Introduction
With the incidence of cancer and mortality rates
rising rapidly worldwide, cancer is anticipated to become both the
most significant obstacle to extending life expectancy, and the
major cause of death worldwide. Cancer has already become the first
or second main cause of mortality in 91 of 172 countries for
persons aged 70 or less, and is ranked third or fourth in a further
22 countries, according to data published by the World Health
Organization (WHO) in 2015 (1).
During 2018, over 18.1 million new cases of cancer and 9.6 million
cancer deaths were estimated to have occurred worldwide. For both
the sexes, in 2018 Asia accounted for nearly one-half of all cancer
cases, and more than one-half of cancer-associated deaths globally
(1). Despite improvements in
radiotherapy, chemotherapy and surgical treatment technologies,
various types of cancer remain very difficult to treat, such as
gastric cancer (GC), non-small cell lung cancer (NSCLC) and bladder
cancer (BC) (2,3). Therefore, study of the pathogenesis
and therapeutic targets of cancer is imperative.
The transcriptional background of all organisms has
been shown to be much more complicated than was at first
envisioned, since various protein-coding RNAs and noncoding RNAs
(ncRNAs) are extensively transcribed from large portions of genomic
sequences (4,5). Thus, to resolve these problems, a
clear understanding of the intracellular transcriptional
environment must be obtained. Benefiting from the development of
new generation sequencing technologies, it has been shown that, in
humans, over 70% of the genome sequence is transcribed. Among these
transcripts, less than 2% of the transcripts contain protein-coding
RNAs, whereas the remaining transcripts produce ncRNAs (6–9).
Previously, it was considered that the products of most
non-protein-coding genes were without any function, simply produced
during the process of gene transcription. However, with the
development of research in this area, there is a new understanding
that the products of these non-protein-coding genes are involved in
the regulation of a variety of biological processes in cells
(6–9). At present, ncRNAs are a hot research
topic in the life sciences.
NcRNAs comprise regulatory RNAs (for instance,
miRNAs) and structural RNAs [such as small nuclear RNAs (snRNAs),
ribosomal RNAs (rRNAs) and transfer RNAs (tRNAs)] (10). Thanks to recent advances in the
acquisition of genome and transcriptome sequencing data, the
catalogue of regulatory molecules now contains numerous long
noncoding RNAs (lncRNAs), a loosely classified group of long RNA
transcripts that lack protein-coding function (11,12).
As early as 1991, it was found that X-inactive specific transcript
(XIST) is involved in regulating X chromosome inactivation,
providing an initial insight into lncRNA functionality (13,14).
At present, 96,308 lncRNA genes as well as 172,216 lncRNA
transcripts, have been identified in humans [data from NONCODE, the
ncRNA database including sequence, documentation and analysis
(http://www.bioinfo.org/noncode/)], which
influence a great variety of cellular biological processes. lncRNAs
were undervalued for a long time, being considered as merely
transcriptional ‘garbage’ across the entire length of the genome
(15). However, the expression of
lncRNAs is now known to be dysregulated in a great majority of
tumor types, including lung cancer (16), hepatocellular carcinoma (HCC)
(17), glioma (18), colorectal cancer (CRC) (19), breast cancer (BC) (20), osteosarcoma (21), ovarian cancer (22), gastric cancer (GC) (23), and esophageal squamous cell
carcinoma (ESCC) (24). Emerging
studies have now shown that lncRNA has a vital role in a wide
variety of cellular biological processes, including cellular
differentiation, cell cycle, proliferation, migration, metabolism,
as well as apoptosis, especially in tumor cells (25,26).
These findings suggest that lncRNA functions as an oncogene or
tumor suppressor through interaction with other biomolecules or via
chromatin modifications (27).
lncRNAs are able to interfere in numerous cellular
processes, including chromatin organization, transcription complex
recruitment, post-transcriptional regulation, translation, and
post-translational processes (12,28,29).
Several of the main mechanisms of lncRNAs are discussed below.
Chromatin organization
The dosage compensation effect of the X chromosome
in mammals provides a striking instance of lncRNA-mediated
chromatin regulation. Xist, one of the first functionally annotated
lncRNAs, regulates dosage compensation in female mammals by
localizing to the X chromosome, and recruiting various factors
directly or indirectly to accomplish X chromosome inactivation
(XCI) (30). In brief, dosage
compensation refers to the process by which the gene expression
level of the two X chromosomes in female cells is made equivalent
to the one X chromosome in male cells. The expression of lncRNA
Xist in female cells is from one of the two X chromosomes, which
subsequently changes the chromatin structure of the whole
chromosome, whereby most of the genes in the inactive X chromosome
are silenced during transcription (31–33).
Significantly, Xist can interact with polycomb repressive complex 2
(PRC2) via a structural domain called Repeat A, resulting in PRC2
and its cognate histone marker histone H3 lysine 27 trimethylation
(H3K27me3) being located on the inactive X chromosome. lncRNAs can
also recruit PRC2 to modulate distal genes throughout the genome
(34). In addition, lncRNAs
expressed only in embryonic stem cells are able to directly
interact with chromatin to regulate gene expression and maintain
pluripotency (35). The interaction
between DNA and lncRNA is achieved either via sequence
complementation or via combination in helical structures (28). Therefore, lncRNAs are associated
with organization of the nuclear architecture and the general
structuring of the genome, thereby affecting the expression of
related genes (36).
Transcriptional regulation
At the transcriptional level, lncRNA is often used
as a molecular scaffold to recruit two or more proteins to the
promoter regions of their target genes, and modulates the
transcription of the target genes (37–39).
These proteins include zinc-finger proteins, DNA
methyltransferases, transcription factors (TFs), and other
transcriptional regulators (40).
Furthermore, lncRNA can act as an adaptor to recruit associated
proteins into discrete complexes (41). For instance, the lncRNA HOTAIR
synchronously binds both PRC2 and LSD1-CoREST complexes through
specific structural domains of the RNA (42). This interaction effectively
harmonizes the methylation of H3K27 and demethylation of H3K4me2,
guaranteeing that gene silencing occurs. Functioning as a molecular
scaffold is among the primary mechanisms by which HOXA11-AS
functions.
Post-transcriptional regulation
At the post-transcriptional level, lncRNAs operate
in diverse ways to affect miRNAs and mRNAs (43,44).
First, lncRNAs are hypothesized to act as competing endogenous RNA
(ceRNA), or as ‘RNA sponges’, which are able to interact with
miRNAs, thereby blocking the interaction between miRNAs and mRNAs,
thus decreasing their regulatory impact on target mRNAs (45,46).
For example, HOXA11-AS serves as a ceRNA to modulate the expression
level of the transcription factor Sp1 by sequestering miR-124.
Findings demonstrated that improvements in cell invasion and
proliferation mediated by HOXA11-AS were reversed by miR-124
(47). Secondly, lncRNAs can be
precursors of miRNAs, which are able to modulate different
processes in miRNA production and have microprocessor activity to
complete primary transcripts via a mechanism independent of
polyadenylation (48). For
instance, lncRNA LOC554202 was shown to be the putative precursor
of miR-31, exerting an important role in preventing metastasis of
BC (49). In addition, the
maturation of miR-145 can be hindered by lncRNA colon
cancer-associated transcript 2 (CCAT2) via the repression of Dicer
cleavage and cytoplasmic export (50). Moreover, lncRNAs also affect
alternative splicing and the stability of mRNA. For example, first
apoptosis signal (Fas)-antisense lncRNA-SAF complex is able to
combine with the human splicing factor SPF45, resulting in removal
of exon 6 during Fas splicing and the production of a soluble Fas
protein that serves to inhibit Fas/Fas ligand (FASL)-mediated
apoptosis in different human cell lines (51). In addition, lncRNA PDCD4-AS1
stabilizes programmed cell death 4 (PDCD4) RNA via formation of an
RNA duplex that dictates the mutual effect between RNA decay
promoting factors and PDCD4 RNA, in human BC (52).
Recently, numerous studies (19,53)
have reported that exosomes can function as autocrine or paracrine
factors to influence the significant biological functions that
mediate intercellular interactions. An increasing amount of
evidence has shown that cancer cells are able to release exosomes,
and exosome-transmitted lncRNAs are able to promote tumor
metastasis, angiogenesis and drug resistance. For example,
lncRNA-APC1 is able to inhibit the production of exosomes, and
reduce their stability by directly binding Rab5b mRNA, thereby
inhibiting the growth, metastasis and tumor angiogenesis of CRC
cells (19). Additionally, Qu et
al (53) found that bioactive
lncARSR [lncRNA activated in renal cell carcinoma (RCC) with
sunitinib resistance] can be integrated into exosomes and delivered
to sensitive cells in RCC. When exosomes containing lncARSR reached
the sensitive RCC cells, lncARSR was released into the cytoplasm.
The expression of the receptor tyrosine kinases AXL and c-MET in
RCC cells was promoted by lncARSR competitively binding
miR-34/miR-449, thus promoting sunitinib resistance. AXL and c-MET
are responsible for lncARSR-mediated sunitinib resistance in RCC
(53).
Translational process
In addition to the abovementioned effects, lncRNAs
are also able to promote or inhibit the translational process. For
example, dopaminergic neurons specifically express ubiquitin
carboxy-terminal hydrolase L1 (Uchl1). Uchl1 can be modulated by
its antisense transcript (AS Uchl1), which binds polysomes through
its repetitive domain termed ‘SINEB2’ to facilitate cap-independent
translation (54). Furthermore,
lncRNA-p21, a post-transcriptional modulator, passively modulates
translation of the transcripts of the transcription factor JUNB and
β-catenin through incomplete complementary base pairing at diverse
sites in the coding and non-coding regions [both 5′- and
3′-untranslated regions (UTRs)] of JUNB (8 sites) and β-catenin (15
sites) mRNA, leading to the formation of an lncRNA-p21-mRNA
complex. The communication between mRNAs and Fragile X mental
retardation protein (FMRP), as well as the translational repressor
RCK, may be improved by the lncRNA-p21-mRNA complex, resulting in
suppression of the translation target transcripts via the reduction
of ribosome drop-off and polysome sizes (54–56).
Post-translational modification
lncRNAs not only regulate the translational process,
but also modify proteins produced after translation via mechanisms
such as phosphorylation and ubiquitination. The activity and
stability of a protein can be altered through these modifications
(57). For instance, the lncRNA
SLCO4A1-AS1 interacts with β-catenin to improve its stability
through weakening the communication between glycogen synthase
kinase β (GSKβ) and β-catenin, restricting its phosphorylation and
leading to Wnt/β-catenin signaling activation in CRC (58). In addition, the lncRNA SNHG15 can
sustain Slug stability by inhibiting the interaction between Slug
and β-transducin repeat containing (BTRC) E3 ubiquitin protein
ligase, blocking BTRC-mediated Slug ubiquitination in colon cancer
(59).
It may be concluded that, from the chromatin level
to transcription, post-transcription, translation and
post-translational regulation, lncRNAs fulfill important roles in
all aspects of cell physiology.
