Introduction
Colorectal cancer (CRC) includes malignant tumors
that occur in the colon, rectum and anus. With its high morbidity
and mortality, CRC is among the most malignant tumors worldwide. It
has been estimated that there were >1.8 million new CRC cases
and 880,000 CRC-associated deaths in 2018, accounting for
approximately one-tenth of all cancer cases and deaths. Among all
cancers worldwide, CRC ranks third in terms of morbidity and second
in terms of mortality (1). In China,
CRC has high incidence and mortality, and is one of the top five
most commonly diagnosed tumors (2).
The leading cause of mortality for patients with CRC is metastasis.
The 5 year overall survival (OS) rate of patients with primary CRC
can be as high as 80–90%, but this may be reduced to 5–10% in
patients with metastatic tumors (3,4). Like many
other cancers, CRC is a heterogeneous disease in which genetic
variation, cellular context and environmental effects have an
impact on the initiation, progression and metastasis of tumors
(5). Accordingly, it is highly
crucial to locate biomarkers and prognostic indicators for the
early detection of malignant cell transformation.
The use of whole-genome data to screen for markers
of tumors, which can be applied to diagnosis and prognosis, is
efficient and effective and can be used to guide the exploration of
prospective mechanisms. The Gene Expression Omnibus (GEO) is the
most comprehensive, well known and largest international public
database for the storage and query of expression data; it is
developed and maintained by the National Center for Biotechnology
Information. Its purpose is to provide a good platform for
post-data mining and information promotion by collecting a large
amount of high-throughput experimental data (6).
The gene C-X-C motif chemokine ligand 3
(CXCL3), a member of the CXC chemokine family, encodes a
secreted growth factor that signals through the G protein-coupled
receptor CXC receptor 2, and thereby serves a role in inflammation
and acts as a chemoattractant of neutrophils (7,8). Previous
studies have investigated the prognostic relationship between
CXCL3 and CRC. The study by Doll et al (9) identified no significant correlation
between CXCL3 expression and CRC survival, whereas the
findings of Xiong et al (10)
suggested that CRC patients with high CXCL3 expression
levels had a shorter OS time. More than 50% of CRCs are colon
cancer (CC) (1); CC and rectal cancer
have different causes (11,12), and their pathogenesis and histological
types also differ. In the study conducted by Xiong et al
(10), patients with colon and rectal
cancer from a TCGA dataset were combined for the prognostic
analysis of CXCL3; however, as patients with colon and
rectal cancer are two separate cohorts, the results require further
investigation. Furthermore, the study lacked analysis at the
protein level. Therefore, the aim of the present study was to use a
patient cohort from Guangxi Medical University and a GEO dataset to
investigate and validate CXCL3 for the diagnosis and
prognosis of CC, and to explore its prospective molecular
mechanism.
Materials and methods
Reverse transcription-quantitative PCR
(RT-qPCR) of CXCL3 expression in CC tissue
Patient tissue samples and ethical
approval
From April to June 2018, cancer and adjacent normal
tissues were continuously collected during the resective surgery of
patients with CC in the Department of Colorectal and Anal Surgery,
First Affiliated Hospital of Guangxi Medical University (Nanning,
Guangxi). Immediately after surgery, the tissue was smeared in RNA
protection solution and stored in refrigerator at −80°C. The
inclusion criteria for patients were as follows: i) Without
restrictions of age and sex; ii) underwent resection of colon
tumor; and iii) with a pathological diagnosis of colon cancer. The
exclusion criteria include: i) Complicated with other known tumors;
ii) received radiotherapy or chemotherapy prior to surgery; iii)
refused to provide written informed consent; iv) the tumor was too
small for a specimen to be acquired. The study was conducted in
accordance with the Declaration of Helsinki, all patients signed an
informed consent form, and the Ethics Committee of the First
Affiliated Hospital of Guangxi Medical University approved the
experimental protocol [Ethics no.: 2019(KY-E-001)].
RNA extraction and RT-qPCR
Total RNA was extracted from the patients' tissues
using TRIzol reagent (cat. no. 15596026; Invitrogen, Thermo Fisher
Scientific, Inc.). Then, PrimeScript™ RT Reagent kit with gDNA
Eraser (cat. no. RR047A; Takara Bio, Inc.) was used to transform
the total RNA into first-strand cDNA. The reverse transcription
reaction conditions were as follows: 42°C for 60 min, 70°C for 5
min, and 4°C until required. qPCR was then conducted using
FastStart Universal SYBR Green Master (ROX) (Roche Diagnostics
GmbH) in an Applied Biosystems QuantStudio™ 6 Real-PCR System
(Thermo Fisher Scientific, Inc.). All procedures were conducted in
accordance with the manufacturer's instructions. The expression
level of CXCL3 was calculated using the 2∆∆Cq
method (13,14), and was normalized to GAPDH expression.
The primer sequences were as follows: CXCL3 forward,
CCAAACCGAAGTCATAGCCAC and reverse, TGCTCCCCTTGTTCAGTATCT; GAPDH
forward, GTCAGCCGCATCTTCTTT and reverse, CGCCCAATACGACCAAAT.
Immunohistochemistry (IHC) of CXCL3
expression in CC tissue
Patient tissue samples and ethical
approval
Tumor tissue and adjacent normal tissue (slice
thickness, 4 µm), fixed with 10% neutral formalin at room
temperature for 16 h and embedded in paraffin wax blocks, were
retrospectively collected from patients who had undergone colonic
tumor resection in the First Affiliated Hospital of Guangxi Medical
University between May 2012 and May 2013. The inclusion criteria
for patients were as follows: i) Without restrictions of age and
sex; ii) received resection of colon tumor; and iii) with a
pathological diagnosis of colon cancer. The exclusion criteria
include: i) Complicated with other known tumors; ii) received
radiotherapy or chemotherapy prior to surgery; iii) refused to sign
informed consent; iv) the tumor was too small for a specimen to be
acquired. Tumors were identified and categorized according to the
tumor node metastasis (TNM) staging system of the American Joint
Committee on Cancer (8th edition, 2017) (15). Information about the patients was
recorded as follows: Sex, age, preoperative carcinoembryonic
antigen levels, TNM stage, tumor location, general classification,
tumor differentiation, tumor thrombus, tumor size, tumor number,
lymph node status, radical resection, tumor metastasis, nerve
infiltration and postoperative chemotherapy. The study was
conducted in accordance with the Declaration of Helsinki. Prior to
the study, all patients received informed consent and ethical
approval for the study was provided [Ethics no.:
2019(KY-E-001)].
Evaluation of IHC
IHC was applied for evaluation of the expression of
CXCL3. A CXCL3 antibody (cat. no. #35751) supplied by Signalway
Antibody LLC, IHC staining reagents (DAB) and Secondary Antibody,
HRP (cat. no. D-3004-15) from Shanghai ChangDao Biotech Co., Ltd.
were used. Antigen retrieval was conducted using sodium citrate
buffer for 2.5 min at high pressure, followed by cooling for 5 min,
and washing with PBS buffer for 3 min three times. The IHC
procedure and steps were performed strictly following the
manufacturers' protocols (incubation with primary antibody
incubation at 1:100 dilution, 37°C for 2.5 h; incubation with
ready-to-use secondary antibody for 30 min at room temperature).
The slides were observed under an Olympus upright microscope, white
light (magnification ×400). Two independent pathologists scored the
average percentage of positive cells as follows: 0 (0%); 1 (1–25%);
2 (26–50%); 3 (51–75%); and 4 (76–100%). The intensity of staining
was scored as follows: 0 (negative); 1 (weak); 2 (moderate) and 3
(strong). The positive cell percentage was multiplied by the
staining intensity score as previously described to provide the
final pathological score, and a score >2 was considered to
indicate a positive staining result (16).
Validation of CXCL3 expression in
normal colon and colon tumor tissues
The expression level of CXCL3 in normal human
tissues was obtained from Human Protein Atlas (HPA: https://www.proteinatlas.org, accessed December 22,
2018) (17). Expression levels of the
CXCL3 gene in normal colon and primary tumor tissues were
determined using the online tool GEPIA (http://gepia.cancer-pku.cn/detail.php?gene=cxcl3,
accessed February 17, 2019) (18).
Validation cohort for the prognosis
value of CXCL3 from the GEO database
A dataset of CXCL3 gene expression values and
corresponding clinical data was downloaded from the GEO database
(https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE40967,
accessed December 23, 2018) (19).
The data were chosen according to the following inclusion criteria:
i) Histopathological diagnosis of colon cancer; ii) primary tumor
that could be surgically removed; iii) complete postoperative
follow-up data; iv) all patients did not receive preoperative
chemotherapy and/or radiation therapy; and v) number of cases
>500. The exclusion criteria include: i) Complicated with other
known tumors; ii) the subject of the study was not colon cancer;
iii) sample size ≤500. Since these datasets were obtained from
public databases, their use did not need require ethical
approval.
