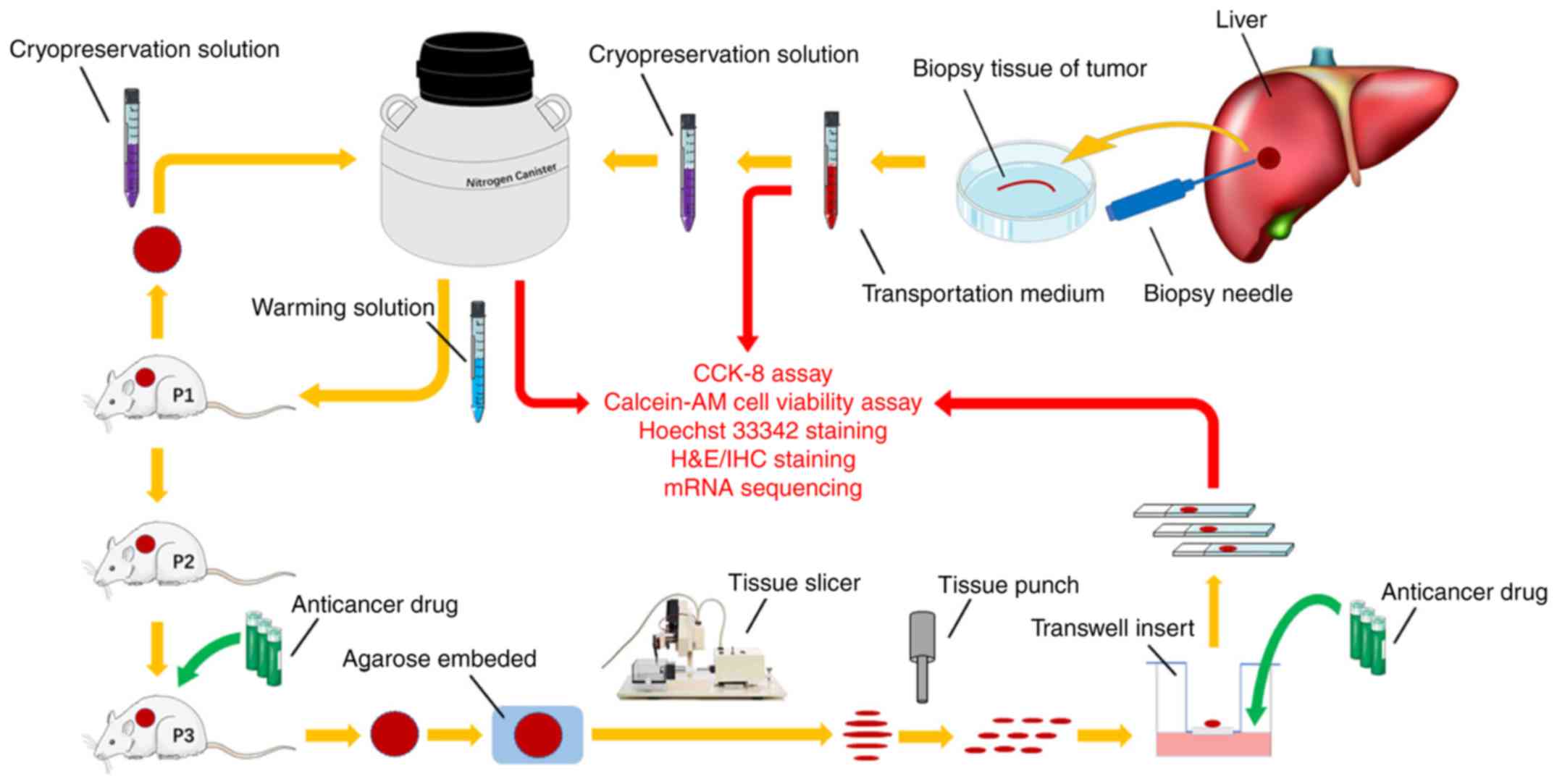
Introduction
Colorectal cancer, including rectal cancer, is the
third most prevalent malignancy worldwide and a leading cause of
cancer-related deaths in the developed world (1). Synchronous distant metastases are
diagnosed in approximately 15–20% of rectal cancer patients, most
commonly involving the liver (2).
Surgery remains the standard of care for rectal cancer liver
metastasis (RLM). However, 85% of patients with RLM are considered
unresectable (3). Those patients
require systemic chemotherapy, in the form of a fluoropyrimidine
doublet (FOLFOX/CAPOX or FOLFIRI/CAPIRI) combined with a biologic
targeting either angiogenesis, or epidermal growth factor receptor
in patients with RAS wild-type tumors (4). Unfortunately, many patients do not
respond to first-line chemotherapy, or develop resistance.
Consequently, further cancer research in drug testing and optimized
personalization strategies for RLM treatment are urgently
needed.
In previous years, established cell lines have been
widely used in many aspects of medical research, and particularly
as in vitro models in preclinical drug testing (5). However, these are an imperfect model
because the three-dimensional architecture of the tumor tissue and
cell-cell communications that clearly exist in vivo are
lost, and therefore they do not faithfully reflect the tumor of
origin (6). Recently, various novel
strategies have been applied to maintain or reconstitute an
environment closely resembling the tumor tissue, such as
two-dimensional culture of dissociated tumor cells and
three-dimensional spheroid cultures (7). However, these still cannot imitate the
intricate tissue architecture and the high degree of variability
seen in individual tumors. It is becoming increasingly clear that
the progression of cancer and the response to chemotherapeutic
drugs mainly depends on specific intercommunications between tumor
cells and surrounding tissue components (8). Therefore, an ideal model is required
for the accurate characterization of chemotherapy sensitivity in
RLM tumors from patients.
A potentially desirable model was provided by
precision-cut slicing, which maintains the complete tissue
architecture and full heterogeneity of the tumor in vivo
(9,10). However, there is no standard
approach to cultivating tumor slices in vitro. Besides,
slice cultivation derived from fresh biopsy tissue, which is
convenient to obtain and preserve, is rarely reported. Another
problem is that no preservation method has thus far been applied to
maintain living fresh tissue, to the best of our knowledge.
Currently, the conventional preservation techniques for fresh tumor
tissue, either as formalin-fixed paraffin embedded samples or by
flash freezing in liquid nitrogen, lead to the absolute
inactivation of the fresh tissue and can only provide morphological
and genetic information, which has led to the infrequent
utilization of fresh specimens. Recently, a standardized
vitrification-based cryopreservation method has been developed to
preserve fresh tumor tissue, by which the biological
characteristics of the original tumor can be retained and the
utilization of specimens may be markedly improved.
The present study combined the vitrification-based
cryopreservation method with the precision-cut slicing technique,
using available tumor tissues from a mouse model of human RLM
biopsy to assess anticancer drug responses.
