Introduction
Distant metastasis is the leading cause of
cancer-associated mortality in patients with colorectal cancer
(CRC); ≤25% of patients have synchronous colorectal liver
metastases (CLM) upon diagnosis (1).
Such synchronous presentation has been associated with poor
survival outcome (2). Thus, it is
important to understand the molecular mechanisms underlying distant
metastasis to aid developments in effective therapeutic strategies
for treating metastatic CRC or preventing metastasis with adjuvant
chemotherapy. A key event in the process of distant metastasis is
epithelial-mesenchymal transition (EMT), which induces the invasive
and metastatic abilities of tumor cells in various types of cancer,
such as ovarian cancer (3), breast
cancer (4), CRC (5) and is associated with poor prognosis
(6). During the EMT process, cancer
cells lose their expression of cellular adhesion proteins,
including E-cadherin and γ-catenin, and acquire the expression of
mesenchymal markers, such as vimentin and N-cadherin (7). Loss of E-cadherin expression has been
reported to be a hallmark of the EMT process (8).
Insulin-like growth factor (IGF) is a potent mitogen
involved in normal growth and development. The growth-promoting and
metabolic activities of IGFs are modulated by IGF factor binding
proteins (IGFBPs) and their receptors (9). IGFBP7 has been reported as a tumor
suppressor in carcinomas. For example, IGFBP7 exhibits certain
tumor-suppressive functions in colon cancer as well as
hepatocellular carcinoma (10,11).
Furthermore, low IGFBP7 expression was determined to be associated
with poorly differentiated breast cancer tumors and higher-stage
disease (12) when
immunohistochemical and microarray analyses were used to identify
IGFBP7 expression in tumor cells. The mechanism underlying the
putative tumor-suppressor function of IGFBP7 requires further
investigation. Previous findings suggested that epithelial IGFBP7
acts as an IGF-1/2 antagonist that inhibits IGF-1 receptor
activation by binding to the receptor, thereby suppressing cell
growth and survival (13). Although
mounting evidence indicates the importance of IGFBP7 in various
cancer types, systematic investigations into the role of IGFBP7 in
the metastasis of human CRC to organs, such as the liver, are
required.
The aim of the present study was to investigate the
expression of IGFBP7 in colon cancer with liver metastasis (LM),
and compare the expression of IGFBP7 between primary CRC (PC) and
matched LM tissues. Furthermore, the potential mechanism was
determined by analyzing the effects of IGFBP7 overexpression and
knockdown on the expression of EMT-associated proteins, including
E-cadherin, N-cadherin and Vimentin protein levels.
Materials and methods
Patients and tissue samples
A tissue microarray was purchased from Shanghai
Xinchao Industry Co., Ltd. (Shanghai). The microarray contained
formalin-fixed, paraffin-embedded colon adenocarcinoma tissues (90
CRC and 90 adjacent tumor tissues). Of these 81 pairs of colon
cancer tissues and adjacent normal tissues were included, as
medical record data were lacking for 9 subjects and were therefore
excluded. In addition, 24 PC (primary colon cancer) and matched
corresponding LM (liver metastasis) tissues were obtained from the
Shanxi Tumor Hospital (Taiyuan, China) and The First Clinical
Hospital of Shanxi Medical University (Taiyuan, China). All cases
were diagnosed according to the Union for International Cancer
Control (UICC) TNM Classification of Malignant Tumors. Two
pathologists were blinded to the samples and analyzed all samples
to confirm diagnosis. Written informed consent was obtained from
all patients prior to enrolment. All the procedures were conducted
in accordance with standard guidelines for the Study of Humans and
were approved by the Research Ethics Committee of First Hospital of
Shanxi Medical University.
Cell lines
HCT116, SW480, SW620, RKO, LοVο and Caco2 CRC cell
lines were purchased from the American Type Culture Collection
(Manassas) to analyze the expression of IGFBP7. Cells were cultured
in Dulbecco's modified Eagle's medium (DMEM; Gibco; Thermo Fisher
Scientific, Inc.) containing 10% heat-inactivated fetal bovine
serum (Thermo Fisher Scientific, Inc.), penicillin (100 U/ml),
streptomycin (100 U/ml) and sodium bicarbonate (1.5 g/l) at 37°C in
a humidified atmosphere of 5% CO2.
Immunohistochemistry (IHC)
Paraffin-embedded sections (4 µm) were
deparaffinized and treated with 0.3% (v/v) hydrogen peroxide to
block endogenous peroxidase activity. Then, heat-induced antigen
retrieval was performed using 0.01 M sodium citrate (pH 6.0) and
10% (v/v) bovine serum albumin (Sigma-Aldrich; Merck KGaA) in PBS
for 10 min at room temperature. Sections were then incubated
overnight at 4°C with mouse monoclonal antibodies (1:50) against
IGFBP7 (cat. no. ab74169; Abcam, Cambridge, UK), E-cadherin (cat.
no. ab1416; Abcam), N-cadherin (cat. no. ab18203; Abcam) and
Vimentin (cat. no. ab8069; Abcam) overnight at 4°C. A
streptavidin-biotin-peroxidase complex kit (SABC kit; Zymed) was
used to detect the protein conjugates, according to the
manufacturer's protocol. The sections were developed using a
diaminobenzidine substrate (Sigma-Aldrich; Merck KGaA) and
counterstained with hematoxylin. Negative controls (NCs) were run
in parallel; however, PBS or mouse IgG1 (AMS/Immunokontact) was
applied.
Evaluation of immunostaining
Subjective visual scoring of staining was used to
assess the immunoreactivity of samples; scores were based on the
proportion of positive tumor cells to total tumor cells
(positivity, %) as follows: 0, negative or <5%; 1, 5–25%; 2,
26–50%; 3, 51–75% and 4, >75% positive cells. The staining
intensity was evaluated as 0, negative; 1, weak; 2, moderate and 3,
intense. Providing the staining intensity was heterogeneous, and
scoring was based on the greatest degree of intensity (14). As >95% of tumor cells stained
positively for IGFBP7 in all specimens, only immunostaining
intensity scores were analyzed. For the analysis of E-cadherin
expression, staining intensity scores were categorized as negative
(<5%) or positive (≥5%). For statistical analysis, scores of 0–7
were considered to indicate low expression and scores of 8–12 were
considered to indicate high expression. Staining analysis was
conducted by two researchers independently without knowledge of
patient clinical data and the presence of LM.
RNA preparation and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from cultured cells or fresh
frozen colon cancer tissues using TRIzol® reagent
(Thermo Fisher Scientific, Inc.) according to the manufacturer's
protocol. Total RNA (1 µg) was reverse transcribed using Reverse
ace qPCR RT Kit (Toyobo). The primers used in the present study are
presented in Table I. A volume of 2.0
µl of each diluted cDNA (1:20) was subjected to RT-qPCR in a final
volume of 20 µl containing 100 nM of each specific primer and
SYBR-Green Mix (Takara Biotechnology, Co., Ltd.). The thermocycling
conditions as follows: Initial enzyme activation at 95°C for 2 min,
followed by 40 cycles of 95°C for 15 sec and 60°C for 20 sec, and
elongation at 72°C for 20 sec. The experiment was performed in
triplicate, and the relative expression of IGFBP7 was calculated
using the 2−ΔΔCq method (15) and normalized to a-tubulin.
 | Table I.Sequence primers used for
RT-qPCR. |
Table I.
Sequence primers used for
RT-qPCR.
| Gene | Primer
sequence |
|---|
| h-a-tubulin-5′ |
5′-ATATGTGGCCAGAGGGAAGT-3′ |
| h-a-tubulin-3′ |
5′-GGCTGTGTTTGTAGACTTGG-3′ |
| h
E-cadherin-5′ |
5′-AATGCCGCCATCGCTTAC-3′ |
| hE-cadherin-3′ |
5′-TCAGGCACCTGACCCTTGTA-3′ |
| hN-cadherin-5′ |
5′-GTGCCATTAGCCAAGGGAATTCAGC-3′ |
| hN-cadherin-3′ |
5′-GCGTTCCTGTTCCACTCATAGGAGG-3′ |
| hvimentin-5′ |
5′-GTCCACTGAGTACCGGAGACA-3′ |
| hvimentin-3′ |
5′-CGAAGGTGACGAGCCATTT-3′ |
| hIGFBP7-5′ |
5′-TGGAACAAGGTAAAAAGGGGT-3′ |
| hIGFBP7-3 |
5′-TGGTATTTCATGTAAGGCATAC-3′ |
Generation of stable IGFBP7 knockdown
or overexpression
For short hairpin (sh)RNA-mediated IGFBP7 knockdown,
the constructs (pLenti-1#miR-EGFP) developed by The RNAi Consortium
(Open Biosystems; Thermo Fisher Scientific, Inc.) were packaged
into lentiviral particles and used to infect the LοVο cell line.
