Introduction
Hepatocellular carcinoma (HCC) is the fifth most
prevalent and the third most lethal type of cancer, making it a
highly aggressive malignancy. Over six million new cases and a
similar number of deaths are reported worldwide each year (1,2). Although
surgical resection and transplantation are effective for HCC
treatment, patients who have undergone tumor resection have a high
rate of recurrence primarily, due to intrahepatic spread and
extrahepatic metastasis (3). Indeed,
metastasis remains the primary obstacle to survival in patients
with HCC. Understanding the essential molecular pathways for tumor
metastasis is essential in order to improve therapeutic strategies.
However, there is limited knowledge regarding the molecular and
environmental mechanisms mediating the metastatic recurrence of
HCC, and therapeutic choices for HCC treatment remain scarce. Thus,
elucidating the mechanisms of HCC metastasis may provide a solid
basis for the clinical prediction and prevention of HCC
metastasis.
MicroRNAs (miRNAs) are small non-coding RNAs of ~22
nucleotides in length that negatively regulate the expression of
various genes, primarily through interactions with the 3′
untranslated regions (3′UTRs) of their mRNAs (4,5). miRNAs
can post-transcriptionally modulate ~30% of human genes, indicating
that they may serve as crucial regulators of human biological
processes, including cancer development, cell proliferation,
differentiation, and apoptosis (6,7). Several
dozen miRNAs have been demonstrated to be aberrantly expressed in
various human cancers. For example, miR-96, miR-128, and miR-193b
are associated with pancreatic cancer (8–10);
miR-200, miR-183, and miR-22 are associated with gastric cancer
(11–13); and miR-185, miR-143, and miR-145 are
associated with breast cancer (14,15).
However, the role of miRNAs in HCC metastasis remains poorly
understood. A more thorough understanding of deregulated miRNAs in
HCC may elucidate the mechanisms of HCC development and may result
in improvements of HCC diagnosis and treatment.
In a previous study from our group, a tumor tissue
microarray was used to identify that miR-141 was significantly
downregulated in renal cell carcinoma (RCC). That previous study
also revealed that high expression of miR-141 dramatically
inhibited the proliferation and metastasis of RCC cells both in
vitro and in nude mouse models (16). Recently, miR-141 was reported to be
correlated with metastasis in various types of cancer, including
gastric cancer (17,18), non-small cell lung cancer (19), pancreatic cancer (20), and bladder cancer (21). Several studies have also demonstrated
that miR-141 functions as a metastasis suppressor by targeting E2F
transcription factor 3 (E2F3), zinc-finger E-box binding homeobox 2
(ZEB2), and T cell lymphoma invasion and metastasis 1 (Tiam1) in
HCC (22–24). However, these studies were not
in-depth analyses of the clinical value of miR-141 in HCC or its
functional roles.
The present study demonstrated for the first time
that miR-141 directly targeted transforming growth factor β
receptor 1 (TGFβR1), which is an important factor in HCC growth,
migration and invasion. These findings suggest that miR-141 may
have clinical applications owing to its effects on proliferation
and invasion.
Materials and methods
Patients and tissue samples
Clinical HCC tissues and adjacent non-tumor tissues
(≥3 cm from the tumor), used for reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) and western blotting, were
collected from 19 patients undergoing hepatectomy at Union
Hospital, Tongji Medical College, Huazhong University of Science
and Technology (Wuhan, China). These specimens were obtained from
patients with HCC from April to August, 2013. Patients aged 50–60
years and diagnosed with only primary hepatocellular carcinoma were
selected as a late stage study. Informed consent was obtained from
all patients prior to enrollment in the study. The Ethics Committee
of Tongji Medical College approved the present study. Clinical
pathological reports were collected. Tissue samples were
immediately snap-frozen in liquid nitrogen at the time of surgery
and stored at −80°C.
Cell culture and materials
The human hepatocellular carcinoma cell lines Huh7
and SK-HEP-1 and the human liver cell line LO2 (HL-7702) were
purchased from the China Center for Type Culture Collection (Wuhan,
China). Cells were cultured in Dulbecco's modified Eagle's medium
(DMEM; HyClone; GE Healthcare Life Sciences, Logan, UT, USA),
supplemented with 10% fetal bovine serum (FBS; HyClone; GE
Healthcare Life Sciences), 100 U/ml penicillin sodium, and 0.1
mg/ml streptomycin sulfate in a humidified incubator at 37°C with
an atmosphere containing 5% CO2.
RNA extraction and RT-qPCR
Total RNA was extracted from the patient liver
tissues or cultured cells using TRIzol reagent (Invitrogen; Thermo
Fisher Scientific, Inc., Waltham, MA, USA) according to the
manufacturer's protocol. The RNA concentrations and A260/A280
ratios were measured with a plate reader (BioTek Instruments, Inc.,
Winooski, VT, USA). Subsequently, reverse transcription was
performed using a RevertAid First Strand cDNA Synthesis kit (Thermo
Fisher Scientific, Inc.). qPCR was performed using a SYBR Green
qPCR Supermix UDG kit (Invitrogen; Thermo Fisher Scientific Inc.).
The primers for miRNA detection were purchased from Ribobio Co.,
Ltd. (Guangzhou, China), and the primers for mRNA detection were
purchased from Genewiz, Inc. (Jiangsu, China; Table I). U6 small nuclear RNA and GAPDH were
used as internal controls for miRNA and mRNA, respectively. Primer
sequences were as follows: TGFβR1, forward
5′-CACAGAGTGGGAACAAAAAGGT-3′ and reverse
5′-CCAATGGAACATCGTCGAGCA-3′; GAPDH, forward
5′-GGTGAAGGTCGGAGTCAACGG-3′ and reverse
5′-GAGGTCAATGAAGGGGTCATTG-3′; miR-141, forward
5′-GGCGTCACAGGCATTCACAGTC-3′ and reverse
5′-TCTTCCTCCTTCCTTCTCCG-3′; and U6, forward
5′-CTCGCTTCGGCAGCACATATACT-3′ and reverse 5′-CGGCTGCAGATGAGATAG-3′.
qPCR was performed on a Roche Light 480 system (LightCycler 480;
Roche Applied Science, Penzberg, Germany). All reactions were
performed in triplicate. Relative fold changes in mRNA expression
were calculated using the formula 2−ΔΔCT.
 | Table I.Primers for qRT-PCR. |
Table I.
Primers for qRT-PCR.
| Primers | Forward
(5′-3′) | Reverse
(5′-3′) | Product size
(bp) |
|---|
| TGFβR1 |
CACAGAGTGGGAACAAAAAGGT |
CCAATGGAACATCGTCGAGCA | 143 |
| GAPDH |
GGTGAAGGTCGGAGTCAACGG |
GAGGTCAATGAAGGGGTCATTG | 112 |
Protein isolation and
immunoblotting
Cells or tissues were washed twice with ice-cold
PBS, scraped into RIPA buffer (50 mM Tris-HCl, pH 7.4, 1% v/v
Triton X-100, 1 mM ethylenediaminetetraacetic acid, 0.1% m/v SDS,
1% m/v glycodeoxycholate, and 150 mM NaCl) with freshly added
protease inhibitor cocktail and phenylmethylsulfonyl fluoride for
30 min on ice, and then centrifuged at 12,000 × g at 4°C for 10
min. The supernatants were then collected. Protein (50 µg) was
separated on 10% SDS-polyacrylamide gels and then transferred to
polyvinylidene difluoride membrane (0.45 µm; EMD Millipore,
Billerica, MA, USA). The membranes were blocked with 5% non-fat
milk, and the primary antibody was added at 4°C overnight. The
membranes were then incubated with horseradish
peroxidase-conjugated secondary antibody for 1 h at room
temperature. The following antibodies were used: rabbit polyclonal
anti-TGFβR1 (sc-399; 1:200; Santa Cruz Biotechnology, Inc., Dallas,
TX, USA), rabbit polyclonal anti-SMAD family member 2 (ab40855;
1:1,000; Abcam, Cambridge, MA, USA), rabbit monoclonal
anti-phosphorylated (p-) Smad2 (ab188334; 1:1,000; Abcam), mouse
monoclonal anti-β-actin (sc-81178; 1:200; Santa Cruz Biotechnology,
Inc.), anti-mouse secondary horseradish peroxidase (HRP)-conjugated
antibodies, and anti-rabbit secondary HRP-conjugated antibodies
(ab205718; 1:2,000; Abcam). Reactive bands were visualized with ECL
reagent (Pierce; Thermo Fisher Scientific, Inc.) on a Bio-Rad Image
Analysis System (Bio-Rad Laboratories, Inc., Hercules, CA, USA).
