Introduction
Breast cancer is one of the most common types of
cancer in females, with 2.1 million newly diagnosed cases in 2018
alone, accounting for ~25% of all female cancer diagnoses (1). Approximately 15% of all female
cancer-associated mortalities in the United States of America were
attributed to breast cancer (2).
Current treatment therapies include surgical treatment,
chemotherapy, radiotherapy and endocrine therapy (3,4). However,
treatment may be ineffective, particularly due to the development
of chemotherapy and endocrine therapy resistance. There is
therefore a requirement for the identification of novel therapeutic
strategies for the treatment of breast cancer.
X-linked ribosomal S6 kinase 4 (RSK4) is a
serine-threonine protein kinase (5)
that participates in the regulation of cellular senescence
(6) and in tumor protein
P53-dependent cell growth arrest signaling, acting as an inhibitor
during embryogenesis (7). The
distribution RSK4 in normal and tumor tissues has not been
systematically investigated. Aberrant RSK4 expression has been
reported in several types of cancer, including breast cancer
(8), colorectal cancer (9), acute myeloid leukemia (10), non-small cell lung carcinoma (11), endometrial cancer (12) and ovarian cancer (13,14).
Several studies have demonstrated that RSK4 is downregulated in
breast cancer and may therefore act as a tumor suppressor gene
(8,15–17).
Additionally, downregulation of RSK4 in breast cancer is associated
with hypermethylation of the RSK4 promoter (8,15).
The sex hormone estrogen (E2) is involved in the
growth, development and physiology of the mammary glands. E2 binds
to the E2 receptor (ER) α or β and regulates cellular proliferation
and differentiation. ERα upregulation occurs in ~70% of breast
cancer cases (18,19) and is associated with tumor progression
(20,21). ER upregulation is the most commonly
used clinical biomarker for breast cancer and ER-positive
(ER+) breast cancer is amenable to endocrine therapy
(22). E2 stimulation promotes the
expression of RSK4 in normal mammary epithelial cells and the
expression of RSK4 in breast cancer cells is closely associated
with RSK4 methylation (15). ER
signaling may therefore affect the expression of RSK4 or the
methylation of the RSK4 promoter.
The present study explored the association between
ER status, RSK4 expression and promoter methylation in breast
cancer tissues. Additionally, the role of RSK4 and the effect of ER
signaling on RSK4 promoter methylation in ER+ breast
cancer cells were investigated.
Materials and methods
Tumor tissue specimens
A total of 60 female patients who underwent breast
surgery at the Guangxi Medical University Cancer Hospital between
January 2013 to December 2014 were included in the current study.
The clinical characteristics of the patients are presented in
Table I. The patients had a mean age
of 45.3 years (range, 25–71 years). The inclusion criteria were as
follows: i) Pathological diagnosis of luminal A or luminal type B
invasive breast cancer; and ii) no family history of breast cancer.
Patients who had received tumor-associated treatment (including
radiotherapy and chemotherapy) prior to admission were excluded
from the current study.
 | Table I.Characteristics of 60 patients with
breast cancer, stratified by molecular subtype. |
Table I.
Characteristics of 60 patients with
breast cancer, stratified by molecular subtype.
| Characteristic | Total | Luminal A | Luminal B1 | Luminal B2 | P-value |
|---|
| n (%) | 60 (100) | 21 (35) | 19 (32) | 20 (33) |
|
| Age (years), n |
|
|
|
| 0.858a |
|
<45 | 24 | 9 | 8 | 10 |
|
|
≥45 | 36 | 12 | 11 | 10 |
|
| Median (range) |
| 45 (30–65) | 45 (28–71) | 46 (25–69) |
|
| Menopausal status,
n |
|
|
|
| 0.859a |
|
Premenopausal | 28 | 11 | 10 | 12 |
|
|
Postmenopausal | 32 | 10 | 9 | 8 |
|
| Pathology type,
n |
|
|
|
|
|
|
Invasive ductal carcinoma | 60 | 21 | 19 | 20 |
|
| T stage, n |
|
|
|
| 0.744b |
| T1 | 8 | 2 | 4 | 2 |
|
| T2 | 23 | 8 | 8 | 7 |
|
| T3 | 29 | 11 | 7 | 11 |
|
| N stage, n |
|
|
|
| 0.922b |
| N0 | 6 | 2 | 3 | 1 |
|
| N1 | 28 | 10 | 8 | 10 |
|
| N2 | 26 | 9 | 8 | 9 |
|
| TNM stage, n |
|
|
|
| 0.783b |
| IA | 3 | 1 | 2 | 0 |
|
|
IIA | 9 | 3 | 4 | 2 |
|
|
IIB | 25 | 8 | 7 | 10 |
|
|
IIIA | 23 | 9 | 6 | 8 |
|
| Hormone receptor
status, n |
|
|
|
|
|
|
Positive | 60 | 21 | 19 | 20 |
|
|
Negative | – | – | – | – |
|
| Tumor nuclear
grade, n |
|
|
|
| 0.387b |
| I | 18 | 5 | 6 | 7 |
|
| II | 20 | 6 | 9 | 5 |
|
|
III | 22 | 10 | 4 | 8 |
|
Surgical resection specimens were obtained in the
Department of Breast Surgery at the Guangxi Medical University
Cancer Hospital. Breast cancer tissue and non-cancerous glandular
tissue samples (at a distance of ≥2 cm from the tumor edge) were
obtained from all patients. All samples were immediately placed in
liquid nitrogen, stored at 80°C and subsequently embedded in
paraffin. The present study was approved by the Research Ethics
Committee of the Guangxi Medical University Cancer Hospital and
patients provided written informed consent.
Cell culture
The ER+ human breast cancer cell line
MCF-7 and the ER− human breast cancer cell lines
MDA-MB-231 and MDA-MB-453 were purchased from the Chinese Academy
of Sciences. The breast cancer cell lines were cultured in
Dulbecco's Modified Eagles medium (DMEM; HyClone; GE Life Sciences)
containing 10% fetal bovine serum (FBS; Sigma-Aldrich; Merck KGaA).
293T and human umbilical vein endothelial cells (HUVECs) were
purchased from the Chinese Academy of Sciences. All cells were
maintained at 37°C and 5% CO2.
Lentiviral packaging and cell
transfection
The RSK4 lentiviral expression vector carrying the
RSK4 coding sequence (pLenti-RSK4; 650 ng/µl; Thermo Fisher
Scientific, Inc.) was co-transfected into 293T cells together with
the packaging plasmids (Invitrogen; Thermo Fisher Scientific,
Inc.). Medium containing lentiviral particles was subsequently
harvested 48 h following transfection. Lentivirus stock was stored
at −80°C and used to infect MCF-7 cells in subsequent
experiments.
MCF-7 cells were seeded in 6-well plates at a
density of 3×105 cells/well. Upon reaching 70%
confluence, the cells were transfected with the lenti-RSK4-eGFP
virus (RSK4-OE) or a lenti-eGFP virus as a negative control (NC) at
a multiplicity of infection of 30. Nontransfected MCF-7 cells
served as additional controls. Transfections were performed in
triplicate. Cells were observed 24, 48 and 72 h following
transfection using bright field/fluorescence microscopes
(magnification, ×100; three fields), and the transfection
efficiency was further assessed by western blotting.
Immunohistochemistry
Paraffin blocks containing fixed tissues were sliced
into 4-µm sections, placed on a glass slide, dewaxed with xylene
and dehydrated using an alcohol gradient (100, 95, 70, 50%).
Endogenous peroxidase activity was blocked by incubating the
sections in 3% hydrogen peroxide at 37°C for 10 min. Samples were
microwaved at high power for 4 min and low power for 20 min and
allowed to reach room temperature to allow antigen recovery.
Sections were immersed in normal goat serum (Abcam) for 10 min and
subsequently incubated overnight at 4°C with primary antibodies
against ER (1:500; Abcam, Ab75635) and RSK4 (1:1,000; Abcam,
Ab76117). Following primary antibody incubation, samples were
incubated with rabbit secondary antibody (1:200; Jackson
ImmunoResearch Laboratories Inc., cat. no. 117360), stained with
DAB (Beijing Solarbio Science & Technology Co., Ltd.) for 2 min
and counterstained with hematoxylin for 5 min at room temperature.
Samples were visualized using a DM750 microscope (Leica,
Germany).
