.
S1A). Immunoprecipitation of A549 miCDCP1 cells with anti-FLAG
and anti-HA antibodies showed the suppression of CDCP1 homophilic
complex formation on addition of the CUB2 (10 µg/ml), but not the
CUB3 domain (Fig. 4E). The two
control IgG antibodies did not co-precipitate CDCP1res-FLAG or
CDCP1res-HA (Fig. S1B). These
results suggested that the extracellular CUB2 domain of CDCP1
regulates homophilic complex formation at the plasma membrane.
The CUB2 domain of CDCP1 regulates
cancer cell migration via SFK activation
Since rMBP-CUB2 effectively inhibited CDCP1
homophilic complex formation, the effect of rMBP-CUB2 on
intracellular SFK-CDCP1-PKCδ signaling in parental A549 cells was
investigated. Neither rMBP protein affected cell survival (Fig. S2), though SFK activation was reduced
in A549 miCDCP1 cells, whose CDCP1 expression was suppressed
(Fig. 5A). Notably, Tyr416
(Y416) phosphorylation of SFK, which is an indicator of SFK
activation, was suppressed following the addition of rMBP-CUB2
(Fig. 5B; SFK, lane 3), but not that
of rMBP or rMBP-CUB3 (Fig. 5B, SFK,
lanes 2 and 4). In addition, phosphorylation of PKCδ, a downstream
signal molecule of SFK, was reduced by the addition of rMBP-CUB2
(Fig. 5B PKCδ lane 3), but not rMBP
or rMBP-CUB3 (Fig. 5B, PKCδ, lanes 2
and 4). Comparable results were observed in the pancreatic cancer
cell line BxPC3 (Fig. S3).
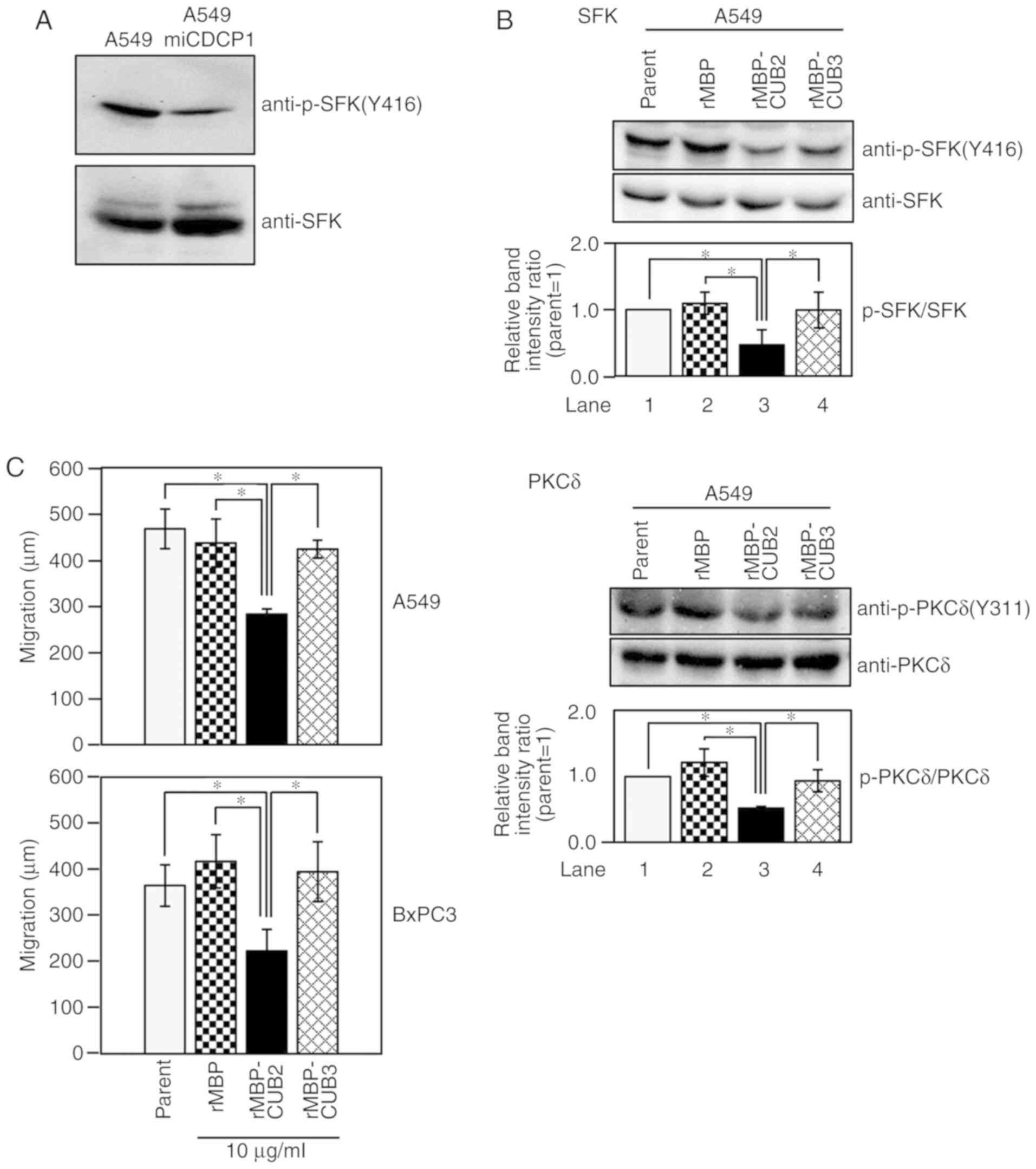 | Figure 5.CDCP1 CUB2 domain attenuates SFK
activation and inhibits cancer cell migration. (A) Reduced CDCP1
expression attenuates SFK activation. A549 and A549 miCDCP1 cells
(1×106 each) were cultured in RPMI for 24 h. Cell
lysates were subjected to western blotting using anti-p-SFK(Y416)
and anti-SFK antibodies. (B) Effects of each rMBP fusion protein
(rMBP, rMBP-CUB2 and rMBP-CUB3) on CDCP1 signaling in A549 cells.
Cells were cultured with each rMBP protein for 24 h. Lysates were
subjected to immunoblotting with the indicated antibodies:
Anti-p-SFK (Y416), anti-SFK, anti-p-PKCδ (Y311) and anti-PKCδ.
Adding rMBP-CUB2 alone inhibits SFK and PKCδ phosphorylation.
Relative intensity ratios are shown; mean ± standard deviation; n=3
for each cell; *P<0.05, tukey's test. (C) Effects of the CUB2
domain on wound-healing in A549 and BxPC3 cells. After scratching
the cells and exchanging the medium, 10 µg/ml MBP proteins were
added, and images were immediately captured (0 h). The scratched
cells were incubated for 12 (BxPC3 cells) or 24 h (A549 cells) in