Materials and methods
Collection of human RLM biopsy
tissues
Fresh sterile biopsy tissues were obtained from
patients with RLM at the Department of Interventional Oncology,
Renji Hospital Affiliated to Shanghai Jiaotong University School of
Medicine (Shanghai, China). Samples were collected between June
2016 and December 2016. There were a total of 20 patients,
including 10 men and 10 women, with a mean age of 60 years. The
pathological diagnosis of all patients was RLM and none had
received any prior treatment. The fresh biopsy tissues were then
directly transported to the laboratory within 2 h. All specimens
were kept at 4°C on ice and transported in preservation medium
(Tissue Mate™; Celliver Biotechnology, Inc.) which is composed of
energy substrate, iron chemicals and antioxidants, without protein.
This investigation was approved by the ethics committee of Renji
Hospital and all patients provided written informed consent.
Details are illustrated in Fig.
1.
Cryopreservation and warming
procedures
All cryopreservation solutions (cat. no. LT2601;
LiveTissue™) and warming solutions (cat. no. LT2602; LiveTissue™)
were provided by Celliver Biotechnology, Inc. For tissue
cryopreservation, vitrification solution 1 (V1), vitrification
solution 2 (V2) and vitrification solution 3 (V3) were pre-warmed
in a 26°C water bath. Fresh RLM biopsy tissues were cleaned twice
with sterile PBS and transferred into 5 ml V1, 5 ml V2 and 5 ml V3
for 3, 3 and 6 min, respectively. Tissues were then placed onto a
thin metal strip and submerged into liquid nitrogen for at least 5
min. Finally, the strips with tissues were placed into frozen
storage tubes and preserved in the nitrogen canister. The tissue
samples were stored in the liquid nitrogen until warming. For
tissue warming, the frozen storage tubes were removed from the
nitrogen canister and the strips with the cryopreserved biopsy
tissues were quickly transferred into 10 ml warming solution 1, and
incubated for 3 min in a 37°C water bath. The tissues were then
transferred into 5 ml warming solution 2 and 5 ml warming solution
3 for 5 and 10 min, respectively, at 26°C. Warmed tissues were
cleaned twice with sterile PBS and kept on ice until use.
Tissue slice preparation and
cultivation
Warmed tissues were used to establish the first
generation of patient-derived xenografts (PDXs), in order to
produce ample tumor tissues for further experiments. Subcutaneous
xenografts were cut into 300-µM-thick precision-cut slices using a
microtome for slice preparation [slices of 300 µM were considered
to be the most suitable thickness for RLM after conducting several
slicing experiments with different thicknesses (data not
shown)].
For precision-cut slice preparation, tissues were
embedded in 2% low temperature gelling agarose (Sigma-Aldrich;
Merck KGaA) and 300-µm-thick slices were prepared using a VF-300
Microtome (Bio-Gene Technology, Ltd.). Parameter settings, such as
the frequency and amplitude of vibration slicing, were determined
by the specific tumor texture. Tissue slices (diameter, 2 mm) were
then prepared using a hand-held coring tool, and all the procedures
were performed under sterile conditions.
Precision-cut slices were maintained on Transwell
inserts (pore size, 0.4 µm; Corning, Inc.), with two slices to each
insert. Cultivation was performed in 24-well plates containing 450
µl RPMI-1640 medium (BasalMedia) with 10% fetal bovine serum
(Biological Industries), penicillin (100 U/ml; BasalMedia) and
streptomycin (100 U/ml; BasalMedia), and kept at 37°C in a
humidified incubator with 5% CO2. For drug testing of
slices in vitro, oxaliplatin (OXA; MedChemExpress, LLC) was
used and tested at a concentration of 20 µM. Drug testing commenced
after 24 h of slice culture and was performed for an additional 72
h. Medium changes were performed every 24 h.
Experimental methods for mRNA
sequencing
RNA purity was checked using the
kaiaoK5500® Spectrophotometer (Beijing Kaiao Technology
Development Co., Ltd). RNA integrity and concentration was assessed
using the RNA Nano 6000 Assay kit and the Bioanalyzer 2100 system
(Agilent Technologies, Inc.). A total amount of 2 µg RNA/sample was
used as input material for the RNA sample preparations. Sequencing
libraries were generated using NEBNext® Ultra™ RNA
Library Prep kit for Illumina® (cat. no. E7530L; New
England BioLabs, Inc.), following the manufacturer's
recommendations, and index codes were added to attribute sequences
to each sample. Briefly, mRNA was purified from the total RNA using
poly-T oligo-attached magnetic beads. Fragmentation was carried out
using divalent cations under elevated temperature in NEBNext First
Strand Synthesis Reaction Buffer (5X). First strand cDNA was
synthesized using random hexamer primer and RNase H. Second strand
cDNA synthesis was subsequently performed using buffer, dNTPs, DNA
polymerase I and RNase H. The library fragments were purified with
QiaQuick PCR kits (Qiagen, Inc.) and elution with EB buffer, then
terminal repair, A-tailing and adapter adding were implemented. The
products were retrieved and PCR was performed, then the library was
completed. The RNA concentration of the library was measured using
a Qubit® RNA Assay kit in Qubit® 3.0 (Thermo
Fisher Scientific, Inc.)for preliminary quantification, and then
diluted to 1 ng/µl. Insert size was assessed using the Agilent
Bioanalyzer 2100 system (Agilent Technologies, Inc.), and qualified
insert size was accurately quantified using the StepOnePlus™
Real-Time PCR System (Thermo Fisher Scientific, Inc.; library valid
concentration, >10 nM). The clustering of the index-coded
samples was performed on a cBot cluster generation system using a
HiSeq PE Cluster kit v4-cBot-HS (Illumina, Inc.) according to the
manufacturer's instructions. After cluster generation, the
libraries were sequenced on an Illumina, Inc. platform and 150 bp
paired-end reads were generated. The variations in gene expression
could be detected by different colors in the heat map. Kyoto
Encyclopedia of Genes and Genomes (KEGG) enrichment exhibited the
main types and functions of the detected differential genes. Kyoto
Encyclopedia of Genes and Genomes (KEGG) pathway analysis was
processed using KOBAS 3.0 (http://kobas.cbi.pku.edu.cn/).
Establishment of xenograft model
A total of 20 female NOD/SCID mice of 6–8 weeks old
were housed and treated under specific pathogen-free conditions at
the Experimental Animal Center of Shanghai Jiaotong University
School of Medicine. Mice were housed in a 12-h light/dark cycle
with food and water available at all times. The room temperature
was maintained at 24±1°C and relative humidity at 50%. The average
weight of the animals was 18 g. All mice were purchased from the
Shanghai Experimental Center of Chinese Academy of Science. All the
experimental procedures performed on the animals were approved by
the Shanghai Medical Experimental Animal Care Commission.
