Introduction
Chronic myeloid leukemia (CML) arises from
pluripotent hematopoietic stem cells by a reciprocal translocation
between chromosomes 9 and 22, forming Philadelphia chromosome (Ph)
(1). The product of this genetic
rearrangement consists of BCR-ABL fusion protein with deregulated
tyrosine kinase activity, leading to an endless proliferation of
immune precursors (2). Over the past
two decades, BCR-ABL tyrosine kinase inhibitors (TKIs) have
markedly improved the treatment of CML (3–5). Despite
these great advances, therapy-resistant leukemic stem cells (LSCs)
still persist after TKI treatment, presenting an obstacle on the
road to long-term progression-free survival (PFS) in CML patients
(6). Recent findings have revealed
that BCR-ABL fusion protein still had leukemogenic ability without
tyrosine kinase activity (7,8), and the survival of CML stem cells was
independent of tyrosine kinase activity of this fusion protein
(9). These findings strongly call for
new targeting strategies of BCR-ABL for CML treatment.
It is well established that downregulation of the
BCR-ABL protein level could suppress the growth of CML cells, and
many attempts have been made to achieve this purpose. Goussetis
et al revealed that autophagic degradation of BCR-ABL fusion
protein was critical for the potent exhibition of the anti-leukemic
effects of As2O3 (10). According to previous studies, genetic
knockdown technologies, such as antisense oligonucleotides and
small interfering RNA, have been used for the downregulation of the
protein levels of BCR-ABL in CML cells (11–13).
Recently, potent degraders against BCR-ABL have been developed by
conjugating dasatinib to ligands for E3 ubiquitin ligases,
revealing a more sustained inhibition of CML cell growth than ABL
kinase inhibitor (14).
Homoharringtonine (HHT), a plant alkaloid with
antitumor properties, was originally identified nearly 40 years ago
(15), and has been revealed to play
an important role in the treatment of CML, both before and after
the clinical use of TKIs (16,17). In
2010, the use of HHT for relapsed/refractory CML was approved by
the FDA (18). HHT is an alternative
treatment for patients with CML who exhibit resistance or
intolerance to TKIs. The drug involves a distinct mechanism and may
be able to inhibit leukemic stem cells, which play a key role in
the progression of CML (19). There
are different theories regarding the mechanisms of HHT. First, it
is widely considered that HHT exerts a role in the treatment of
leukemia. This is mainly achieved by inhibiting the synthesis of
proteins associated with cell apoptosis and cell survival, such as
Mcl-1, XIAP, and Myc (20–22). Recently, it was reported that HHT is
involved in other mechanisms. For example, it can regulate
alternative splicing of Bcl-x and caspase-9 and regulate Smad3
protein tyrosine kinase phosphorylation (23,24).
Therefore, understanding the mechanism of HHT is very complex and
has not been fully elucidated yet. In particular, it is unclear as
to how HHT regulates the oncoprotein BCR-ABL. Although it has been
reported that HHT could downregulate the BCR-ABL protein by
inhibiting protein synthesis (25),
it is speculated that there may be more specific mechanisms
involved. In the present study, it was reported that HHT promoted
oncoprotein BCR-ABL degradation and cell apoptosis in the
imatinib-resistant CML K562G cell line, and the p62-mediated
autophagy-lysosome pathway was involved in BCR-ABL protein
degradation induced by HHT. Depletion of p62 or inhibition of
autophagy not only reversed HHT-induced BCR-ABL protein
degradation, but also affected cytotoxicity of HHT on CML
cells.
Materials and methods
Cell culture and treatment
K562 and K562G cells were provided by Professor Jie
Jin (The First Affiliated Hospital of Zhejiang University). The
cells were maintained in RPMI-1640 medium (Gibco; Thermo Fisher
Scientific, Inc.) with 10% fetal bovine serum (FBS; Gibco; Thermo
Fisher Scientific, Inc.) at 37°C in 5% CO2 atmosphere.
Imatinib-resistant K562G cells was established by culturing cells
in gradually increasing concentrations of imatinib and gained
resistance to imatinib over several months of culture (Appendix I).
However, K562G cells do not contain mutations within the TK domain.
Autophagic inhibitor Bafilomycin A1 (Baf-A1) was purchased from
Abcam, autophagic inhibitor chloroquine (CQ), proteasome inhibitor
MG132 and cycloheximide (CHX) were purchased from Sigma-Aldrich
(Merck KGaA). HHT was purchased from Selleckchem (cat. no. s9015).
Protein A/G plus agarose beads were obtained from Thermo Fisher
Scientific, Inc. (cat. no. 20423). Compounds were dissolved in
dimethyl sulfoxide (DMSO) and added to culture media until it
reaches a final concentration of 0.1% DMSO. For the vehicle
control, 0.1% DMSO alone was used.
Cell viability assay
K562, K562G, and siRNA-K562G cells were seeded in
384-well plates at a density of 8,000 cells/well, and then treated
with 0.1% DMSO (vehicle control) or HHT at concentrations of 100,
50, 25, 12.5, 6.2, 3.1, 1.56, 0.78 and 0.39 ng/ml for 48 h. Cell
proliferation was assayed using an MTS assay (Promega Corp.)
according to the manufacturer's instructions and the absorbance was
measured at 490 nm using the EnVision microplate reader
(PerkinElmer).
Western blotting and antibodies
Cells were lysed using RIPA buffer (pH 7.4)
containing a protease inhibitor cocktail (Roche Diagnostics).