Gene set enrichment analysis
(GSEA)
For investigation of the prospective molecular
mechanism of CXCL3 in patients with a prognosis CC,
differential metabolic pathways and biological processes at the
transcriptome level between high and low CXCL3 gene
expression, which was based on the 75% cut-off values, were
analyzed using GSEA (http://software.broadinstitute.org/gsea/index.jsp,
accessed December 24, 2018) v3.0 (20). GSEA was used with reference to gene
sets from the Molecular Signatures Database, namely c5 GO gene sets
for biological process, cellular component and molecular function
(c5.bp.v6.2.symbols.gmt, c5.cc.v6.2.symbols.gmt and
c5.mf.v6.2.symbols.gmt) and c2 KEGG gene sets
(c2.cp.kegg.v6.2.symbols.gmt). The number of permutations was set
at 1,000. Enrichment results with one nominal P-value <0.05 and
one false discovery rate (FDR) <0.25 were considered
statistically significant.
Statistical analysis
The paired t-test was used to analyze the difference
in the mRNA expression of CXCL3 between tumors and adjacent
non-tumor tissues. χ2 test was used to compare the
distribution of IHC staining scores between tumors and adjacent
non-tumor tissues. The Kaplan-Meier method was performed for
survival analysis. Cox proportional hazards regression analysis was
applied to calculate the crude and adjusted hazard ratio (HR) and
95% confidence interval (CI) in uni- and multivariate analyses. The
FDR in GSEA was adjusted for multiple testing with the
Benjamini-Hochberg procedure (21,22). A
scatter plot, receiver operating characteristic (ROC) curves and
Kaplan-Meier survival curves were drawn using GraphPad Prism 7.0
(GraphPad Software, Inc.). P<0.05 was considered statistically
significant. SPSS v.24.0 software (IBM Corp.) was used to conduct
the data analysis.
Results
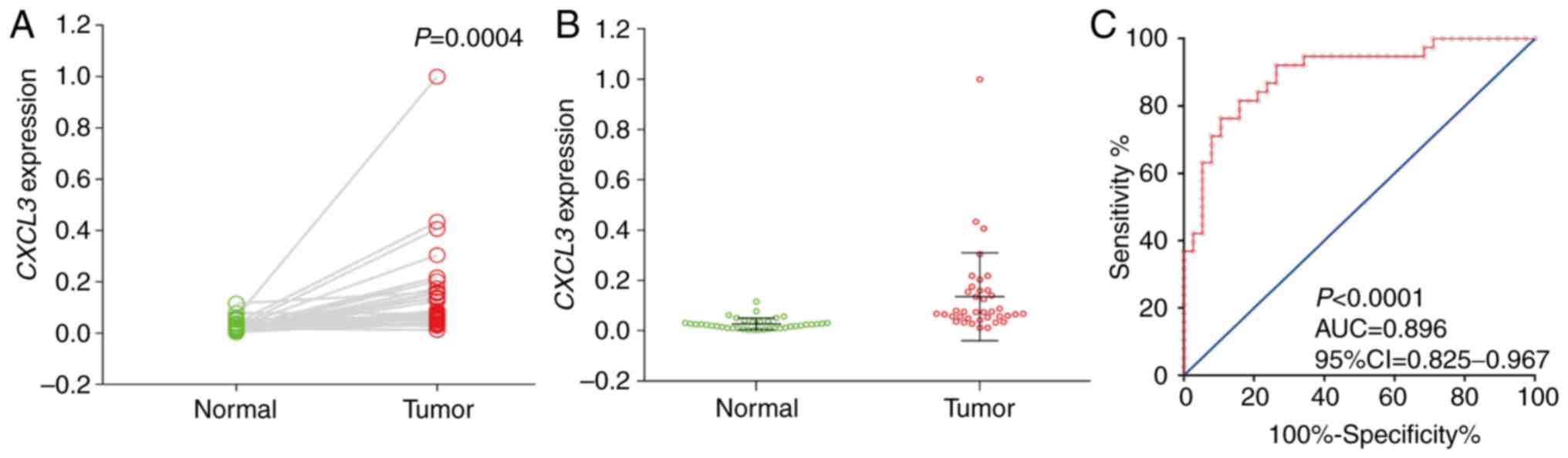
RT-qPCR analysis of CXCL3 expression
in CC tissue
RT-qPCR was performed on the CC and adjacent normal
tissue samples of 38 patients with CC. These CC patients ranged in
age from 35 to 85 years, and included 25 men and 13 women. Analysis
using a paired t-test demonstrated that the expression of
CXCL3 in cancer tissues was significantly higher than that
in adjacent normal tissues (P=0.0004, 95% CI=0.052–0.164; Fig. 1A and B). In addition, diagnostic ROC
curve analysis indicated that CXCL3 has a high diagnostic
value for CC (P<0.0001, AUC=0.896, 95% Cl=0.825–0.967; Fig. 1C).
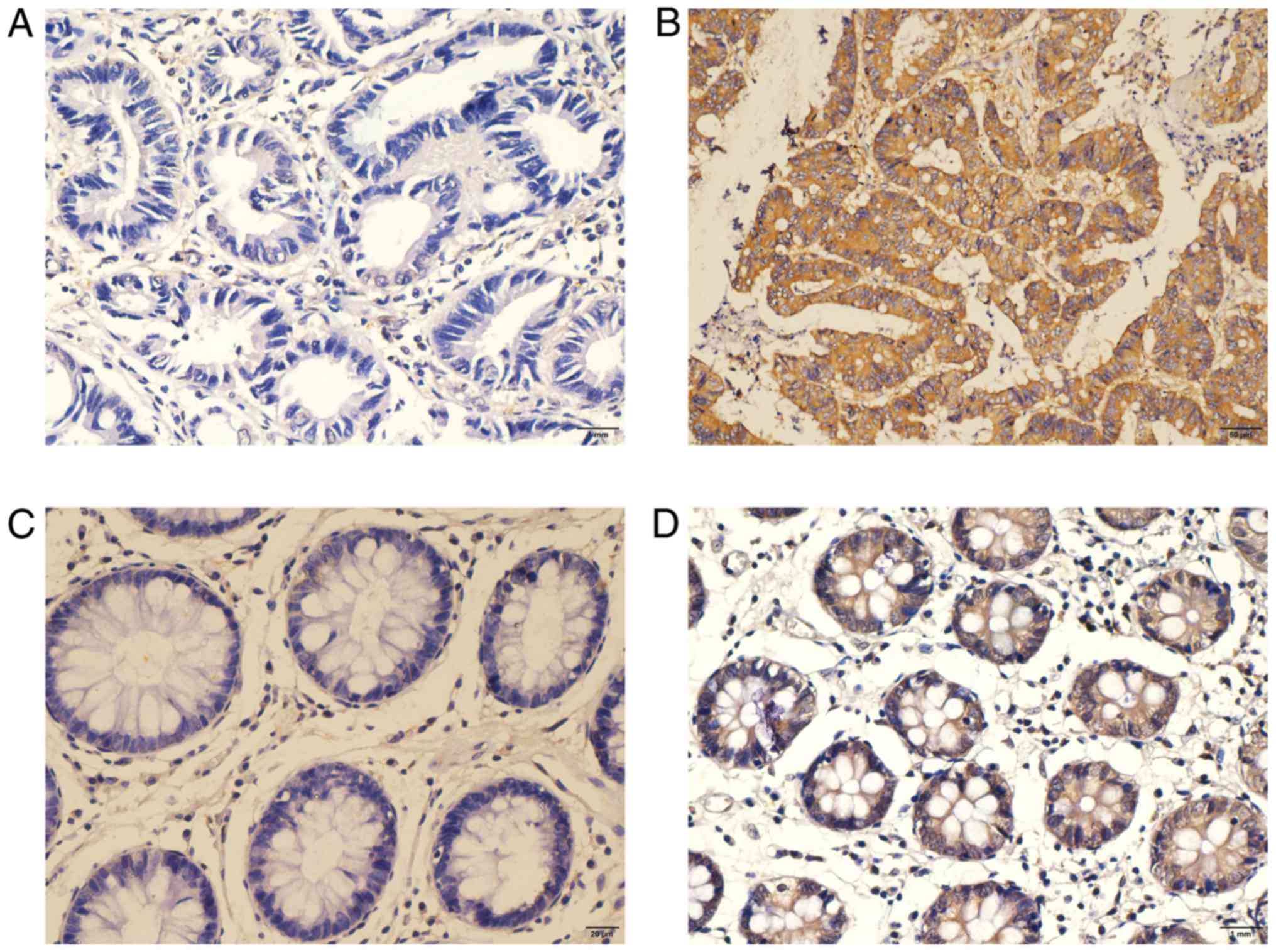
IHC of CXCL3 expression in CC
tissue
IHC testing was performed on another 212 tumor and
46 adjacent normal tissue samples, preserved in wax blocks, that
were acquired from 212 patients with CC. The positive signaling of
CXCL3, located in the cytoplasm of CC cells or adjacent normal
colonic epithelium cells, was shown by the formation of a diffuse
brown-yellow or dark-brown color following immunohistochemical
staining (Fig. 2). Among the 212
cases of CC, 90 cases were CXCL3-positive (42.5%), while positive
CXCL3 expression was observed in only 4/46 (8.7%) of the adjacent
normal tissues.