Mice were implanted subcutaneously with warmed
biopsy tissues on the right flank, and tumor growth was monitored
twice a week. When the tumor burden was ~500 mm3 [tumor
volume was measured with calipers and calculated using the formula
V=1/2 (length × width2)], the mice were sacrificed and
the tumors were used for continuous passaging to the third
generation. The subcutaneous xenograft tumors of PDXs derived from
warmed biopsy tissues were cut into 1-mm-thick slices in a metal
mold before cryopreservation. When the tumor burden was 100–150
mm3, mice with xenografts derived from fresh tissue and
warmed tissue were injected intraperitoneally with OXA in 200 µl
PBS at a dose of 5 mg/kg (twice a week for 4 weeks). All the mice
in the PBS group were injected intraperitoneally with 200 µl PBS.
The mice in the control group were injected with nothing. The mice
were sacrificed 4 weeks later and the tumors were used for
hematoxylin and eosin (H&E)/immunohistochemical (IHC)
staining.
Cell Counting Kit-8 (CCK-8) assay
A CCK-8 assay (Dojindo Molecular Technologies, Inc.)
was used to evaluate the viability of tissue slices at each time
point (24, 48, 72 and 96 h) during the culturing process in
vitro. RPMI-1640 medium (50 µl/well) and CCK-8 solution (10
µl/well) were added into 96-well plates. The tissue slices were
added one slice/well. The plates were kept at 37°C in a humidified
incubator with 5% CO2 for 2 h. The slices were removed
from the 96-well plates and the plates were transferred to the
Thermo Fisher Scientific, Inc. microplate reader (Multiskan GO).
The absorbance at 450 nm was measured and three wells were tested
for each sample at each time point. In addition, during preliminary
work, the cell viability following different lengths of
preservation time in liquid nitrogen were compared by CCK-8
assay.
Acetoxymethyl ester of calcein
(calcein-AM) cell viability assay and Hoechst 33342 staining
The Live/Dead® Viability Assay kit
(Nanjing KeyGen Biotech Co., Ltd.) and Hoechst 33342 (Beyotime
Institute of Biotechnology) were stored at −20°C and allowed to
warm to room temperature prior to experimentation. The viability
assay stock reagents (calcein-AM, 4 mM) were diluted to 1 µM in
physiological solution and mixed with 2 µg/ml Hoechst 33342 stock
reagents at room temperature for 30 min. Representative images were
captured with the Leica TCS SP8 confocal microscope (Leica
Microsystems GmbH). The ratio of living cells in the calcein-AM
cell viability assay/Hoechst 33342 staining was calculated based on
manual counting within ten random microscopic fields.
H&E/IHC staining
Tissue slices were fixed in 10% phosphate-buffered
formalin for at least 24 h at room temperature and subsequently
paraffin-embedded. For the examination of histopathology, paraffin
sections (4 µm) were stained with H&E at room temperature.
Before incubating with antibodies, samples were blocked with 20%
goat serum (cat. no. Ab138478; Abcam) for 15 min at room
temperature. For Ki67 staining, sections were incubated with the
primary antibody (cat. no. Ab15580; Abcam; 1:1,000 dilution) and a
horseradish peroxidase (HRP)-conjugated secondary antibody (cat.
no. Ab205718; Abcam; 1:20,000 dilution). For caspase-3 staining,
sections were incubated with the primary antibody (cat. no.
Ab184787; Abcam; 1:1,000 dilution) and an HRP-conjugated secondary
antibody (cat. no. Ab97051; Abcam; 1:500 dilution). Sections were
dewaxed in xylene and rehydrated in an ethanol gradient of 100, 95
and 80%, and heat-mediated antigen retrieval of the tissue sections
was carried out at a temperature of 96–98°C before they were
allowed to cool. Paraffin-embedded sections (4 µm) were incubated
with primary antibodies overnight at 4°C. The secondary antibody
was used to detect the primary for 1 h at room temperature.
Confocal laser scanning microscopy was performed using an Olympus
Corporation BX51 instrument.
Statistical analysis
Statistical evaluations were performed using a
Student's t-test and one-way ANOVA with post hoc least significant
difference test by IBM SPSS Statistics 22.0 (IBM Corp). P<0.05
was considered to indicate a statistically significant difference.
Three repeats were performed.
Results
Biological characteristics of RLM
biopsy tissues are retained by vitrification-based
cryopreservation
By conducting several CCK-8 assays during
preliminary work, it was found that no difference was induced by
different lengths of preservation time in liquid nitrogen (data not
shown). All the fresh RLM biopsies were obtained from Department of
Tumor Interventional Oncology, Renji Hospital, School of Medicine.
The study was performed using standard procedures, as depicted in
Fig. 1. The freshly collected RLM
biopsy tissues were cryopreserved and warmed according to the
schedule in Fig. 2A, using
cryopreservation solutions (Fig.
2B, left) and warming solutions (Fig. 2B, right). Fresh biopsy tissues
(Fig. 2C) were soft and fragile,
and required gentle manipulation in the cryopreservation and
warming procedures. The cryopreserved tissues (Fig. 2D, top) were found to be translucent
and sclerotic, while the warmed tissues (Fig. 2D, bottom) recovered their softness
and luster. Xenograft models were successfully established with
both fresh biopsy tissues (Fig. 2E,
left) and warmed biopsy tissues (Fig.
2E, right). Fresh and warmed tumor biopsies from 20 patients
with RLM were implanted into NOD-SCID mice. From these, 11
×enografts derived from 20 fresh RLM biopsies and 10 ×enografts
derived from 20 warmed RLM biopsies were successfully established
(take rates, 55 and 50%, respectively).