Protein concentrations of the extracts were measured by BCA™
protein assay kit (Pierce; Thermo Fisher Scientific, Inc.) and
equal amounts (20 µg) of total protein from each sample was
separated by SDS-PAGE and then were transferred onto NC
(nitrocellulose) membrane (Millipore, Minneapolis, MN, USA). NC
membranes were blocked with 10% skim milk powder dissolved in TBST
buffer at room temperature for 1 h. Then, NC membranes were washed
with TBST, and then incubated with a primary antibody (dilution of
1:1,000) in TBST containing 5% bovine serum albumin (BSA) overnight
at 4°C. Next, membranes were washed with TBST three times and
incubated with an HRP-conjugated anti-rabbit (dilution 1:10,000;
cat. no. A16096) or anti-mouse secondary antibody (dilution
1:10,000; cat. no. 31430) (Thermo Fisher Scientific, Inc.) for 1 h
at room temperature. The signals were detected using an ECL reagent
(Pierce; Thermo Fisher Scientific, Inc.). All western blots were
replicated in three times and the intensity of the western blot
signals was analyzed using ImageJ 1.8.0 analysis software (NIH;
National Institutes of Health, Bethesda, MD, USA). The primary
antibodies were as follows: Antibody against C-Abl (cat. no.
sc-887) and antibody against ubiquitin (Ub; cat. no. sc-166553;
both from Santa Cruz Biotechnology, Inc.), antibodies against p62
(cat. no. PM045; MBL Beijing Biotech Co., Ltd.), antibodies against
LC3B (cat. no. 2775), antibodies against ATG5 (cat. no. 2630) and
antibodies against Beclin-1 (cat. no. 3495; all from Cell Signaling
Technology, Inc.), antibodies against β-tubulin (cat. no. HC101-02)
and antibodies against β-actin (cat. no. HC201-02; both from
Beijing Transgen Biotech Co., Ltd.), antibodies against poly
adenosine diphosphate (ADP)ribose polymerase (PARP; cat. no 9542)
and antibodies against cleave caspase3 (cat. no. 9664; both from
Cell Signaling Technology, Inc.).
Flow cytometry for apoptotic
analysis
The K562G cells were treated with concentrations of
0, 20, 50 and 80 ng/ml HHT for 24 h. Subsequently, the cells
underwent centrifugation at 100 × g centrifugation for 5 min, and
then were washed twice with cold PBS. The cells were resuspended
with 100 µl 1X binding buffer, followed by the addition of 5 µl
Annexin V-FITC and 10 µl PI staining solution (cat. no 556547; BD
Biosciences) for 15 min. Next, cell apoptosis was analyzed by flow
cytometry (BD FACSCalibur™).
Immunoprecipitation (IP)
K562G cells were treated with HHT and harvested at
different time-points (0, 2, 4, 8, 16 and 24 h). The cells were
then washed once and lysed in IP buffer for 30 min (1% NP40, 150 mM
NaCl, 5 mM KCl, 2 mM MgCl2, 1 mM EDTA, 50 mM Tris-HCl, pH 7.4) with
proteases (cat. no. 04693116001; Roche Diagnostics) and phosphatase
inhibitors. Lysates were cleared by centrifugation at 13,500 × g
and a fraction was saved as a loading input control. The remaining
lysates were incubated with p62 or c-Abl antibodies overnight at
4°C. Protein A/G beads (cat. no. 20423; Thermo Fisher Scientific,
Inc.) were added for another 4 h at 4°C. The beads were then washed
5–7 times in IP buffer, and the bound proteins were eluted by
directly adding SDS loading buffer with 2 mM DTT. Beads and the
saved lysates were analyzed by immunoblotting.
Fluorescence microscopy
Cells transfected with Ad-mcherry-GFP-LC3 (cat. no
C3011; Beyotime Institute of Biotechnology) were seeded in 96-well
plates at a density of 1×105/ml, and then were treated
with DMSO (vehicle control) or 50 ng/ml HHT for 8 h followed by
staining with 1 nM Hoechst 33342 (nuclear stain) for 10 min at
37°C. The cells were visualized using the Operetta CLS (Perkin
Elmer) at an ×40 magnification under excitation/emission filters
587/610 nm (mcherry) and 350/461 nm (Hoechst 33342). The resulting
images were then analyzed using Harmony software 4.6 (Perkin Elmer)
and autophagosomes were quantified by counting the number of spots
per mCherry-positive cells. Data represent the mean values of
puncta per cell for mCherry-positive cells, and error bars
represent standard deviations. Statistical analysis was performed
by Kruskal-Wallis test assuming equal variances. Representative
confocal microscopy images were captured using an Olympus
fluorescence microscope at an ×40 magnification [Olympus IX71
(fluorescence microscope); Olympus DP71 (CCD; Olympus Corp.].
Lentiviral infection and generation of
stable cell lines
K562G cells were transfected with a lentiviral
expression system for siRNA. LV-NC siRNA, LV-ATG5 siRNA,
LV-Beclin-1 siRNA and LV-p62 siRNA lentivirus stocks were obtained
from Shanghai GeneChem Co., Ltd. K562G cells were transfected with
lentiviruses targeting ATG5, Beclin-1 or p62 with 8 µg/ml of
Polybrene (cat. no. H9268; Sigma-Aldrich, Merck KGaA). Two days
after transfection, the medium was changed and 2 µg/ml of puromycin
was added to select stable knockdown of ATG5, Beclin-1 or p62 gene
K562G cells. Cells with stable reduction of ATG5, Beclin-1 or p62
were used to assess BCR-ABL levels and analyze the apoptotic
sensitivity to HHT. The targeting sequences were as follows: ATG5,
GAAGTTTGTCCTTCTGCTA; Beclin-1, CTCAGGAGAGGAGCCATTT; p62,
GCATTGAAGTTGATATCGAT.
Statistical analysis
One-way analysis of variance (ANOVA) and
Student-Newman-Keuls test were used to analyze the differences in
the target protein level of western blotting data from ImageJ
analysis. For the cell viability assay, data were analyzed using
GraphPad Prism 5.0 1 (GraphPad Software, Inc.). The results are
expressed as the means ± standard deviation. Differences were
considered to be statistically significant at P<0.05, P<0.01
and P<0.001 (*P<0.05, **P<0.01 and ***P<0.001 as
indicated in the figures and legends).
Results
HHT induces cell apoptosis and
downregulates BCR-ABL in imatinib-resistant K562G cells
The K562G cell line was transformed from K562 cells
and is widely used for leukemia research. To qualify the K562G cell
line in the present study, the sensitivity of K562G cells to
imatinib was verified. K562G exhibited high resistance to imatinib
with a 10-fold difference compared to K562 cells (IC50,
320 nM vs. 38 nM; P<0.05; Fig.