Protein expression was quantified using QuantityOne v4.6.2 software
(Bio-Rad Laboratories, Inc.).
Analysis of cell viability, colony
formation, and cell cycle distribution
Dimethyl thiazolyl diphenyl tetrazolium (MTT;
Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) assays were
performed to determine cell viability in 96-well plates according
to standard procedures. Cells were seeded at a density of 3,000
cells/well containing 100 µl culture medium. At 24-h intervals, 20
µl of 5 mg/ml MTT was added, and plates were incubated at 37°C for
4 h. The medium was then removed, and 150 µl dimethyl sulfoxide was
added per well to dissolve the precipitate. The optical density was
tested at a wavelength of 490 nm. Wells with 100 µl culture medium
and no cells were used as blanks.
For colony formation assays, single-cell suspensions
(2,000 cells/well) were plated in each well of 6-well plates. After
1 week of culture, the surviving colonies were fixed with 95%
methanol for 10 min, stained with 0.1% crystal violet for 20 min,
imaged, and counted. The experiments were repeated at least three
times. For cell cycle assays, the cells were fixed in 70% ethanol,
stored at −20°C for at least 24 h, and then stained with propidium
iodide prior to flow cytometry (ZE5; Bio-Rad Laboratories,
Inc.).
In vitro migration and invasion
assay
Twenty-four-well Transwell plates (8 µm pore size;
Corning Inc., Corning, NY, USA) were used to measure the migratory
and invasive abilities of the cells. Cells were serum starved for
24 h, trypsinized, and suspended in serum-free medium at
5×104 cells/200 µl. For Transwell migration assays,
5×104 Huh7-NC or Huh7-141 cells were added into the
upper of Transwell chambers containing uncoated membranes. For
invasion assays, a similar number of cells was seeded into the
upper chamber of the Transwell that were precoated with 200 mg/ml
Matrigel (BD Biosciences). Then, 600 µl DMEM supplemented with
10–20% FBS was added to the lower of the Transwells. After
incubation at 37°C for 20 h, the cells were removed and washed with
PBS three times. Non-migratory or non-invasive cells on the top
chambers were gently removed using cotton wool. The cells on the
underside of the membranes were fixed with 95% methanol for 10 min,
air-dried, stained with 0.1% crysal violet for 15 min, and counted
under an inverted light microscope (magnification, ×200).
miRNA and small interfering RNA
(siRNA) transfection
For establishment of cell lines, cells were infected
with lentivirus (GV369 vector; GeneChem Co., Ltd., Shanghai, China)
after reaching 30–50% confluence, according to the manufacturer's
recommendations. After 48 h of incubation, the infection efficiency
was detected by evaluating green fluorescent protein (GFP)
expression under a fluorescence microscope; infection of at least
90% of cells was considered effective. Successful infection was
also confirmed using RT-qPCR. After identification, the cells were
cultured and harvested for subsequent experiments.
siRNA against TGFβR1 and negative control
oligonucleotides and siRNA against miR-141 and negative control
oligonucleotides were synthesized by GenePhama Co., Ltd. (Shanghai,
China). The oligonucleotides used in these studies were as follows:
TGFβR1 siRNA, 5′-AGAAAUGAAGUGGCAUUAATT-3′ (sense) and
5′-UUAAUGCCACUUCAUUUCUTT-3′ (antisense). siRNA against miR-141 and
the negative control oligonucleotides were ready-made by GenePhama
Co., Ltd. Oligonucleotide transfection was performed using
Lipofectamine 2000 Reagent (Invitrogen; Thermo Fisher Scientific,
Inc.) according to the manufacturer's protocol. At 48 h
post-transfection, the cells were harvested for subsequent
experiments.
Bioinformatic analysis
The prediction of miRNA targets was performed using
these publicly available algorithms: TargetScan (http://www.targetscan.org; Whitehead Institute,
Cambridge, MA, USA), MiRanda (http://cbio.mskcc.org/mirnaviewer; Memorial
Sloan-Kettering Cancer Center, New York, NY, USA) and StarBase
(http://starbase.sysu.edu.cn; Sun Yat-sen
University, Guangzhou, China).
Vector construction and dual
luciferase reporter assays
The wild-type (wt) 3′ untranslated region (UTR) of
the human TGFβR1 gene, which was predicted to interact with
miR-141, and a mutant (mut) variant, were constructed and cloned
into the GV272 vector (GeneChem, Inc., Daejeon, Korea). For
luciferase reporter activity assays, cells were seeded in 24-well
plates and allowed to reach 70% confluence prior to transfection.
Next, wt TGFβR1 3′UTR reporter vector, mut TGFβR1 3′UTR reporter
vector, or the negative control vector were cotransfected with the
Renilla firefly luciferase-expressing vector into the cells
using Lipofectamine 2000 reagent. Cell lysates were collected 24 h
later, and luciferase activity was determined with a
Dual-Luciferase Reporter System (Promega Corporation, Madison, WI,
USA). Relative Renilla luciferase activities were used as an
internal control for transfection efficiency.
Statistical analysis
All values are presented as means ± standard
deviations. MTT assays, RT-qPCR, dual luciferase reporter assays,
and cell cycle analysis were performed in triplicate. Statistical
analyses were performed with GraphPad Prism (version 5.01 for
Windows; GraphPad, Inc., La Jolla, CA, USA). Differences between
two groups were assessed by Student's t-tests. Differences among
multiple groups were assessed with one-way analysis of variance,
and Tukey's post hoc test. The correlation between levels of
miR-141 and TGFβR1 mRNA in HCC tissues was analyzed with Pearson's
correlation analysis. P<0.05 was considered to indicate a
statistically significant difference.
