Introduction
Sentinel lymph node (SN) biopsy is widely used to
determine axillary lymph node (LN) status in clinically
node-negative breast cancer patients (1,2). In
practice, SN metastasis is detected by intraoperative
histopathological examination of frozen section(s) or cytological
observation of touch imprints, and is confirmed by postoperative
pathological examination of permanent sections (3,4). One-step
nucleic acid amplification (OSNA) can be used to detect SN
metastasis through the amplification of cytokeratin 19 (CK19) mRNA
(which is expressed in tumor cells, but not normal cells of LNs)
with the same accuracy as routine pathological examination
(5). OSNA is also used to determine
total tumor load (TTL) in SNs as the sum of CK19 mRNA copies, which
is reportedly useful for predicting non-SN metastatic status
(6,7),
as well as patient prognosis (8).
However, TTL determination by OSNA does not always
accurately reflect the total number of tumor cells in the SN, since
the copy number of CK19 mRNAs per tumor cell varies considerably.
In fact, it is reported that OSNA predicts a 30-fold difference in
CK19 mRNA copies among tumors of the same size (9). By contrast, the amount of DNA per tumor
cell is thought to be less variable, thus the detection of SN tumor
cells from tumor-derived DNA is considered to more accurately
determine TTL.
Ras association domain-containing protein 1
(RASSF1A) promoter methylation is one of the most frequently
observed epigenomic changes in breast cancer (10,11).
Methylation-specific PCR (MSP) following bisulfite treatment is
widely used to quantify methylated DNA. However, bisulfite
treatment often results in considerable DNA loss (12,13), and
requires specialized optimization for digital PCR (dPCR) (14). A novel dPCR technique for the
detection of methylated DNA was recently reported, using a
methylation-specific restriction enzyme without bisulfite treatment
(15–17). The aim of the present study was to
develop a highly sensitive dPCR assay to detect RASSF1A methylation
following restriction enzyme digestion (RE-dMSP), for the detection
of tumor-derived methylated RASSF1A in SN lysates.
Materials and methods
Patients and samples
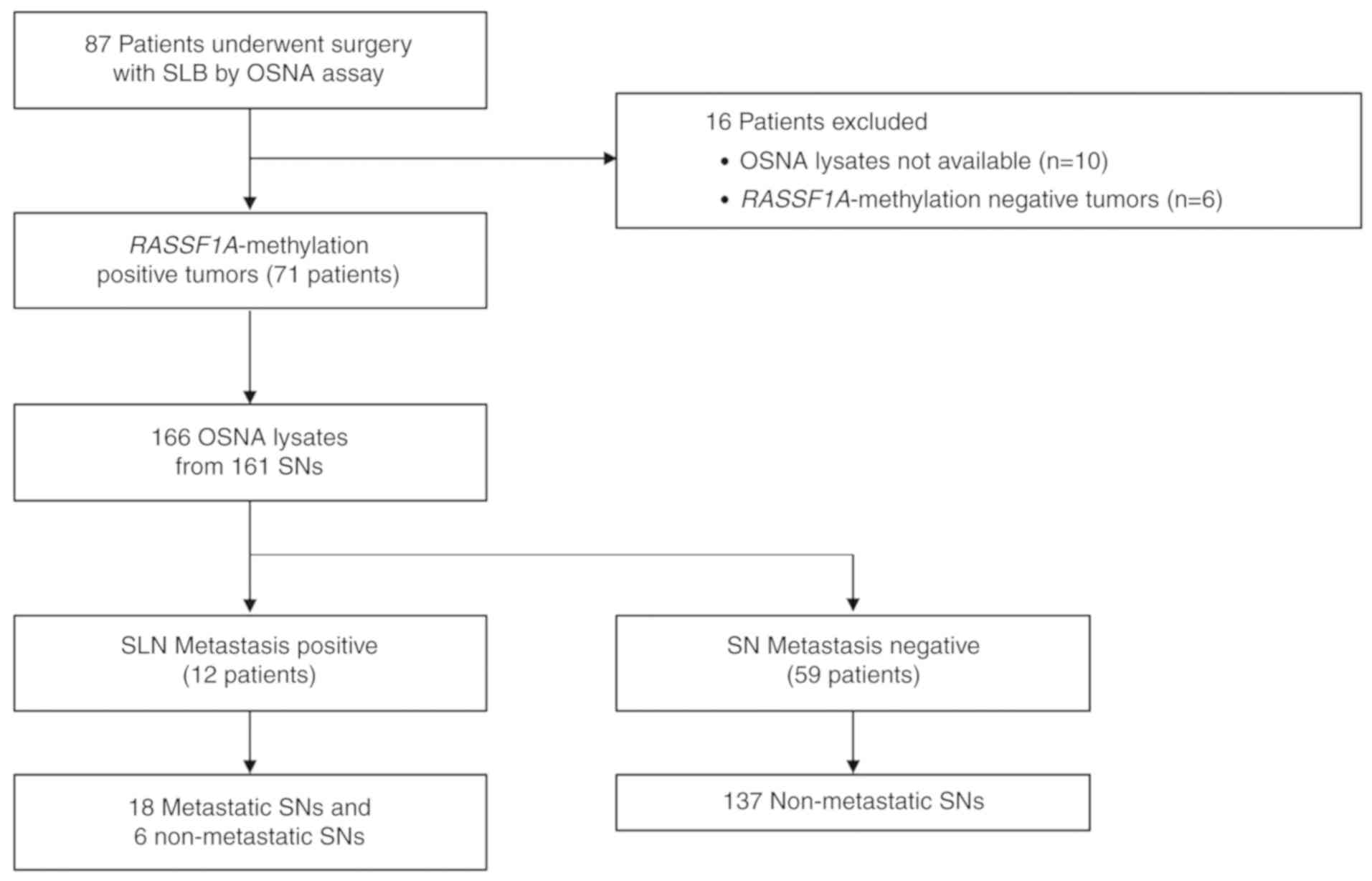
A total of 87 patients with breast cancer who
underwent surgery with sentinel lymph node biopsy (SNB), and whose
SNs were examined by OSNA at Osaka University between November 2015
and April 2017, were retrospectively included in this study
(Fig. 1). The study was approved by
the Ethical Review Board of Osaka University Hospital (approval
date/number: 14 Aug 2014/#14111), and informed consent was obtained
from each patient. Of the 87 patients, 10 were excluded due to a
lack of OSNA lysates, and six were excluded due to the lack of
RASSF1A methylation in their primary tumors. Ultimately, 161 LNs
from 71 patients were included, and 166 lysates were analyzed (the
LN was separated into two lysates in three SNs, and three lysates
in one SN, due to its large size). SNB was performed with a
combination of dye (patent blue and/or indocyanine green) and
radiocolloid (technetium-99m tin colloid) or dye alone. A
1-mm-thick slice was cut from the center of each SN and
intraoperatively subjected to frozen section analysis. The
remaining LN tissue was used for OSNA, where the SN was homogenized
in 4 ml Lynorhag solution (Sysmex Corporation), of which 20 µl
lysate was used. The remaining lysate was stored at −80°C until
use. The CK19 copy number per assay was classified as follows:
>5,000, (++); >250 and ≤5,000, (+); >0 and ≤250, (−); and
0, (N.D.). OSNA (++) and (+) were considered to be positive, and
isolated tumor cells (ITCs) were considered negative for SN
metastasis.