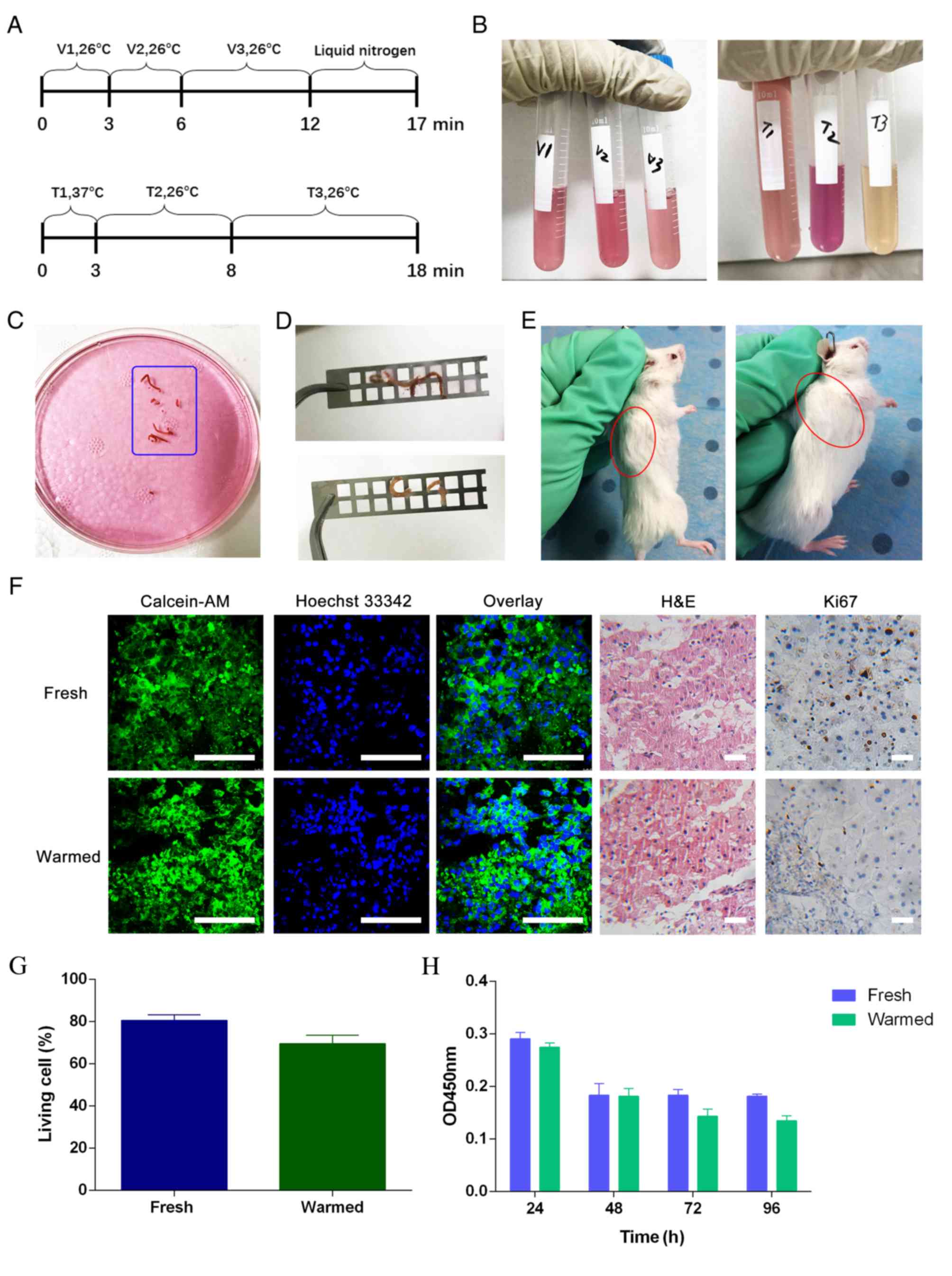 | Figure 2.Cryopreservation and warming of RLM
biopsy tissue. (A) The time schemes of the cryopreservation (top)
and warming procedures (bottom). (B) Cryopreservation solutions
(left) and warming solutions (right). (C) Fresh RLM biopsy. (D)
Cryopreserved tissues (top) and warmed tissues (bottom). (E)
Xenograft model derived from a fresh RLM biopsy tissue (left, n=11)
and warmed RLM biopsy tissue (right, n=10). (F) Calcein-AM cell
viability assay/Hoechst 33342 staining and H&E/IHC staining.
Blue nuclei comprise both living and dead nuclei, while green
sections represent the cytoplasm of living cells. IHC staining
indicated the percentage of cells expressing the viability marker
Ki67. Scale bars, 100 µm. (G) The living cell ratios of fresh and
warmed tissues. The ratios were found to be not statistically
different by Student's t-test (P=0.38). (H) Cell Counting Kit-8
assay of in vitro slice cultures before and after
cryopreservation. (I) Heat map of mRNA sequencing. The color change
in the heat map is defined as the difference in gene expression
between warmed and fresh tissues. Each row represents one gene, and
each column represents one type of sample. Compared to the areas of
blue color, red color represents increased gene expression. The
deeper the red, the more greatly increased the gene expression. The
deeper the blue, the lesser the gene expression. (J) KEGG
enrichment analysis. In the KEGG diagram, the size of each dot
represents the number of differential genes in the corresponding
pathway. The different colors of each dot indicate the different
degrees of KEGG enrichment. Red represents the highest KEGG
enrichment degree. V, vitrification solution; T, warming solution;
calcein-AM, acetoxymethyl ester of calcein; H&E, hematoxylin
and eosin; OD, optical density; KEGG, Kyoto Encyclopedia of Genes
and Genomes; IHC staining, immunohistochemical staining. |
Calcein-AM cell viability assay/Hoechst 33342
staining and H&E/IHC staining indicated that no obvious
difference was detectable between fresh tissues and warmed tissues
(Fig. 2F). Tissue viability was
assessed by calcein-AM cell viability assay/Hoechst 33342 staining.
The living cell ratio was 76.5% in fresh tissues and 75% in warmed
tissues, which confirmed that cryopreservation had little influence
on the biological viability of the tissue (Fig. 2G). H&E staining showed that the
morphological features remained in the warmed tissues, and were
similar to those of fresh tissues. The proliferative rate, as
assessed by Ki67, was maintained at almost the same level between
fresh and warmed tissues, indicating that little damage was induced
by the cryopreservation and warming procedures to the tissue
proliferative capacity. The CCK-8 assay illustrated that both
warmed and fresh tissues could be cultured in vitro for at
least 96 h (Fig. 2H). From the heat
map, it was apparent that the color of left column, representing
the warmed tissue, was mostly consistent with that of the fresh
tissue on the right. Only a small part of the heat map in the left
column was different from the right column, which corresponded to
genes with metabolic functions. Therefore, limited variation in
gene expression between fresh and warmed tissues was identified by
the heat map (Fig. 2I). According
to the KEGG enrichment analysis, it was found that many of the
differential genes, which were represented by large red dots, were
closely related to metabolic processes (Fig. 2J). These results confirmed that the
RLM biopsies were applicable to the establishment of PDX models,
and that the vitrification-based cryopreservation method was able
to largely maintain the biological activity and histological
features of the RLM biopsy tissues.
