Introduction
Hepatocellular carcinoma (HCC) is a common malignant
tumor with high lethality, and infection with hepatitis B virus
(HBV) is the leading cause of HCC (1). HBV can accelerate HCC via multiple
mechanisms. First, HBV induces immune reactions that lead to
repeated hepatic inflammation, fibrosis and a deficient immune
microenvironment. Subsequently, HBV can modify host genes near the
insertion point through DNA integration to cause host cell genome
instability and to generate carcinogenic fusion proteins (2,3).
However, the approaches for HCC screening in HBV-infected
individuals, the risk factors of HCC, the possible mechanisms
leading to HCC and the potential therapeutic approaches of
HBV-related HCC have not been systematically reviewed. Thus, it is
urgent to characterize the pathogenic mechanisms of HBV-related HCC
in order to identify novel targets for its treatment.
The lncRNA plasmacytoma variant translocation 1
(PVT1) has 1,716 nucleotides and is located in the chr8q24.21
region, which is 57 kb downstream of the MYC gene on human
chromosome 8q24 (4,5). PVT1 produces a wide variety of spliced
non-coding RNAs as well as a cluster of six annotated microRNAs:
miR-1205, miR-1204, miR-1206, miR-1207-5p, miR-1208, miR-1207-3p,
as a P53-inducible target gene (6).
Several researchers have observed that lncRNA PVT1 plays an
important role in tumorigenesis in non-small cell lung cancer,
ovarian and breast cancer, gastric cancer, and high expression of
PVT1 predicts poor patient prognosis (7–10). As
to liver cancer, lncRNA PVT1 has been reported to participate in
tumor progression by promoting cell proliferation, invasion and
metabolism of HCC cells (11,12),
and these studies indicate the important roles of PVT1 in HCC
carcinogenesis, yet the mechanisms remain unclear (13). lncRNA-PVT1 participates in many
physiological and pathological processes by modulating gene
expression at the transcriptional, post-transcriptional and
epigenetic levels (14,15). lncRNAs recruit the PRC2 complex to
chromatin, such as HOTAIR, Kcnq 1ot1, and Braveheart, suggesting
the specific association between PRC2 and lncRNAs (16). RNA immunoprecipitation with
microarray analysis revealed that 20% of the lncRNAs in human cells
are associated with the PRC2 complex (17). Polycomb repressive complex-2 (PRC2)
is a highly conserved histone methyltransferase for H3K27
methylation, and contains the subunits Enhancer of Zeste 2 (EZH2),
Embryonic Ectoderm Development (EED), and Suppressor of Zeste 12
(SUZ12) (18). EZH2 is detectable
in various HCC cell lines, and was found to play a critical role in
HCC tumorigenesis in vivo and may serve as a powerful
diagnostic biomarker (19).
Although the important roles of PVT1 in many cancer have been
reported, the mechanism of PVT1 in the occurrence and development
of hepatitis B virus-positive HCC remains unclear.
In the present study, it was demonstrated that
lncRNA PVT1 is highly expressed in hepatitis B virus-positive HCC
tissues and regulates cell proliferation, migration, and invasion
in liver cancer cell lines. Importantly, we present evidence that
lncRNA PVT1 interferes with the recruitment of EZH2 onto the
MYC promoter, which contributes to the suppression of
H3K37me3 modification and promotes c-Myc expression in hepatitis B
virus-positive HCC cells. Moreover, EZH2 protein was found to be
negatively correlated with lncRNA PVT1 expression. Taken together,
our study may provide a new approach and facilitate
lncRNA-PVT1-directed diagnostic and therapeutic strategies for
hepatitis B virus-positive liver cancer at the early stages.
Materials and methods
Patient tissue samples
A total of 24 HCC tissues were obtained from
patients undergoing routine hepatic resection at Tianjin Second
People's Hospital. Fifteen of the patients presented with
HBV-positive HCC. Patients included 14 males and 10 females, with
ages ranging from 33 to 74 years, with a median age of 46 years.
Pathological progression was classified as TNM I (n=5), TNM II
(n=9), and TNM III (n=10) according to the International Union
Against Cancer TNM classification system (20). These patients did not receive any
form of chemotherapy or radiotherapy before surgery. The tissues
were immediately snap-frozen and stored in liquid nitrogen after
resection. Written informed consent was obtained from all patients
enrolled in this study, and the present study protocol was approved
by the Ethics Committee of Tianjin Second People's Hospital.
Isolation of primary human hepatocytes
(PHHs)
Primary hepatocyte isolation was prepared according
to a previous report (21).
Briefly, tissues obtained from resected human liver of patients
with benign local liver diseases were digested with
EDTA/collagenase perfusion technique, and the cell suspension was
centrifuged at 50 × g for 5 min at 4°C to separate the
non-parenchymal cell (NPC) fraction, and the cell pellet was used
to isolate the PHHs. The PHH fraction was subjected to a 25%
Percoll density gradient centrifugation at 1,250 × g for 20 min at
4°C without stopping to remove non-viable cells, and then the cells
were washed with phosphate-buffered saline (PBS) and re-suspended
and cultured in Dulbecco's modified Eagle's medium (DMEM) with 15%
fetal bovine serum (FBS) (both from Gibco; Thermo Fisher
Scientific, Inc.) in a humidified atmosphere of 5% CO2
at 37°C.
Cell culture
The human liver cancer cell lines Hep3B and HepG2
were purchased from the American Type Culture Collection (ATCC).
Cells were maintained in Dulbecco's modified Eagle's medium (DMEM)
with 10% fetal bovine serum (FBS) (both from Gibco; Thermo Fisher
Scientific, Inc.), 100 U/ml penicillin-streptomycin, and cultured
in a humidified atmosphere of 5% CO2 at 37°C.
Quantitative real-time PCR
Total RNA was extracted from the frozen tissues or
cell lines with TRIzol™ reagent (Thermo Fisher Scientific, Inc.)
according to the manufacturer's instructions. Total RNA (2 µg) was
reverse transcribed with SuperScript™ IV Reverse Transcriptase
(Thermo Fisher Scientific, Inc.). The quantitative real-time PCR
was performed with FastStart Universal SYBR Green Master (Rox)
(Roche) on QuantStudio 3 Real-Time PCR System (Applied Biosystems;
Thermo Fisher Scientific, Inc.). The amplification conditions
consisted of 95°C for 10 min; 95°C for 15 sec, 60°C for 60 sec; 40
cycles. The relative expression of PVT1 was calculated using
the 2−ΔΔCq method (22).
The gene-specific primers were as follows: PVT1 forward,
5′-GGGAATAACGCTGGTGGAAC-3′ and reverse, 5′-GATCTCAACCCTCTCAGCCA-3′;
GAPDH forward, 5′-AATGGCAGCCGTTAGGAAA-3′ and reverse,
5′-GCCCAATACGACCATCAGAG-3′.