Detection of RASSF1A methylation using
RE-dMSP
DNA was extracted from 100 µl SN lysate using the
QIAamp Circulating Nucleic Acid Kit (Qiagen GmbH) and eluted in 50
µl desalted water. DNA solution (6.6 µl) was incubated for 16 h at
37°C in a final volume of 20 µl, containing 1X ddPCR Supermix for
probes (Bio-Rad Laboratories, Inc.), 900 nM each primer, 250 nM
probe and 10 U HhaI, HpaII (New England BioLabs,
Inc.) and BstUI (Thermo Fisher Scientific, Inc.) each. These
three methylation-sensitive restriction enzymes were selected since
they can be used at the same incubation temperature (37°C). The
reaction time was set to 16 h (16,17) to
allow for the complete digestion of unmethylated DNA. Methylation
analysis was performed using three wells per assay. As a control to
confirm the presence of DNA, the DNA solution (2.0 µl) was also
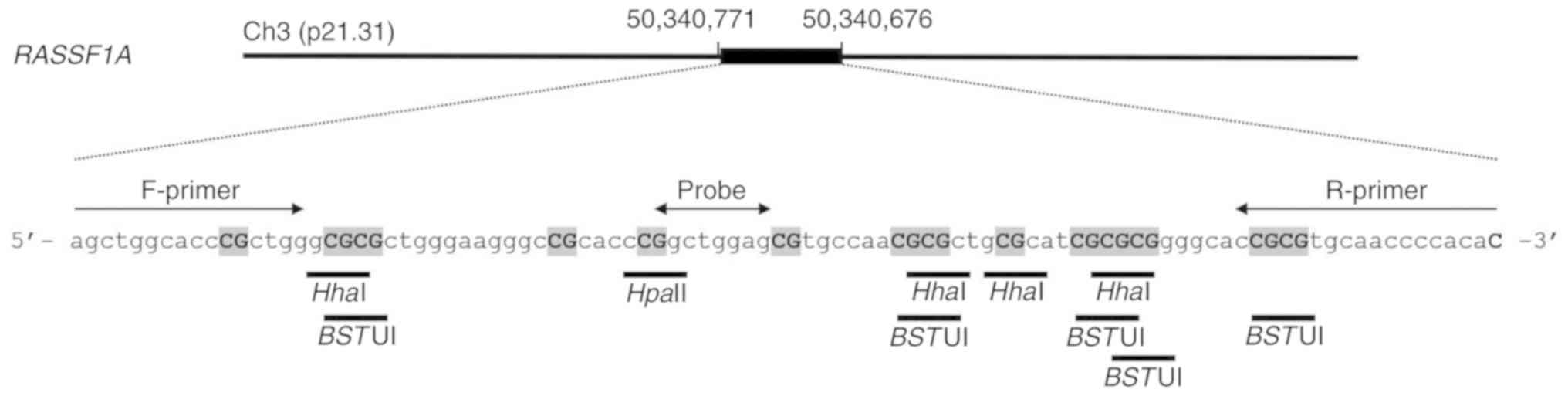
incubated without restriction enzymes. The primers (18) and probe (Universal ProbeLibrary #19;
cat. no. 04686926001; Roche Diagnostics BmbH) are presented in
Fig. 2 and Table SI. After incubation, droplet
generation oil was added, and the mixture was loaded onto a QX100
droplet generator (Bio-Rad Laboratories, Inc.). Then, 40 µl
emulsified mixture was subjected to PCR using a T100 thermal cycler
(Bio-Rad Laboratories, Inc.) under the following conditions: 95°C
for 10 min, followed by 40 cycles at 94°C for 30 sec and 60°C for 1
min, and 98°C for 10 min. The data were analyzed using the QX100
droplet reader and QuantaSoft software version 1.7.4 (both Bio-Rad
Laboratories, Inc.). The presence of ≥2 dots/well was regarded as a
positive result, and the copy numbers of three positive wells were
totaled. The results for each SN divided into multiple lysates were
summed.
For methylation analysis of primary breast tumors,
DNA was extracted from three 10-µm formalin-fixed paraffin-embedded
(FFPE) tumor sections using the QIAamp DNA FFPE kit (Qiagen GmbH),
and RE-dMSP was performed. For sensitivity analysis of RE-dMSP, 0,
1, 3, 10, 30 and 100 copies of methylated DNA template
(EpiScope® Methylated HeLa gDNA; Takara Bio, Inc.)
spiked in 10,000 copies of unmethylated DNA from the peripheral
blood leukocytes of a healthy individual were subjected to RE-dMSP
with or without restriction enzymes. Conventional MSP with
real-time PCR after bisulfite modification (qMSP) was performed as
previously reported (19). The
initial amount of DNA before bisulfite treatment was adjusted so
that the input DNA copy number per well was the same as that of
RE-dMSP. The sensitivity and positive detection rates were compared
between the RE-dMSP and qMSP assays over eight wells.
Mutational analysis of PIK3CA in SNs
and primary tumors
For the mutational analysis of primary breast
tumors, DNA extracted from the FFPE tissue sections was subjected
to real-time PCR analysis to detect the PIK3CA-H1047R mutation, as
previously reported (20). For
PIK3CA-mutation detection in SNs, DNA was extracted from 100 µl SN
lysate and eluted in 50 µl desalted water. Then, 9 µl DNA solution
was subjected to QuantStudio™ 3D dPCR (Thermo Fisher Scientific,
Inc.) (20). The sequences of the
primers and probes are displayed in Table SI.
Estimation of DNA fragment size of
methylated RASSF1A in SN tissues
DNA was extracted from 100 µl SN lysate and eluted
with 50 µl desalted water. To estimate the fragment size of the
methylated DNA, 14 µl DNA from each SN lysate was electrophoresed
on a 2% agarose gel, and subsequently separated into short (<500
bp) and long (>500 bp) fragments. The DNA was extracted from
each fraction using the QIAquick Gel Extraction Kit (Qiagen GmbH)
and RE-dMSP was performed.