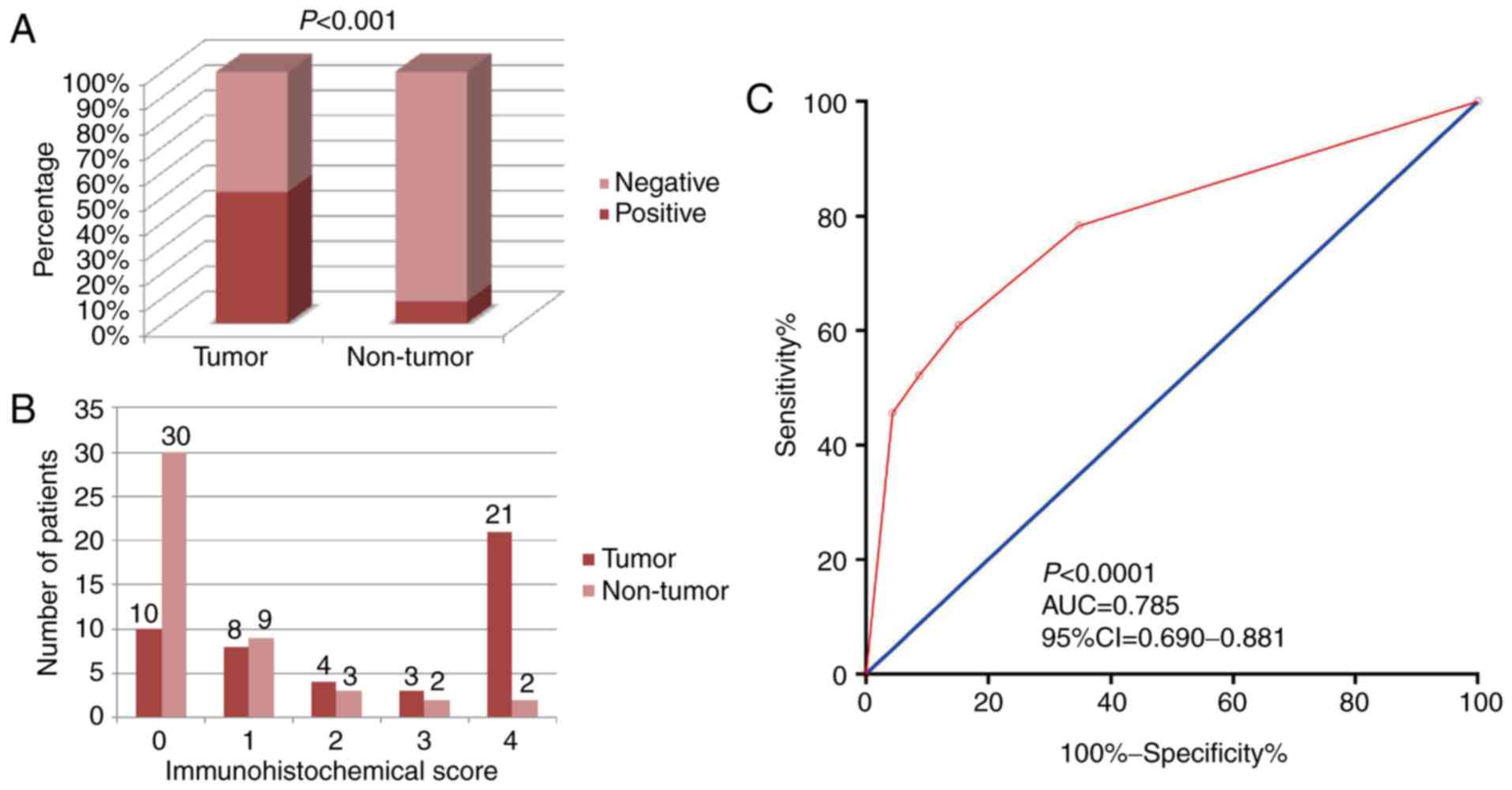
Clinical and pathological factors that may be
associated with prognosis were evaluated (Table I). A total of 137 male and 75 female
patients, with an average age of 58 years were included in the
evaluation. The median follow-up time after surgery was 1,934 days
(range, 36–2,236 days); 10 patients were lost to follow-up. The
positive rate of CXCL3 in cancer tissues was significantly higher
than that in adjacent normal tissues (χ2=20.536,
P<0.001; Fig. 3A) in the 46 CC
patients for which both types of tissue were available. The number
of patients with IHC scores are shown in Fig. 3B. Diagnostic ROC curve analysis of
CXCL3 revealed a moderate diagnostic value for CC (P<0.0001,
AUC=0.785, 95% Cl=0.690–0.881; Fig.
3C).
 | Table I.Clinical and pathological parameters
of 212 patients with colon cancer. |
Table I.
Clinical and pathological parameters
of 212 patients with colon cancer.
| Variable | No. of
patients | MST (days) | OSa, HR (95% CI)b | Log rank
P-valuec |
|---|
| Sex |
|
|
| 0.801 |
|
Male | 137 | NA | 1 |
|
|
Female | 75 | NA | 0.934
(0.552–1.582) |
|
| Age (years) |
|
|
| 0.536 |
|
≤65 | 137 | NA | 1 |
|
|
>65 | 75 | NA | 1.174
(0.707–1.950) |
|
| CEA (ng/ml) |
|
|
| 0.169 |
|
1–5 | 113 | NA | 1 |
|
|
>5 | 93 | NA | 1.424
(0.858–2.363) |
|
|
Missing | 6 |
|
|
|
| TNM stage |
|
|
| <0.0001 |
|
I–II | 88 | NA | 1 |
|
|
III–IV | 124 | NA | 5.049
(2.563–9.945) |
|
| Location |
|
|
| 0.806 |
|
Right | 102 | NA | 1 |
|
|
Left | 109 | NA | 0.929
(0.565–1.529) |
|
|
Both | 1 | NA | 0
(0–2.209×10211) |
|
| General
classification |
|
|
| 0.691 |
|
Invasive | 11 | NA | 1 |
|
|
Ulcerative | 153 | NA | 1.511
(0.367–6.221) |
|
|
Mass | 42 | NA | 1.203
(0.267–5.428) |
|
|
Missing | 6 |
|
|
|
| Tumor
differentiation |
|
|
| 0.019 |
|
Well | 10 | NA | 1 |
|
|
Moderately | 160 | NA | 1.451
(0.352–5.993) |
|
|
Poor | 42 | NA | 3.076
(0.710–13.318) |
|
| Tumor thrombus |
|
|
| <0.0001 |
| No | 185 | NA | 1 |
|
|
Yes | 26 | 660 | 4.571
(2.568–8.134) |
|
|
Missing | 1 |
|
|
|
| Tumor size
(cm) |
|
|
| 0.236 |
|
<5 | 90 | NA | 1 |
|
| ≥5 | 116 | NA | 0.739
(0.447–1.221) |
|
|
Missing | 6 |
|
|
|
| Tumor number |
|
|
| 0.138 |
|
One | 205 | NA | 1 |
|
|
Two | 7 | 1,917 | 2.119
(0.768–5.844) |
|
| Lymph node |
|
|
| <0.0001 |
|
Negative | 120 | NA | 1 |
|
|
Positive | 91 | NA | 3.546
(2.075–6.061) |
|
|
Missing | 1 |
|
|
|
| Radical
resection |
|
|
| <0.0001 |
|
Yes | 175 | NA | 1 |
|
| No | 37 | 481 | 11.536
(6.836–19.469) |
|
| Tumor
metastasis |
|
|
| <0.0001 |
| No | 179 | NA | 1 |
|
|
Yes | 33 | 401 | 14.344
(8.376–24.565) |
|
| Nerve
infiltration |
|
|
| 0.173 |
| No | 207 | NA | 1 |
|
|
Yes | 4 | 1,079 | 2.572
(0.628–10.540) |
|
|
Missing | 1 |
|
|
|
| Postoperative
chemotherapy |
|
|
| 0.833 |
| No | 69 | NA | 1 |
|
|
Yes | 124 | NA | 1.061
(0.610–1.846) |
|
|
Missing | 19 |
|
|
|
| CXCL3 |
|
|
| 0.730 |
|
Negative | 122 | NA | 1 |
|
|
Positive | 90 | NA |
0.914(0.548–1.524) |
|
