Introduction
Predicting the prognosis of patients with estrogen
receptor-positive/human epidermal growth factor receptor
2-negative/node-negative (ER+/HER2−/N0)
breast cancer with a high accuracy is critical to decide the
indication for adjuvant chemotherapy. Accordingly, several
multigene assays (MGAs) that have been developed based on the
expression of multiple genes in breast cancer tissues, such as
Oncotype DX and MammaPrint (1–4), are
widely used in clinical practice.
We previously developed Curebest 95GC Breast (Sysmex
Co., Kobe, Japan), a 95-gene classifier (95GC) using DNA microarray
(GeneChip Human Genome U 133 Plus 2.0 Array). The 95GC helps
classify patients with ER+/HER2-/N0 breast cancer types
into high- and low-risk groups. In addition, intermediate risk
breast cancer types [recurrence score (RS), 18–30] classified using
21GC (21GCRS were calculated using Recurrence Online)
can be further dichotomized using a 95GC into low- and high-risk
groups (5). This dichotomization is
expected to lead to a significant difference in disease prognosis,
suggesting the use of 95GC in predicting the prognosis of
intermediate-risk breast cancers.
Although 95GC dichotomizes breast cancer into high-
and low-risk groups by using a defined RS cutoff, the recurrence
risk is thought to increase in proportion to the RS, as was clearly
shown by the correlation between recurrence rate and RS in Oncotype
DX (1,2,6,7). Predicting the recurrence risk using an
RS for each patient could enable better decision-making for
adjuvant chemotherapy indication than using 95GC information of
high or low-risk patients. The present study primarily aimed to
develop a 95GC-based RS (95GCRS) and demonstrate a
correlation with recurrence rate in
ER+/HER2−/N0 breast cancer types. In
addition, the study aimed to apply 95GC, originally developed using
fresh-frozen (FF) tissues, to FFPE tissues, because FFPE tissues
are routinely prepared and are readily available. Although we
previously reported the applicability of 72GC to FFPE tissues
(8), the present study aimed to
improve the accuracy of 95GC for FFPE tissues using the reference
robust multiarray average (refRMA) method, optimized for FFPE
tissues. Therefore, a 95GCRS was first developed and
then the accuracy of the newly developed 95GC algorithm for FFPE
tissues was evaluated using the 95GCRS.
Materials and methods
Breast cancer tissues
Development of 95GCRS
A total of 257 patients with ER+/HER2-/N0
breast cancer who underwent breast-conserving surgery or mastectomy
at Osaka University Hospital (Suita, Japan) and were treated using
only adjuvant hormonal therapy were retrospectively included in
this study (Table I). FF tumor
tissues were obtained from surgical specimens and stored at −80°C
until use. The median follow-up period was 86 months (range, 12–190
months). Of the 257 patients, 106 were treated postoperatively with
goserelin (3.75 mg/4 weeks) plus tamoxifen (20 mg/day) and 151 were
treated with anastrozole (1 mg/day). Tamoxifen and anastrozole were
administered for 5 years or until recurrence, whichever occurred
earlier, whereas goserelin was administered for 2 years. Informed
consent to participate in the study was obtained from all patients
before surgery. The present study was approved by the Ethics
Committee of Osaka University Hospital.
 | Table I.Clinicopathological parameters of
patients with estrogen receptor-positive/human epidermal growth
factor receptor 2-negative/node-negative breast cancer included in
the study for correlation between 95GCRS and distant
recurrence rate. |
Table I.
Clinicopathological parameters of
patients with estrogen receptor-positive/human epidermal growth
factor receptor 2-negative/node-negative breast cancer included in
the study for correlation between 95GCRS and distant
recurrence rate.