MTS cell proliferation assay
Cells were seeded in 96-well microplates at a
density of 3,000 cells per well. Cell viability was assessed using
the CellTiter 96 AQueous One Solution Reagent (Promega) according
to the manufacturer's instructions.
Colony formation assay
Single-cell suspension of cells at the concentration
of 1×104 cells in DMEM with 10% FBS were plated on the
bottom layer containing 0.8% agarose in a 6-well plate. The cells
were photographed on day 14 after plating (Motic AE2000;
Xiamen).
Cell mobility analysis
Cell migration and invasion were examined by
wound-healing and Transwell assay. For the wound-healing assay,
Hep3B and HepG2 liver cancer cells were seeded with a defined cell
number of 3×103 cells/cm2 in 24-well plates.
A scratch was made using a pipette tip as a cross through the whole
well after reaching a uniform confluence of 95%. The scratch area
was monitored and measured with Fiji software (ImageJ V1.48;
National Institutes of Health, Bethesda, MD, USA) at time-points 0,
6, 12 and 24 h.
A Transwell chamber (8-µm pore size; Millipore) with
Matrigel (BD Biosciences) matrix was used to determine cell
invasion. Two hundred microliters of the cell suspension
(1×104 cells) in serum-free medium was plated in the top
chamber, whereas the lower chamber contained 600 µl. The cells on
the top surface of the membrane were removed by a cotton swab after
24 h. Cells on the bottom surface of the membrane were stained for
15 min at room tempreture with crystal violet solution and image
were captured (Motic AE2000; Xiamen).
Cell cycle and apoptosis analysis
Hep3B and HepG2 cells were cultured as described in
the above section. Subsequently, cells were trypsinized and fixed
in 70% ethanol and incubated at 4°C overnight followed by
incubation with DNAse-free RNAse A (Sigma-Aldrich; Merck KGaA)
containing propidium iodide (PI)/Triton X-100 staining solution for
20 min at room tempreture. The analysis was performed according to
the manufacturer's instructions using a FACS Calibur
instrument.
Cell apoptosis was detected with the Annexin V-FITC
Apoptosis Detection Kit (Sigma-Aldrich; Merck KGaA) according to
the instruction manual. Briefly, the cells were washed with PBS and
incubated with Dead Cell Apoptosis Kit (Thermo Fisher Scientific,
Inc.) with Annexin V FITC and PI, for flow cytometry at 4°C for 30
min. The flow cytometry data were analyzed with CellQuest 3.0
software (BD Biosciences).
Construction of vectors and cell
transfection
The lncRNA- PVT1 small hairpin RNA (shRNA) oligos
and the non-targeting scramble (NC Ctrl) were synthesized by
Shanghai Invitrogen Biotechnology (Shanghai, China), and the
sequence tested with the highest efficacy was
GGACATGAGAAGGACAGAATA. The lncRNA-PVT1 overexpression (OE) vector
and the vector control (Vector) were constructed by GeneChem Co.
(Shanghai, China). Vector transfections were all performed by using
FuGENE® HD Transfection Reagent (Promega) following the
manufacturer's instructions.
Western blot analysis
Total protein was extracted using RIPA protein
extraction reagent containing protease inhibitors (cOmplete™, Mini,
EDTA-free Protease Inhibitor Cocktail Tablets, Roche Diagnostics),
and the concentration was measured using a BCA protein assay kit
(Thermo Fisher Scientific, Inc.). The protein lysates were analyzed
with 10% SDS-PAGE and subsequently transferred to a PVDF membrane.
Antibodies for EZH2 (cat. no. 5246), c-Myc (cat. no. 9402), SUZ12
(cat. no. 3737) and EED (cat. no. 51673) were purchased from Cell
Signaling Technology (all at 1:1,000 dilution), and anti-GAPDH
(cat. no. G9545) was purchased from Sigma-Aldrich/Merck KGaA
(1:5,000 dilution). The HRP-conjugated secondary antibody (cat. no.
A0545) was purchased from Sigma-Aldrich; Merck KGaA. The signal was
detected with Amersham ECL Prime Western Blotting Detection Reagent
(GE Healthcare Life Sciences) and exposed using the Gel Doc™ EZ
System (Bio-Rad Laboratories).
RNA pull-down assay
RNA pull-down assays were performed using a Magnetic
RNA Protein Pull-Down kit (Thermo Fisher Scientific, Inc.)
according to the kit manual. Biotinylated PVT1 RNA was synthesized
by RiboBio. For each assay, 50 pM biotinylated RNA was incubated
with 50 µl prewashed streptavidin-agarose beads for 1 h at 4°C.
Then, RNA-bound beads were incubated with lysates from Hep3B cell
cytosolic/nuclear extracts, and eluted proteins were detected by
western blot analysis.
RNA immunoprecipitation assay
The EZ Magna RNA Immunoprecipitation Kit (Millipore)
was used following the manufacturer's guidelines. Briefly, Hep3B
cells were lysed in RIPA lysis buffer. Magnetic beads were
pre-incubated with antibodies for 30 min at room temperature, and
the cell lysates were immunoprecipitated with beads for 6 h at 4°C.
Then, RNA was purified and detected by RT-qPCR.
Chromatin-immunoprecipitation-qPCR
(ChIP-qPCR)
Cells (4×107) were washed in PBS and
cross-linked with 1% formaldehyde for 10 min at room temperature
and then quenched by addition of glycine for 5 min. Chromatin was
fragmented to 200–500 bp using 14 cycles using the Vibra-Cell
Ultrasonic Liquid Processors (Sonics and Materials, Inc.). For each
IP, chromatin was immunoprecipitated with 2 mg antibody, EZH2 (cat.
no. 5246) and H3K27me3 (cat. no. 9733) (Cell Signaling Technology),
in IP dilution buffer (1% Triton X-100, 2 mM EDTA, 150 mM NaCl, 20
mM Tris-HCl, pH 8.0) at 4°C overnight. Chromatin was precleared for
2 h each with protein G agarose beads before immunoprecipitation.
The samples were removed from the beads, reversed cross-linked
overnight at 65°C and DNA was isolated using QIAquick PCR
Purification Kit (Qiagen). Precipitated DNA was analyzed by qPCR
using upstream primer of the c-Myc gene,
5′-CCTACCCTCTCAACGACAGC-3′ and downstream primer,
5′-CTTGTTCCTCCTCAGAGTCGC-3′.
Statistical analysis
Data are shown as mean ± SD for at least three
independent experiments. Differences between groups were determined
using paired two-tailed Student's t-test for matched data in two
groups or one-way ANOVA for three or more groups, and the Least
Significant Difference (LSD) was used for post hoc analysis. A
P-value <0.05 was considered statistically significant.