Immunohistochemistry analysis of
CK19
The expression of CK19 protein in primary breast
tumors was assessed using immunohistochemistry with 4-µm FFPE
tissue sections. Immunohistochemical staining of each section was
performed as previously described (21) with mouse monoclonal anti-CK19 primary
antibody (clone, RCK 108; 1:50; Dako; Agilent Technologies, Inc.)
and a peroxidase-conjugated secondary antibody [cat. no. 414131F;
Histofine Simple Stain MAX PO (M); Nichirei Biosciences, Inc.].
Finally, the sections were visualized with 3,3-diaminobenzidine
tetrahydrochloride (Wako Pure Chemical Industries, Ltd.) and
counterstained with hematoxylin.
Quantification of methylated RASSF1A
and CK19 mRNA in breast cancer cell lines
A total of six breast cancer cell lines (MCF7,
MDA-MB-361, BT474, MDA-MB-453, MDA-MB-231 and BT20) were cultured
in DMEM (Sigma-Aldrich; Merck KGaA), and five (ZR75-1, T47D,
ZR75-30, SKBR3 and AU565) were cultured in RPMI-1640
(Sigma-Aldrich; Merck KGaA) at 37°C (5% CO2) in a
humidified atmosphere. DNA and mRNA were extracted from 10E+6 cells
from each cell line using the DNeasy Blood & Tissue Kit and the
RNeasy Mini Kit (both Qiagen GmbH), respectively. The DNA was
subjected to RE-dMSP, and the copy number of methylated RASSF1A per
cell was obtained. Briefly, 1 µg total RNA was reverse-transcribed
into cDNA using the ReverTra Ace® qPCR RT kit (Toyobo
Life Science), and CK19 mRNA expression was assessed using the
Light Cycler 480 Real-time PCR System (Roche Applied Science) with
the following conditions: 95°C for 10 min, followed by 50 cycles at
95°C for 15 sec and 60°C for 60 sec, with a final cycle at 50°C for
10 sec. KRT19 (CK19) TaqMan® Gene Expression Assays
(Hs01051611_gH; Applied Biosystems; Thermo Fisher Scientific, Inc.)
were used to conduct real-time PCR. The relative CK19 mRNA
expression level per cell was obtained, and a PCR product was used
as the standard.
Statistical analysis
Statistical analyses were performed using JMP Pro
11.2.0 (SAS Institute, Inc.) or GraphPad Prism 6 software (GraphPad
Software, Inc.). The association between clinicopathological
parameters and the copy number of methylated DNA or CK19 mRNA in
SNs was evaluated using Fisher's exact test. Associations between
the copy number of methylated DNA and CK19 mRNA in lysates were
evaluated using the Wilcoxon signed-rank sum test. Differences in
the copy number ranges of methylated DNA and CK19 mRNA among breast
cancer cell lines were evaluated using the F-test.
Results
Sensitivity of RE-dMSP
The sensitivity of RE-dMSP was evaluated for the
detection of methylated RASSF1A, using 0, 1, 3, 10, 30 and 100
copies of methylated DNA spiked in 10,000 copies of unmethylated
DNA per well. A linear correlation was observed across all
concentrations of methylated RASSF1A between the input copy number
and the RE-dMSP results (Fig. 3A). No
copies of methylated RASSF1A were detected in the 100% unmethylated
DNA samples (data not shown), indicating that methylated RASSF1A
was completely removed by restriction enzyme digestion. The assay
results without restriction enzymes accurately represented the
total amount of input DNA. The detection sensitivity of RE-dMSP in
eight wells was 37.5, 62.5 and 100% for 1, 3 and ≥10 copies of
methylated DNA, respectively, while that of qMSP was 0, 25 and 100%
for ≤10, 30 and 100 copies of methylated DNA, respectively. This
indicated that RE-dMSP was ≥10 times more sensitive than
conventional qMSP following bisulfite modification (Fig. 3B). Considering the probability
distribution of methylated DNA in the templates (according to the
binominal model), the sensitivity of RE-dMSP was estimated to be
between two and three copies per well.
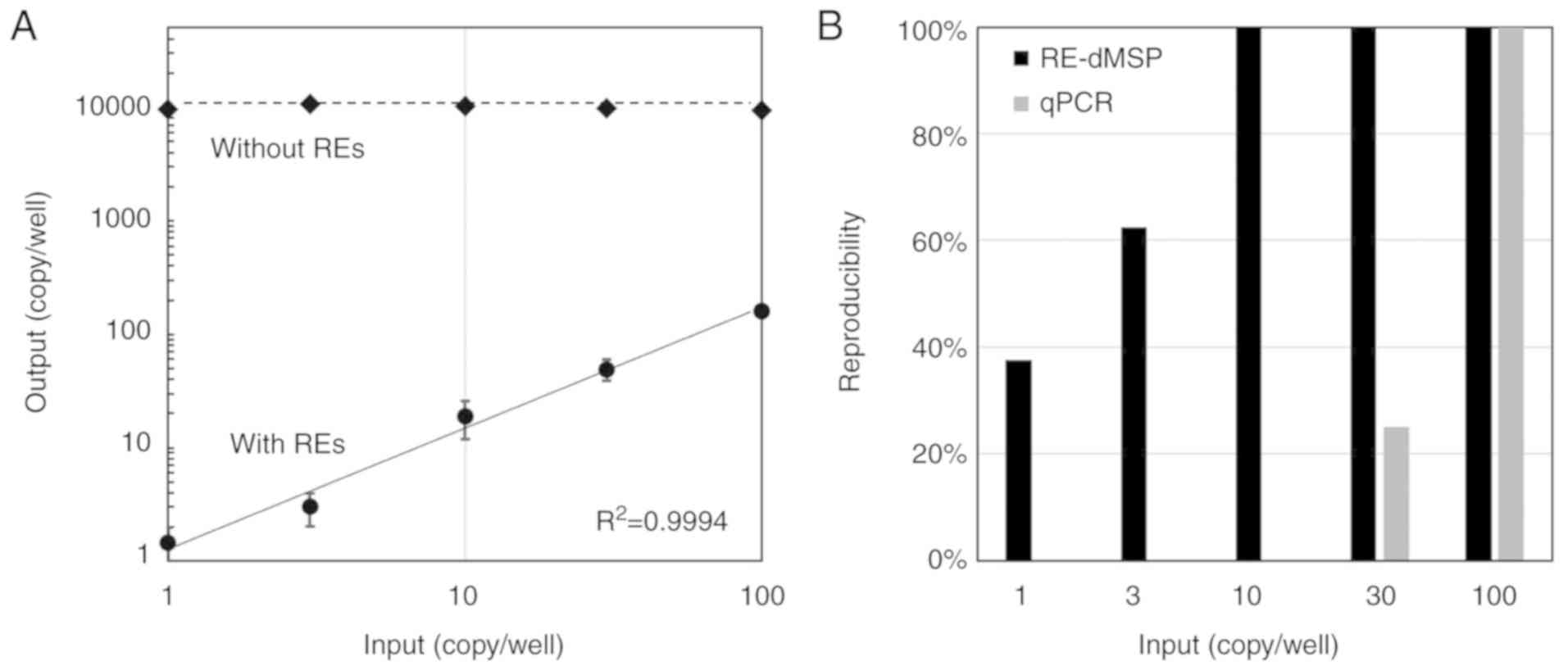 | Figure 3.Sensitivity of RE-dMSP for the
detection of methylated RASSF1A. (A) Detection sensitivity of
RE-dMSP was assessed using 0, 1, 3, 10, 30 and 100 copies of
methylated genomic DNA, spiked in 10,000 copies of unmethylated
genomic DNA extracted from the peripheral blood leukocytes of a
healthy individual. Methylated RASSF1A was quantified by RE-dMSP
with restriction enzymes (solid line, with REs), and the total
inputs of methylated and unmethylated DNA were measured without
restriction enzymes (dotted line, without REs). Error bars indicate
the standard deviation of eight experiments. (B) Positive detection
rate in eight experiments for each sample, compared between RE-dMSP
and qPCR with bisulfite modification. RE-dMSP, dPCR with
methylation-specific restriction enzymes; RASSF1A, Ras association
domain-containing protein 1; RE, restriction enzyme; qPCR,
quantitative PCR. |
RE-dMSP using SN lysates for OSNA
Using the primary tumor samples of 77 breast cancer
patients who underwent SNB and OSNA, RASSF1A methylation was
screened by RE-dMSP; 71 (92.2%) of the samples were revealed to
exhibit RASSF1A methylation (Fig. 1).