Selection was conducted with puromycin (Invitrogen; Thermo Fisher
Scientific, Inc.) at 1 µg/ml. All the experiments were conducted
according to local biosafety regulations. In order to induce IGFBP7
expression, IGFBP7 Lentifect lentiviral particles (GeneCopoeia,
Inc.) were used to overexpress the protein in HT-29 cells. The
infection and selection processes were performed as aforementioned.
Cells without any treatment comprised the blank group, and negative
control was the Lenti-EGFP-infected group.
Western blot analysis
Proteins were extracted from cells in a Triton-X
lysis (1% Triton-X, 50 Mm Tris and 150 mM NaCl) containing protease
inhibitors (pepstatin, phenylmethane sulfonyl fluoride, aprotinin
and leupettin) for 1 h in 4°C. Cell lysates were centrifuged at
12,000 × g for 20 min at 4°C, and the protein concentration in the
supernatant was determined using a bicinchoninic acid protein
assay. Protein extracts (50 µg) were separated by 10% SDS-PAGE and
then transferred to a polyvinylidene difluoride membrane (Merck
KGaA). The membrane was blocked with 5% skim milk and tris-buffered
saline with Tween-20 (25 mM Tris-HCl, pH 7.5, 150 mM NaCl and 0.05%
v/v Tween-20) for 2 h at room temperature. Subsequently, the
membrane was incubated with a goat anti-IGFBP7 antibody (cat. no.
ab74169; Abcam) at 0.1 µg/ml (1:1,000), and antibodies against
E-cadherin (cat. no. ab1416; Abcam), N-cadherin (cat. no. ab18203;
Abcam) and Vimentin (cat. no. ab8069; Abcam) overnight at 4°C.
Tubulin was used for the normalization of protein expression. An
electrochemiluminescent chromogenic substrate was used to visualize
the bands, and Image Lab 5.0 software (Bio-Rad Laboratories, Inc.)
was used for quantitative analysis.
Cell Counting Kit-8 (CCK-8) assay for
the analysis of cell proliferation
Cell lines including HT29, HT-29 miRNA, LοVο and
LοVο cells with IGFBP7 shRNA-mediated knockdown were cultured in
DMEM with 10% FBS containing 1 µg/ml recombinant IGFBP7 for 6 days.
A CCK-8 assay was conducted to analyze cell proliferation at days
0, 1, 2, 3, 4, 5 and 6 following treatment. Briefly,
5×103 cells were seeded into 96-well plates and cultured
overnight in DMEM supplemented with 10% FBS; cells were then
cultured at 37°C with 5% CO2 for 24 h. CCK-8 reagent (10
µl; Dojindo Molecular Technologies, Inc.) was added to the
maintenance cell medium at various time points and incubated at
37°C for an additional 2 h. Absorbance values were determined using
a microplate reader (Multiskan MK3; Thermo Fisher Scientific, Inc.)
at 450 nm.
Invasion assay
The invasion assay was performed using transwell
culture chambers (8 µm pores; Costar, Corning, NK, USA) according
to the manufacturer's instructions. The upper chamber was loaded
with 1×105 cells in 0.2 ml serum-free medium, while 0.6
ml medium containing 10% FBS was loaded to the lower chamber. After
incubation for 48 h at 37°C in a humidified atmosphere of 5%
CO2, the cells on the lower side were fixed in 95%
ethanol and stained with crystal violet at room temperature for 10
min and counted under a microscope (Olympus Corp.). Four
microscopic fields were randomly selected for cell counting. The
images were captured at ×100 magnification. Each experiment was
performed at least three times.
Statistical analysis
Data were analyzed with GraphPad Prism 7.0 software
(GraphPad Software, Inc.). Data were presented as means ± SD.
Paired t-tests and a Mann-Whitney U test were used. To analyze the
results from CRC tissues, comparisons of clinicopathological
parameters and EMT markers between the high- and low-IGFBP7
expression groups were performed via χ2 or Fisher's
exact tests. Correlations were analyzed using Spearman's
coefficient. P<0.05 was considered to indicate a statistically
significant difference.
Results
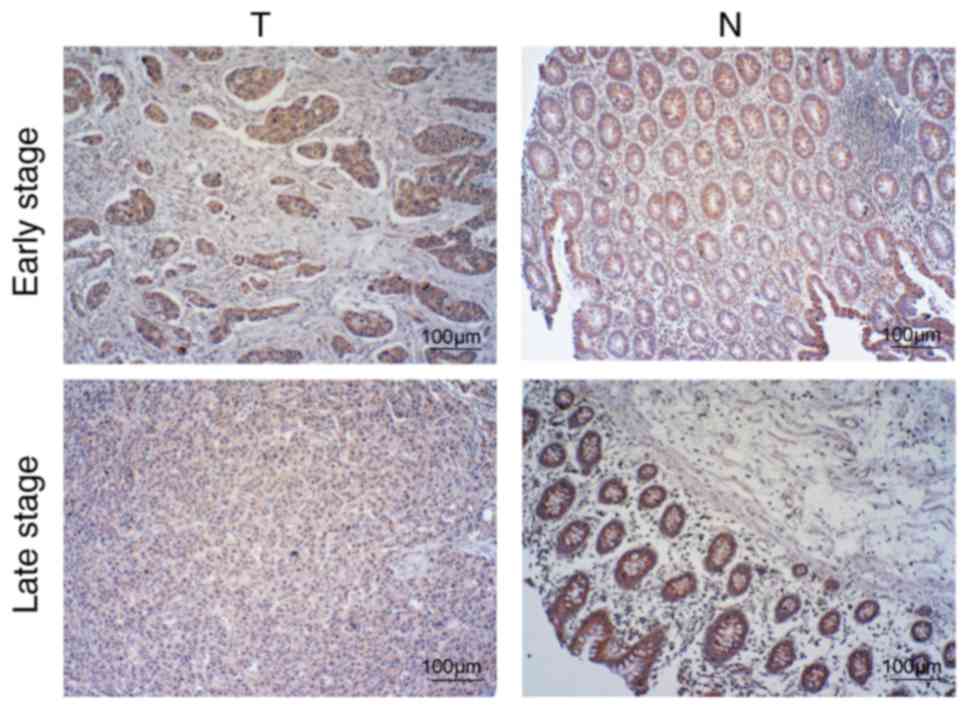
IGFBP7 is significantly downregulated
in LM tissues of patients with CRC
IHC was performed using primary CRC tissues with and
without metastasis, as well as in matched PC and LM tissues. In all
81 pairs of primary CRC and tumor-adjacent tissue samples, the
majority (>95%) of tumor cells and adjacent mucosa stained
positively for IGFBP7. The staining pattern was predominantly
cytoplasmic and nuclear staining was observed in only one sample.
There were no significant differences in the expression of IGFBP7
between early stage (I + II) CRC and adjacent normal colonic mucosa
(P=0.285); however, late stage (III + IV) CRC revealed a
significantly low expression of IGFBP7 compared with adjacent
normal colonic mucosa (P=0.031; Table
II, Fig. 1). Furthermore, in CRC
samples, the low IGFBP7 expression group exhibited significantly
higher lymphatic metastasis (P=0.039), liver metastasis
(P=0.002) and advanced tumor stage (P=0.014) compared with
the high IGFBP7 expression group, which is consistent with a
previous study (7). In addition, no
significant correlations between IGFBP7 expression and other
clinicopathological features, including age, sex, tumor location,
tumor size and histological grade, were reported (P>0.05;
Table III).
 | Table II.Expression of IGFBP7 in colon cancer
and tumor-adjacent tissue by IHC (means ± SD). |
Table II.
Expression of IGFBP7 in colon cancer
and tumor-adjacent tissue by IHC (means ± SD).
| Stage | n | T(OD) | N(OD) | P-value |
|---|
| I+II | 37 | 0.758±0.218 | 0.824±0.112 | 0.105 |
| III+IV | 44 | 0.579±0.231 | 0.682±0.191 | 0.031a |
 | Table III.Association between IGFBP7 expression
and clinicopathological characteristics in colon cancer by IHC. |
Table III.