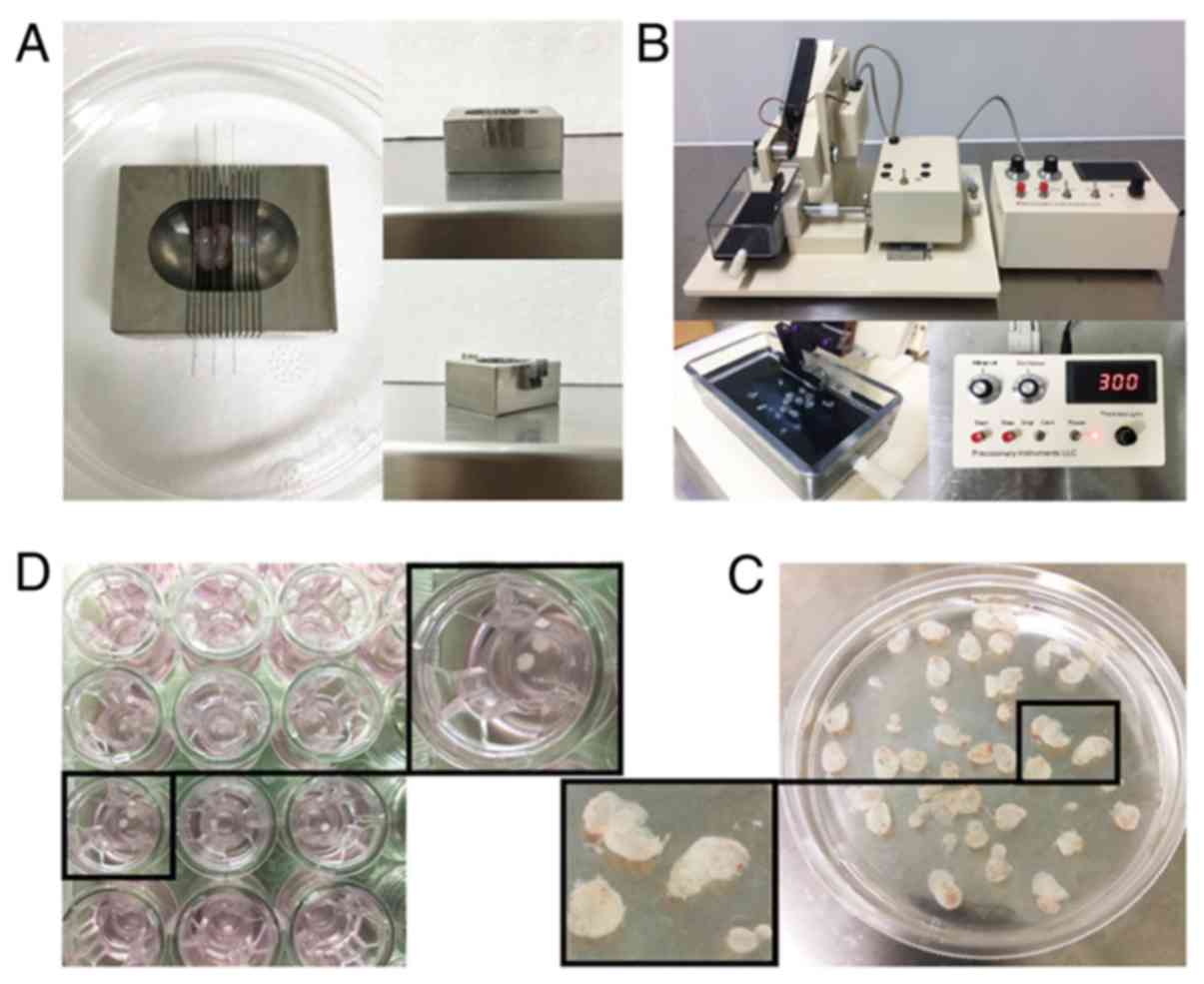
Precision-cut slices provide the
complete three-dimensional architecture and full heterogeneity of
the original tumor
The subcutaneous xenograft tumors of PDXs derived
from warmed biopsy tissues were cut into 1-mm-thick slices in a
metal mold before cryopreservation (Fig. 3A). Precision-cut 300-µM-thick slices
were obtained using a VF-300 microtome (Fig. 3B and C). Tissue slices (2 mm in
diameter) were then prepared using a hand-held coring tool and
maintained on Transwell inserts (pore size, 0.4 µm) (Fig. 3D).
To test the in vitro anticancer responses of
both fresh and warmed slices using OXA, standardized slice
culturing and drug testing were conducted, and repeated three
times. The calcein-AM cell viability assay/Hoechst 33342 staining
and H&E/IHC staining indicated that no obvious differences were
detectable after 24 h of slice culturing before drug testing
(Fig. 4A, row 1; Fig. 4B, rows 1 and 3; Fig. 4C, P>0.05). However, compared with
slices without drug treatment, both fresh and warmed tissue slices
cultured for 96 h showed a marked decrease in cell viability by IHC
staining when treated with drugs (Fig.
4A, row 2). Morphological features visible by H&E staining
and Ki67 staining were also changed in both fresh and warmed slices
treated with OXA, while they were retained in control slices
(Fig. 4B, row 2). Accordingly, a
significant decline in Ki67 was detected by IHC staining in slices
treated with drugs, while no significant change was found in the
control (Fig. 4B, row 4).
Furthermore, tissue slices treated for 96 h with anticancer drugs
showed a specific time-dependent reduction in tissue viability by
CCK-8 assay (Fig. 4C). Besides, it
was found that fresh and warmed tissues both showed evident
responses to OXA (P<0.05), which indicated that the
cryopreservation technique had little influence on the tumor
biology. Above all, the precision-cut slices retained the
three-dimensional architecture and tumor heterogeneity, and may
therefore represent an optimal tool for in vitro drug
testing.