|
| 95GC |
|
|
|
|---|
|
|
|
|
|
|
|---|
| Variable | Low-risk
(n=180) | High-risk
(n=77) | OR | 95% CI |
P-valuea |
|---|
| Menopause, n
(%) |
|
|
| 0.39–1.15 | 0.167 |
|
Premenopausal | 69 | 37 (35) | 1.00 |
|
|
|
Postmenopausal | 111 | 40 (26) | 0.672 |
|
|
| cT, n |
|
|
| 0.61–1.80 | 0.890 |
| 1 | 105 | 44 | 1.00 |
|
|
|
2+3 | 75 | 33 | 1.05 |
|
|
| HG, n |
|
|
| 1.38–9.30 | <0.0001 |
| 1 | 93 | 16 | 1.00 |
|
|
| 2 | 79 | 50 | 1.00 |
|
|
| 3 | 8 | 11 | 3.58 |
|
|
| PR, n |
|
|
| 0.22–0.93 | 0.045 |
|
Negative | 19 | 16 | 1.00 |
|
|
|
Positive | 161 | 61 | 0.45 |
|
|
| Ki67,
nb |
|
|
| 0.81–4.00 | 0.197 |
|
<20% | 94 | 34 | 1.00 |
|
|
|
≥20% | 20 | 13 | 1.80 |
|
|
Correlation between 95GCRS
and response to chemotherapy
A total of 126 patients with ER+ breast
cancer (stage II–III) who were treated with neoadjuvant
chemotherapy (NAC) followed by mastectomy or breast-conserving
surgery at Osaka University Hospital between 2004 and 2012 were
retrospectively included in this study (Table II). NAC consisted of paclitaxel (80
mg/m2) weekly for 12 cycles, followed by a combination
of 5-fluorouracil (500 mg/m2), epirubicin (75
mg/m2) and cyclophosphamide (500 mg/m2) every
3 weeks for four cycles (P-FEC). Before initiating NAC, all
patients underwent tumor biopsy using a vacuum-assisted core-biopsy
instrument (Mammotome 8G HH; Ethicon Endosurgery Inc.) under
ultrasonographic guidance for histological examination and gene
expression analysis. Tumor samples for histological examination
were fixed in 10% buffered formaldehyde, and tumor samples for gene
expression analysis were snap-frozen in liquid nitrogen and stored
at −80°C until use. Informed consent to participate in the study
was obtained from all patients before performing the tumor biopsy.
In addition, 299 patients with ER+ breast cancer who
were treated with neoadjuvant sequential taxane and fluorouracil,
doxorubicin and cyclophosphamide [P-(F)AC] were selected from the
GSE25066 dataset (9) available in the
public database GEO (https://www.ncbi.nlm.nih.gov/geo/).
 | Table II.Clinicopathological parameters of
patients included in the study for correlation between
95GCRS and pCR rate to neoadjuvant chemotherapy in
patients (n=126) at Osaka University Hospital. |
Table II.
Clinicopathological parameters of
patients included in the study for correlation between
95GCRS and pCR rate to neoadjuvant chemotherapy in
patients (n=126) at Osaka University Hospital.
| Parameter | Value |
|---|
| Age, years |
|
|
Median | 47 |
|
Range | 24-76 |
| Postmenopausal,
n | 57 |
| cT, n |
|
| T1 | 8 |
| T2 | 89 |
| T3 | 18 |
| T4 | 11 |
| cN, n |
|
| N0 | 46 |
| N1 | 80 |
| Histological grade,
n |
|
| 1 | 26 |
| 2 | 80 |
| 3 | 20 |
| ER, n |
|
|
Positive | 126 |
|
Negative | 0 |
| PR |
|
|
Positive | 84 |
|
Negative | 42 |
| HER2 |
|
|
Positive | 25 |
|
Negative | 101 |
| Ki67a |
|
|
Positive | 41 |
|
Negative | 47 |
Application of 95GC to FFPE
tissues
Two adjacent tumor specimens were obtained from each
of the 56 ER+/HER2−/N0 breast cancers: One
specimen was stored at −80°C as FF tissue and the other was fixed
in 10% buffered formalin for FFPE tissue preparation (Table SI). Of these 56 breast cancer types,
25 samples were from patients treated at Osaka University Hospital
without NAC and 31 were purchased from East West Biopharma LLC as
paired FF/FFPE specimens (Table
SI).
RNA extraction and DNA microarray assay
in FF and FFPE tissues
FF tissues
RNA was extracted from FF tumor tissues using Qiagen
RNeasy Lipid Tissue Mini kits (Qiagen GmbH). Approximately 100 ng
RNA (RNA integrity number >7) was used to generate second-strand
cDNA, and cRNA was amplified using the oligodeoxynucleotide
ribosylthymine primers, then biotinylated and fragmented using the
Gene Profiling Reagent kit (Affymetrix; Thermo Fisher Scientific,
Inc.), followed by hybridization using U133 Plus 2.0 arrays
overnight (17 h) according to the manufacturer's protocol. Finally,
the hybridized DNA microarray was fluorescently stained with
GeneChip Fluidics Station 450, and scanned using a GeneChip Scanner
3000.
FFPE tissues
RNA was extracted from four consecutive sections (10
µm) of each FFPE tissue using RNeasy FFPE kits (Qiagen GmbH).