Results
lncRNA PVT1 expression is upregulated
in hepatitis B virus-positive HCC tissues and liver cancer
cells
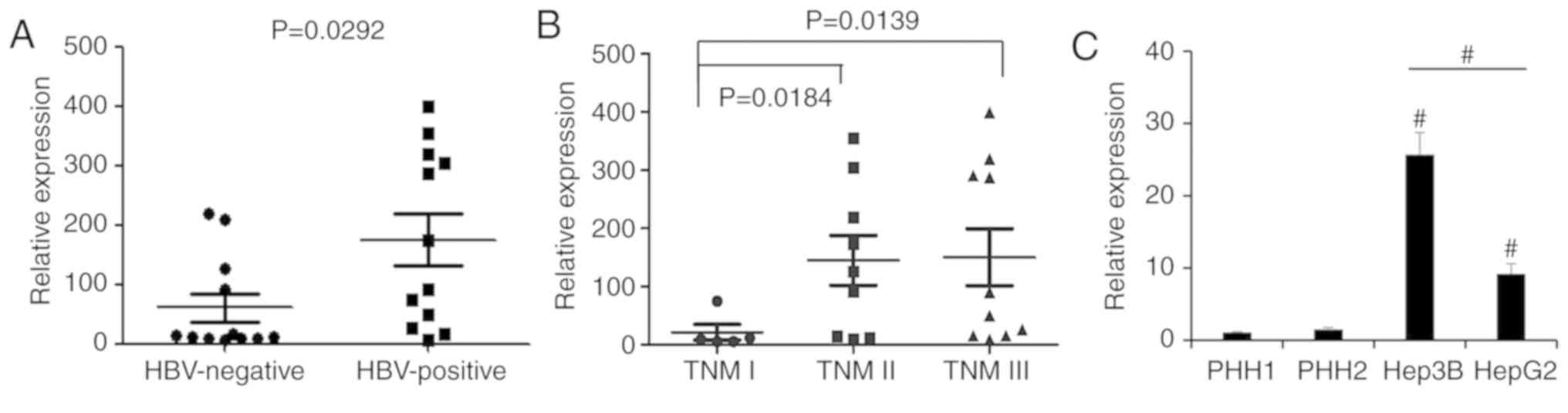
In order to assess the lncRNA-PVT1 expression
pattern associated with HBV virus infection in HCC tissues, we
analyzed lncRNA-PVT1 expression in 24 HCC tissues from HBV-positive
and HBV-negative patients using real-time PCR assay. Our results
showed that HBV-positive HCC tissues showed a significantly higher
PVT1 expression than the HBV-negative HCC patient tissues (Fig. 1A). To investigate the difference
between pathological stages, we further assessed samples of
different stages (TNM I, II and III) by qPCR. The results showed
that PVT1 expression was significantly upregulated in patients with
stage TNM II and III HCC compared with stage TNM I (Fig. 1B). Furthermore, in comparison with
the primary human hepatocytes (PHH) from healthy donors and the
HBV-negative HepG2 liver cancer cell line, PVT1 was highly
expressed in the HBV-positive liver cancer cell line, Hep3B
(Fig. 1C). Our study revealed that
PVT1 was upregulated in HBV-positive HCC tissues, suggesting that
PVT1 may play a specific role in the progression of HBV-positive
liver cancer.
PVT1 promotes HBV-positive liver
cancer cell proliferation
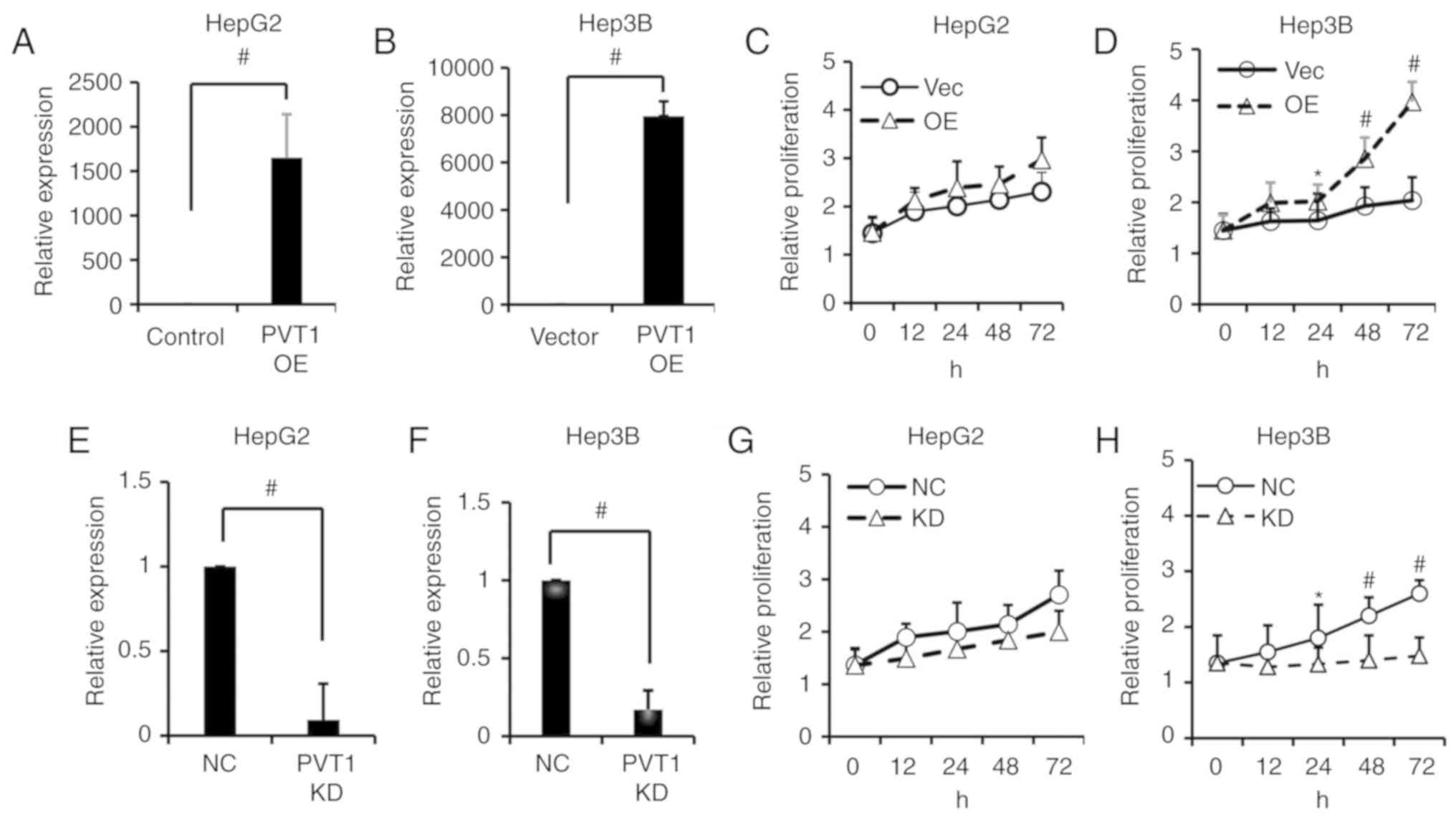
To investigate the biological role of PVT1 in liver
cancer cells, we transfected HepG2 and Hep3B cells with PVT1 shRNA
to knock down PVT1 expression, or the cell lines were transfected
with expression vectors to overexpress the PVT1 gene, and
the non-targeting scramble (NC) or vector control (Vector) were
used as mock controls, respectively. The transfection efficiency
was examined by qPCR. PVT1 was successfully overexpressed in the
Hep3B and HepG2 cells after transfection with synthesized PVT1 RNA
compared with the vector control (Fig.
