Introduction
Oral squamous cell carcinoma (OSCC) accounts for
more than 90% of all oral cancers, and is considered as the most
frequently diagnosed type of cancer in the head and neck region
(1,2).
Currently, the 5 year overall survival rate for OSCC has been
estimated at approximately 50% and little improvement has been
noted, regardless of the many advances made in surgery, radiation
therapy and chemotherapy in the past few decades (3). In addition to the frequent late-stage
diagnosis associated with poor patient prognosis, a combination of
genetic alterations, environmental risk factors and viral infection
results in an increased incidence of OSCC (4). All these factors result in a challenging
task for predicting OSCC prognosis. Therefore, it is important to
better understand the molecular mechanisms associated with OSCC
pathogenesis, so as to develop more effective therapeutic
strategies for OSCC patients.
Circular RNAs (circRNAs) are a novel type of
endogenous non-coding RNA molecules that regulate gene expression
(5). Unlike linear RNA, circRNAs form
a closed continuous loop by back-splicing with covalently joined
3′- and 5′-ends (6), which makes it
highly stable and largely resistant to RNA degradation (7). This closed structure also presents with
advantages for existing as novel molecular biomarkers for many
diseases. Increasing evidence indicates that circRNAs play an
important role in many diseases, including cardiac senescence
(8), atherosclerosis (9) and Alzheimer's disease (10), and especially in human cancers. For
example, the expression of hsa_circ_0023404 has been shown to be
significantly upregulated in cervical cancer (11). CircRNA-Cdr1as exerts anti-oncogenic
functions in bladder cancer (12),
and hsa_circRNA_103809 could serve as a prognostic biomarker for
patients with lung cancer (13).
Recently, the involvement of circRNAs has also been
reported in OSCC. A report has shown that circDOCK1 may target
BIRC3 by inhibiting miR-196a-5p to be involved in OSCC development
(14). Hsa_circ_100290 has been
suggested as a potential prognostic biomarker for OSCC (15). However, these findings are only
preliminary and there is a lack of experimental and clinical
evidence. With the development of microarray technology,
bioinformatic detection tools provide novel insight into a
comprehensive analysis of molecular targets involved in tumor
progression. The predictive value of circRNAs has been explored in
breast cancer by Galasso et al (16) and Nair et al (17).
Hence, we performed circRNA microarray to establish
the circRNA expression profile and identified significantly
differentially expressed circRNAs between OSCC tissues and paired
adjacent non-cancerous tissues. Subsequently, qPCR was used to
confirm these differentially expressed circRNAs detected in the
microarray. We then selected the most upregulated or downregulated
circRNAs to further explore their biological function in OSCC in
vitro. The present study provides a molecular basis for
investigating the regulatory mechanisms of the circRNAs and
highlights their potential as diagnostic and prognostic biomarkers
for OSCC patients.
Materials and methods
Tissue samples and cell culture
Tumor and adjacent tissues (2 cm distant from the
tumor margin) were collected from OSCC patients who underwent
surgical resection of the primary lesions at Hainan General
Hospital (Haikou, Hainan). All of the patients underwent neither
chemotherapy nor radiotherapy, and written informed consent was
provided by all patients prior to participation. After radical
resection, all tissues were snap-frozen in liquid nitrogen and
immediately stored at −80°C until use. The histopathological
quality of all tissue specimens was independently validated by two
licensed pathologists, including the adjacent tissues
(histopathological quality of all adjacent tissues were without
cancer cells). After histopathological vetting, a total of 23
paired tumor and adjacent tissues were finally included in this
study. The microarray chip assay was performed in 3 pairs of OSCC
vs. adjacent tissues. The remaining 20 paired tissues were used for
verification using quantitative real-time PCR (qPCR). The
clinicopathological data of 20 patients is provided in Table I. This study was approved by the
Ethics Committee of Hainan General Hospital.
 | Table I.Correlation between hsa_circRNA_043621
and circRNA_102459 expression and clinicopathological
characteristics of the 20 patients with oral squamous cell
carcinoma. |
Table I.
Correlation between hsa_circRNA_043621
and circRNA_102459 expression and clinicopathological
characteristics of the 20 patients with oral squamous cell
carcinoma.
|
|
| Expression of
hsa_circRNA_043621 |
|
| Expression of
hsa_circRNA_102459 |
|
|---|
|
|
|
|
|
|
|
|
|---|
| Characteristics | Total | High | Low | P-value | Total | High | Low | P-value |
|---|
| Sex |
| Male | 12 | 7 | 5 | 0.325 | 12 | 8 | 4 | 0.085 |
|
Female | 8 | 3 | 5 |
| 8 | 2 | 6 |
|
| Age (years) |
|
|
|
|
|
|
|
|
|
<60 | 9 | 5 | 4 | 0.5 | 9 | 3 | 6 | 0.185 |
|
≥60 | 11 | 5 | 6 |
| 11 | 7 | 4 |
|
| Clinical stage |
|
I+II | 13 | 9 | 4 | 0.029 | 13 | 3 | 10 | 0.017 |
|
III+IV | 7 | 1 | 6 |
| 9 | 7 | 2 |
|
| Tumor
differentiation |
|
Well | 9 | 8 | 1 | 0.02 | 9 | 2 | 7 | 0.011 |
|
Moderately | 5 | 1 | 4 |
| 5 | 2 | 3 |
|
|
Poorly | 6 | 1 | 5 |
| 6 | 6 | 0 |
|
| Lymph node
metastasis |
|
Yes | 12 | 4 | 8 | 0.034 | 12 | 9 | 3 | 0.003 |
| No | 8 | 6 | 2 |
| 8 | 1 | 7 |
|
The human OSCC cell line, TSCC1, was obtained from
the Cell Bank of Type Culture Collection of Chinese Academy of
Sciences (Shanghai, China). TSCC1 cells were cultured in Dulbecco's
modified Eagle's medium (DMEM) supplemented with 10% fetal bovine
serum (FBS) (Invitrogen; Thermo Fisher Scientific, Inc.) and
maintained in a humidified air atmosphere containing 5%
CO2 at 37°C.
Total RNA extraction
Total RNA was extracted from the tissue samples
using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.)
following the manufacturer's instructions. A NanoDrop 2000
spectrophotometer (NanoDrop; Thermo Fisher Scientific, Inc.) was
used to measure the quantity (ng/ml) and purity (260/280 and
260/230 ratios) of the RNAs. Specifically, OD260/OD280 ratios
between 1.8 and 2.1, while OD260/OD230 ratios >1.8 were deemed
acceptable. RNA integrity and DNA contamination were then assessed
through electrophoresis on a denaturing agarose gel.
Sample labeling and hybridization
Sample labeling and array hybridization were
performed according to the manufacturer's protocol (Arraystar
Inc.). Briefly, Rnase R (Epicentre, Inc.) was used to digest total
RNAs in order to degrade any linear RNAs and enrich circular RNAs.
These enriched circular RNAs were amplified and transcribed into
fluorescent cRNA using a random priming method (Arraystar Super RNA
Labeling Kit; Arraystar). The labeled cRNAs underwent purify by
RNeasy Mini Kit (Qiagen) and quantitation by NanoDrop ND-1000
(Thermo Fisher Scientific, Inc.). A total of 1 µg of each labeled
cRNA was fragmented by adding 5 µl 10 X blocking agent and 1 µl of
25X fragmentation buffer, and then the mixture was heated at 60°C
for 30 min, and finally diluted by adding 25 µl 2X hybridization
buffer. Subsequently, 50 µl of hybridization solution was dispensed
into the gasket slide and applied to the circRNA expression
microarray slide. The slides were incubated for 17 h at 65°C in an
Agilent Hybridization Oven (Agilent Technologies, Inc.). The
hybridized arrays were washed, fixed and scanned using the Agilent
Scanner G2505C (Agilent Technologies, Inc.).
Microarray data analysis
The raw data were extracted by importing the scanned
images by Agilent G2565C Microarray Scanner into Agilent Feature
Extraction software (version 11.0.1.1; Agilent Technologies, Inc.).