Association between IGFBP7 expression
and clinicopathological characteristics in colon cancer by IHC.
|
|
| IGFBP7
expression |
|
|
|---|
|
|
|
|
|
|
|---|
| Variable | Total
(n) | Low n (%) | High n (%) | r | P-value |
|---|
| Age (years) |
|
|
| 0.015 | 1.000 |
|
<60 | 33 | 16 (48.5) | 17 (54.5) |
|
|
|
≥60 | 72 | 35 (48.6) | 37 (51.4) |
|
|
| Sex |
|
|
| −0.002 | 0.326 |
|
Male | 57 | 28 (49.1) | 29 (50.9) |
|
|
|
Female | 48 | 29 (60.4) | 19 (39.6) |
|
|
| Histologic
grade |
|
|
| 0.281 | 0.027a |
|
Low | 71 | 24 (33.8) | 47 (66.2) |
|
|
|
High | 29 | 17 (58.6) | 12 (41.4) |
|
|
| Lympho-node
metastasis |
|
|
| 0.34 | 0.039a |
|
Negative | 41 | 21 (51.2) | 20 (48.8) |
|
|
|
Positive | 64 | 46 (71.9) | 18 (28.1) |
|
|
| Liver
metastasis |
|
|
| 0.537 | 0.002a |
|
Negative | 81 | 22 (27.2) | 59 (72.8) |
|
|
|
Positive | 24 | 15 (62.5) | 9 (37.5) |
|
|
| TNM stage |
|
|
| 0.410 | 0.040a |
|
I+II | 37 | 16 (43.2) | 21 (56.8) |
|
|
|
III+IV | 68 | 44 (73.3) | 24 (35.3) |
|
|
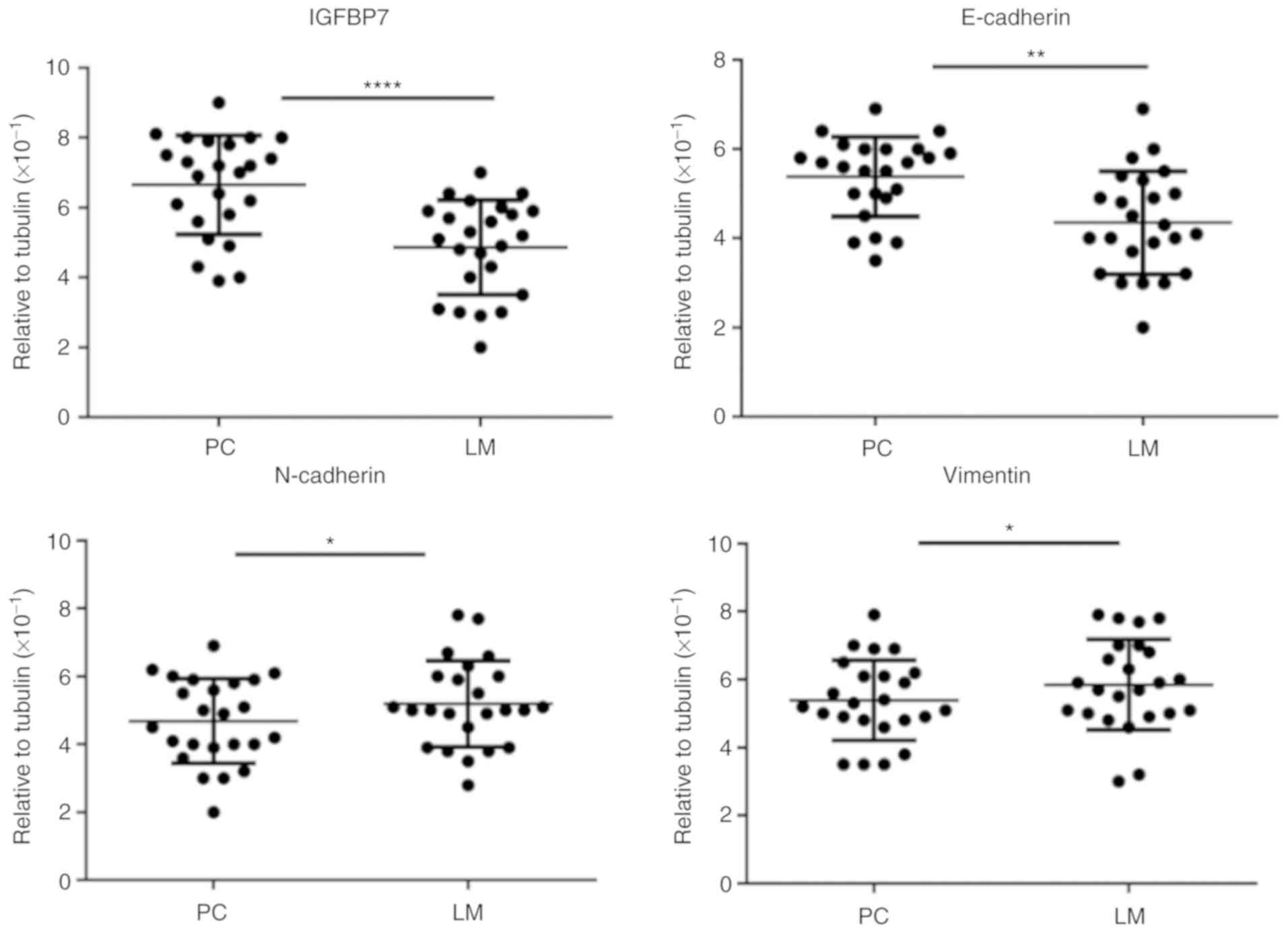
Additionally, 24 of 101 patients had synchronous LM.
Of note, IHC analysis of PC and matched LM tissues revealed very
low or undetectable expression of IGFBP7 in metastasized CRC
tissues compared with matched PC and normal hepatocytes in adjacent
tissues (Fig. 2A and B). Furthermore,
IGFBP7 expression was gradually reduced at the invasive front,
indicating that the suppression of IGFBP7 expression may increase
the metastatic potential of CRC cells at the invasive front
(Fig. 2B).
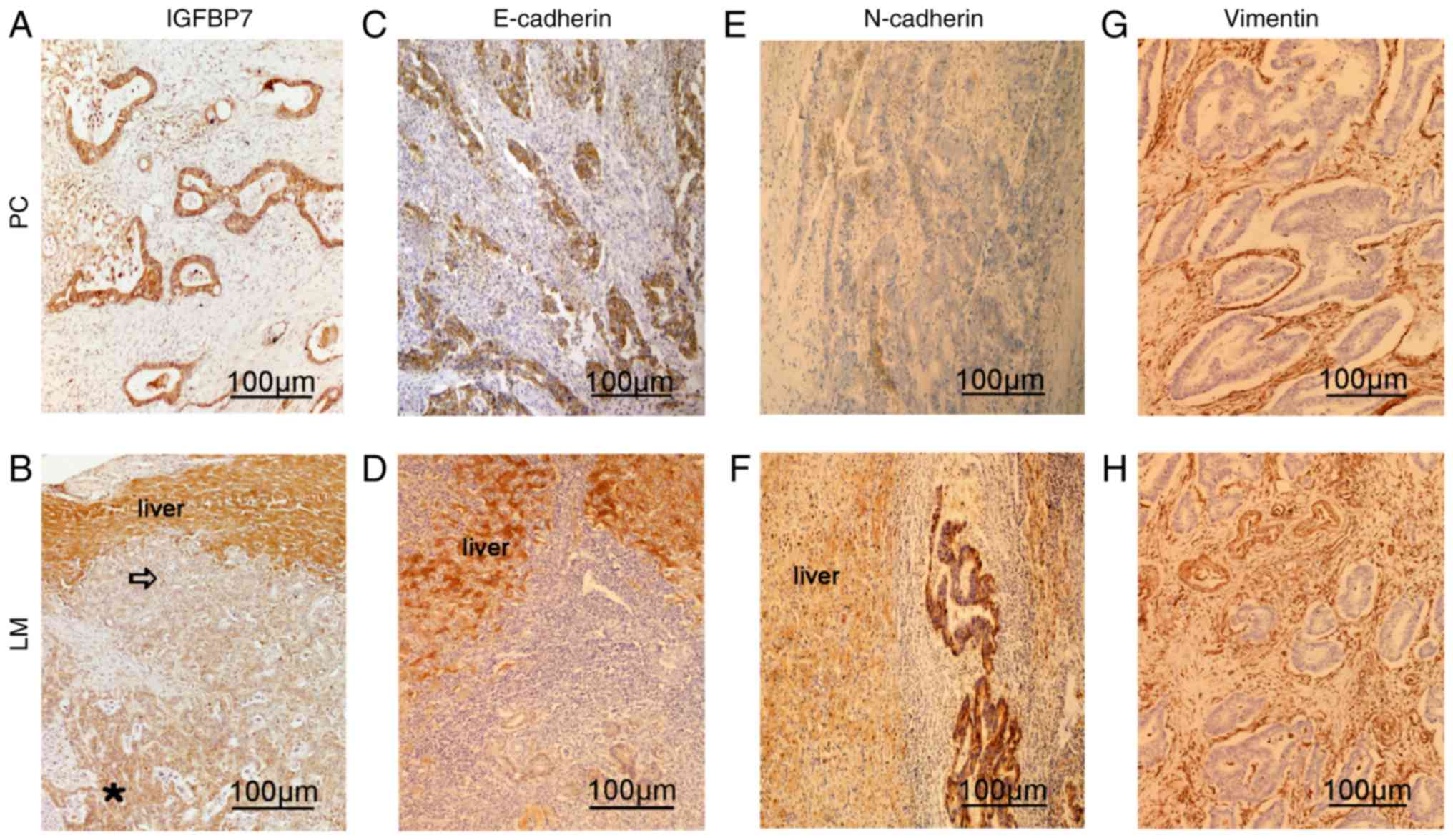 | Figure 2.Expression of IGFBP7, E-cadherin,
N-cadherin and Vimentin in PC and LM tissues as determined by
immunohistochemistry. Expression of (A and B) IGFBP7, (C and D)
E-cadherin, (E and F) N-cadherin and (G and H) Vimentin in PC and
LM tissues. IGFBP7 was downregulated at the invasive front of the
LM site. Scale bar, 100 µm. IGFBP7, insulin-like growth factor
binding protein 7; PC, primary colon cancer; LM, liver metastasis.