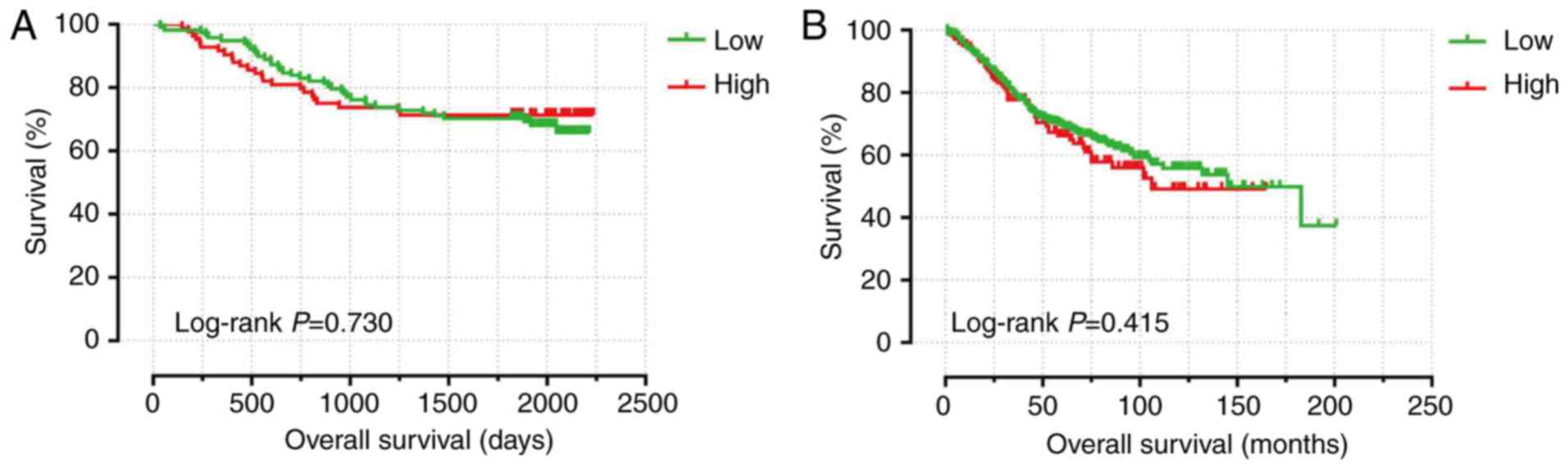
Univariate analysis revealed that advanced TNM
stage, poorer tumor differentiation, tumor thrombus, lymph node
positivity, non-radical resection and tumor metastasis were
associated with poor outcomes (Table
I). Kaplan-Meier analysis indicated that CXCL3 expression was
not relevant to survival (Fig. 4A)
and multivariate analysis showed that CXCL3 positive expression was
not relevant to OS following adjustment for TNM stage, tumor
differentiation, tumor thrombus and radical resection (adjusted
P=0.934, adjusted HR=1.022, 95% CI=0.604–1.729).
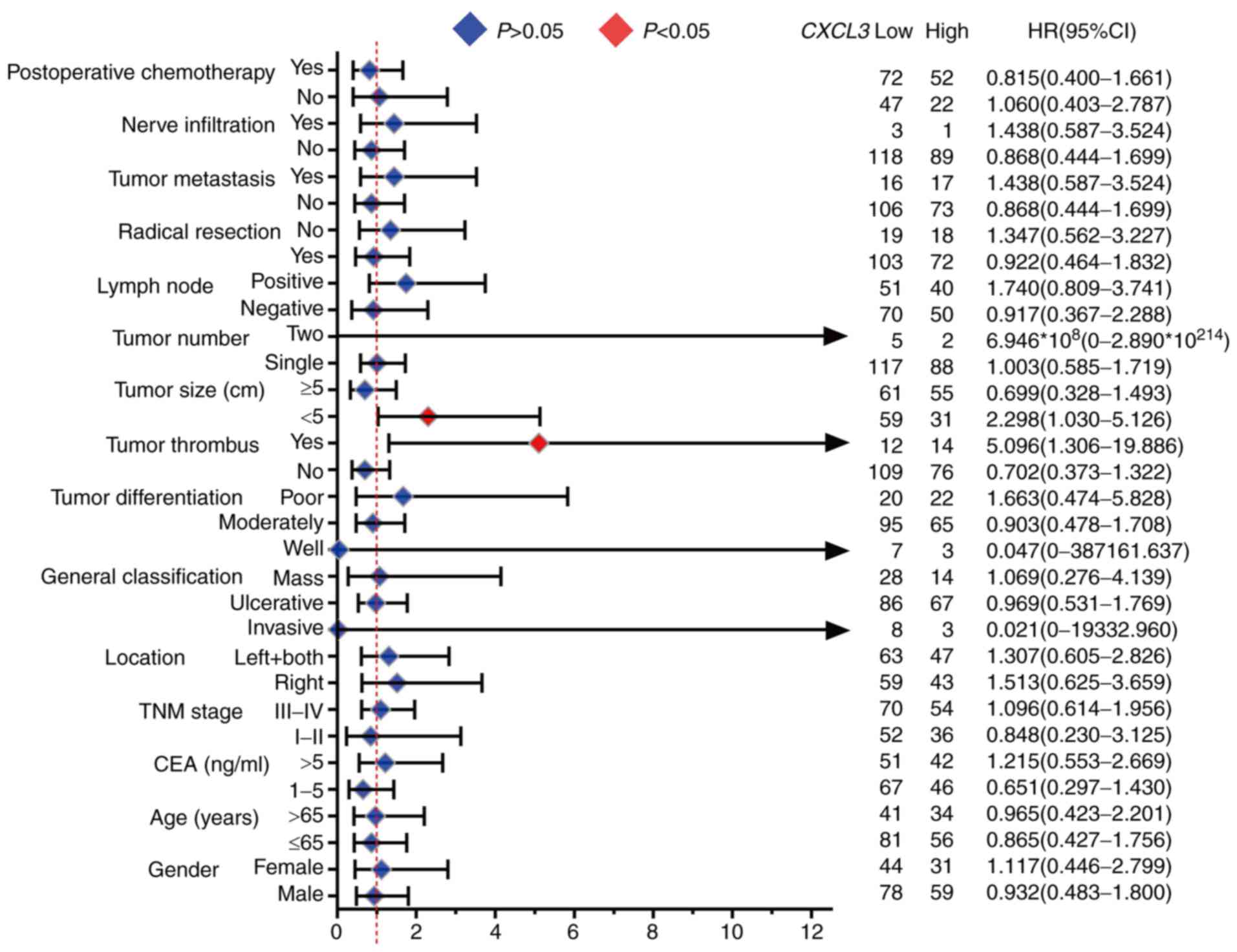
Results of the stratified analysis of the
association of CXCL3 with OS for different stratified clinical
characteristics are displayed in Fig.
5. High expression of CXCL3 was significantly associated with
an increased risk of death in the subgroups of patients with tumor
size <5 cm (adjusted P=0.042, adjusted HR=2.298, 95%
CI=1.030–5.126) and with tumor thrombus (adjusted P=0.019, adjusted
HR=5.096, 95% CI=1.306–19.886).
Validation of CXCL3 expression in
normal colon and colon tumor tissue
The expression level of CXCL3 in normal human
tissues was obtained from the Human Protein Atlas. Data were
extracted from the Functional Annotation of Mammalian Genomes 5
(FANTOM5) project, Genotype-Tissue Expression (GTEx) project and
the HPA RNA-seq dataset (Fig. S1).
Expression level analysis was performed for the CXCL3 gene
in normal colon and colon tumor tissues. The expression of
CXCL3 in colon cancer tissues was significantly higher
compared with that in normal colon tissues (Fig. S2).
Validation of the prognostic value of
CXCL3 using the GEO database
The GPL570 expression profile chip data and clinical
data were downloaded from the GSE40967 dataset. This included data
for 585 patients. The sex, age (years), TNM stage, tumor location,
adjuvant chemotherapy, mismatch repair (MMR) status, CpG island
methylator phenotype status, chromosomal instability status,
TP53 mutation, KRAS mutation, BRAF mutation
and Cartes d'Identité des Tumeurs (CIT) molecular subtype of these
patients were collected.
The patients had a median age of 69 years (range,
22–97 years), and comprised 322 males and 263 females. In the
GSE40967 CC cohort, it was observed that age >65 years, advanced
TNM stage, KRAS mutations and the CIT molecular subtype C4
was associated with a significantly higher risk of CC death
(Table II). Kaplan-Meier survival
analysis with the 75% cut-off values of CXCL3 expression
suggested that CXCL3 expression in the GSE40967 cohort was
not significantly associated with OS (Fig
4B). However, multivariate analysis indicated that high
expression of CXCL3 (adjusted P=0.049, adjusted HR=1.416,
95% CI=1.002–2.003) was closely associated with poor OS in CC,
after adjusting for age, TNM stage, KRAS gene and CIT
subtypes.
 | Table II.Clinical and pathological parameters
of 585 patients with colon cancer from the GSE40967 cohort. |
Table II.