R software package limma package (http://bioconductor.riken.jp/packages/3.0/bioc/html/limma.html)
was used to carry out a series of data processing including
quantile normalization of raw data. Subsequently, differentially
expressed circRNAs between the tumor and normal groups were
evaluated using t-test, and the P-values were corrected for False
Discovery Rate (FDR) by Benjamini-Hochberg (BH) procedure. CircRNAs
exhibiting fold changes ≥1.5 and FDR P-values <0.05 were
considered as significantly differentially expressed.
Hierarchical Clustering was used to show the
distinguishable circRNA expression pattern among samples. Scatter
plots were performed to evaluate differentially expressed circRNAs
with statistical significance between two groups. Volcano Plot
filtering was utilized to visualize the significantly differential
circRNAs between each pair wise comparison. Hierarchical
Clustering, Scatter plots and Volcano Plot filtering were performed
using Agilent Feature Extraction software (version 11.0.1.1;
Agilent Technologies, Inc.)
Bioinformatic analysis
Gene Ontology (GO) enrichment analysis and Kyoto
Encyclopedia of Genes and Functional Categories analysis were
performed with the DAVID online tool (https://david.ncifcrf.gov/). GO analysis was performed
to annotate genes meaningfully in terms of their biological
processes (BP) and molecular functions (MF). The -log10 (P-value)
yields an enrichment score representing the significance of GO term
enrichment among differentially expressed genes. Functional
Categories analysis was performed to determine the involvement of
target genes in the different biological pathway.
Validation of candidate circRNAs by
qPCR
Based on fold change >2, we selected the top 20
including 10 most upregulated circRNAs, and all 10 downregulated
circRNAs for subsequent validation. Briefly, total RNA was
extracted from 20 pairs of OSCC samples and matched adjacent
tissues using TRIzol reagent. The cDNAs were synthesized with the
Prime-Script RT Reagent kit (TakaraBiotechnology) from 500 ng of
total RNA. The qPCR was performed with the ABI 7300 PCR instrument
using the SYBR Green (Takara Bio Inc., Dalian, China) detection
method with divergent primers, as listed in Table II. The thermal cycling parameters
used for amplification were as follows: denaturation at 94°C for 2
min, followed by 40 cycles at 94°C for 20 sec, 58°C for 20 sec and
72°C for 30 sec. The reaction was stopped by incubation at 75°C for
5 min. GAPDH was used as an internal control. The relative gene
expression levels were analyzed by the 2−ΔΔCq method
(18).
 | Table II.Primers used for qPCR analysis of
circRNA and mRNA levels. |
Table II.
Primers used for qPCR analysis of
circRNA and mRNA levels.
| Target ID | Primer sequence
5′-3′ | PS (bp) |
|---|
|
hsa_circRNA_102459 | F:
CATCTGGAGGAACAGGACAGT | 125 |
|
| R:
ATAAGCAACTTCTTCACCAGC |
|
|
hsa_circRNA_102034 | F:
ACAAGTCGATGGATTCCTCTCA | 148 |
|
| R:
TCACTCTGTTCTGCTTCTGAGT |
|
|
hsa_circRNA_043621 | F:
CCTGAAGAAGAACCACGAGGA | 122 |
|
| R:
TCAACTCTGTCTCATACTTGGTG |
|
|
hsa_circRNA_101996 | F:
AAGATCTACTGGAACTGTCTTGC | 121 |
|
| R:
GCCTGTCCGTTTAGTTGTTGT |
|
|
hsa_circRNA_102733 | F:
AGGGGGACATATAACAGCTGAA | 120 |
|
| R:
TGTGCACCAATCATGTACCC |
|
|
hsa_circRNA_058819 | F:
CGCTGGGCAGACATACCA | 121 |
|
| R:
CCTTGAGTGCCGTTCACAC |
|
|
hsa_circRNA_066361 | F:
TGTCCGCCTATGGCACG | 125 |
|
| R:
CATGTAAGGTCCCTCCTGAGA |
|
|
hsa_circRNA_101036 | F:
TCCGTCTGTGGATAATGGGAG | 133 |
|
| R:
GGTCTCATGGTAAGCAGGAA |
|
|
hsa_circRNA_100245 | F:
CCCGGACTTCTTATCGTGGA | 120 |
|
| R:
CCCGGACTTCTTATCGTGGA |
|
|
hsa_circRNA_088200 | F:
TCCGGACCAAAACCATCAGT | 123 |
|
| R:
AGATCCCATCGGTAGCCATC |
|
|
hsa_circRNA_003949 | F:
ATGCCAGAAGACTGTCACCAT | 127 |
|
| R:
AAATGAATGCCAAGAAGGCAG |
|
|
hsa_circRNA_100259 | F:
AGACCTGGAAGAACCATCCT | 143 |
|
| R:
CTGCTTTGATTTGCACCAGT |
|
|
hsa_circRNA_100045 | F:
CTATGCAGGGGTGGTCAAC | 128 |
|
| R:
ACTTCAGGCAAACAGGTGCT |
|
|
hsa_circRNA_101217 | F:
GGTGGACCTGTACCTCAACA | 123 |
|
| R:
TTTCAATCCGGCTTTGACGA |
|
|
hsa_circRNA_005882 | F:
CTCTCTGTGCACGACTCTCA | 155 |
|
| R:
AGGTACTCTCTGTTCTTGGCT |
|
|
hsa_circRNA_102068 | F:
CCACCACCTTTGAGAGGGAC | 120 |
|
| R:
TCGGATGAGGTCATTGCTGT |
|
|
hsa_circRNA_037767 | F:
GTGCTGCAGTTCCTAGTCA | 120 |
|
| R:
AGTCCCAAAGCTCTGGTTGTT |
|
|
hsa_circRNA_402901 | F:
AGTTTGGAAAGAGTTGCTGTTA | 127 |
|
| R:
TGATAACTACATCTCAAAGGCAG |
|
|
hsa_circRNA_404462 | F:
TTCCCAGTGGACCAAGGCT | 144 |
|
| R:
TCCTGGCTCATCCCAAGTC |
|
|
hsa_circRNA_405247 | F:
GAAGTCCTTGCCTTGTTCCTGA | 289 |
|
| R:
AACAGAGTTTAGACTGACTTGGC |
|
| GAPDH | F:
TGTTCGTCATGGGTGTGAAC | 154 |
|
| R:
ATGGCATGGACTGTGGTCAT |
|
Cell transfection
To construct the circRNA_102459 overexpression
plasmid, the full-length circRNA_102459 cDNA sequence was amplified
and cloned into the pcircRNA 1.2 vector (Invitrogen; Thermo Fisher
Scientific, Inc.) and sequenced, named as pcRNA-circRNA_102459
(circRNA_102459), while the mock plasmid served as the control
vector. The specific small interfering RNAs (siRNAs) targeting
circRNA_043621(si-circRNA_043621) and scrambled siRNA control were
designed and synthesized by RiboBio Co., Ltd. (Guangzhou, China).
All of these plasmids and oligonucleotides were transfected into
TSCC1 cells using Lipofectamine 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's instructions. At
48 h after transfection, cells were harvested and used for the
in vitro experiments.
CCK-8 assay
Cell growth status was measured using Cell Counting
Kit-8 (CCK-8) reagent (Beyotime, China) according to the
manufacturer's instructions. Briefly, 1×105 cells were
seeded into the wells of a 96-well plate, each group of TSCC1 cells
was collected at 48 h after transfection. Then, 10 µl CCK-8
solution was added into each well and incubation was carried out
for 2 h at 37°C in darkness. Finally, the absorbance in each well
at 450 nm was measured using a microplate reader (Bio-Tek, USA).
The experiment was conducted in three separate wells for each
sample, and performed in triplicate.