S1). The anti-proliferative effects of HHT in
imatinib-sensitive K562 cells and imatinib-resistant K562G cells
were compared. The two cell lines were treated with different
concentrations of HHT for 48 h, respectively, and cell
proliferation was assessed using MTS assay. The results revealed
that the 50% inhibitory concentration (IC50) of HHT in
K562 and K562G cell lines was 27 and 24 ng/ml, respectively
(Fig. 1A). HHT revealed similar
cytotoxicity results in both cell lines. Subsequently, it was then
investigated whether HHT could induce apoptosis in
imatinib-resistant K562G cells. As revealed in Fig. 1B, K562G cells were treated with
different concentrations of HHT at 0, 20, 50, 80 ng/ml for 24 h,
and the apoptotic rates were 4.55, 12.6, 14.5 and 49.6%,
respectively. Furthermore, it was revealed that HHT triggered a
dose-dependent cleavage of caspase-3 and PARP in K562G cells
(Fig. 1C), confirming the induction
of cell apoptosis by HHT in imatinib-resistant K562G cells. BCR-ABL
is the main pathogenic factor of CML, and thus the effects of HHT
on BCR-ABL oncoprotein were examined. K562G cells were treated with
different concentrations of HHT and the protein level of BCR-ABL
was examined by western blotting. As anticipated, the levels of the
BCR-ABL protein revealed a significant reduction by HHT treatment
in a dose- and time-dependent manner (Fig. 1D and E). In addition, HHT induced
downregulation of BCR-ABL in K562 cells (Fig. S2). It is worth mentioning that HHT
exhibited no significant effect on the c-Abl protein. These results
indicated that HHT downregulated BCR-ABL oncoprotein and induced
cell apoptosis in imatinib-resistant K562G CML cells.
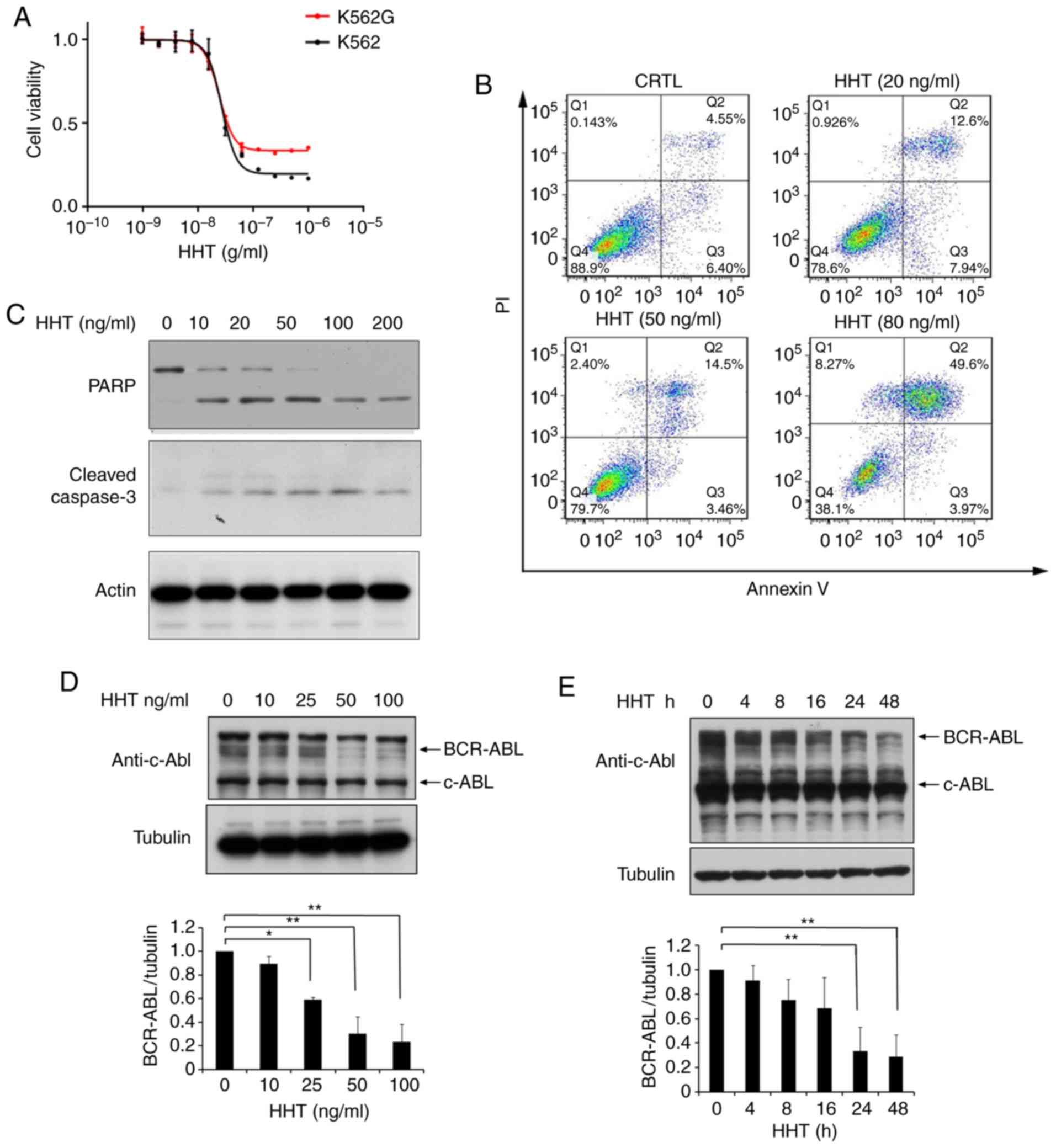 | Figure 1.HHT induces cell apoptosis and
downregulates BCR-ABL in CML cells. (A) HHT inhibited cell
proliferation in K562 cells and K562G cells. K562 and K562G cells
were treated with HHT at indicated concentrations for 48 h,
respectively. Cell viability was determined by MTS assay. Values