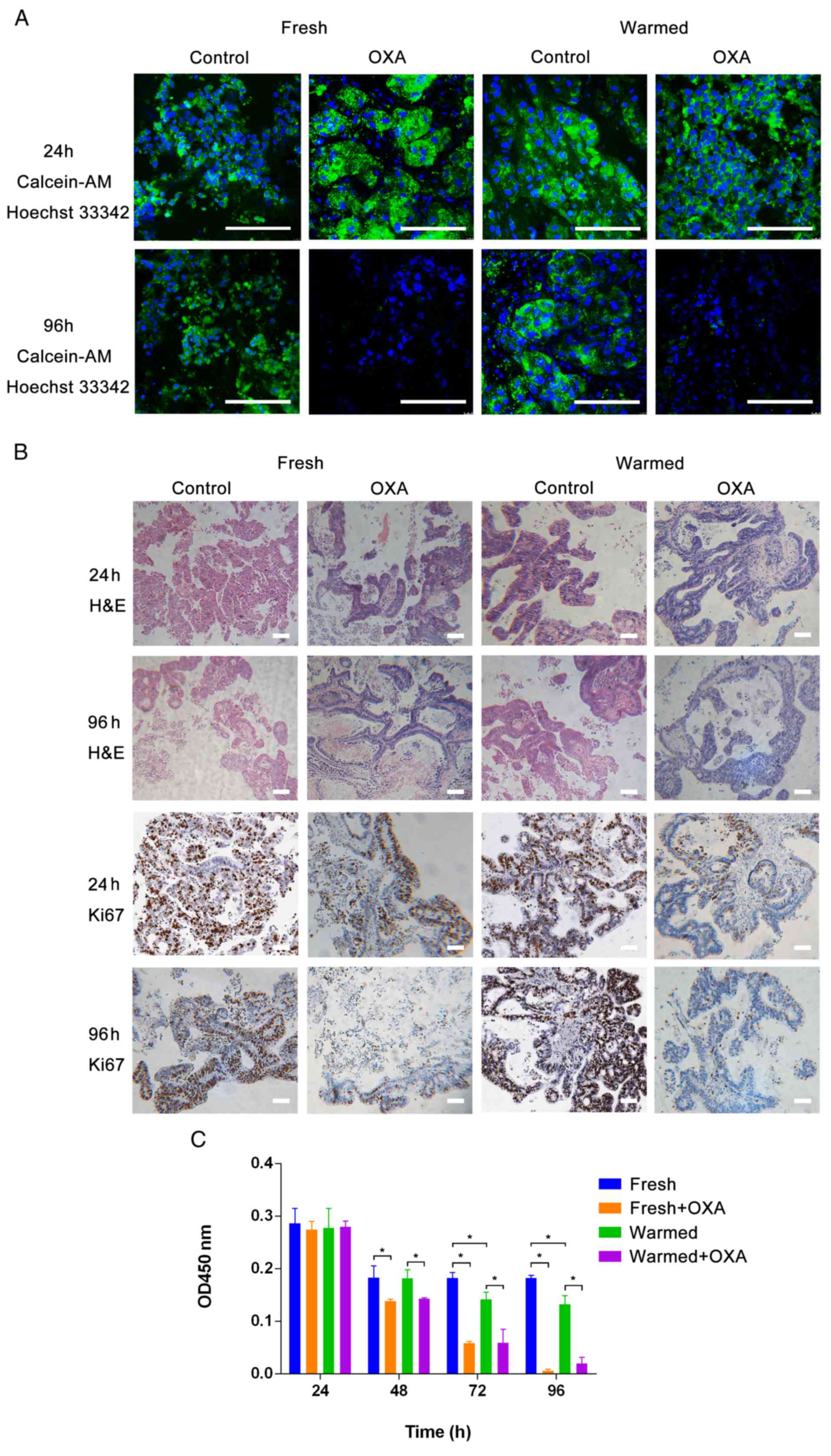 | Figure 4.Drug testing in precision-cut slice
cultures. (A) Calcein-AM cell viability assay/Hoechst 33342
staining. Scale bars, 100 µm. (B) H&E/immunohistochemical
staining. Scale bars, 100 µm. (C) CCK-8 assay of fresh and warmed
tissue slice cultures treated with OXA. No obvious difference was
detected after 24 h of slice culturing. After 48 h, the OD values
of the fresh group were similar to those in the warmed group, while
the OD values of the fresh group were different from those of the
fresh + OXA group. Also, the OD values of the warmed group were
different from those of the warmed + OXA group. After 72 h, the OD
values of the fresh group were different to those of the warmed
group. Also, the OD values of the no drug group were different from
those of the drug group. After 96 h, the result was the same as at
72 h. *P<0.05. Calcein-AM, acetoxymethyl ester of calcein;
H&E, hematoxylin and eosin; OXA, oxaliplatin; OD, optical
density. |
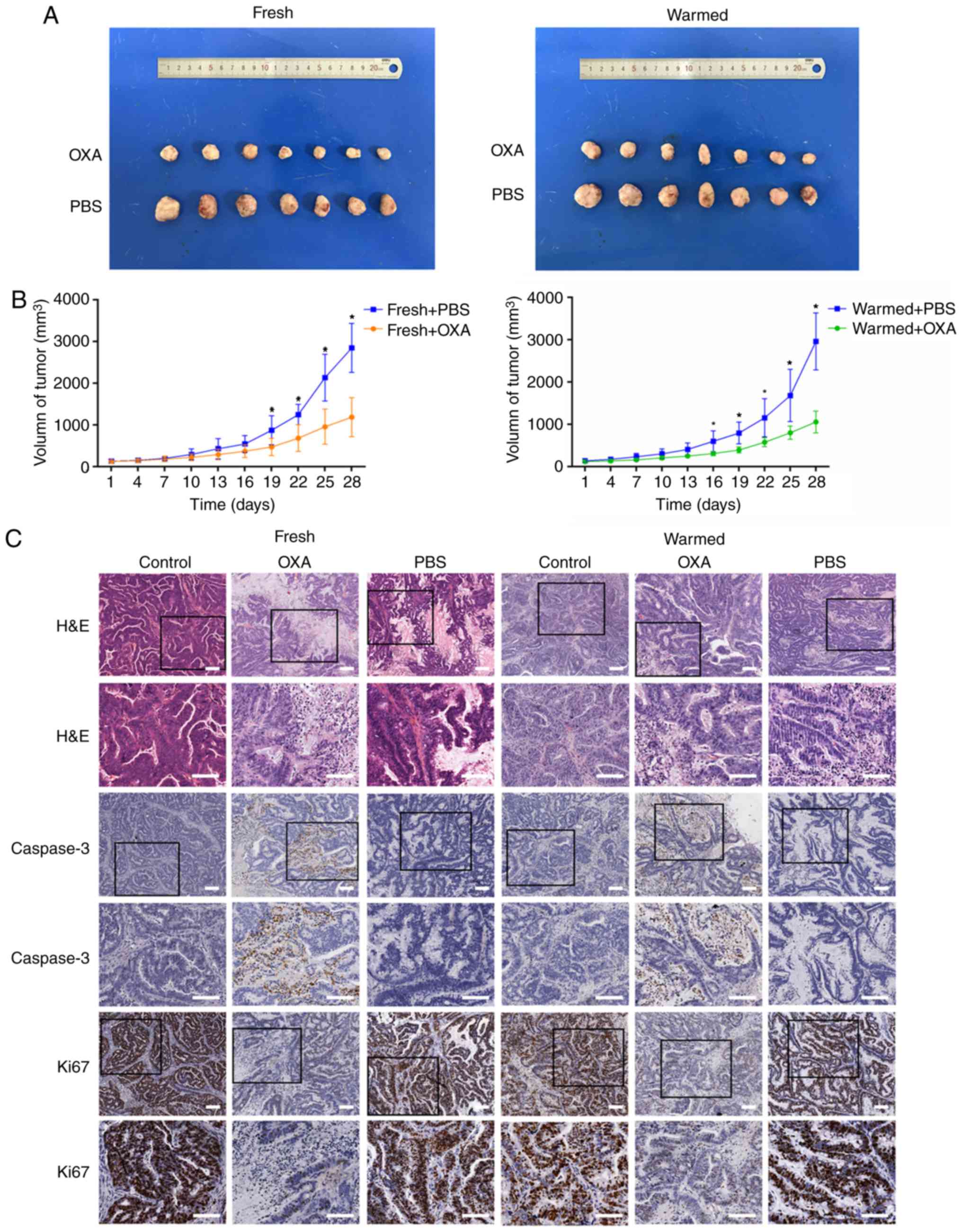
Positive drug responses in xenograft
models are consistent with those in slice cultures in vitro
In order to examine the concordance of anticancer
activity of OXA in vivo and in vitro, PDX models were
established. The PDX mice treated with drugs showed a significant
difference in tumor size, compared with the mice treated with PBS
(Fig. 5A and B; P<0.05). H&E
staining showed an obvious difference in the morphological
characteristics of the tumors between mice treated with drugs and
control mice (Fig. 5C, rows 1 and
2). Accordingly, IHC staining showed increased expression of
cleaved caspase-3 and decreased expression of the viability marker
Ki-67 in warmed slices treated with OXA, compared to those treated
with PBS (Fig. 5C, rows 3–6).