2A and B). The effect of PVT1 on cell proliferation was
investigated by MTS assay and colony formation assay. As shown in
Fig. 2C and D, the cell
proliferation was significantly increased at 12, 24, 48 and 72 h
following PVT1 overexpression in the Hep3B cell line but not in the
HepG2 cell line. Compared with non-target scrambles (NC), the shRNA
of PVT1 had a significant knockdown (KD) efficacy in the HepG2 and
Hep3B cell lines (Fig. 2E and F).
Following PVT1 KD, the proliferation of Hep3B cells was
significantly suppressed at 24, 48 and 72 h when compared with the
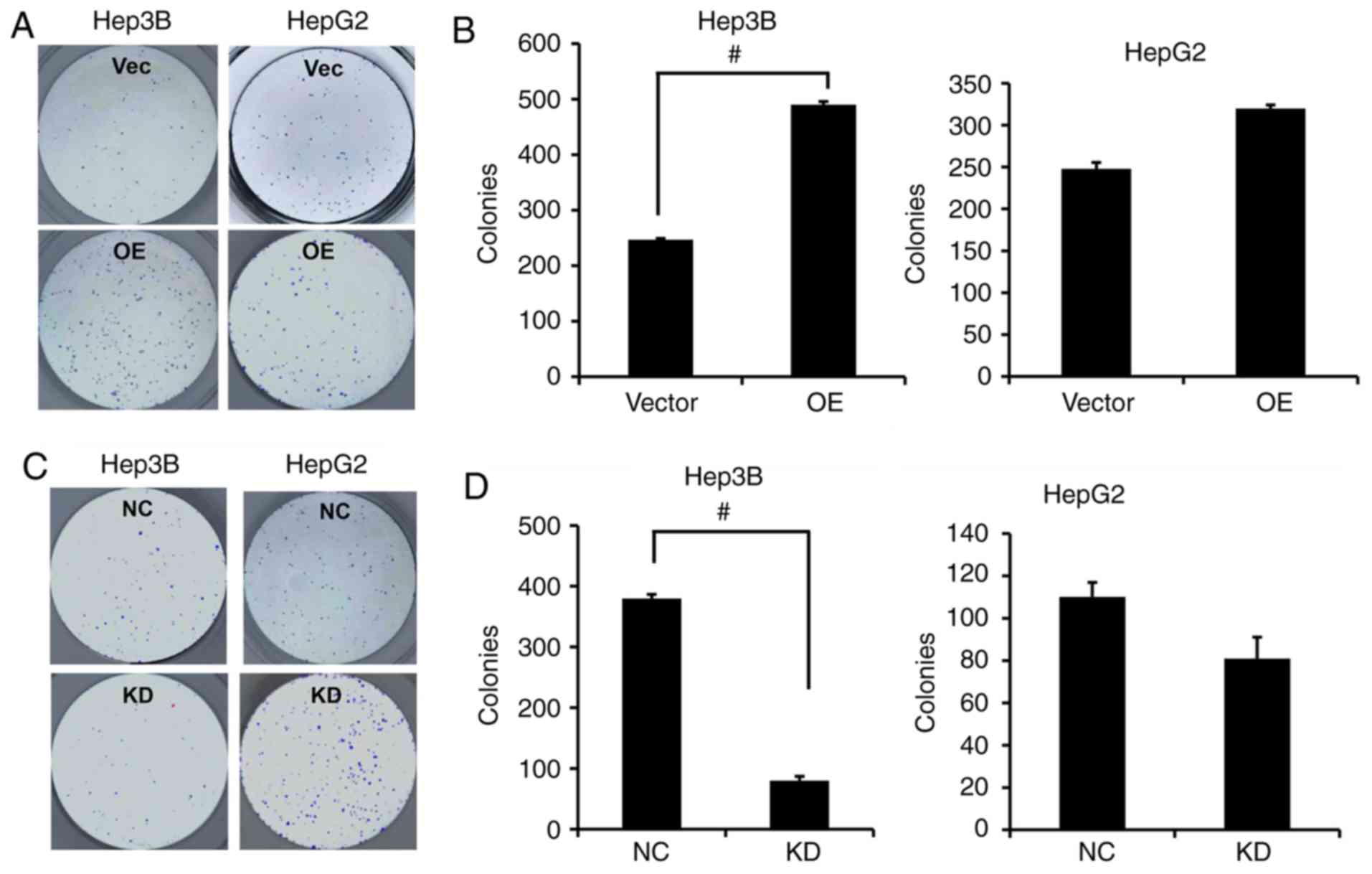
HepG2 cell line which showed no significant decrease (Fig. 2G and H). In addition, the colony
formation capacity was significantly enhanced following
overexpression of PVT1 in the Hep3B cells while the colony
formation capacity in the HepG2 cells was only markedly increased
without significance (Fig. 3A and
B); in contrast, significantly fewer colonies were formed in
the Hep3B cells rather than the HepG2 cells when PVT1 was KD by
shRNA compared with the NC group (Fig.
3C and D).