ER and RSK4 staining was independently assessed by
two senior pathologists. Three randomly selected fields were scored
under 100× magnification using a light microscope. A total of 200
cells were scored per field, with ≥600 cells scored per section. ER
and RSK4 staining was scored as follows: i) <10% Positively
stained cells, 0 points; ii) 10–30% positively stained cells, 1
point; iii) 31–60% positively stained cells, 2 points; and iv)
>61% positively stained cells, 3 points. ER and RSK4 staining
was additionally scored for color intensity as follows: i)
Colorless, 0 points; ii) light yellow, 1 point; iii) brown, 2
points; and iv) tan, 3 points. The overall level of ER and RSK4
expression was determined by multiplying the two scores as follows:
i) 0–1 Points, negative; ii) 1.1–2.0 points, weakly positive; iii)
2.1–3.0, moderately positive; and iv) 3.1–5.0 staining, strongly
positive.
RNA extraction and
reverse-transcription quantitative polymerase chain reaction
(RT-qPCR)
Total RNA was extracted from patient tissue samples
and transduced cells using TRIzol® reagent (Thermo
Fisher Scientific, Inc.) according to the manufacturer's protocol.
The RNA was reverse-transcribed into cDNA using the RevertAid First
Strand cDNA Synthesis kit (Thermo Fisher Scientific, Inc.) in
accordance with the manufacturer's instructions. qPCR was
subsequently performed using SYBR® Green mix (Thermo
Fisher Scientific, Inc.) according to the manufacturer's protocol
at Mx3000P Real-time fluorescence quantification PCR instrument
(Agilent Technologies, Inc.). The thermocycling conditions were as
follows: 95°C for 2 min, followed by 40 cycles of 95°C for 15 sec,
60°C for 20 sec and 72°C for 20 sec and a final extension step
60.5°C for 30 sec. mRNA levels were quantified using the
2−ΔΔCq method (23) and
normalized to β-actin. Primer sequences were as follows: RSK4
forward, 5′-AGATTCTCCCGGTTTGCC-3′ and reverse,
5′-AAGGGTCTCGCTTACTTTTGT-3′; β-actin forward,
5′-GGCACTCTTCCAGCCTTCC-3′ and reverse, 5′-GAGCCGCCGATCCACAC-3′.
Western blotting
Cells were lysed with cell lysis buffer (Beyotime
Institute of Biotechnology) and the protein concentration was
determined using a Pierce™ BCA Protein Assay kit (Thermo Fisher
Scientific, Inc.). Protein aliquots (30 µg/lane) were separated via
SDS-PAGE on a 10% gel. The separated proteins were subsequently
transferred onto a polyvinylidene difluoride membrane and blocked
with 5% skim milk powder in Tris-buffered saline with Tween-20
(TBST) for 1 h at room temperature. The membranes were incubated
with primary antibodies against RSK4 (1:1,000; Abcam), ER (1:400;
Abcam) and GAPDH (1:1,000; Sungene), β-actin (1:2,000; Boster
Biological Technology) or tubulin (1:2,000; Sungene) overnight at
4°C. Following three washes with TBST, the membranes were incubated
with rabbit HRP secondary antibodies against RSK4 and ER (1:3,000;
Jackson ImmunoResearch Laboratories Inc.), and GAPDH, β-actin and
tubulin) (1:3,000; Jackson ImmunoResearch Laboratories Inc.) for 1
h at room temperature. The membranes were washed three times with
TBST and protein bands were visualized using the Tanon-4200
automatic chemiluminescence image analysis system (Tanon-4200 Fully
Automatic Chemiluminescence Image Analyzer, Tianneng).
DNA extraction and bisulfite
sequencing
Genomic DNA was extracted from patient tissues and
cells and treated with bisulfite using the EZ DNA Methylation-Gold™
kit (Zymo Research Corp.) according to the manufacturer's
protocols. The bisulfite-treated DNA was subsequently amplified by
PCR using the following primer pair: RSK4 forward,
5′-TGAGAGGGTTTGTTGAGTATGTGTG-3′ and reverse,
5′-CCTCTATACTACCTCTCCAAAAACTAC-3′. The PCR products were cloned
into the pMD19-T vector (Takara Biotechnology Co., Ltd.) according
to manufacturer's protocols. The ligation product (10 µl) was
transformed into DH5α competent cells (Invitrogen; Thermo Fisher
Scientific, Inc.) using the conversion method. The cells were
subsequently plated onto Luria broth (LB; Thermo Fisher Scientific,
Inc.) plates containing 0.1 mg/ml ampicillin and cultured overnight
at 37°C. Eight bacterial colonies were selected from each plate and
the presence of the insert was confirmed by PCR as aforementioned.
Colonies containing the cloned inserts (at least 5 per plate) were
inoculated in 3 ml LB containing 0.1 mg/ml ampicillin and cultured
overnight at 37°C. Plasmids were extracted using the AxyPerp
Plasmid Miniprep kit (Axygen; Corning, Inc.) and analyzed using the
Quantification Tool For Methylation Analysis (quma.cdb.riken.jp).
Five clones were sequenced per locus.
Cell proliferation assay
The proliferation of transduced cells was determined
using the Cell Counting Kit-8 (CCK-8; Dojindo Molecular
Technologies, Inc.). Cells were seeded in a 96-well plate at a
density of 3×103 cells per well and cultured at 37°C. A
total of 10 µl CCK-8 solution was added following 1, 2, 3, 4 and 5
days of incubation, and cells were incubated at 37°C for a further
2 h. Cell proliferation was subsequently measure at a wavelength of
450 nm a microplate reader (Thermo Fisher Scientific, Inc.).
Samples were analyzed in triplicate.
Clone formation assay
A total of 2 ml transfected cell suspension
(1×103 cells/ml) were added to each well of a 6-well
plate and cultured until 30–50 cell clusters had formed, ~5–7 days
later. The spent cell culture medium was discarded and the cells
were fixed in methanol (0.1%, 37°C, 15 min) and stained with
crystal violet (0.1%, 37°C, 20 min). Clones were counted using an
inverted fluorescence microscope (Olympus Corporation;
magnification, ×100). Clone formation was calculated as the number
of clones formed/number of cells seeded ×100%.
Cell migration assay
A total of 100 µl transfected cell suspension was
pipetted into the upper chamber of 24-well Transwell inserts at a
density of 4×104 cells per well. The lower chamber was
filled with DMEM supplemented with 10% FBS. Cells were incubated at
37°C and 5% CO2 for 24 h, after which the cells in the
upper chamber that had not migrated through the polycarbonate
membrane were wiped off. Cells in the lower chamber were fixed with
4% paraformaldehyde (Beijing Solarbio Science & Technology Co.,
Ltd.) for 30 min at 37°C and stained with 0.1% crystal violet
(Beijing Solarbio Science & Technology Co., Ltd.) for 20 min at
37°C. The cells were subsequently washed three times with PBS.
Stained cells were counted in five randomly selected fields using
an inverted fluorescence microscope (Olympus Corporation;
magnification, ×100). Each well was counted in triplicate.
Flow cytometry
After 48 h following transduction, cells were washed
with PBS, trypsinized and suspended in 500 µl binding buffer which
was included in the kit. Cells were subsequently incubated with 5
µl Annexin V-allophycocyanin (APC) and 5 µl propidium iodide (PI;
Annexin V-APC-PI Apoptosis Analysis kit; Sungene) in the dark at
room temperature for 10–15 min. Annexin V fluorescence was measured
using a FACSAria Cell Sorter c6 flow cytometer (Accuri C6; BD
Biosciences). The experiment was performed in triplicate.
Angiogenesis assay
Matrigel (BD Biosciences) was pipetted into a
96-well plate (50 µl/well), incubated for 30 min at 4°C and allowed
to set for 30 min at 37°C. A total of 5×104 HUVECs
suspended in 50 µl serum-free conditioned medium taken from MCF-7
cells cultured for 24 h were seeded on Matrigel-coated wells.
Endothelial tube formation was assessed following 5 h of culture
and the mean number of tubes was counted in three randomly selected
fields per well using an inverted fluorescence microscope (Olympus
Corporation; magnification, ×100).
E2 induction of MCF-7 cells
MCF-7 cells were plated into a 6-well plate at a
density of 3×105 cells/well and incubate at 37°C and 5%
CO2. E2 (Sigma-Aldrich; Merck KGaA) was diluted in
dimethyl sulfoxide (Beijing Solarbio Science & Technology Co.,
Ltd.) to prepare a stock solution of 10 mM and added to MCF-7 cells
at final concentrations of 1, 5 or 10 nM. Cells were subsequently
incubated with E2 for 48 h at 37°C. Untreated MCF-7 cells served as
controls. Cells were imaged prior and following treatment using an
inverted fluorescence microscope (Olympus Corporation;
magnification, ×100; three random fields were selected).