Therefore, it was concluded that the drug responses in vivo
were in accordance with those of the slice cultures in
vitro, indicating the effectiveness of the precision-cut slice
model for drug response assessment.
Discussion
Tumors are intricate and heterogeneous pathological
‘organs’, which have a close connection with their hosts (11). Various models of cancer in
vitro have been applied to the research of cancer biology and
drug testing (12–16). However, ~95% of new anticancer drugs
eventually fail in clinical trials (9), because these models fail to maintain
the essential features of the heterogeneous architecture of the
original tumor. Therefore, innovative models that better capture
the key characteristics of heterogeneous tumors, and mimic the
interactions between tumor cells and their microenvironment, are
required. In the present study, human RLM biopsy tissues were used
for the first time, to the best of our knowledge, to establish
models in vitro and in vivo to evaluate anticancer
drug responses, combining vitrification-based cryopreservation
methods with precision-cut slice cultivation. It was specially
found that the standardized vitrification method, as an optimized
cryopreservation technique, could effectively maintain the
biological activity and histological features of the RLM tissues
and substantially improve the efficiency of tumor specimen
utilization. In addition, with the precision-cut slice culture
method in vitro, it was possible to provide
three-dimensional architecture and tumor heterogeneity, which may
lead to an improvement in the positive response rate of drug
testing.
Up to now, only a few studies have reported on the
long-term preservation of living tumor tissues, such as human
embryos and ovaries (17).
Vitrification of biopsy tissues, especially RLM biopsies, has not
been reported in the literature to the best of our knowledge.
Currently, most PDX models have been established from fresh
surgical resections (18–20). In the present study, PDX models were
established successfully using both fresh and warmed RLM biopsy
tissues, indicating that cryopreservation of both fresh and warmed
biopsy tissues is feasible and efficient. By conducting several
CCK-8 assays in the earlier stages of the research, it was found
that no difference was induced by different lengths of preservation
time in liquid nitrogen. However, the whole cryopreservation and
warming procedure should be conducted strictly in accordance with
the time scheme used in the present study. Additionally, the fresh
biopsy should be transported to the laboratory within 2 h, which is
critical to the activity of the living tissue. Since the amount of
biopsy tissues was limited in the present study, it was determined
to first cryopreserve the biopsy tissues and then establish the
xenografts to generate enough tissues for further experiments. The
results of the present study showed that the novel
vitrification-based cryopreservation method is capable of retaining
the proliferative capacity and morphological characteristics of the
original tumor. No obvious difference was detected in the
biological characteristics of the tissues before and after
cryopreservation. According to the heat map generated, it was clear
that the amount of differential gene expression was limited.
Furthermore, the KEGG enrichment analysis revealed that the
majority of the differential genes were associated with metabolism;
it is hypothesized that the large changes in temperature during the
cryopreservation and warming procedures may be the main reason for
this. Compared with conventional specimen preservation methods, the
novel vitrification-based cryopreservation method is likely to
efficiently improve viability after warming. Notably, this method
avoided ice crystallization, by which the cell viability may be
reduced (21). Furthermore, the
required time and basic cost of this cryopreservation method were
lower than those of conventional cryopreservation methods (22).
Precision-cut slices have been applied to set up
micro-tissue culture systems in vitro, which can be used to
perform preclinical and clinical studies on novel therapies or for
basic research (18,23). Slices of 300 µM were thought to be
the most suitable thickness for RLM after conducting several
slicing experiments with different thicknesses. The most prominent
merit of precision-cut slicing was that the full heterogenous
architecture of the tissues was retained, and slice-based drug
testing avoided the potential damaging effects of the process of
tumor cell isolation from the biological environment (24,25).
In the present study, the tissue slices processed by microtome all
showed evident responses to anticancer drugs. Furthermore, drug
responses in the PDXs were consistent with those in the slice
cultures in vitro. Compared with the PDX model, tissue slice
cultivation represented a more convenient and practical model in
light of the shorter culture cycle and simpler operation.
Certainly, the insert support was helpful for maintaining slice
viability, and enabled the implementation of a steady platform and
provided an ample supply of oxygen and nutrition, compared with
floating alone (18,26,27).
On the whole, precision-cut slice cultivation retained the
heterogeneous architecture of the original tumor and thus might
provide a basis for in-depth study of pharmacokinetic mechanisms in
pharmacology and gene pathways in tumor biology.
The present study standardized a workflow for the
cryopreservation and warming of RLM biopsy tissues, combined with
precision-cut slice cultivation to assess anticancer drug
responses. The present study demonstrated that biopsy tissues from
RLM could be effectively cryopreserved, and that the tumor
biological characteristics were well retained. In addition,
precision-cut slices derived from the warmed tissues provided an
efficient tool to assess anticancer drug responses in vitro.
Above all, the cryopreserved biopsy tissues combined with precision
slice culture represented an optimal method of investigating
personalized therapeutics. Further studies to evaluate this model
for novel preclinical and clinical drug trials are warranted.
Acknowledgements
The authors would like to thank Dr Tao Wang for
technical assistance.
Funding
This study was funded by the National Natural
Science Foundation of China (grant no. 81472845).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
YZ conceived and designed the study, conducted the
experiments and drafted the article. WJH performed the H&E/IHC
staining of tissue and was involved in drafting the manuscript.
QRY, HDZ and XJZ performed the statistical analysis. XZ and MZ made
substantial contributions to the conception and design. ZYW, WJL,
HSJ, XBZ, YPS, HH and HXY were involved in drafting the manuscript
and revising it critically for important intellectual content. ZHL
made substantial contributions to the design and agreed to be
accountable for all aspects of the work in ensuring that questions
related to the accuracy or integrity of any part of the work are
appropriately investigated and resolved. BZ was involved in
drafting and revising the manuscript, made substantial
contributions to the design, and approved the final version to be
submitted. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
The investigation was approved by ethics committee
of Renji Hospital and all patients provided written informed
consent. All the experimental procedures performed on the animals
were approved by the Shanghai Medical Experimental Animal Care
Commission. Informed consent was obtained from all individual
participants included in the study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
RLM
|
rectal cancer liver metastasis
|
|
PDX
|
patient-derived xenograft
|
|
H&E
|
hematoxylin and eosin
|
|
IHC
|
immunohistochemistry
|
|
OXA
|
oxaliplatin
|
|
KEGG
|
Kyoto Encyclopedia of Genes and
Genomes
|
References
|
1
|
Ozyurt H, Ozden AS, Ozgen Z, Gemici C and
Yaprak G: Pre- and post-surgery treatments in rectal cancer: A
long-term single-centre experience. Curr Oncol. 24:e24–e34. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kim KH, Shin SJ, Cho MS, Ahn JB, Jung M,
Kim TI, Park YS, Kim H, Kim NK and Koom WS: A phase II study of
preoperative mFOLFOX6 with short-course radiotherapy in patients
with locally advanced rectal cancer and liver-only metastasis.