Clinical and pathological parameters
of 585 patients with colon cancer from the GSE40967 cohort.
| Variable | No. of
patients | MST (months) | OSa, HR (95% CI)b | Log-rank
P-valuec |
|---|
| Sex |
|
|
| 0.066 |
|
Male | 322 | 112 | 1 |
|
|
Female | 263 | 183 | 0.765
(0.573–1.020) |
|
| Age (years) |
|
|
| 0.010 |
|
≤65 | 228 | NA | 1 |
|
|
>65 | 356 | 105 | 1.479
(1.094–1.999) |
|
|
Missing | 1 |
|
|
|
| TNM stage |
|
|
| <0.0001 |
|
0-II | 313 | 183 | 1 |
|
|
III–IV | 270 | 105 | 1.774
(1.335–2.358) |
|
|
Missing | 2 |
|
|
|
| Location |
|
|
| 0.584 |
|
Distal | 351 | 145 | 1 |
|
|
Proximal | 232 | NA | 1.084
(0.812–1.447) |
|
|
Missing | 2 |
|
|
|
| Chemotherapy
adjuvant |
|
|
| 0.607 |
| No | 326 | 183 | 1 |
|
|
Yes | 240 | 145 | 0.926
(0.690–1.243) |
|
|
Missing | 19 |
|
|
|
| MMR status |
|
|
| 0.397 |
|
dMMR | 77 | NA | 1 |
|
|
pMMR | 459 | NA | 1.227
(0.762–1.977) |
|
|
Missing | 49 |
|
|
|
| CIMP status |
|
|
| 0.589 |
|
Negative | 420 | 145 | 1 |
|
|
Positive | 93 | NA | 1.115
(0.751–1.656) |
|
|
Missing | 72 |
|
|
|
| CIN status |
|
|
| 0.170 |
|
Negative | 112 | NA | 1 |
|
|
Positive | 369 | 145 | 0.770
(0.529–1.121) |
|
|
Missing | 104 |
|
|
|
| TP53
mutation |
|
|
| 0.312 |
|
Mutant | 190 | 105 | 1 |
|
| Wild
type | 161 | NA | 0.836
(0.590–1.185) |
|
|
Missing | 234 |
|
|
|
| KRAS
mutation |
|
|
|
|
|
Mutant | 217 | 132 | 1 | 0.037 |
| Wild
type | 328 | 145 | 0.736
(0.551–0.983) |
|
|
Missing | 40 |
|
|
|
| BRAF
mutation |
|
|
| 0.689 |
| M | 51 | NA | 1 |
|
| WT | 460 | 145 | 0.900
(0.538–1.508) |
|
|
Missing | 74 |
|
|
|
| CIT molecular
subtype |
|
|
| 0.002 |
| C1 | 116 | 86 | 1 |
|
| C2 | 104 | NA | 0.722
(0.447–1.165) |
|
| C3 | 74 | NA | 0.639
(0.360–1.137) |
|
| C4 | 59 | 46 | 1.790
(1.125–2.850) |
|
| C5 | 152 | 145 | 0.855
(0.567–1.288) |
|
| C6 | 60 | 105 | 1.001
(0.602–1.665) |
|
|
Missing | 20 |
|
|
|
| CXCL3 |
|
|
| 0.415 |
|
Low | 439 | 145 | 1 |
|
|
High | 146 | 106 | 1.139
(0.829–1.566) |
|
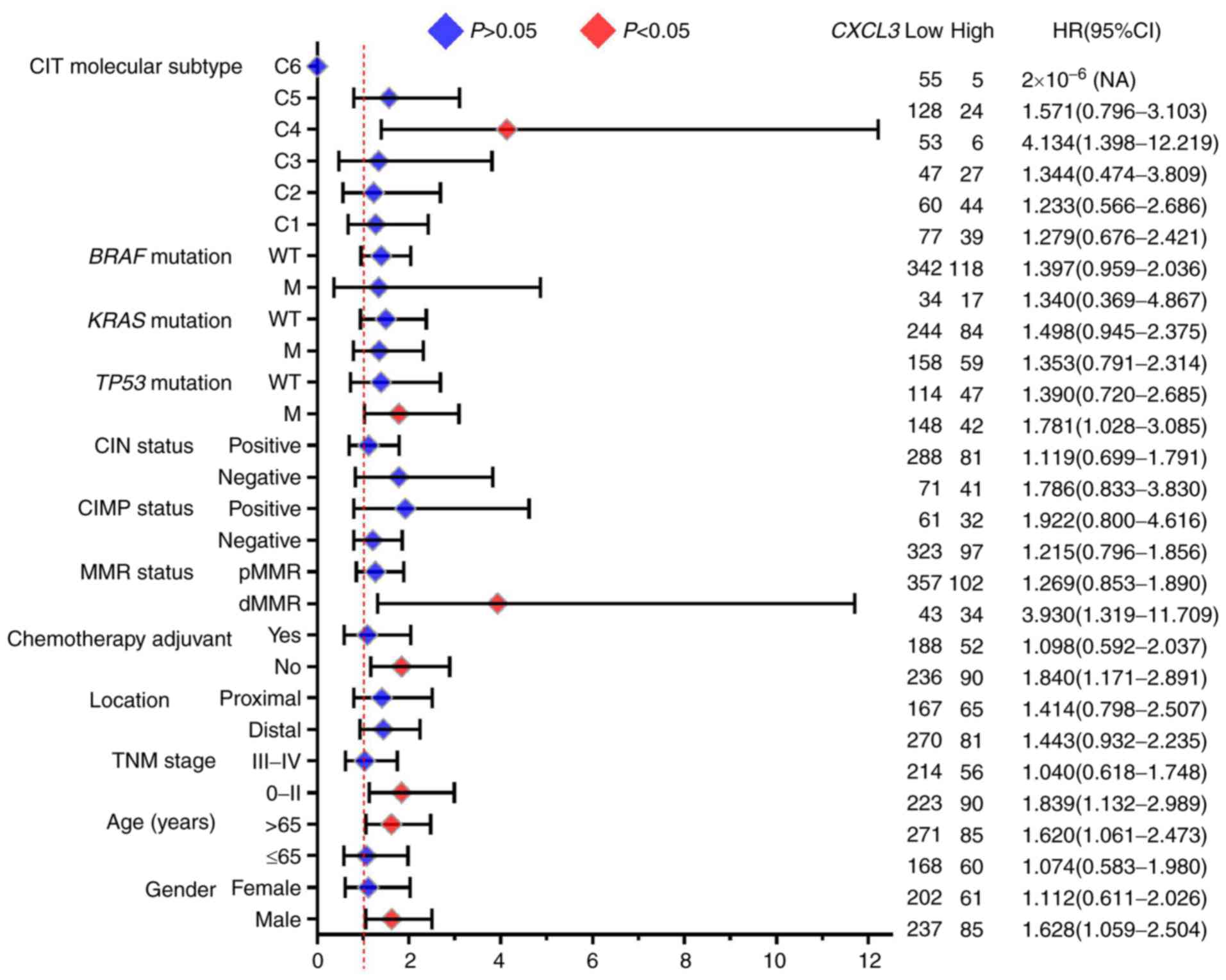
Furthermore, the results of the stratified analysis
of the association of CXCL3 with OS for different stratified
characteristics are presented in Fig.
6. High expression of CXCL3 was associated with a
significantly increased risk of death in the following patient
subgroups: Age >65 years (adjusted P=0.025, adjusted HR=1.620,
95% CI=1.061–2.473), TNM stage 0-II (adjusted P=0.014, adjusted
HR=1.839, 95% CI=1.132–2.989), deficient MMR status (adjusted
P=0.014, adjusted HR=3.930, 95% CI=1.319–11.709), TP53
mutation (adjusted P=0.039, adjusted HR=1.781, 95% CI=1.028–3.085),
CIT molecular subtype C4 (adjusted P=0.010, adjusted HR=4.134, 95%
CI=1.398–12.219) and male sex (adjusted P=0.026, adjusted HR=1.628,
95% CI=1.059–2.504).
GSEA of CXCL3
GSEA of CXCL3 was also conducted in the
GSE40967 cohort. The genome-wide expression profile dataset of the
GSE40967 cohort was assorted into two categories in accordance with
the 75% cut-off values of CXCL3 gene expression. GSEA
results of the GSE40967 cohort are displayed in Figs. 7 and 8
and Tables SI and SII, and indicate that the high expression
of CXCL3 exhibited appreciable relevance to DNA repair, cell
cycle process, cell apoptosis process and the P53 regulation
pathway.