Second-strand cDNA was generated using 70–100 ng of RNA, and cDNA
was amplified using the oligodeoxynucleotide ribosylthymine and
random primers using an Ovation FFPE whole-transcriptome
amplification system (NuGEN Technologies, Inc.) according to the
manufacturer's protocol. The cDNA was then biotinylated and
fragmented using the Encore Biotin Module (NuGEN Technologies,
Inc.), followed by hybridization on U133 Plus 2.0 arrays overnight
(17–20 h) according to the manufacturer's protocol. Finally, the
hybridized DNA microarray was fluorescently stained using a
GeneChip Fluidics Station 450, and scanned using a GeneChip Scanner
3000 (both Affymetrix; Thermo Fisher Scientific, Inc.).
Histological examination
Pathological response to NAC was evaluated using
surgical specimens obtained during surgery. Specimens were cut into
5-mm slices, and hematoxylin and eosin-stained sections were
prepared to determine the presence or absence of tumor cells. A
complete absence of invasive tumor cells in the breast and lymph
nodes was defined as pathological complete response (pCR),
irrespective of the presence or absence of non-invasive breast
cancer cells. ER, PR, and Ki67 levels in the tumor biopsy samples
were immunohistochemically determined as previously described
(10–12). The cut-off values were 10% for ER, 10%
for PR and 20% for Ki67. HER2 amplification was determined using
fluorescence in situ hybridization (FISH) using the
PathVysion HER-2 DNA Probe kit (Vysis/Abbott Molecular Inc.)
according to the manufacturer's instructions. Tumors were
classified as HER2-amplified if the FISH ratio was ≥2.0.
Statistical analysis
Gene expression datasets obtained by DNA microarray
were normalized using the refRMA procedure, followed by analysis