The patient clinicopathological characteristics are presented in
Table I. Of these 71 patients, 12
(16.9%) possessed SN metastases. In total, 161 SNs from these 71
patients were analyzed using OSNA, including 18 positive and 143
negative SNs. Among the 161 SNs, RASSF1A-methylation analysis was
performed by RE-dMSP, and methylation was detected in 22 SNs from
14 patients. The amount of total DNA in the SN lysates ranged from
1,600 to 1,593,000 copies per 100 µl, confirming successful DNA
extraction from all samples. Methylated RASSF1A was observed
significantly more frequently in patients with large tumors that
exhibited positive lymphovascular invasion (Table I). The expression levels of CK19 mRNA
also exhibited a similar trend, although the difference was not
significant. The relationship between the amounts of methylated
RASSF1A and CK19 mRNA in SNs is presented in Fig. 4A. Methylated RASSF1A was detected in
all of the SNs in which CK19 mRNA was highly expressed [OSNA (++)]
(range, 9.8–95,000 copies/assay; n=13), and methylated RASSF1A was
not present in SNs in which CK19 mRNA was not detected [OSNA
(N.D.); n=91] (Fig. 4B). Methylated
RASSF1A was detected in three of the five OSNA (+) SNs (60%; range,
7.4–12,800 copies/assay), and in six of the 52 OSNA (−) SNs (11.5%;
range, 19.8–348 copies/assay). The concordance rate between the
methylated RASSF1A status and the OSNA results was 95.0% (153/161).
In the six RASSF1A-methylation (+) and OSNA (−) SNs from five
patients (one patient possessed two SNs), immunohistochemistry was
used to assess CK19 protein expression in the primary tumors; the
results revealed strong-positive staining in all five patient
samples (Fig. S1).
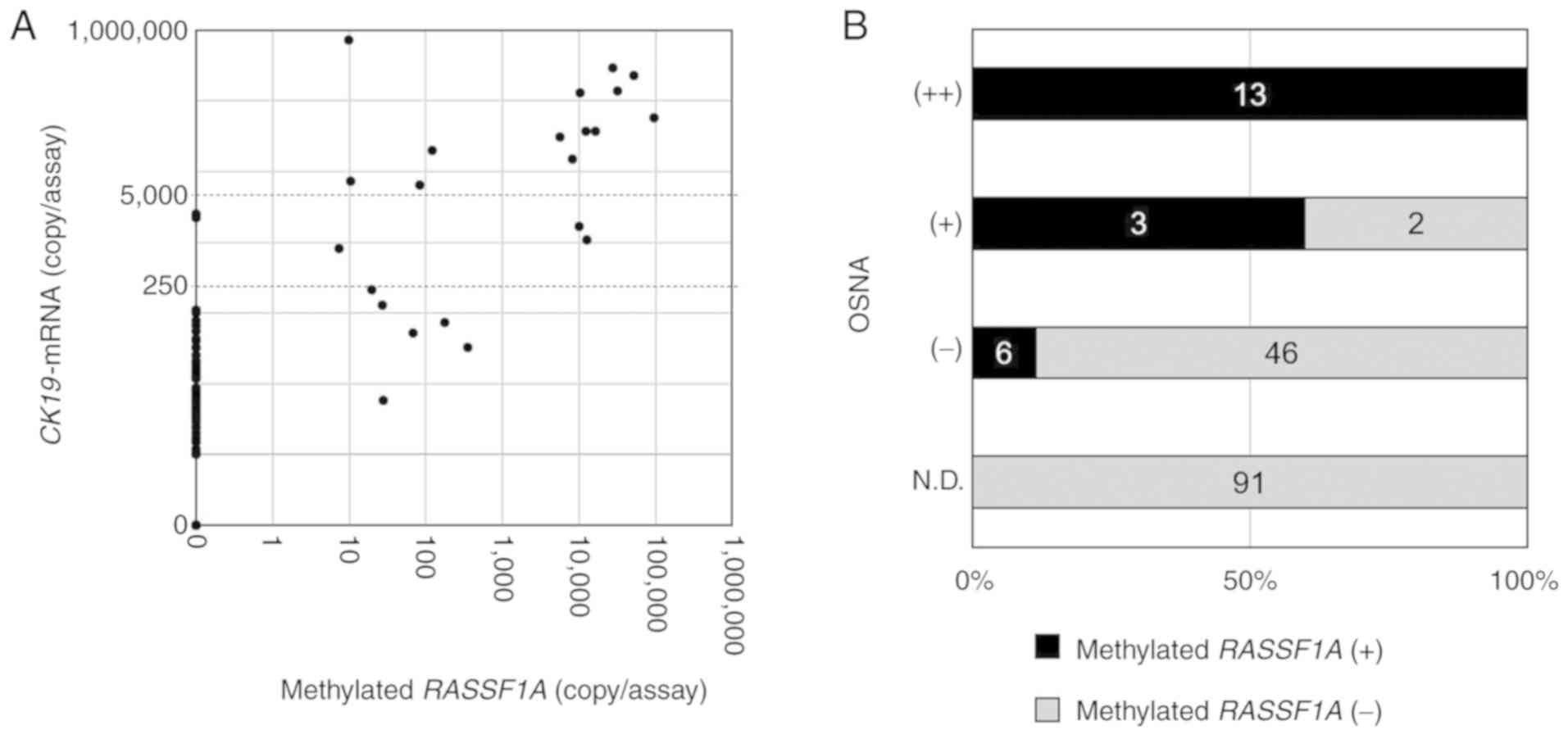 | Figure 4.Association between the methylated
RASSF1A copy number and CK19 mRNA expression in SN lysates of OSNA.