PVT1 enhances HBV-positive liver
cancer cell migration and invasion
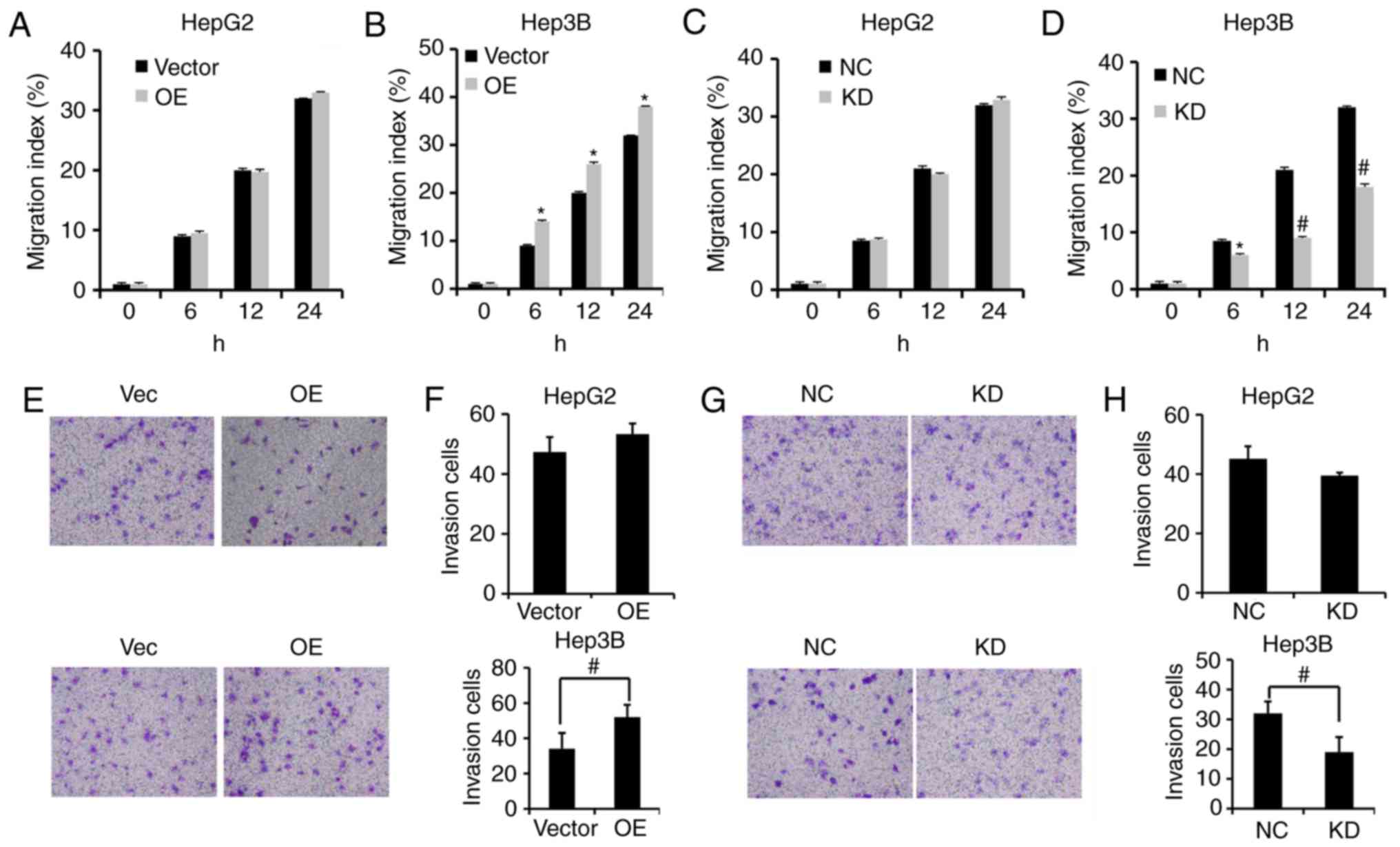
To assess the effects of PVT1 on the migration and
invasion of HBV-positive liver cancer cells, we performed a
wound-healing assay and invasion assay, respectively. Our results
showed that PVT1 overexpression (OE) significantly enhanced the
wound healing capacity in Hep3B cells rather than HepG2 cells
(Fig. 4A and B). Moreover, KD of
PVT1 by shRNA significantly reduced migration capacity of the Hep3B
cells rather than the HepG2 cells compared with the NC group
(Fig. 4C and D). The invasion
ability of HBV-positive Hep3B cells was significantly enhanced
following PVT1 OE (Fig. 4E and F)
and was impeded by PVT1 KD compared with the Vector and NC groups
(Fig. 4G and H).
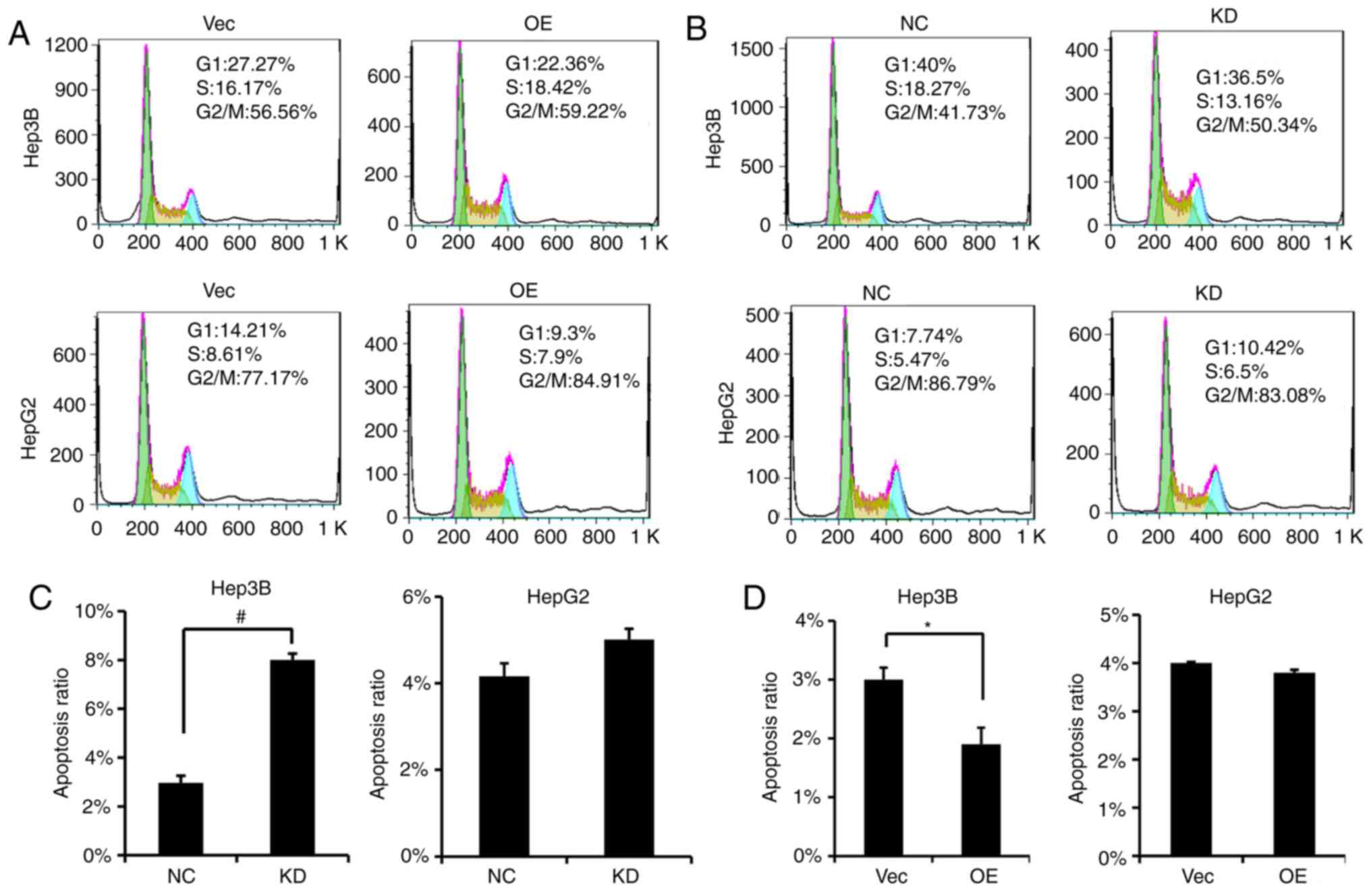
PVT1 causes changes in cell cycle
distribution and induces apoptosis of HBV-positive HCC cells
Based on our results, alterations in PVT1 expression
led to cell proliferation, migration and invasion changes.