5-Ethynyl-2′-deoxyuridine (EdU)
staining
Cell proliferation was determined by the Cell-Light
EdU DNA Cell Proliferation Kit (RiboBio Co., Ltd.) according to the
manufacturer's instructions. Briefly, the transfected cells were
seeded in triplicate in 96-well plates at a density of
3×103 cells per well, and then exposed to 50 mM EdU for
2 h at 37°C. After collection, the cells were fixed with 4%
formaldehyde at room temperature for 20 min and incubated with 100
µl of 1X Apollo® reaction (Ribobio, Guangzhou, China)
for 30 min. The cells were subsequently incubated with DAPI
detecting liquid for 10 sec followed by microscopic observation
using a fluorescent microscope (magnification ×200; Olympus Corp.,
Tokyo, Japan). The percentage of the EdU-positive relative to the
DAPI-positive cells was calculated by ImageJ software (ImageJ
bundled with 64-bit Java 1.8.0 112; National Institutes of Health,
Bethesda, MD, USA). The experiment was conducted in three separate
wells for each sample, and performed in triplicate.
Cell cycle and apoptosis assay
For cell cycle analysis, 5×105
transfected cells were harvested by trypsinization, washed two
times with PBS, and then fixed in 75% ice-cold ethanol overnight at
4°C. The fixed cells were resuspended in PBS and stained with
propidium iodide (PI) (50 µg/ml) containing RNase (0.1 µg/ml). Cell
cycle distribution analysis was performed using FACSan flow
cytometry (BD Biosciences, San Jose, CA, USA). Data are presented
as the percentages of cells in the G0/G1, S and G2/M stages.
Apoptosis analysis was performed using Annexin
V-FITC Kit (Abcam, Cambridge, UK). In brief, transfected cells were
harvested by trypsinization and resuspended in binding buffer. Then
the cells were stained with Annexin V-FITC and PI for 15 min at
room temperature. Cell apoptotic rates were assessed by FACSan flow
cytometry (BD Biosciences).
Hoechst 33258 staining
Following cell transfection, cells at the density of
3×105 were plated in 6-well plates and further cultured
for 12 h. Then cells were washed twice with pre-cooled PBS after
the upper culture solution was carefully discarded. Subsequently,
cells were fixed with 4% paraformaldehyde for 10 min and washed
with distilled water, followed by staining with Hoechst 33258
(Thermo Fisher Scientific, Inc.) for 20 min in the dark. Then, the
cells were washed with distilled water, after which the morphology
of cells was observed under a fluorescence microscope
(magnification ×200; Olympus Corp.).
Western blot analysis
Total protein was isolated from cells using RIPA
Lysis Buffer (Thermo Fisher Scientific, Inc.). After collecting the
supernatants, the protein concentration was determined with a BCA
protein assay kit (Bio-Rad Laboratories, Inc.). Equal amounts (20
µg) of total protein were separated by 12% sodium dodecyl
sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and
transferred onto PVDF membranes (Millipore). Then, the membranes
were blocked with 5% non-fat dry milk in 1X TBS containing 0.05%
Tween-20 at room temperature for 1 h. Next, the membranes were
incubated with primary antibodies against MAPK (dilution 1:1,000,
ab205926; Abcam), PI3K (dilution 1:500; cat. no. ab70912; Abcam),
p-PI3K (dilution 1:800, ab182651; Abcam), AKT (dilution 1:1,000;
cat. no. ab8805; Abcam), p-AKT (dilution 1:800; cat. no. ab38449;
Abcam), Bcl-2 (dilution 1:1,000; cat. no. ab32124; Abcam), Bax
(dilution 1:1000; cat. no. ab32503; Abcam) at 4°C overnight and
GAPDH (dilution 1:10,000; Cell Signaling Technology) used as
internal control. Afterwards, the membranes were incubated with
HRP-conjugated secondary antibodies (cat. nos. sc-516102 or
sc-2357; Santa Cruz Biotechnology) for 2 h at room temperature. The
protein signal was detected and visualized using an enhanced
chemiluminescence reagent (Thermo Fisher Scientific, Inc.).
Densitometric quantification was determined using Bio-Rad Quantity
One software v.4.6.3 (Bio-Rad Laboratories, Hercules, CA, USA).
Statistical analysis
The fold-change of each circRNA was computed from
the profile difference between the cancer and control groups, and
the significance was analyzed with a paired t-test. All the in
vitro data are expressed as the mean ± standard deviation (SD)
from at least triplicate trials. Statistical analyses were
performed by GraphPad Prism 5 (GraphPad Software, Inc., La Jolla,
CA, USA). The statistical significance for multi-group comparisons
was assessed using one-way ANOVA test with Tukey's test. A P-value
<0.05 was considered to indicate statistical significance.
Results
CircRNA expression profiles in
OSCC
To study the potential involvement of circRNAs in
OSCC, a high throughput circRNA microarray assay was performed in
the three pairs of human OSCC and matched normal tissue. Fig. 1 is a hierarchical cluster displaying
the circRNA expression pattern in the OSCC and adjacent tissues.
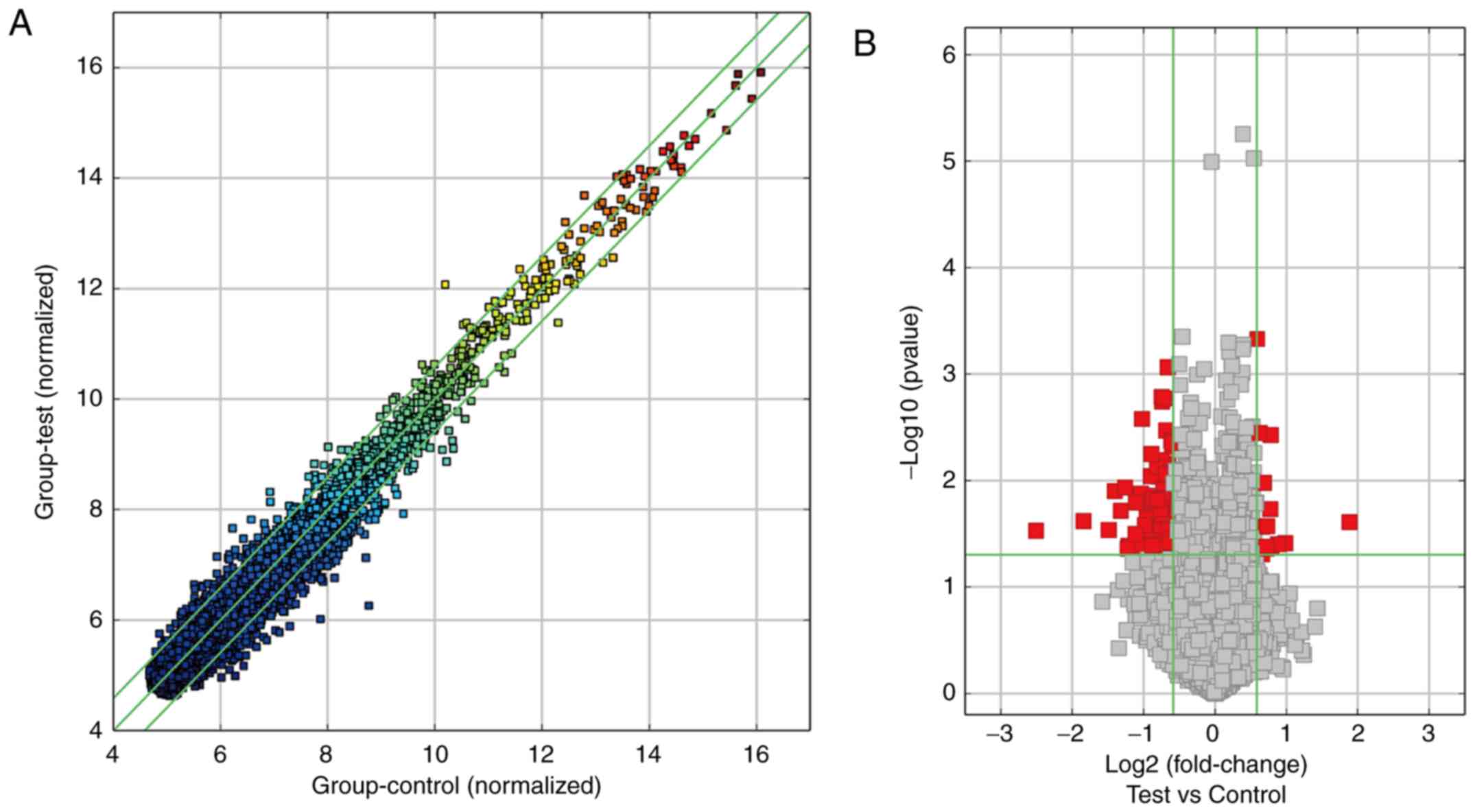
The scatter plot of circRNA expression profile was used to describe
the variation in circRNA expression between OSCC and adjacent
tissues (Fig. 2A). The statistical
significance of the differentially expressed circRNAs between the
OSCC and adjacent tissues was additionally visualized using volcano
plot filtering (Fig. 2B). Based on
the filter criteria (fold-change ≥1.5 and a P-value <0.05), a
total of 213 significantly differentially expressed circRNAs were
identified, including 124 upregulated and 89 downregulated circRNAs
in the OSCC tissues compared with adjacent tissues. Considering
that false positives can be caused by multi-comparisons, we used
the FDR method to adjust the P-values. After FDR correction, the
top 20 differentially expressed circRNAs, including 10
significantly upregulated and 10 significantly downregulated
circRNAs were identified, which are listed in Table III.
