Introduction
Globally, gastric cancer (GC) is the fifth most
commonly diagnosed cancer and the third leading cause of
cancer-related mortality (1,2).
Although the incidence and mortality of GC in China are declining,
GC still ranks second in both incidence and mortality, with an
estimated 679,100 new cases and 498,0000 deaths reported in 2015
(3). The high mortality is largely
due to the late diagnosis of the disease; at present, the majority
of newly diagnosed cases have locally advanced or metastatic
disease. Therefore, identification of factors that participate in
the development and progression of GC will help establish optimal
prevention, early diagnosis, and treatment strategies for GC.
Homeobox genes, which encode homeodomain
transcription factors, have been demonstrated to play critical
roles in embryo patterning along the anterior-posterior axis and
maintaining patterns in adult tissues (4,5).
Studies have demonstrated that some homeobox genes are upregulated
whereas others are downregulated in cancers, and some homeobox
genes exhibit both tumor-promoting and tumor-suppressing activities
depending on the specificity of the tissues and cells (6–9).
BARX2, also known as BarH-like homeobox 2, is
located at 11q24-q25 and encodes af 254-amino acid homeodomain
transcription factor (10).
BARX2 plays a key role during embryonic development
(11,12) and participates in cytoskeletal
organization, growth factor signaling, cell adhesion, and
transcriptional regulation (11,13–15).
Several studies have shown that BARX2 downregulation is
associated with ovarian cancer, breast cancer, primary
hepatocellular carcinoma (HCC), colorectal cancer, lung cancer, and
GC (16), along with poor patient
prognosis (16–21). In addition, BARX2 promotes
myogenic differentiation, regulates muscle-specific gene
expression, and regulates cell adhesion and cytoskeleton remodeling
during muscle cell fusion and cartilage formation (10). BARX2 regulates various
cellular adhesion molecules and promotes tissue differentiation
(14). Moreover, BARX2
functions as a tumor suppressor, with anti-oncogenic effects, as
shown in an in vitro study (16). However, the underlying mechanisms by
which BARX2 expression is downregulated and by which
BARX2 exerts anti-oncogenic effects remain to be
elucidated.
Several mechanisms, such as loss of heterozygosity,
histone deacetylation, gene amplification, and especially CpG
island promoter hypermethylation are involved in the aberrant
expression of homeobox genes (22–24).
DNA methylation within the promoter of tumor-suppressor genes,
which is commonly found in cancer cells, leads to transcriptional
silencing, and subsequently promotes cancer development (25). DNA methyltransferase (DNMT) is
responsible for DNA methylation (26). Promoter hypermethylation and
decreased expression of various homeobox genes, such as CDX1
(22), CDX2 (23) and PDX1 (24), have been reported in cancers such as
squamous esophageal cancer, GC, and colorectal cancers. Whether CpG
island promoter hypermethylation is responsible for the
downregulation or loss of BARX2 expression is unclear.
Therefore, the present study aimed to determine whether DNA
methylation downregulates BARX2 expression and whether
BARX2 is associated with suppression of gastric
carcinogenesis.
Materials and methods
Tissue microarray chips, cell lines,
and animals
The tissue microarray chips containing
formalin-fixed, paraffin-embedded specimens surgically taken from
gastric malignancies of 208 patients and endoscopically taken from
normal gastric mucosa of 8 individuals were provided by Xi'an Alena
Biotechnology Company (Xi'an, China) and used for
immunohistochemical BARX2 detection. The clinical and histological
characteristics of the patients and normal controls are listed in
Table I.
 | Table I.Associations of BARX2 protein
expression with demographic and pathological characteristics of the
patients with gastric cancer (n=208) and normal controls (n=8). |
Table I.
Associations of BARX2 protein
expression with demographic and pathological characteristics of the
patients with gastric cancer (n=208) and normal controls (n=8).
| Variable | Group | Cases (n) | BARX2
expressiona | Positive percentage
(%) |
|---|
| Age (years) | Gastric cancer
patients |
|
|
|
|
|
<60 | 103 | 0.469
(0.018, 1.859) | 53.40 (55/103) |
|
|
60–79 | 98 | 0.171
(0.018, 2.037) | 55.10 (54/98) |
|
|
≥80 | 7 | 0.017
(0.016, 2.868) | 42.86 (3/7) |
|
| Normal
controls |
|
|
|
|
|
<60 | 8 | 6.085
(4.032, 13.049) | 100 (8/8) |
| Sex | Gastric cancer
patients |
|
|
|
|
|
Male | 156 | 0.351
(0.018, 1.854) | 51.92 (81/156) |
|
|
Female | 52 | 0.241
(0.018, 2.631) | 59.61 (31/52) |
|
| Normal
controls |
|
|
|
|
|
Male | 5 | 6.640±2.780 | 62.5 (5/8) |
|
|
Female | 3 | 10.287±2.330 | 37.5 (3/8) |
| Pathological
types | Normal | 8 | 6.085
(4.032, 13.049) | 100 (8/8) |
|
| Mucinous
adenocarcinoma | 12 | 0.06
(0.018, 1.952) | 41.67 (5/12) |
|
| Signet-ring cell
carcinoma | 6 | 0.052
(0.025, 0.491) | 33.33 (2/6) |
|
| Undifferentiated
carcinoma | 5 | 0.891
(0.243, 5.463) | 100 (5/5) |
|
| Carcinoid | 3 | 0.19
(0.021, 4.499) | 0.00 (0/3) |
|
| Malignant
interstitialoma | 9 | 0.0189 (0.016,
0.018) | 100 (9/9) |
|
| Squamous cell
carcinoma | 1 | 0.092 | 100
(1/1)d |
| TNM
stageb | Normal | 8 | 6.085
(4.032, 13.049) | 100 (8/8) |
|
| I | 116 | 0.641
(0.018, 2.318) | 59.48 (69/116) |
|
| II | 41 | 0.039
(0.018, 0.927) | 36.59 (15/41) |
|
|
III | 12 | 0.022
(0.017, 1.509) | 41.67 (5/12) |
|
| IV | 3 | 0.021
(0.016, 5.867)f | 33.33
(1/3)e |
| Pathological
grading | Normal | 8 | 6.085
(4.032, 13.049) | 100.00 (8/8) |
|
|
Well-differentiated | 8 | 0.659
(0.079, 2.825) | 62.50 (5/8) |
|
| Moderately
differentiated | 40 | 0.459 (0.017,
2.42) | 47.5 (19/40) |
|
| Poorly
differentiated | 110 | 0.444
(0.019, 2.002) | 53.64 (59/110) |
|
|
Undifferentiated | 8 | 0.143
(0.016, 1.758)e | 50 (4/8) |
|
| Not
reportedc | 5 | 0.039 (0.018,
9.03) | 40 (2/5) |
| Lymph node
metastasis | No | 150 | 0.426
(0.019, 2.028) | 54.67 (82/150) |
|
| Yes | 24 | 0.02
(0.017, 1.174) | 36.36 (8/22) |
To observe the correlation between the expression of
BARX2 and DNM-1 (a commonly used marker of DNA methylation), a
separate batch of tissue microarray chips containing specimens from
22 cases of gastric adenocarcinoma and 8 normal controls were used
for immunohistochemical detection of BARX2 and DNMT-1.
Human GC cell lines including AGS, MGC803 (both
derived from primary human gastric adenocarcinoma), MKN7
(metastatic gastric tubular adenocarcinoma), MKN74 (metastatic
gastric tubular adenocarcinoma), and HGC27 (metastatic gastric
carcinoma) were purchased from the Type Culture Collection of the
Chinese Academy of Sciences (Shanghai, China). GES1 (a normal
gastric mucosal cell) was kindly provided by Dr Sui Peng (The First
Affiliated Hospital, Sun Yat-sen University, Guangzhou, China).