using a between-group analysis (BGA) classifier model for 95GC as
previously reported by our group (5)
for classifying patients into low- and high-risk groups. All
statistical analyses were performed using R statistical software
(version 3.5.1; http://www.r-project.org/), apart from the comparison
of 95GCRS between FF and FFPE tissues, as shown in
Fig. 3, which was performed in
Microsoft Excel 2010 (Microsoft Corporation) using the CORREL
function. Fisher's exact test was used to compare 2×2 groups. All
statistical analyses were two-sided, and P<0.05 was considered
to indicate a statistically significant difference.
Results
Development of 95GCRS
BGA was used to separate low- and high-risk groups
in 95GC (5). BGA assigns a value to
each tumor, where valueS >0 are considered high-risk tumors and
valueS ≤0 are considered low-risk tumors. These values were then
converted to 95GCRS 0–100 using the following
formula:
95GC score: round {(1000⁄3) × original
value+50}
95GCRS was calculated for 257 patients
with ER+/HER2−/N0 breast cancer receiving
adjuvant hormonal therapy alone as an independent validation set.
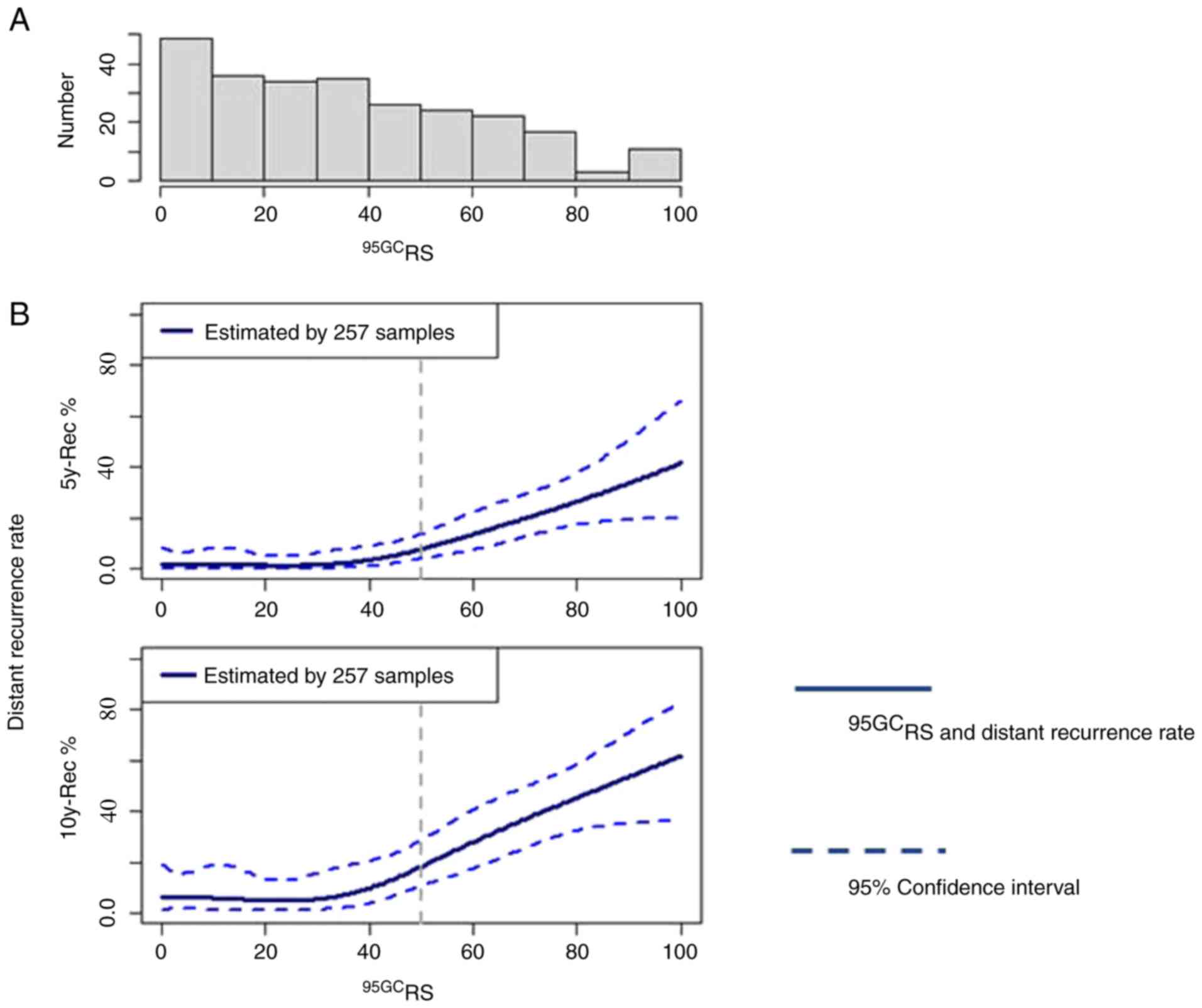
The histogram of patients according to 95GCRS is shown
in Fig. 1A and the correlation
between 95GCRS and distant recurrence rates at 5 and 10
postoperative years is shown in Fig.
1B. Distant recurrence rates were significantly low in patients
with 95GCRS ≤50 (low-risk) and increased in proportion
to 95GCRS in patients with 95GCRS >50
(high-risk).
95GCRS and response to
NAC
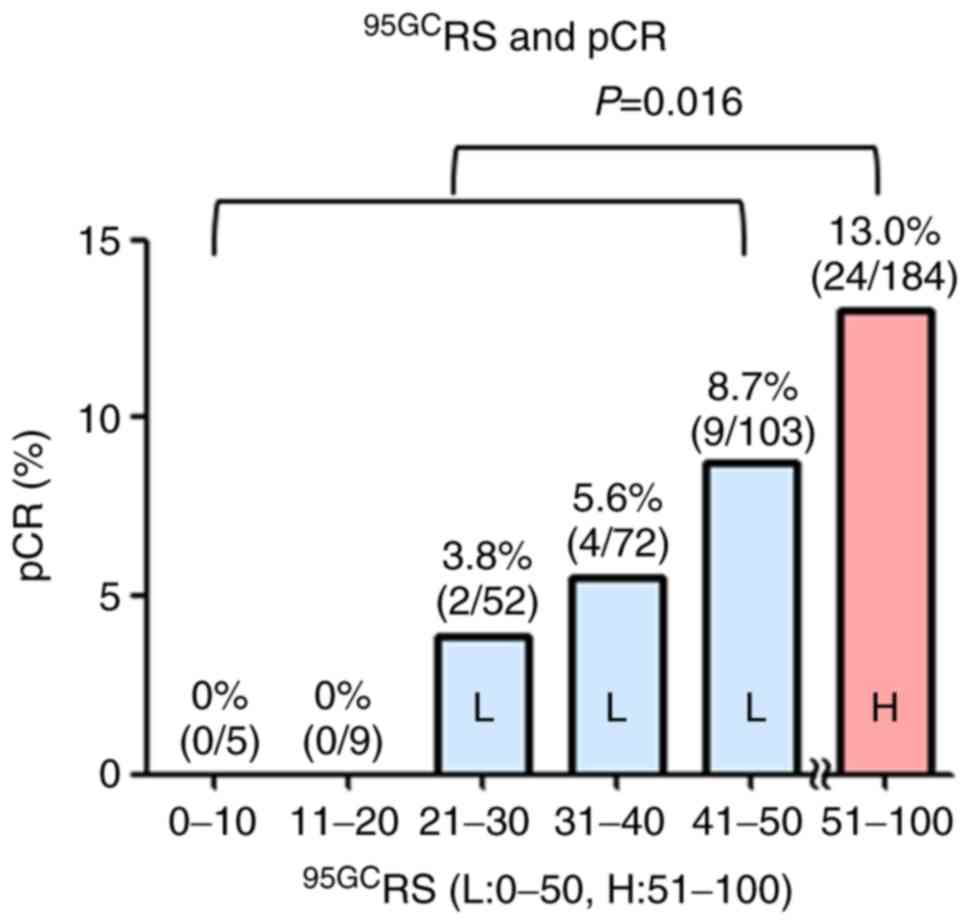
The correlation between 95GCRS and
response (pCR) to NAC was examined in 425 patients with
ER+ breast cancer treated with NAC [P-FEC or P-(F)AC].