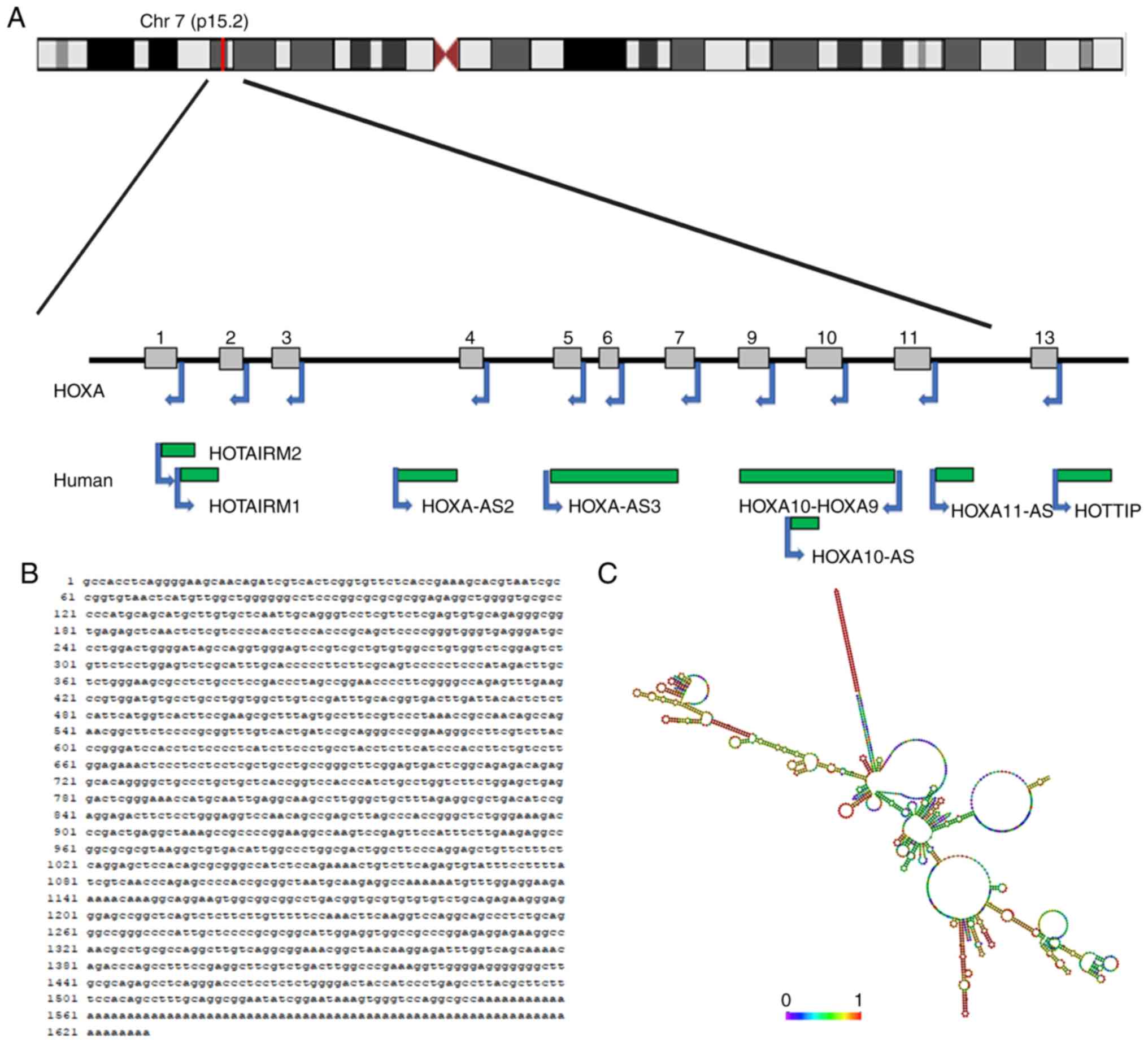
Discovery and description of HOXA11-AS
lncRNA HOXA11-AS, located in the HOXA gene cluster,
has been reported to exert an impact on the occurrence of variety
of human diseases and their subsequent development (60). HOXA11-AS is located on chromosome
7p15.2, and is referred to as HOXA11AS, HOXA11-AS1, HOXA11S,
HOXA-AS5 or NCRNA00076. The chromosomal localization and secondary
structure of HOXA11-AS are shown in Fig. 1. The length of the HOXA11-AS gene is
3,885 bp, whereas the HOXA11-AS transcript is 1628 bp in length
(61). HOX genes are organized into
four clusters (A, B, C and D) on four diverse chromosomes, and HOXA
is a member of the homeobox (HOX) family in the human genome
(62,63). The HOXA gene has a sense strand and
an antisense strand that include protein-coding genes and ncRNA
genes, respectively. The 5′-region of the HOXA gene refers to the
direction of the sense strand relative to the direction of protein
coding genes, and the most abundant protein-coding gene of the
5′-region is HOXA13. Furthermore, a further three protein-coding
genes, HOXA9, HOXA10 and HOXA11, and 3 lncRNAs, HOXA10-AS,
HOXA11-AS and HOTTIP, are located in the 5′-region (64,65).
HOXA11-AS is a novel lncRNA that functions as an oncogene or
tumor-suppressor gene in diverse types of tumor. For example,
HOXA11-AS can serve as an oncogene in non-small cell lung cancer
(NSCLC), HCC, glioma, BC, GC, renal cancer (RC), uveal melanoma
(UM), laryngeal squamous cell carcinoma (LSCC), cervical cancer
(CC), ESCC and osteosarcoma. By contrast, HOXA11-AS functions as a
tumor suppressor in epithelial ovarian cancer (EOC) (7,66,67).
The subsequent sections of this review provide an
overview of the clinical significance, biological functions, and
molecular mechanisms of HOXA11-AS in tumors and several other
diseases types, with the aim of intuitively understanding the role
of HOXA11-AS in the occurrence and development of human
disease.
HOXA11-AS in cancer
Nonsmall-cell lung cancer (NSCLC)
Lung cancer is the primary cause of cancer mortality
and the most commonly occurring type of cancer worldwide (68,69).
No fewer than 2.1 million people are diagnosed with lung cancer
annually (1). Histologically, lung
cancer can be divided into small cell lung cancer (SCLC) and NSCLC.
Approximately 80–85% of newly diagnosed cases of lung cancer belong
to the NSCLC type (70,71). Numerous lncRNAs, including
HOXA11-AS, have been shown to have significant roles in NSCLC.
Zhang et al (73,74) reported that the expression of
HOXA11-AS is higher in both squamous cell carcinoma (SCC) and lung
adenocarcinoma (LUAD) compared with that in normal lung tissues,
and HOXA11-AS knockdown suppresses tumorigenesis, angiogenesis,
proliferation, migration and invasion of NSCLC cells, inducing
apoptosis by impeding the cell cycle at the G0/G1 or the G2/M
phase. Moreover, Chen et al (75) identified that high levels of
HOXA11-AS predict poor prognosis in patients with NSCLC.
Furthermore, Yu et al (48)
demonstrated that the expression level of HOXA11-AS is associated
with lymph node metastasis and tumor size. Zhao et al
(76) also reported that high
levels of HOXA11-AS are associated with poor prognosis.
Mechanistically, Zhang et al (72–74)
revealed that HOXA11-AS expression is negatively correlated with
dedicator of cytokinesis 8 (DOCK8) in SCC and LUAD. Those authors
conjectured that HOXA11-AS could have an oncogenic role in the
development and progression of NSCLC by modulating various
pathways, such as the transforming growth factor (TGF)-β pathway,
the phosphoinositide 3-kinase (PI3K)-Akt pathway and the Hippo
signaling pathway, and genes, such as DOCK8 gene. In other
studies, the same authors demonstrated that the expression of
HOXA11-AS co-expressed genes in NSCLC may be partly regulated by
the NSCLC pathway and that HOXA11-AS could affect various
biological processes of NSCLC via regulation of the expression of
miR-642b-3p by targeting the expression of phosphodiesterase 4D
(PDE4D) or other target genes (72–74).
Additionally, Chen et al (75) found that HOXA11-AS interacts with
DNA (cytosine-5)-methyltransferase 1 (DNMT1) and enhancer of zeste
homolog 2 (EZH2), recruiting these proteins to the promoter regions
of miR-200b and mediating methylation silencing of miR-200b in
NSCLC cells, a process that promotes both NSCLC cell
epithelial-mesenchymal transition (EMT) via regulation of the
protein levels of E-cadherin, N-cadherin, Snail1/2 and ZEB1/2 and
tumor progression. Yu et al (48) revealed that HOXA11-AS acts as a
ceRNA to positively modulate the expression of transcription factor
Sp1 by sequestering miR-124, a process that can inhibit cell
proliferation and the invasion-promoting effects of HOXA11-AS.
Futhermore, those authors found that HOXA11-AS can serve as an
oncogene by promoting EMT via regulation of the protein levels of
E-cadherin, β-catenin, vimentin and the EMT-mediating transcription
factors Slug and Snail in NSCLC (48). Zhao et al (76) discovered that, in human LUAD cells,
HOXA11-AS can function as a ceRNA to facilitate cisplatin tolerance
through the miR-454-3p/Stat3 pathway (the abovementioned mechanisms
are shown in Fig. 2A). Recent
findings (77) have shown that
membrane-bound extracellular vesicles, especially exosomes, serve
significant roles as mediators for communication among different
tissues and organs. Exosomes have a vital role in signal
transduction among cells, and a wide range of biological functions.
Wu et al (77) reported that
high expression levels of HOXA11-AS in exosomes are closely
associated with smoking and NSCLC in lung tissues. These findings
indicate that HOXA11-AS has several functions associated with
oncogenesis, regulating various physiological activities in NSCLC.
These findings shed light upon the effects of HOXA11-AS on the
progression of NSCLC and indicate that HOXA11-AS is a potential
target for future treatment of NSCLC.
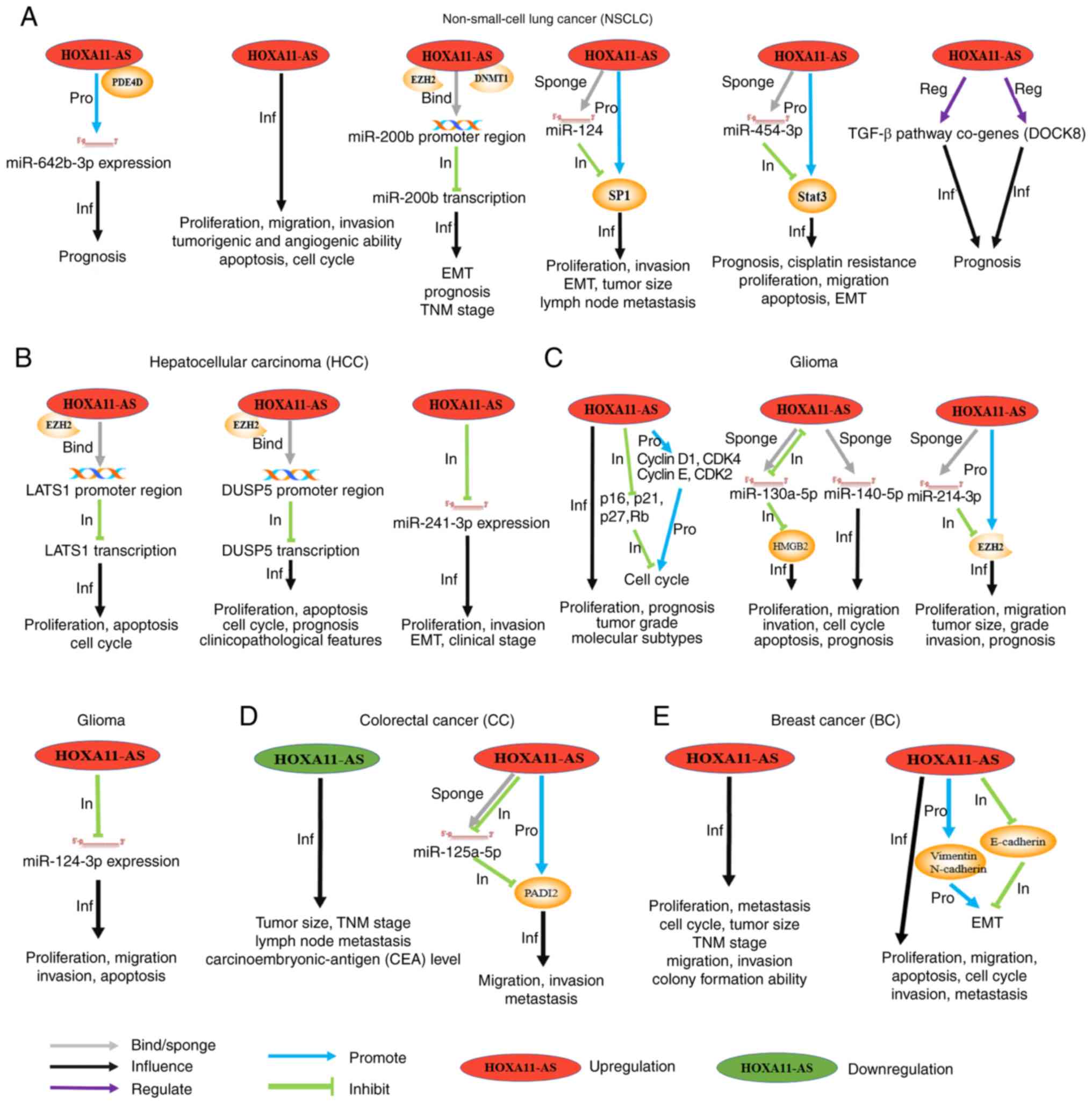 | Figure 2.Schematic diagram that introduces in
detail the molecular mechanisms, clinical values and cell
phenotypes affected by HOXA11-AS in different tumor types,
including (A) NSCLC, (B) HCC, (C) glioma, (D) CC, and (E) BC.
‘Upregulation’/‘downregulation’ indicates that the expression level
of HOXA11-AS in such a tumor was increased/decreased compared with
normal tissues. The gray arrows (‘bind’/‘sponge’) indicate that
HOXA11-AS binds to the promoter region of a gene or, as a sponge,
binds to a miRNA. The black arrows (‘Inf’) indicate that HOXA11-AS
affects the phenotypes and the clinical characteristics of the
cancer under consideration. The blue (‘Pro’)/green (‘In’) arrows
indicate the promoting or inhibitory effects on the downstream
genes, respectively. The purple arrows (‘Reg’) indicate the
regulatory effects on the downstream pathway. NSCLC, non-small cell
lung cancer; HCC, hepatocellular carcinoma; CRC, colorectal cancer;
BC, breast cancer; HOXA11-AS, HOXA11 antisense RNA. |
Hepatocellular carcinoma
Hepatocellular carcinoma (HCC), which accounts for
90% of cases of liver cancer, is the fifth most frequent cancer in
men and the ninth in women worldwide (78,79).