were presented as the means ± SD, of the three independent
experiments. (B) HHT induced apoptosis in K562G cells. K562G cells
were treated with 0, 20, 50, and 80 ng/ml HHT for 24 h, and cell
apoptosis was determined by flow cytometric analysis with
Annexin-V/PI staining. (C) K562G cells were treated with different
concentrations of HHT for 24 h. Cleaved PARP and caspase-3 were
examined by western blotting. (D) BCR-ABL protein was downregulated
by HHT in K562G cells. K562G cells were treated with HHT at
indicated concentrations for 24 h. The concentrations were 0, 10,
25, 50 and 100 ng/ml. (E) K562G cells were treated with 20 ng/ml
HHT at indicated time-points. Cell lysates were analyzed to
determine BCR-ABL protein levels by western blotting. Densitometric
values were quantified using the ImageJ software and normalized to
the control. The values of the control were set to 1. Error bars
represent the standard deviations for n=3 experiments and western
blotting revealed representative data (*P<0.05, **P<0.01
compared with the vehicle control). CML, chronic myelogenous
leukemia; HHT, homoharringtonine. |
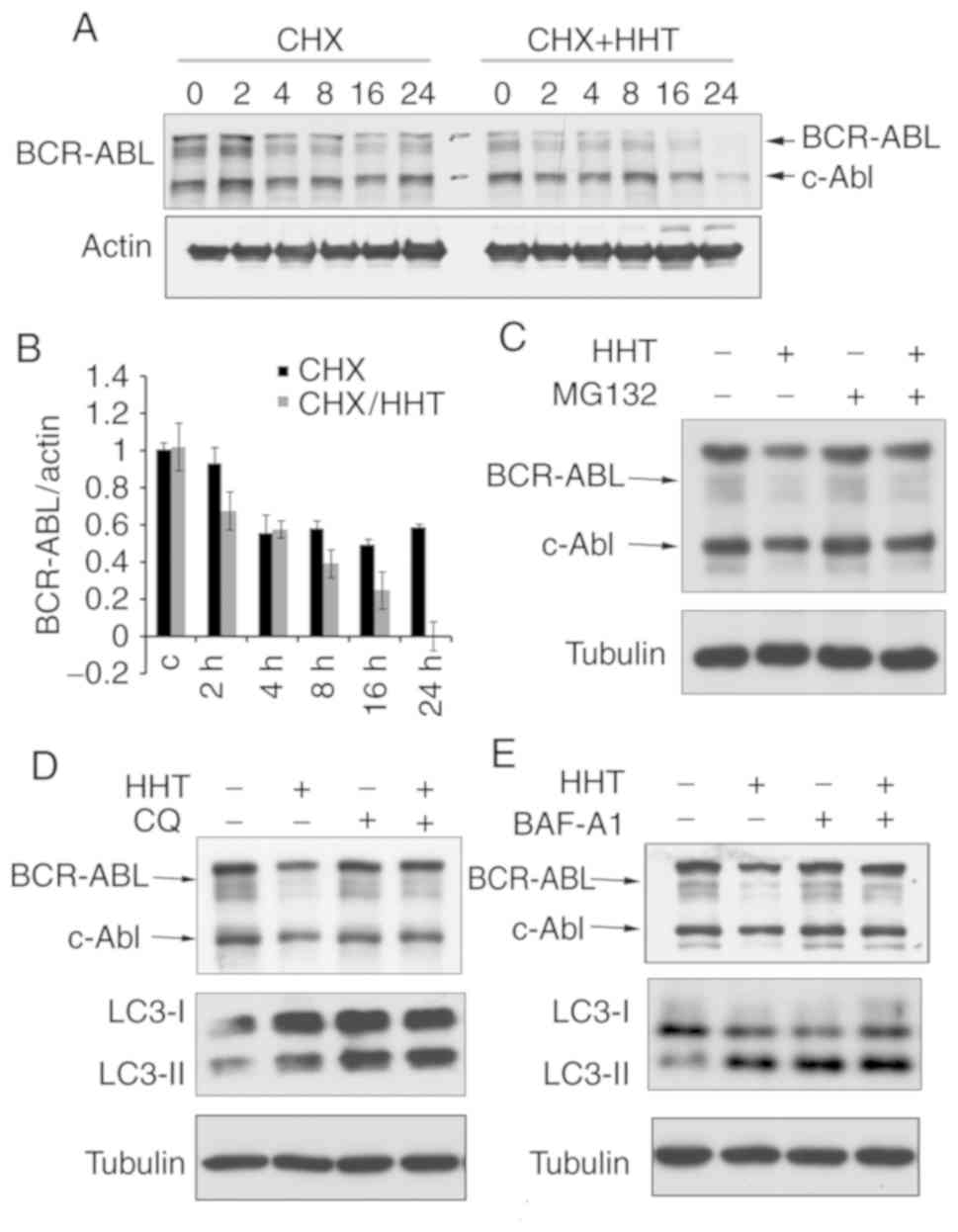
HHT promotes BCR-ABL degradation
though the autophagy pathway
Since HHT could downregulate BCR-ABL oncoprotein,
the next question was whether the downregulation was caused by
inhibiting protein synthesis or by promoting its degradation. Some
evidence has indicated that HHT downregulates BCR-ABL by inhibiting
the synthesis of BCR-ABL (26).
However, little attention has been paid to whether HHT affects
protein degradation or not. To study the effect of HHT on BCR-ABL
protein degradation, cycloheximide (CHX) chase assay was performed
on HHT-treated K562G cells. The results revealed that the BCR-ABL
protein levels began to decline at 2 h and were markedly decreased
at 24 h in HHT-treated K562G cells, whereas in control cells, the
BCR-ABL protein levels began to decline at 4 h and were maintained
until 24 h (Fig. 2A and B). These
results confirmed that HHT could promote BCR-ABL degradation.