Results
miR-141 is frequently downregulated in
human HCC tissues and cell lines
Based on published reports (25–29) and
our previous studies (16,30) of miR-200 family members in several
types of cancer, the present study attempted to clarify the role of
the miR-200 family member miR-141 in human HCC progression. To
investigate the clinical implications of miR-141 in HCC
progression, first the expression levels of miR-141 were determined
in 19 pairs of primary HCC tissues and their matched adjacent
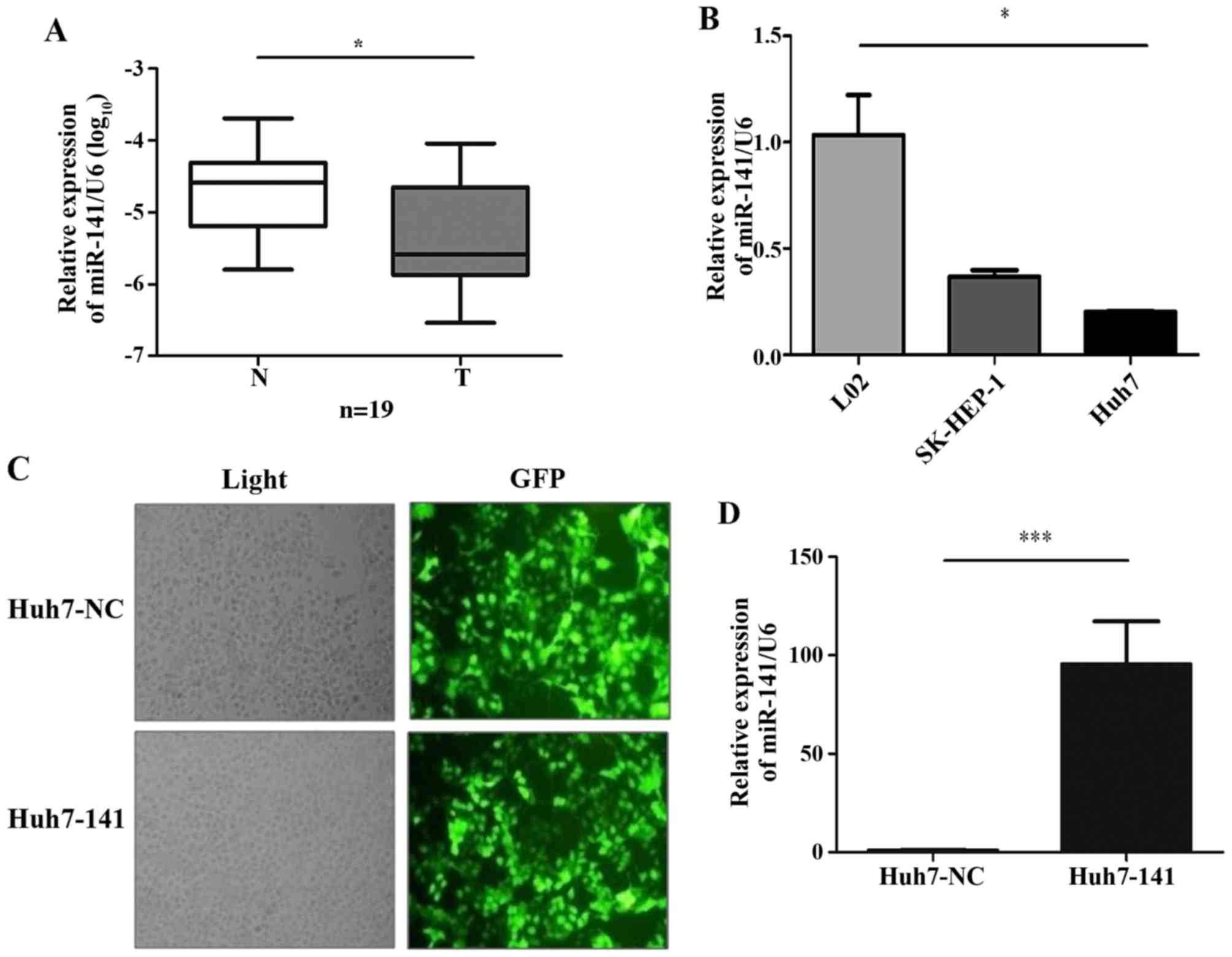
non-tumor tissues, using miRNA-based RT-qPCR. The results
demonstrated that miR-141 was significantly downregulated in 89%
(17 of 19; P=0.012) of HCC tissues compared with their matched
non-tumor tissues (Fig. 1A),
suggesting that decreased expression of miR-141 occurred frequently
in HCC and that miR-141 may function as a suppressor of HCC
development.
To investigate the role of miR-141 in HCC in
vitro, the expression levels of miR-141 were detected in three
human liver cell lines. The results revealed that miR-141
expression was substantially lower in two liver cancer cell lines,
SK-HEP-1 and Huh7, compared with the normal liver LO2 cells
(Fig. 1B). Next, stable cell lines
were established via infection of the Huh7 cells with either a
lentivirus harboring an miR-141-overexpressing vector (cells termed
Huh7-141) or a control empty vector (cells termed Huh7-NC). The
efficiency of infection was >90%, as evidenced by the expression
of GFP encoded in the lentiviral vector (Fig. 1C). The efficiency of miR-141
overexpression was further confirmed by RT-qPCR (Fig. 1D).
miR-141 negatively regulates HCC
proliferation, migration, and invasion in vitro
Tumor cell proliferation, migration, and invasion
are crucial processes of HCC progression. The significant
downregulation of miR-141 in HCC tissues and cell lines, compared
with normal tissues and cells, indicated that miR-141 may have a
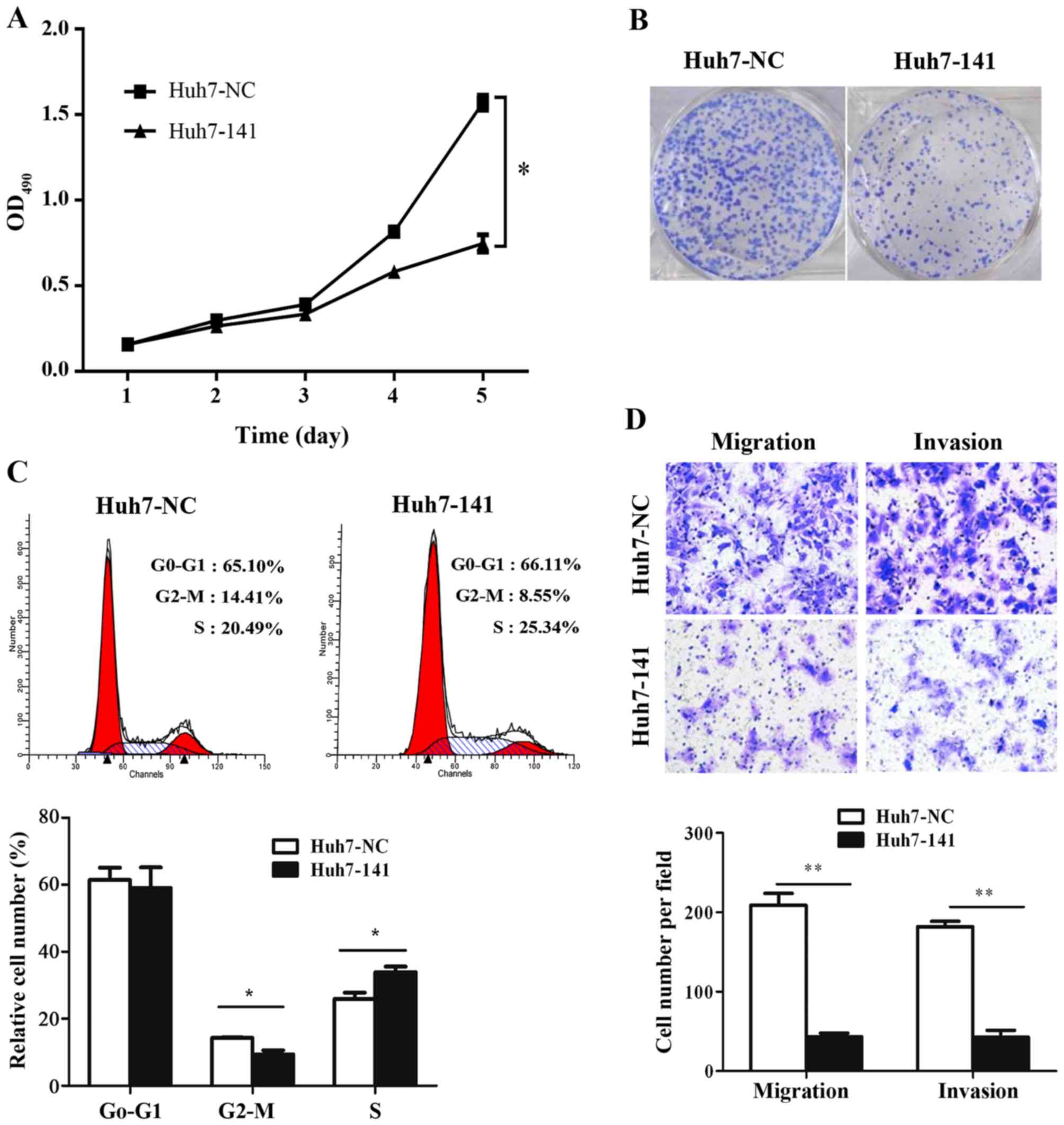
biologically significant role in HCC tumorigenesis. First, MTT
assays, colony formation assays, and flow cytometry were used to
investigate whether miR-141 affects the growth of HCC cells in
vitro. MTT assays demonstrated that overexpression of miR-141
significantly reduced the numbers of viable Huh7 cells over time
(Fig. 2A). The viability of Huh7-141
cells decreased by 52.37% by day 5 compared with Huh7-NC cells
(Fig. 2A). These data implied that
miR-141 suppressed HCC cell growth. Colony formation assays were
employed to confirm these results. As illustrated in Fig. 2B, far fewer colonies were formed
following miR-141 overexpression in HCC cells, compared with the
control cells. In addition, cell cycle analysis revealed that
overexpression of miR-141 increased cell cycle arrest at the S
phase by 4.85%, with 5.86% fewer cells entering G2/M phase
(Fig. 2C; P<0.05). Taken together,
these observations suggested that miR-141 suppressed the
proliferation of Huh7 cells by inducing cell cycle arrest.