As shown in Fig. 2, pCR rates
increased in proportion to 95GCRS, indicating the
increased sensitivity of breast cancers with high 95GCRS
to chemotherapy.
Application of 95GCRS to
FFPE tissues
When 95GC was calculated for FF tissues, gene
expression data were normalized using refRMA constructed for FF
tissues (5). Since gene expression in
FFPE tissues is significantly affected by mRNA degradation during
FFPE tissue preparation, it is necessary to construct a refRMA
specific to FFPE tissues. A refRMA was constructed for FFPE tissues
using the GSE47109 (13) and GSE51450
(14) datasets available in the GEO
public database (https://www.ncbi.nlm.nih.gov/geo/) (comprising gene
expression data from FFPE breast cancer tissues) to optimize the
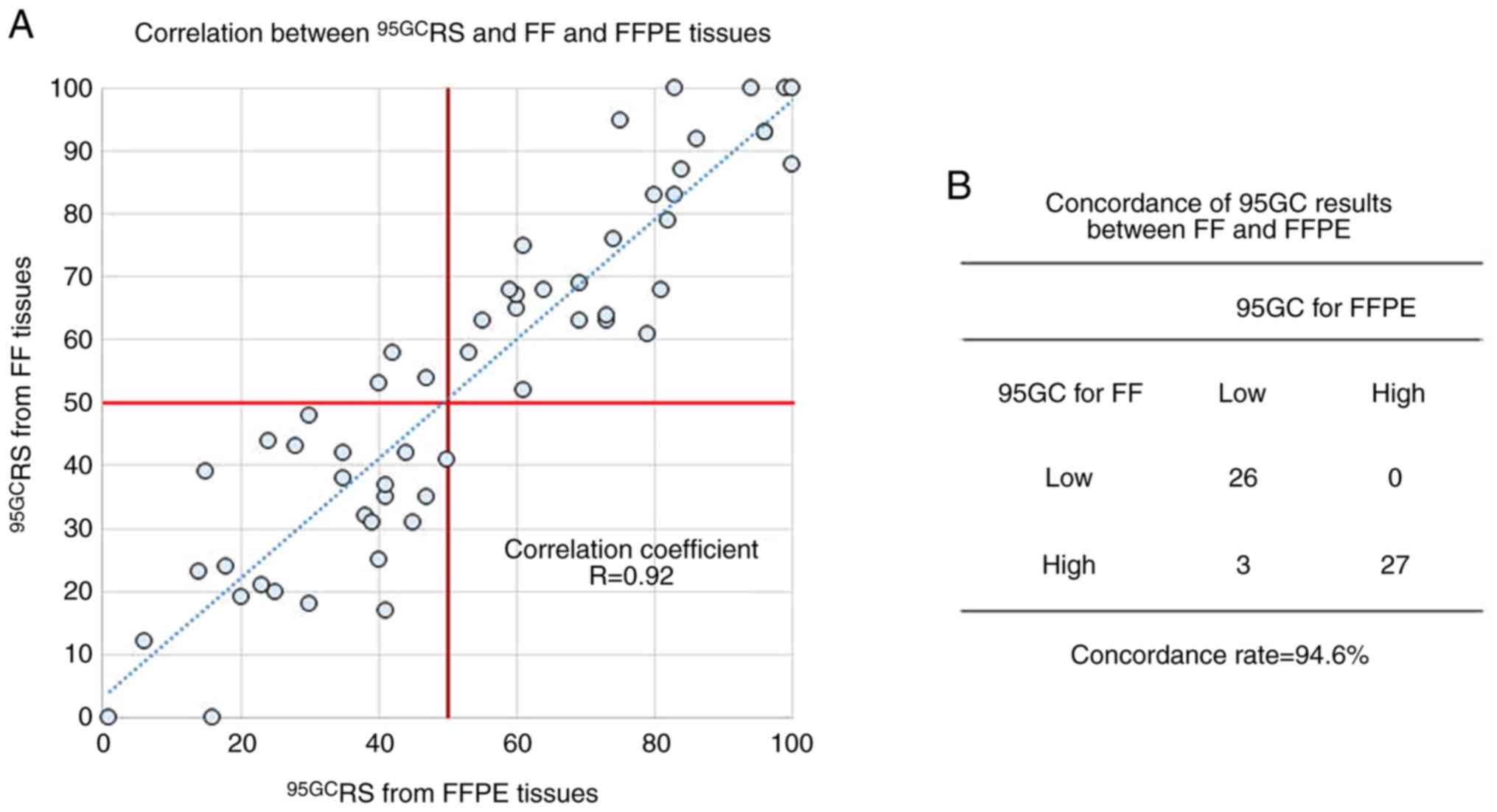
concordance of 95GC results between FF and FFPE tissues (Fig. S1). Subsequently, 56 pairs of FF and
FFPE breast cancer tissues were subjected to 95GC assay and
95GCRS values were calculated (FF tissues were analyzed
using refRMA for FF tissues and FFPE tissues were analyzed using
refRMA for FFPE tissues) (Fig. 3).
The correlation coefficient was significantly high (R=0.92) between
95GCRS obtained from the FF and FFPE tissues, and the
concordance rate (94.6%) of the high- and low-risk groups was also
notably high between the tissues (Fig.
3).
Discussion
In the present study, 95GCRS ranging from
0 to 100 was first developed, which correlated well with distant
recurrence. Breast cancer with 95GCRS ≤50 had a
significantly low recurrence rate, whereas that with
95GCRS >50 had a high recurrence rate. In addition,
the recurrence rate increased in proportion to 95GCRS.
Similar results were previously reported for 21GCRS
(1,2,6,7). Information on recurrence risk using
95GCRS for individual patients could enable better
decision-making in a clinical setting for adjuvant chemotherapy
indication than binary results (high- or low-risk groups).
We previously reported a correlation between risk
groups determined by 95GC and response to NAC (5,15). Breast
cancers in the 95GC high-risk group exhibited a significantly