Protein degradation usually involves two pathways: The
ubiquitin-proteasome pathway and the autophagy-lysosome pathway. To
further investigate the mechanism of HHT-induced BCR-ABL
degradation, K562G cells were treated with proteasome inhibitors
and autophagolysosome inhibitors, respectively and then their
effects were observed on HHT-induced BCR-ABL degradation.
Proteasome inhibitor MG132 exhibited no obvious effect on
HHT-induced BCR-ABL degradation (Fig.
2C), however the autophagolysosome inhibitor CQ and Baf-A1
reversed the HHT-induced reduction of the protein levels of BCR-ABL
(Fig. 2D and E). Similar results were
also obtained in K562 cells (Fig.
S3). These results indicated that the autophagy-lysosome
pathway was involved in HHT-induced BCR-ABL degradation.
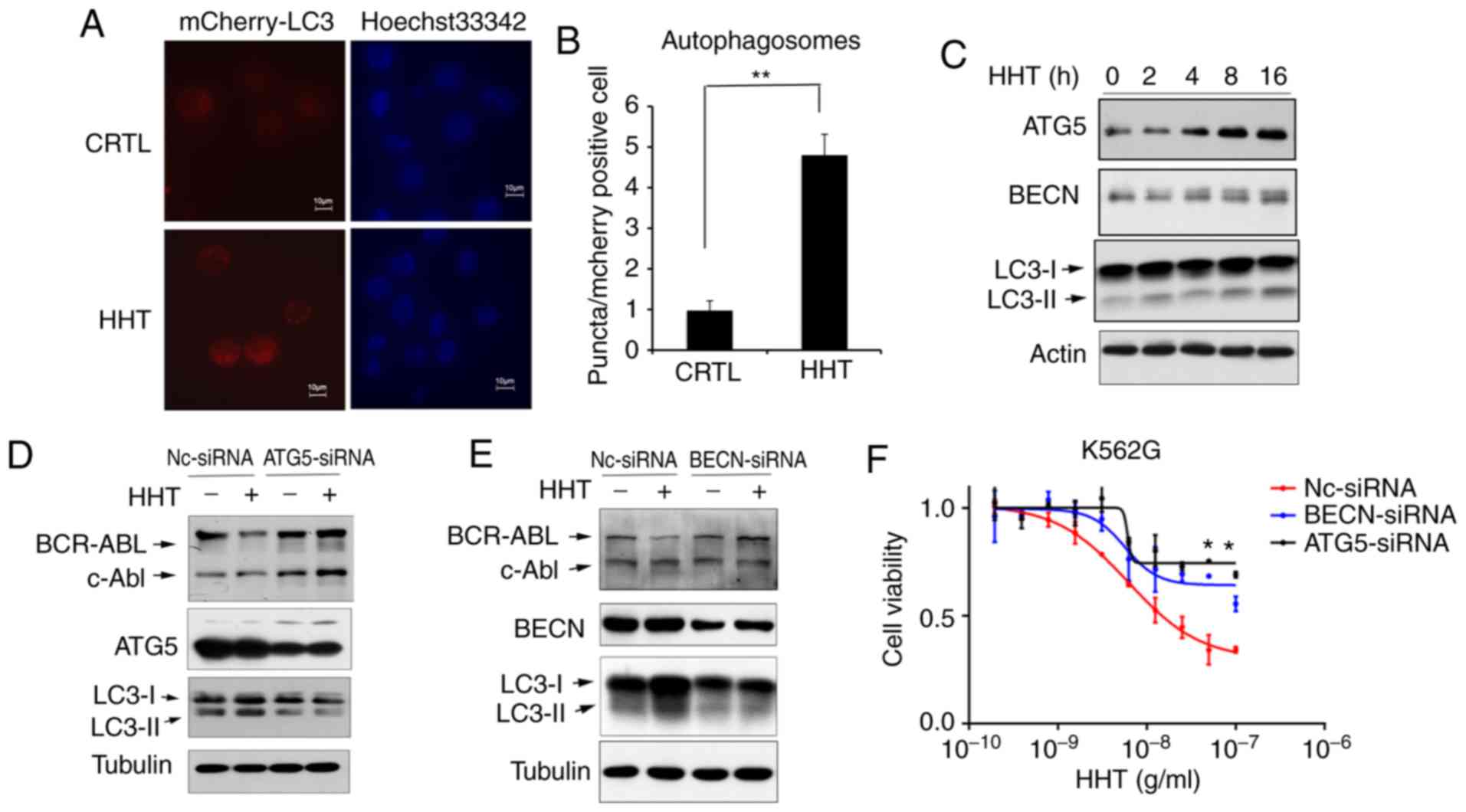
Inhibition of the autophagy pathway reversed the
effect of HHT on BCR-ABL degradation and cell apoptosis in K562G
cells. To confirm HHT-induced autophagy, K562G cells were
transfected with Ad-mCherry-GFP-LC3B, which is considered as a
well-documented model for monitoring the conversion of MAP1LC3B
from a cytosolic MAP1LC3B-I form to a phagophore- and
autophagosome-bound MAP1LC3B-II form. Under normal conditions,
mCherry-GFP-LC3B fluorescence was diffused in the cytoplasm, while
it converted to a punctate pattern when autophagy was activated.
Next, treatment with 50 ng/ml of HHT for 8 h markedly increased the
average number of fluorescent spots in mCherry-GFP-LC3B-positive
cells (Fig. 3A and B), indicating
that HHT induced autophagy in K562G cells. In addition, the levels
of autophagic key proteins were detected by western blotting. The
results revealed that treatment of K562G cells with HHT resulted in
the upregulation of the LC3II/LC3I ratio and autophagic key factors
ATG5 and Beclin-1 (Fig. 3C).
Furthermore, the key molecules of autophagic machinery were
targeted using specific siRNAs and assessed their effects on
HHT-induced downregulation of the BCR-ABL protein. Selective
knockdown of either Beclin-1 or ATG5 (Fig. 3D and E) reversed the suppressive
effects of HHT treatment on the levels of BCR-ABL. In addition,
siRNA-mediated silencing of autophagic machinery inhibited
cytotoxicity of HHT on K562G cells (Fig.