 | Table III.Top 20 differentially expressed
circRNAs in the microarray data in oral squamous cell
carcinoma. |
Table III.
Top 20 differentially expressed
circRNAs in the microarray data in oral squamous cell
carcinoma.
| CircRNAs | P-value |
Log2FC | CircRNA_type | Chrom | Strand | Gene symbol |
|---|
| Upregulated
circRNAs |
|
hsa_circRNA_043621 | 0.0245729 | 3.6875525 | Exonic | chr17 | – | KRT14 |
|
hsa_circRNA_101996 | 0.0386753 | 1.9776622 | Exonic | chr17 | + | SPECC1 |
|
hsa_circRNA_102068 | 0.0396215 | 1.8445549 | Exonic | chr17 | – | TNS4 |
|
hsa_circRNA_088200 | 0.0422007 | 1.7255309 | Exonic | chr9 | – | TNC |
|
hsa_circRNA_100045 | 0.0037498 | 1.7117976 | Exonic | chr1 | – |
CTNNBIP1 |
|
hsa_circRNA_101217 | 0.0185312 | 1.7076297 | Exonic | chr12 | – | ANKLE2 |
|
hsa_circRNA_404462 | 0.0268066 | 1.6557805 | Intronic | chr1 | + |
BC016143 |
|
hsa_circRNA_102733 | 0.0416789 | 1.6436409 | Exonic | chr2 | – | XPO1 |
|
hsa_circRNA_058819 | 0.0436044 | 1.6430251 | Exonic | chr2 | – | COL6A3 |
|
hsa_circRNA_066361 | 0.0104826 | 1.6047044 | Exonic | chr3 | + | FLNB |
| Downregulated
circRNAs |
|
hsa_circRNA_102459 | 0.0296076 | −5.6804226 | Exonic | chr19 | + | MAST1 |
|
hsa_circRNA_101036 | 0.0239938 | −3.591020 | Exonic | chr12 | – | TMTC1 |
|
hsa_circRNA_102034 | 0.0290761 | −2.8047351 | Exonic | chr17 | + | RHOT1 |
|
hsa_circRNA_005882 | 0.0125354 | −2.6437297 | Exonic | chr2 | – | STK39 |
|
hsa_circRNA_037767 | 0.0191608 | −2.4977462 | Exonic | chr16 | – | PPL |
|
hsa_circRNA_003949 | 0.0116305 | −2.3968409 | Exonic | chr7 | – | PTN |
|
hsa_circRNA_402901 | 0.0435932 | −2.3361872 | Exonic | chr3 | – | EIF4E3 |
|
hsa_circRNA_100259 | 0.0408402 | −2.3186901 | Exonic | chr1 | – | WDR78 |
|
hsa_circRNA_405247 | 0.0317352 | −2.1636677 | Intronic | chr14 | + | FUT8 |
|
hsa_circRNA_100245 | 0.0160437 | −2.1563291 | Exonic | chr1 | + | FGGY |
GO enrichment and functional category
analysis
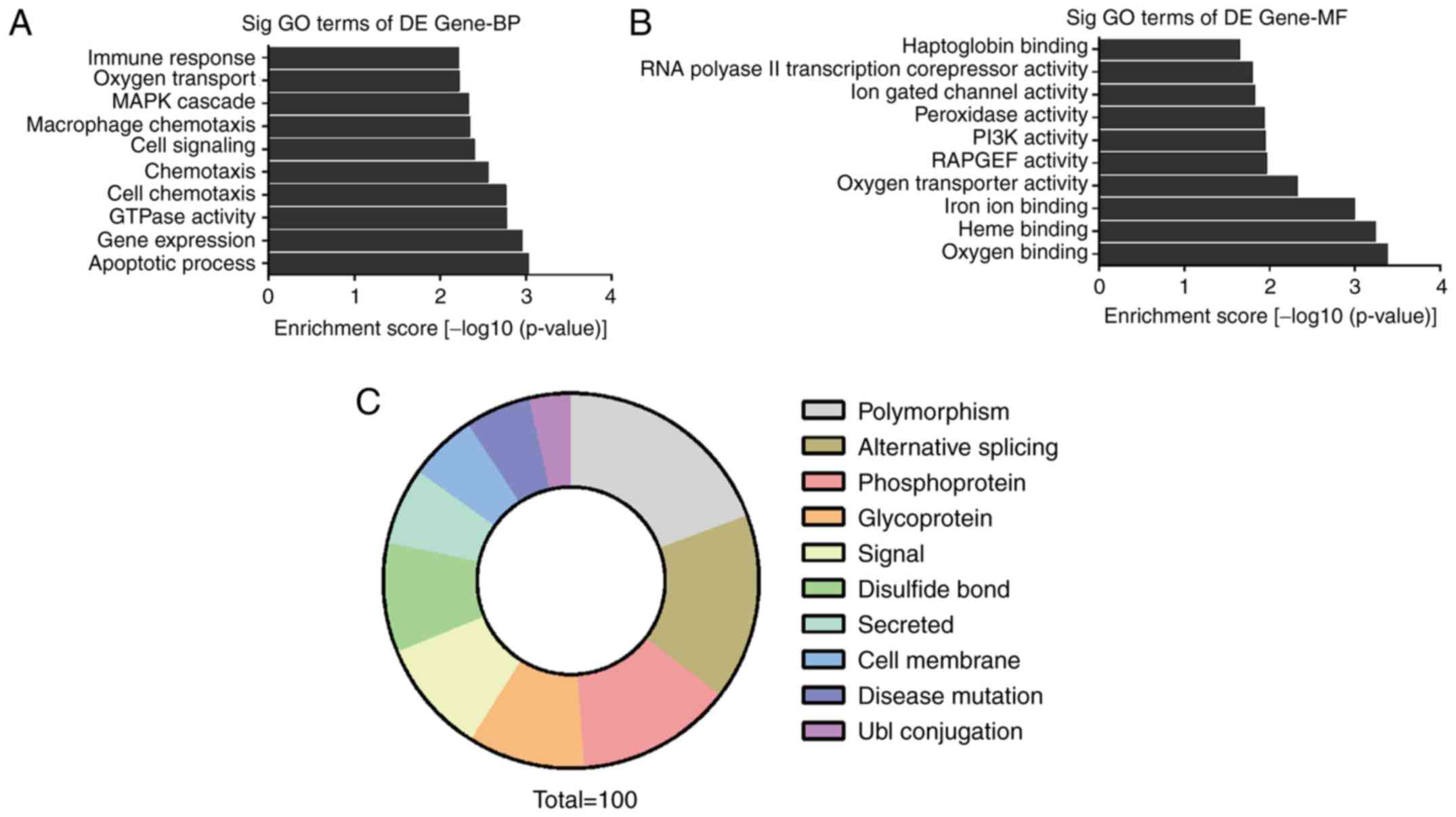
To explore the expression of circRNAs on a more
functional level, we performed GO enrichment analysis for the genes
involved with the circRNAs that were found to be differentially
expressed in the circRNA microarray results. The count number >2
and FDR <0.05 were chosen as cut-off criteria. The results
showed that the most significant enriched GO terms in the
biological processes (BP) included ‘MAPK cascade’, ‘cell signaling’
and ‘apoptotic process’ (Fig. 3A).