The mortality rate of HCC ranks second in the world (80,81).
Recently, cancer-associated studies focused on lncRNAs have
demonstrated that a considerable number of lncRNAs are involved in
the progression of HCC (82–84).
Previous studies revealed that HOXA11-AS is overexpressed in HCC
cells and tissues (82–84). Liu et al (82) found that high expression levels of
HOXA11-AS are significantly correlated with vascular invasion,
cirrhosis, tumor size and Edmondson grade. The overall survival
(OS) rate of patients with high levels of HOXA11-AS expression is
markedly shorter compared with those with lower levels of HOXA11-AS
expression. Overexpression of HOXA11-AS promotes cell cycle
progression, proliferation, invasion, and EMT, as well as
repressing apoptosis in HCC cells.
Yu et al (83) reported that HOXA11-AS recruits PRC2
to the promoter region of large tumor suppressor 1 (LATS1) to
obstruct its transcription, thereby promoting HCC growth and
inhibiting apoptosis and cell cycle progression at the G0/G1 phase.
The results from flow cytometry experiments indicated that the cell
cycle progression of HCC cells was stalled at the G0/G1 phase when
HOXA11-AS RNAi was transfected into HCC cells (83). Zhan et al (84) showed that the overexpression of
HOXA11-AS is able to facilitate HCC proliferation and invasion, and
induce EMT by repressing the expression of miR-214-3p. In addition,
these authors demonstrated that the expression of miR-214-3p in
early clinical stages (I–II) was higher than that in advanced
clinical stages (III–IV). Furthermore, Liu et al (82) reported that HOXA11-AS recruits EZH2
to the promoter region of dual specificity protein phosphatase 5
(DUSP5), thereby inhibiting the transcription of DUSP5, which is a
downstream target of HOXA11-AS and can function as a tumor
suppressor gene. HOXA11-AS performs an oncogenic role in HCC by
interacting with PRC2 (the abovementioned mechanisms are shown in
Fig. 2B). HOXA11-AS may function as
an oncogene in HCC development. The interaction between HOXA11-AS
and LATS1, DUSP5 or miR-214-3p may supply novel prognostic markers
and therapeutic targets for HCC.
Glioma
Glioma, accounting for approximately 80% of primary
malignant brain tumors, is the most aggressive primary tumor of the
nervous system. Effective treatment of gliomas is very difficult,
particularly glioblastomas (GBMs, also known as grade IV
astrocytomas), with a median survival time for patients of less
than 15 months using standard therapy (85–88).
To date, although many studies have been focused on seeking
improvements in diagnosis and treatment, the trend of poor
prognosis has not been reversed (89). Among the numerous biomolecules
involved in the occurrence and development of glioma, lncRNAs have
attracted sustained attention due to their abnormal expression
during tumorigenesis. Wang et al (90) discovered that high levels of
HOXA11-AS expression are closely correlated with OS in high-grade
glioma, and HOXA11-AS may be an independent prognostic factor for
GBM. HOXA11-AS overexpression might occur during initial
gliomagenesis and increased levels might be maintained in higher
grades of gliomas. Those authors reported that the expression
levels of HOTTIP, HOXA9, HOXA10, and HOXA13 are prominently
correlated with HOXA11-AS expression, and HOXA11-AS can alter the
expression of cell cycle-associated proteins, thus regulating cell
cycle progression. Moreover, Cui et al (91) and Xu et al (92) found that high expression levels of
HOXA11-AS are correlated with decreased survival time and poorer
prognosis compared with patients with lower HOXA11-AS expression.
In addition, Xu et al (93)
found that the overexpression of HOXA11-AS is associated with
advanced stages of glioma and poor prognosis. Authors of that study
reported that knocking down the expression of HOXA11-AS leads to
suppression of the proliferation, migration, and invasion rates of
glioma cells in vitro, with the consequent further
enhancement of cell cycle arrest at the G0/G1 stage and improved
apoptotic responses.
Cui et al (91) showed that miR-140-5p is able to
directly target the 3′-UTR of HOXA11-AS, and the effects of
HOXA11-AS knockdown are shown to be reversed by an miR-140-5p
inhibitor, as determined by its effects on cell cycle arrest,
proliferation and apoptosis. Xu et al (92) demonstrated that HOXA11-AS functions
as a ceRNA by sponging miR-214-3p, which can directly target EZH2
and suppress its mRNA transcriptional level. It was thereby
confirmed that HOXA11-AS may serve in an oncogenic role by
regulating the miR-214-3p/EZH2 pathway. Yang et al (94) found that HOXA11-AS leads to a marked
increase in proliferation, invasion and migration rates, while
inhibiting apoptosis by absorbing miR-124-3p in glioma cells.
Furthermore, Xu et al (93)
found that HOXA11-AS can exert its oncogenic role by directly
binding to miR-130a-5p as a ceRNA, which inhibits the inhibitory
effect of miR-130a-5p on HMGB2 expression. HMGB2 has been
demonstrated to be involved in several diseases, such as sepsis,
arthritis and cancer (the abovementioned mechanisms are shown in
Fig. 2C). The above results suggest
that HOXA11-AS may be an oncogene participating in the prognosis
and treatment response of specific GBM subtypes.
Colorectal cancer
Colorectal cancer (CRC) ranks third among the most
common types of cancer in men, and second among women worldwide,
with ~55% of CRC cases occurring in developed countries (95,96).
The incidence of CRC has rapidly increased in China, and in 2011,
it was shown to be the second most common cause of
cancer-associated mortality (97).
Although recent advances have been made in the diagnosis and
treatment of CRC in recent years, the prognosis of patients
diagnosed with CRC remains poor; in fact, the 5-year survival rate
in patients with metastatic CRC has been shown to be less than 20%
(98,99). Li et al (100) reported that HOXA11-AS is
downregulated in CRC cell lines and tissues, and low expression
levels of HOXA11-AS are associated with advanced tumor-lymph
node-metastasis (TNM) stages, lymphatic metastasis, large tumor
size and elevated levels of carcinoembryonic antigen (CEA). Authors
of that study also showed that the lncRNA HOXA11-AS may distinguish
CRC tissue from noncancerous tissue, and they further explored the
correlation between HOXA11-AS and lymph node metastasis. Moreover,
Chen et al (101) reported
that the level of HOXA11-AS was markedly upregulated in CRC
patients with liver metastasis, and the migration and invasion of
CRC cells was facilitated by HOXA11-AS. In addition, Chen et
al (101) found that HOXA11-AS
can act as a ceRNA sequestering miR-125a-5p to modulate the
expression of protein-arginine deiminase type-2 (PADI2), which was
shown to facilitate metastasis and the invasion of CRC (the
abovementioned mechanisms are shown in Fig. 2D). Consequently, a novel
HOXA11-AS/miR-125a-5p/PADI2 pathway, involved in CRC liver
metastasis, was identified. Taken together, these results
demonstrated that HOXA11-AS is a potential biomarker and molecular
therapy target in CRC.
Breast cancer
Breast cancer (BC) is one of the most frequent
causes of cancer-associated mortality in women worldwide (102,103). Although great progress has been
made in earlier diagnosis of this disease and in the effective of
systemic therapies, the overall prognosis of BC remains
unsatisfactory. In particular, distant metastasis is a barrier for
successful treatment (104–106).
Su and Hu (107) discovered that
lncRNA HOXA11-AS is overexpressed in human BC, and the expression
of HOXA11-AS is markedly associated with metastasis, tumor size and
TNM staging. Clinical statistics revealed that the expression of
HOXA11-AS was correlated with Ki-67 protein and human epidermal
growth factor receptor (HER2), but not with estrogen receptor (ER)
or progesterone receptor (PR). Their results indicated that
decreased expression levels of HOXA11-AS in human BC led to
suppression in the rates of cell proliferation, migration and
invasion, and cell cycle arrest at the G0/G1 phase. Li et al
(108) also reported that low
levels of HOXA11-AS expression inhibited cell proliferation and
induced tumor cell apoptosis by inducing cell cycle arrest at the
G0/G1 stage. By contrast, high levels of HOXA11-AS expression
facilitated metastasis and invasion both in vitro and in
vivo by exerting an influence on EMT in BC (the abovementioned
mechanisms are shown in Fig. 2E).
Note that the above articles (107,108) only determined the involvement of
HOXA11-AS in these processes; the specific mechanisms have yet to
be properly elucidated. These studies, however, have confirmed that
HOXA11-AS exerts carcinogenic effects in BC, thereby providing some
novel insights for even earlier diagnosis in the future, and for
the therapy of BC.
Gastric cancer. Gastric cancer (GC) ranks the fifth
among the most common types of cancer, and is the third leading
cause of cancer mortality (109).
GC is more common in men: The mortality rate in men is 2-fold
higher compared with women (1). In
men, GC is the most commonly occurring gastrointestinal malignancy
in East Asia, the most common cancer in several Western Asian
countries and the main cause of cancer deaths (110,111). Despite advances in surgical
techniques and the successful development of targeted drugs, the
5-year OS rate, however, remains unsatisfactory, and the majority
of patients are diagnosed with advanced cancer along with lymphatic
metastasis (98). Sun et al
(112) reported that HOXA11-AS is
significantly upregulated in GC tissues. High levels of HOXA11-AS
expression are closely related to poor differentiation, larger
tumor size, lymph node metastasis and advanced pathological stage
in GC. Researchers also found that progression-free survival (PFS)
and OS rates in excess of 3 years in the group with high levels of
HOXA11-AS expression were lower compared with the group with low
levels of HOXA11-AS expression. HOXA11-AS overexpression leads to
increased rates of cell growth, migration and invasion, and
inhibition of apoptosis in GC. By contrast, Liu et al
(113) reported that HOXA11-AS
knockdown induces G0/G1 phase arrest in GC cells and decreases GC
cell metastasis, migration and invasion, both in vitro and
in vivo.
In terms of the underlying mechanism, Sun et
al (112) demonstrated that
HOXA11-AS acts as a protein scaffold to recruit EZH2, accompanied
by DNMT1 or LSD1. In addition, HOXA11-AS functions as a miR-1297
sponge, thereby regulating the translation of EZHZ, a direct
miR-1297 target. Knockdown of HOXA11-AS increases the expression of
the tumor suppressors, serine protease 8 (PRSS8) and Krüppel-like
Factor 2 (KLF2), leading to the promotion of cell proliferation and
invasion and inhibiting apoptosis in GC cells. Those authors
reported that EZH2 can directly bind to the promoter regions of
PRSS8 and KLF2 and DNMT1 can directly bind to the promoter region
of KLF2 to stimulate H3K27me3 modifications. LSD1 is also able to
directly bind to the promoter region of PRSS8 to mediate H3K4
demethylation. Knockdown of HOXA11-AS suppresses the binding
capability of DNA with several chromatin modification factors
(namely, PRC2, LSD1 and DNMT1). Moreover, Sun et al
(112) identified the potential
regulator E2F1, which can directly bind to the promoter region of
HOXA11-AS and regulate the transcription of HOXA11-AS. In addition,
HOXA11-AS can function as an oncogene by regulating the
miR-1297/EZH2 and HOXA11-AS-EZH2/DNMT1/LSD1-KLF2/PRSS8 pathways to
promote GC cell proliferation, migration and invasion and inhibit
apoptosis. Liu et al (113)
identified that HOXA11-AS enhances β-catenin transcription by
interacting with WD repeat-containing protein 5 (WDR5), and p21
transcription is subsequently inhibited via binding with EZH2. In
addition, HOXA11-AS can interact with double-stranded RNA-binding
protein staufen homolog 1 (STAU1) to promote KLF2 mRNA degradation.
These findings revealed that HOXA11-AS may serve as an oncogene by
modulating the HOXA11-AS-WDR5/EZH2/STAU1-β-catenin/p21/KLF2 pathway
(Fig. 3A). Taken together, these
results have revealed that HOXA11-AS can function as an oncogene in
GC cells, further elucidating the role of HOXA11-AS in promoting
better diagnosis of the disease, and developing further how lncRNA
may be used in treatments of GC.