3F). These results demonstrated that autophagic inhibition
reversed the effect of HHT on BCR-ABL degradation and inhibited the
cytotoxicity of HHT in K562G CML cells, further confirming that HHT
induced BCR-ABL degradation via the autophagy pathway.
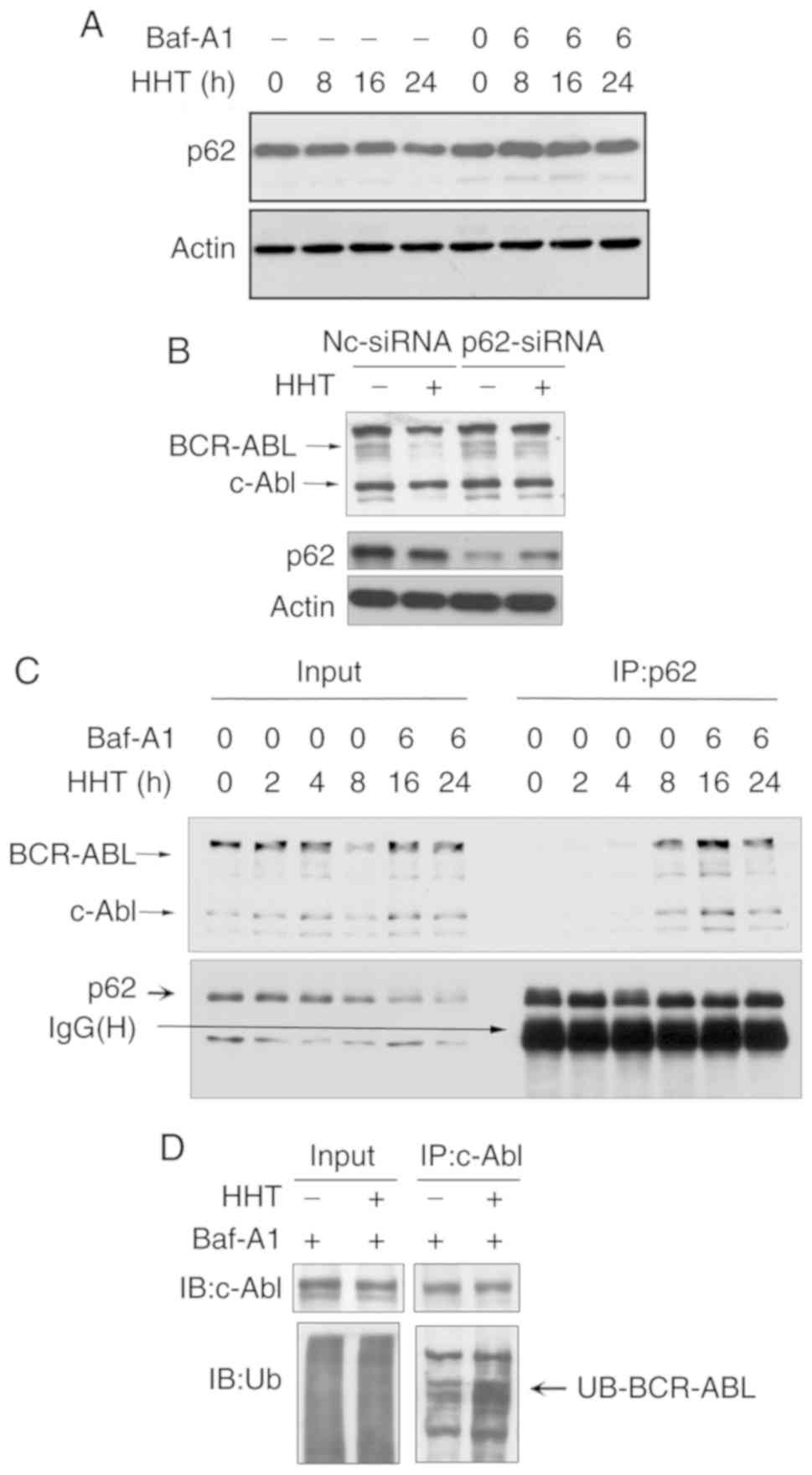
p62 is involved in HHT-induced BCR-ABL
degradation via the autophagy pathway
p62 plays an important role in the
autophagy-lysosome pathway and was revealed to target
polyubiquitinated proteins to autophagosomes for degradation by
directly interacting with LC3 (27).
Next, it was determined whether the classical autophagic receptor
p62 was involved in HHT-induced BCR-ABL degradation. Western
blotting assays revealed that HHT induced a time-dependent decrease
of p62 levels, whereas the autophagy inhibitor Baf-A1 inhibited the
reduction of p62 (Fig. 4A). These
results indicated that p62 may be related to autophagic degradation
induced by HHT. Furthermore, siRNAs were designed to knockdown
endogenous p62. As revealed in Fig.
4B, endogenous p62 expression was effectively inhibited with
p62-siRNA, and p62 knockdown significantly reversed the decrease of
BCR-ABL protein levels induced by HHT. These results confirmed that
p62 was necessary for HHT-induced autophagic degradation of the
BCR-ABL protein. Next, whether HHT promoted BCR-ABL to interact
with p62 was assessed. K562G cells were treated with HHT for 0, 2,
4, 8, 16 and 24 h and then were lysed, followed by
immunoprecipitation with an anti-p62 antibody and immunoblotting
with an anti-ABL antibody. To prevent the degradation of BCR-ABL
and p62 from HHT treatment for 16 and 24 h, Baf-A1 (20 nM) was
added during the last 6 h. An HHT-inducible time-dependent increase
was observed during the interaction between BCR-ABL and p62
(Fig. 4C). Finally, the results
revealed that ubiquitination levels of BCR-ABL were significantly
increased in HHT-treated K562G cells when compared with the control
group (Fig. 4D). These results
indicated that HHT promoted the interaction of p62 with
ubiquitinated BCR-ABL, and p62 was involved in the autophagic
degradation of the BCR-ABL protein.