Disease-free survival and overall
survival
The 60 patients with breast cancer enrolled in the
current study were followed-up by telephone or by electronic
medical records until November 30, 2018. The median follow-up time
was 52 months (range, 10–66 months). Local recurrence was defined
as recurrence in the ipsilateral chest wall or regional lymph nodes
detected by imaging or histology. Distant metastasis was defined as
distant metastatic lesions detected by imaging. Disease-free
survival was defined as time from the date of surgery to recurrence
or metastasis. Overall survival was defined as time from the date
of surgery to death. Patients with RSK4 methylation levels above or
equal to the mean methylation value of 60 patients were defined as
hypermethylated, while those with RSK4 methylation below the mean
were defined as hypomethylated. Patient characteristics are
presented in Table I.
Statistical analysis
Data were presented as mean ± standard deviation.
Data were analyzed using SPSS software (version 19; IBM Corp.). A
Student's t-test was used for the comparison of two groups while
one-way ANOVA was used for the comparison of three or more groups
followed by a Least Significant Difference post-hoc test. Pearson's
correlation analysis was used to assess the correlation of ER
status with RSK4 expression. Patient survival was assessed using
the Kaplan-Meier method, and survival curves were compared using
the log-rank test. R × C bidirectional disordered count data were
analyzed via a Pearson's χ2 test. Grade data were
analyzed via a Mann-Whitney U test. P<0.05 was considered to
indicate a statistically significant difference. Three independent
experiments were performed.
Results
ER+ status is associated
with higher RSK4 expression and lower promoter methylation in
breast cancer tissue compared with ER− status
The current study examined whether there was an
association between RSK4 expression and breast cancer by comparing
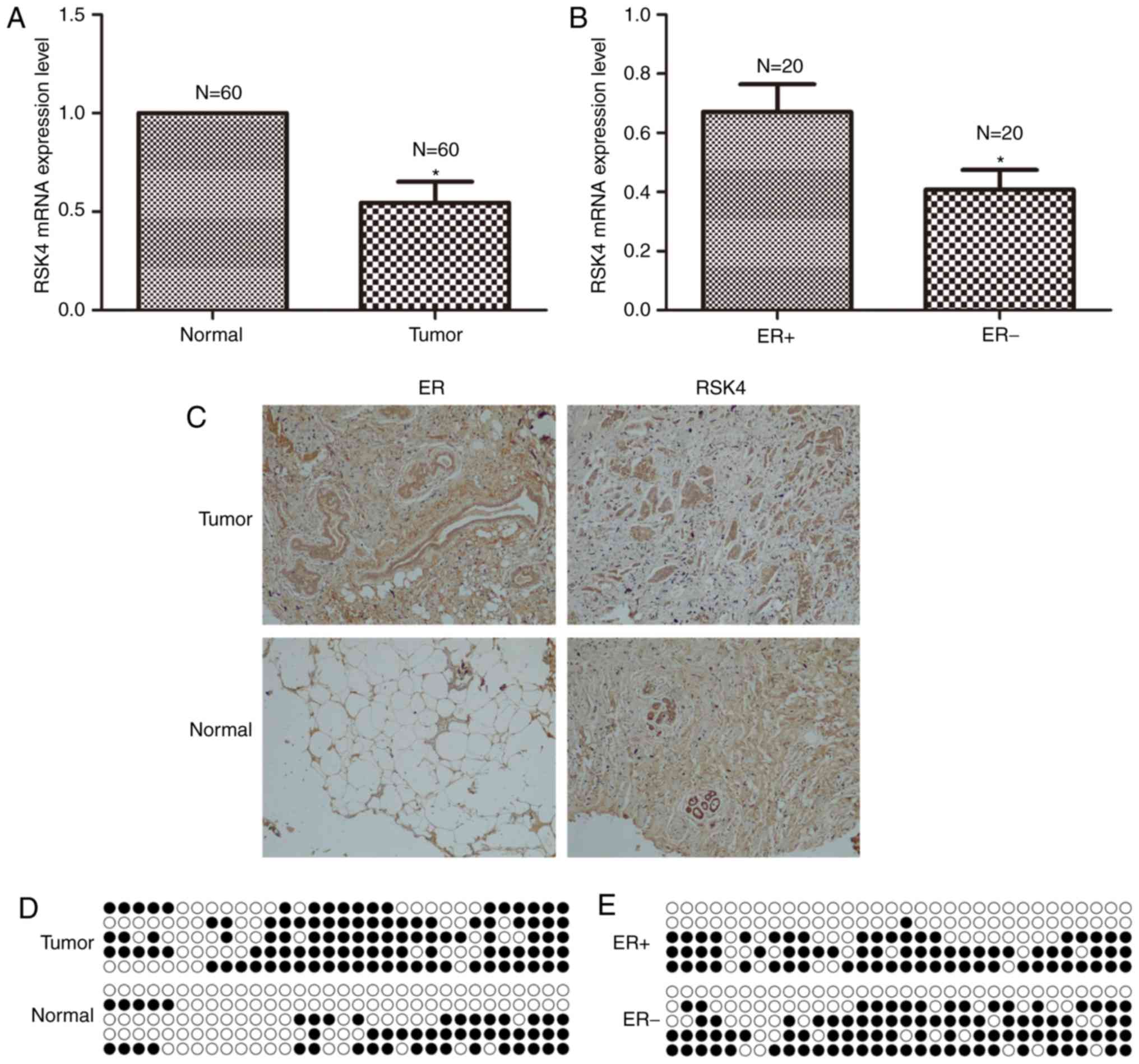
breast cancer and adjacent normal tissues. RSK4 mRNA expression was
significantly decreased in breast cancer tissues compared with
adjacent normal tissues (P=0.003; Fig.
1A). Similarly, immunohistochemical analysis revealed that RSK4
protein expression was markedly decreased in breast cancer tissues
compared with adjacent normal tissues (Fig. 1C). ER protein expression was notably
increased in breast cancer tissues compared with adjacent normal
tissues (Fig. 1C). Bisulfite
sequencing revealed that RSK4 promoter methylation was increased in
breast cancer tissues compared with adjacent normal tissues (66.2
vs. 31.2%; Fig. 1D).
The association between RSK4 expression and ER
status in breast cancer was investigated by analyzing
ER+ and ER− tumor tissues. RSK4 mRNA
expression was significantly decreased in ER− breast
cancer tissues compared with ER+ breast cancer tissues
(P=0.037; Fig. 1B). Bisulfite
sequencing demonstrated that RSK4 promoter methylation was
decreased in ER+ breast cancer tissues compared with
ER− breast cancer tissues (45 vs. 56.9%; Fig. 1E).