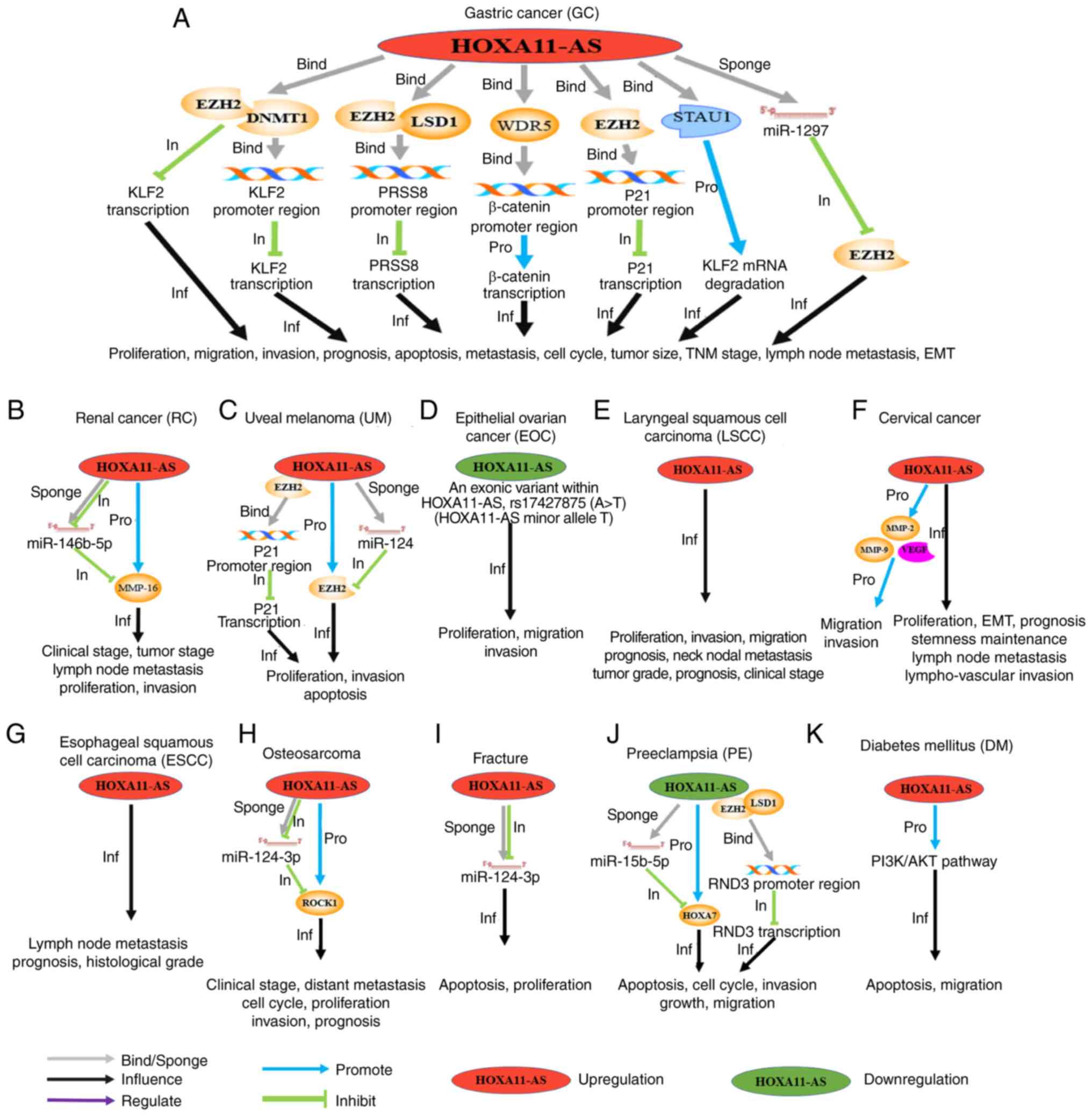 | Figure 3.Schematic diagram, introducing in
detail the molecular mechanisms, clinical values and cell
phenotypes affected by HOXA11-AS in different types of tumors,
including (A) GC, (B) RC, (C) UM, (D) EOC, (E) LSCC, (F) CC, (G)
ESCC, (H) osteosarcoma, (I) fracture, (J) PE and (K) DM.
‘Upregulation’/‘downregulation’ indicates that the expression level
of HOXA11-AS in such a tumor was increased/decreased compared with
normal tissues. The gray arrows (‘bind/sponge’) indicate that
HOXA11-AS binds to the promoter region of a gene or, as a sponge,
binds to a miRNA. The black arrows (‘Inf’) indicate that HOXA11-AS
affects the phenotypes and the clinical characteristics of the
cancer under consideration. The blue (‘Pro’) and green (‘In’)
arrows indicate the promoting or inhibitory effects on the
downstream genes, respectively. The purple arrows (‘Reg’) indicate
the regulatory effects on the downstream pathway. GC, gastric
cancer; RC, renal cancer; UM, uveal melanoma; EOC, epithelial
ovarian cancer; LSCC, laryngeal squamous cell carcinoma; ESCC,
esophageal squamous cell carcinoma; PE, pre-eclampsia; DM, diabetes
mellitus; HOXA11-AS, HOXA11 antisense RNA. |
Renal cancer
Clear cell renal cell carcinoma (ccRCC) is a common
malignant tumor of the urinary system, accounting for >80% of
all types of RC (114). No fewer
than 209,000 new cases of ccRCC are reported each year, and among
these cases, approximately 25–30% of the patients present with
distant metastasis and 40% of patients have local recurrence after
the initial diagnosis (115,116). The biological behavior of ccRCC is
extremely complex and the underlying molecular mechanisms have yet
to be fully elucidated, factors which explain how prognosis is both
poor and difficult to predict (117). Recently, Yang et al
(118) found that HOXA11-AS was
clearly upregulated in ccRCC cell lines and tissues, and high
HOXA11-AS expression levels were very closely correlated with lymph
node metastasis, tumor stage and advanced clinical stage.
Downregulation of the HOXA11-AS transcript led to a marked decrease
in the growth, proliferation, invasion, and EMT of the ccRCC cells.
Regarding the mechanism, Yang et al (118) discovered that HOXA11-AS functions
as a ceRNA to suppress the expression of miR-146b-5p, which
subsequently modulates the expression of its downstream target,
matrix metalloproteinase-16 (MMP16) in RC. Therefore, HOXA11-AS
promotes RC cell invasion and proliferation by regulating the
miR-146b-5p/MMP16 pathway (Fig.
3B). During this process, HOXA11-AS may function as an oncogene
in ccRCC and may therefore represent an efficient therapeutic
target for RCC.
Uveal melanoma. Uveal melanoma (UM) ranks first
among primary intraocular malignant tumors in adults, and the uvea
ranks second among the common sites of primary melanoma (119,120). The metastatic rate of UM is very
high, and UM ultimately spreads to the liver in up to 50% of
patients (121). Lu et al
(122) discovered that HOXA11-AS
is overexpressed in UM, and HOXA11-AS is shown to enhance the rates
of UM cell proliferation and invasion, while repressing apoptosis.
Authors of that study also reported that HOXA11-AS recruits EZH2 to
the promoter region of p21 and mediates H3K27me3 to suppress its
transcription. Furthermore, HOXA11-AS acts as a ceRNA for miR-124,
which directly targets EZH2, and the cell proliferation and
invasion-increasing effects of HOXA11-AS were attenuated upon
miR-124 overexpression (Fig. 3C).
Taken together, these studies indicate that HOXA11-AS has an
oncogenic role through regulating the HOXA11-AS/EZH2/p21 and
miR-124/EZH2 pathways in UM tumorigenesis, and HOXA11-AS may also
represent a potential therapeutic target for treatment of UM.
Epithelial ovarian cancer
Ovarian malignancies are among the most commonly
occurring malignancies in female reproductive organs. The most
frequent ovarian malignancy type is cutaneous carcinoma, followed
by malignant germ cell tumor. Among these malignant gynecological
tumors, the mortality rate of epithelial ovarian cancer (EOC) is
the highest, posing a serious threat to women's health (1). A large number of studies have shown
that not only HOXA11-AS, but also many other lncRNAs act as
oncogenes or tumor suppressor genes in different cancers. Ignarski
et al (123) reported that
the expression pattern of lncRNA genes is far more tissue- and
cell-type specific than is the case for protein coding genes.
HOXA11-AS exerts opposing effects in different types of cancer.
Such expressional and functional discrepancies of HOXA11-AS in
different types of cancer could be caused by distinct gene
expression backgrounds in different tumors (124). For example, in EOC, HOXA11-AS can
act as a tumor suppressor gene, which is different from the tumor
mentioned above.
It has been reported that common germline genetic
variants or single nucleotide polymorphisms (SNPs) affecting
lncRNAs are conducive to the development of various types of cancer
(125), such as EOC (126). Richards et al (126) reported that overexpression of
HOXA11-AS results in a clear reduction in cell proliferation and
survival, which are two main cellular processes associated with EOC
development in both common and minor allele constructs. In
particular, the existence of minor allele constructs markedly
reduced the proliferation and survival of EOC cells compared with
the common allele, and the expression of both alleles served to
diminish cell migration and invasion. However, compared with the
common allele, the minor allele exerted a more pronounced
suppressive effect in both assays. Richards et al (126) found that the minor allele
repressed the carcinogenic phenotypes to a greater extent compared
with the common allele in EOC cells. The finding has identified a
greatly decreased risk of EOC among women who have the HOXA11-AS
rs17427875 T allele (126). In EOC
tumor tissue, HOXA11-AS expression was reduced by over 60% on
average compared with normal ovarian tissue. Richards et al
(126) also found HOXA11-AS cannot
exert any influence upon HOXA11 and HOXA13 (Fig. 3D). Thus, these studies suggest that
HOXA11-AS may function as a tumor suppressor gene in EOC.
Laryngeal squamous cell carcinoma
Laryngeal squamous cell carcinoma (LSCC) is a
commonly occurring malignant tumor of the upper respiratory tract.
Laryngeal carcinoma is ranked second in terms of the most
frequently occurring malignant tumors in the head and neck
(127). Qu et al (128) found that the expression levels of
HOXA11-AS are closely correlated with pathological grade, T (tumor)
grade, clinical stage, neck nodal metastasis and advanced tumors of
grade T3 to T4 in LSCC. Furthermore, advanced clinical stages, poor
prognosis, poor differentiation and lymph node metastasis were
identified in patients who expressed higher levels of HOXA11-AS.
Authors of that study also found that HOXA11-AS was able to promote
LSCC cell proliferation, invasion and migration (Fig. 3E). Notably, only the general effects
attributable to HOXA11-AS were identified in the study of Qu et
al (128), and the specific
mechanism involved requires further investigation. However, their
results have demonstrated that HOXA11-AS may function as an
oncogene, suggesting that it is a potential novel biomarker and an
efficient therapeutic target in LSCC.
Cervical cancer
It is estimated that CC is the fourth leading cause
of cancer mortality, and ranks fourth out of the most commonly
diagnosed cancers among women. In 2018, there were approximately
570,000 CC cases and 311,000 cancer-associated deaths worldwide
(1). In fact, in terms of its
incidence and the mortality rates, CC is ranked second behind BC
(1). Kim et al (129) reported that the expression of
HOXA11-AS is specifically upregulated in CC, and OS and 5-year
survival rates are both reduced in CC patients with HOXA11-AS
overexpression. Cox multivariate proportional hazards analysis
revealed that nodal metastasis, tumor stage, and HOXA11-AS are
independent prognosticators of OS. Those authors found that
HOXA11-AS is associated with enhanced cell proliferation, also
leading to increased rates of cell migration and invasion in CC. In
terms of the underlying mechanism, HOXA11-AS promotes CC cell
migration and invasion by upregulating the levels of MMP-9, MMP-2,
and vascular endothelial growth factor (VEGF), and disordering of
the EMT-associated genes suggested that HOXA11-AS may be involved
in cell migration and invasion in CC (Fig. 3F). Kim et al (129) also showed that HOXA11-AS can
promote activation of the genetic program that supports the cancer
stem cell (CSC) phenotype and improves EMT, suggesting that
HOXA11-AS may function as an oncogene in CC. Thus, HOXA11-AS may be
a therapeutic target in the search for improved treatments for
CC.
Esophageal squamous cell
carcinoma
Among the different types of cancer, ESCC is ranked
seventh in terms of its incidence and sixth in terms of overall
mortality, which means that 1 out of every 20 cases of
cancer-associated mortality in 2018 was estimated to have been
caused by ESCC (1). Sun et
al (130) found that high
expression levels of HOXA11-AS are correlated with lymph node
metastasis and histological grade in patients with ESCC. Compared
with those patients with HOXA11-AS overexpression, patients with
low HOXA11-AS expression exhibited both increased median
disease-free survival (DFS) and median OS. Sun et al
(130) found that lymph node
metastasis and the expression of HOXA11-AS were independent poor
prognosis factors in patients with ESCC (Fig. 3G). Taking all these findings into
consideration, HOXA11-AS may have an oncogenic role in ESCC, and
therefore HOXA11-AS may also represent a predictive marker in
postoperative ESCC patients.
Osteosarcoma
Osteosarcoma is ranked the first among the most
commonly occurring types of primary malignant bone cancer, and is
placed second in terms of cancer-associated deaths in pediatrics
(131). lncRNAs are well known to
be involved in the development of most tumors. Cui et al
(60) found that a high level of
HOXA11-AS expression is correlated with distant metastasis, reduced
OS and advanced clinical stage in patients with osteosarcoma. Their
study suggested that HOXA11-AS knockdown in osteosarcoma induces
cell cycle arrest at the G0/G1 phase, and HOXA11-AS overexpression
led to a substantial improvement in cell invasion and growth rates
in osteosarcoma cells via the competitive binding of miR-124-3p,
which targets Rho-associated, coiled-coil-containing protein kinase
1 (ROCK1) (Fig. 3H). In this
manner, HOXA11-AS may exert oncogenic functions by regulating the
miR-124-3p/ROCK1 pathway. Investigating the underlying mechanistic
roles of the HOXA11-AS/miR-124-3p/ROCK1 pathway may be an important
step in developing novel osteosarcoma therapeutic strategies.