(A) Correlation between the copy number (copies/assay) of
methylated RASSF1A as determined by RE-dMSP, and CK19 mRNA
expression as determined by OSNA. (B) The number of methylated
RASSF1A-positive and -negative SNs is presented according to OSNA
diagnoses; CK19 mRNA >5,000, (++); >250 and ≤5,000, (+);
>0 and ≤250, (−); 0, (N.D.). RASSF1A, Ras association
domain-containing protein 1; SN, sentinel node; CK19, cytokeratin
19; OSNA, one-step nucleic acid amplification; RE-dMSP, dPCR with
methylation-specific restriction enzymes. |
 | Table I.Association between patient
clinicopathological parameters and RASSF1A methylation or CK19 mRNA
expression in SNs. |
Table I.
Association between patient
clinicopathological parameters and RASSF1A methylation or CK19 mRNA
expression in SNs.
|
|
| RASSF1A methylation
in SN |
| CK19 mRNA in
SN |
|
|---|
|
|
|
|
|
|
|
|---|
| Variables | n | Positive | Negative | P-value | Positive | Negative | P-value |
|---|
| Total patients |
| 71 | 14 | 57 |
| 12 | 59 |
| Age |
|
<50 | 23 | 7 | 16 | 0.107 | 5 | 18 | 0.332 |
|
≥50 | 48 | 7 | 41 |
| 7 | 41 |
|
| Tumor size
(mm) |
|
<20 | 40 | 4 | 36 | 0.033 | 3 | 37 | 0.025 |
|
≥20 | 31 | 10 | 21 |
| 9 | 22 |
|
| Histological
grade |
| 1,
2 | 53 | 11 | 42 | 0.332 | 11 | 42 | 0.128 |
| 3 | 18 | 3 | 15 |
| 1 | 17 |
|
| LVI |
|
Positive | 7 | 5 | 2 | 0.003 | 3 | 4 | 0.089 |
|
Negative | 64 | 9 | 55 |
| 9 | 55 |
|
| ER/PgR |
|
Positive | 65 | 13 | 52 | 0.663 | 11 | 54 | 0.685 |
|
Negative | 6 | 1 | 5 |
| 1 | 5 |
|
| HER2 |
|
Positive | 5 | 0 | 5 | 0.322 | 0 | 5 | 0.385 |
|
Negative | 66 | 14 | 52 |
| 12 | 54 |
|
| Recurrence |
|
Positive | 0 | 0 | 0 | – | 0 | 0 | – |
|
Negative | 71 | 14 | 57 |
| 12 | 59 |
|
DNA fragment size of methylated RASSF1
in SNs
It has previously been reported that circulating
tumor DNA (ctDNA) generated in primary tumors can be detected in
SNs (22). ctDNA is segmented into
<180 bp fragments by apoptosis, and can therefore be detected by
the short amplicon dPCR product (96 bp) of RE-dMSP (23,24). By
contrast, metastatic tumor cells in SNs can produce long DNA
fragments of methylated RASSF1A. In the present study, DNA fragment
size was assessed to determine whether the methylated RASSF1A
detected in SNs was derived from tumor cells, or from methylated
RASSF1A fragments from the primary tumor migrating through the
lymphatic vessels. A total of six methylated RASSF1A-positive, CK19
mRNA-negative SNs were selected. The SN lysates were available from
three of these SNs and subjected to the following experimental
procedures: Total DNA extracted from the lysates was separated into
short (<500 bp) and long (>500 bp) DNA fractions by agarose
gel electrophoresis, and evaluated by RE-dMSP (Fig. S2A). Methylated RASSF1A was detected
in the long DNA fractions of all three SNs, and in the short
fractions of two SNs (Fig. S2B).
Given that all methylated RASSF1A-positive SNs contained long DNA
fragments, SN-associated methylated RASSF1A was considered to
originate from tumor cells in the SN, and not from the primary
tumor.
Detection of SN metastasis by dPCR for
the PIK3CA mutation
To further investigate whether the methylated
RASSF1A was derived from unexpected methylation in non-tumor cells
of the SN (including lymphocytes), mutational analysis of the SN
lysates was performed, targeting a mutation specific to primary
tumors (which does not occur in non-cancerous cells) (25). The PIK3CA H1047R mutation was used in
this study, since it is one of the most frequently observed
mutations in breast cancer (26,27). A
total of 71 tumors were screened using real-time PCR, and 22 were
revealed to possess the mutation. A total of 59 SN lysates from
these 22 patients were subjected to dPCR analysis for the PIK3CA
mutation, which was detected in 11 SNs (18.6%; range, 6.5–6,106.9
copies/assay; Fig. S3A). Methylated
RASSF1A was detected in all of the 11 PIK3CA mutation-positive SNs,
but not in the remaining 48 mutation-negative SNs, indicating
complete agreement between mutation and methylation status
(Fig. S3B).
Association between methylated RASSF1A
and CK19 mRNA expression in breast cancer cells
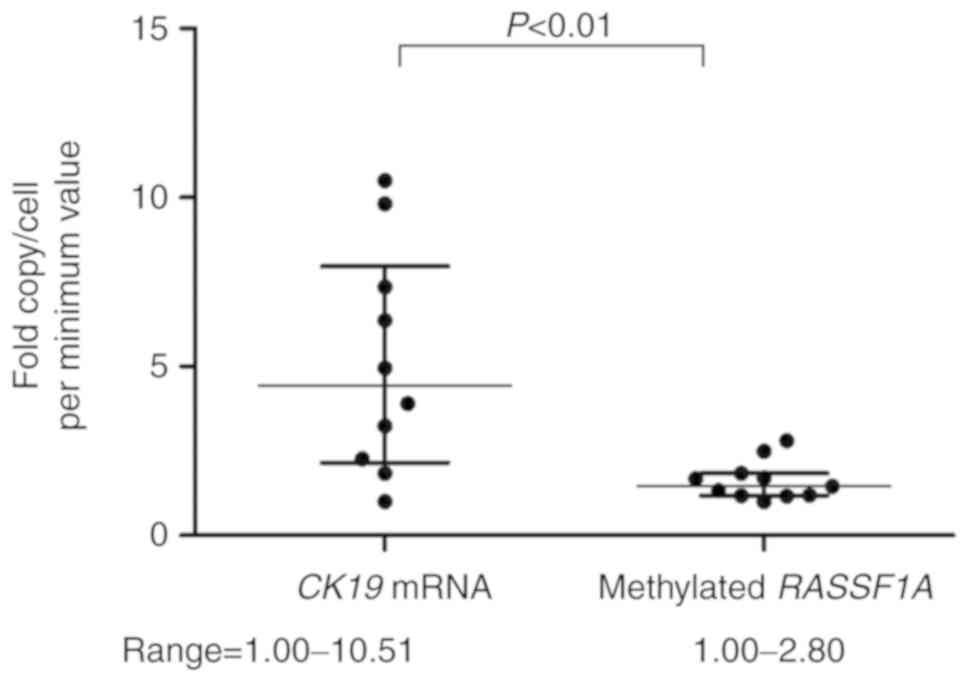
CK19 mRNA expression and the presence of methylated
RASSF1A alleles were analyzed in 11 breast cancer cell lines. An
extremely low level of CK19 mRNA expression was detected in
MDA-MB-231 cells (20 copies/cell), which were considered to be CK19
negative. In the other 10 cell lines, the expression level ranged
from 3,224 to 33,877 copies/cell, which equated to a 10.5-fold
difference (Fig. 5). By contrast, the
copy numbers of methylated RASSF1A alleles were in the range of
0.52–1.44 copies/cell, exhibiting a 2.80-fold difference. The fold
difference in copy number per cell was significantly lower for
RASSF1A methylation than for CK19 mRNA, indicating that methylated
DNA alleles more precisely reflect the number of tumor cells.