Discussion
BCR-ABL is the main oncogene of CML that causes
malignant proliferation of hematopoietic stem cells and progenitor
cells. Inhibitors of ABL tyrosine kinase (TKIs) have been revealed
to exhibit marked therapeutic effects on CML (28), however, mutations of several kinase
domains have conferred high-level resistance to TKIs. Furthermore,
TKIs cannot eliminate leukemic stem cells, as the survival of CML
stem cells does not depend on the activity of BCR-ABL kinase
(29). Therefore, it is necessary to
explore novel therapeutic strategies to improve CML treatment.
HHT has been used in China for the treatment of
hematological malignancies for over 40 years, and was approved by
the FDA in the US for its use in refractory/relapsed CML in 2010.
Despite its clinical efficacy, the exact underlying mechanisms are
yet to be revealed. In the present study, HHT directly inhibited
the protein levels of BCR-ABL by promoting the degradation of
fusion protein, which was different from the previously reported
mechanisms, i.e., inhibition of protein synthesis. This is
considered a novel angle to investigate HHT and develop strategies
to downregulate the levels of BCR-ABL, which in turn may shed new
insights in the treatment of CML.
There are two major mechanisms involved in protein
degradation, including the ubiquitin-proteasome system and the
autophagy-lysosome pathway (30). HHT
activated autophagy in K562G cells in the present study. When
autophagy was inhibited by ATG5 or Beclin-1 siRNA, HHT induced
BCR-ABL protein degradation and apoptosis was also inhibited. In
addition, autophagic lysosome inhibitors Baf-A1 and CQ reversed the
degradation of BCR-ABL in K562G cells induced by HHT, however, the
proteasome inhibitor MG132 did not reverse the degradation.
Therefore, it is surmised that degradation of the BCR-ABL protein
induced by HHT was mainly achieved through the autophagy-lysosome
pathway.
Conventionally, autophagy requires membrane
trafficking and remodeling for the formation of autophagosomes and
delivers its internal contents to the lysosome for degradation.
Although autophagy has been regarded as a random cytoplasmic
degradation process, the involvement of ubiquitin as a targeting
signal for selective autophagy is rapidly emerging (31,32). In
addition, the selection of autophagy is mediated by autophagic
receptors, such as SQSTM1/p62 and NBR1 (NBR1, autophagy cargo
receptor). These autophagic receptors interact with both ubiquitin
conjugated to target proteins and autophagosome-specific
MAP1LC3/LC3, promoting protein autophagic degradation (33). As is well known, ubiquitin is a
universal degradation signal used for protein degradation via
proteasomes and autophagy (34). In
the present study, HHT enhanced ubiquitination of the BCR-ABL
protein, which may in turn provide signal recognition by autophagic
receptor p62, and then lead to subsequent autophagic degradation.
As anticipated, HHT treatment significantly facilitated the
interaction of p62 with BCR-ABL. Moreover, knockdown of p62 in
K562G cells reversed the effect of HHT on the degradation of
BCR-ABL. These results demonstrated p62-mediated HHT-induced
autophagic degradation of BCR-ABL. However, how HHT induces BCR-ABL
ubiquitination and how p62 interacts with ubiquitin BCR-ABL require
further study.
Collectively, these results reported that HHT
induced autophagy in CML cells and promoted BCR-ABL ubiquitination.
The ubiquitinated BCR-ABL was bound to p62 forming a BCR-ABL-p62
complex followed by autophagic degradation. The present study
elucidated how HHT downregulated CML oncoprotein BCR-ABL, which may
subsequently be a potential new strategy to overcome the resistance
to TKIs.
Supplementary Material
Supporting Data
Acknowledgements
We are grateful to Professor Jie Jin for providing
K562 and K562G cells.
Funding
The present study was supported by the National
Natural Science Foundation under grant no. 81670139 to YT; and the
Natural Science Foundation of Shanghai under grant no. 16ZR1427800
to YT.
Availability of data and materials
The datasets used during the present study are
available from the corresponding author upon reasonable
request.
Authors' contributions
YT and CW designed the research. SL, ZB and YJ
performed the experiments. SL and XS analyzed the data and wrote
the manuscript. All authors read and approved the final manuscript
and agree to be accountable for all aspects of the research in
ensuring that accuracy or integrity of any parts of the work are
appropriately investigated and resolved.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Shtivelman E, Lifshitz B, Gale RP and
Canaani E: Fused transcript of abl and bcr genes in chronic
myelogenous leukaemia. Nature. 315:550–554. 1985. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Druker BJ: Translation of the philadelphia
chromosome into therapy for CML. Blood. 112:4808–4817. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chopade P and Akard LP: Improving outcomes
in chronic myeloid leukemia over time in the era of tyrosine kinase
inhibitors. Clin Lymphoma Myeloma Leuk. 18:710–723. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Jabbour E, Kantarjian H and Cortes J: Use
of second- and third-generation tyrosine kinase inhibitors in the
treatment of chronic myeloid leukemia: An evolving treatment
paradigm. Clin Lymphoma Myeloma Leuk. 15:323–334. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Sawyers CL, Hochhaus A, Feldman E, Goldman
JM, Miller CB, Ottmann OG, Schiffer CA, Talpaz M, Guilhot F,
Deininger MW, et al: Imatinib induces hematologic and cytogenetic
responses in patients with chronic myelogenous leukemia in myeloid
blast crisis: Results of a phase II study. Blood. 99:3530–3539.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Larson RA: Is there a best TKI for chronic
phase CML? Blood. 126:2370–2375. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Warmuth M, Bergmann M, Priess A, Hauslmann
K, Emmerich B and Hallek M: The Src family kinase Hck interacts
with Bcr-Abl by a kinase-independent mechanism and phosphorylates
the Grb2-binding site of Bcr. J Biol Chem. 272:33260–33270. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wertheim JA, Forsythe K, Druker BJ, Hammer
D, Boettiger D and Pear WS: BCR-ABL-induced adhesion defects are
tyrosine kinase-independent. Blood. 99:4122–4130. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Inoue A, Kobayashi CI, Shinohara H,
Miyamoto K, Yamauchi N, Yuda J, Akao Y and Minami Y: Chronic
myeloid leukemia stem cells and molecular target therapies for
overcoming resistance and disease persistence. Int J Hematol.