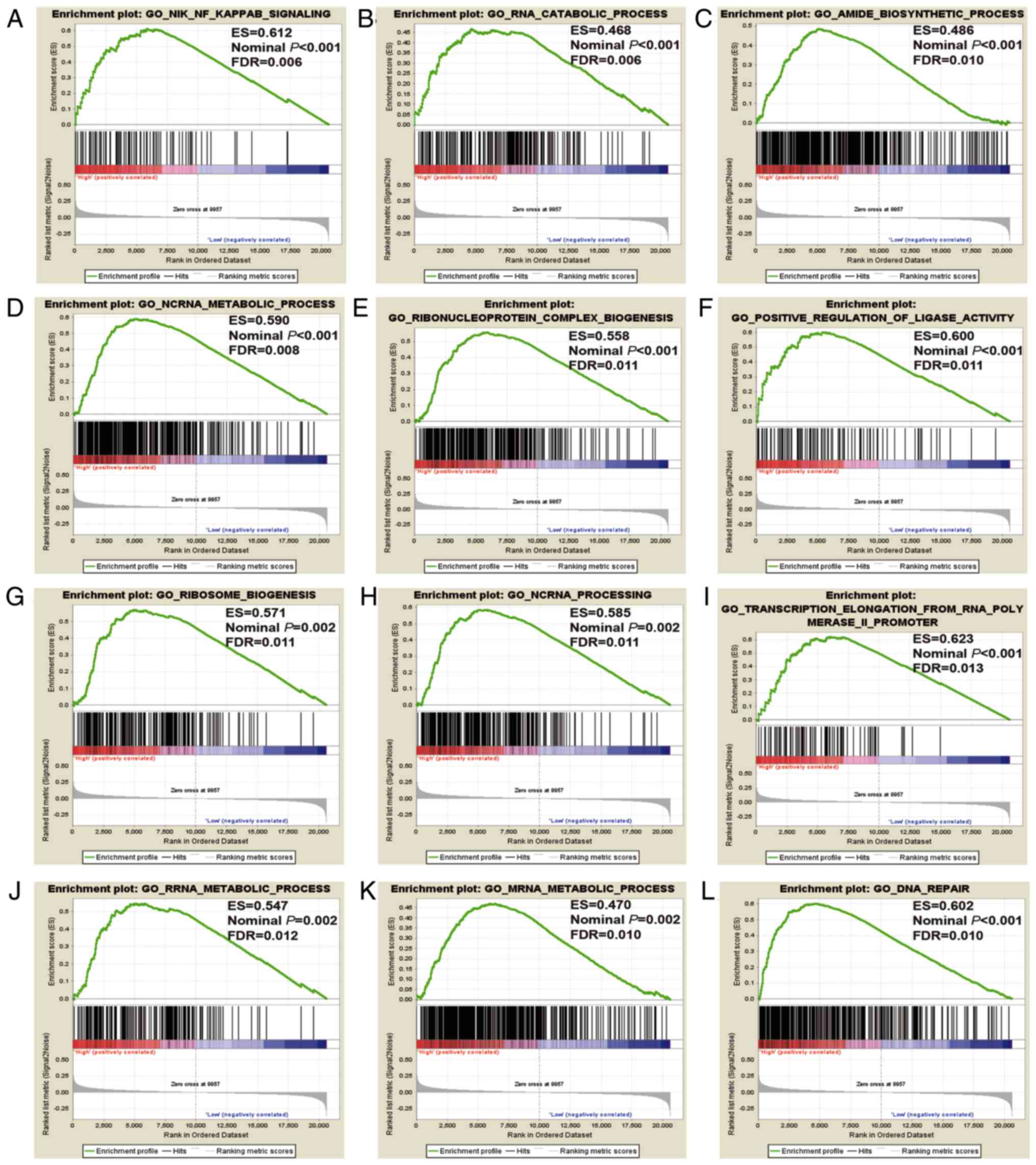 | Figure 7.GSEA results of CXCL3 in the
GSE40967 cohort, based on a GO dataset. (A) NIK NF κB signaling,
(B) RNA catabolic process, (C) amide biosynthetic process (D) ncRNA
metabolic process, (E) ribonucleoprotein complex biogenesis, (F)
positive regulation of ligase activity, (G) ribosome biogenesis,
(H) ncRNA processing, (I) transcription elongation from RNA
polymerase II promoter, (J) rRNA metabolic process, (K) mRNA
metabolic process and (L) DNA repair. GSEA, gene set enrichment
analysis; CXCL3, C-X-C motif chemokine ligand 3; ES,
enrichment score; FDR, false discovery rate; ncRNA, non-coding RNA;
rRNA, ribosomal RNA; mRNA, messenger RNA. |
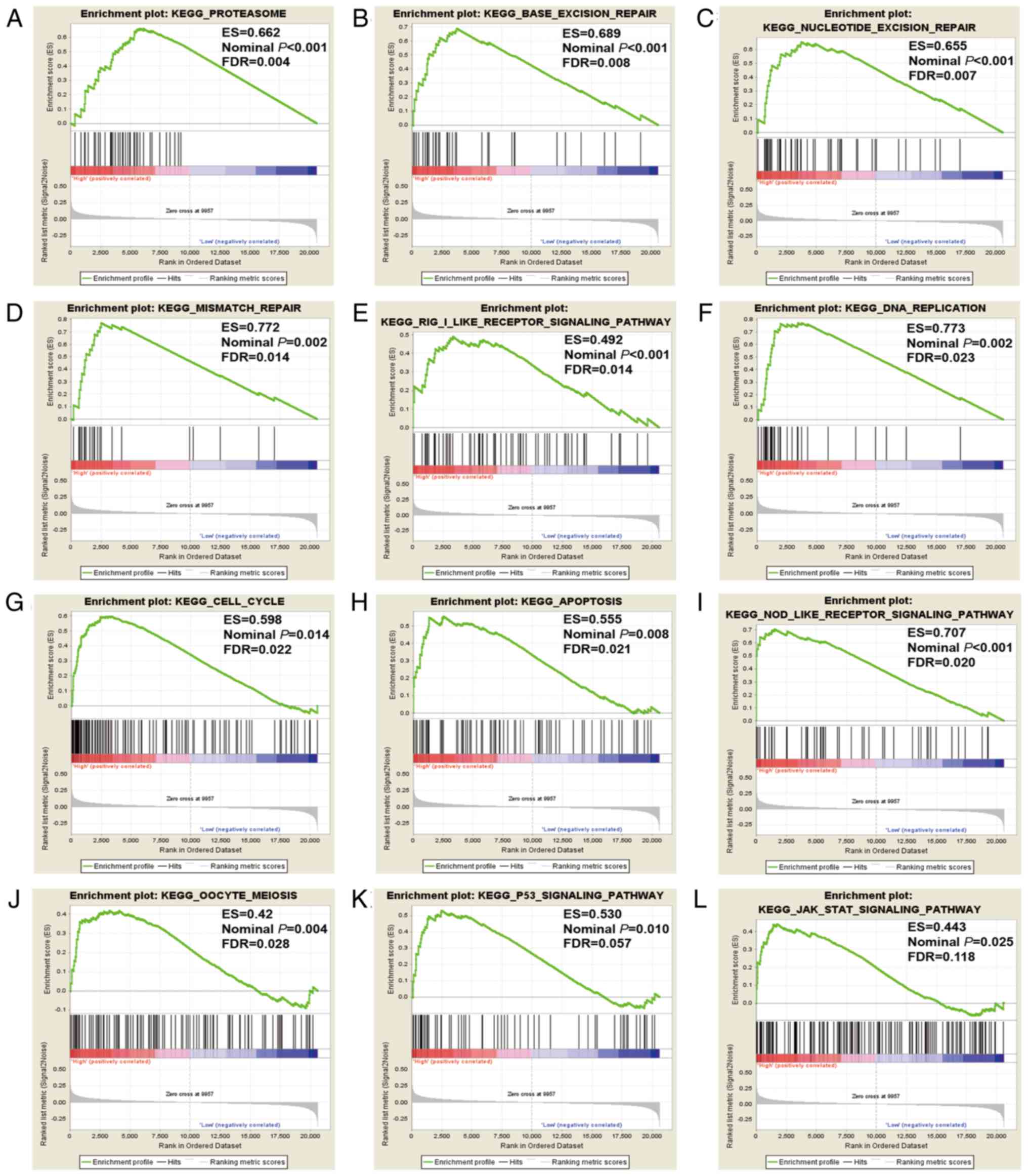 | Figure 8.GSEA results of CXCL3 in the
GSE40967 cohort, based on a KEGG dataset. (A) proteasome, (B) base
excision repair, (C) nucleotide excision repair, (D) mismatch
repair, (E) RIG I-like receptor signaling pathway, (F) DNA
replication, (G) cell cycle, (H) apoptosis, (I) NOD-like receptor
signaling pathway, (J) oocyte meiosis, (K) P53 signaling pathway,
(L) JAK STAT signaling pathway. GSEA, gene set enrichment analysis;
CXCL3, C-X-C motif chemokine ligand 3; RIG I, retinoic
acid-inducible gene-I; NOD, nucleotide-binding oligomerization
domain; ES, enrichment score; FDR, false discovery rate. |
Discussion
The CXCL3 gene is located in a cluster of
other CXC chemokines on chromosome 4 (23). It is a small cytokine belonging to the
CXC chemokine family, and is also known as GRO3 oncogene, GRO
protein gamma and macrophage inflammatory protein-2-beta (7,8). CXC
chemokines have a heparin-binding domain at the C-terminus of the
molecule, that serve different roles in the regulation of
angiogenesis (24). Simpson et
al (25) reported that
CXCL3 is widely expressed in the liver, and is involved in
liver injury and inflammation; Luan et al (26) reported that CXCL3 is an
important mediator of tumor initiation in human melanoma. Recent
studies have shown that CXCL3 has significant functions in
the progression and metastasis of malignant tumors. See et
al (27) reported that
CXCL3 is involved in breast cancer metastasis and may be a
potential target for cancer treatment. Gui et al (28) suggested that CXCL3 is
overexpressed in prostate cancer and might play various roles in
prostate cancer progression and metastasis. However, Li et
al (29) found no significant
difference in CXCL3 expression in non- and low-metastatic colon
cancer cells, compared with highly metastatic colon cancer cells.