Therefore, we investigated whether PVT1 would disrupt the cell
cycle profile of HBV-positive liver cancer cells. We found that in
contrast to the Vector group, the percentage of cells in the S
phase was increased following PVT1 OE in the Hep3B cells (Fig. 5A), and the percentage of cell in the
S phase was decreased following PVT1 KD in the Hep3B cells
(Fig. 5B). Furthermore, the cell
apoptosis rate was significantly increased in the PVT1 KD Hep3B
cells (Fig. 5C); on the other hand,
following PVT1 OE, the apoptotic ratio of the Hep3B cells was
significanly decreased (Fig.
5D).
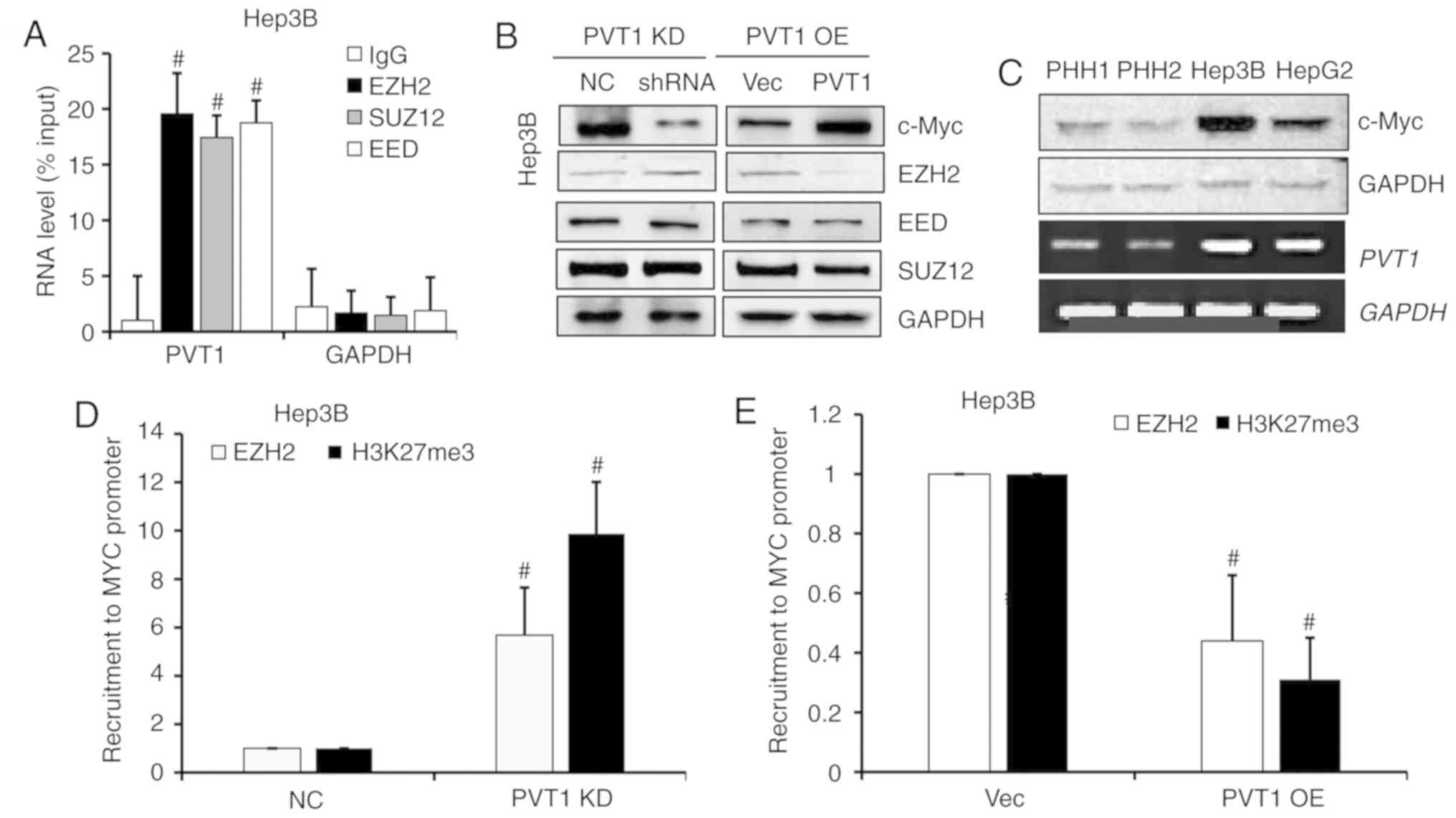
PVT1 negatively regulates EZH2
expression and dysregulates recruitment of EZH2 to the MYC
promoter
To further study the regulatory role of PVT1 in
HBV-positive HCC cells, RNA immunoprecipitation assay was
performed. As shown in Fig. 6A, we
detected a strong interaction between PVT1 with polycomb repressive
xomplex 2 (PRC2) subunits, such as EZH2, SUZ12 and EED (Fig. 6A). Next, to confirm this regulated
interaction, we determined the protein levels of PRC2 complex
members in PVT1 OE and KD Hep3B cells by western blot analysis.
When the PVT1 expression was KD or OE, the c-Myc protein level was
markedly downregulated or upregulated respectively, and EZH2 showed
a negative correlation with the PVT1 expression, but no obvious
changes in EED and SUZ12 were observed (Fig. 6B). Moreover, the c-Myc protein level
was positively correlated with PVT1 expression level in the PHH
cells and liver cancer cells, where in the HBV-positive Hep3B cells
the c-Myc protein level and PVT1 mRNA level were obviously high
than that in the HBV-negative HepG2 and PHH cells (Fig. 6C). To investigate a potential effect
on the recruitment ability of PVT1, we designed another experiment
which assessed the EZH2 protein and H3K27me3 level of the MYC
promotor by ChIP-qPCR. Lower PVT1 expression in the Hep3B cell line
induced EZH2 recruitment to the specific target gene, MYC that
contributed to H3K37me3 alteration (Fig. 6D). PVT1 OE significantly inhibited
EZH2 recruitment to the MYC promoter and decreased the H3K27me3
level of this region (Fig. 6E).