Discussion
In order to detect SN tumor-derived DNA, an RE-dMSP
assay was developed to accurately measure RASSF1A methylation using
dPCR following restriction enzyme digestion. RE-dMSP was able to
detect as few as three copies of methylated RASSF1A by complete
digestion of unmethylated DNA, that corresponds to 150 tumor cells
per node, showing a sensitivity >10 times greater than that of
the bisulfite method. A highly linear correlation between the
RE-dMSP results and the amount of input DNA also ensured accurate
and quantitative measurement of SN tumor cells.
The RE-dMSP assay, which was conducted with 161 SN
lysates, demonstrated a high concordance of 95% (153/161 SNs) with
OSNA; eight discordant cases were found, including six OSNA
(−)/methylation (+) and two OSNA (+)/methylation (−) SNs. The fact
that the PIK3CA mutation status in the SN lysates revealed complete
agreement with the RASSF1A methylation status indicates that
non-tumorous cells in the lymph nodes do not exhibit RASSF1A
methylation, because, if they did, a considerable amount of RASSF1A
methylation would have been detected in PIK3CA mutation-negative
SNs. Therefore, it is surmised that the six OSNA (−)/methylation
(+) SNs are unlikely to have been RE-dPCR false-positives, and were
more likely to be OSNA false-negatives. Since CK19 protein
expression was confirmed in the primary tumors of all patients,
most of these false-negatives are unlikely to be attributable to
low CK19 mRNA expression within the tumor cells, although the
possibility of low CK19 mRNA expression in CK19 protein-positive
tumors still remains (9).
In addition, the DNA fragment size of methylated
RASSF1A in the OSNA (−)/methylation (+) SN group was analyzed, in
order to determine whether methylated RASSF1A originated from
metastatic tumor cells in the SNs, or from primary tumors via the
lymphatic vessels. Taking advantage of the fact that methylated
RASSF1A from primary tumors has a short DNA fragment size (<500
bp; as it is generated from apoptosis), while that from metastatic
tumor cells may be either short or long (>500 bp), the origin of
the methylated RASSF1A was distinguished by analyzing DNA fragment
size. In the present study, the presence of long DNA fragments was
indicated in all three of the analyzed SNs, confirming the presence
of tumor cells in SNs. Thus, it is highly likely that OSNA
(−)/methylation (+) SNs reflect tumor metastases, and thus
represent false-negatives from OSNA.
Only two of the SNs were OSNA (+)/methylation (−),
suggesting the possibility of false-negatives from RE-dMSP.
However, in addition to being negative for RASSF1A methylation,
these two SNs were also PIK3CA mutation-negative, although the
corresponding primary tumors were positive for the PIK3CA mutation.
Since RE-dMSP is sensitive enough to detect only a few copies per
assay, it is unlikely that tumor cells in the SNs were missed by
RE-dMSP; it is more likely that OSNA resulted in
false-positives.
The TTL in SNs has been reported to correlate with
the extent of non-SN metastases (28), and CK19 mRNA copies measured by OSNA
have been used to estimate TTL in several predictive models
(6,7).
However, there is an ~30-fold difference in CK19 mRNA expression
among breast tumors (9). In line with
this, the present study also demonstrated a 10.8-fold difference in
CK19 mRNA expression per cell in 10 breast cancer cell lines (plus
one cell line that was CK19-negative). By contrast, the fold
difference in the methylated RASSF1A copy number was as low as 2.8
among these 11 cell lines. This difference was assumed to result
from a loss of heterozygosity (29)
or aneuploidy (30–32). These results indicate that methylated
RASSF1A can predict TTL more accurately than CK19 mRNA expression
levels. At present, SN micrometastases (equivalent to ITCs) are
considered to have little significance in prognosis (33). However, in these previous studies, SN
metastases were usually evaluated by histological examination of a
few representative sections of each SN, rather than a series of
sections from each SN. On the other hand, RE-dMSP can detect
metastases in each entire SN, thus quantification of TTL in each SN
is considered to be more accurate than histological examination.
Thus, it is possible that future RE-dMSP studies may disclose a new
prognostic value for small SN metastases which are not detectable
by histological examination.
A possible limitation of the present study is that
RASSF1A methylation is not observed in all breast cancers, and its
expression in the primary tumor is a prerequisite for RE-dMSP. In
the present study, as many as 92.2% of breast tumors were RASSF1A
methylation-positive; this was consistent with previous studies
reporting frequencies of 90.4–97.8% (10,19),
suggesting that RE-dMSP is applicable for use in >90% of breast
tumors. Moreover, we had already reported that ≥1 of the three
RASSF1A, GSTP1 and RARB2 genes is methylated in 98% of breast
tumors, indicating that for RE-dMSP, the addition of GSTP1 and
RARB2 to RASSF1A would enhance its applicability to nearly all
breast tumors (10,34). OSNA has been repeatedly revealed to be
as accurate as routine histological examination for the detection
of SN metastases in unselected breast tumors (5), and is used in clinical practice in
numerous countries. A lack of CK19 mRNA expression (a target of
OSNA) has been reported in 1.6–3.0% of breast tumors (35,36). These
results suggest that RE-dMSP targeting methylated GSTP1, RARB2 and
RASSF1A genes may also be applicable to unselected breast tumors,
much like OSNA. Further studies to pursue this possibility would be
worthwhile.