Other diseases
Fracture
Given the ever-increasing aging population,
fractures have become a serious health problem, constituting the
most common injuries sustained worldwide. Despite the body being
capable of healing fractures, many risk factors have substantially
delaying effects on the process of fracture healing, including
advanced age, smoking, diabetes mellitus (DM) and anti-cancer drugs
(132). Therefore, it is
imperative to explore the mechanisms underlying fracture healing,
notably in patients with these factors, is an urgent requirement.
Numerous studies have been published that demonstrate how lncRNAs
are able to participate in the occurrence and development of many
diseases, including fracture healing (133). Wang et al (133) found that HOXA11-AS overexpression
led to a suppression of OS-732 osteoblast proliferation and
improved apoptosis. In addition, HOXA11-AS can act as a ceRNA by
sequestering miR-124-3p to inhibit cell proliferation and enhance
apoptosis (Fig. 3I). These results
may provide a novel perspective for deciphering the mechanism of
fracture healing.
Preeclampsia
Preeclampsia (PE) is the leading cause of
pregnancy-associated death and fetal defects (134). PE is characterized as having a
blood pressure exceeding 140/90 mmHg from the 20th week of
pregnancy onwards. Between 3 and 5% of pregnant women experience
PE, especially in developing countries (135). Xu et al (136) found that HOXA11-AS expression was
markedly downregulated in pre-eclamptic placental tissues, and
reducing HOXA11-AS expression led to a clear inhibition of
trophoblast cell growth and migration. Mechanistically, those
authors revealed that HOXA11-AS recruits LSD1 and EZH2 proteins to
the RND3 gene promoter region in the nucleus to repress its
expression in trophoblast cells. When the level of HOXA11-AS
expression was reduced in trophoblast cells, its ability to bind to
LSD1 and EZH2 was also reduced, leading to decreased LSD1 and EZH2
binding to the RND3 gene promoter region and decreasing the
inhibitory effect on RND3, which suppresses cell growth and
proliferation. Furthermore, HOXA11-AS facilitated the expression of
HOXA7 in the cytoplasm via sequestration of miR-15b-5p, thus
exerting an influence on trophoblast proliferation (Fig. 3J). These studies revealed that
HOXA11-AS may have an oncogenic function via modulating the
LSD1/EZH2-RND3 and HOXA11-AS/miR-15b-5p/HOXA7 pathways.
Consequently, these results have verified that abnormal expression
of HOXA11-AS is involved in the occurrence and development of PE,
and that this lncRNA may function as a putative target for
diagnosis and treatment in PE.
Diabetes mellitus (DM)
With rapid economic development and aging of the
population, the number of patients with DM is increasing annually.
Persistent hyperglycemia induces hyperglycemia-associated
complications that pose severe medical risks, such as diabetic
arteriosclerosis (DAA), atherosclerosis (AS) and cardiomyopathy
(137). Studies have revealed that
the incidence of cardio-cerebrovascular diseases in DM patients is
markedly higher than in non-DM patients. In view of the role of
lncRNAs in a variety of different tumor types and diseases,
researchers have also examined the role of lncRNA in diabetes. Jin
et al (138) found that
expression levels of HOXA11-AS and pro-inflammatory genes were
substantially increased in carotid endarterectomy specimens of DM
patients, and HOXA11-AS expression was also significantly increased
in the carotid arteries of DM mice. Mechanistically, HOXA11-AS
knockdown reduces the expression of proliferation-associated gene
(PCNA), the cell cycle-related genes p21 and
p53, and platelet-derived growth factor
(PDGF)-induced growth and migration of vascular smooth
muscle cells (VSMCs) is repressed, significantly downregulating the
expression of inflammation-associated genes in VSMCs induced by
tumor necrosis factor-α (TNF-α). Moreover, in vascular endothelial
cells (VECs), low expression levels of HOXA11-AS were shown to
suppress the expression of TNF-α-induced pro-inflammatory genes and
PDGF-induced vascular inflammation-related genes. Low expression
levels of HOXA11-AS inhibited the PDGF-induced stimulation of the
PI3K/AKT pathway by inhibiting the phosphorylation of PI3K and AKT
in VSMCs and VECs (Fig. 3K). The
biological functions of HOXA11-AS in DAA-induced inflammation
should be further explored to identify potential new effective
treatments for DAA.
Conclusion
In conclusion, an increasing number of studies have
shown that lncRNAs are dysregulated in various types of cancer, and
aberrant expression of lncRNAs is involved in the occurrence,
development, and metastasis of cancer (139). lncRNAs function mainly by
interacting with other DNA, RNA or protein molecules to exert their
pre-transcriptional or post-transcriptional regulatory functions.
At present, emerging in-depth studies are elucidating the
regulatory effects of HOXA11-AS on a majority of different tumor
types, including NSCLC, HCC and glioma, although its mechanisms and
targets are generally found not to be similar when comparing among
the different malignancies. Table I
summarizes HOXA11-AS expression patterns and its functional and
clinical value in different types of human cancer. Additionally,
functional characteristics and molecular mechanisms of HOXA11-AS in
diverse types of cancer and other diseases are summarized in
Fig. 4. Although many studies have
investigated HOXA11-AS, much remains to be determined before we are
in a position to fully understand the mechanism of HOXA11-AS in
diseases including BC, ESCC and LSCC. However, with further
research, HOXA11-AS is likely to gain in importance as a novel
target and guide for the prevention, diagnosis and treatment of
tumors and additional diseases.
 | Table I.HOXA11-AS expression pattern,
function and clinical value in human. |
Table I.
HOXA11-AS expression pattern,
function and clinical value in human.
| Cancer types | Expression
level | Gene types | Number of
cases | Cell lines
involved | Genes, RNAs and
proteins interact with HOXA11-AS | Signaling
pathways | HOXA11-AS molecular
mechanisms | HOXA11-AS binding
sites (5′→3′) | Affected
phenotypes | Association with
patients' outcome | (Refs.) |
|---|
| Lung cancer | Up | Oncogene | Three pairs NSCLC
tissues (HOXA11-AS and HOXA11-AS RNAi) | A549 | DOCK8 | TGF-beta
pathway |
|
|
|
| Zhang et al
(2016) (72) |
|
| Up | Oncogene | 287 lung
adenocarcinoma cases vs. 12 non-cancerous lung cases and 463 lung
squamous cell carcinoma cases vs. 12 non-cancerous lung cases | PC9, A549, H460 and
H1299 |
| ErbB, MAPK,
Calcium, PI3K-Akt and P53 signaling pathways |
|
| Proliferation,
migration, invasion, apoptosis, tumorigenic angiogenic ability |
| Zhang et al
(2017) (73) |
|
| Up | Oncogene | 78 pairs NSCLC
tissues and corresponding adjacent normal tissues | A549, H1299 | miR-124 |
HOXA11-AS/miR-124/Sp1 pathway | ceRNA | 509–515
(NR_002795.2) (GTGCCTT) | Proliferation,
invasion, EMT | Tumor size, lymph
node metastasis | Yu et al
(2017) (47) |
|
| Up | Oncogene | 78 pairs NSCLC
tissues and adjacent normal tissue samples | A549, H1299, 95D,
16HBE | EZH2, DNMT1,
miR-200b |
| Scaffold |
| Invasion, EMT | Prognosis, lymph
node metastasis, TNM stage | Chen et al
(2017) (75) |
|
| Up | Oncogene | Cell line
study | A549, H157,
A549-CR, H157-CR (cisplatin-Resistantcells), HEK-293T | miR-454-3p |
HOXA11-AS/miR-454-3p/Stat3 pathway | ceRNA | 1057–1075
(ENST00000520395.2) (CTAXXXXXXTATT-GCACT) | Proliferation,
migration, apoptosis, EMT | Prognosis | Zhao et al
(2018) (76) |
|
| Up | Oncogene | Cell line
study | A549 | miR-642b-3p,
PDE4D |
HOXA11-AS/miR-642b-3p/PDE4D axis |
|
|
ENST00000-520395.2 |
| Zhang et al
(2018) (74) |
| Liver cancer | Up | Oncogene | 72 pairs HCC
tissues and its adjacent tissue specimens | HepG2, Hep3B,
MHCC-97H | LAST1, EZH2 |
| Scaffold |
| Proliferation,
apoptosis, cell cycle progression, tumor formation ability |
| Yu et al
(2017) (83) |
|
| Up | Oncogene | 66 pairs HCC
tissues and adjacent normal tissues | HepG2, Hep3B | DUSP5, EZH2 |
| Scaffold |
| Proliferation,
apoptosis, cell cycle | Vascular invasion,
cirrhosis, tumor size and edmindson grade, prognosis | Liu et al
(2017) (82) |
|
| Up | Oncogene | 40 pairs of HCC
samples and the adjacent noncancerous samples | Hep3B,
MHCC97-H | miR-214-3p |
HOXA11-AS/miR-214-3p axis |
|
| Proliferation,
invasion, EMT |
| Zhan et al
(2018) (84) |
| Glioma | Up | Oncogene | 220 glioma cases
and 5 normal brain samples | U87 (ATCC), LN229,
U251, HA |
|
|
|
| Proliferation, cell
cycle | Grade, prognosis,
molecular, subtypes | Wang et al
(2016) (90) |
|
| Up | Oncogene | 43 cases of glioma
tissues and normal brain tissues | SHG44, U251,
HA | miR-140-5p |
HOXA11-AS/miR-140-5p axis | ceRNA | (HGNC 24957)
(AAACCACT) | Proliferation,
apoptosis, cell cycle | Prognosis | Cui et al
(2017) (91) |
|
| Up | Oncogene | 45 pairs glioma
tissues and the adjacent normal brain tissues | U251, U87 (ATCC),
LN229, A172, NHA, HEK-293T, SHG-44 | miR-214-3p,
EZH2 |
HOXA11-AS/miR-214-3p/EZH2 axis | ceRNA | 502–515
(NR_002795.2) (CXCXXXXGT-GCCTT) | Proliferation,
migration, invasion, tumor growth | Prognosis, tumor
size, grade | Xu et al
(2017) (92) |
|
| Up | Oncogene | 43 pairs of glioma
tissues and noncancerous tissues | U251, U87
(ATCC) | miR-130a-5p, HMGB2
pathway |
miR-130a-5p/HMGB2 | ceRNA | 1487–1494
(ENST00000520395.2) (ATTGCACT) | Proliferation,
migration, invasion, apoptosis | Prognosis, advanced
stage | Xu et al
(2019) (93) |
| Colorectal
cancer | Down | Suppressor
gene | 84 CRC tissues and
adjacent non-cancerous tissues, in addition to 3 CRC cell lines and
1 human normal colorectal cell line. | CCD-18Co, HCT8,
HCT116, RKO |
|
|
|
|
ENST0000-0520395.2 | Tumor size, TNM
stage, lymph node metastasis, carcinoem-bryonic antigen level | Li et al
(2016) (100) |
|
| Up | Oncogene | 30 primary CRC
samples (15 patients with CRC) and liver metastasis and 15 patients
with CRC without metastasis) | Colo205, Lovo,
HCT116, SW620, Caco-2, SW480 | miR-125a-5p,
PADI2 |
HOXA11-AS/miR-125a-5p/PADI2 regulatory
network | ceRNA | 1482–1487
(NR_002795.2) (CCTGAG) | Migration,
invasion | Liver
metastasis | Chen et al
(2017) (101) |
| Breast cancer | Up | Oncogene | 100 pairs BC
tissues and adjacent non-cancerous tissues | MCF10A, MDA-MB-231,
MDA-MB-436, MCF7, T47D |
|
|
|
| Proliferation,
migration, invasion, colony formation, cell cycle progression | Tumor size,
metastasis, TNM stage, molecular subtypes | Su and Hu (2017)
(107) |
|
| Up | Oncogene | 68 pairs breast
cancer tissues and adjacent normal tissue specimens | MDA-MB-468,
MDA-MB-231, SKBR3, MCF-10A |
|
|
|
| Migration,
invasion, apoptosis, cell cycle progression, EMT | Metastasis | Li et al
(2017) (108) |
| Gastric cancer | Up | Oncogene | 85 pairs gastric
cancer and adjacent nontumor tissues | GES1, AGS | miR-1297, EZH2,
LSD1, DNMT1 | Cell-cell adhesion
pathways, HOXA11-AS/miR-1297/EZH2 cross-talk | Scaffold ceRNA | 457–469
(NR_002795.2) (CXXXXTXACTTGA) 5′ region bind with EZH2 3′ region
bind with LSD1 | Growth,
proliferation, migration, invasion, apoptosis, tumorigenesis, tumor
progression | Prognosis, tumor
size, differentiation, pathologic stage, lymph node metastasis | Sun et al
(2016) (112) |
|
| Up | Oncogene | Cell line
study | AGS | WDR5, STAU1,
EZH2 |
| Scaffold |
| Proliferation,
migration, invasion, cell cycle progression, EMT | Metastasis | Liu et al
(2017) (113) |
| Renal cancer | Up | Oncogene | 103 pairs ccRCC
specimens and adjacent nontumor tissues | ACHN, 786-O, A498,
OSRC- 2, HK-2 | miR-146b-5p | miR-146b-5p/MMP16
axis. | ceRNA | 15–42
(ENST00000522674.1) (AGCXXXXGAXXXTCAXXXXXXGTTCTCA) | Growth,
proliferation, invasion, EMT | Advanced clinical
stage, tumor stage, lymph | Yang et al
(2018) (118) |
| Uveal melanoma | Up | Oncogene | Five primary UM
samples | C918, MUM-2B and
D78 | EZH2, miR-124 |
| Scaffold ceRNA | 509–515
(NR_002795.2) (GTGCCTT) | Growth,
proliferation, invasion, apoptosis |
| Lu et al
(2017) (122) |
| Epithelial ovarian
cancer | Down | Suppressor
gene | Case control
study |
| HOXA11-AS minor
allele T and common allele A |
|
|
| Proliferation,
growth migration, invasion | Prognosis | Richards et
al (2015) (126) |
| Laryngeal squamous
cell carcinoma | Up | Oncogene | 25 pairs cancerous
and adjacent noncancerous tissues | AMC-HN-8 |
|
|
|
| Proliferation,
growth migration, invasion | Grade, neck nodal
metastasis, clinical stage, prognosis | Qu et al
(2018) (128) |
| Cervical
cancer | Up | Oncogene | 92 cervical cancer
tissues and 30 normal cervix samples | SiHa, HeLa, CaSki,
ME-180, C33A and HOSE | MMP-9, MMP-2, and
VEGF |
|
|
| Proliferation,
growth, migration, invasion, sphere formation, EMT, stemness
maintenance | Prognosis | Kim et al
(2016) (129) |
| Esophageal squamous
cell carcinoma | Up | Oncogene | 73 pairs ESCC
tissues and adjacent tissue samples | HET-1A, EC109,
EC9706 |
|
|
|
|
| Histological grade,
lymph node metastasis, prognosis | Sun et al
(2018) (130) |
| Osteosarcoma | Up | Oncogene | 51 pairs OS tissues
and adjacent tissue samples | U2OS, MG-63, KHOS
and NHost | ROCK1,
miR-124-3p |
HOXA11-AS/miR-124-3p/ROCK1 signaling
axis | ceRNA | 502–515
(NR_002795.2) (CXCXXXXGTGCCTT) | Proliferation,
invasion, cell cycle progression | Clinical stage,
distant metastasis, prognosis | Cui et al
(2017) (60) |
Acknowledgements
Not applicable.