Discussion
The present study investigated the function of long
non-coding RNA (lncRNA) PVT1 in HBV-positive liver cancer
development and progression. We found that lncRNA PVT1 was
upregulation in HBV-positive HCC tissues and liver cancer cell
lines. We present evidence that the cell proliferation, migration
and invasion of the liver cancer cells was regulated by PVT1.
Knockdown of PVT1 significantly arrested HBV-positive liver cancer
cell cycle and induced apoptosis. More importantly, the protein
expression and recruitment of the core subunit of PRC2, EZH2, was
negatively regulated by PVT1. These results indicated that PVT1
expression could be an independent prognostic and diagnostic factor
for HBV-positive HCC and is critical for the tumorigenesis and
progression of HBV-positive HCC.
lncRNA PVT1 has been studied in various human
cancers and has been identified to play an oncogenic role.
Increased PVT1 expression was significantly associated with
histological grade, lymph node metastasis, and poor overall
survival in non-small cell lung cancer (NSCLC) (10). The PVT1 locus nearby the
normal-MML-network encompassed tumor-related genes indicating a
possible role of surveillance to tumorigenesis (23). PVT1 was found to increase FOXM1
post-translation by directly binding FOXM1 protein, and FOXM1 also
could bind to the PVT1 promoter to activate its transcription in
gastric cancer (24). PVT1 is also
highly expressed in the tissues of cisplatin-resistant gastric
patients and cisplatin-resistant cells, and upregulation of PVT1
was found to increase the expression of MDR1, and exhibit an
anti-apoptotic effect (25). Our
results showed that lncRNA PVT1 was upregulated in HBV-positive HCC
tissues and liver cancer cells, and PVT1 was significantly
associated with the proliferation, migration, and invasion of
HBV-positive liver cancer cells rather than HBV-negative liver
cancer cells.
Enhancer of zeste homolog 2 (EZH2), a member of the
polycomb group (PcG) protein family, suppresses many
tumor-suppressor genes, including miRNAs by modified transcription
at the epigenetic level (26,27).
PVT1 binding to EZH2 epigenetically regulates P15 and P16 causing
G1 arrest in gastric cancer (28).
Zhang et al demonstrated that PVT1 could combine with EZH2
and recruit EZH2 to the miR-200b promoter, thereby increasing
histone H3K27 trimethylation level of the miR-200b promoter, and
inhibiting miR-200b expression in cervical cancer progression
(29). RNA immunoprecipitation and
chromatin immunoprecipitation assays demonstrated that PVT1
recruits EZH2 to the promoter of large tumor suppressor kinase2
(LATS2) and represses LATS2 transcription in NSCLC (30). The location of PVT1 is recognized as
a cancer risk locus that is shared with the well-known MYC
oncogene. The gain of PVT1 expression was found to be required for
MYC protein upregulation in human cancer cells and was co-increased
in more than 98% of MYC-copy-increase cancers (31). Guan et al found that
inhibition of PVT1 induced apoptosis and suppression of MYC
expression contributing independently to ovarian and breast cancer
(7). In this study, we provide
evidence that PVT1 binds EZH2 and interferes with the recruitment
of EZH2 onto MYC promoter in HBV-positive liver cancer cells
by RNA immunoprecipitation and chromatin immunoprecipitation
assays. The important role of PVT1 in many types of cancer has been
studied, and the present study explored its functional role in
HBV-positive HCC. Nevertheless, whether the upregulation of lncRNA
PVT1 is a consequence of HBV infection or tumorigenesis remains
unelaborated in our study, and lack of comparison between normal
liver tissue and HBV infected tissue is the main limitation of the
present study. Our study clearly showed that expression of lncRNA
PVT1 was higher in the HBV-positive HCC tissues when compared with
the HBV-negative HCC tissues, and the expression was elevated
gradually with the progression of HCC. Moreover, PVT1expression was
higher in liver cancer cells compared with normal liver cells.
These data indicate that, at least partially, high expression of
PVT1 is associated with HBV infection and tumor development, but
this conclusion should be tested in HBV-positive and HBV-negative
normal liver tissues. Since virus infection is crucial for
activation of gene transcription initiation and elongation
(32), the lncRNA PVT1 is involved
in HBV-related HCC development. Thus, the present study may provide
a new approach and facilitate lncRNA PVT1-directed diagnostic and
therapeutic strategies for HBV-positive HCC at the early
stages.
Acknowledgements
Not applicable.
Funding
This research was supported by the Foundation of
Tianjin Second People's Hospital (YS-0008 to BJ), the Tianjin
Natural Science Foundation (18JCQNJC11800 to BJ), the Tianqing
Foundation (TQGB20190021 to BJ), the National Natural Science
Foundation of China (30870583 to QW), the Youth Teachers Fund of
Peking Union Medical College (2014zlgc0755 to QW) and the Natural
Science Foundation of Tianjin (16JCQNJC12100 to BY).
Availability of data and materials
The datasets used during the present study are
available from the corresponding author upon reasonable
request.
Authors' contributions
BJ and BY contributed to the writing of the
manuscript. BJ, QW, BY and XZ contributed to performing of the
experiments and the statistical analyses. YG and XZ provided the
patient samples and performed the clinical statistics. WL
contributed to the design of the experiments. All authors have
approved the final version of the publication and agree to be
accountable for all aspects of the research in ensuring that the
accuracy or integrity of any part of the work are appropriately
investigated and resolved.
Ethics approval and consent to
participate
Written informed consent was obtained from all
patients enrolled in this study, and the present study protocol was
approved by the Ethics Committee of Tianjin Second People's
Hospital.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Ringelhan M, Pfister D, O'Connor T,
Pikarsky E and Heikenwalder M: The immunology of hepatocellular
carcinoma. Nat Immunol. 19:222–232. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Liu J, Yang HI, Lee MH, Lu SN, Jen CL,
Batrla-Utermann R, Wang LY, You SL, Hsiao CK, Chen PJ, et al:
Spontaneous seroclearance of hepatitis B seromarkers and subsequent
risk of hepatocellular carcinoma. Gut. 63:1648–1657. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Levrero M and Zucman-Rossi J: Mechanisms
of HBV-induced hepatocellular carcinoma. J Hepatol. 64 (1
Suppl):S84–S101. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Shtivelman E, Henglein B, Groitl P, Lipp M
and Bishop JM: Identification of a human transcription unit
affected by the variant chromosomal translocations 2;8 and 8;22 of
Burkitt lymphoma. Proc Natl Acad Sci USA. 86:3257–3260. 1989.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Huppi K, Siwarski D, Skurla R, Klinman D
and Mushinski JF: Pvt-1 transcripts are found in normal tissues and
are altered by reciprocal(6;15) translocations in mouse
plasmacytomas. Proc Natl Acad Sci USA. 87:6964–698. 1990.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Barsotti AM, Beckerman R, Laptenko O,
Huppi K, Caplen NJ and Prives C: p53-Dependent induction of PVT1
and miR-1204. J Biol Chem. 287:2509–2519. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Guan Y, Kuo WL, Stilwell JL, Takano H,
Lapuk AV, Fridlyand J, Mao JH, Yu M, Miller MA, Santos JL, et al:
Amplification of PVT1 contributes to the pathophysiology of ovarian
and breast cancer. Clin Cancer Res. 13:5745–5755. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Takahashi Y, Sawada G, Kurashige J, Uchi
R, Matsumura T, Ueo H, Takano Y, Eguchi H, Sudo T, Sugimachi K, et
al: Amplification of PVT-1 is involved in poor prognosis via
apoptosis inhibition in colorectal cancers. Br J Cancer.