higher response rate to NAC than those in the low-risk group.
Similar reports have also been reported following the use of
Oncotype DX and MammaPrint, which have shown the increased
sensitivity of high-risk breast cancers to NAC (16–20). In
the present study, breast cancers were further categorized using
95GCRS and demonstrated the gradual increase of pCR in
proportion to 95GCRS, indicating greater
chemosensitivity in breast cancers with high 95GCRS.
Altogether, the results suggest the remarkable ability of 95GC to
categorize patients at high-risk for relapse who would likely
benefit from adjuvant chemotherapy.
The 95GC was originally developed using gene
expression data from FF tissues; however, this needs to be
applicable to FFPE tissues prepared in routine practice to enhance
its clinical use. Thus, the present study attempted to modify and
apply 95GC to FFPE tissues. As preparation of FFPE tissues leads to
mRNA degradation, refRMA was first developed for FFPE tissues and
then 95GCRS was calculated. A significantly high
correlation coefficient (R=0.92) was demonstrated between
95GCRS and FF and FFPE tissues, as well as a markedly
high concordance rate (94.6%) between the high- and low-risk
groups, demonstrating the potential use of 95GC for FFPE
tissues.
In conclusion, in the present study, a
95GCRS was developed that correlated well with
recurrence rate, and it was demonstrated that 95GC is applicable to
FFPE tissues. These preliminary results need to be confirmed in
future studies.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by a Grant-in-Aid
for Scientific Research (grant no. 16K10456) from the Ministry of
Education, Culture, Sports, Science and Technology.
Availability of data and materials
The datasets generated and/or analyzed during the
current study are not publicly available due to the patients'
consent for provision to other facilities, but are available from
the corresponding author on reasonable request. Datasets GSE25066,
GSE47109 and GSE51450 are available from the GEO public
database.