The most significant enriched GO terms in molecular functions (MF)
included ‘oxygen binding’, ‘PI3K activity’ and ‘Heme binding’
(Fig. 3B). Functional categories
analysis revealed that pathways such as ‘phosphoprotein’ and
‘alternative splicing’ were related to the differentially expressed
circRNAs (Fig. 3C).
Validation of the deregulated
circRNAs
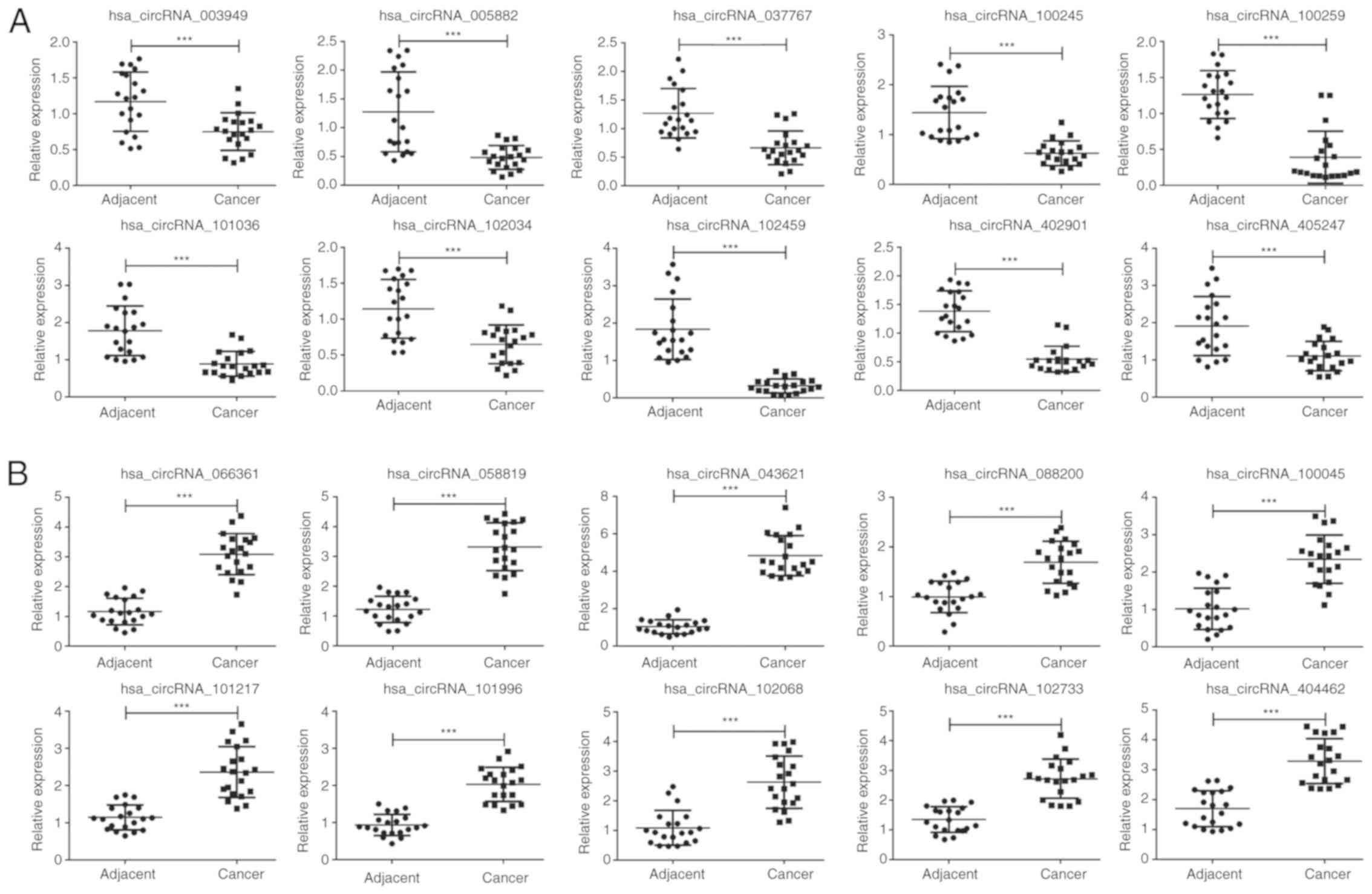
To verify that the differentially expressed circRNAs
discovered through the microarray were accurately identified, we
selected 20 potentially significant circRNAs involved in the MAPK
and PI3K signaling pathways, including 10 upregulated and 10
downregulated circRNAs for validation by qPCR in 20 pairs of human
OSCC and matched normal tissues. As expect, the expression patterns
of the 10 downregulated (Fig. 4A,
P<0.001) and 10 upregulated circRNAs (Fig. 4B, P<0.001) were consistent with the
microarray data. Notably, hsa_circRNA_102459 and hsa_circRNA_043621
displayed the lowest and highest expression in the 20 OSCC tissues
relative to the normal tissues, respectively, which may play a
crucial role in the development and progression of OSCC. In
addition, circRNA-miRNA network prediction was conducted, and the
results revealed that there are numerous miRNAs that are able to
interact with hsa_circRNA_102459 and hsa_circRNA_043621 (Fig. S1).
Table I shows the
correlation between circRNA_ 043621/circ_102459 and the
clinicopathological characteristics of the 20 OSCC patients.
Specifically, there were no obviously significant correlation
between expression and patient age and sex (P>0.05). However,
clinicopathological characteristics, including clinical stage,
tumor differentiation and lymph node metastasis presented
significant difference in regards to the expression of
circRNA_043621 and circRNA_102459 (P<0.05, Table I).