Cells were grown in F-12K nutrient mixture containing 10% fetal
bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.) in a cell
culture incubator with 5% CO2 at 37°C for more than 24
h, and then lysed with 0.05% of trypsin-EDTA (Thermo Fisher
Scientific, Inc.). All cell lines were used for reverse
transcription polymerase chain reaction (RT-PCR) or real-time
quantitative PCR (qPCR), and AGS cells were used for western blot
analysis, qPCR, and lentivirus (LV) transfection.
Ten male BALB/c-nu/nu mice (weighing 16–18 g) were
provided by Guangdong Medical Laboratory Animal Center (Guangzhou,
China) and used to determine the effect of BARX2 on
tumorigenicity. Mice were housed at room temperature with 40–60%
humidity, and with a light cycle of 10-h light/14-h dark under
pathogen-free conditions. All animal protocols were approved by the
Guangdong General Hospital Ethics Committee.
Immunohistochemical staining
After deparaffinizing and rehydration, the chips
were incubated with mouse anti-BARX2 (dilution 1:50; cat. no.
sc-53177; Santa Cruz Biotechnology) and rabbit anti-DNMT-1
(dilution 1:50; product code ab19905; Abcam) primary antibodies
overnight at 4°C. The chips were then incubated with
peroxidase-conjugated anti-mouse secondary antibody (dilution
1:100; cat. no. 7076; Cell Signaling Technology) and
peroxidase-conjugated anti-rabbit secondary antibody (dilution
1:100; cat. no. 7074; Cell Signaling Technology) respectively. The
chips were visualized with 3,3′-diaminobenzidine (1 mg/ml) and then
counterstained with hematoxylin. Finally, BARX2 expression was
analyzed using a Leica DM2500 system microscope (magnification,
×100; Meyer Instruments). The percentage of the area with
positively stained cells, defined as the area ratio, was determined
using ImageJ 1.52a software (National Institutes of Health,
Bethesda, MD, USA) (http://imagej.net/Downloads) to represent quantitative
expression. The percentage of positively stained cells (i.e. cells
with BARX2 signal among GC cells) was also calculated.
Western blot analysis
AGS cells were washed with ice-cold
phosphate-buffered saline (PBS; Thermo Fisher Scientific, Inc.) and
scraped using a 10-cm cold plastic cell scraper. After
centrifugation at 2,000 × g at 4°C for 5 min, the cell pellet was
added into RIPA lysate buffer (Sigma-Aldrich; Merck KGaA)
containing protease inhibitors for protein extraction. The AGS cell
lysates were electrophoresed on sodium dodecyl
sulfate-polyacrylamide 5% gel (SDS-PAGE) and transferred to a
polyvinylidene difluoride membrane. The membrane was probed with
primary antibodies against BARX2 (dilution 1:1,000; cat. no.
sc-53177; Santa Cruz Biotechnology), proliferating cell nuclear
antigen (PCNA) (dilution 1:1,000; product code ab146970), Ki-67
(dilution 1:500, product code; ab254123), matrix
metalloproteinase-7 (MMP7) (dilution 1:1,000; product code
ab207299), MMP9 (dilution 1:2,000; product code ab73734; all from
Abcam), E-cadherin (dilution 1:1,000; cat. no. 3195) and MMP3
(dilution 1:1,000; cat. no. 14351; both from Cell Signaling
Technology), and subsequently with horseradish
peroxidase-conjugated secondary antibodies (dilution 1:2,500; cat.
no. BA1055; Boster Biological Technology, Ltd.). β-actin (dilution
1:1500; Abcam) served as an internal control. An enhanced
chemiluminescence system (Amersham) was used to visualize the
antigen-antibody complex. The ImageJ 1.52a software (National
Institutes of Health) was used for quantification.
Total RNA extraction, RT-PCR and
qPCR
Total RNA was extracted using TRIzol reagent
(Invitrogen; Thermo Fisher Scientific, Inc.) after the cells were
harvested. RNA concentrations were measured using a NanoDrop
2000/2000c spectrophotometer (Thermo Fisher Scientific, Inc.). RNA
was reverse transcribed to complementary DNA using the PrimeScript™
RT reagent Kit (Perfect Real Time; Takara Bio Inc.). The PCR
program was performed in a 20-µl reaction mixture containing 2 µl
complementary DNA and 0.2 U Hot Start Taq DNA polymerase
(cat. no. M0495S; New England Biolabs, Inc.) and run for 30 cycles
of denaturation (at 94°C for 30 sec), annealing (at 56°C for 30
sec) and elongation (at 72°C for 45 sec). The primer sequences for
RT-PCR are shown in Table II.
Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as the
internal control.
 | Table II.Primer sequences for BARX2
mRNA expression, MSP and BSP analysis. |
Table II.
Primer sequences for BARX2
mRNA expression, MSP and BSP analysis.
| Gene | Primer
sequence | Product size
(bp) |
|---|
| BARX2
(RT-PCR) | Forward:
5′-GCAGCGAGTCAGAGACGGAACA-3′ | 424 |
|
| Reverse:
5′-GCCATCTCTAAGGGGACATCACG-3′ |
|
| GAPDH
(RT-PCR) | Forward:
5′-GTCAACGGATTTGGTCGTATTG-3′ | 200 |
|
| Reverse:
5′-CTCCTGGAAGATGGTGATGGG-3′ |
|
| BARX2
(qPCR) | Forward:
5′-CGGAGTCGCACCATCTTCAC-3′ | 100 |
|
| Reverse:
5′-GAGCCAAGTCCAACCTGTCT-3′ |
|
| β-actin (qPCR) | Forward:
5′-GGCACCACACCTTCTACAATGAG-3′ | 167 |
|
| Reverse:
5′-GGATAGCACAGCCTGGATAGCA-3′ |
|
| MSP | Forward:
5′-AAGAGTAATGTAAAGTCGGGGTTTCGA-3′ | 208 |
|
| Reverse:
5′-ACCGCCAATAAACTAAATACTTACGAACG-3′ |
|
| USP | Forward:
5′-AAGAGTAATGTAAAGTTGGGGTTTTGA-3′ | 267 |
|
| Reverse:
5′-CCACCAATAAACTAAATACTTACAAACAAT-3′ |
|
| BSP | Forward:
5′-GGGGAGGGGGAGGAGAGTTAAA-3′ | 267 |
|
| Reverse:
5′-AAACCCACCCRCAAATCAACATCTTC-3′ |
|
AGS cells were used for qPCR using the ABI PRISM
7000 Fluorescent Quantitative PCR System (Applied Biosystems;
Thermo Fisher Scientific, Inc.). The PCR program was carried out in
a 20 µl mixture containing the Power SYBR Green PCR Master Mix
(Applied Biosystems; Thermo Fisher Scientific, Inc.), 500 nmol of
the primers and 300 ng of complementary DNA templates. An initial
denaturation at 95°C for 5 min was followed by 50 cycles of
denaturation (at 94°C for 20 sec), annealing (at 60°C for 20 sec),
and elongation (at 72°C for 40 sec), with a final extension at 72°C
for 5 min. The melting curves and the generated Ct values, which
were calculated using the ΔΔCq-method and expressed as
2−ΔΔCq (27), were used
to quantify BARX2 mRNA. The primer sequences for RT-qPCR are
shown in Table II.
Methylation-specific PCR (MSP) and
bisulfite DNA sequencing PCR (BSP)
The TRED database (28) and Methprimer software (29) were used to search a 1,000-bp genomic
sequence that included the ATG translation starting codon (−1,000
nt to 0 nt) of BARX2 to predict the promoter. MSP and BSP were used
to examine the methylation status of the CpG islands in the five GC
cell lines.