Authors' contributions
SN and YN conceived and designed the study, drafted
and revised the paper critically for important intellectual
content, and approved the final version of the manuscript. SN, YN,
YS, KK, MS, NK, TM, TT, KS and SJK were responsible for the
acquisition, analysis or interpretation of data. All authors read
and approved the final manuscript.
Ethics approval and consent to
participate
The present study complied with the current relevant
laws and guidelines for Japan. Informed consent to participate in
the study was obtained from all patients before surgery. Approval
was obtained from the Ethics Committee of Osaka University
Hospital. Patient consent was obtained from all patients before
surgery.
Patient consent for publication
Not applicable.
Competing interests
SN has been an advisor for Taiho, AstraZeneca and
Novartis, and received research funding for other studies from
Sysmex, AstraZeneca, Novartis, Chugai, Daiichi-Sankyo, Kyowa-Kirin,
Takeda, Pfizer, Ono, Taiho, and Eisai, and honoraria from
AstraZeneca, Novartis, Pfizer, Chugai, Takeda, Sysmex, Nippon
Kayaku. YN received research funding from Sysmex and AstraZeneca.
NK received honoraria from AstraZeneca and Novartis. MS received
research funding from Novartis and AstraZeneca, and honoraria from
Chugai, Eisai, Novartis, and Takeda. KS received honoraria from
AstraZeneca, Chugai, and Sysmex. SJK received honoraria from
AstraZeneca, Chugai, Eisai, Kyowa-Kirin, Novartis, Pfizer,
Shimadzu, Taiho, and Takeda. YS and KK are the agents of Sysmex
Corporation. SN and YN are patent holders about 95GC.
Glossary
Abbreviations
Abbreviations:
|
ER
|
estrogen receptor
|
|
FF
|
fresh-frozen
|
|
FFPE
|
formalin-fixed paraffin-embedded
|
|
HER2
|
human epidermal growth factor receptor
2
|
|
NAC
|
neoadjuvant chemotherapy
|
|
pCR
|
pathological complete response
|
|
95GCRS
|
95-gene classifier recurrence
score
|
|
P-FEC
|
paclitaxel followed by a combination
of 5-fluorouracil, epirubicin and cyclophosphamide
|
|
P-[F]AC
|
taxane and fluorouracil, doxorubicin
and cyclophosphamide
|
References
|
1
|
Paik S, Shak S, Tang G, Kim C, Baker J,
Cronin M, Baehner FL, Walker MG, Watson D, Park T, et al: A
multigene assay to predict recurrence of tamoxifen-treated,
node-negative breast cancer. N Engl J Med. 351:2817–2826. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Paik S, Tang G, Shak S, Kim C, Baker J,
Kim W, Cronin M, Baehner FL, Watson D, Bryant J, et al: Gene
expression and benefit of chemotherapy in women with node-negative,
estrogen receptor-positive breast cancer. J Clin Oncol.
24:3726–3734. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
van't Veer LJ, Dai H, van de Vijver MJ, He
YD, Hart AA, Mao M, Peterse HL, van der Kooy K, Marton MJ,
Witteveen AT, et al: Gene expression profiling predicts clinical
outcome of breast cancer. Nature. 415:530–536. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
van de Vijver MJ, He YD, van't Veer LJ,
Dai H, Hart AA, Voskuil DW, Schreiber GJ, Peterse JL, Roberts C,
Marton MJ, et al: A gene-expression signature as a predictor of
survival in breast cancer. N Engl J Med. 347:1999–2009. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Naoi Y, Kishi K, Tsunashima R, Shimazu K,
Shimomura A, Maruyama N, Shimoda M, Kagara N, Baba Y, Kim SJ and
Noguchi S: Comparison of efficacy of 95-gene and 21-gene classifier
(Oncotype DX) for prediction of recurrence in ER-positive and
node-negative breast cancer patients. Breast Cancer Res Treat.
140:299–306. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Habel LA, Shak S, Jacobs MK, Capra A,
Alexander C, Pho M, Baker J, Walker M, Watson D, Hackett J, et al:
A population-based study of tumor gene expression and risk of
breast cancer death among lymph node-negative patients. Breast
Cancer Res. 8:R252006. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kim C, Tang G, Pogue-Geile KL, Costantino
JP, Baehner FL, Baker J, Cronin MT, Watson D, Shak S, Bohn OL, et
al: Estrogen receptor (ESR1) mRNA expression and benefit from
tamoxifen in the treatment and prevention of estrogen
receptor-positive breast cancer. J Clin Oncol. 29:4160–4167. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Nishio M, Naoi Y, Tsunashima R, Nakauchi
C, Kagara N, Shimoda M, Shimomura A, Maruyama N, Shimazu K, Kim SJ
and Noguchi S: 72-gene classifier for predicting prognosis of
estrogen receptor-positive and node-negative breast cancer patients
using formalin-fixed, paraffin-embedded tumor tissues. Clin Breast
Cancer. 14:e73–e80. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hatzis C, Pusztai L, Valero V, Booser DJ,
Esserman L, Lluch A, Vidaurre T, Holmes F, Souchon E, Wang H, et
al: A genomic predictor of response and survival following
taxane-anthracycline chemotherapy for invasive breast cancer. JAMA.