Blank arrow, front of tumor cells invading the liver. Star, IGFBP7
staining of liver metastases. |
Correlation of IGFBP7 and
EMT-associated proteins in CRC
To further investigate the association between
IGFBP7 expression and EMT-associated proteins, including
E-cadherin, N-cadherin and Vimentin, the distribution of the
aforementioned proteins was analyzed in PC and matched LM tissues
by IHC (Fig. 2C-H). Immunostaining of
IGFBP7 and E-cadherin (r=0.451, P=0.015) revealed a positive
correlation. Conversely, a negative correlation was determined
between IGFBP7 and N-cadherin (r=−0.381, P=0.035) and Vimentin
(r=−0.314, P=0.035) in PC and matched LM tissues (Table IV). In addition, the expression
levels of E-cadherin, N-cadherin and Vimentin were investigated by
RT-qPCR. The expression levels of IGFBP7 and E-cadherin in LM
tissues were significantly decreased compared with PC tissues
(P<0.001 and P=0.016, respectively). By contrast, the expression
levels of N-cadherin and Vimentin were significantly increased in
matched LM tissues (P=0.041 and P=0.027, respectively; Fig. 3).
 | Table IV.Correlation between IGFBP7 expression
and EMT markers in colon cancer by IHC. |
Table IV.
Correlation between IGFBP7 expression
and EMT markers in colon cancer by IHC.
|
|
| IGFBP7
expression |
|
|
|---|
|
|
|
|
|
|
|---|
| Variable | Total (n=24) | Low n (%) | High n (%) | r | P-value |
|---|
| E-cadherin |
|
|
| 0.451 | 0.015a |
|
Negative | 5 | 2 (40.0) | 3 (60.0) |
|
|
|
Postive | 19 | 3 (15.8) | 16 (84.2) |
|
|
| N-cadherin |
|
|
| −0.381 | 0.035a |
|
Negative | 4 | 1 (25.0) | 3 (75.0) |
|
|
|
Positive | 20 | 17 (85.0) | 3 (15.0) |
|
|
| Vimentin |
|
|
| −0.314 | 0.035a |
|
Negative | 6 | 3 (50.0) | 3 (50.0) |
|
|
|
Positive | 18 | 17 (94.4) | 1 (5.6) |
|
|
Association between the expression of
IGFBP7 and markers of EMT in colon cancer cells
To verify the aforementioned findings from CRC
tissues and investigate the potential role of the IGFBP7 in colon
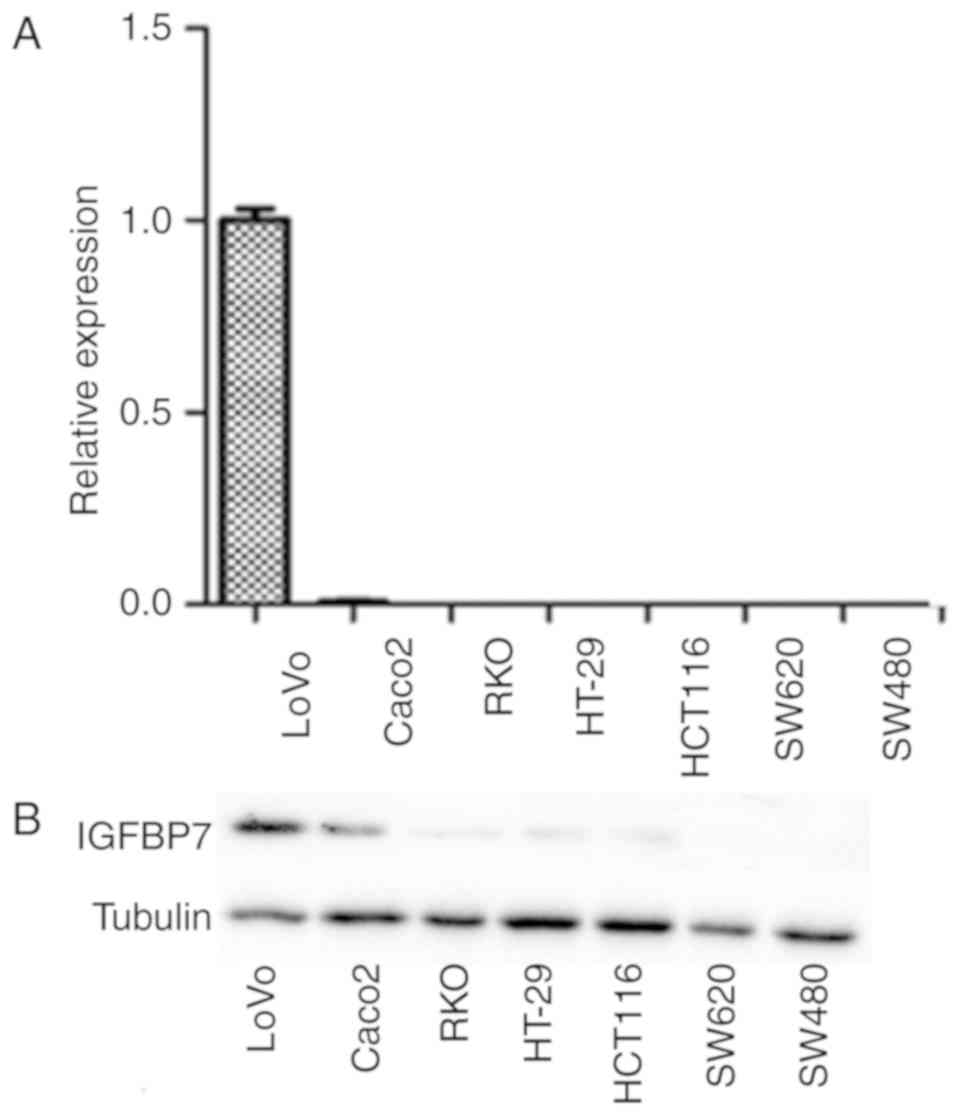
cancer, the expression of IGFBP7 in CRC cells lines was analyzed by
RT-qPCR. LοVο cells exhibited robust IGFBP7 expression, whereas
downregulated expression was observed in Caco2 cells. The
expression of IGFBP7 in RKO, HT-29, HCT116, SW620 and SW480 cells
was undetectable (Fig. 4A). Similarly
the expression of protein by western blotting was confirmed
(Fig. 4B).