ER+ status is associated
with increased RSK4 protein levels and decreased promoter
methylation in breast cancer cell lines compared with
ER− status
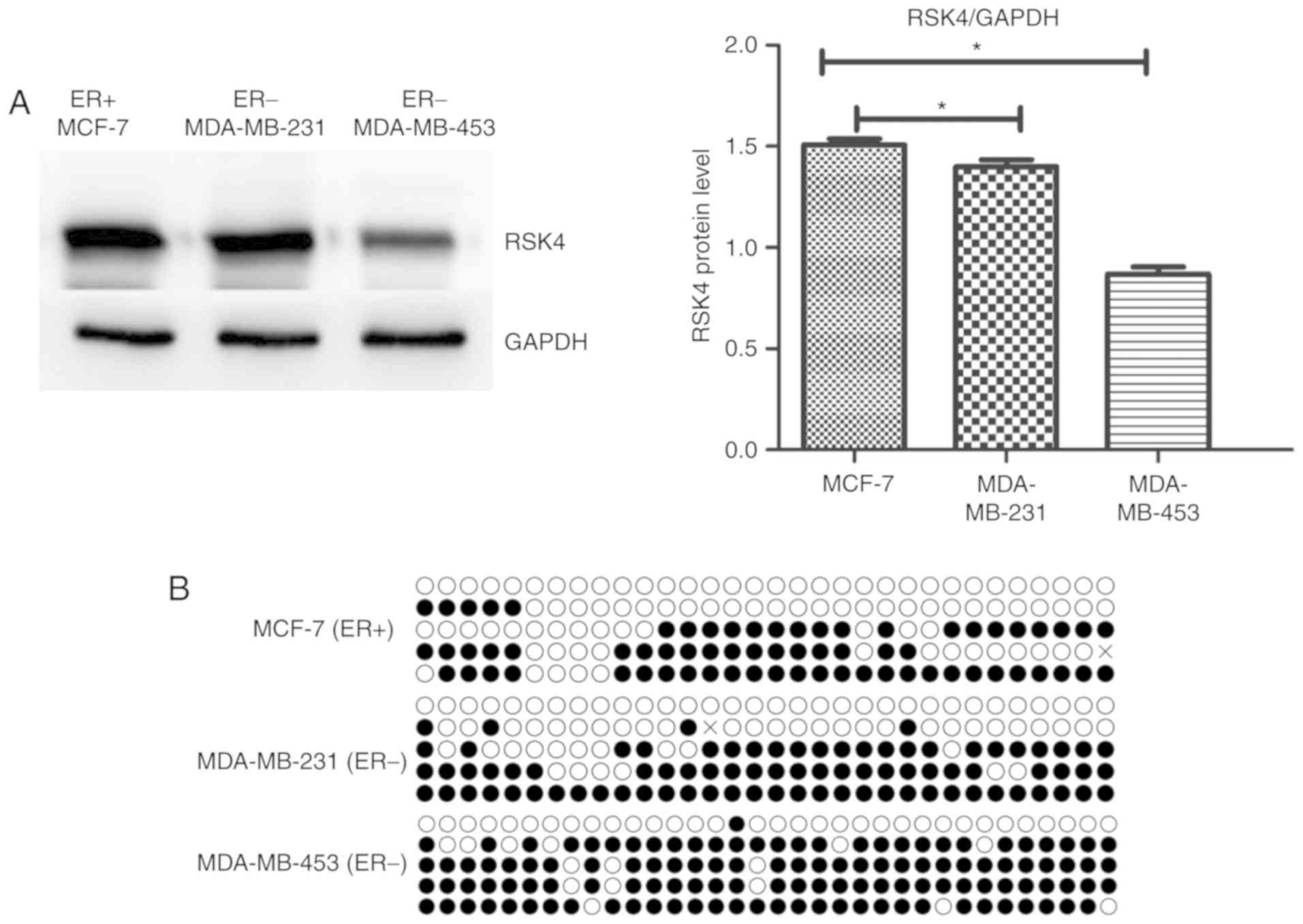
The RSK4 protein levels in breast cancer cell lines
were assessed using western blotting. Similarly to the mRNA data
obtained from patient tissue samples, RSK4 protein expression was
significantly increased in the ER+ breast cancer cell
line MCF-7 compared with the ER− breast cancer cell
lines MDA-MB-231 (P=0.037) and MDA-MB-453 (P<0.001; Fig. 2A). The aforementioned results
suggested that RSK4 expression decreased with increasing tumor
malignancy as MDA-MB-231 and MDA-MB-453 cells have a higher degree
of malignancy than MCF-7 cells (24).
Furthermore, the ER+ breast cancer cell line MCF-7
exhibited decreased RSK4 promoter methylation (compared with the
ER− breast cancer cell lines MDA-MB-231 and MDA-MB-453
(42.8 vs. 52.8% and 71.8%, respectively; Fig. 2B).
RSK4 overexpression inhibits the
proliferation and clone formation in ER+ breast cancer
cells
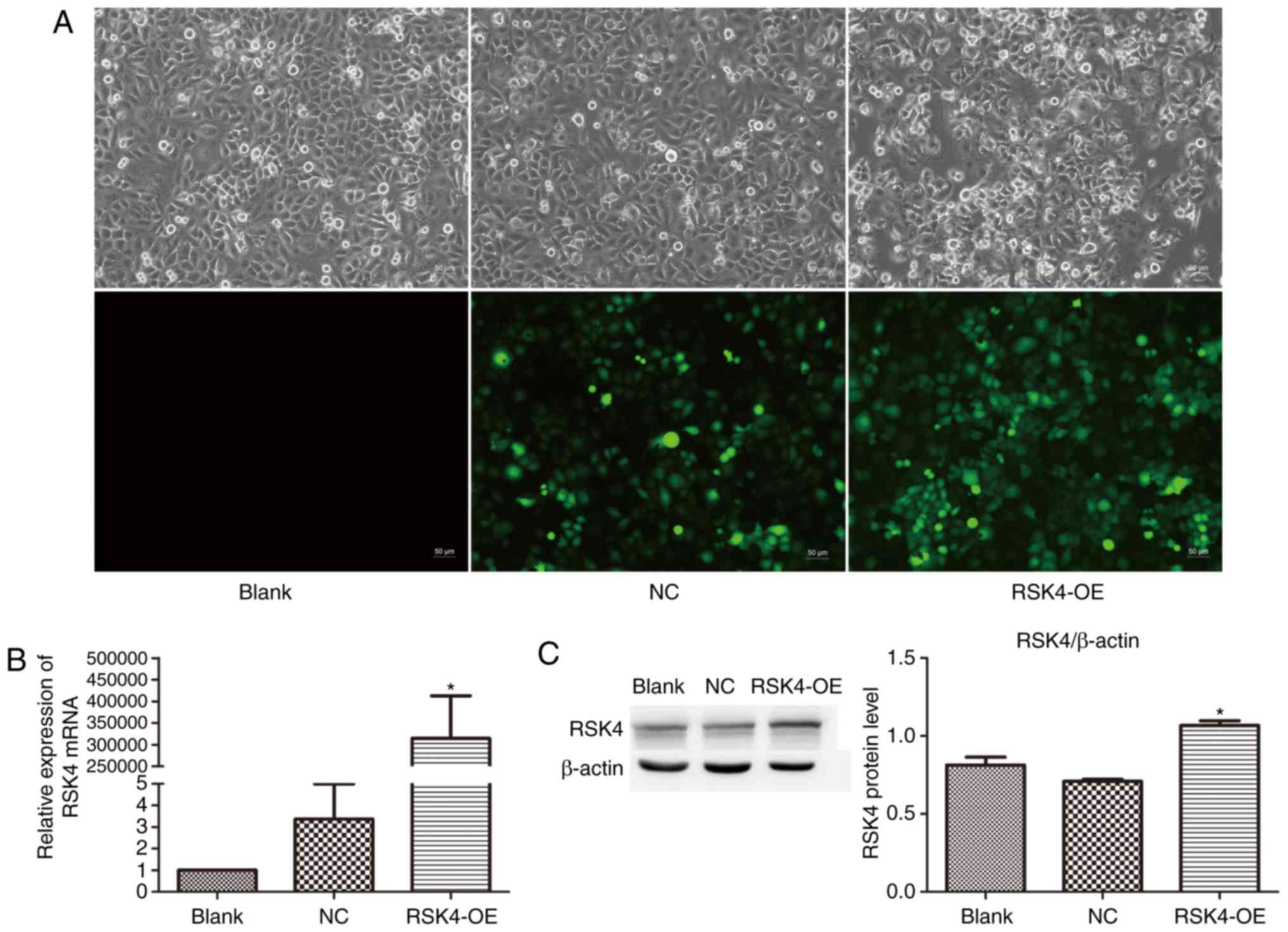
To investigate the effects of RSK4 overexpression on
breast cancer cells, MCF-7 breast cancer cells were transfected
with RSK4-OE or Lenti-EGFP (NC) vectors. MCF-7 cells were selected
since they are ER+ breast cancer cells. Cells
transfected with RSK4-OE exhibited a notable increase in EGFP
fluorescence over 72 h; the transfection efficiency was over 80%
(Fig. 3A). Quantitation of RSK4 mRNA
and protein by RT-qPCR and western blotting revealed that RSK4-OE
cells exhibited a significant increase in RSK4 expression compared
with NC cells (RSK4 mRNA, P=0.008; RSK4 protein, P=0.002) and
nontransfected cells (RSK4 mRNA, P=0.008; RSK4 protein, P<0.001;
Fig. 3B and C). No significant
differences were observed between NC and nontransfected cells.