5% CO2, and images were captured (Fig. S4). Wound widths were measured, and
migration distances were calculated by subtracting the width at 12
or 24 h from that at 0 h. Mean ± standard deviation; n=3 for each
cell line; *P<0.05, tukey's test. CUB, complement C1r/C1s,
urchin embryonic growth factor, bone morphogenetic protein 1; SFK,
Src family kinase; CDCP1, CUB domain-containing protein 1; p-,
-phosphorylated; rMBP, recombinant maltose binding protein; PKCδ,
protein kinase C δ. |
Finally, the effect of rMBP-CUB2 on cell migration
was examined using a scratch wound-healing assay (Fig. S4). Cell migration was reduced by
treatment with rMBP-CUB2 in both the A549 and BxPC3 cell lines; by
contrast, rMBP and rMBP-CUB3 did not affect cell migration
(Fig. 5C). These results indicated
that the extracellular CUB2 domain of CDCP1 regulates cell
migration by promoting SFK activation.
Discussion
In the present study, the formation of a CDCP1
homophilic complex via the CUB2 domain was demonstrated at the cell
surface. This formation allows for the tighter molecular
arrangement of signaling molecules at the cell surface. CDCP1
mediates the phosphorylation of PKCδ by active SFK, thus, it is
conceivable that one of the CDCP1 molecules in the complex acts as
an active SFK receptor, and that the other molecule serves as a
PKCδ receptor. In the present study, co-expression of CDCP1res-F
with CDCP1res-HA demonstrated that the formation of the CDCP1
complex (Fig. 1) may generate an
intracellular signal (Fig. 5B, lane
1). Previous reports have indicated that cleavage between the CUB1
and CUB2 domains of the CDCP1 ECD is important for dimerization,
and that the CUB2 domain may be a key region for homophilic complex
formation (18,21). In the present study, rMBP-CUB2
prevented CDCP1 homophilic complex formation (Fig. 4E) and decreased PKCδ phosphorylation
(Fig. 5B, PKCδ lane 3); therefore,
the interaction between CDCP1 molecules via the CUB2 domain is most
likely a mechanism for initiating SFK-CDCP1-PKCδ signaling by
clustering the receptor for active SFK and PKCδ at the cell
membrane. In addition, Hooper et al (5) revealed that CDCP1 possessed 14 putative
N-glycosylation sites. The released CUB1 region of CDCP1 (30–368
residues) has a greater number of putative glycosylation sites than
the remaining ECD (Fig. S5). Thus,
it is speculated that the CUB1-containing region possessing these
glycosylation sites may be a regulator for SFK-CDCP1-PKCδ
signaling.