Upregulation of circRNA_102459 or
downregulation of circRNA_043621 significantly suppresses OSCC cell
proliferation
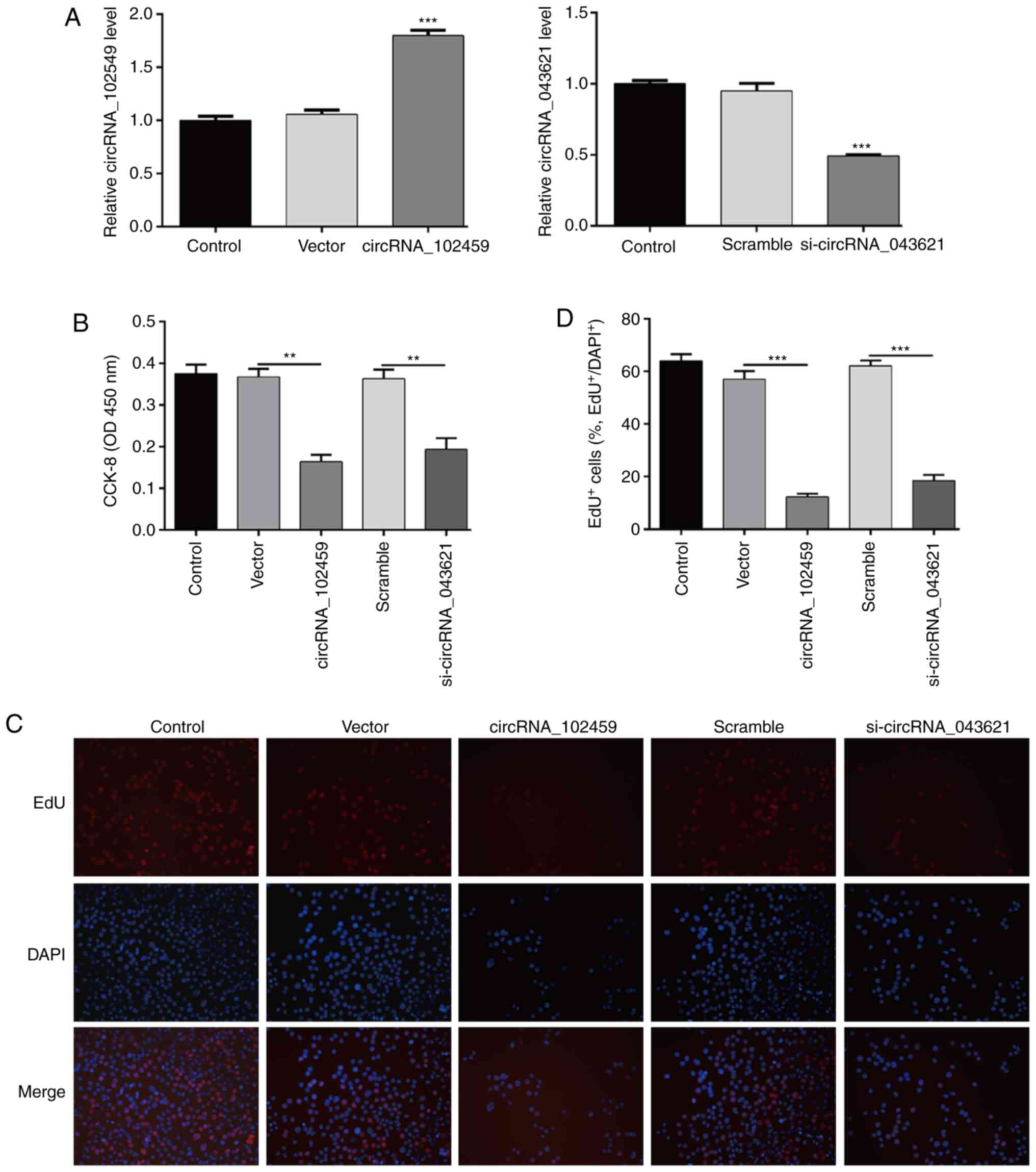
Next, we performed gain-of-function and
loss-of-function assays to evaluate the biological functions of
circRNA_102459 and circRNA_043621 in OSCC cells, respectively. As
shown in Fig. 5A, qPCR analysis
demonstrated that following circRNA_102459 transfection the
expression of circRNA_102459 was significantly elevated in the
TSCC1 cells (P<0.001), while si-circRNA_043621 transfection
remarkably reduced the expression of circRNA_043621 (P<0.001) in
the TSCC1 cells. Subsequent CCK-8 assay revealed that upregulation
of circRNA_102459 and downregulation of circRNA_043621
significantly suppressed the growth of TSCC1 cells (Fig. 5B, P<0.01). In addition, the linear
RNA expression levels of circRNA_102459 and circRNA_043621 did not
change various to overexpression and knockdown of circRNA_102459
and circRNA_043621 (Fig. S2).
Furthermore, 5-ethynyl-2′-deoxyuridine (EdU) staining assay showed
that the proliferation potential of TSCC1 cells was impaired upon
upregulation of circRNA_102459 or downregulation of circRNA_043621
(Fig. 5C and D, P<0.001).
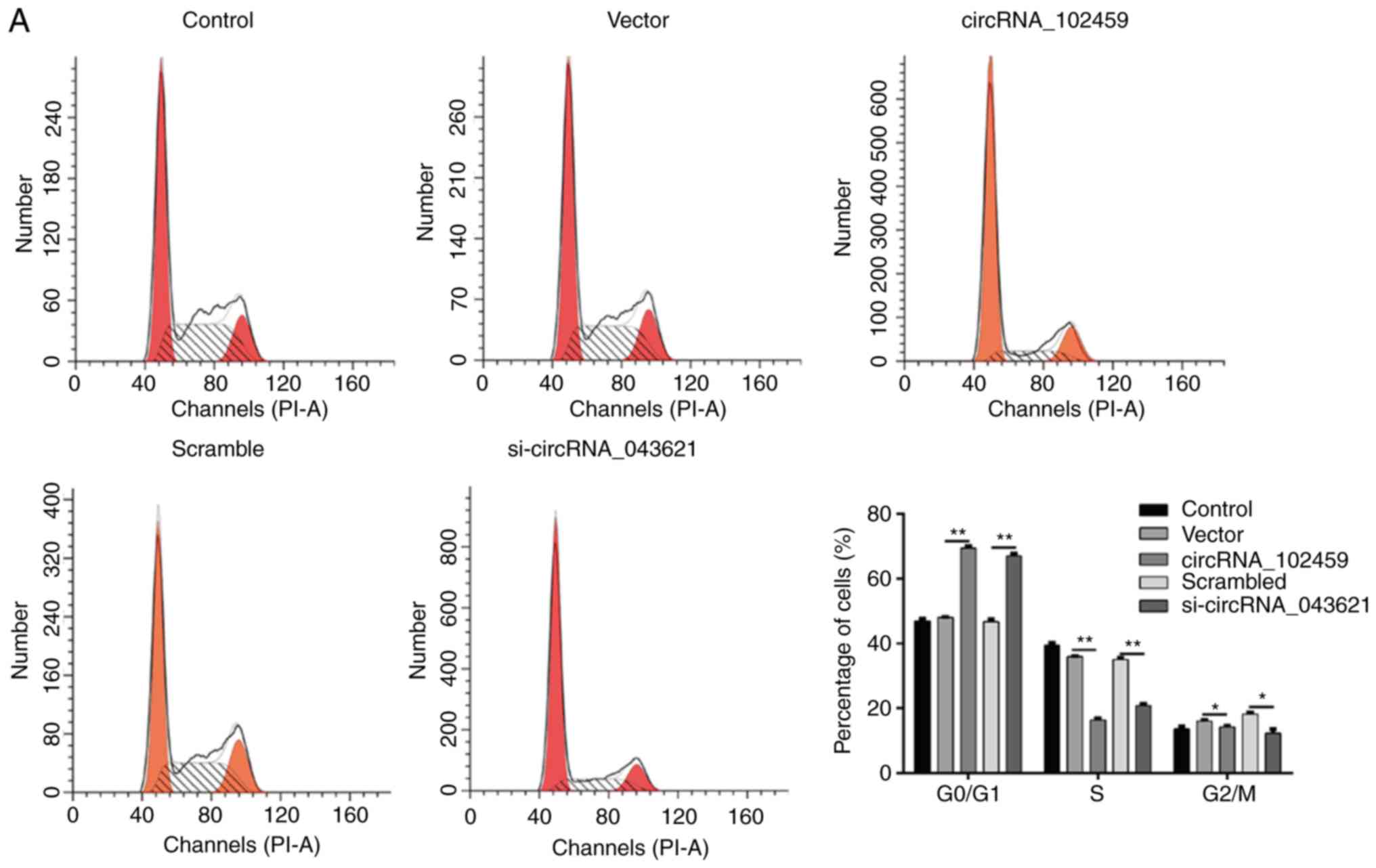
Upregulation of circRNA_102459 or
downregulation of circRNA_043621 causes cell cycle G0/G1 phase
arrest and promotes apoptosis in OSCC cells
To determine whether the effects of circRNA_102459
or circRNA_043621 on the proliferation of OSCC cells are mediated
by inhibition of cell cycle progression, flow cytometry was used to
evaluate the cell cycle distribution in TSCC1 cells. As shown in
Fig. 6A, circRNA_102459
overexpression or circRNA_043621 knockdown led to the significant
accumulation of cells in the G0/G1-phase (P<0.01) and a
significant decrease in cells in the S-phase (P<0.01) and
G2/M-phase (P<0.05) compared with the corresponding control.