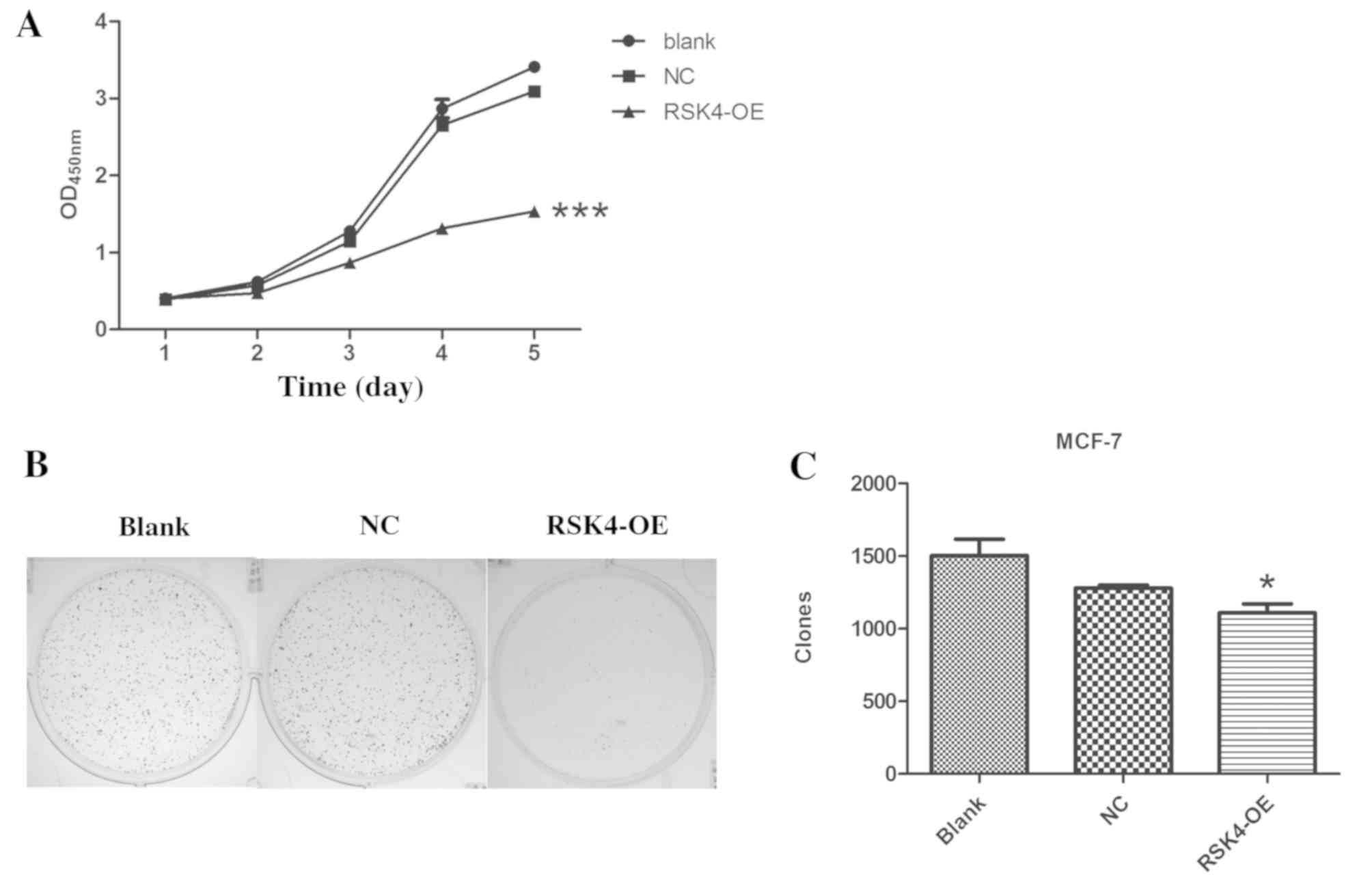
The proliferation of transfected cells was assessed
using a CCK-8 assay. RSK4-OE cells exhibited significant decreases
in the proliferation compared with NC (P<0.001) and
nontransfected groups (P<0.001; Fig.
4A). Furthermore, RSK4-OE cells formed significantly fewer
clones than NC cells (P=0.001) and nontransfected cells
(P<0.001; Fig. 4B and C). There
was no difference between NC cells and nontransfected cells.
RSK4 overexpression reduces the
migration of ER+ breast cancer cells
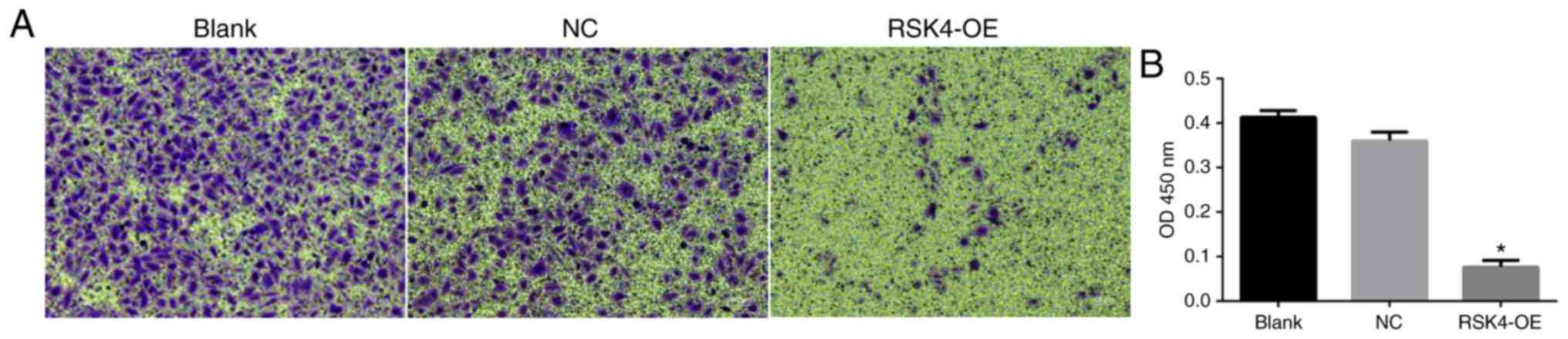
The effects of RSK4 overexpression on the migration
of transfected MCF-7 cells was assessed using a Transwell assay.
RSK4-OE cells exhibited significantly reduced migration compared
with NC cells (P<0.001) and nontransfected cells (P<0.001;
Fig. 5A and B). There was no
significant difference in migration between NC and nontransfected
cells.
RSK4 overexpression promotes the
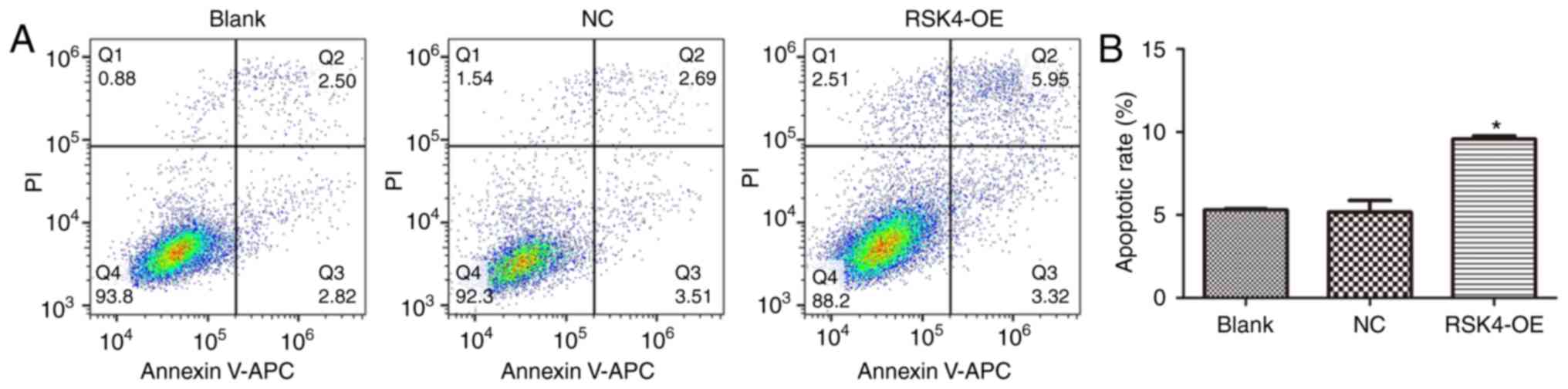
apoptosis in ER+ breast cancer cells
Flow cytometry analysis of cells stained with
Annexin V-APC and PI revealed that RSK4-OE cells exhibited a
significant increase in the number of apoptotic cells compared with
the NC (P<0.001) and nontransfected cells (P<0.001; Fig. 6). There was no significant difference
in apoptosis between NC cells and nontransfected cells
(P=0.822).