First, DNA was isolated from the cells using the
DNeasy Blood and Tissue Kit (Qiagen China Co. Ltd.). Each genomic
DNA sample (1.0 µg) was denatured with NaOH (2 mol/l) at 37°C for
10 min, and incubated with sodium bisulfate (3 mol/l, pH 5.0;
Sigma-Aldrich; Merck KGaA) at 50°C for 16 h. The bisulfite-treated
DNA was amplified with methylation-specific or
unmethylation-specific primers (Table
II). For MSP analysis, the PCR program was carried out in a
25-µl reaction mixture containing 10 pmol/l primers, 25 µmol/l
deoxynucleoside triphosphates, 2 µl of bisulfate-treated DNA, and
0.5 U of Hot-Start Taq polymerase. The hot start at 95°C for
20 min was followed by 40 cycles of denaturation (at 94°C for 30
sec), annealing (at 52°C for 30 sec), and elongation (at 72°C for
45 sec), with a final extension at 72°C for 10 min. The PCR
products were visualized on a 2% agarose gel and stained with
ethidium bromide.
For BSP analysis, the bisulfite-treated DNA sample
(2 µl) was amplified in 20 µl reaction mixture, using the same PCR
program for MSP, except that the primers used were the
bisulfate-treated DNA sequencing PCR primers (Table II). For GC cell lines, the fragment
containing 27 CpG sites, as identified by the Methprimer software,
was amplified using bisulfite-modified DNA as a template and
inserted into the pGEM-T4 vector after purification
(Promega). Ten white clones were selected for each sample and were
then sequenced to determine the aberrant methylation of each CpG
site of the wild-type and modified sequences of the BARX2
promoter fragment in MGC803, MKN74 and HGC27 cells.
Lentiviral (LV) transfection of AGS
cells
LV specifically targeting BARX2 for gene
overexpression and sequences of the controls were purchased from
Biolink Biotechnology Co. (Shanghai, China). LV-BARX2 or
LV-empty vectors were transfected into AGS cells following the
manufacturer's protocol. Puromycin (5 µg/ml) was used for 1 week to
eradicate untransfected cells, and then the transfected cells
(AGS-LV-BARX2 and AGS-LV cells) were passaged at a ratio of 1:15
(vol/vol), and cultured for 4 weeks in F-12K nutrient mixture
containing puromycin (5 µg/ml). Finally, stably transfected clones
were selected for immunohistochemical detection of BARX2 expression
and maintained in the F-12K nutrient mixture prior to the
subsequent experiments.
Cell proliferation assays
Cell proliferation was detected using the Cell
Counting Kit-8 (CCK-8, Dojindo Laboratories). Briefly, AGS cells
transfected with LV-BARX2 or LV-empty were seeded into
96-well plates at 2.0×103 or 4.0×103 cells
per well. The relative ratio of absorbance at 490 nm of the
transfected cells (i.e. AGS-BARX2 and AGS-LV cells) was recorded
with a microplate reader (Bio-Rad Laboratories, Inc.) at 24, 48, 72
and 96 h, and expressed as a proliferation index. Each experiment
was performed in quadruplicate.
Cell proliferation was also detected using
5′-ethynyl-2′-deoxyuridine (EdU) (Thermo Fisher Scientific, Inc.).
Briefly, the transfected cells were seeded into 12-well-plates at
4.0×105 cells per well, stained with EdU and
4′,6-diamidino-2-phenylindole (DAPI) (Thermo Fisher Scientific,
Inc.) 48 h later, and randomly photographed under four high-power
fields with a fluorescence microscope.
Colony formation assay
Transfected cells were seeded into 6-well plates at
three different cell densities (0.5×103,
1.0×103 and 2.0×103 cells per well) and
cultured for 4 weeks. Cell colonies were fixed with methyl alcohol
for 15 min and stained with 0.1% crystal violet solution for 15 min
and were then counted under the Leica DM2500 system microscope
(magnification, ×40; Meyer Instruments). Three independent
experiments were performed, each in triplicate.
Cell migration assay
Cell migration was assayed using the Transwell
24-well Boyden chamber with a 8-µm polycarbonate membrane (Corning,
Inc.). Briefly, 3.0×104 cells were plated in the upper
chamber containing 200 µl serum-free media, while the bottom
chamber contained 500 µl media supplemented with 10% FBS as a
chemoattractant. After 48 h, the migrated cells were fixed by 4%
paraformaldehyde, stained with 0.1% crystal violet, and finally
eluted with 1 ml of 33% acetic acid. Absorbance of the migrated
cells was measured at 460 nm and expressed as an A460 value. Three
independent experiments were performed.
Tumorigenicity in nude mice
Ten male BALB/c-nu/nu mice were divided into two
groups (n=5 per group). The monoclonal LV-BARX2 or LV-empty
transfectants of AGS cells (i.e. AGS cells stably
transfected with LV-BARX2 or LV-empty vectors,
1.0×107 cells in 150 µl PBS) were inoculated
subcutaneously into the right flank of the mice. Tumor formation
was observed for 4 weeks, and then the mice were euthanized. The
xenografted tumors were dissected and the volume (V) was calculated
according to the following formula: V (mm3) = length
(mm) × width2 (mm2). Whole proteins were
extracted to detect Ki-67, PCNA, E-cadherin, and MMPs using western
blot analysis as described above.
Statistical analysis
Numerical data are expressed as mean ± standard
error of the mean (SEM) or median (25, 75%). The Student's t-test
was used for numeric data with normal distribution, and the
Mann-Whitney U-test was used for numeric data with abnormal
distribution. The Chi-square or Fisher's exact test, where
appropriate, was used for categorical data, with odds ratio (OR)
and 95% confidence interval (CI). Correlation was performed using
Pearson linear correlation. All statistical analyses were performed
using SPSS 16.0 software (SPSS, Inc.). A P-value of <0.05 was
assigned as indicative of statistical significance.
Results
BARX2 expression is downregulated in
GC tissues and cell lines
Immunohistochemical analysis showed that BARX2 was
expressed in the nuclei and cytoplasm of glandular epithelial cells
in the normal gastric tissues (Fig. 1Ag
and H). In contrast, BARX2 expression was low or even
undetectable in the gastric malignant tissues (Fig. 1Aa-F). BARX2 was expressed in all 8
(100%) normal gastric mucosal samples, and 112 (53.85%) cases with
gastric malignancy (χ2=4.163, P=0.041). Quantitative
analysis showed that the positive staining area was significantly
larger in the normal group (n=8) compared to the gastric malignant
group (n=208, 8.01±1.95 vs. 1.29±0.14, P<0.001) (Fig. 1B); the area was significantly larger
in the normal group than in the subgroup with gastric
adenocarcinoma (n=172, 8.01±1.95, vs. 1.23±0.14, P<0.001).
BARX2 mRNA levels were also low in AGS, MKN74, and MKN7
cells, MGC803 and HGC27 cells compared to GES1, as detected by qPCR
(vs. GES1, P<0.001) (Fig.
1C).