To determine the effects of miR-141 on the migration
and invasion abilities of HCC cells, Transwell assays were
performed in vitro. In accordance with our hypothesis,
ectopic miR-141 expression significantly suppressed HCC cell
migration and invasion compared with control cells (Fig. 2D). These data revealed an obvious
negative correlation between miR-141 expression and HCC migration
and invasion.
miR-141 directly targets the 3′UTR of
TGFβR1
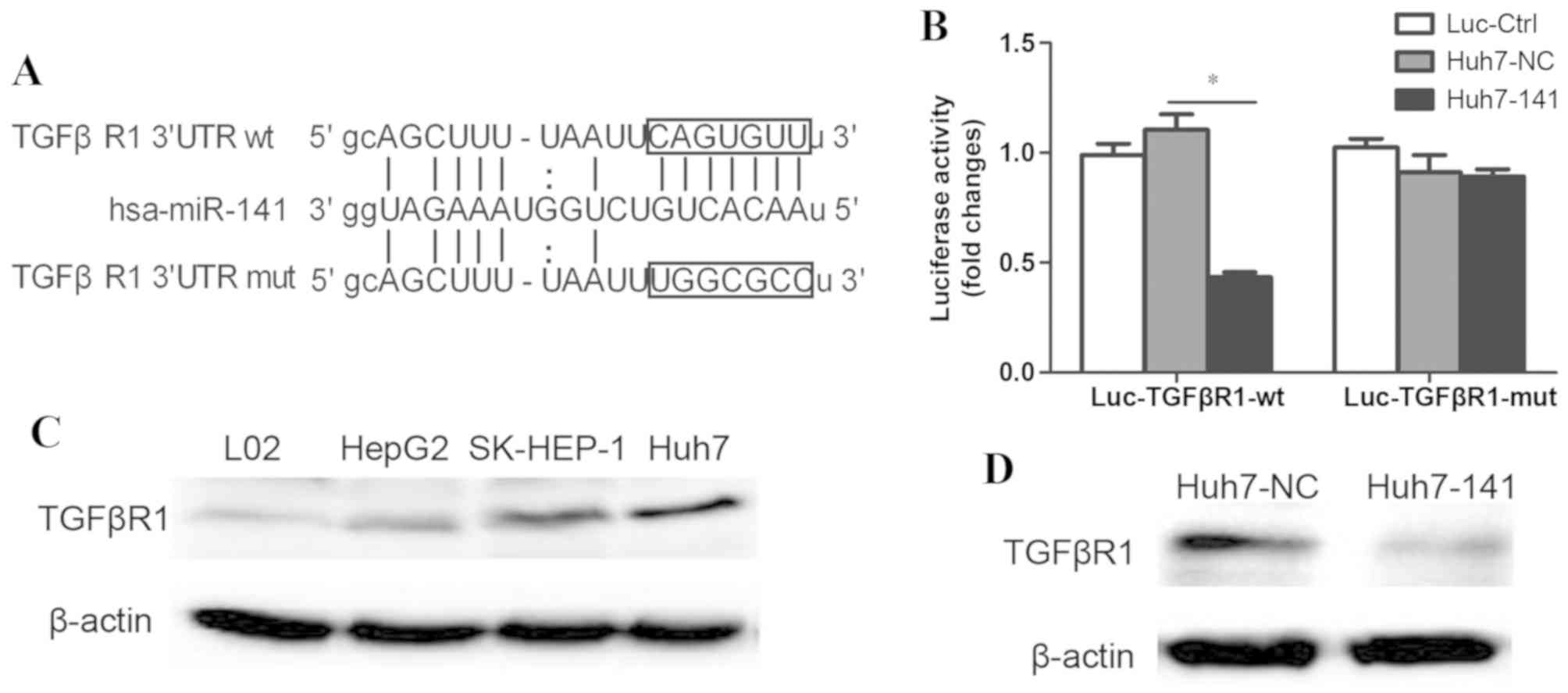
To explore the molecular mechanisms through which
miR-141 suppressed HCC migration, a search was performed to
identify candidate gene targets of miR-141 that are involved in HCC
pathogenesis. With a combination of miRNA target prediction
programs, such as TargetScan and MiRanda, and through review of the
literature, the 3′UTR of TGFβR1 was identified as complementary to
the miR-141 sequence; thus, miR-141 may exert its biological
functions by targeting TGFβR1 mRNA (Fig.
3A). To test this hypothesis, a luciferase-reporter plasmid
(Luc-TGFβR1-wt) was constructed in which the nucleotides of the
wild-type (wt) TGFβR1 3′UTR (complementary to miR-141) were
inserted into a GV238 vector downstream of the luciferase reporter
gene. A mutant reporter (Luc-TGFβR1-mut; in which the first six
nucleotides in the miR-141 seed region were changed; Fig. 3A), as well as a negative control
reporter (Luc-Ctrl) were also constructed. Luc-TGFβR1-wt,
Luc-TGFβR1-mut, or Luc-Ctrl was transfected into Huh7 cells
overexpressing miR-141. Luciferase assays revealed that the
luciferase activity decreased significantly only in
miR-141-overexpressing Huh7 cells that were transfected with
Luc-TGFβR1-wt (Fig. 3B). However, no
effect was observed following transfection with Luc-TGFβR1-mut
(Fig. 3B).
To further investigate whether miR-141 reduced
endogenous TGFβR1, the expression of TGFβR1 was detected in various
cell lines by western blotting. The results demonstrated that the
protein expression levels of TGFβR1 were higher in Huh7 cells
compared with the other cell lines (Fig.
3C), and appeared to be inversely correlated with the
expression levels of miR-141 (Fig.
1B). Additionally, overexpression of miR-141 in Huh7 cells
significantly reduced the protein expression levels of TGFβR1
protein (Fig. 3D). These data
demonstrated that miR-141 specifically bound to the 3′UTR of wt
TGFβR1 (but not the mutant), and reduced the expression of TGFβR1.
Therefore, the present results indicated that TGFβR1 was a specific
target of miR-141.