110:164–171. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ding J, Li D, Gong M, Wang J, Huang X, Wu
T and Wang C: Expression and clinical significance of the long
non-coding RNA PVT1 in human gastric cancer. OncoTargets Ther.
7:1625–1630. 2014. View Article : Google Scholar
|
|
10
|
Yang YR, Zang SZ, Zhong CL, Li YX, Zhao SS
and Feng XJ: Increased expression of the lncRNA PVT1 promotes
tumorigenesis in non-small cell lung cancer. Int J Clin Exp Pathol.
7:6929–6935. 2014.PubMed/NCBI
|
|
11
|
Xu Y, Luo X, He W, Chen G, Li Y, Li W,
Wang X, Lai Y and Ye Y: Long non-coding RNA PVT1/miR-150/HIG2 axis
regulates the proliferation, invasion and the balance of iron
metabolism of hepatocellular carcinoma. Cell Physiol Biochem.
49:1403–1419. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Guo J, Hao C, Wang C and Li L: Long
noncoding RNA PVT1 modulates hepatocellular carcinoma cell
proliferation and apoptosis by recruiting EZH2. Cancer Cell Int.
18:982018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Derderian C, Orunmuyi AT, Olapade-Olaopa
EO and Ogunwobi OO: PVT1 signaling is a mediator of cancer
progression. Front Oncol. 9:5022019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Cui M, You L, Ren X, Zhao W, Liao Q and
Zhao Y: Long non-coding RNA PVT1 and cancer. Biochem Biophys Res
Commun. 471:10–4. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chen L, Sun L, Dong L, Cui P, Xia Z, Li C
and Zhu Y: The role of long noncoding RNA-LET in cell proliferation
and invasion of nasopharyngeal carcinoma and its mechanism.
OncoTargets Ther. 10:2769–2778. 2017. View Article : Google Scholar
|
|
16
|
Davidovich C and Cech TR: The recruitment
of chromatin modifiers by long noncoding RNAs: Lessons from PRC2.
RNA. 21:2007–2022. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Khalil AM, Guttman M, Huarte M, Garber M,
Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van
Oudenaarden A, et al: Many human large intergenic noncoding RNAs
associate with chromatin-modifying complexes and affect gene
expression. Proc Natl Acad Sci USA. 106:11667–116672. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
He A, Shen X, Ma Q, Cao J, von Gise A,
Zhou P, Wang G, Marquez VE, Orkin SH and Pu WT: PRC2 directly
methylates GATA4 and represses its transcriptional activity. Genes
Dev. 26:37–42. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chen Y, Lin MC, Wang H, Chan CY, Jiang L,
Ngai SM, Yu J, He ML, Shaw PC, Yew DT, et al: Proteomic analysis of
EZH2 downstream target proteins in hepatocellular carcinoma.
Proteomics. 7:3097–3104. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Villanueva A: Hepatocellular Carcinoma. N
Engl J Med. 380:1450–1462. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Pfeiffer E, Kegel V, Zeilinger K,
Hengstler JG, Nüssler AK, Seehofer D and Damm G: Featured article:
Isolation, characterization, and cultivation of human hepatocytes
and non-parenchymal liver cells. Exp Biol Med (Maywood).
240:645–656. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Colombo T, Farina L, Macino G and Paci P:
PVT1: A rising star among oncogenic long noncoding RNAs. Biomed Res
Int. 2015:3042082015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Xu MD, Wang Y, Weng W, Wei P, Qi P, Zhang
Q, Tan C, Ni SJ, Dong L, Yang Y, et al: A positive feedback loop of
lncRNA-PVT1 and FOXM1 facilitates gastric cancer growth and
invasion. Clin Cancer Res. 23:2071–2080. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhang XW, Bu P, Liu L, Zhang XZ and Li J:
Overexpression of long non-coding RNA PVT1 in gastric cancer cells
promotes the development of multidrug resistance. Biochem Biophys
Res Commun. 462:227–232. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ezhkova E, Pasolli HA, Parker JS, Stokes
N, Su IH, Hannon G, Tarakhovsky A and Fuchs E: Ezh2 orchestrates
gene expression for the stepwise differentiation of tissue-specific
stem cells. Cell. 136:1122–1135. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Gao SB, Zheng QF, Xu B, Pan CB, Li KL,
Zhao Y, Zheng QL, Lin X, Xue LX and Jin GH: EZH2 represses target
genes through H3K27-dependent and H3K27-independent mechanisms in
hepatocellular carcinoma. Mol Cancer Res. 12:1388–1397. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kong R, Zhang EB, Yin DD, You LH, Xu TP,
Chen WM, Xia R, Wan L, Sun M, Wang ZX, et al: Long noncoding RNA
PVT1 indicates a poor prognosis of gastric cancer and promotes cell
proliferation through epigenetically regulating p15 and p16. Mol
Cancer. 14:822015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhang S, Zhang G and Liu J: Long noncoding
RNA PVT1 promotes cervical cancer progression through
epigenetically silencing miR-200b. APMIS. 124:649–658. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wan L, Sun M, Liu GJ, Wei CC, Zhang EB,
Kong R, Xu TP, Huang MD and Wang ZX: Long noncoding RNA PVT1
promotes non-small cell lung cancer cell proliferation through
epigenetically regulating LATS2 expression. Mol Cancer Ther.
15:1082–1094. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Tseng YY, Moriarity BS, Gong W, Akiyama R,
Tiwari A, Kawakami H, Ronning P, Reuland B, Guenther K, Beadnell
TC, et al: PVT1 dependence in cancer with MYC copy-number increase.
Nature. 512:82–86. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Clark DN and Hu J: Hepatitis B virus
reverse transcriptase-target of current antiviral therapy and
future drug development. Antiviral Res. 123:132–137. 2015.
View Article : Google Scholar : PubMed/NCBI
|