To further analyze the association between the
expression of EMT markers and the potential role of IGFBP7 in colon
cancer cells, two colon cancer cell lines, including LοVο cells
with robust IGFBP7 expression and HT29 cells with undetectable
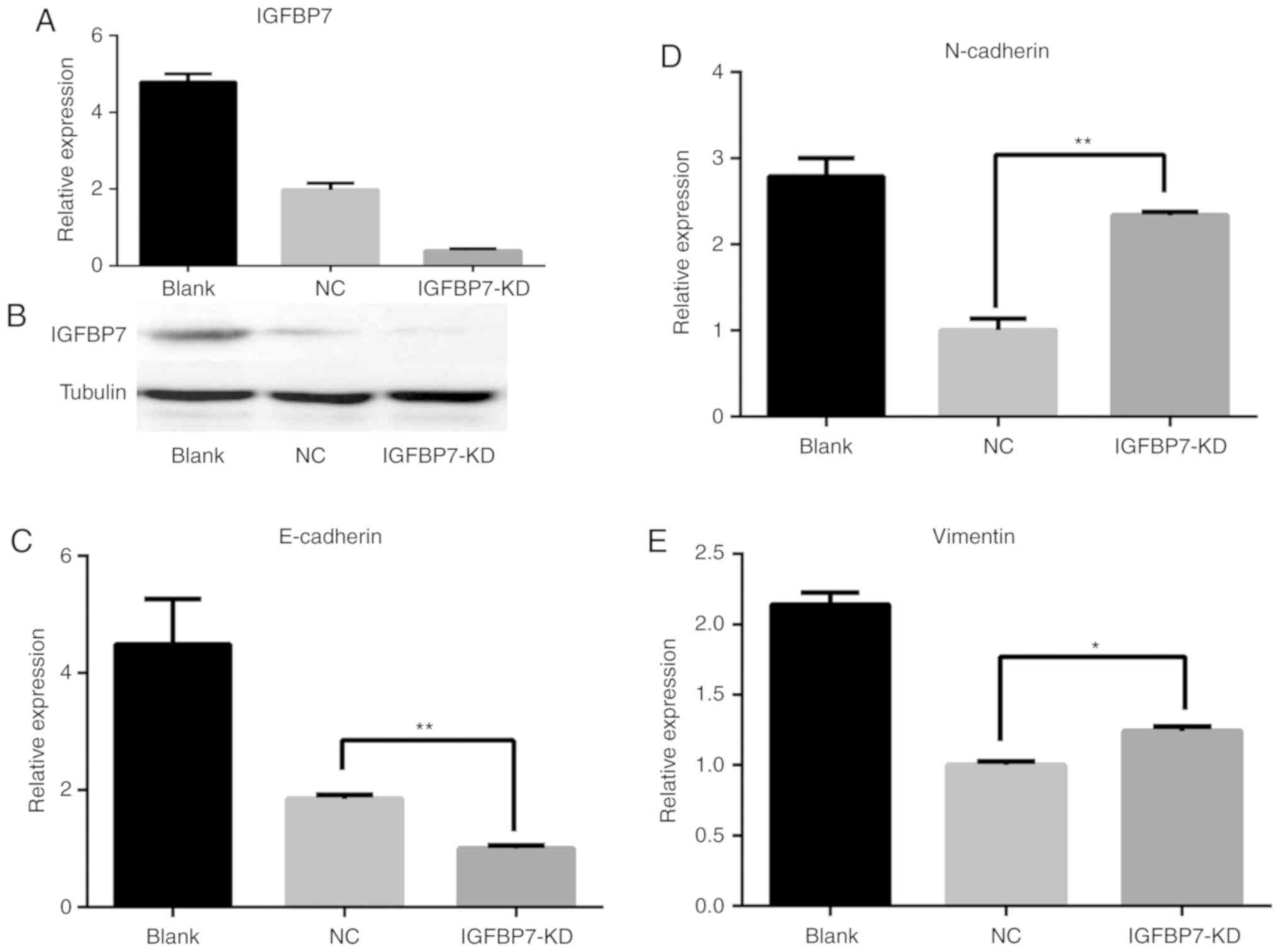
expression were selected. Then, IGFBP7 was knocked down in LοVο
cells and overexpressed in HT-29 cells to examine the functional
role of IGFBP7 in EMT. Specifically, LοVο cells were transfected
with IGFBP7 shRNA via lentivirus for stable knockdown
(IGFBP7-shRNA), which resulted in a >90% reduction in IGFBP7
expression by RT-qPCR (Fig. 5A),
confirmed in the protein level by western blotting (Fig. 5B). Cells of the control group were
transfected with a scrambled shRNA via lentivirus to establish an
NC. The expression of E-cadherin was significantly downregulated
following IGFBP7-knockdown in LοVο cells (P=0.007; Fig. 5C), while that of N-cadherin and
Vimentin was upregulated (P=0.006 and P=0.019, respectively)
compared with the NC (Fig. 5D and E).
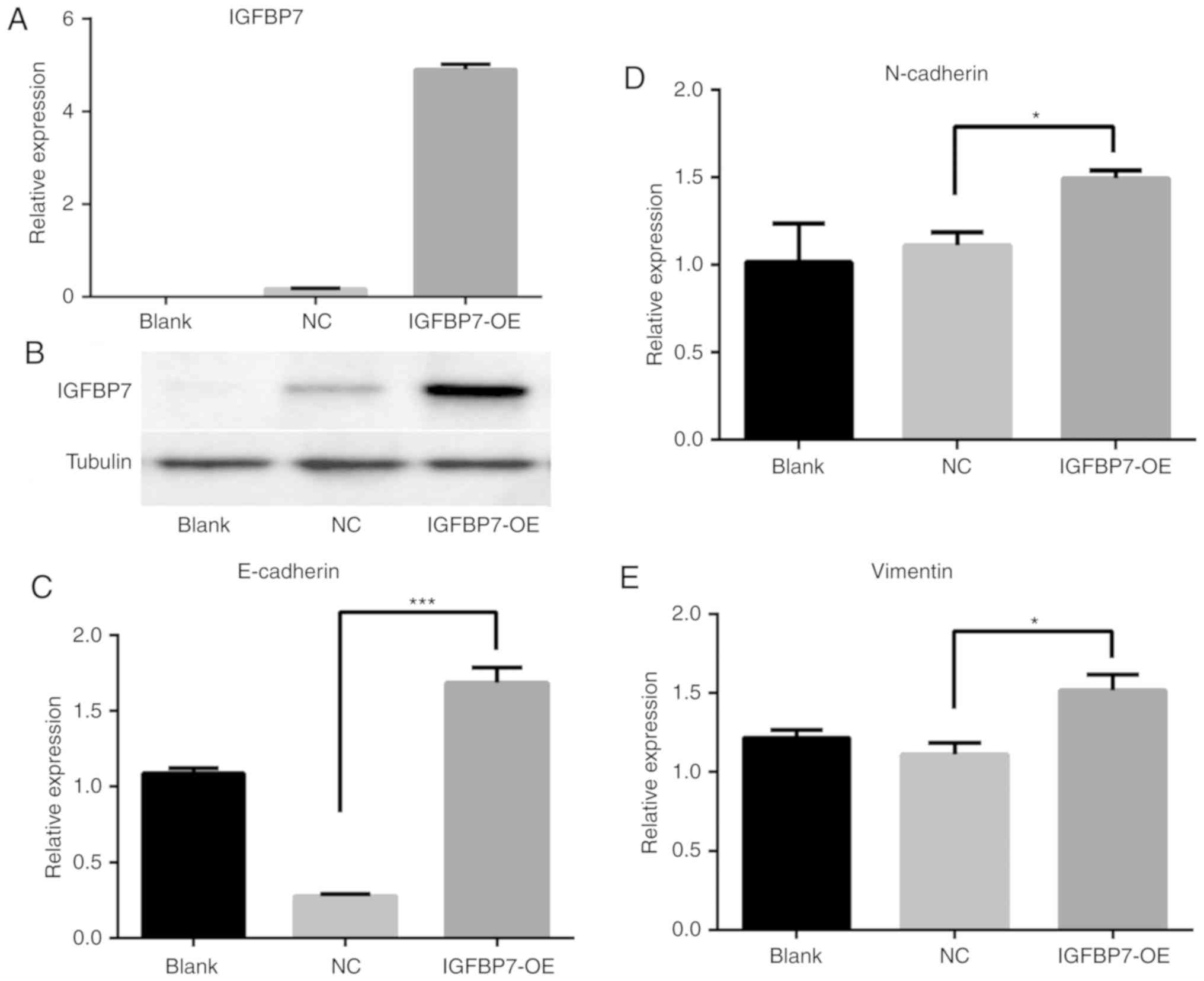
By contrast, overexpression of IGFBP7 in HT-29 cells by RT-qPCR
(Fig. 6A) and western blotting
(Fig. 6B) were associated with
upregulated E-cadherin expression (P<0.001; Fig. 6C), whereas the expression levels of
N-cadherin and Vimentin were downregulated (P=0.031 and P=0.029,
respectively; Fig. 6D and E) compared
with the NC. As expected, knockdown of IGFBP7 in LοVο cells
demonstrated downregulated E-cadherin expression, while N-cadherin
and Vimentin were upregulated compared with the NC (P=0.007,
P=0.003 and P=0.019, respectively; Fig.