Funding
This study was supported by grants (nos. 81773187
and 81572496) from the National Nature Science Foundation of China.
Support was also received from the Tianjin High School Program for
Young and Middle-aged Talents Backbone and the Tianjin Young
Medical Talents Program.
Availability of data and materials
Not applicable.
Authors' contributions
LH and YZ conceived the review. CW and LZ drafted
the manuscript and revised it before submission. CW and HL
collected the references. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Li J, Li Z, Zheng W, Li X, Wang Z, Cui Y
and Jiang X: PANDAR: A pivotal cancer-related long non-coding RNA
in human cancers. Mol Biosyst. 13:2195–2201. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hu Q, Tai S and Wang J: Oncogenicity of
lncRNA FOXD2-AS1 and its molecular mechanisms in human cancers.
Pathol Res Pract. 215:843–848. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Adams BD, Parsons C, Walker L, Zhang WC
and Slack FJ: Targeting noncoding RNAs in disease. J Clin Invest.
127:761–771. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Djebali S, Davis CA, Merkel A, Dobin A,
Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F,
et al: Landscape of transcription in human cells. Nature.
489:101–118. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhu S, Li W, Liu J, Chen CH, Liao Q, Xu P,
Xu H, Xiao T, Cao Z, Peng J, et al: Genome-scale deletion screening
of human long non-coding RNAs using a paired-guide RNA CRISPR-Cas9
library. Nat Biotechnol. 34:1279–1286. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Xue JY, Huang C, Wang W, Li HB, Sun M and
Xie M: HOXA11-AS: A novel regulator in human cancer proliferation
and metastasis. Onco Targets Ther. 11:4387–4393. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhao W, Ma X, Liu L, Chen Q, Liu Z, Zhang
Z, Ma S, Wang Z, Li H, Wang Z and Wu J: SNHG20: A vital lncRNA in
multiple human cancers. J Cell Physiol. Jan 15–2019.(Epub ahead of
print). doi: 10.1002/jcp.28143.
|
|
9
|
Schmitt AM and Chang HY: Long noncoding
RNAs in cancer pathways. Cancer Cell. 29:452–463. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Anastasiadou E, Jacob LS and Slack FJ:
Non-coding RNA networks in cancer. Nat Rev Cancer. 18:5–18. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Atianand MK, Caffrey DR and Fitzgerald KA:
Immunobiology of long noncoding RNAs. Annu Rev Immunol. 35:177–198.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Khorkova O, Hsiao J and Wahlestedt C:
Basic biology and therapeutic implications of lncRNA. Adv Drug
Deliv Rev. 87:15–24. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Borsani G, Tonlorenzi R, Simmler MC,
Dandolo L, Arnaud D, Capra V, Grompe M, Pizzuti A, Muzny D,
Lawrence C, et al: Characterization of a murine gene expressed from
the inactive X chromosome. Nature. 351:325–329. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Gendrel AV and Heard E: Noncoding RNAs and
epigenetic mechanisms during X-chromosome inactivation. Annu Rev
Cell Dev Biol. 30:561–580. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Li ZX, Zhu QN, Zhang HB, Hu Y, Wang G and
Zhu YS: MALAT1: A potential biomarker in cancer. Cancer Manag Res.
10:6757–6768. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Seiler J, Breinig M, Caudron-Herger M,
Polycarpou-Schwarz M, Boutros M and Diederichs S: The lncRNA VELUCT
strongly regulates viability of lung cancer cells despite its
extremely low abundance. Nucleic Acids Res. 45:5458–5469. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ma M, Xu H, Liu G, Wu J, Li C, Wang X,
Zhang S, Xu H, Ju S, Cheng W, et al: Metabolism-induced tumor
activator 1 (MITA1), an Energy Stress-inducible long noncoding RNA,
promotes hepatocellular carcinoma metastasis. Hepatology.
70:215–230. 2019.PubMed/NCBI
|
|
18
|
Lu YF, Cai XL, Li ZZ, Lv J, Xiang YA, Chen
JJ, Chen WJ, Sun WY, Liu XM and Chen JB: lncRNA SNHG16 functions as
an oncogene by sponging MiR-4518 and Up-regulating PRMT5 expression
in Glioma. Cell Physiol Biochem. 45:1975–1985. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wang FW, Cao CH, Han K, Zhao YX, Cai MY,
Xiang ZC, Zhang JX, Chen JW, Zhong LP, Huang Y, et al:
APC-activated long noncoding RNA inhibits colorectal carcinoma
pathogenesis through reduction of exosome production. J Clin
Invest. 129:727–743. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kim J, Piao HL, Kim BJ, Yao F, Han Z, Wang
Y, Xiao Z, Siverly AN, Lawhon SE, Ton BN, et al: Long noncoding RNA
MALAT1 suppresses breast cancer metastasis. Nat Genet.
50:1705–1715. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhu C, Cheng D, Qiu X, Zhuang M and Liu Z:
Long noncoding RNA SNHG16 promotes cell proliferation by sponging
MicroRNA-205 and upregulating ZEB1 expression in osteosarcoma. Cell
Physiol Biochem. 51:429–440. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yan H, Li H, Li P, Li X, Lin J, Zhu L,
Silva MA, Wang X, Wang P and Zhang Z: Long noncoding RNA MLK7-AS1
promotes ovarian cancer cells progression by modulating
miR-375/YAP1 axis. J Exp Clin Cancer Res. 37:2372018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhuo W, Liu Y, Li S, Guo D, Sun Q, Jin J,
Rao X, Li M, Sun M, Jiang M, et al: Long noncoding RNA GMAN,
up-regulated in gastric cancer tissues, is associated with
metastasis in patients and promotes translation of Ephrin A1 by
competitively binding GMAN-AS. Gastroenterology. 156:676–691.e11.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhang E, Han L, Yin D, He X, Hong L, Si X,
Qiu M, Xu T, De W, Xu L, et al: H3K27 acetylation activated-long
non-coding RNA CCAT1 affects cell proliferation and migration by
regulating SPRY4 and HOXB13 expression in esophageal squamous cell
carcinoma. Nucleic Acids Res. 45:3086–3101. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Peng WX, Koirala P and Mo YY:
lncRNA-mediated regulation of cell signaling in cancer. Oncogene.
36:5661–5667. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wang Z, Yang B, Zhang M, Guo W, Wu Z, Wang
Y, Jia L, Li S; Cancer Genome Atlas Research Network, ; Xie W and
Yang D: lncRNA epigenetic landscape analysis identifies EPIC1 as an
oncogenic lncRNA that interacts with MYC and promotes Cell-cycle
progression in cancer. Cancer Cel. 33:706–720.e9. 2018. View Article : Google Scholar
|
|
27
|
Bhan A, Soleimani M and Mandal SS: Long
noncoding RNA and cancer: A new paradigm. Cancer Res. 77:3965–3981.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Chen YG, Satpathy AT and Chang HY: Gene
regulation in the immune system by long noncoding RNAs. Nat
Immunol. 18:962–972. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kopp F and Mendell JT: Functional
classification and experimental dissection of long noncoding RNAs.
Cell. 172:393–407. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Chu C, Zhang QC, da Rocha ST, Flynn RA,
Bharadwaj M, Calabrese JM, Magnuson T, Heard E and Chang HY:
Systematic discovery of Xist RNA binding proteins. Cell.
161:404–416. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Samata M and Akhtar A: Dosage compensation
of the X chromosome: A complex epigenetic assignment involving
chromatin regulators and long noncoding RNAs. Annu Rev Biochem.
87:323–350. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Vallot C, Patrat C, Collier AJ, Huret C,
Casanova M, Liyakat AT, Tosolini M, Frydman N, Heard E, Rugg-Gunn
PJ and Rougeulle C: XACT noncoding RNA competes with XIST in the
control of X Chromosome activity during human early development.
Cell Stem Cell. 20:102–111. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Furlan G, Gutierrez Hernandez N, Huret C,
Galupa R, van Bemmel JG, Romito A, Heard E, Morey C and Rougeulle
C: The Ftx noncoding locus controls X chromosome inactivation
independently of its RNA products. Mol Cell. 70:462–472.e8. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Sarma K, Cifuentes-Rojas C, Ergun A, Del
Rosario A, Jeon Y, White F, Sadreyev R and Lee JT: ATRX directs
binding of PRC2 to Xist RNA and Polycomb targets. Cell.
159:869–883. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Guttman M, Donaghey J, Carey BW, Garber M,
Grenier JK, Munson G, Young G, Lucas AB, Ach R, Bruhn L, et al:
lincRNAs act in the circuitry controlling pluripotency and
differentiation. Nature. 477:295–300. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Khalil AM, Guttman M, Huarte M, Garber M,
Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van
Oudenaarden A, et al: Many human large intergenic noncoding RNAs
associate with chromatin-modifying complexes and affect gene
expression. Proc Natl Acad Sci USA. 106:11667–11672. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Engreitz JM, Haines JE, Perez EM, Munson
G, Chen J, Kane M, McDonel PE, Guttman M and Lander ES: Local
regulation of gene expression by lncRNA promoters, transcription
and splicing. Nature. 539:452–455. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Sun TT, He J, Liang Q, Ren LL, Yan TT, Yu
TC, Tang JY, Bao YJ, Hu Y, Lin Y, et al: lncRNA GClnc1 promotes
gastric carcinogenesis and may act as a modular scaffold of WDR5
and KAT2A complexes to specify the histone modification pattern.