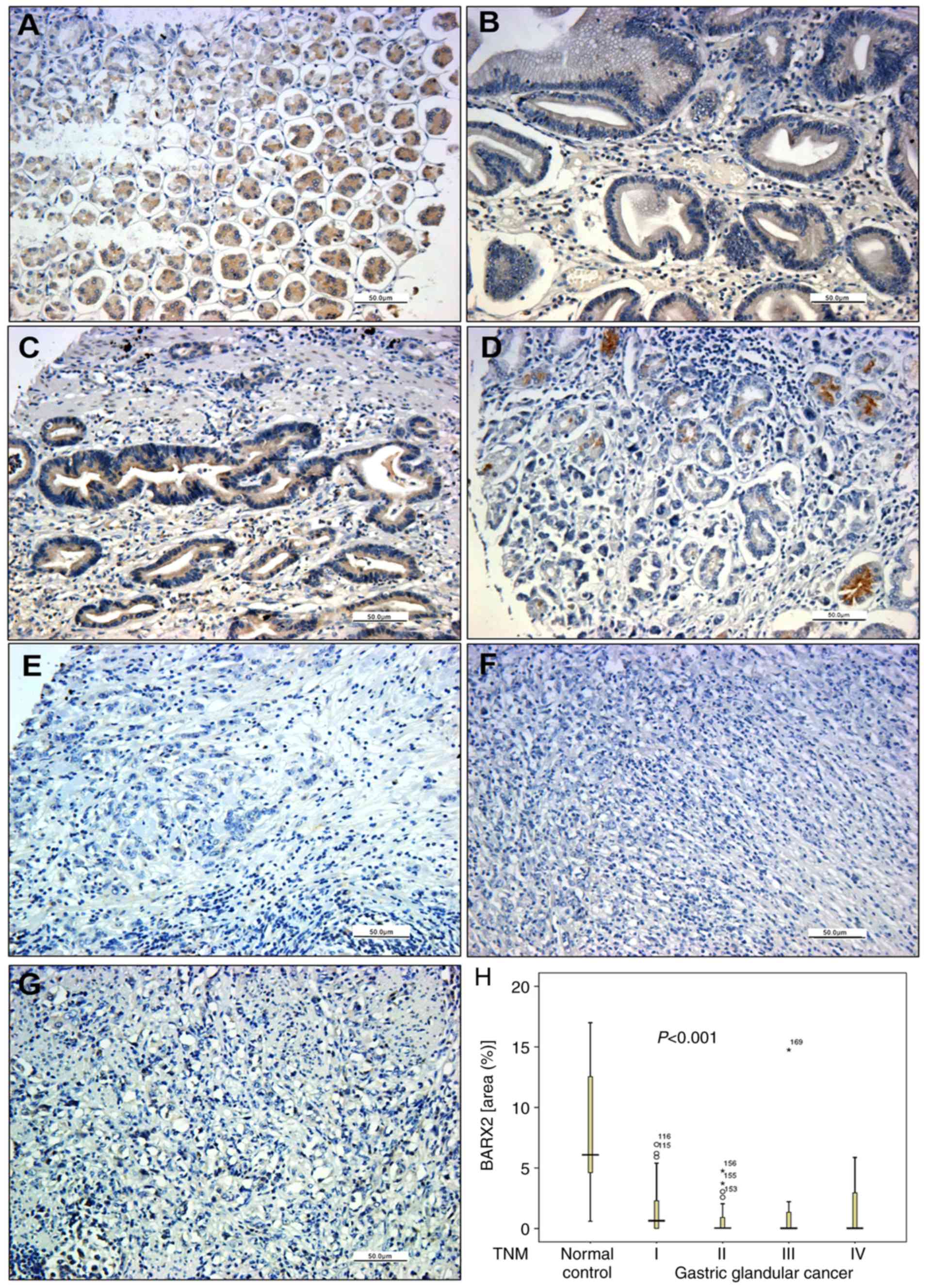
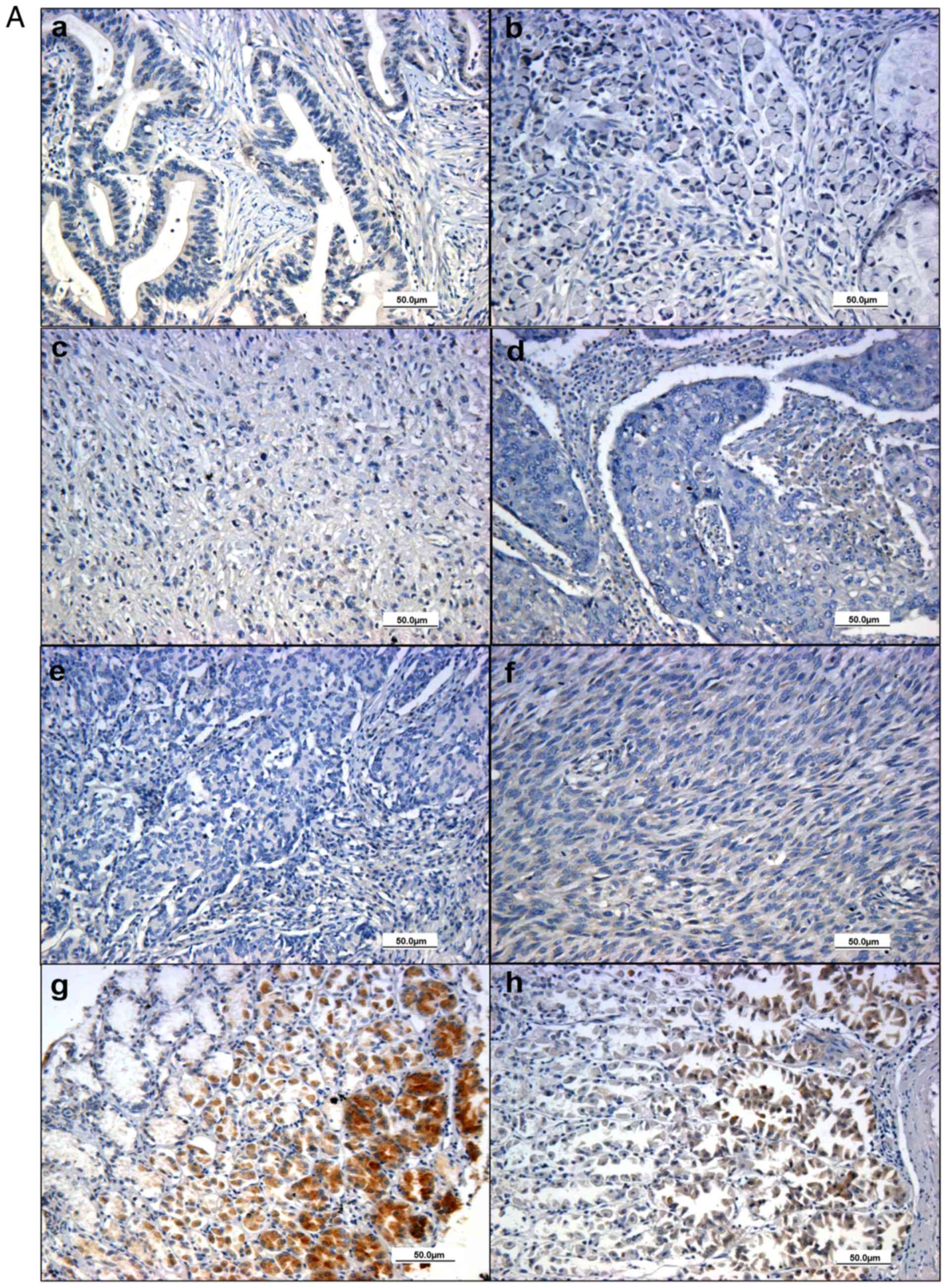 | Figure 1.BARX2 expression in normal and GC
tissues. BARX2 protein expression was weak and even absent in
gastric malignant samples, including adenocarcinoma (T2N0M0) (Aa),
signet-ring cell carcinoma (T2N0M0) (Ab), undifferentiated
carcinoma (T2N0M0) (Ac), squamous cell carcinoma (T2N0M0) (Ad),
carcinoid (T2N0M0) (Ae) and malignant interstitialoma (T2N0M0)
(Af). BARX2 stained as a yellowish brown and was commonly expressed
in the cytoplasm and nucleus of the glandular epithelial cells in
normal gastric mucosa (Ag and Ah), especially in the proliferative
glandular lumens (Ah) as detected by immunohistochemistry. (B)
Quantitative analysis shows that the positive staining area of
BARX2 was lower in the gastric malignant group (n=208, right bar),
compared to normal controls (n=8, left bar, *P<0.001). (C) qPCR
analysis showed that BARX2 mRNA levels were decreased and
even lost in the gastric carcinoma cell lines MGC803, AGS, MKN74,
MKN7, and HGC27, compared with the gastric mucosal cell line GES1
(*P<0.001). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was
used as the internal control. The Student's t-test was used for
statistical analysis. BARX2, BarH-like homeobox 2; GC,
gastric cancer. |
The associations of BARX2 expression with
demographical, clinical, and pathological characteristics are
summarized in Table I. There was a
significant difference in the positive percentage of BARX2
(χ2=4.748, P=0.029), but not in the area ratio, among
the different pathological types of gastric malignant tumors. As
most patients had gastric adenocarcinoma, we further investigated
the associations between BARX2 expression with pathological TNM
stages and grading of gastric adenocarcinoma (Table I). BARX2 protein expression was
positive in normal gastric tissues (Fig. 2A), but began to decline in gastric
adenocarcinoma from TNM stage I to IV (Fig. 2B-G) in terms of both percentage and
area ratio (Table I,
χ2=22.496, P<0.001). Additionally, there was a
gradual decline among the different pathological grades of
differentiation (well-differentiated, moderately differentiated,
poorly differentiated, and undifferentiated) in terms of the
percentage and area ratio (χ2=18.255, P=0.001) (Table I).