Furthermore, the effects of circRNA_102459 or circRNA_043621 on
apoptosis were assessed in TSCC1 cells. The percentage of cells
undergoing apoptosis, including early apoptosis and late apoptosis,
was significantly increased in the circRNA_102459 overexpression or
circRNA_043621 knockdown group compared with the corresponding
control (Fig. 6B, P<0.05).
Apoptosis could also be observed when cells were stained with
Hoechst 33258. Here, circRNA_102459 overexpression or
circRNA_043621 knockdown induced morphological changes
characteristic of apoptosis such as chromatin condensation and
nuclear blebbing (Fig. 6C). Taken
together, these data suggest that circRNA_102459 overexpression or
circRNA_043621 knockdown induces G0/G1 phase arrest and enhances
apoptosis in OSCC cells.
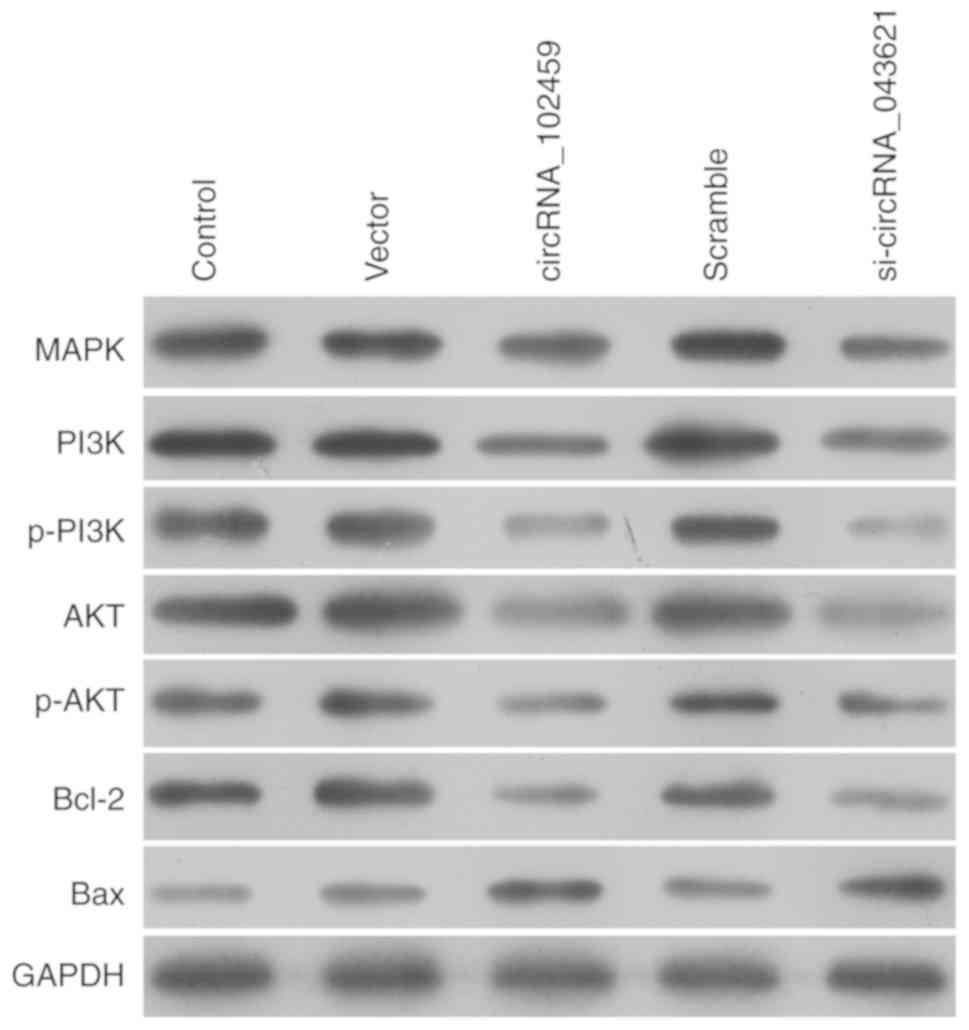
CircRNA_102459 and circRNA_043621
mediate the expression of molecules associated with MAPK, PI3K/Akt
and apoptotic signaling in OSCC cells
To explore the potential mechanism underlying the
role of circRNA_102459 or circRNA_043621 in cell proliferation, the
activation of the MAPK and PI3K/Akt pathways was assessed using
western blotting. As shown in Fig. 7,
circRNA_102459 overexpression or circRNA_043621 knockdown
downregulated the expression of p38 MAPK. The PI3K/Akt pathway was
suppressed by circRNA_102459 overexpression or circRNA_043621
knockdown, as revealed by decreased PI3K and Akt phosphorylation.
In addition, we found that circRNA_102459 overexpression or
circRNA_043621 knockdown obviously reduced anti-apoptotic Bcl-2
expression, but elevated pro-apoptotic Bax expression in the TSCC1
cells.
Discussion
It is well known that the initiation and progression
of OSCC is a complex pathological process. Studies suggest that
noncoding circRNAs are potential new diagnostic markers for
diseases, especially cancer. To date, several studies have reported
that circRNAs function as important regulators in OSCC (14,15).
However, the global circRNA expression profile in OSCC has not been
fully uncovered.
In the present study, we performed circRNA
microarray analysis to identify a number of aberrantly expressed
circRNAs in OSCC compared with adjacent tissues. Through GO and
Functional categories analysis, a total of 20 significantly
differentially expressed circRNAs were screened to be involved in
MAPK and PI3K signaling pathways. Accumulating evidence indicates
that simultaneous activation of the MAPK and PI3K/AKT pathways is
frequently observed to promote the progression of cancers (19,20). We
thus utilized qPCR to validate these 20 differentially expressed
circRNAs, including 10 upregulated and 10 downregulated circRNAs in
another 40 OSCC tissues and paired adjacent normal tissues. As
expect, the results of qPCR analysis were consistent with the
microarray data, confirming that the microarray data were
reliable.
Importantly, hsa_circRNA_102459 and
hsa_circRNA_043621 were further screened to study their biological
function in TCSS1 cells as they displayed the lowest and highest
expression in 20 OSCC tissues relative to normal tissues,
respectively, which might play a crucial role in the development
and progression of OSCC. Hsa_circRNA_102459 (MAST1) is a member of
the MAST family genes and its recurrent gene rearrangements play
key roles in subsets of carcinomas (21). Hsa_circRNA_043621 (KRT14) is a
valuable hub gene involved in the immune response and tumor cell
development in tumorigenesis (22,23).
Considering their potential correlation with the progression of
tumors, we performed loss-of-function and rescue assays to evaluate
the biological functions of circRNA_102459 and circRNA_043621 in
OSCC cells, respectively. The results demonstrated that
upregulation of circRNA_102459 and downregulation of circRNA_043621
significantly suppressed TSCC1 cell proliferation, induced cell
cycle G0/G1 phase arrest and promoted apoptosis, suggesting that
circRNA_102459 and circRNA_043621 are closely associated with tumor
cell proliferation. However, in the present study, we used only one
cell line TSCC1 for our research, and thus this is a limitation of
the present study. Two or more cell lines will be used to validate
these findings in future research by our group.
The present study further investigated the
activation of the MAPK and PI3K/Akt pathways by measuring the
phosphorylation of proteins in the MAPK and PI3K/Akt pathways,
including p38 MAPK, p-PI3K and p-AKT, in
circRNA_102459-overexpressing and circRNA_043621-knockdown TSCC1
cells. When circRNA_102459 was overexpressed, or circRNA_043621 was
silenced, the MAPK and PI3K/Akt pathways were suppressed, as
presented by decreased p38 MAPK, p-PI3K and p-Akt levels.