The second limitation is that RE-dMSP cannot be used
for the intraoperative diagnosis of SN metastasis, since it
requires an overnight assay procedure. We believe that OSNA and
RE-dMSP are complementary to each other for the detection of SN
metastasis: OSNA is quicker than RE-dMSP and thus suitable for
intraoperative analysis, while RE-dMSP provides a more accurate
assessment of TTL, and is thus more suitable for postoperative
analyses. The association between OSNA and RE-dMSP is analogous to
that between intraoperative frozen and postoperative FFPE section
analyses. Therefore, if the clinical significance of TTL determined
by RE-dMSP is confirmed in the future, RE-dMSP may potentially be
used alongside OSNA in daily practice, replacing the need for
histological analysis. The third limitation was our limited sample
size, which included only 161 LNs from 71 patients. We are
currently working with other institutions to increase the sample
population and hope to corroborate the findings in the future.
In conclusion, the present study demonstrated the
development of an RE-dMSP assay to precisely detect RASSF1A
methylation by methylation-specific restriction enzyme digestion,
followed by dPCR. RE-dMSP was indicated to detect SN metastasis
more accurately, and to estimate TTL more precisely than OSNA.
However, the clinical utility of RE-dMSP requires further
validation, including future studies with a greater number of
patients.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
No specific grants were received from funding
agencies in the public, commercial or not-for-profit sectors. SN
received honoraria and research funding from Sysmex for research
not related to the present study, and holds joint patents with
Sysmex on subjects not related to the present study. YN also
received honoraria from Sysmex, and holds joint patents with Sysmex
on subjects not related to the present study.
Availability of data and materials
The datasets used during the present study are
available from the corresponding author upon reasonable
request.
Authors' contributions
MA performed the experiments and analyzed the data.
TM, TT, YN, MS, KS, and SJK collected the clinical samples. MA, NK
and SN designed and drafted the manuscript. All authors read and
approved the final manuscript and agree to be accountable for all
aspects of the research in ensuring that the accuracy or integrity
of any part of the work are appropriately investigated and
resolved.
Ethics approval and consent to
participate
The study was approved by the Ethical Review Board
of Osaka University Hospital (approval date/number: 14 Aug
2014/#14111), and informed consent was obtained from each
patient.
Patient consent for publication
Not applicable.
Competing interests
This study was supported in part by the research
funding from Novartis, Pfizer, and Sysmex. Shinzaburo Noguchi has
received research grant for other study from AstraZeneca and
honoraria from AsstraZeneca, Novartis, Pfizer, and Sysmex, and has
been an advisor for AstraZeneca and Novartis. Naofumi Kagara
received honoraria from AstraZeneca and Novartis. Yasuto Naoi
received honoraria from AstraZeneca and Sysmex and research grant
from AstraZeneca for other study. Masafumi Shimoda received
honoraria from Novartis. Kenzo Shimazu received honoraria from
AstraZeneca. Seung Jin Kim received honoraria from AstraZeneca,
Novartis and Pfizer. The other authors declare that they do not
have a financial relationship with the organizations that sponsored
the research.
References
|
1
|
Lyman GH, Giuliano AE, Somerfield MR,
Benson AB III, Bodurka DC, Burstein HJ, Cochran AJ, Cody HS III,
Edge SB, Galper S, et al: American society of clinical oncology
guideline recommendations for sentinel lymph node biopsy in
early-stage breast cancer. J Clin Oncol. 23:7703–7720. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lyman GH, Temin S, Edge SB, Newman LA,
Turner RR, Weaver DL, Benson AB III, Bosserman LD, Burstein HJ,
Cody H III, et al: Sentinel lymph node biopsy for patients with
early-stage breast cancer: American Society of Clinical Oncology
clinical practice guideline update. J Clin Oncol. 32:1365–1383.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Veronesi U, Paganelli G, Viale G, Luini A,
Zurrida S, Galimberti V, Intra M, Veronesi P, Maisonneuve P, Gatti
G, et al: Sentinel-lymph-node biopsy as a staging procedure in
breast cancer: Update of a randomised controlled study. Lancet
Oncol. 7:983–990. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Krag DN, Anderson SJ, Julian TB, Brown AM,
Harlow SP, Ashikaga T, Weaver DL, Miller BJ, Jalovec LM, Frazier
TG, et al: Technical outcomes of sentinel-lymph-node resection and
conventional axillary-lymph-node dissection in patients with
clinically node-negative breast cancer: Results from the NSABP B-32
randomised phase III trial. Lancet Oncol. 8:881–888. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Tamaki Y: One-step nucleic acid
amplification assay (OSNA) for sentinel lymph node biopsy. Breast
Cancer. 22:230–234. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Shimazu K, Sato N, Ogiya A, Sota Y,
Yotsumoto D, Ishikawa T, Nakamura S, Kinoshita T, Tsuda H, Ohi Y,
et al: Intraoperative nomograms, based on one-step nucleic acid
amplification, for prediction of non-sentinel node metastasis and
four or more axillary node metastases in breast cancer patients
with sentinel node metastasis. Ann Surg Oncol. 25:2603–2611. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Rubio IT, Espinosa-Bravo M, Rodrigo M,
Amparo Viguri Diaz M, Hardisson D, Sagasta A, Dueñas B and Peg V:
Nomogram including the total tumoral load in the sentinel nodes
assessed by one-step nucleic acid amplification as a new factor for
predicting nonsentinel lymph node metastasis in breast cancer
patients. Breast Cancer Res Treat. 147:371–380. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Osako T, Iwase T, Ushijima M, Yonekura R,
Ohno S and Akiyama F: A new molecular-based lymph node staging
classification determines the prognosis of breast cancer patients.
Br J Cancer. 117:1470–1477. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Tsujimoto M, Nakabayashi K, Yoshidome K,
Kaneko T, Iwase T, Akiyama F, Kato Y, Tsuda H, Ueda S, Sato K, et
al: One-step nucleic acid amplification for intraoperative
detection of lymph node metastasis in breast cancer patients. Clin
Cancer Res. 13:4807–4816. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yamamoto N, Nakayama T, Kajita M, Miyake
T, Iwamoto T, Kim SJ, Sakai A, Ishihara H, Tamaki Y and Noguchi S:
Detection of aberrant promoter methylation of GSTP1, RASSF1A and
RARβ2 in serum DNA of patients with breast cancer by a newly
established one-step methylation-specific PCR assay. Breast Cancer
Res Treat. 132:165–173. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Park SY, Kwon HJ, Lee HE, Ryu HS, Kim SW,
Kim JH, Kim IA, Jung N, Cho NY and Kang GH: Promoter CpG island
hypermethylation during breast cancer progression. Virchows Arch.