Upregulation of TGFβR1 was inversely
correlated with miR-141 expression in vivo
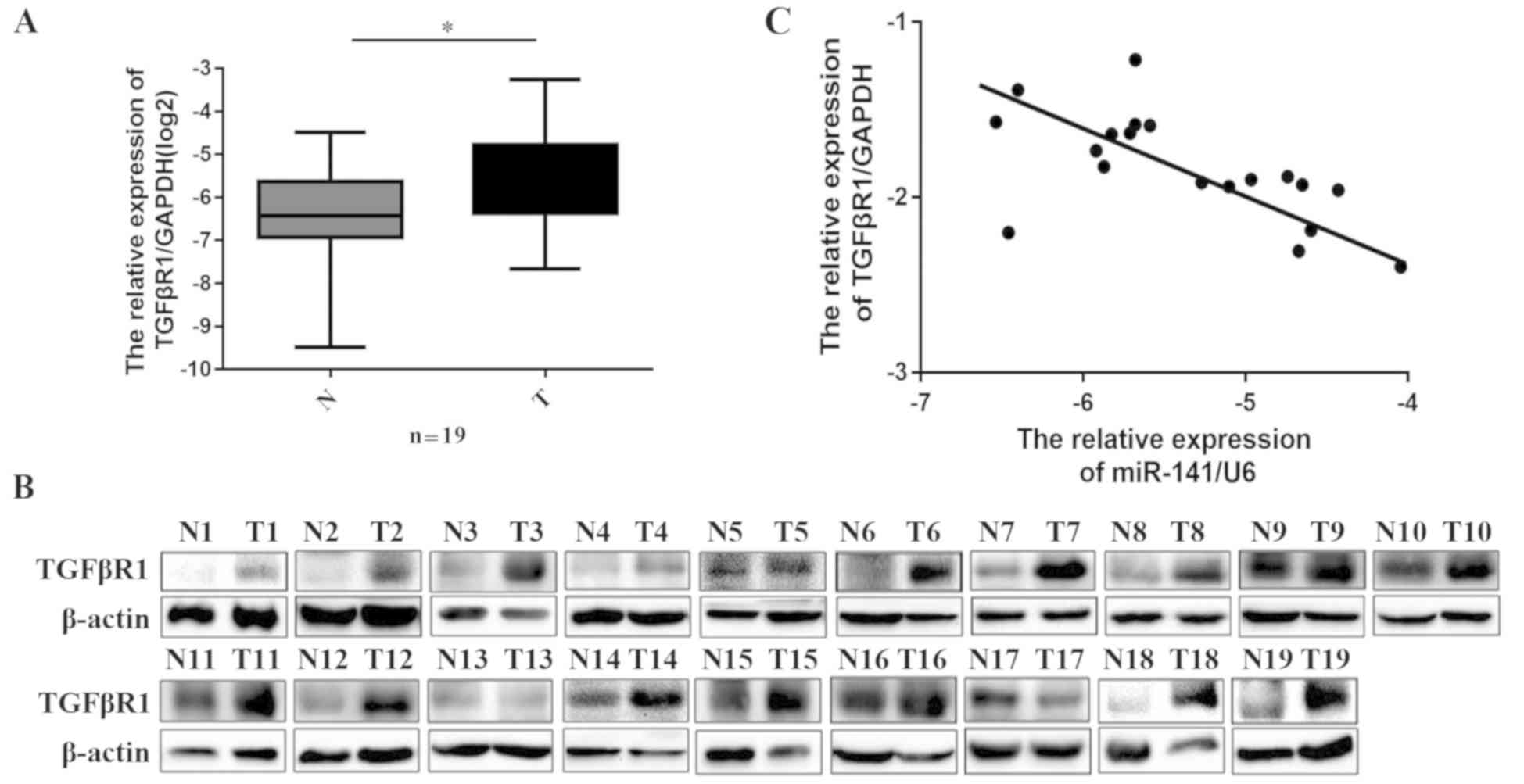
Because miR-141 was downregulated in HCC patient
samples and because miR-141 was demonstrated to specifically target
TGFβR1, we next investigated whether the expression of TGFβR1 was
negatively associated with miR-141 levels in patient tissues. The
mRNA expression levels of TGFβR1 were determined by RT-qPCR in the
20 pairs of HCC tissues and matched adjacent non-tumor tissues
collected in the present study. The results demonstrated that
TGFβR1 mRNA expression was significantly higher in HCC tissues
compared with adjacent non-tumor tissues (Fig. 4A; P=0.027). In addition, western
blotting revealed that the protein levels of TGFβR1 in HCC were
significantly higher compared with adjacent non-tumor tissues
(Fig. 4B). In Fig. 1A, miR-141 expression was demonstrated
to be downregulated in HCC tissues. Indeed, the expression of
miR-141 was significantly inversely correlated with TGFβR1
expression (Fig. 4C; P=0.048,
R2=0.21). Taken together, these findings suggested that TGFβR1 was
a functional target of miR-141 in HCC.
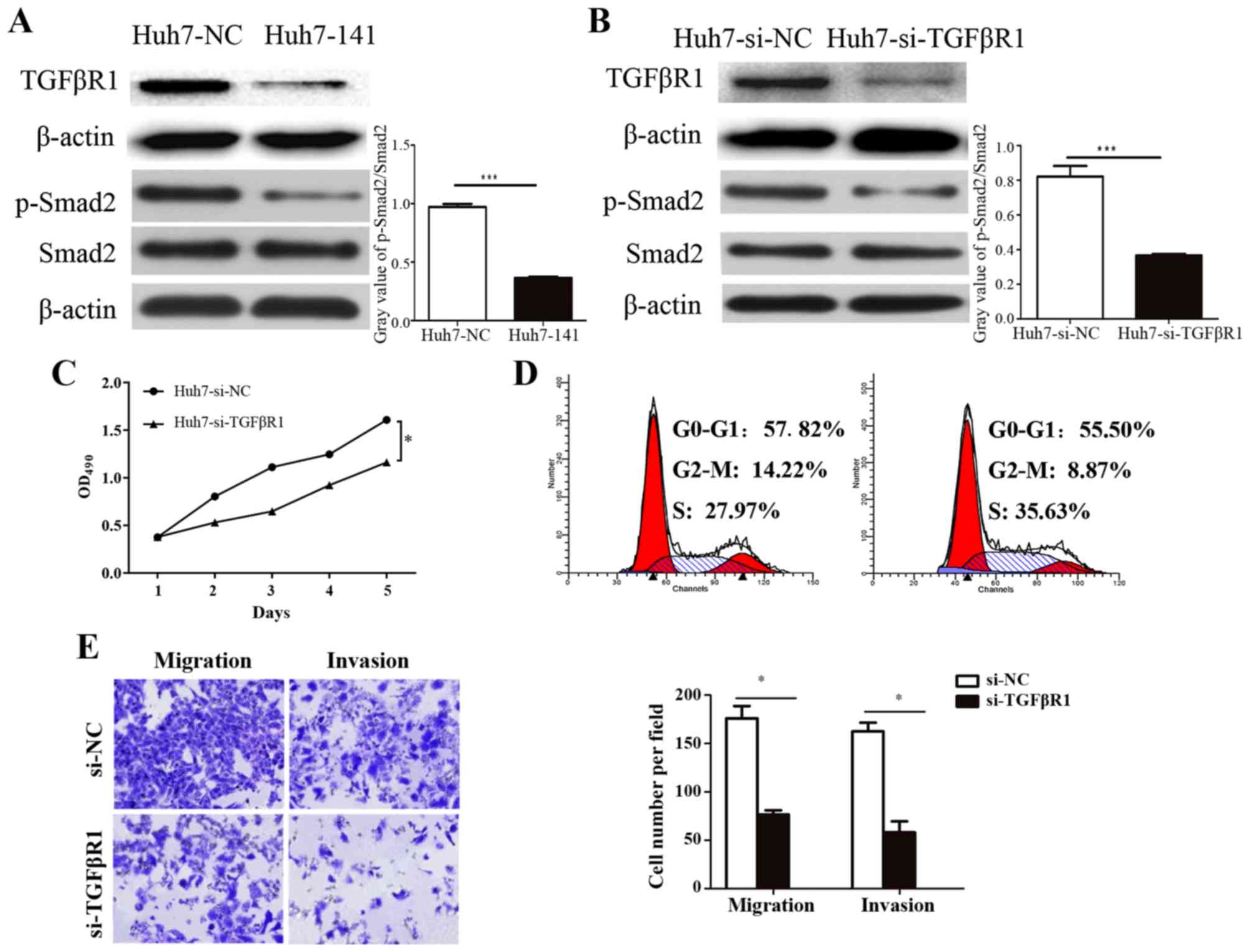
Knockdown of TGFβR1 has similar
effects to miR-141 overexpression in vitro
To further investigate the specificity of miR-141
and TGFβR1, TGFβR1 expression was silenced using siRNA, and the
effects were compared to the effects of miR-141 overexpression.