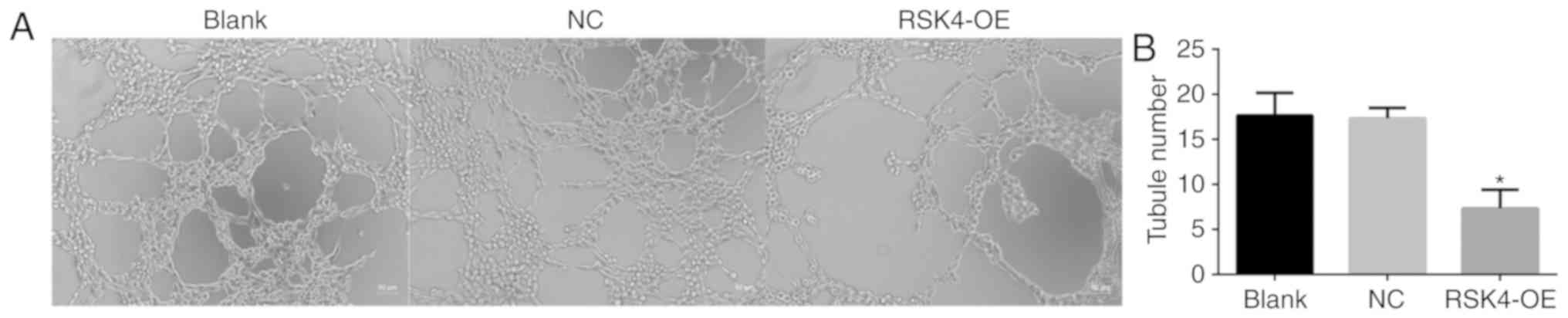
RSK4 overexpression reduces HUVEC
tubule formation in vitro
The effects of RSK4 on the ability of HUVECs to form
tubes in vitro was assessed. HUVECs were incubated with
conditioned medium from nontransfected MCF-7 cells, and those
transfected with RSK4-OE or NC cultured for 24 h. Cultured medium
from RSK4-OE cells induced significantly fewer tubules compared
with NC (P=0.001) and nontransfected controls (P=0.001; Fig. 7). There was no significant difference
in tubule formation between conditioned medium from NC and
nontransfected cells.
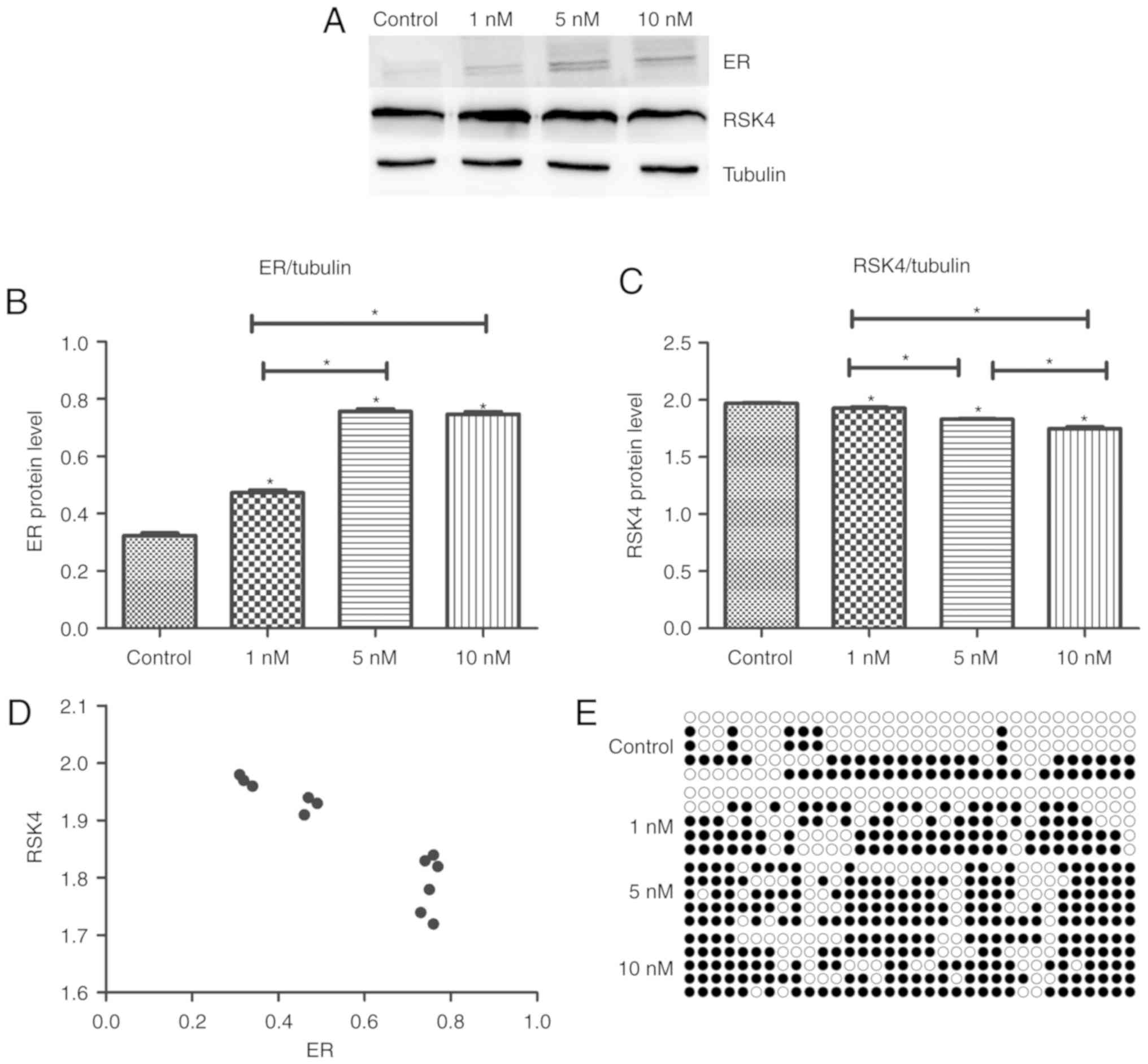
E2 stimulation of MCF-7 cells
increases the expression of ER and RSK4 promoter methylation, but
decreases the expression of RSK4
To investigate the effects of E2 signaling on ER and
RSK4 expression, MCF-7 cells were treated with E2 for 48 h. ER
protein levels were significantly increased in treated cells
compared with untreated controls (P<0.001). Furthermore, ER
protein levels were increased in the 5 and 10 nM groups compared
with the 1 nM-treated group (P<0.001). There was no significant
difference between the 5 and 10 nM groups (P=0.446; Fig. 8A and B). RSK4 protein levels were
decreased in the treated cells compared with the untreated controls
(P<0.05). RSK4 protein levels were significantly decreased in
the 5 and 10 nM groups compared with the 1 nM group (P<0.001),
and it was lower in the 10 nM group compared with the 5 nM-treated
group (P=0.001; Fig. 8A and C). ER
and RSK4 expression exhibited a negative correlation (r=−0.914;
P<0.001; Fig. 8D). Bisulfite
sequencing demonstrated that RSK4 promoter methylation was markedly
increased in the 10 nM-treated group (75.3%) compared with the 5 nM
(71.2%) and 1 nM-treated groups, (53.8%) and the control group
(36.9%; Fig. 8E).