7A and B). E-cadherin was upregulated, while N-cadherin and
Vimentin were downregulated in HT-29 following IGFBP7
overexpression (P=0.008, P=0.003 and P=0.029, respectively;
Fig. 7A and C). The data suggested
that the expression of IGFBP7 may serve a crucial role to in the
induction of the epithelial phenotype and the simultaneous loss of
mesenchymal features in CRC cells.
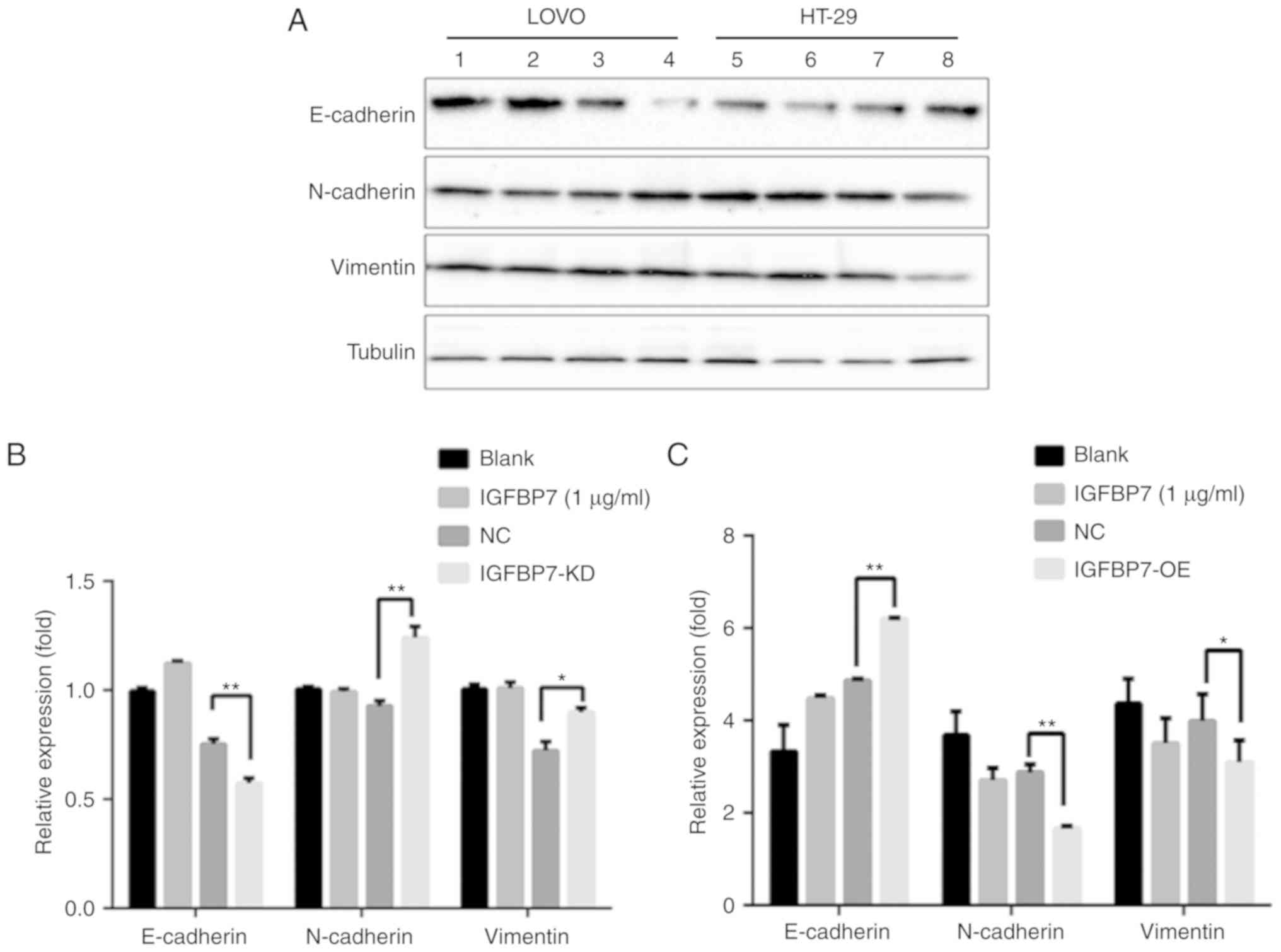 | Figure 7.Effects of IGFBP7 knockdown or
overexpression in LοVο cells and HT-29 respectively as determined
by western blotting. (A) E-cadherin was downregulated, while
N-cadherin and Vimentin were significantly upregulated compared
with the NC in LοVο cells. In addition, the IGFBP7-treated group (1
µg/ml) exhibited upregulated E-cadherin compared with the blank
group; however, no notable variations in the expression of
N-cadherin and Vimentin between the blank and IGFBP7-treated groups
were observed. In HT-26 cells, E-cadherin was upregulated following
IGFBP7 overexpression, while the expression of N-cadherin and
Vimentin was downregulated compared with the NC. Lane 1, Blank 2,
IGFBP7 3, NC 4, IGFBP7-KD 5, Blank 6, IGFBP7 7, NC 8, IGFBP7-OE.
(B) Quantification of protein band intensity in LοVο cells. (C)
Quantification of protein band intensity in HT-29 cells. *P<0.05
and **P<0.01. IGFBP7, insulin-like growth factor binding protein
7; Blank, cells without any treatment; NC, negative control. |
Expression of IGFBP7 affects
proliferation and invasive activity
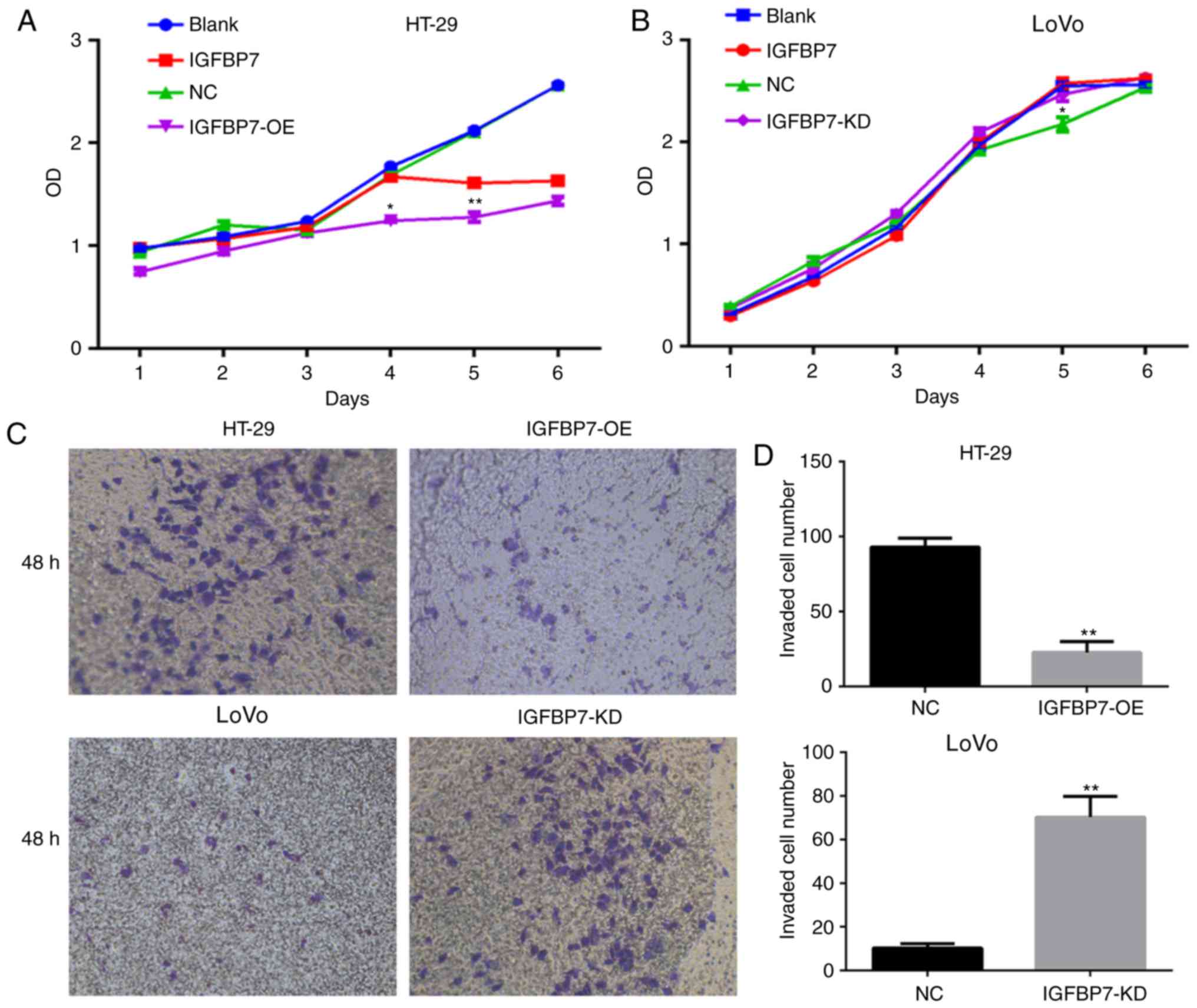
A CCK-8 assay was performed to analyze the effects
of IGFBP7 overexpression on the proliferation of HT-29 cells.