Furthermore, Farquharson et al (30) demonstrated that insulin and
adiponectin can participate in the occurrence of colon cancer
through the regulation of CXCL3.
The study by Doll et al (9) showed that when CXCL3 mRNA
expression was tested by RT-qPCR in the CRC tissues of 97 patients
and normal colon tissues of 16 patients, CXCL3 gene
expression was significantly increased in CRC compared with normal
colon tissue. In the study by Xiong et al (10), the analysis of 695 RNA results from
645 CRC patients from the TCGA showed that the expression of
CXCL3 in cancer tissues was considerably higher than that in
adjacent normal tissues, which was verified by the RT-qPCR testing
of 25 pairs of fresh CRC and adjacent noncancerous tissues
collected from 25 patients at the First Affiliated Hospital of
Chongqing Medical University. Similar results have also been found
in other cancer studies; for example, one study found that
CXCL3 was higher in early stage non-small cell lung cancer
tissue as compared with the matched normal tissue (31). A meta-analysis also obtained
comparable results for CXCL3 in breast cancer (27). In the present study, the analysis of
CXCl3 mRNA in the paired cancer and adjacent tissues of 38
CC patients revealed that CXCl3 was overexpressed in CC; the
IHC scores of cancer and adjacent normal tissues in 46 patients
revealed that the CXCL3 score for cancer tissues were higher
than that for the adjacent tissues. These results are consistent
with the results obtained using GEPIA. Therefore, the present study
verified the overexpression of CXCL3 in CC tissues at both
the genetic and protein levels, which indicates that CXCL3
may be a potential marker for the diagnosis of CC.
Previous studies have found that overexpression of
CXCL3 indicates poor prognosis, Specifically, hepatocellular
carcinoma patients with higher CXCL3 expression have been
observed to have a shorter survival time (32). In addition, shorter OS was observed in
CRC patients with increased CXCL3 expression (10). In the current study of CC, similar
results were obtained. In the multivariate analysis of the Guangxi
Medical University cohort of 212 CC patients, although CXCL3
expression was not closely and directly connected with OS time,
further subgroup analysis revealed that CXCL3 positive
expression in patients who had a tumor diameter <5 cm or a tumor
embolus indicated poorer prognosis. A subsequent multivariate
analysis of prognosis in the GEO cohort, which was performed to
verify the results obtained from Guangxi Medical university cohort,
found that CXCL3 gene expression was notably relevant to
overall patient survival, and patients with high CXCL3 gene
expression had shorter survival times. These results also suggest
that CXCL3 might be a candidate prognostic biomarker for
CC.
CXCL3 is considered to serve a major role in
tumor initiation and invasion. The expression of CXCL3 in
normal colon tissue is high, indicating that it plays a certain
role in the physiological function of normal intestinal tissues,
but is dysregulated in cancer, indicating that expression disorder
of CXCL3 may be involved in the tumorigenesis of CC
(33). To examine the potential
mechanism of CXCL3 in CC, a genome-wide RNA sequencing
dataset in GSEA was analyzed in the present study. The results
indicated that the mechanism by which CXCL3 affects CC
prognosis may involve biological processes and signaling pathways
connected with DNA repair, cell cycle, apoptosis and P53
signaling. Previous studies have suggested an association between
DNA repair and CRC development (34–36).
Soreide et al (37) reported
that cell cycle and apoptosis are associated with the prognosis of
CRC. Numerous studies have reported a relationship between
P53 and the development of CRC (38–40).
However, to the best of our knowledge, the functional correlations
of DNA repair, cell cycle, apoptosis and P53 with
CXCL3 have not been previously reported. The GSEA of
CXCL3 in the present study supported the conclusion that
CXCL3 might affect CC via DNA repair, cell cycle, apoptosis
and the P53 pathway. However, these hypotheses require
further research for confirmation.
The present study used GSE40967 and Guangxi cohorts
to analyze the prognostic value of CXCL3 in CC at the mRNA and
protein levels. These two cohorts belong to retrospective cohort
studies with a level of evidence of four, as defined on the basis
of the Oxford Centre for Evidence-based Medicine-Levels of Evidence
(41). However, the present study has
certain limitations. The clinical information from the GEO database
was incomplete, and information such as tumor size, histology,
tumor differentiation, lymphatic invasion and venous invasion were
unattainable from the GEO website. The results of this study also
require validation in a larger sample population and in a
multi-center, multi-regional and multi-ethnic population.
Furthermore, in vitro and in vivo functional trials
are needed to further explore the roles of CXCL3 in CC
initiation, development, metastasis, proliferation and
angiogenesis. However, to the best of our knowledge, the current
study is the first to discover the value of CXCL3 in the
diagnosis and prognosis of CC, rather than CRC. Another advantage
of this study is that, in addition to identifying the prognostic
value of CXCL3 in CC in large samples, a GEO genome-wide
dataset was also used to explore prospective molecular mechanisms
through the GSEA approach.
In conclusion, the present study demonstrated that
CXCL3 is not only considerably upregulated in tumor tissue
but also has potential diagnostic value in patients with CC.
Survival analysis in Guangxi Medical University and GEO cohorts
suggested that CXCL3 may serve as a potential prognostic
biomarker in CC. The prospective molecular mechanism identified by
GSEA suggested that CXCL3 may influence the prognosis of CC
through involvement in the regulation of DNA repair, cell cycle
process, cell apoptosis process and P53 regulation pathways.
However, these results require further verification using in
vivo and in vitro experiments in future studies.
Supplementary Material
Supporting Data
Supporting Data
Supporting Data
Acknowledgements
The authors would like to acknowledge the support
(experimental environments and equipment) provided by the National
Key Clinical Specialty Programs (General Surgery and Oncology) and
Key Laboratory of Early Prevention and Treatment for Regional
High-Incidence-Tumor (Guangxi Medical University), Ministry of
Education, China. In addition, the authors also like to acknowledge
the helpful comments on this paper received from the reviewers.
Funding
This study was supported in part by the 2018
Innovation Project of Guangxi Graduate Education (grant no.
YCBZ2018036). This study was also supported by the Graduate Course
Construction Project of Guangxi Medical University (grant nos.
YJSB2017008 and YJSA2017014).
Availability of data and materials
The analyzed datasets generated during the study are
available from Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/), and the datasets
for the colon cancer cohort from the First Affiliated Hospital of
Guangxi Medical University used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
GTR and YZG wrote the manuscript. GTR and FG made
substantial contributions to the conception, design and
intellectual content of the study. GTR, YZG, XWL, SW, WH, XKW, GZZ
and CL made key contributions to the analysis and interpretation of
data. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
All patients signed an informed consent form, and
the experimental protocol was approved by the Ethics Committee of
the First Affiliated Hospital of Guangxi Medical University [No.
2019(KY-E-001)].
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Garborg K: Colorectal cancer screening.
Surg Clin North Am. 95:979–989. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Pita-Fernández S, González-Sáez L,
López-Calviño B, Seoane-Pillado T, Rodríguez-Camacho E,
Pazos-Sierra A, González-Santamaría P and Pértega-Díaz S: Effect of
diagnostic delay on survival in patients with colorectal cancer: A
retrospective cohort study. BMC Cancer. 16:6642016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Aran V, Victorino AP, Thuler LC and
Ferreira CG: Colorectal Cancer: Epidemiology disease mechanisms and
interventions to reduce onset and mortality. Clin Colorectal
Cancer. 15:195–203. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Clough E and Barrett T: The gene
expression omnibus database. Methods Mol Biol. 1418:93–110. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ahuja SK and Murphy PM: The CXC chemokines
growth-regulated oncogene (GRO) alpha, GRObeta, GROgamma,
neutrophil-activating peptide-2, and epithelial cell-derived
neutrophil-activating peptide-78 are potent agonists for the type
B, but not the type A, human interleukin-8 receptor. J Biol Chem.