SFK activation is important for SFK-CDCP1-PKCδ
signaling (9). CDCP1 phosphorylation
is increased in metastatic cancer cells; however, the mechanism of
enhanced phosphorylation is not clear. Substrates such as p130Cas
and Ossa can activate SFK by associating with their phosphorylated
regulatory domains (22,23). CDCP1 can also activate SFK, possibly
via a similar mechanism (24). In the
present study, rMBP-CUB2 prevented CDCP1 homophilic complex
formation (Fig. 4E) and SFK
activation in cancer cells (Fig. 5B
SFK lane 3). Therefore, CDCP1 accumulation at the cell membrane via
the essential CUB2 domain may be required for SFK activation. The
CUB2-mediated interaction of CDCP1 most likely promotes the
activation of SFK by clustering inactive SFK molecules and CDCP1
within the cell membrane. It is not clear whether dimer formation
is sufficient for effective signal transduction, or if
oligomerization would enhance signaling activity. Furthermore,
since SFK may be activated by the homophilic complex of
phosphorylated CDCP1, analysis of the initial activation mechanism
of SFK is required. Nonetheless, the CUB2 domain interaction
between CDCP1 molecules appears to be critical for SFK
activation.
In the present study, a regulatory mechanism of
CDCP1 in cancer cell migration was revealed. Consistent with a
previous report by Miyazawa et al (13), the CDCP1 ECD was revealed to regulate
cell migration (Fig. 3). Furthermore,
a positive correlation was observed between CDCP1 complex formation
via the CUB2 domain and cell migration in different cancer cell
lines (Figs. 5, S3 and S4).
SFK-mediated CDCP1 phosphorylation is a requirement for cell
migration (13). Thus, the mechanisms
of CDCP1 homophilic complex formation-induced SFK activation via
CUB2 may regulate cell migration mediated by intracellular
SFK-CDCP1-PKCδ signaling.
The extracellular CUB domain is thought to mediate
interactions between various proteins (15). A number of integrins have been
reported to influence CDCP1 phosphorylation (25). Moreover, a possible association
between N-cadherin (which is involved in epithelial-mesenchymal
transition) and CDCP1 has also been reported (26). In addition to these previous findings,
the present study delineated a role for the CUB2 domain in CDCP1
homophilic complex formation, and in the regulation of cell
migration. Therefore, the CUB2 domain may be broadly involved in
molecular interactions that mediate biological functions such as
cell migration.
Kollmorgen et al (20) suggested that CDCP1 may form homophilic
dimer complexes via its transmembrane and cytoplasmic domains. In
the present study, the interaction between CDCP1res-HA and ΔECD-F
was observed (Fig. 1); however, the
motility of cancer and A549 miCDCP1 cells was markedly reduced
(Fig. 3). This suggests that dimer
formation by transmembrane and cytoplasmic domains may regulate
cancer cell functions other than migration. Therefore, further
studies with CDCP1 variants are required to investigate the effects
of this additional binding site on cancer cell function.
The present study suggested that the CDCP1 CUB2
domain may act as a therapeutic target. Recombinant proteins
containing the CUB2 and CUB3 domains of CDCP1 are able to suppress
cell migration (18), but the
required domain was previously unknown. Herein, it was demonstrated
that rMBP-CUB2 decreased SFK-CDCP1-PKCδ signaling, and the
migration of A549 and BxPC3 cell (Figs.
5, S3 and S4). Also, an interaction between rMBP-CUB3
and CDCP1res-HA was detected (Fig.
4B); however, rMBP-CUB3 did not effectively inhibit homophilic
complex formation (Fig. 4E) or cancer
cell migration (Fig. 5C). In
addition, the comparison of amino acid sequences of each CUB domain
showed low rates of homology (Fig.