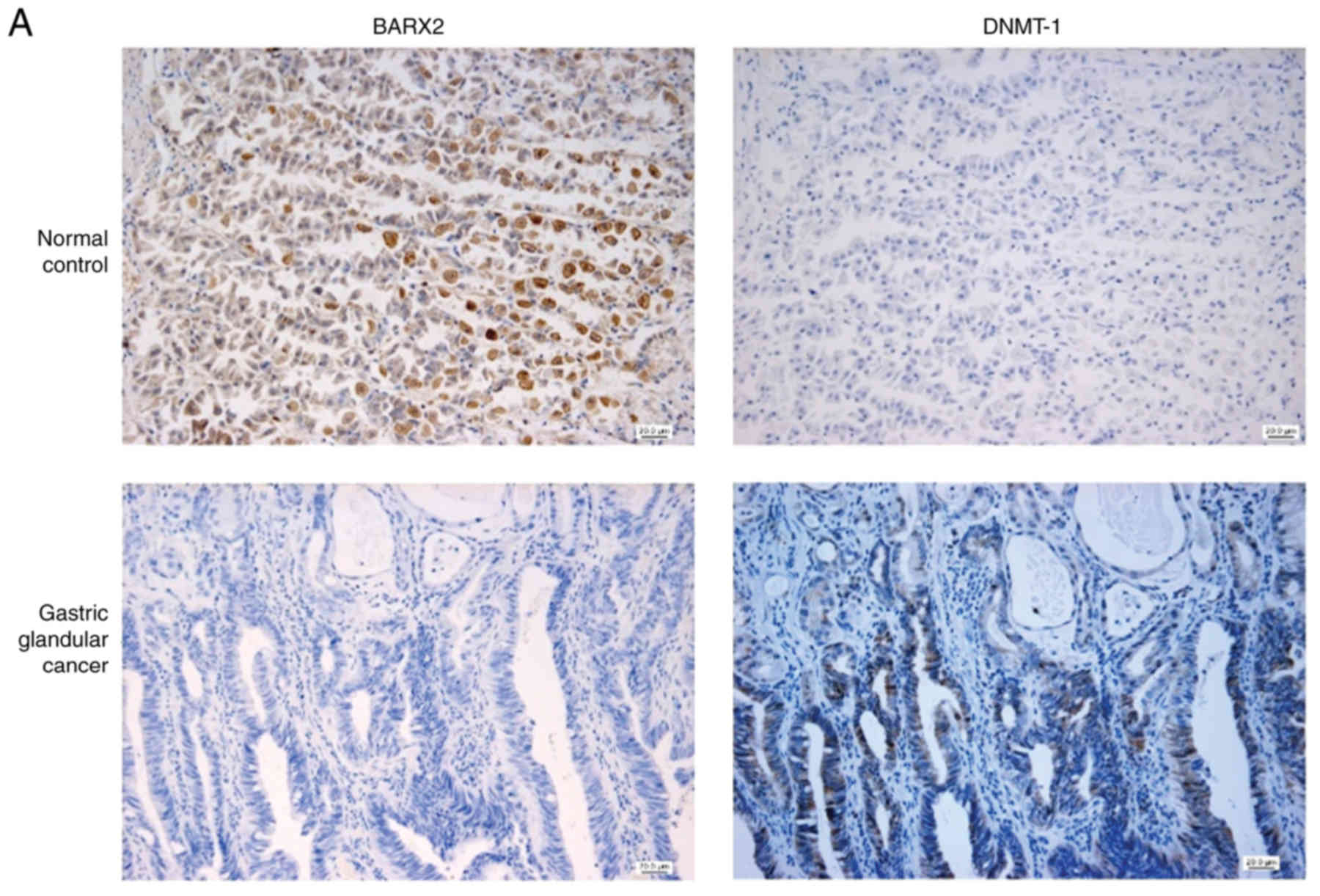
BARX2 expression is negatively
correlated with DNMT-1 expression
BARX2 expression was commonly present in normal
gastric mucosal glands, while DNMT-1 expression was often positive
in gastric adenocarcinoma cells (Fig.
3A). The quantitative protein expression of BARX2 in gastric
adenocarcinoma was lower than that of the normal mucosa [0.05
(0.021, 2.121) vs. 7.67 (4.657, 13.282), P=0.001], whereas the
expression pattern of DNMT-1 was reversed [0.018 (0.016, 0.166)
(control) vs. 3.3395 (1.312, 6.007) (adenocarcinoma), P<0.001]
(Fig. 3B). BARX2 expression was
negatively correlated with DNMT-1 expression (Pearson correlation
r=−0.369, P=0.045) (Fig. 3C).
BARX2 promoter is hypermethylated in
GC cell lines
Locations of two putative CpG islands were
identified using the Methprimer software (Fig. 4A). The methylation status of these
CpG islands was determined using MSP and BSP analyses in the five
GC cell lines. MSP showed partial methylation of BARX2 in
MGC803, MNK7, MKN74, and AGS cells and complete methylation in
HGC27 cells (Fig. 4B). BSP analysis
demonstrated 27 candidate CpG sites for methylation in the
BARX2 promoter fragment as none of these sites was altered
into T (Fig. 4C). Among the cell
lines studied, MGC803 cells displayed a high level of methylation
in most of the 27 CpG sites, while MKN74 and HGC27 cells only
showed partial methylation of the 27th CpG site (Fig. 4D). Both MSP and BSP showed that DNA
hypermethylation was present at the 5′flanking conserved promoter
region of BRAX2 in GC cells.