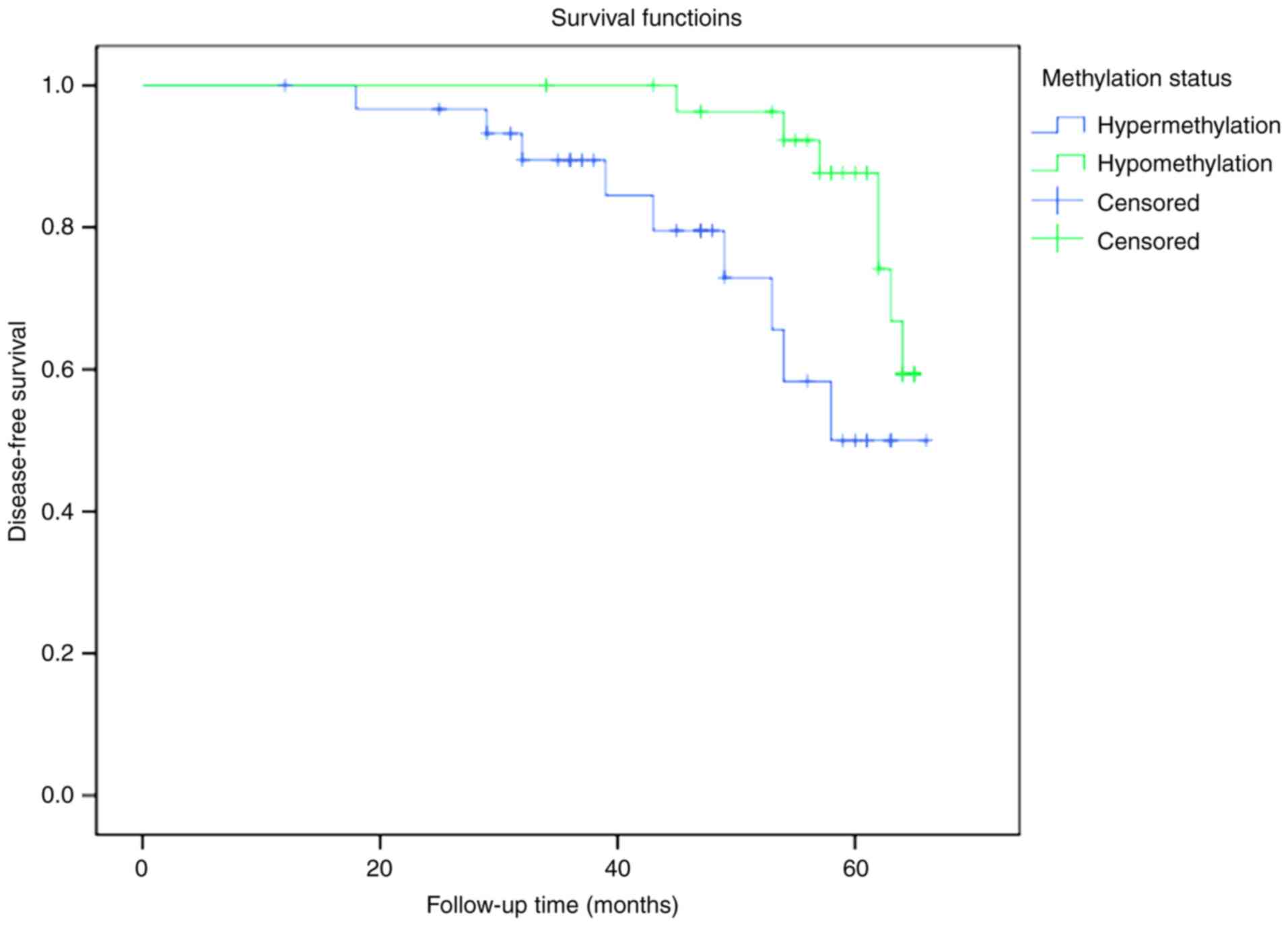
RSK4 hypomethylation correlates with
longer disease-free survival in patients
The correlation between RSK4 promoter methylation
and survival was investigated in the 60 patients with breast cancer
enrolled in the current study. Patients with RSK4 methylation
levels above or equal to the mean methylation value (n=31) were
defined as hypermethylated, while those with RSK4 methylation below
the mean (n=29) were defined as hypomethylated. The disease-free
survival rate was significantly increased in the hypomethylated
group compared with the hypermethylated group (P=0.026; Fig. 9). No patient succumbed due to breast
cancer during the follow-up period, so the overall survival could
not be compared.
Discussion
The current study revealed that breast cancer
tissues expressed significantly decreased levels of RSK4 and
increased RSK4 promoter methylation compared with adjacent normal
tissues. Furthermore, RSK4 expression was negatively correlated
with ER expression in breast cancer cell lines. In addition,
ER+ status was associated with increased RSK4 expression
and decreased promoter methylation compared with ER−
status. RSK4 overexpression inhibited ER+ breast cancer
cell proliferation, migration and clone formation, and promoted
apoptosis. Additionally, conditioned medium from an ER+
cell line overexpressing RSK4 inhibited HUVEC tubule formation
in vitro. The aforementioned results suggested that RSK4
acts as a tumor suppressor, and its downregulation through the
hypermethylation of the RSK4 promoter may be associated with breast
cancer. Consistent with this hypothesis, the disease-free survival
in the current study was significantly longer among patients with
RSK4 promoter hypomethylation than among those with
hypermethylation.
Previous studies have revealed that the E2-ER
signaling pathway regulates mammary gland growth, development and
apoptosis through genomic and non-genomic effects, which alter the
expression of genes in normal breast epithelial cells (25,26).
Dysregulation of the E2-ER signaling pathway alters the expression
of genes, including cell surface heparan sulfate proteoglycans, and
function of proteins such as the receptors for insulin-like growth
factor and epidermal growth factor (27–31), which
in turn dysregulates cell proliferation and mammary gland apoptosis
(32–35). Additionally, alterations of the E2-ER
signaling pathway may promote the conversion of E2-dependent tumors
to non-dependent tumors, which are much more resistant to therapy
(36–38). However, few studies have investigated
the regulatory kinases and upstream/downstream molecules of the
estrogen-ER signaling pathway. The results of the present study
suggested a mechanistic link between ER signaling and RSK4 in
breast cancer.
The present study revealed that RSK4 expression was
reduced in breast cancer tissues compared with adjacent normal
tissues, and that RSK4 expression decreased with increasing tumor
malignancy. A previous study using a smaller number of breast
cancer tissues reported a negative correlation between RSK4 mRNA
expression and breast cancer tumor size, and clinical stage
(8).
The results obtained in the present study revealed
that RSK4 expression may correlate with ER upregulation in breast
cancer tissues and cell lines, suggesting a mechanistic link
between ER signaling and RSK4. The results revealed that the
expression of ER was enhanced with increasing E2 concentration.
Additionally, RSK4 protein levels decreased and the methylation of
the RSK4 promoter increased with increasing E2 concentration.
Furthermore, the expression of the was negatively correlated with
the expression of RSK4, suggesting that the E2/ER signaling pathway
may regulate the methylation of the RSK4 promoter. Inducing E2/ER
signaling in breast cancer cells may therefore increase the
methylation of the RSK4 promoter, thereby reducing the expression
of RSK4 and affecting the development of breast cancer.
In the present study, patients with breast cancer
and RSK4 hypomethylation had longer disease-free survival than
patients with RSK4 hypermethylation. RSK4 methylation status may
thus serve as an independent prognostic marker in breast cancer.
However, an association between methylation status and overall
survival was not observed in the current study. In summary, our
results suggested that altered estrogen-ER signaling may be
associated with decreased RSK4 expression and increased RSK4
methylation in breast cancer, leading to enhanced cell
proliferation that may accelerate tumor development.
Acknowledgements
Not applicable.
Funding
The present study was supported by a grant from the
National Natural Science Foundation of China (grant no.
81560431).
Availability of data and materials
The datasets used during the present study are
available from the corresponding author upon reasonable
request.
Authors' contributions
QL designed, analyzed and revised the experiment. YJ
designed and analyzed the experiment. HH conducted experiments,
data analysis and article writing. XY conducted data processing and
collected specimens. HY carried out specimen collection and
follow-up. All authors read and approved the final manuscript and
agree to be accountable for all aspects of the research in ensuring
that the accuracy or integrity of any part of the work are
appropriately investigated and resolved.
Ethics approval and consent to
participate
The present study was approved by the Research
Ethics Committee of the Guangxi Medical University Cancer Hospital
and patients provided written informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2019. CA Cancer J Clin. 69:7–34. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Huang X and Yin YM: Updates of Chinese
society of clinical oncology (CSCO) guideline for breast cancer in
2018. Zhonghua yi xue za zhi (In Chinese). 98:1213–1217. 2018.
|
|
4
|
Waks AG and Winer EP: Breast cancer
treatment: A review. JAMA. 321:288–300. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Houles T and Roux PP: Defining the role of
the RSK isoforms in cancer. Seminars Cancer Biol. 48:53–61. 2018.
View Article : Google Scholar
|
|
6
|
Yin Z, Fan L, Huang G, Wang H and Wang Z:
The possible role of ribosomal protein S6 kinase 4 in the
senescence of endothelial progenitor cells in diabetes mellitus.
Cardiovasc Diabetol. 11:122012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Dummler BA, Hauge C, Silber J, Yntema HG,
Kruse LS, Kofoed B, Hemmings BA, Alessi DR and Frödin M: Functional
characterization of human RSK4, a new 90-kDa ribosomal S6 kinase,
reveals constitutive activation in most cell types. J Biol Chem.
280:13304–13314. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Li Q, Jiang Y, Wei W, Ji Y, Gao H and Liu
J: Frequent epigenetic inactivation of RSK4 by promoter methylation
in cancerous and non-cancerous tissues of breast cancer. Med Oncol.