S6). Although, the possibility that CUB3 is critical to CDCP1
homophilic complex formation cannot be excluded, as the current
data support cell migration primarily via the extracellular CUB2
domain. A previous report detailed the homophilic complex of MMP14
formed by the hemopexin-like (PEX) domain. Expression of the
exogenous PEX domain also resulted in dose-dependent inhibition of
cancer cell invasion via cell surface activation of proMMP-2
(27). Therefore, the present study
supported that the CUB2 domain may present a novel target for the
inhibition of CDCP1 homophilic complex formation and cell
migration, thus contributing to the suppression of cancer cell
invasion.
The present study indicated that the CUB2 domain of
CDCP1 stimulates SFK activation through homophilic binding, and
that it also regulates cancer cell migration. In addition,
molecules that inhibit CDCP1 homologous complex formation, such as
rMBP-CUB2, may be viable therapeutic candidates to combat invasive
cancer.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by a Grant-in-Aid
for Scientists by the Ministry of Education, Culture, Sports,
Science and Technology of Japan [JSPS KAKENHI, grant no. 15K06850
(TU)] and the National Cancer Center Research Development Fund
[grant no. 23-B-24 (TU)].
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
TS, KN, TI, RS and TU made substantial contributions
to conception and design or analysis and interpretation of the
data. TS and TU analyzed and interpreted the data regarding CDCP1
homophilic complex formation via CUB2 domain. TS performed the
examination of cell migration and SFK activation and was a primary
contributor to the manuscript writing. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
CDCP1
|
CUB domain-containing protein 1
|
|
CDCP1res-F
|
FLAG-tagged CDCP1 rescue
|
|
CDCP1res-HA
|
HA-tagged CDCP1 rescue
|
|
CUB
|
complement C1r/C1s, urchin embryonic
growth factor, bone morphogenetic protein 1
|
|
ECD
|
extracellular domain
|
|
∆ECD-F
|
extracellular domain deletion mutant
of CDCP1 with a FLAG tag at the C-terminus
|
|
rMBP-CUB2/3
|
recombinant CUB2 or CUB3 domain
proteins combined with maltose binding protein
|
|
SFK
|
Src family kinase
|
|
SH2
|
Src homology 2
|
References
|
1
|
Hu G, Place AT and Minshall RD: Regulation
of endothelial permeability by Src kinase signaling: Vascular
leakage versus transcellular transport of drugs and macromolecules.
Chem Biol Interact. 171:177–189. 2010. View Article : Google Scholar
|
|
2
|
Brown MT and Cooper JA: Regulation,
substrates and functions of src. Biochim Biophy Acta. 1287:121–149.
1996.
|
|
3
|
Irby RB and Yeatman TJ: Role of Src
expression and activation in human cancer. Oncogene. 19:5636–5642.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Summy JM and Gallick GE: Src family
kinases in tumor progression and metastasis. Cancer Metastasis Rev.
22:337–358. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hooper JD, Zijlstra A, Aimes RT, Liang H,
Claassen GF, Tarin D, Testa JE and Quigley JP: Subtractive
immunization using highly metastatic human tumor cells identifies
SIMA135/CDCP1, a 135 kDa cell surface phosphorylated glycoprotein
antigen. Oncogene. 22:1783–1794. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Brown TA, Yang TM, Zaitsevskaia T, Xia Y,
Dunn CA, Sigle RO, Knudsen B and Carter WG: Adhesion or plasmin
regulates tyrosine phosphorylation of a novel membrane glycoprotein
p80/gp140/CUB domain-containing protein 1 in epithelia. J Biol
Chem. 279:14772–14783. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bhatt AS, Erdjument-Bromage H, Tempst P,
Craik CS and Moasser MM: Adhesion signaling by a novel mitotic
substrate of src kinase. Oncogene. 24:5333–5343. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Scherl-Mostageer M, Sommergruber W,
Abseher R, Hauptmann R, Ambros P and Schweifer N: Identification of
a novel gene, CDCP1, overexpressed in human colorectal cancer.
Oncogene. 20:4402–4408. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Uekita T, Jia L, Narisawa-Saito M, Yokota
J, Kiyono T and Sakai R: CUB domain-containing protein 1 is a novel
regulator of anoikis resistance in lung adenocarcinoma. Mol Cell
Biol. 27:7649–7660. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Benes CH, Wu N, Elia AE, Dharia T, Cantley
LC and Soltoff SP: The C2 domain of PKCdelta is a phosphotyrosine
binding domain. Cell. 121:271–280. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wortmann A, He Y, Christensen ME, Linn M,
Lumley JW, Pollock PM, Waterhouse NJ and Hooper JD: Cellular
settings mediating Src Substrates switching between focal adhesion
kinase tyrosine 861 and CUB-domain-containing protein 1 (CDCP1)
tyrosine 734. J Biol Chem. 286:42303–42315. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Dong Y, He Y, de Boer L, Stack MS, Lumley
JW, Clements JA and Hooper JD: The cell surface glycoprotein CUB
domain-containing protein 1 (CDCP1) contributes to epidermal growth
factor receptor-mediated cell migration. J Biol Chem.