Consistently, circRNA_043621 (KRT14) was found to be increased upon
mortalin overexpression, which is involved in PI3K/Akt signaling
contributing to EMT of metastatic cancer (24). In addition, it was found that
circRNA_102459 overexpressing and circRNA_043621 silencing
significantly upregulated Bcl-2, but downregulated Bax expression
in the TSCC1 cells. Apoptosis is a process of programmed cell death
and its activation provides a new direction for the treatment of
cancer, among which the Bcl-2 family plays a pivotal role in either
inhibiting Bcl-2 or promoting Bax expression (25,26). Thus,
we infer that circRNA_102459 and circRNA_043621 affect the process
of cell apoptosis by influencing changes in Bcl-2 family
members.
In summary, the present study provided a profile of
circRNAs in OSCC and adjacent tissues. We discovered that
circRNA_102459 and circRNA_043621 may function as a tumor
suppressor and promoter, respectively, of OSCC carcinogenesis, and
thus may be valuable diagnostic biomarkers of OSCC. However, some
limitations must be considered in the interpretation of ours
results, including the limited sample size and in vivo
experimental validation. Therefore, further specific studies are
still needed to decipher whether circRNA_102459 and circRNA_043621
participate in cancer-related pathways and sequester miRNAs.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
This research was supported by the National Natural
Science Foundation of China (no. 81360172).
Availability of data and materials
The datasets used during the present study are
available from the corresponding author upon reasonable
request.
Authors' contributions
SZ contributed to the conception and design of the
study. WD and WP performed the experiments. TW and JC completed
data analysis and interpretation. XQ and LF contributed to writing
the manuscript. All authors read and approved the final manuscript.
All authors read and approved the final manuscript and agree to be
accountable for all aspects of the research in ensuring that the
accuracy or integrity of any part of the work are appropriately
investigated and resolved.
Ethics approval and consent to
participate
This study was approved by the Ethics Committee of
Hainan General Hospital (Haikou, Hainan, China). Written informed
consent was provided by all patients prior to participation.
Patient consent for publication
Not applicable.
Competing interests
The authors state that they have no competing
interests.
References
|
1
|
Mignogna MD, Fedele S and Lo Russo L: The
world cancer report and the burden of oral cancer. Eur J Cancer
Prev. 13:139–142. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
van Zyl A and Bunn BK: Clinical features
of oral cancer. SADJ. 67:566–569. 2012.PubMed/NCBI
|
|
3
|
Blatt S, Kruger M, Ziebart T, Sagheb K,
Schiegnitz E, Goetze E, Al-Nawas B and Pabst AM: Biomarkers in
diagnosis and therapy of oral squamous cell carcinoma: A review of
the literature. J Craniomaxillofac Surg. 45:722–730. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhao X, Sun S, Zeng X and Cui L:
Expression profiles analysis identifies a novel three-mRNA
signature to predict overall survival in oral squamous cell
carcinoma. Am J Cancer Res. 8:450–461. 2018.PubMed/NCBI
|
|
5
|
Wilusz JE and Sharp PA: Molecular biology.
A circuitous route to noncoding RNA. Science. 340:440–441. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hentze MW and Preiss T: Circular RNAs:
Splicing's enigma variations. EMBO J. 32:923–925. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Memczak S, Jens M, Elefsinioti A, Torti F,
Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer
M, et al: Circular RNAs are a large class of animal RNAs with
regulatory potency. Nature. 495:333–338. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Du WW, Yang W, Chen Y, Wu ZK, Foster FS,
Yang Z, Li X and Yang BB: Foxo3 circular RNA promotes cardiac
senescence by modulating multiple factors associated with stress
and senescence responses. Eur Heart J. 38:1402–1412.
2017.PubMed/NCBI
|
|
9
|
Burd CE, Jeck WR, Liu Y, Sanoff HK, Wang Z
and Sharpless NE: Expression of linear and novel circular forms of
an INK4/ARF-associated non-coding RNA correlates with
atherosclerosis risk. PLoS Genet. 6:e10012332010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lukiw WJ: Circular RNA (circRNA) in
Alzheimer's disease (AD). Front Genet. 4:3072013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhang J, Zhao X, Zhang J, Zheng X and Li
F: Circular RNA hsa_circ_0023404 exerts an oncogenic role in
cervical cancer through regulating miR-136/TFCP2/YAP pathway.
Biochem Biophys Res Commun. 22:428–433. 2018. View Article : Google Scholar
|
|
12
|
Li P, Yang X, Yuan W, Yang C, Zhang X, Han
J, Wang J, Deng X, Yang H, Li P, et al: CircRNA-Cdr1as exerts
anti-oncogenic functions in bladder cancer by sponging
microRNA-135a. Cell Physiol Biochem. 46:1606–1616. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Liu W, Ma W, Yuan Y, Zhang Y and Sun S:
Circular RNA hsa_circRNA_103809 promotes lung cancer progression
via facilitating ZNF121-dependent MYC expression by sequestering
miR-4302. Biochem Biophys Res Commun. 500:846–851. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wang L, Wei Y, Yan Y, Wang H, Yang J,
Zheng Z, Zha J, Bo P, Tang Y, Guo X, et al: CircDOCK1 suppresses
cell apoptosis via inhibition of miR196a5p by targeting BIRC3 in
OSCC. Oncol Rep. 39:951–966. 2018.PubMed/NCBI
|
|
15
|
Chen L, Zhang S, Wu J, Cui J, Zhong L,
Zeng L and Ge S: CircRNA_100290 plays a role in oral cancer by
functioning as a sponge of the miR-29 family. Oncogene.
36:4551–4561. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Galasso M, Costantino G, Pasquali L,
Minotti L, Baldassari F, Corrà F, Agnoletto C and Volinia S:
Profiling of the predicted circular RNAs in ductal in situ and
invasive breast cancer: A pilot study. Int J Genomics.
2016:45038402016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Nair AA, Niu N, Tang X, Thompson KJ, Wang
L, Kocher JP, Subramanian S and Kalari KR: Circular RNAs and their
associations with breast cancer subtypes. Oncotarget. 7:809672016.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hou P, Liu D, Shan Y, Hu S, Studeman K,
Condouris S, Wang Y, Trink A, El-Naggar AK and Tallini G: Genetic
alterations and their relationship in the phosphatidylinositol
3-kinase/Akt pathway in thyroid cancer. Clin Cancer Res.
13:11612007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Li S, Ma YM, Zheng PS and Zhang P: GDF15
promotes the proliferation of cervical cancer cells by
phosphorylating AKT1 and Erk1/2 through the receptor ErbB2. J Exp
Clin Cancer Res. 37:802018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Robinson DR, Kalyana-Sundaram S, Wu YM,
Shankar S, Cao X, Ateeq B, Asangani IA, Iyer M, Maher CA, Grasso
CS, et al: Functionally recurrent rearrangements of the MAST kinase
and Notch gene families in breast cancer. Nat Med. 17:1646–1651.
2011. View
Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wang LX, Li Y and Chen GZ: Network-based
co-expression analysis for exploring the potential diagnostic
biomarkers of metastatic melanoma. PLoS One. 13:e01904472018.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zheng Y, Zhao G, Xu B, Liu C, Li C, Zhang
X and Chang X: PADI4 has genetic susceptibility to gastric
carcinoma and upregulates CXCR2, KRT14 and TNF-α expression levels.
Oncotarget. 7:62159–62176. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Na Y, Kaul SC, Ryu J, Lee JS, Ahn HM, Kaul
Z, Kalra RS, Li L, Widodo N, Yun CO and Wadhwa R: Stress chaperone
mortalin contributes to epithelial-mesenchymal transition and
cancer metastasis. Cancer Res. 76:2754–2765. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yu C, Liu SL, Qi MH, Zou X, Wu J and Zhang
J: Herbal medicine guan chang fu fang enhances 5-fluorouracil
cytotoxicity and affects drug-associated genes in human colorectal
carcinoma cells. Oncol Lett. 9:701–708. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Brunelle JK and Letai A: Control of
mitochondrial apoptosis by the Bcl-2 family. J Cell Sci.
122:437–441. 2009. View Article : Google Scholar : PubMed/NCBI
|