458:73–84. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Okamoto A: Chemical approach toward
efficient DNA methylation analysis. Org Biomol Chem. 7:21–26. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Grunau C, Clark SJ and Rosenthal A:
Bisulfite genomic sequencing: Systematic investigation of critical
experimental parameters. Nucleic Acids Res. 29:E65–E75. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Li M, Chen WD, Papadopoulos N, Goodman SN,
Bjerregaard NC, Laurberg S, Levin B, Juhl H, Arber N, Moinova H, et
al: Sensitive digital quantification of DNA methylation in clinical
samples. Nat Biotechnol. 27:858–863. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wu Z, Bai Y, Cheng Z, Liu F, Wang P, Yang
D, Li G, Jin Q, Mao H and Zhao J: Absolute quantification of DNA
methylation using microfluidic chip-based digital PCR. Biosens
Bioelectron. 96:339–344. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Suehiro Y, Zhang Y, Hashimoto S, Takami T,
Higaki S, Shindo Y, Suzuki N, Hazama S, Oka M, Nagano H, et al:
Highly sensitive faecal DNA testing of TWIST1 methylation in
combination with faecal immunochemical test for haemoglobin is a
promising marker for detection of colorectal neoplasia. Ann Clin
Biochem. 55:59–68. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Suehiro Y, Hashimoto S, Higaki S, Fujii I,
Suzuki C, Hoshida T, Matsumoto T, Yamaoka Y, Takami T, Sakaida I
and Yamasaki T: Blood free-circulating DNA testing by highly
sensitive methylation assay to diagnose colorectal neoplasias.
Oncotarget. 9:16974–16987. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kristiansen S, Nielsen D and Sölétormos G:
Detection and monitoring of hypermethylated RASSF1A in serum from
patients with metastatic breast cancer. Clin Epigenetics. 8:352016.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Takahashi H, Kagara N, Tanei T, Naoi Y,
Shimoda M, Shimomura A, Shimazu K, Kim SJ and Noguchi S:
Correlation of methylated circulating tumor DNA with response to
neoadjuvant chemotherapy in breast cancer patients. Clin Breast
Cancer. 17:61–69.e3. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Oshiro C, Kagara N, Naoi Y, Shimoda M,
Shimomura A, Maruyama N, Shimazu K, Kim SJ and Noguchi S: PIK3CA
mutations in serum DNA are predictive of recurrence in primary
breast cancer patients. Breast Cancer Res Treat. 150:299–307. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Morimoto K, Kim SJ, Tanei T, Shimazu K,
Tanji Y, Taguchi T, Tamaki Y, Terada N and Noguchi S: Stem cell
marker aldehyde dehydrogenase 1-positive breast cancers are
characterized by negative estrogen receptor, positive human
epidermal growth factor receptor type 2 and high Ki67 expression.
Cancer Sci. 100:1062–1068. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Miyamura Y, Kagara N, Miyake T, Tanei T,
Naoi Y, Shimoda M, Shimazu K, Kim SJ and Noguchi S: Drainage of
tumor-derived DNA into sentinel lymph nodes in breast cancer
patients. Pathol Oncol Res. Feb 25–2019.(Epub ahead of print).
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Jahr S, Hentze H, Englisch S, Hardt D,
Fackelmayer FO, Hesch RD and Knippers R: DNA fragments in the blood
plasma of cancer patients: Quantitations and evidence for their
origin from apoptotic and necrotic cells. Cancer Res. 61:1659–1665.
2001.PubMed/NCBI
|
|
24
|
Heitzer E, Ulz P and Geigl JB: Circulating
tumor DNA as a liquid biopsy for cancer. Clin Chem. 61:112–123.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Broderick DK, Di C, Parrett TJ, Samuels
YR, Cummins JM, McLendon RE, Fults DW, Velculescu VE, Bigner DD and
Yan H: Mutations of PIK3CA in anaplastic oligodendrogliomas,
high-grade astrocytomas and medulloblastomas. Cancer Res.
64:5048–5050. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Cancer Genome Atlas Network, .
Comprehensive molecular portraits of human breast tumours. Nature.
490:61–70. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Pereira B, Chin SF, Rueda OM, Vollan HK,
Provenzano E, Bardwell HA, Pugh M, Jones L, Russell R, Sammut SJ,
et al: The somatic mutation profiles of 2,433 breast cancers
refines their genomic and transcriptomic landscapes. Nat Commun.
7:114792016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Teramoto A, Shimazu K, Naoi Y, Shimomura
A, Shimoda M, Kagara N, Maruyama N, Kim SJ, Yoshidome K, Tsujimoto
M, et al: One-step nucleic acid amplification assay for
intraoperative prediction of non-sentinel lymph node metastasis in
breast cancer patients with sentinel lymph node metastasis. Breast.
23:579–585. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Maitra A, Wistuba II, Washington C,
Virmani AK, Ashfaq R, Milchgrub S, Gazdar AF and Minna JD:
High-resolution chromosome 3p allelotyping of breast carcinomas and
precursor lesions demonstrates frequent loss of heterozygosity and
a discontinuous pattern of allele loss. Am J Pathol. 159:119–130.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Cornelisse CJ, van de Velde CJ, Caspers
RJ, Moolenaar AJ and Hermans J: DNA ploidy and survival in breast
cancer patients. Cytometry. 8:225–234. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kimmig R, Wimberger P, Kapsner T and
Hillemanns P: Flow cytometric DNA analysis using cytokeratin
labeling for identification of tumor cells in carcinomas of the
breast and the female genital tract. Anal Cell Pathol. 22:165–178.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Dayal JH, Sales MJ, Corver WE, Purdie CA,
Jordan LB, Quinlan PR, Baker L, ter Haar NT, Pratt NR and Thompson
AM: Multiparameter DNA content analysis identifies distinct groups
in primary breast cancer. Br J Cancer. 108:873–880. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Galimberti V, Cole BF, Viale G, Veronesi
P, Vicini E, Intra M, Mazzarol G, Massarut S, Zgajnar J, Taffurelli
M, et al: Axillary dissection versus no axillary dissection in
patients with breast cancer and sentinel-node micrometastases
(IBCSG 23-01): 10-year follow-up of a randomised, controlled phase
3 trial. Lancet Oncol. 19:1385–1393. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Fujita N, Nakayama T, Yamamoto N, Kim SJ,
Shimazu K, Shimomura A, Maruyama N, Morimoto K, Tamaki Y and
Noguchi S: Methylated DNA and total DNA in serum detected by
one-step methylation-specific PCR is predictive of poor prognosis
for breast cancer patients. Oncology. 83:273–282. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Chu PG and Weiss LM: Keratin expression in
human tissues and neoplasms. Histopathology. 40:403–439. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Vilardell F, Novell A, Martin J, Santacana
M, Velasco A, Díez-Castro MJ, Cuevas D, Panadés MJ, González S
Llombart A, et al: Importance of assessing CK19 immunostaining in
core biopsies in patients subjected to sentinel node study by OSNA.
Virchows Arch. 460:569–575. 2012. View Article : Google Scholar : PubMed/NCBI
|