Similar to the effects of miR-141 overexpression (Fig. 5A), western blotting results
demonstrated that TGFβR1 siRNA transfection clearly reduced the
expression of TGFβR1 and decreased the phosphorylation of its
downstream protein Smad2 (Fig. 5B).
MTT assays revealed that TGFβR1 knockdown significantly inhibited
Huh7 cell growth (Fig. 5C), while
cell cycle analysis revealed that TGFβR1 knockdown reduced cell
cycle arrest at the G2/M phase by 5.35%, and increased the ratio of
cells entering the S phase by 7.66% (Fig.
5D; P<0.05). Finally, silencing of TGFβR1 significantly
impaired migration and invasion in Huh7 cells (Fig. 5E). These results further confirmed
that TGFβR1 was a key factor involved in miR-141-induced
suppression of HCC cell proliferation, migration, and invasion, by
blocking TGFβ signaling.
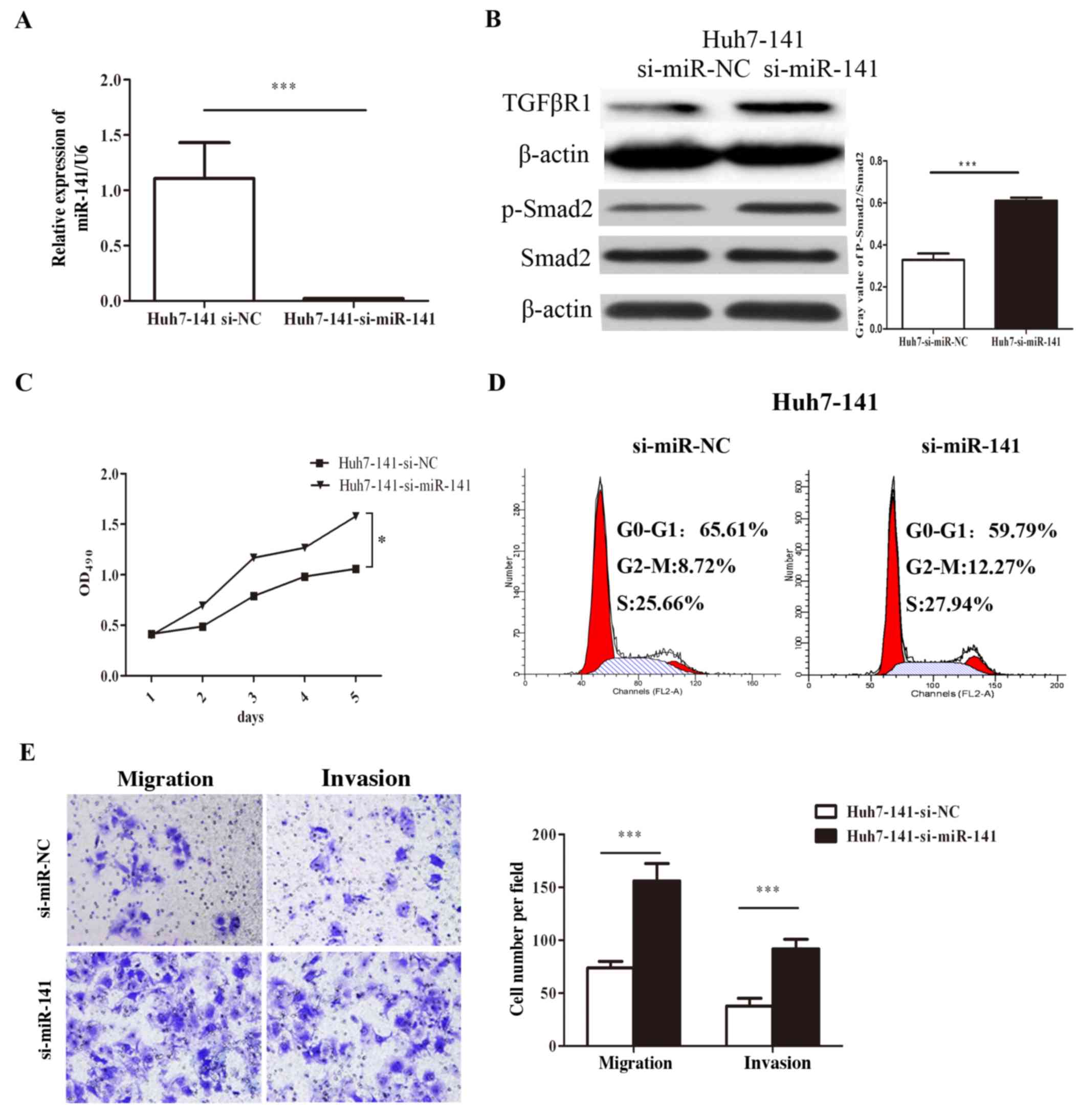
Inhibition of miR-141 promotes HCC
cell proliferation, migration and invasion in vitro
A rescue experiment was performed, and the results
revealed that inhibition of miR-141 restored HCC proliferation,
migration, and invasion, as well as the expression of TGFβR1 and
p-Smad2 (Fig. 6). First, RT-qPCR
results confirmed that specific siRNA against miR-141 (si-miR-141)
significantly inhibited the expression of miR-141 in the stable
Huh-141 cells (Fig. 6A). Silencing
miR-141 in Huh7-141 cells increased the expression of TGFβR1 and
the p-Smad2/Smad2 ratio (Fig. 6B).
Additionally, MTT assays revealed that inhibition of miR-141
enhanced HCC cell growth. The viability of Huh7-141 cells
transfected with si-miR-141 increased by 49.09% on day 5 compared
with the cells transfected with si-miR-NC (Fig. 6C). Cell cycle analysis indicated that
silencing of miR-141 increased the number of cells in G2/M phase by
3.55% (Fig. 6D). Finally, Transwell
assays revealed that silencing of miR-141 restored HCC cell
migration and invasion capacity (Fig.
6E). These data suggested that inhibition of miR-141 promoted
HCC cell proliferation, migration, and invasion.
Discussion
Metastasis is a complex, multistep process through
which primary tumor cells invade adjacent tissues and proliferate
from microscopic growth to macroscopic secondary tumors (31). Many studies have described numerous
genes and gene products that can drive the metastatic process. In
addition to alterations in protein-encoding genes, a class of
non-coding small RNAs also contributes to cancer metastasis. For
example, miRNAs can lead to silencing of their cognate target genes
by cleaving mRNA molecules or by inhibiting their translation
(32). Emerging evidence has
suggested that miRNAs regulate diverse cellular pathways. Although
miRNAs can function as oncogenes or tumor suppressors, the roles of
miRNAs in mediating cancer metastasis remain unclear.
Hepatocarcinogenesis is a highly complicated process
due to changes in signaling induced by mutation of tumor
suppressors, chromosomal amplifications and deletions, and
epigenetic alterations (33).
Signaling cascades related to HCC proliferation and metastasis are
activated as a result. Consequently, miRNAs may to some extent
regulate the expression of key oncogenes (34). The altered expression of miRNA
profiling is common in HCC; thus, research on these small RNAs has
become a hotspot in attempts to control the crucial steps of cancer
progression (35,36).
A previous study from our group demonstrated that
miR-141 suppresses proliferation and metastasis in RCC in
vitro and in vivo (16).
The miR-200 family, including miR-141, miR-200c, miR-200b, miR-200a
and miR-429, has been reported to be downregulated in HCC (22–24) and
other tumor types (16,37,38).
Although the importance of miR-141 in HCC development has
previously been highlighted, studies focusing on its functions
remain limited. Therefore, based on our previous discovery that
miR-141 downregulation is associated with RCC metastasis (16), the present study selected to examine
the functions of miR-141 in HCC progression.