31:7932014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Cai J, Ma H, Huang F, Zhu D, Zhao L, Yang
Y, Bi J and Zhang T: Low expression of RSK4 predicts poor prognosis
in patients with colorectal cancer. Int J Clin Exp Pathol.
7:4959–4970. 2014.PubMed/NCBI
|
|
10
|
Rafiee M, Keramati MR, Ayatollahi H,
Sadeghian MH, Barzegar M, Asgharzadeh A and Alinejad M:
Down-regulation of ribosomal S6 kinase RPS6KA6 in acute myeloid
leukemia patients. Cell J. 18:159–164. 2016.PubMed/NCBI
|
|
11
|
Li A, Liu D, Liu Y, Zhou Y, Du Z and Song
J: A pilot study of RSK4 expression in patients with human
non-small cell lung carcinoma. Ann Clin Lab Sci. 48:484–489.
2018.PubMed/NCBI
|
|
12
|
Banno K, Yanokura M, Iida M, Masuda K and
Aoki D: Carcinogenic mechanisms of endometrial cancer: Involvement
of genetics and epigenetics. J Obstet Gynaecol Res. 40:1957–1967.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Niskakoski A, Kaur S, Staff S,
Renkonen-Sinisalo L, Lassus H, Jarvinen HJ, Mecklin JP, Bützow R
and Peltomäki P: Epigenetic analysis of sporadic and
Lynch-associated ovarian cancers reveals histology-specific
patterns of DNA methylation. Epigenetics. 9:1577–1587. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Arechavaleta-Velasco F, Zeferino-Toquero
M, Estrada-Moscoso I, Imani-Razavi FS, Olivares A, Perez-Juarez CE
and Diaz-Cueto L: Ribosomal S6 kinase 4 (RSK4) expression in
ovarian tumors and its regulation by antineoplastic drugs in
ovarian cancer cell lines. Med Oncol. 33:112016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Jiang Y, Ye X, Ji Y, Zhou X, Yang H, Wei W
and Li Q: Aberrant expression of RSK4 in breast cancer and its role
in the regulation of tumorigenicity. Int J Mol Med. 40:883–890.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhu J, Li QY, Liu JL, Wei W, Yang HW and
Tang W: RSK4 knockdown promotes proliferation, migration and
metastasis of human breast adenocarcinoma cells. Oncol Rep.
34:3156–3162. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhu L, Zhang S, Huan X, Mei Y and Yang H:
Down-regulation of TRAF4 targeting RSK4 inhibits proliferation,
invasion and metastasis in breast cancer xenografts. Biochem
Biophys Res Commun. 500:810–816. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Harvey JM, Clark GM, Osborne CK and Allred
DC: Estrogen receptor status by immunohistochemistry is superior to
the ligand-binding assay for predicting response to adjuvant
endocrine therapy in breast cancer. J Clin Oncol. 17:1474–1481.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Musgrove EA and Sutherland RL: Biological
determinants of endocrine resistance in breast cancer. Nat Rev
Cancer. 9:631–643. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
20
|
Leary AF, Drury S, Detre S, Pancholi S,
Lykkesfeldt AE, Martin LA, Dowsett M and Johnston SR: Lapatinib
restores hormone sensitivity with differential effects on estrogen
receptor signaling in cell models of human epidermal growth factor
receptor 2-negative breast cancer with acquired endocrine
resistance. Clin Cancer Res. 16:1486–1497. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Nguyen PL, Taghian AG, Katz MS, Niemierko
A, Abi Raad RF, Boon WL, Bellon JR, Wong JS, Smith BL and Harris
JR: Breast cancer subtype approximated by estrogen receptor,
progesterone receptor, and HER-2 is associated with local and
distant recurrence after breast-conserving therapy. J Clin Oncol.
26:2373–2378. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Srinivasan S, Nwachukwu JC, Bruno NE,
Dharmarajan V, Goswami D, Kastrati I, Novick S, Nowak J, Cavett V,
Zhou HB, et al: Full antagonism of the estrogen receptor without a
prototypical ligand side chain. Nat Chem Biol. 13:111–118. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Jiang G, Dong S, Yu M, Han X, Zheng C, Zhu
X and Tong X: Influence of gap junction intercellular communication
composed of connexin 43 on the antineoplastic effect of adriamycin
in breast cancer cells. Oncol Lett. 13:857–866. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Acconcia F, Fiocchetti M and Marino M:
Xenoestrogen regulation of ERalpha/ERbeta balance in
hormone-associated cancers. Mol Cell Endocrinol. 457:3–12. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Tsonis AI, Afratis N, Gialeli C, Ellina
MI, Piperigkou Z, Skandalis SS, Theocharis AD, Tzanakakis GN and
Karamanos NK: Evaluation of the coordinated actions of estrogen
receptors with epidermal growth factor receptor and insulin-like
growth factor receptor in the expression of cell surface heparan
sulfate proteoglycans and cell motility in breast cancer cells.
FEBS J. 280:2248–2259. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Hsieh DJ, Kuo WW, Lai YP, Shibu MA, Shen
CY, Pai P, Yeh YL, Lin JY, Viswanadha VP and Huang CY:
17β-estradiol and/or estrogen receptor β attenuate the autophagic
and apoptotic effects induced by prolonged hypoxia through
HIF-1α-mediated BNIP3 and IGFBP-3 signaling blockage. Cell Physiol
Biochem. 36:274–284. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Panic A, Stanimirovic J, Obradovic M,
Zafirovic S, Sudar-Milovanovic E, Petrovic N and Isenovic ER:
17β-estradiol inhibits hepatic iNOS via the activation of the
estrogen receptor ERα and inhibition of erk1/2-mir-221 axis. J Biol
Regul Homeost Agents. 32:1369–1377. 2018.PubMed/NCBI
|
|
29
|
Go RE, Hwang KA, Kim CW, Byun YS, Nam KH
and Choi KC: Effect of dioxin and 17β-estradiol on the expression
of cytochrome P450 1A1 gene via an estrogen receptor dependent
pathway in cellular and xenografted models. Environ Toxicol.
32:2225–2233. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Lee SH and Nam HS: TNF alpha-induced
down-regulation of estrogen receptor alpha in MCF-7 breast cancer
cells. Mol Cells. 26:285–290. 2008.PubMed/NCBI
|
|
31
|
Boerner JL, Gibson MA, Fox EM, Posner ED,
Parsons SJ, Silva CM and Shupnik MA: Estrogen negatively regulates
epidermal growth factor (EGF)-mediated signal transducer and
activator of transcription 5 signaling in human EGF family
receptor-overexpressing breast cancer cells. Mol Endocrinol.
19:2660–2670. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Huang J, Li X, Yi P, Hilf R, Bambara RA
and Muyan M: Targeting estrogen responsive elements (EREs): Design
of potent transactivators for ERE-containing genes. Mol Cell
Endocrinol. 218:65–78. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Bartucci M, Morelli C, Mauro L, Ando S and
Surmacz E: Differential insulin-like growth factor I receptor
signaling and function in estrogen receptor (ER)-positive MCF-7 and
ER-negative MDA-MB-231 breast cancer cells. Cancer Res.
61:6747–6754. 2001.PubMed/NCBI
|
|
34
|
Zhou Y, Eppenberger-Castori S, Marx C, Yau
C, Scott GK, Eppenberger U and Benz CC: Activation of nuclear
factor-kappaB (NFkappaB) identifies a high-risk subset of
hormone-dependent breast cancers. Int J Biochem Cell Biol.
37:1130–1144. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Singh RR and Kumar R: Steroid hormone
receptor signaling in tumorigenesis. J Cell Biochem. 96:490–505.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Song RX, McPherson RA, Adam L, Bao Y,
Shupnik M, Kumar R and Santen RJ: Linkage of rapid estrogen action
to MAPK activation by ERalpha-Shc association and Shc pathway
activation. Mol Endocrinol. 16:116–127. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zhou Y, Yau C, Gray JW, Chew K, Dairkee
SH, Moore DH, Eppenberger U, Eppenberger-Castori S and Benz CC:
Enhanced NF kappa B and AP-1 transcriptional activity associated
with antiestrogen resistant breast cancer. BMC Cancer. 7:592007.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Filardo EJ, Quinn JA, Frackelton AR Jr and
Bland KI: Estrogen action via the G protein-coupled receptor,
GPR30: Stimulation of adenylyl cyclase and cAMP-mediated
attenuation of the epidermal growth factor receptor-to-MAPK
signaling axis. Mol Endocrinol. 16:70–84. 2002. View Article : Google Scholar : PubMed/NCBI
|