287:9792–9803. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Miyazawa Y, Uekita T, Hiraoka N, Fujii S,
Kosuge T, Kanai Y, Nojima Y and Sakai R: CUB domain-containing
protein 1, a prognostic factor for human pancreatic cancers,
promotes cell migration and extracellular matrix degradation.
Cancer Res. 70:5136–5146. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Uekita T and Sakai R: Roles of CUB
domain-containing protein 1 signaling in cancer invasion and
metastasis. Cancer Sci. 102:1943–1948. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Bork P and Beckmann G: The CUB domain. A
widespread module in developmentally regulated proteins. J Mol
Biol. 231:539–545. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lee HX, Mendes FA, Plouhinec JL and De
Robertis EM: Enzymatic regulation of pattern: BMP4 binds CUB
domains of Tolloids and inhibits proteinase activity. Genes Dev.
23:2551–2562. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Romero A, Varela PF, Sanz L,
Töpfer-Petersen E and Calvete JJ: Crystallization and preliminary
X-ray diffraction analysis of boar seminal plasma spermadhesin
PSP-I/PSP-II, a heterodimer of two CUB domains. FEBS Lett.
382:15–17. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wright HJ, Arulmoli J, Motazedi M, Nelson
LJ, Heinemann FS, Flanagan LA and Razorenova OV: CDCP1 cleavage is
necessary for homodimerization-induced migration of triple-negative
breast cancer. Oncogene. 35:4762–4772. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yang L, Dutta SM, Troyer DA, Lin JB, Lance
RA, Nyalwidhe JO, Drake RR and Semmes OJ: Dysregulated expression
of cell surface glycoprotein CDCP1 in prostate cancer. Oncotarget.
6:43743–43758. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kollmorgen G, Bossenmaier B, Niederfellner
G, Häring HU and Lammers R: Structual requirements for cub domain
containing protein 1 (CDCP1) and Src dependent cell transformation.
PLoS One. 7:e530502012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Casar B, He Y, Iconomou M, Hooper JD,
Quigley JP and Deryugina EI: Blocking of CDCP1 cleavage in vivo
prevents Akt-dependent survival and inhibits metastatic
colonization through PARP1-medited apoptosis of cancer cells.
Oncogene. 31:3924–3938. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Burnham MR, Bruce-Staskal PJ, Harte MT,
Weidow CL, Ma A, Weed SA and Bouton AH: Regulation of c-SRC
activity and function by the adapter protein CAS. Mol Cell Biol.
20:5865–5878. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Tanaka M, Sasaki K, Kamata R, Hoshino Y,
Yanagihara K and Sakai R: A novel RNA-binding protein,
Ossa/C9orf10, regulates activity of Src kinase to protect cells
from oxidative stress-induced apoptosis. Mol Cell Biol. 22:402–413.
2009. View Article : Google Scholar
|
|
24
|
Liu H, Ong SE, Badu-Nkansah K, Schindler
J, White FM and Hynes RO: CUB-domain-containing protein 1 (CDCP1)
activates Src to promotes melanoma metastasis. Proc Natl Acad Sci
USA. 108:1379–1384. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Xia Y, Gil SG and Carter WG: Anchorage
mediated by integrin alpha6beta4 to laminin 5 (epiligrin) regulates
tyrosine phosphorylation of a membrane-associated 80-kD protein. J
Cell Biol. 134:724–740. 1996.
|
|
26
|
Miura S, Hamada S, Masamune A, Satoh K and
Shimosegawa T: CUB-domain containing protein 1 represses the
epithelial phenotype of pancreatic cancer cell. Exp Cell Res.
321:209–218. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Itoh Y, Takamura A, Ito N, Maru Y, Sato H,
Suenaga N, Aoki T and Seiki M: Homophilic complex formation of
MT1-MMP facilitates proMMP-2 activation on the cell surface and
promotes tumor cell invasion. EMBO J. 20:4782–4793. 2001.
View Article : Google Scholar : PubMed/NCBI
|