Overexpression of IGFBP7 in HT-29 cells resulted in decreased cell
proliferation compared with the empty vector control over a period
of 6 days. Its cell viability was significantly lower than that of
the NC group at days 4 and 5 (P=0.016 and P=0.002) (Fig. 8A). As IGFBP7 is a secreted protein,
cells were treated with 1 µg/l IGFBP7 recombinant protein to
evaluate the effects of IGFBP7 on cell proliferation. The results
revealed that cell proliferation was slowly inhibited and the
effects of inhibition were noted until the fifth day compared with
the blank control group (P=0.019) (Fig.
8A). Similarly, the proliferation of LοVο cells with IGFBP7
shRNA-mediated knockdown increased until day 5 compared with the NC
groups (P=0.023) (Fig. 8B).
Conversely, the proliferation of LοVο cells treated with 1 µg/l
IGFBP7 markedly increased compared with the blank group but there
was no significant difference. The invasive ability of the
IGFBP7-overexpressing HT-29 cells was significantly weaker than
that of the negative control cells (P=0.008) (Fig. 8C), while the invasive ability of LοVο
cells with IGFBP7-KD was stronger than that of the negative control
cells (P=0.006) (Fig. 8D).
Discussion
Approximately 90% of cancer-associated mortalities
are characterized by the metastatic ability of cancer cells to
spread from the site of origin and colonize distant organs or nodes
(16). The metastatic process has
been investigated; the metastasis of various types of cancer has a
heterogeneous biology, and may be dependent on certain factors
associated with the tissue of origin and region of metastasis
(17). Thus, it is important to
determine the biology of metastasis in various types of cancer.
IGFBP7 is a secreted protein that is diffusely expressed in
gastrointestinal tract tissues (18),
the ovaries (19) and liver (20). The present study investigated the role
of IGFBP7 in the progression of CRC from a primary state to the
development of metastatic disease, and highlights the importance of
IGFBP7 downregulation in LMs originating from CRC. To the best of
our knowledge, the present study is the first to directly analyze
the expression of IGFBP7 in PC and matched LM tissues. The results
revealed IGFBP7 downregulation in LM tissues compared with PC
tissues. Of note, IGFBP7 was significantly downregulated in the
invasive front of LM tissues, which may promote cell behaviors that
facilitate dissemination from the tumor. Furthermore, the potential
mechanism by which IGFBP7 serves a pivotal role in the process of
LM was investigated.
In the process of metastasis, CRC cells initially
lose their epithelial phenotype while simultaneously acquiring the
mesenchymal characteristics required for EMT. Numerous studies have
highlighted the vital role of EMT in the progression of cancer due
to its invasive and metastatic behaviors (21–23). To
the best of our knowledge, only one study has investigated the
association between IGFBP7 with EMT, in which IGFBP7 inhibited EMT
and tumor metastasis by suppressing transforming growth
factor-β-mediated EMT via the Smad signaling cascade (24). However, the differential expression of
IGFBP7 between PC and LM was not determined.
In the present study, it was demonstrated that the
downregulation of IGFBP7 from PC to LM tissue was correlated with
decreased E-cadherin, and increased N-cadherin and Vimentin
expression, suggesting a negative role for IGFBP7 in regulating
colon cancer cell invasion via EMT. In addition, several studies
have reported the effects of IGFBP7 overexpression on suppressing
the growth, invasion and migration of cancer cells. Of note, one
study revealed IGFBP7 upregulation during the process of homing
into the liver from as early as 3 days; however, the expression
thereof returned to basal levels thereafter in a rat model
(25). Those findings are
inconsistent with the observations of the present study as IGFBP7
was significantly downregulated in LM tissues compared with in PC
tissues. This suggests that the expression of IGFBP7 is tightly
regulated by its microenvironment. In addition, we noted blood
vessels, fibroblasts in stromal tissues, showing IGFBP7-positive
staining. IGFBP7 has been studied as a modulator of angiogenesis
and was found to inhibit tumor angiogenesis in several contexts
(11,26). To a certain degree, it is supposed
that IGFBP7 plays a role as mediator for tumor-stroma
interactions.
Notably, the majority of colon cancer cell lines
used in the present study, including HCT116, SW620, RKO and Caco2
revealed undetectable IGFBP-7 expression by RT-qPCR. By contrast,
the LοVο cell line exhibited the highest expression levels of
IGFBP7 compared with the aforementioned cell lines. One possible
reason is that IGFBP7 is completely or strongly methylated in those
cells except LοVο cells, where relatively weak methylation was
found (27). Furthermore,
inconsistent with the findings in the colon cancer cell lines in
this study, the expression of IGFBP7 has been detected in the
majority of colon cancer and adjacent normal tissues, though its
expression varies across these cells. In addition, tumor xenografts
generated by direct intravenous injection of prostate cell lines,
LNCaP or C4-2, into the bone marrow space resulted in tumors that
stained positively for IGFBP-7; however, IGFBP7 was undetected in
these cells lines in vitro (28). This indicated that the expression of
IGFBP-7 may be induced in vivo within an appropriate host
environment. Alterations in the expression of IGFBP7 were reported
in the present study. Upregulated expression at early Union for
International Cancer Control (UICC) stages was observed, followed
by a plateau and a decrease at the most advanced tumor stage IV,
which suggests the variable role of the IGFBP7 in CRC.
As the varying expression of IGFBP7 at different
UICC stages and the role of IGFBP7 in LM in CRC remain unknown, the
present study investigated a possible mechanism underlying the
regulation of IGFBP7 expression in metastasis. The expression of
IGFBP7 was positively associated with that of E-cadherin, but was
negatively associated with N-cadherin and Vimentin in colon cancer
tissues compared with matching LM tissues. Of note, cells at the
invasive tumor front in LM tissues exhibited downregulated IGFBP7
expression, which was accompanied with the loss of epithelial
markers, such as E-cadherin and cell-cell junctions, and a gain in
the expression of mesenchymal markers.
It has been suggested that cancer cells undergo EMT
in the PC site to facilitate the invasion and dissemination of a
tumor; this process is then reversed and is termed the
mesenchymal-epithelial transition (MET) in LM, for clonal outgrowth
at metastatic sites (29). It is
difficult to determine whether EMT or MET occurs during the
development of LM from primary tumors. In the present study, the
expression of E-cadherin was downregulated, while that of
N-cadherin and Vimentin were upregulated in matched LM tissues
compared with in PC samples. This is inconsistent with the findings
reported by Hur et al (30).
This may be accounted for by the use of synchronous LM tissues in
the present study, which are considered to have poorer prognosis
compared with patients with metachronous LM. In addition, MET is
not required in the process of tumor cell migration from the PC
site to the liver due failed cell cycle arrest upon the induction
of EMT, leading to genomic instability (31). This instability may result in highly
metastatic tumors that are resistant to next-line therapies
(32,33). Thus, the results of the present study
may provide insight into the behavior of tumors in synchronous LM
tissues; however, the specific mechanism remains unknown (34). Analysis of LM tissues indicated the
enhanced invasion and dissemination of tumors compared with that in
metachronous LM. Furthermore, these properties may be associated
with the varied expression of E-cadherin, N-cadherin and Vimentin
in the same sample cohort observed in the present study. Thus,
minor differences in the expression of E-cadherin, N-cadherin and
Vimentin between PC and LM tissues may occur as it is difficult to
determine which regions of tissue express EMT-associated proteins
prior to collection.
In summary, the findings of the present study
indicated that IGFBP7 overexpression leads to the direct targeting
of EMT-associated genes, which in turn regulates the metastatic
behavior of colon cancer cells. These data suggest the biological
and clinical importance of IGFBP7 in CRC; however, the role of
IGFBP7 in CRC-associated LM requires further investigation. In
addition, downregulation of IGFBP7 in PC associated with the
development of LM may facilitate the proliferation and expansion of
CRC cells.
Acknowledgements
The authors wish to thank Professor Shao Lee (School
of Life Sciences, Shanxi University, China) for critically
reviewing the manuscript in advance of submission.
Funding
No funding was received.