271:20545–20550. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Smith DF, Galkina E, Ley K and Huo Y: GRO
family chemokines are specialized for monocyte arrest from flow. Am
J Physiol Heart Circ Physiol. 289:H1976–H1984. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Doll D, Keller L, Maak M, Boulesteix AL,
Siewert JR, Holzmann B and Janssen KP: Differential expression of
the chemokines GRO-2, GRO-3, and interleukin-8 in colon cancer and
their impact on metastatic disease and survival. Int J Colorectal
Dis. 25:573–581. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Xiong Y, You W, Wang R, Peng L and Fu Z:
Prediction and validation of hub genes associated with colorectal
cancer by integrating PPI network and gene expression data. Biomed
Res Int. 2017:24214592017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Magalhaes B, Peleteiro B and Lunet N:
Dietary patterns and colorectal cancer: Systematic review and
meta-analysis. Eur J Cancer Prev. 21:15–23. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Rebbeck TR, Devesa SS, Chang BL, Bunker
CH, Cheng I, Cooney K, Eeles R, Fernandez P, Giri VN, Gueye SM, et
al: Global patterns of prostate cancer incidence, aggressiveness,
and mortality in men of african descent. Prostate Cancer.
2013:5608572013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Rong M, He R, Dang Y and Chen G:
Expression and clinicopathological significance of miR-146a in
hepatocellular carcinoma tissues. Ups J Med Sci. 119:19–24. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Dai J, Wu H, Zhang Y, Gao K, Hu G, Guo Y,
Lin C and Li X: Negative feedback between TAp63 and Mir-133b
mediates colorectal cancer suppression. Oncotarget. 7:87147–87160.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Amin MB, Edge S, Greene F, Byrd DR,
Brookland RK, Washington MK, Gershenwald JE, Compton CC, Hess KR,
Sullivan DC, et al: AJCC Cancer Staging Manual. 8th. Springer;
Chicago, IL: pp. 202017
|
|
16
|
Zhang Y, Luo J, He R, Huang W, Li Z, Li P,
Dang Y, Chen G and Li S: Expression and clinicopathological
implication of DcR3 in lung cancer tissues: A tissue microarray
study with 365 cases. Onco Targets Ther. 9:4959–4968. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Uhlen M, Fagerberg L, Hallstrom BM,
Lindskog C, Oksvold P, Mardinoglu A, Sivertsson A, Kampf C,
Sjostedt E, Asplund A, et al: Proteomics. Tissue-based map of the
human proteome. Science. 347:12604192015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Tang Z, Li C, Kang B, Gao G and Zhang Z:
GEPIA: A web server for cancer and normal gene expression profiling
and interactive analyses. Nucleic Acids Res. 45:W98–W102. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Marisa L, de Reynies A, Duval A, Selves J,
Gaub MP, Vescovo L, Etienne-Grimaldi MC, Schiappa R, Guenot D,
Ayadi M, et al: Gene expression classification of colon cancer into
molecular subtypes: Characterization, validation, and prognostic
value. PLoS Med. 10:e10014532013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Subramanian A, Tamayo P, Mootha VK,
Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub
TR, Lander ES, et al: Gene set enrichment analysis: A
knowledge-based approach for interpreting genome-wide expression
profiles. Proc Natl Acad Sci USA. 102:15545–15550. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Benjamini Y and Hochberg Y: Controlling
the false discovery rate: A practical and powerful approach to
multiple testing. J Royal Stat Soc Series B (Methodological).
57:289–300. 1995. View Article : Google Scholar
|
|
22
|
Reiner A, Yekutieli D and Benjamini Y:
Identifying differentially expressed genes using false discovery
rate controlling procedures. Bioinformatics. 19:368–375. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
O'Donovan N, Galvin M and Morgan JG:
Physical mapping of the CXC chemokine locus on human chromosome 4.
Cytogenet Cell Genet. 84:39–42. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Airoldi I and Ribatti D: Regulation of
angiostatic chemokines driven by IL-12 and IL-27 in human tumors. J
Leukoc Biol. 90:875–882. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Simpson KJ, Henderson NC, Bone-Larson CL,
Lukacs NW, Hogaboam CM and Kunkel SL: Chemokines in the
pathogenesis of liver disease: So many players with poorly defined
roles. Clin Sci (Lond). 104:47–63. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Luan J, Shattuck-Brandt R, Haghnegahdar H,
Owen JD, Strieter R, Burdick M, Nirodi C, Beauchamp D, Johnson KN
and Richmond A: Mechanism and biological significance of
constitutive expression of MGSA/GRO chemokines in malignant
melanoma tumor progression. J Leukoc Biol. 62:588–597. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
See AL, Chong PK, Lu SY and Lim YP: CXCL3
is a potential target for breast cancer metastasis. Curr Cancer
Drug Targets. 14:294–309. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Gui SL, Teng LC, Wang SQ, Liu S, Lin YL,
Zhao XL, Liu L, Sui HY, Yang Y, Liang LC, et al: Overexpression of
CXCL3 can enhance the oncogenic potential of prostate cancer. Int
Urol Nephrol. 48:701–709. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Li A, Varney ML and Singh RK: Constitutive
expression of growth regulated oncogene (gro) in human colon
carcinoma cells with different metastatic potential and its role in
regulating their metastatic phenotype. Clin Exp Metastasis.
21:571–579. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Farquharson AJ, Steele RJ, Carey FA and
Drew JE: Novel multiplex method to assess insulin, leptin and
adiponectin regulation of inflammatory cytokines associated with
colon cancer. Mol Biol Rep. 39:5727–5736. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kowalczuk O, Burzykowski T, Niklinska WE,
Kozlowski M, Chyczewski L and Niklinski J: CXCL5 as a potential
novel prognostic factor in early stage non-small cell lung cancer:
Results of a study of expression levels of 23 genes. Tumour Biol.
35:4619–4628. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhang L, Li H, Ge C, Zhao F, Tian H, Chen
T, Jiang G, Xie H, Cui Y, Yao M, et al: CXCL3 contributes to
CD133(+) CSCs maintenance and forms a positive feedback regulation
loop with CD133 in HCC via Erk1/2 phosphorylation. Sci Rep.
6:274262016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Fagerberg L, Hallstrom BM, Oksvold P,
Kampf C, Djureinovic D, Odeberg J, Habuka M, Tahmasebpoor S,
Danielsson A, Edlund K, et al: Analysis of the human
tissue-specific expression by genome-wide integration of
transcriptomics and antibody-based proteomics. Mol Cell Proteomics.
13:397–406. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Dörsam B, Seiwert N, Foersch S, Stroh S,
Nagel G, Begaliew D, Diehl E, Kraus A, McKeague M, Minneker V, et
al: PARP-1 protects against colorectal tumor induction, but
promotes inflammation-driven colorectal tumor progression. Proc
Natl Acad Sci USA. 115:E4061–E4070. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
AlDubayan SH, Giannakis M, Moore ND, Han
GC, Reardon B, Hamada T, Mu XJ, Nishihara R, Qian Z, Liu L, et al:
Inherited DNA-repair defects in colorectal cancer. Am J Hum Genet.
102:401–414. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Aggarwal N, Donald ND, Malik S, Selvendran
SS, McPhail MJ and Monahan KJ: The association of low-penetrance
variants in DNA repair genes with colorectal cancer: A systematic
review and meta-analysis. Clin Transl Gastroenterol. 8:e1092017.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Soreide K, Buter TC, Janssen EA,
Gudlaugsson E, Skaland I, Korner H and Baak JP: Cell-cycle and
apoptosis regulators (p16INK4A, p21CIP1, beta-catenin, survivin,
and hTERT) and morphometry-defined MPECs predict metachronous
cancer development in colorectal adenoma patients. Cell Oncol.
29:301–313. 2007.PubMed/NCBI
|
|
38
|
Noda M, Okayama H, Kofunato Y, Chida S,
Saito K, Tada T, Ashizawa M, Nakajima T, Aoto K, Kikuchi T, et al:
Prognostic role of FUT8 expression in relation to p53 status in
stage II and III colorectal cancer. PLoS One. 13:e02003152018.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Wu Y, Li Y, Zhao X, Dong D, Tang C, Li E
and Geng Q: Combined detection of the expression of Nm23-H1 and p53
is correlated with survival rates of patients with stage II and III
colorectal cancer. Oncol Lett. 13:129–136. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Katkoori VR, Manne U, Chaturvedi LS,
Basson MD, Haan P, Coffey D and Bumpers HL: Functional consequence
of the p53 codon 72 polymorphism in colorectal cancer. Oncotarget.
8:76574–76586. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Howick J, Chalmers I, Glasziou P,
Greenhalgh T, Heneghan C, Liberati A, Moschetti I, Phillips B and
Thornton H: The 2011 Oxford CEBM Levels of Evidence (Introductory
Document). Oxford Centre for Evidence-Based Medicine. http://www.cebm.net/index.aspx?o=5653
|