Cancer Discov. 6:784–801. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Li D, Liu X, Zhou J, Hu J, Zhang D, Liu J,
Qiao Y and Zhan Q: Long noncoding RNA HULC modulates the
phosphorylation of YB-1 through serving as a scaffold of
extracellular signal-regulated kinase and YB-1 to enhance
hepatocarcinogenesis. Hepatology. 65:1612–1627. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Long Y, Wang X, Youmans DT and Cech TR:
How do lncRNAs regulate transcription? Sci Adv. 3:eaao21102017.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Hu WL, Jin L, Xu A, Wang YF, Thorne RF,
Zhang XD and Wu M: GUARDIN is a p53-responsive long non-coding RNA
that is essential for genomic stability. Nat Cell Biol. 20:492–502.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Bhan A and Mandal SS: Estradiol-induced
transcriptional regulation of long non-coding RNA, HOTAIR. Methods
Mol Biol. 1366:395–412. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Grelet S, Link LA, Howley B, Obellianne C,
Palanisamy V, Gangaraju VK, Diehl JA and Howe PH: Addendum: A
regulated PNUTS mRNA to lncRNA splice switch mediates EMT and
tumour progression. Nat Cell Biol. 19:14432017. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Wu Q, Guo L, Jiang F, Li L, Li Z and Chen
F: Analysis of the miRNA-mRNA-lncRNA networks in ER+ and ER-breast
cancer cell lines. J Cell Mol Med. 19:2874–2887. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Song YX, Sun JX, Zhao JH, Yang YC, Shi JX,
Wu ZH, Chen XW, Gao P, Miao ZF and Wang ZN: Non-coding RNAs
participate in the regulatory network of CLDN4 via ceRNA mediated
miRNA evasion. Nat Commun. 8:2892017. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Shen L, Wang Q, Liu R, Chen Z, Zhang X,
Zhou P and Wang Z: lncRNA lnc-RI regulates homologous recombination
repair of DNA double-strand breaks by stabilizing RAD51 mRNA as a
competitive endogenous RNA. Nucleic Acids Res. 46:717–729. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Yu W, Peng W, Jiang H, Sha H and Li J:
lncRNA HOXA11-AS promotes proliferation and invasion by targeting
miR-124 in human non-small cell lung cancer cells. Tumour Biol.
39:10104283177214402017. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Dhir A, Dhir S, Proudfoot NJ and Jopling
CL: Microprocessor mediates transcriptional termination of long
noncoding RNA transcripts hosting microRNAs. Nat Struct Mol Biol.
22:319–327. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Augoff K, McCue B, Plow EF and
Sossey-Alaoui K: miR-31 and its host gene lncRNA LOC554202 are
regulated by promoter hypermethylation in triple-negative breast
cancer. Mol Cancer. 11:52012. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Yu Y, Nangia-Makker P, Farhana L and
Majumdar A: A novel mechanism of lncRNA and miRNA interaction:
CCAT2 regulates miR-145 expression by suppressing its maturation
process in colon cancer cells. Mol Cancer. 16:1552017. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Villamizar O, Chambers CB, Riberdy JM,
Persons DA and Wilber A: Long noncoding RNA Saf and splicing factor
45 increase soluble Fas and resistance to apoptosis. Oncotarget.
7:13810–13826. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Jadaliha M, Gholamalamdari O, Tang W,
Zhang Y, Petracovici A, Hao Q, Tariq A, Kim TG, Holton SE, Singh
DK, et al: A natural antisense lncRNA controls breast cancer
progression by promoting tumor suppressor gene mRNA stability. PLoS
Genet. 14:e10078022018. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Qu L, Ding J, Chen C, Wu ZJ, Liu B, Gao Y,
Chen W, Liu F, Sun W, Li XF, et al: Exosome-Transmitted lncARSR
promotes sunitinib resistance in renal cancer by acting as a
competing endogenous RNA. Cancer Cell. 29:653–668. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Rashid F, Shah A and Shan G: Long
non-coding RNAs in the cytoplasm. Genomics Proteomics
Bioinformatics. 14:73–80. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Yoon JH, Abdelmohsen K, Srikantan S, Yang
X, Martindale JL, De S, Huarte M, Zhan M, Becker KG and Gorospe M:
LincRNA-p21 suppresses target mRNA translation. Mol Cell.
47:648–655. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Wang KC, Yang YW, Liu B, Sanyal A,
Corces-Zimmerman R, Chen Y, Lajoie BR, Protacio A, Flynn RA, Gupta
RA, et al: A long noncoding RNA maintains active chromatin to
coordinate homeotic gene expression. Nature. 472:120–124. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
57
|
He RZ, Luo DX and Mo YY: Emerging roles of
lncRNAs in the post-transcriptional regulation in cancer. Genes
Dis. 6:6–15. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Yu J, Han Z, Sun Z, Wang Y, Zheng M and
Song C: lncRNA SLCO4A1-AS1 facilitates growth and metastasis of
colorectal cancer through β-catenin-dependent Wnt pathway. J Exp
Clin Cancer Res. 37:2222018. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Jiang H, Li T, Qu Y, Wang X, Li B, Song J,
Sun X, Tang Y, Wan J, Yu Y, et al: Long non-coding RNA SNHG15
interacts with and stabilizes transcription factor Slug and
promotes colon cancer progression. Cancer Lett. 425:78–87. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Cui M, Wang J, Li Q, Zhang J, Jia J and
Zhan X: Long non-coding RNA HOXA11-AS functions as a competing
endogenous RNA to regulate ROCK1 expression by sponging miR-124-3p
in osteosarcoma. Biomed Pharmacother. 92:437–444. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Zheng LL, Li JH, Wu J, Sun WJ, Liu S, Wang
ZL, Zhou H, Yang JH and Qu LH: deepBase v2.0: Identification,
expression, evolution and function of small RNAs, lncRNAs and
circular RNAs from deep-sequencing data. Nucleic Acids Res.
44:D196–D202. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Spencer DH, Young MA, Lamprecht TL, Helton
NM, Fulton R, O'Laughlin M, Fronick C, Magrini V, Demeter RT,
Miller CA, et al: Epigenomic analysis of the HOX gene loci reveals
mechanisms that may control canonical expression patterns in AML
and normal hematopoietic cells. Leukemia. 29:1279–1289. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Garcia-Fernández J: The genesis and
evolution of homeobox gene clusters. Nat Rev Genet. 6:881–892.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Kelly ZL, Michael A, Butler-Manuel S,
Pandha HS and Morgan RG: HOX genes in ovarian cancer. J Ovarian
Res. 4:162011. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Cheng W, Liu J, Yoshida H, Rosen D and
Naora H: Lineage infidelity of epithelial ovarian cancers is
controlled by HOX genes that specify regional identity in the
reproductive tract. Nat Med. 11:531–537. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Lu S, Jiang X, Su Z, Cui Z, Fu W and Tai
S: The role of the long non-coding RNA HOXA11-AS in promoting
proliferation and metastasis of malignant tumors. Cell Biol Int.
42:1596–1601. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Lu CW, Zhou DD, Xie T, Hao JL, Pant OP, Lu
CB and Liu XF: HOXA11 antisense long noncoding RNA (HOXA11-AS): A
promising lncRNA in human cancers. Cancer Med. 7:3792–3799. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Xu YJ, Du Y and Fan Y: Long noncoding RNAs
in lung cancer: What we know in 2015. Clin Transl Oncol.
18:660–665. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Kang CG, Lee HJ, Kim SH and Lee EO:
Zerumbone suppresses Osteopontin-induced cell invasion through
inhibiting the FAK/AKT/ROCK pathway in human Non-small cell lung
cancer A549 cells. J Nat Prod. 79:156–160. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Osmani L, Askin F, Gabrielson E and Li QK:
Current WHO guidelines and the critical role of immunohistochemical
markers in the subclassification of non-small cell lung carcinoma
(NSCLC): Moving from targeted therapy to immunotherapy. Semin
Cancer Biol. 52:103–109. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Li Y, Xuan J, Song Y, Qi W, He B, Wang P
and Qin L: Nanoporous Glass integrated in volumetric Bar-chart chip
for Point-of-Care diagnostics of non-small cell lung cancer. Acs
Nano. 10:1640–1647. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Zhang Y, He RQ, Dang YW, Zhang XL, Wang X,
Huang SN, Huang WT, Jiang MT, Gan XN, Xie Y, et al: Comprehensive
analysis of the long noncoding RNA HOXA11-AS gene interaction
regulatory network in NSCLC cells. Cancer Cell Int. 16:892016.
View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Zhang Y, Chen WJ, Gan TQ, Zhang XL, Xie
ZC, Ye ZH, Deng Y, Wang ZF, Cai KT, Li SK, et al: Clinical
significance and effect of lncRNA HOXA11-AS in NSCLC: A study based
on bioinformatics, in vitro and in vivo verification. Sci Rep.
7:5567–18. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Zhang Y, Luo J, Wang X, Wang HL, Zhang XL,
Gan TQ, Chen G and Luo DZ: A comprehensive analysis of the
predicted targets of miR-642b-3p associated with the long
non-coding RNA HOXA11-AS in NSCLC cells. Oncol Lett. 15:6147–6160.
2018.PubMed/NCBI
|
|
75
|
Chen JH, Zhou LY, Xu S, Zheng YL, Wan YF
and Hu CP: Overexpression of lncRNA HOXA11-AS promotes cell
epithelial-mesenchymal transition by repressing miR-200b in
non-small cell lung cancer. Cancer Cell Int. 17:642017. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Zhao X, Li X, Zhou L, Ni J, Yan W, Ma R,
Wu J, Feng J and Chen P: lncRNA HOXA11-AS drives cisplatin
resistance of human LUAD cells via modulating miR-454-3p/Stat3.
Cancer Sci. 109:3068–3079. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Wu F, Yin Z, Yang L, Fan J, Xu J, Jin Y,
Yu J, Zhang D and Yang G: Smoking induced extracellular vesicles
release and their distinct properties in Non-small cell lung
cancer. J Cancer. 10:3435–3443. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Vogel A, Cervantes A, Chau I, Daniele B,
Llovet J, Meyer T, Nault JC, Neumann U, Ricke J, Sangro B, et al:
Hepatocellular carcinoma: ESMO clinical practice guidelines for
diagnosis, treatment and follow-up. Ann Oncol. 29 (Suppl
4):iv238–iv255. 2018. View Article : Google Scholar
|
|
79
|
Koeberle D, Dufour JF, Demeter G, Li Q,
Ribi K, Samaras P, Saletti P, Roth AD, Horber D, Buehlmann M, et
al: Sorafenib with or without everolimus in patients with advanced
hepatocellular carcinoma (HCC): A randomized multicenter,
multinational phase II trial (SAKK 77/08 and SASL 29). Ann Oncol.
27:856–861. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Tai WM, Yong WP, Lim C, Low LS, Tham CK,
Koh TS, Ng QS, Wang WW, Wang LZ, Hartano S, et al: Ann A phase Ib
study of selumetinib (AZD6244, ARRY-142886) in combination with
sorafenib in advanced hepatocellular carcinoma (HCC). Ann Oncol.
27:2210–2215. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Ciuleanu T, Bazin I, Lungulescu D, Miron
L, Bondarenko I, Deptala A, Rodriguez-Torres M, Giantonio B, Fox
NL, Wissel P, et al: A randomized, double-blind, placebo-controlled
phase II study to assess the efficacy and safety of mapatumumab
with sorafenib in patients with advanced hepatocellular carcinoma.
Ann Oncol. 27:680–687. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Liu B, Li J, Liu X, Zheng M, Yang Y, Lyu Q
and Jin L: Long non-coding RNA HOXA11-AS promotes the proliferation
HCC cells by epigenetically silencing DUSP5. Oncotarget.
8:109509–109521. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Yu J, Hong JF, Kang J, Liao LH and Li CD:
Promotion of lncRNA HOXA11-AS on the proliferation of
hepatocellular carcinoma by regulating the expression of LATS1. Eur
Rev Med Pharmacol Sci. 21:3402–3411. 2017.PubMed/NCBI
|
|
84
|
Zhan M, He K, Xiao J, Liu F, Wang H, Xia
Z, Duan X, Huang R, Li Y, He X, et al: lncRNA HOXA11-AS promotes
hepatocellular carcinoma progression by repressing miR-214-3p. J
Cell Mol Med. May 15–2018.(Epub ahead of print). doi:
10.1111/jcmm.13633. View Article : Google Scholar
|
|
85
|
Gusyatiner O and Hegi ME: Glioma
epigenetics: From subclassification to novel treatment options.
Semin Cancer Biol. 51:50–58. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Cohen ZR, Ramishetti S, Peshes-Yaloz N,
Goldsmith M, Wohl A, Zibly Z and Peer D: Localized RNAi
therapeutics of chemoresistant grade IV glioma using
hyaluronan-grafted lipid-based nanoparticles. ACS Nano.
9:1581–1591. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Zhang KL, Han L, Chen LY, Shi ZD, Yang M,
Ren Y, Chen LC, Zhang JX, Pu PY and Kang CS: Blockage of a
miR-21/EGFR regulatory feedback loop augments anti-EGFR therapy in
glioblastomas. Cancer Lett. 342:139–149. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Wang X, Wang K, Han L, Zhang A, Shi Z,
Zhang K, Zhang H, Yang S, Pu P, Shen C, et al: PRDM1 is directly
targeted by miR-30a-5p and modulates the Wnt/β-catenin pathway in a
Dkk1-dependent manner during glioma growth. Cancer Lett.