Availability of data and materials
All data generated or analyzed during this study are
included in this article or are available from the corresponding
author on reasonable request.
Authors' contributions
YLi and LL contributed to the study design and wrote
the manuscript. YX, GZ, JJ and HH performed the sample collection
and data analysis. YLi, YLiu and YG performed the experiments. All
authors read and approved the final manuscript.
Ethics approval and consent to
participate
Written informed consent was obtained from all
patients prior to enrolment. All procedures were conducted in
accordance with standard guidelines for the Study of Humans and
were approved by the Research Ethics Committee of First Hospital of
Shanxi Medical University.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Miller KD, Siegel RL, Lin CC, Mariotto AB,
Kramer JL, Rowland JH, Stein KD, Alteri R and Jemal A: Cancer
treatment and survivorship statistics, 2016. CA Cancer J Clin.
66:271–289. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wale A, Van Cutsem E, Rao S, Cunningham D
and Brown G: Session 2: Synchronous metastatic disease-liver first
or primary first? The oncologist decides. Colorectal Dis. 20 (Suppl
1):S52–S55. 2018. View Article : Google Scholar
|
|
3
|
Loret N, Denys H, Tummers P and Berx G:
The role of epithelial-to-mesenchymal plasticity in ovarian cancer
progression and therapy resistance. Cancers (Basel). 11:E8382019.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Blick T, Widodo E, Hugo H, Waltham M,
Lenburg ME, Neve RM and Thompson EW: Epithelial mesenchymal
transition traits in human breast cancer cell lines. Clin Exp
Metastasis. 25:629–642. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bastid J: EMT in carcinoma progression and
dissemination: Facts, unanswered questions, and clinical
considerations. Cancer Metastasis Rev. 31:277–283. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Gomes LR, Terra LF, Sogayar MC and
Labriola L: Epithelial-mesenchymal transition: Implications in
cancer progression and metastasis. Curr Pharm Biotechnol.
12:1881–1890. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Brabletz T, Kalluri R, Nieto MA and
Weinberg RA: EMT in cancer. Nat Rev Cancer. 18:128–134. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Brabletz T: To differentiate or not-routes
towards metastasis. Nat Rev Cancer. 12:425–436. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yamanaka Y, Wilson EM, Rosenfeld RG and Oh
Y: Inhibition of insulin receptor activation by insulin-like growth
factor binding proteins. J Biol Chem. 272:30729–30734. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ruan W, Xu E, Xu F, Ma Y, Deng H, Huang Q,
Lv B, Hu H, Lin J, Cui J, et al: IGFBP7 plays a potential tumor
suppressor role in colorectal carcinogenesis. Cancer Biol Ther.
6:354–359. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Tamura K, Hashimoto K, Suzuki K, Yoshie M,
Kutsukake M and Sakurai T: Insulin-like growth factor binding
protein-7 (IGFBP7) blocks vascular endothelial cell growth factor
(VEGF)-induced angiogenesis in human vascular endothelial cells.
Eur J Pharmacol. 610:61–67. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Benatar T, Yang W, Amemiya Y, Evdokimova
V, Kahn H, Holloway C and Seth A: IGFBP7 reduces breast tumor
growth by induction of senescence and apoptosis pathways. Breast
Cancer Res Treat. 133:563–573. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Evdokimova V, Tognon CE, Benatar T, Yang
W, Krutikov K, Pollak M, Sorensen PH and Seth A: IGFBP7 binds to
the IGF-1 receptor and blocks its activation by insulin-like growth
factors. Sci Signal. 5:ra922012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zlobec I, Terracciano L, Jass JR and Lugli
A: Value of staining intensity in the interpretation of
immunohistochemistry for tumor markers in colorectal cancer.
Virchows Arch. 451:763–769. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Gupta GP and Massagué J: Cancer
metastasis: Building a framework. Cell. 127:679–695. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Joyce JA and Pollard JW:
Microenvironmental regulation of metastasis. Nat Rev Cancer.
9:239–252. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
18
|
Takeno A, Takemasa I, Doki Y, Yamasaki M,
Miyata H, Takiguchi S, Fujiwara Y, Matsubara K and Monden M:
Integrative approach for differentially overexpressed genes in
gastric cancer by combining large-scale gene expression profiling
and network analysis. Br J Cancer. 99:1307–1315. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Gambaro K, Quinn MC, Cáceres-Gorriti KY,
Shapiro RS, Provencher D, Rahimi K, Mes-Masson AM and Tonin PN: Low
levels of IGFBP7 expression in high-grade serous ovarian carcinoma
is associated with patient outcome. BMC Cancer. 15:1352015.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Tomimaru Y, Eguchi H, Wada H, Kobayashi S,
Marubashi S, Tanemura M, Umeshita K, Kim T, Wakasa K, Doki Y, et
al: IGFBP7 downregulation is associated with tumor progression and
clinical outcome in hepatocellular carcinoma. Int J Cancer.
130:319–327. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Liao TT and Yang MH: Revisiting
epithelial-mesenchymal transition in cancer metastasis: The
connection between epithelial plasticity and stemness. Mol Oncol.
11:792–804. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Cao H, Xu E, Liu H, Wan L and Lai M:
Epithelial-mesenchymal transition in colorectal cancer metastasis:
A system review. Pathol Res Pract. 211:557–569. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Heerboth S, Housman G, Leary M, Longacre
M, Byler S, Lapinska K, Willbanks A and Sarkar S: EMT and tumor
metastasis. Clin Transl Med. 4:62015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhu S, Zhang J, Xu F, Xu E, Ruan W, Ma Y,
Huang Q and Lai M: IGFBP-rP1 suppresses epithelial-mesenchymal
transition and metastasis in colorectal cancer. Cell Death Dis.
6:e16952015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Georges RB, Adwan H, Hamdi H, Hielscher T,
Linnemann U and Berger MR: The insulin-like growth factor binding
proteins 3 and 7 are associated with colorectal cancer and liver
metastasis. Cancer Biol Ther. 12:69–79. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Hooper AT, Shmelkov SV, Gupta S, Milde T,
Bambino K, Gillen K, Goetz M, Chavala S, Baljevic M, Murphy AJ, et
al: Angiomodulin is a specific marker of vasculature and regulates
vascular endothelial growth factor-A-dependent neoangiogenesis.
Circ Res. 105:201–208. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Suzuki H, Igarashi S, Nojima M, Maruyama
R, Yamamoto E, Kai M, Akashi H, Watanabe Y, Yamamoto H, Sasaki Y,
et al: IGFBP7 is a p53-responsive gene specifically silenced in
colorectal cancer with CpG island methylator phenotype.
Carcinogenesis. 31:342–349. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Long TJ, Sprenger CC, Plymate SR and
Ratner BD: Prostate cancer xenografts engineered from 3D
precision-porous poly(2-hydroxyethyl methacrylate) hydrogels as
models for tumorigenesis and dormancy escape. Biomaterials.
35:8164–8174. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Thiery JP: Epithelial-mesenchymal
transitions in tumour progression. Nat Rev Cancer. 2:442–454. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Hur K, Toiyama Y, Takahashi M, Balaguer F,
Nagasaka T, Koike J, Hemmi H, Koi M, Boland CR and Goel A:
MicroRNA-200c modulates epithelial-to-mesenchymal transition (EMT)
in human colorectal cancer metastasis. Gut. 62:1315–1326. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Comaills V, Kabeche L, Morris R, Buisson
R, Yu M, Madden MW, LiCausi JA, Boukhali M, Tajima K, Pan S, et al:
Genomic instability is induced by persistent proliferation of cells
undergoing epithelial-to-mesenchymal transition. Cell Rep.
17:2632–2647. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Sun Y, Campisi J, Higano C, Beer TM,
Porter P, Coleman I, True L and Nelson PS: Treatment-induced damage
to the tumor microenvironment promotes prostate cancer therapy
resistance through WNT16B. Nat Med. 18:1359–1368. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Creighton CJ, Li X, Landis M, Dixon JM,
Neumeister VM, Sjolund A, Rimm DL, Wong H, Rodriguez A,
Herschkowitz JI, et al: Residual breast cancers after conventional
therapy display mesenchymal as well as tumor-initiating features.
Proc Natl Acad Sci USA. 106:13820–13825. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Adam R, de Gramont A, Figueras J, Kokudo
N, Kunstlinger F, Loyer E, Poston G, Rougier P, Rubbia-Brandt L,
Sobrero A, et al: Managing synchronous liver metastases from
colorectal cancer: A multidisciplinary international consensus.
Cancer Treat Rev. 41:729–741. 2015. View Article : Google Scholar : PubMed/NCBI
|