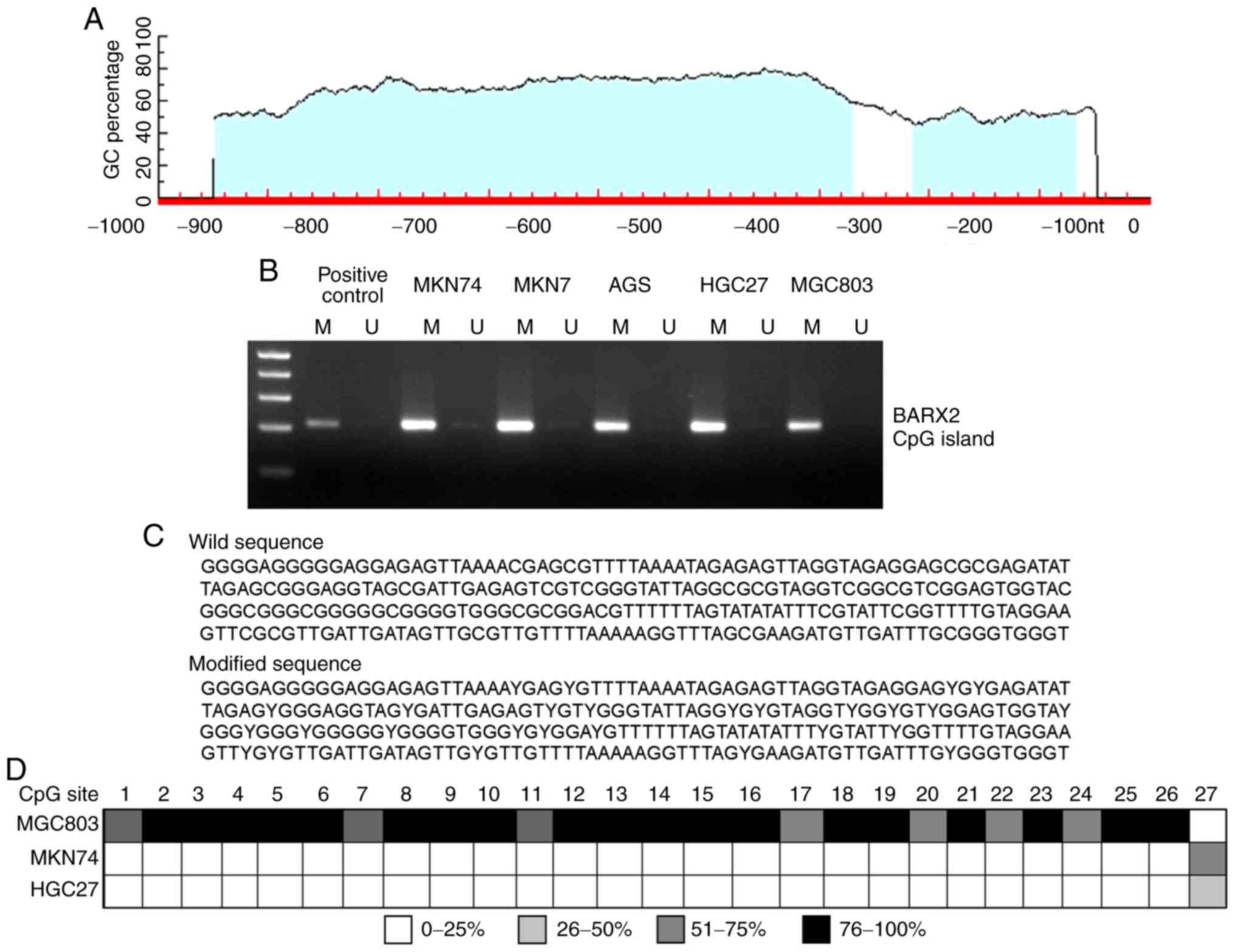 | Figure 4.DNA methylation of the BARX2
promoter was identified in GC cells. (A) Schematic diagram of
putative CpG islands within the BARX2 promoter identified as
determined using bioinformatics analysis. (B) Putative CpG islands
in five GC cell lines, as detected using methylation-specific
polymerase chain reaction (MSP). M, methylated; U, unmethylated.
(C) Methylation status of CpG sites within the functional promoter
fragment, as detected using bisulfite DNA sequencing PCR analysis
(BSP). The methylated CpG dinucleotides that could not be altered
by bisulfite modification are replaced by letter Y. (D) Methylation
status of the 27 CpG dinucleotides within the BARX2 promoter
fragment in three GC cell lines, including MGC803, MKN74, and
HGC27, as detected using BSP. DNA hypermethylation was mostly
observed in MGC803 cells. BARX2, BarH-like homeobox 2; GC,
gastric cancer. |
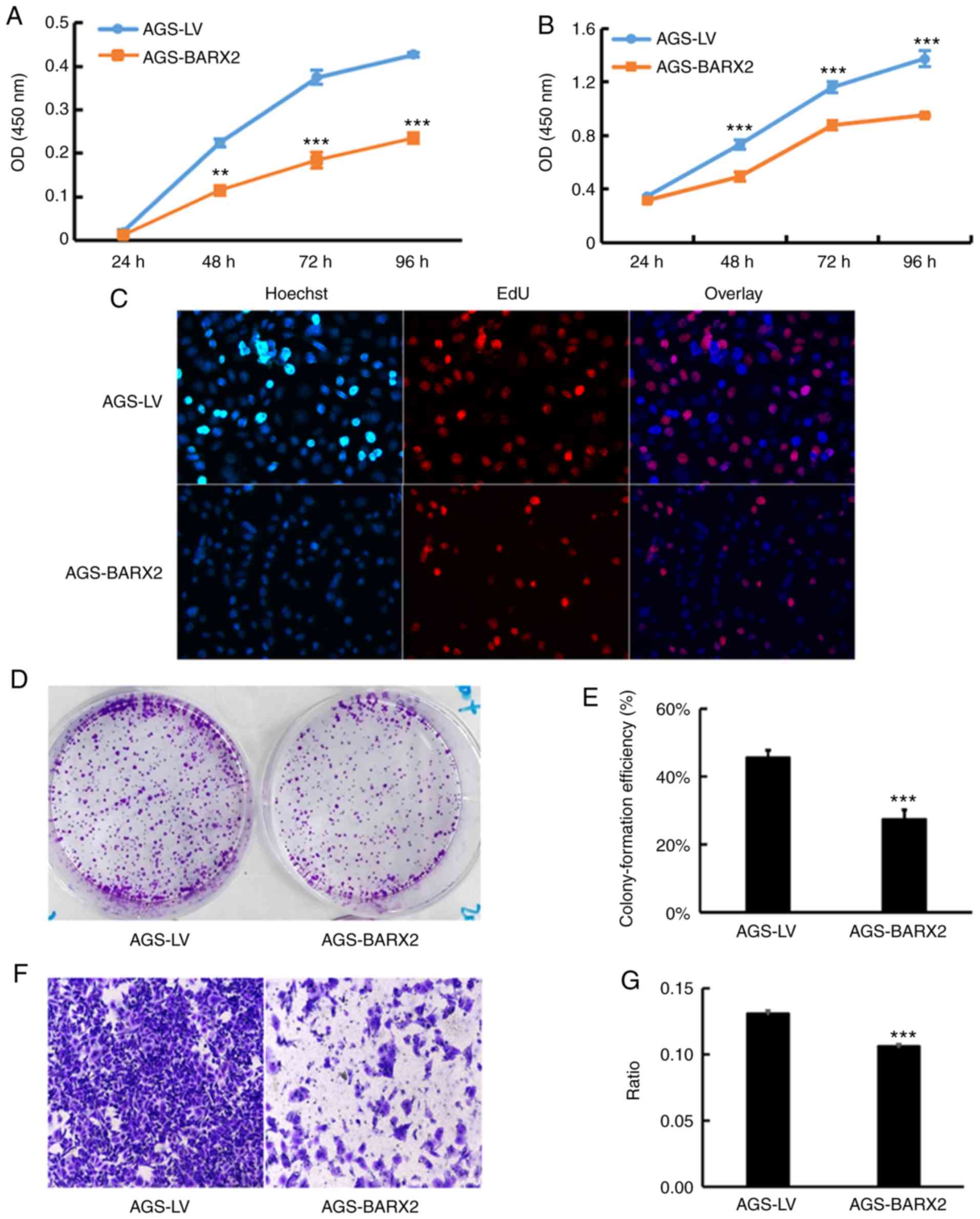
Overexpression of BARX2 is associated
with suppression of GC cell proliferation, colony formation, and
migration
Stable LV-BARX2 transfectants and LV-empty
controls were established in AGS cells to observe the effects on
the biological properties of GC cells in vitro. Three
BARX2-overexpressing single clones detected by qPCR were
selected and named AGS-BARX2.3, AGS-BARX2.5 and AGS-BARX2.6
(Fig. S1). A polyclonal
transfectant of AGS-BARX2 had higher expression of BARX2
compared to the AGS-LV cells (Fig.