305:1873–1881. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Naoi Y, Kishi K, Tanei T, Tsunashima R,
Tominaga N, Baba Y, Kim SJ, Taguchi T, Tamaki Y and Noguchi S:
Development of 95-gene classifier as a powerful predictor of
recurrences in node-negative and ER-positive breast cancer
patients. Breast Cancer Res Treat. 128:633–641. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sota Y, Naoi Y, Tsunashima R, Kagara N,
Shimazu K, Maruyama N, Shimomura A, Shimoda M, Kishi K, Baba Y, et
al: Construction of novel immune-related signature for prediction
of pathological complete response to neoadjuvant chemotherapy in
human breast cancer. Ann Oncol. 25:100–106. 2013. View Article : Google Scholar
|
|
12
|
Tsunashima R, Naoi Y, Kagara N, Shimoda M,
Shimomura A, Maruyama N, Shimazu K, Kim SJ and Noguchi S:
Construction of multi-gene classifier for prediction of response to
and prognosis after neoadjuvant chemotherapy for estrogen receptor
positive breast cancers. Cancer Lett. 365:166–173. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
D'Alfonso TM, van Laar RK, Vahdat LT,
Hussain W, Flinchum R, Brown N, John LS and Shin SJ: BreastPRS is a
gene expression assay that stratifies intermediate-risk Oncotype DX
patients into high- or low-risk for disease recurrence. Breast
Cancer Res Treat. 139:705–715. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Musella V, Callari M, Di Buduo E, Scuro M,
Dugo M, Miodini P, Bianchini G, Paolini B, Gianni L, Daidone MG and
Cappelletti V: Use of formalin-fixed paraffin-embedded samples for
gene expression studies in breast cancer patients. PLoS One.
10:e01231942015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Tsunashima R, Naoi Y, Kishi K, Baba Y,
Shimomura A, Maruyama N, Nakayama T, Shimazu K, Kim SJ, Tamaki Y
and Noguchi S: Estrogen receptor positive breast cancer identified
by 95-gene classifier as at high risk for relapse shows better
response to neoadjuvant chemotherapy. Cancer Lett. 324:42–47. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Pease AM, Riba LA, Gruner RA, Tung NM and
James TA: Oncotype DX® recurrence score as a predictor
of response to neoadjuvant chemotherapy. Ann Surg Oncol.
26:366–371. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Bear HD, Wan W, Robidoux A, Rubin P,
Limentani S, White RL Jr, Granfortuna J, Hopkins JO, Oldham D,
Rodriguez A and Sing AP: Using the 21-gene assay from core needle
biopsies to choose neoadjuvant therapy for breast cancer: A
multicenter trial. J Surg Oncol. 115:917–923. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chang JC, Makris A, Gutierrez MC,
Hilsenbeck SG, Hackett JR, Jeong J, Liu ML, Baker J, Clark-Langone
K, Baehner FL, et al: Gene expression patterns in formalin-fixed,
paraffin-embedded core biopsies predict docetaxel chemosensitivity
in breast cancer patients. Breast Cancer Res Treat. 108:233–240.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Gianni L, Zambetti M, Clark K, Baker J,
Cronin M, Wu J, Mariani G, Rodriguez J, Carcangiu M, Watson D, et
al: Gene expression profiles in paraffin-embedded core biopsy
tissue predict response to chemotherapy in women with locally
advanced breast cancer. J Clin Oncol. 23:7265–7277. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Whitworth P, Stork-Sloots L, de Snoo FA,
Richards P, Rotkis M, Beatty J, Mislowsky A, Pellicane JV, Nguyen
B, Lee L, et al: Chemosensitivity predicted by BluePrint 80-gene
functional subtype and MammaPrint in the prospective neoadjuvant
breast registry symphony trial (NBRST). Ann Surg Oncol.
21:3261–3267. 2004. View Article : Google Scholar
|