108:365–370. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Goussetis DJ, Gounaris E, Wu EJ, Vakana E,
Sharma B, Bogyo M, Altman JK and Platanias LC: Autophagic
degradation of the BCR-ABL oncoprotein and generation of
antileukemic responses by arsenic trioxide. Blood. 120:3555–3562.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Valencia-Serna J, Aliabadi HM, Manfrin A,
Mohseni M, Jiang X and Uludag H: SiRNA/lipopolymer nanoparticles to
arrest growth of chronic myeloid leukemia cells in vitro and in
vivo. Eur J Pharm Biopham. 130:66–70. 2018. View Article : Google Scholar
|
|
12
|
Davidson BL and McCray PB Jr: Current
prospects for RNA interference-based therapies. Nat Rev Genet.
12:329–340. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kim DH and Rossi JJ: Strategies for
silencing human disease using RNA interference. Nat Rev Genet.
8:173–184. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
14
|
Shibata N, Shimokawa K, Nagai K, Ohoka N,
Hattori T, Miyamoto N, Ujikawa O, Sameshima T, Nara H, Cho N and
Naito M: Pharmacological difference between degrader and inhibitor
against oncogenic BCR-ABL kinase. Sci Rep. 8:135492018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lu S and Wang J: Homoharringtonine and
omacetaxine for myeloid hematological malignancies. J Hematol
Oncol. 7:22014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Quintas-Cardama A and Cortes J:
Therapeutic options against BCR-ABL1 T315I-positive chronic
myelogenous leukemia. Clin Cancer Res. 14:4392–4399. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
O'Brien S, Kantarjian H, Keating M, Beran
M, Koller C, Robertson LE, Hester J, Rios MB, Andreeff M and Talpaz
M: Homoharringtonine therapy induces responses in patients with
chronic myelogenous leukemia in late chronic phase. Blood.
86:3322–3326. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kantarjian HM, O'Brien S and Cortes J:
Homoharringtonine/omacetaxine mepesuccinate: The long and winding
road to food and drug administration approval. Clin Lymphhoma
Myeloma Leuk. 13:530–533. 2013. View Article : Google Scholar
|
|
19
|
Al Ustwani O, Griffiths EA, Wang ES and
Wetzler M: Omacetaxine mepesuccinate in chronic myeloid leukemia.
Exp Opin Pharmacother. 15:2397–2405. 2014. View Article : Google Scholar
|
|
20
|
Gurel G, Blaha G, Moore PB and Steitz TA:
U2504 determines the species specificity of the A-site cleft
antibiotics: The structures of tiamulin, homoharringtonine, and
bruceantin bound to the ribosome. J Mol Biol. 389:146–156. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Huang MT: Harringtonine, an inhibitor of
initiation of protein biosynthesis. Mol Pharm. 11:511–519.
1975.
|
|
22
|
Kuroda J, Kamitsuji Y, Kimura S, Ashihara
E, Kawata E, Nakagawa Y, Takeuichi M, Murotani Y, Yokota A, Tanaka
R, et al: Anti-myeloma effect of homoharringtonine with concomitant
targeting of the myeloma-promoting molecules, Mcl-1, XIAP, and
beta-catenin. Int J Hematol. 87:507–515. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Sun Q, Li S, Li J, Fu Q, Wang Z, Li B, Liu
SS, Su Z, Song J and Lu D: Homoharringtonine regulates the
alternative splicing of Bcl-x and caspase 9 through a protein
phosphatase 1-dependent mechanism. BMC Complement Altern Med.
18:1642018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Chen J, Mu Q, Li X, Yin X, Yu M, Jin J, Li
C, Zhou Y, Zhou J, Suo S, et al: Homoharringtonine targets Smad3
and TGF-β pathway to inhibit the proliferation of acute myeloid
leukemia cells. Oncotarget. 8:40318–40326. 2017.PubMed/NCBI
|
|
25
|
Chen R, Gandhi V and Plunkett W: A
sequential blockade strategy for the design of combination
therapies to overcome oncogene addiction in chronic myelogenous
leukemia. Cancer Res. 66:10959–10966. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Novotny L, Al-Tannak NF and Hunakova L:
Protein synthesis inhibitors of natural origin for CML therapy:
Semisynthetic homoharringtonine (Omacetaxine mepesuccinate).
Neoplasma. 63:495–503. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zaffagnini G, Savova A, Danieli A, Romanov
J, Tremel S, Ebner M, Peterbauer T, Sztacho M, Trapannone R,
Tarafder AK, et al: P62 filaments capture and present ubiquitinated
cargos for autophagy. EMBO J. 37:e983082018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Rossari F, Minutolo F and Orciuolo E:
Past, present, and future of Bcr-Abl inhibitors: From chemical
development to clinical efficacy. J Hematol Oncol. 11:842018.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Talati C and Pinilla-Ibarz J: Resistance
in chronic myeloid leukemia: Definitions and novel therapeutic
agents. Curr Opin Hematol. 25:154–161. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Dikic I: Proteasomal and autophagic
degradation systems. Ann Rev Biochem. 86:193–224. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kraft C, Peter M and Hofmann K: Selective
autophagy: Ubiquitin-mediated recognition and beyond. Nat Cell
Biol. 12:836–841. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Lamark T, Svenning S and Johansen T:
Regulation of selective autophagy: The p62/SQSTM1 paradigm. Essays
Biochem. 61:609–624. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Moscat J, Karin M and Diaz-Meco MT: P62 in
Cancer: Signaling adaptor beyond autophagy. Cell. 167:606–609.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Kocaturk NM and Gozuacik D: Crosstalk
between mammalian autophagy and the ubiquitin-proteasome system.
Front Cell Dev Biol. 6:1282018. View Article : Google Scholar : PubMed/NCBI
|