S1). The CCK-8 assay showed that the proliferation of AGS cells
transfected with LV-BARX2 was significantly decreased at 48,
72, and 96 h with either 2.0×103 cells (P<0.01,
Fig. 5A) or 4.0×103
cells (P<0.001, Fig. 5B) seeded
per well. The EdU assay also showed that ectopic expression of
BARX2 suppressed cell proliferation (Fig. 5C). There were fewer proliferating
cells in the AGS-BARX2 cells when compared to that of the AGS-LV
cells. AGS-BARX2 transfectants lost colony-forming capacity by
nearly 40% compared to the AGS-LV controls cells; the percentages
of colonies formed by AGS-BARX2 and AGS-LV cells were 27.37±1.65
and 45.57±1.29%, respectively (P<0.001, Fig. 5D and E). The migration ability of
the AGS-BARX2 cells was significantly lower compared to the AGS-LV
cells (A460 value 0.106±0.001 vs. 0.131±0.001, P<0.001, Fig. 5F and G). These observations indicate
that overexpression of BARX2 in GC cells is associated with
suppression of cell proliferation, colony formation, and
migration.
Overexpression of BARX2 inhibits the
tumorigenesis of GC cells in vivo
Western blot analysis confirmed the overexpression
of BARX2 in the two stable transfectants (Fig. S1). Tumors formed in all 5 nude mice
inoculated with AGS-LV cells and in 4 mice inoculated with
AGS-BARX2 cells 4 weeks after inoculation (Fig. 6A and B). The tumors from mice
inoculated with AGS-BARX2 cells were significantly smaller compared
to tumors from mice inoculated with AGS-LV cells 4 weeks after
inoculation (48.17±14.93 vs. 100.83±5.29 mm3, P=0.011,
Fig. 6C). In mice inoculated with
AGS-BARX2 cells, the tumors expressed higher levels of BARX2
compared to mice inoculated with AGS-LV cells (P=0.032) (Fig. 6D and E). Additionally, the
expression of Ki-67, PCNA, MMP3, MMP7, and MMP9 was decreased,
while E-cadherin expression was increased in tumors formed by
AGS-BARX2 cells compared to expression in those formed by AGS-LV
cells (Fig. 6D and E).
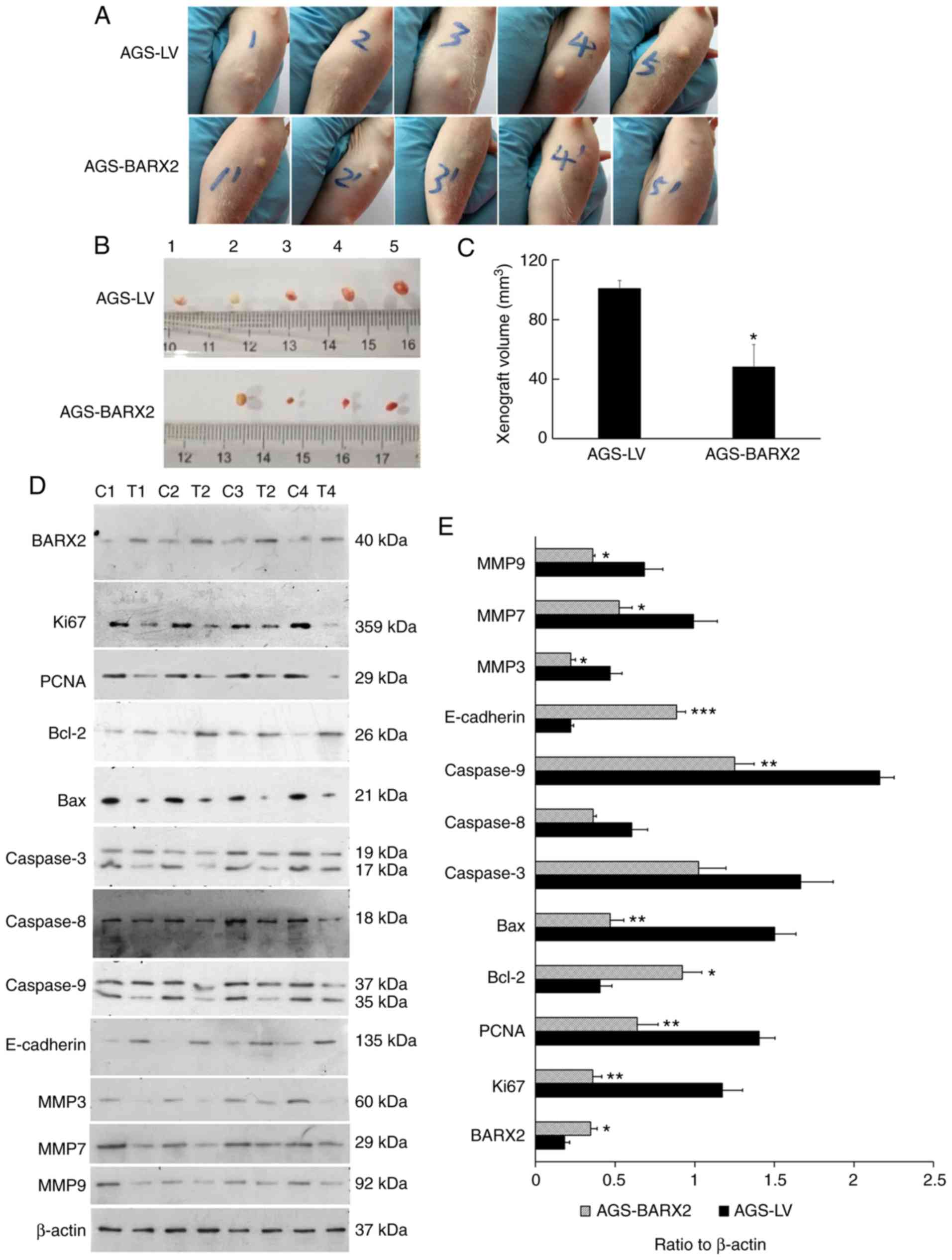 | Figure 6.Overexpression of BARX2
inhibits in vivo tumorigenesis. (A) BALB/c-nu/nu mice were
inoculated subcutaneously with AGS-BARX2 and AGS-LV mono-colony
transfectants (1.0×107) for 4 weeks. (B and C) Xenograft
volume was calculated after euthanization of the mice. The numbers
1–5 indicate the serial numbers of nude mice in both groups
inoculated with AGS-LV cells or AGS-BARX2 cells. Compared to the
nude mice inoculated with AGS-LV cells, the AGS-BARX2
cell-inoculated mice developed smaller transplanted tumors. (D and
E) All xenografts were collected for western blot analysis.
Upregulation of BARX2 suppressed expression of Ki-67, PCNA
and MMPs (MMP3, MMP7 and MMP9), while upregulation of BARX2
stimulated the expression of E-cadherin in the tumor tissues.
Upregulation of BARX2 also stimulated the expression of
Bcl-2 but suppressed the expression of Bax and caspase-9 in the
tumor tissues. *P<0.05, **P<0.01, ***P<0.001, compared
with AGS-LV. The Student's t-test was used for statistical
analysis. C, xenografts inoculated with AGS-LV cells; T, xenografts
inoculated with AGS-BARX2 cells. BARX2, BarH-like homeobox
2; PCNA, proliferating cell nuclear antigen; MMP,
metalloproteinase. |
Discussion
In the present study, BarH-like homeobox 2
(BARX2) expression was lower in gastric malignant tissues,
especially gastric adenocarcinomas, compared to that noted in the
normal gastric mucosa. The aberrant pattern of BARX2 expression was
accompanied by gradual aggravation of pathological stage and tissue
differentiation. We found a negative correlation between BARX2 and
DNA methyltransferase 1 (DNMT-1) (a key marker of DNA methylation)
expression and DNA methylation in the promoter region of
BARX2. Further in vivo experiments demonstrated that
BARX2 suppressed xenograft tumor formation and inhibited
tumor cell proliferation and invasion in nude mice. Overexpression
of BARX2 inhibited gastric cancer (GC) cell proliferation,
invasion, and migration in vitro and xenograft tumor
formation in nude mice. These findings indicate that BARX2
could be a novel tumor suppressor that may play an important role
in gastric carcinogenesis.