Radiother Oncol. 118:369–374. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
De Greef K, Rolfo C, Russo A, Chapelle T,
Bronte G, Passiglia F, Coelho A, Papadimitriou K and Peeters M:
Multisciplinary management of patients with liver metastasis from
colorectal cancer. World J Gastroenterol. 22:7215–7225. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Loree JM and Kopetz S: Recent developments
in the treatment of metastatic colorectal cancer. Ther Adv Med
Oncol. 9:551–564. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Burdall SE, Hanby AM, Lansdown MR and
Speirs V: Breast cancer cell lines: Friend or foe? Breast Cancer
Res. 5:89–95. 2003. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Naipal KA, Verkaik NS, Sanchez H, van
Deurzen CH, den Bakker MA, Hoeijmakers JH, Kanaar R, Vreeswijk MP,
Jager A and van Gent DC: Tumor slice culture system to assess drug
response of primary breast cancer. BMC Cancer. 16:782016.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Usui T, Sakurai M, Enjoji S, Kawasaki H,
Umata K, Ohama T, Fujiwara N, Yabe R, Tsuji S, Yamawaki H, et al:
Establishment of a novel model for anticancer drug resistance in
three-dimensional primary culture of tumor microenvironment. Stem
Cells Int. 2016:70538722016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
van der Kuip H, Murdter TE, Sonnenberg M,
McClellan M, Gutzeit S, Gerteis A, Simon W, Fritz P and Aulitzky
WE: Short term culture of breast cancer tissues to study the
activity of the anticancer drug taxol in an intact tumor
environment. BMC Cancer. 6:862006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hickman JA, Graeser R, de Hoogt R, Vidic
S, Brito C, Gutekunst M and van der Kuip H; IMI PREDECT Consortium,
: Three-dimensional models of cancer for pharmacology and cancer
cell biology: Capturing tumor complexity in vitro/ex vivo.
Biotechnol J. 9:1115–1128. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Holliday DL, Moss MA, Pollock S, Lane S,
Shaaban AM, Millican-Slater R, Nash C, Hanby AM and Speirs V: The
practicalities of using tissue slices as preclinical organotypic
breast cancer models. J Clin Pathol. 66:253–255. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Egeblad M, Nakasone ES and Werb Z: Tumors
as organs: Complex tissues that interface with the entire organism.
Dev Cell. 18:884–901. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Barretina J, Caponigro G, Stransky N,
Venkatesan K, Margolin AA, Kim S, Wilson CJ, Lehár J, Kryukov GV,
Sonkin D, et al: The cancer cell line encyclopedia enables
predictive modelling of anticancer drug sensitivity. Nature.
483:603–607. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Fiebig HH, Maier A and Burger AM:
Clonogenic assay with established human tumour xenografts:
Correlation of in vitro to in vivo activity as a basis for
anticancer drug discovery. Eur J Cancer. 40:802–820. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lee JM, Mhawech-Fauceglia P, Lee N,
Parsanian LC, Lin YG, Gayther SA and Lawrenson K: A
three-dimensional microenvironment alters protein expression and
chemosensitivity of epithelial ovarian cancer cells in vitro. Lab
Invest. 93:528–542. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lin RZ and Chang HY: Recent advances in
three-dimensional multicellular spheroid culture for biomedical
research. Biotechnol J. 3:1172–1184. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chen HC and Hu YC: Bioreactors for tissue
engineering. Biotechnol Lett. 28:1415–1423. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhou XH, Zhang D, Shi J and Wu YJ:
Comparison of vitrification and conventional slow freezing for
cryopreservation of ovarian tissue with respect to the number of
intact primordial follicles: A meta-analysis. Medicine (Baltimore).
95:e40952016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chadwick EJ, Yang DP, Filbin MG, Mazzola
E, Sun Y, Behar O, Pazyra-Murphy MF, Goumnerova L, Ligon KL, Stiles
CD and Segal RA: A brain tumor/organotypic slice co-culture system
for studying tumor microenvironment and targeted drug therapies. J
Vis Exp. e533042015.PubMed/NCBI
|
|
19
|
Unger F, Bentz S, Kruger J, Rosenbrock C,
Schaller J, Pursche K, Sprüssel A, Juhl H and David KA: Precision
cut cancer tissue slices in anti-cancer drug testing. J Mol
Pathophysiol. 4:108–121. 2015. View Article : Google Scholar
|
|
20
|
Bruna A, Rueda OM, Greenwood W, Batra AS,
Callari M, Batra RN, Pogrebniak K, Sandoval J, Cassidy JW,
Tufegdzic-Vidakovic A, et al: A biobank of breast cancer explants
with preserved intra-tumor heterogeneity to screen anticancer
compounds. Cell. 167:260–274 e22. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Elder E, Chen Z, Ensley A, Nerem R,
Brockbank K and Song Y: Enhanced tissue strength in cryopreserved,
collagen-based blood vessel constructs. Transplant Proc.
37:4625–4629. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zeng M, Yang QR, Fu GB, Zhang Y, Zhou X,
Huang WJ, Zhang HD, Li WJ, Wang ZY, Yan HX and Zhai B: Maintaining
viability and characteristics of cholangiocarcinoma tissue by
vitrification-based cryopreservation. Cryobiology. 78:41–46. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Bourke S, Mason HS, Borok Z, Kim KJ,
Crandall ED and Kemp PJ: Development of a lung slice preparation
for recording ion channel activity in alveolar epithelial type I
cells. Respir Res. 6:402005. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Pietras K and Ostman A: Hallmarks of
cancer: Interactions with the tumor stroma. Exp Cell Res.
316:1324–1331. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Davies EJ, Dong M, Gutekunst M, Närhi K,
van Zoggel HJ, Blom S, Nagaraj A, Metsalu T, Oswald E,
Erkens-Schulze S, et al: Capturing complex tumour biology in vitro:
Histological and molecular characterisation of precision cut
slices. Sci Rep. 5:171872014. View Article : Google Scholar
|
|
27
|
Lima D, Silva T, Morais GB, Aquino-Cortez
A, Evangelista J, Xavier Júnior F, Viana DA and Silva L: Different
associations of cryoprotectants for testicular tissue of
prepubertal cats submitted to vitrification. Reprod Domest Anim. 52
(Suppl 2):S235–S241. 2017. View Article : Google Scholar
|