331:211–219. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Gonzalez-Quarante LH, Fernández Carballal
C, Agarwal V, Vargas Lopez AJ, Gil de Sagredo Del Corral OL and
Sola Vendrell E: Angiocentric Glioma in an elderly patient: Case
report and review of the literature. World Neurosurg.
97:755.e5–755.e10. 2017. View Article : Google Scholar
|
|
90
|
Wang Q, Zhang J, Liu Y, Zhang W, Zhou J,
Duan R, Pu P, Kang C and Han L: A novel cell cycle-associated
lncRNA, HOXA11-AS, is transcribed from the 5-prime end of the HOXA
transcript and is a biomarker of progression in glioma. Cancer
Lett. 373:251–259. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Cui Y, Yi L, Zhao J and Jiang Y: Long
Noncoding RNA HOXA11-AS functions as miRNA sponge to promote the
glioma tumorigenesis through targeting miR-140-5p. DNA Cell Biol.
36:822–828. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Xu C, He T, Li Z, Liu H and Ding B:
Regulation of HOXA11-AS/miR-214-3p/EZH2 axis on the growth,
migration and invasion of glioma cells. Biomed Pharmacother.
95:1504–1513. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Xu CH, Xiao LM, Liu Y, Chen LK, Zheng SY,
Zeng EM and Li DH: The lncRNA HOXA11-AS promotes glioma cell growth
and metastasis by targeting miR-130a-5p/HMGB2. Eur Rev Med
Pharmacol Sci. 23:241–252. 2019.PubMed/NCBI
|
|
94
|
Yang JX, Liu B, Yang BY and Meng Q: Long
non-coding RNA homeobox (HOX) A11-AS promotes malignant progression
of glioma by targeting miR-124-3p. Neoplasma. 65:505–514. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: Sources, methods and major
patterns in GLOBOCAN 2012. Int J Cancer. 136:E359–E386. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Rychahou P, Haque F, Shu Y, Zaytseva Y,
Weiss HL, Lee EY, Mustain W, Valentino J, Guo P and Evers BM:
Delivery of RNA nanoparticles into colorectal cancer metastases
following systemic administration. ACS Nano. 9:1108–1116. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Zhang L, Cao F, Zhang G, Shi L, Chen S,
Zhang Z, Zhi W and Ma T: Trends in and predictions of colorectal
cancer incidence and mortality in china from 1990 to 2025. Front
Oncol. 9:982019. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Siegel R, Ma J, Zou Z and Jemal A: Cancer
statistics, 2014. CA Cancer J Clin. 64:9–29. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Lai Y, Wang C, Civan JM, Palazzo JP, Ye Z,
Hyslop T, Lin J, Myers RE, Li B, Jiang B, et al: Effects of cancer
stage and treatment differences on racial disparities in survival
from colon cancer: A united states population-based study.
Gastroenterology. 150:1135–1146. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Li T, Xu C, Cai B, Zhang M, Gao F and Gan
J: Expression and clinicopathological significance of the lncRNA
HOXA11-AS in colorectal cancer. Oncol Lett. 12:4155–4160. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Chen D, Sun Q, Zhang L, Zhou X, Cheng X,
Zhou D, Ye F, Lin J and Wang W: The lncRNA HOXA11-AS functions as a
competing endogenous RNA to regulate PADI2 expression by sponging
miR-125a-5p in liver metastasis of colorectal cancer. Oncotarget.
8:70642–70652. 2017.PubMed/NCBI
|
|
102
|
Anastasiadi Z, Lianos GD, Ignatiadou E,
Harissis HV and Mitsis M: Breast cancer in young women: An
overview. Updates Surg. 69:313–317. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Zhang Y, Leonard M, Shu Y, Yang Y, Shu D,
Guo P and Zhang X: Overcoming Tamoxifen resistance of human breast
cancer by targeted gene silencing using multifunctional pRNA
nanoparticles. Acs Nano. 11:335–346. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Lemler DJ, Lynch ML, Tesfay L, Deng Z,
Paul BT, Wang X, Hegde P, Manz DH, Torti SV and Torti FM: DCYTB is
a predictor of outcome in breast cancer that functions via
iron-independent mechanisms. Breast Cancer Res. 19:252017.
View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Borin TF, Arbab AS, Gelaleti GB, Ferreira
LC, Moschetta MG, Jardim-Perassi BV, Iskander AS, Varma NR, Shankar
A, Coimbra VB, et al: Melatonin decreases breast cancer metastasis
by modulating Rho-associated kinase protein-1 expression. J Pineal
Res. 60:3–15. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
van Roosmalen W, Le Dévédec SE, Golani O,
Smid M, Pulyakhina I, Timmermans AM, Look MP, Zi D, Pont C, de
Graauw M, et al: Tumor cell migration screen identifies SRPK1 as
breast cancer metastasis determinant. J Clin Invest. 125:1648–1664.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Su J and Hu X: Long non-coding RNA
HOXA11-AS promotes cell proliferation and metastasis in human
breast cancer. Mol Med Rep. 16:4887–4894. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Li W, Jia G, Qu Y, Du Q and Liu B and Liu
B: Long Non-coding RNA (lncRNA) HOXA11-AS promotes breast cancer
invasion and metastasis by regulating Epithelial-mesenchymal
transition. Med Sci Monit. 23:3393–3403. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Wang K, Liang Q, Li X, Tsoi H, Zhang J,
Wang H, Go MY, Chiu PW, Ng EK, Sung JJ and Yu J: MDGA2 is a novel
tumour suppressor cooperating with DMAP1 in gastric cancer and is
associated with disease outcome. Gut. 65:1619–1631. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Sugano K: Screening of gastric cancer in
Asia. Best Pract Res Clin Gastroenterol. 29:895–905. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2013. CA Cancer J Clin. 63:11–30. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Sun M, Nie F, Wang Y, Zhang Z, Hou J, He
D, Xie M, Xu L, De W, Wang Z and Wang J: lncRNA HOXA11-AS promotes
proliferation and invasion of gastric cancer by scaffolding the
chromatin modification factors PRC2, LSD1, and DNMT1. Cancer Res.
76:6299–6310. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Liu Z, Chen Z, Fan R, Jiang B, Chen X,
Chen Q, Nie F, Lu K and Sun M: Over-expressed long noncoding RNA
HOXA11-AS promotes cell cycle progression and metastasis in gastric
cancer. Mol Cancer. 16:822017. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Grignon DJ and Che M: Clear cell renal
cell carcinoma. Clin Lab Med. 25:305–316. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Escudier B, Eisen T, Stadler WM, Szczylik
C, Oudard S, Siebels M, Negrier S, Chevreau C, Solska E, Desai AA,
et al: Sorafenib in advanced clear-cell renal-cell carcinoma. N
Engl J Med. 356:125–134. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Ridge CA, Pua BB and Madoff DC:
Epidemiology and staging of renal cell carcinoma. Semin Intervent
Radiol. 31:3–8. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Yang FQ, Zhang JQ, Jin JJ, Yang CY, Zhang
WJ, Zhang HM, Zheng JH and Weng ZM: HOXA11-AS promotes the growth
and invasion of renal cancer by sponging miR-146b-5p to upregulate
MMP16 expression. J Cell Physiol. 233:9611–9619. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Robertson AG, Shih J, Yau C, Gibb EA, Oba
J, Mungall KL, Hess JM, Uzunangelov V, Walter V, Danilova L, et al:
Integrative analysis identifies four molecular and clinical subsets
in Uveal melanoma. Cancer Cell. 32:204–220.e15. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Moore AR, Ceraudo E, Sher JJ, Guan Y,
Shoushtari AN, Chang MT, Zhang JQ, Walczak EG, Kazmi MA, Taylor BS,
et al: Recurrent activating mutations of G-protein-coupled receptor
CYSLTR2 in uveal melanoma. Nat Genet. 48:675–680. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Coupland SE, Lake SL, Zeschnigk M and
Damato BE: Molecular pathology of uveal melanoma. Eye (Lond).
27:230–242. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Lu Q, Zhao N, Zha G, Wang H, Tong Q and
Xin S: lncRNA HOXA11-AS exerts oncogenic functions by repressing
p21 and miR-124 in Uveal melanoma. DNA Cell Biol. 36:837–844. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Ignarski M, Islam R and Müller RU: Long
Non-coding RNAs in Kidney disease. Int J Mol Sci. 20:E32762019.
View Article : Google Scholar : PubMed/NCBI
|
|
124
|
Dong D, Mu Z, Zhao C and Sun M: ZFAS1: A
novel tumor-related long non-coding RNA. Cancer Cell Int.
18:1252018. View Article : Google Scholar : PubMed/NCBI
|
|
125
|
Cheetham SW, Gruhl F, Mattick JS and
Dinger ME: Long noncoding RNAs and the genetics of cancer. Br J
Cancer. 108:2419–2425. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
126
|
Richards EJ, Permuth-Wey J, Li Y, Chen YA,
Coppola D, Reid BM, Lin H, Teer JK, Berchuck A, Birrer MJ, et al: A
functional variant in HOXA11-AS, a novel long non-coding RNA,
inhibits the oncogenic phenotype of epithelial ovarian cancer.
Oncotarget. 6:34745–34757. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Steuer CE, El-Deiry M, Parks JR, Higgins
KA and Saba NF: An update on larynx cancer. CA Cancer J Clin.
67:31–50. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
128
|
Qu L, Jin M, Yang L, Sun C, Wang P, Li Y,
Tian L, Liu M and Sun Y: Expression of long non-coding RNA
HOXA11-AS is correlated with progression of laryngeal squamous cell
carcinoma. Am J Transl Res. 10:573–580. 2018.PubMed/NCBI
|
|
129
|
Kim HJ, Eoh KJ, Kim LK, Nam EJ, Yoon SO,
Kim KH, Lee JK, Kim SW and Kim YT: The long noncoding RNA HOXA11
antisense induces tumor progression and stemness maintenance in
cervical cancer. Oncotarget. 7:83001–83016. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Sun XY, Wang XF, Cui YB, Cao XG, Zhao RH,
Wei HY, Cao W and Wu W: Expression level and clinical significance
of lncRNA HOXA11-AS in esophageal squamous cell carcinoma patients.
Zhonghua Zhong Liu Za Zhi. 40:186–190. 2018.(In Chinese).
PubMed/NCBI
|
|
131
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
132
|
Wang C, Inzana JA, Mirando AJ, Ren Y, Liu
Z, Shen J, O'Keefe RJ, Awad HA and Hilton MJ: NOTCH signaling in
skeletal progenitors is critical for fracture repair. J Clin
Invest. 126:1471–1481. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
133
|
Wang XN, Zhang LH, Cui XD, Wang MX, Zhang
GY and Yu PL: lncRNA HOXA11-AS is involved in fracture healing
through regulating mir-124-3p. Eur Rev Med Pharmacol Sci.
21:4771–4776. 2017.PubMed/NCBI
|
|
134
|
Mirzakhani H, Litonjua AA, McElrath TF,
O'Connor G, Lee-Parritz A, Iverson R, Macones G, Strunk RC,
Bacharier LB, Zeiger R, et al: Early pregnancy vitamin D status and
risk of preeclampsia. J Clin Invest. 126:4702–4715. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
135
|
Powe CE, Levine RJ and Karumanchi SA:
Preeclampsia, a disease of the maternal endothelium: The role of
antiangiogenic factors and implications for later cardiovascular
disease. Circulation. 123:2856–2869. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
136
|
Xu Y, Wu D, Liu J, Huang S, Zuo Q, Xia X,
Jiang Y, Wang S, Chen Y, Wang T and Sun L: Downregulated lncRNA
HOXA11-AS affects trophoblast cell proliferation and migration by
regulating RND3 and HOXA7 expression in PE. Mol Ther Nucleic Acids.
12:195–206. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
137
|
Hu L, Chang L, Zhang Y, Zhai L, Zhang S,
Qi Z, Yan H, Yan Y, Luo X, Zhang S, et al: Platelets express
activated P2Y12 receptor in patients with diabetes mellitus.
Circulation. 136:817–833. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
138
|
Jin QS, Huang LJ, Zhao TT, Yao XY, Lin LY,
Teng YQ, Kim SH, Nam MS, Zhang LY and Jin YJ: HOXA11-AS regulates
diabetic arteriosclerosis-related inflammation via PI3K/AKT
pathway. Eur Rev Med Pharmacol Sci. 22:6912–6921. 2018.PubMed/NCBI
|
|
139
|
Grelet S, Link LA, Howley B, Obellianne C,
Palanisamy V, Gangaraju VK, Diehl JA and Howe PH: A regulated PNUTS
mRNA to lncRNA splice switch mediates EMT and tumour progression.
Nat Cell Biol. 19:1105–1115. 2017. View Article : Google Scholar : PubMed/NCBI
|