In accordance with previous reports (22–24), the
present results demonstrated the downregulation of miR-141 in HCC
tumor tissues compared with matched adjacent non-tumor tissues. The
present in vitro results suggested that enhanced miR-141
expression markedly suppressed HCC cell migration and invasion,
blocked cell growth, and arrested cells at the G2/M phase. Both
previous reports and our current findings implied that miR-141 was
involved in HCC progression and may serve as a negative regulator
of HCC metastasis. The present study, however, has several
limitations. First, only one cell line was used in vitro for
functional experiments. Second, a relatively small number of
tissues was used to confirm the protein expressions in patients.
Further studies will be required in the future to confirm these
results in additional cell lines and larger patient cohorts.
Of note, the present study identified TGFβR1, an
important receptor in the TGFβ signaling pathway, as a novel direct
target of miR-141. TGFβ has emerged as a potent driver of multiple
aspects of cancer progression, particularly induction of the
epithelial-mesenchymal transition. Several reports have indicated
that TGFβ acts as a tumor suppressor by inhibiting epithelial cell
proliferation. However, alterations in TGFβ signaling components
could inactivate the tumor-suppressive abilities of TGFβ and thus
enable cancer cells to survive, escape the primary tumor, and form
metastases, via the TGFβ signaling pathway (39). TGFβ signals through type I and type II
receptors to form a heterotetrameric receptor complex. TGFβR1, one
of the type II receptors of the TGFβ ligand, is a transmembrane
receptor serine-threonine kinase. Using computational prediction,
TGFβR1 was identified as a potential target of miR-141. The present
in vitro findings confirmed that the negative effects
induced by miR-141 were significantly associated with the
expression of TGFβR1 mRNA and protein. Loss-of-function assays also
demonstrated that silencing TGFβR1 by siRNA dramatically suppressed
the migration and invasion of Huh7 cells, in a manner similar to
the miR-141 overexpression. A recent study reported that miR-140-5p
suppressed HCC cell growth and metastasis by targeting TGFβR1
(40), indicating that TGFβR1 was
likely an important modulator of HCC progression and that it could
be simultaneously regulated by multiple miRNAs, such as miR-141 and
miR-140-5p. Taken together, these results suggested that the
downregulation of TGFβR1 may be a mechanism through which miR-141
modulates the growth and metastasis of HCC cells.
Three other important downstream proteins, including
matrix metalloproteinases (MMPs), Jagged1, and focal adhesion
kinase (FAK), have been reported to be activated by the TGFβ
signaling pathway. MMPs, particularly MMP2 and MMP9, are prominent
enzymes in the degradation of the basement membrane and
extracellular matrix (41) and can be
enhanced by the TGFβ signaling pathway (42,43).
Additionally, MMP9 has been identified as a major contributor and a
crucial factor in tumor-induced angiogenesis, due to its effects on
induction of quiescent vasculature formation (44). Preliminary experiments from our group
demonstrated that, although neither MMP2 nor MMP9 were predicted as
potential targets of miR-141, a significant change in their
expression was associated with miR-141 (data not shown). Further
studies will be needed to elucidate the underlying mechanisms
through which miR-141 may regulate MMPs. Jagged1 and FAK are also
activated by the TGFβ/Smad pathway (45–47).
Additionally, FAK has been demonstrated to be a target gene of
miR-151, which promotes HCC cell migration and invasion (46). Taken together, these findings
suggested that miR-141 may prevent HCC cell metastasis and invasion
by attenuating the TGFβ/Smad/MMP/Jagged1/FAK signaling
cascades.
Collectively, the current study indicated that
miR-141 partially inhibited HCC proliferation and invasion by
directly targeting TGFβR1 and blocking the TGFβ signaling pathway.
However, the regulatory network of miRNAs involved in HCC is
complex; the present study on the miR-141/TGFβR1 pathway provides
only preliminary information on the overall signaling networks
affecting HCC progression. Notably, one single miRNA may target
multiple genes and thus control multiple signaling pathways.
Nonetheless, the present study of miR-141 and TGFβR1 improved the
current understanding of the mechanisms underlying HCC development
and suggested that miR-141 may represent a novel therapeutic target
for the treatment of HCC.
Acknowledgements
Not applicable.
Funding
This study was supported by the National Natural
Science Foundation of China (grant nos. 81272560, 31070142,
30872924, 81072095 and 81372760), the Open Research Foundation of
the State Key Laboratory of Virology of Wuhan University (grant no.
2014KF007), the Program for New Century Excellent Talents in
University from the Department of Education of China (grant no.
NCET-08-0223), and the National High Technology Research and
Development Program of China (863 Program; grant no.
2012AA021101).
Availability of data and materials
The datasets used during the present study are
available from the corresponding author upon reasonable
request.
Authors' contributions
YZ, ZX, JZ, HY contributed to the conception and
design of the study. YZ, ZX, JZ contributed to the acquisition,
analysis, and interpretation of the data. YZ drafted the
manuscript. HY revised and approved the final version of the
manuscript. All authors read and approved the manuscript and agree
to be accountable for all aspects of the work ensuring integrity
and accuracy.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of Tongji Medical College and informed consent was
obtained from all patients prior to enrollment in the study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Center MM and Jemal A: International
trends in liver cancer incidence rates. Cancer Epidemiol Biomarkers
Prev. 20:2362–2368. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Parkin DM, Bray F, Ferlay J and Pisani P:
Global cancer statistics, 2002. CA Cancer J Clin. 55:74–108. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chaffer CL and Weinberg RA: A perspective
on cancer cell metastasis. Science. 331:1559–1564. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Baek D, Villén J, Shin C, Camargo FD, Gygi
SP and Bartel DP: The impact of microRNAs on protein output.
Nature. 455:64–71. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ebert MS and Sharp PA: Roles for microRNAs
in conferring robustness to biological processes. Cell.
149:515–524. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lewis BP, Burge CB and Bartel DP:
Conserved seed pairing, often flanked by adenosines, indicates that
thousands of human genes are microRNA targets. Cell. 120:15–20.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
He H, Hao SJ, Yao L, Yang F, Di Y, Li J,
Jiang YJ, Jin C and Fu DL: MicroRNA-218 inhibits cell invasion and
migration of pancreatic cancer via regulating ROBO1. Cancer Biol
Ther. 15:1333–1339. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Huang X, Lv W, Zhang JH and Lu DL: miR-96
functions as a tumor suppressor gene by targeting NUAK1 in
pancreatic cancer. Int J Mol Med. 34:1599–1605. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Li J, Kong F, Wu K, Song K, He J and Sun
W: miR-193b directly targets STMN1 and uPA genes and suppresses
tumor growth and metastasis in pancreatic cancer. Mol Med Rep.
10:2613–2620. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ahn SM, Cha JY, Kim J, Kim D, Trang HT,
Kim YM, Cho YH, Park D and Hong S: Smad3 regulates E-cadherin via
miRNA-200 pathway. Oncogene. 31:3051–3059. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Xu L, Li Y, Yan D, He J and Liu D:
MicroRNA-183 inhibits gastric cancer proliferation and invasion via
directly targeting Bmi-1. Oncol Lett. 8:2345–2351. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Tang Y, Liu X, Su B, Zhang Z, Zeng X, Lei
Y, Shan J, Wu Y, Tang H and Su Q: microRNA-22 acts as a metastasis
suppressor by targeting metadherin in gastric cancer. Mol Med Rep.
11:454–460. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Tang H, Liu P, Yang L and Xie X, Ye F, Wu
M, Liu X, Chen B, Zhang L and Xie X: miR-185 suppresses tumor
proliferation by directly targeting E2F6 and DNMT1 and indirectly
upregulating BRCA1 in triple-negative breast cancer. Mol Cancer
Ther. 13:3185–3197. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yan X, Chen X, Liang H, Deng T, Chen W,
Zhang S, Liu M, Gao X, Liu Y, Zhao C, et al: miR-143 and miR-145
synergistically regulate ERBB3 to suppress cell proliferation and
invasion in breast cancer. Mol Cancer. 13:2202014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chen X, Wang X, Ruan A, Han W, Zhao Y, Lu
X, Xiao P, Shi H, Wang R, Chen L, et al: miR-141 is a key regulator
of renal cell carcinoma proliferation and metastasis by controlling
EphA2 expression. Clin Cancer Res. 20:2617–2630. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhou X, Xia Y, Su J and Zhang G:
Down-regulation of miR-141 induced by helicobacter pylori
promotes the invasion of gastric cancer by targeting STAT4. Cell
Physiol Biochem. 33:1003–1012. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Du Y, Xu Y, Ding L, Yao H, Yu H, Zhou T
and Si J: Down-regulation of miR-141 in gastric cancer and its
involvement in cell growth. J Gastroenterol. 44:556–561. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Mei Z, He Y, Feng J, Shi J, Du Y, Qian L,
Huang Q and Jie Z: MicroRNA-141 promotes the proliferation of
non-small cell lung cancer cells by regulating expression of PHLPP1
and PHLPP2. FEBS Lett. 588:3055–3061. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhao G, Wang B, Liu Y, Zhang JG, Deng SC,
Qin Q, Tian K, Li X, Zhu S, Niu Y, et al: miRNA-141, downregulated
in pancreatic cancer, inhibits cell proliferation and invasion by
directly targeting MAP4K4. Mol Cancer Ther. 12:2569–2580. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wang XL, Xie HY, Zhu CD, Zhu XF, Cao GX,
Chen XH and Xu HF: Increased miR-141 expression is associated with
diagnosis and favorable prognosis of patients with bladder cancer.
Tumour Biol. 36:877–883. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Xue J, Niu YF, Huang J, Peng G, Wang LX,
Yang YH and Li YQ: miR-141 suppresses the growth and metastasis of
HCC cells by targeting E2F3. Tumour Biol. 35:12103–12107. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wu SM, Ai HW, Zhang DY, Han XQ, Pan Q, Luo
FL and Zhang XL: miR-141 targets ZEB2 to suppress HCC progression.
Tumour Biol. 35:9993–9997. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Liu Y, Ding Y, Huang J, Wang S, Ni W, Guan
J, Li Q, Zhang Y, Ding Y, Chen B, et al: miR-141 suppresses the
migration and invasion of HCC cells by targeting Tiam1. PLoS One.
9:e883932014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Goel A, Boland CR and Hur K: Changes in
the expression of mir-200c/141 cluster of micrornas as biomarkers
for epithelial- to-mesenchymal transition in human colorectal
cancer metastasis. World Intellectual Property Organization Patent
WO2012/128902Al. Filed February 29 2012; issued September 27
2012.
|
|
26
|
Bracken CP, Li X, Wright JA, Lawrence DM,
Pillman KA, Salmanidis M, Anderson MA, Dredge BK, Gregory PA,
Tsykin A, et al: Genome-wide identification of miR-200 targets
reveals a regulatory network controlling cell invasion. EMBO J.
33:2040–2056. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Roy SS, Gonugunta VK, Bandyopadhyay A, Rao
MK, Goodall GJ, Sun LZ, Tekmal RR and Vadlamudi RK: Significance of
PELP1/HDAC2/miR-200 regulatory network in EMT and metastasis of
breast cancer. Oncogene. 33:3707–3716. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Feng X, Wang Z, Fillmore R and Xi Y:
miR-200, a new star miRNA in human cancer. Cancer Lett.
344:166–173. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Hung CS, Liu HH, Liu JJ, Yeh CT, Chang TC,
Wu CH, Ho YS, Wei PL and Chang YJ: MicroRNA-200a and −200b mediated
hepatocellular carcinoma cell migration through the epithelial to
mesenchymal transition markers. Ann Surg Oncol. 20 (Suppl
3):S360–S368. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wang X, Chen X, Wang R, Xiao P, Xu Z, Chen
L, Hang W, Ruan A, Yang H and Zhang X: microRNA-200c modulates the
epithelial-to-mesenchymal transition in human renal cell carcinoma
metastasis. Oncol Rep. 30:643–650. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Fidler IJ: The pathogenesis of cancer
metastasis: The ‘seed and soil’ hypothesis revisited. Nat Rev
Cancer. 3:453–458. 2003. View
Article : Google Scholar : PubMed/NCBI
|
|
32
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Villanueva A, Newell P, Chiang DY,
Friedman SL and Llovet JM: Genomics and signaling pathways in
hepatocellular carcinoma. Semin Liver Dis. 27:55–76. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Sun J, Lu H, Wang X and Jin H: MicroRNAs
in hepatocellular carcinoma: Regulation, function, and clinical
implications. ScientificWorldJournal. 2013:9242062013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Farazi TA, Hoell JI, Morozov P and Tuschl
T: MicroRNAs in human cancerMicroRNA Cancer Regulation. Schmitz U,
Wolkenhauer O and Vera J: Springer; Dordrecht: pp. 1–20. 2013,
View Article : Google Scholar
|
|
36
|
Wong CCL, Wong CM, Tung EKK, Au SL, Lee
JM, Poon RT, Man K and Ng IO: The microRNA miR-139 suppresses
metastasis and progression of hepatocellular carcinoma by
down-regulating Rho-kinase 2. Gastroenterology. 140:322–331. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Gregory PA, Bert AG, Paterson EL, Barry
SC, Tsykin A, Farshid G, Vadas MA, Khew-Goodall Y and Goodall GJ:
The miR-200 family and miR-205 regulate epithelial to mesenchymal
transition by targeting ZEB1 and SIP1. Nat Cell Biol. 10:593–601.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Gibbons DL, Lin W, Creighton CJ, Rizvi ZH,
Gregory PA, Goodall GJ, Thilaganathan N, Du L, Zhang Y,
Pertsemlidis A, et al: Contextual extracellular cues promote tumor
cell EMT and metastasis by regulating miR-200 family expression.
Genes Dev. 23:2140–2151. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Ikushima H and Miyazono K: TGFbeta
signalling: A complex web in cancer progression. Nat Rev Cancer.
10:415–424. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Yang H, Fang F, Chang R and Yang L:
MicroRNA-140-5p suppresses tumor growth and metastasis by targeting
transforming growth factor β receptor 1 and fibroblast growth
factor 9 in hepatocellular carcinoma. Hepatology. 58:205–217. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Nelson AR, Fingleton B, Rothenberg ML and
Matrisian LM: Matrix metalloproteinases: Biologic activity and
clinical implications. J Clin Oncol. 18:1135–1149. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Tang J, Cui J, Chen R, Guo K, Kang X, Li
Y, Gao D, Sun L, Xu C, Chen J, et al: A three-dimensional cell
biology model of human hepatocellular carcinoma in vitro. Tumour
Biol. 32:469–479. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Chen R, Cui J, Xu C, Xue T, Guo K, Gao D,
Liu Y, Ye S and Ren Z: The significance of MMP-9 over MMP-2 in HCC
invasiveness and recurrence of hepatocellular carcinoma after
curative resection. Ann Surg Oncol. 19 (Suppl 3):S375–S384. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Bergers G and Benjamin LE: Tumorigenesis
and the angiogenic switch. Nat Rev Cancer. 3:401–410. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Zavadil J, Cermak L, Soto-Nieves N and
Böttinger EP: Integration of TGF-beta/Smad and Jagged1/Notch
signalling in epithelial- to-mesenchymal transition. EMBO J.
23:1155–1165. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Ding J, Huang S, Wu S, Zhao Y, Liang L,
Yan M, Ge C, Yao J, Chen T, Wan D, et al: Gain of miR-151 on
chromosome 8q24. 3 facilitates tumour cell migration and spreading
through downregulating RhoGDIA. Nat Cell Biol. 12:390–399. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Walsh MF, Ampasala DR, Hatfield J, Vander
Heide R, Suer S, Rishi AK and Basson MD: Transforming growth
factor-β stimulates intestinal epithelial focal adhesion kinase
synthesis via Smad- and p38-dependent mechanisms. Am J Pathol.
173:385–399. 2008. View Article : Google Scholar : PubMed/NCBI
|