Human BARX2 shares 100% identity within the
homeodomain murine Barx2, which has been shown to be
strongly expressed in the crypts of the intestine tract and in the
outer cells of gut muscles in rats (30), but is downregulated in many
malignancies including GC, colorectal cancer (20), hepatocellular carcinoma (19), ovarian cancer (17), and non-small cell lung carcinoma
(21). In the present study, we
found reduced expression of BARX2 in gastric malignant tissues
using immunohistochemistry and in GC cell lines using RT-PCR.
Decreased BARX2 expression was also associated with pathological
TNM stage and cell differentiation in GC tissues. Consistently,
BARX2 was not upregulated in the five GC cell lines studied.
Our results support a previous study by Mi et al (16). However, we used GC tissue and
analyzed BARX2 expression using ImageJ, which has never been
previously measured. Interestingly, BARX2 mRNA levels were
decreased in AGS, MKN74 and MKN7 cells and even absent in HGC72 and
BGC803 cells. These findings indicate that BARX2 is
associated with cell differentiation and tumor prognosis as MKN74,
MKN7, AGS, and BGC803 cells are all differentiated GC cell lines,
whereas HGC27 is an undifferentiated gastric carcinoma.
The mechanisms by which BARX2 is
downregulated or lost remain to be elucidated. Mi et al
reported that overexpression of BARX2 reduced nuclear
β-catenin but increased cytoplasmic β-catenin, suggesting that
BARX2 functions as a tumor suppressor in GC cells (16). However, the previous study did not
further explore the molecular mechanism by which BARX2
expression is inhibited. Epigenetic modification, such as DNA
methylation, is known to play an important role in gene
transcription (10). DNA
methyltransferase (DNMT) is a key enzyme that regulates gene
expression during DNA methylation modification, a process that
occurs on some promoters of tumor-suppressor genes in GC (24). In the present study, we first found
an inverse correlation between BARX2 and DNMT-1 protein expression
in GC tissues. In addition, methylation-specific PCR analysis (MSP)
and bisulfite DNA sequencing PCR analysis (BSP) showed that DNA
hypermethylation was present in the putative conserved promoter
region of BARX2 in GC cells. This suggests that DNA
methylation modifications are involved in transcriptional
regulation of the BARX2 promoter, leading to silencing of
BARX2 in GC. However, how DNA methylation regulates
BARX2 transcription and if there are other epigenetic
modifications require further research.
In the present study, we found that the xenograft
volume was significantly decreased in the AGS-BARX2 group compared
to the AGS-LV control group. Both proliferation markers Ki-67 and
PCNA were significantly downregulated, which was consistent with
our in vitro experimental results. In addition, we found
that BARX2 overexpression was associated with changes in the
expression of several apoptotic proteins, including upregulation of
Bcl-2 and downregulation of Bax and caspase-9; however, there was
no change in the expression of caspase 3. It is well known that the
apoptosis process is complex, involving both exogenous and
endogenous pathways (31,32). Although Bcl-2 and Bax play important
roles in regulating apoptosis, they are not the decisive factors in
the occurrence and development of apoptosis, which is primarily
executed by caspase 3 (31,32). Thus, the findings in the present
study suggest that overexpression of BARX2 may regulate the
expression of Bax and Bcl-2, but does not necessarily induce
apoptosis. We speculate that BARX2 overexpression inhibits the
proliferation of GC cells and further induces a compensatory
response that alters expression of apoptotic proteins, but without
substantial induction of apoptosis. More extensive investigation is
required to further reveal the effect of BARX2 on proliferation and
apoptosis.
BARX2 regulates proliferation, migration, invasion,
and metastasis of tumor cells by altering cytoskeletal
rearrangement, cell-matrix interaction, and extracellular matrix
remodeling, all processes that are related to the Wnt/β-catenin
signaling pathway. E-cadherin is a downstream target gene of the
Wnt signaling pathway (33). BARX2
interacts with Wnt to regulate proliferation and differentiation of
embryonic myoblasts (34). Loss of
BARX2 is negatively associated with Ki-67 expression and
epithelial-mesenchymal transition (EMT) markers, including
E-cadherin and vimentin in HCC (19), similar to another Bar homeobox
family genes (35). Recent studies
have found that downregulation of BARX2 in GC is related to
β-catenin (16). Our present study
supports the hypothesis that BARX2 regulates proliferation,
β-catenin expression, and metastasis of GC through the Wnt
signaling pathway. Because BARX2 is essentially a transcription
factor, the downstream effectors of this protein need to be further
identified in future studies. Stevens and Meech (18) found decreased expression of BARX2
and its direct target, estrogen receptor-α (ESR), in breast cancer
cells. BARX2 upregulated the expression of MMP9 and
metalloproteinase inhibitor 4 (TIMP4), which was in response to
extracellular matrix (ECM) signals, and ultimately promoted
invasion of breast cancer cells (18). Mi et al reported that
overexpression of BARX2 was associated with reduced expression of
nuclear β-catenin, but increased expression of cytoplasmic
β-catenin, and that enhanced BARX2 expression reversed the
inhibitory effect of the Wnt signaling pathway in GC (16). Chen et al (21) showed that BARX2 decreased cell
proliferation, migration, and aerobic glycolysis by inhibiting the
Wnt/β-catenin signaling pathway in non-small cell lung
carcinoma.
In the present study, we also explored the effect of
BARX2 on the biological functions of GC cells and the
potential underlying molecular mechanisms. Our experiments showed
that BARX2 overexpression inhibited proliferation and
invasion in vitro and suppressed the growth of transplanted
tumors in vivo. Furthermore, BARX2 overexpression was
associated with altered expression of a series of molecular
proteins. Specifically, BARX2 overexpression downregulated Ki-67,
PCNA, MMP3, MMP7, MMP9 and upregulated E-cadherin in vivo.
Our findings suggest that the silencing of BARX2 promotes
gastric carcinogenesis by responding to ECM and Wnt signals and
regulating genes that are involved in ECM remodeling and GC
invasion. Our study supports some previous studies (16,17,19,21) in
that BARX2 suppressed proliferation, invasion, and migration of
several cancer cell lines, but our data are not consistent with a
previous study (18) that showed
that BARX2 promotes invasion of breast cancer cells by increasing
MMP9 and TIMP4 in the presence of ESR. The discrepancies between
these studies may be explained by the hypothesis that BARX2
bidirectionally regulates carcinogenesis in different organs and
tissues through various target factors, such as estrogen. Indeed,
this bidirectional regulation has been reported for other
homologous genes, such as PDX1 (7,9).
However, this hypothesis and the detailed mechanisms need to be
further explored.
Despite the major exciting findings presented here,
we were not able to fully explain the effects of the epigenetic
modification on BARX2 transcription and, more importantly,
to determine the interactive effects between BARX2 and the
Wnt/β-catenin signaling pathway. We are currently planning to
establish a GC transplantation model to explore the regulatory
effects of BARX2 on the Wnt/β-catenin pathway or vice versa,
using techniques such as RNA-Seq and ChIP-Seq (36–39).
In addition, our preliminary immunohistochemical experiments on the
expression pattern of BARX2 in colorectal cancer,
surprisingly, indicate that BARX2 protein expression is higher in
colorectal cancer compared to normal colon mucosa (data not shown).
This observation suggests that the function of BARX2 in
gastrointestinal tumors is complex and that BARX2 may play
different roles depending on the type of malignancy and tumor
environment and condition. More extensive investigation is required
to elucidate the roles of BARX2.
In conclusion, BARX2 expression is aberrantly
reduced in GC, which is associated with DNA methylation of its
promoter. BARX2 inhibits GC cell proliferation, migration,
and tumor formation. Our findings suggest that BARX2 could act as a
tumor suppressor in gastric carcinogenesis and, more importantly,
BARX2 may be a potential target for GC treatment.
Supplementary Material
Supporting Data
Acknowledgements
We thank Dr Harry H-X Xia for editing the
manuscript.
Funding
The present study was supported by the Natural
Science Foundation of Guangdong Province of China (2016A030313765),
the Medical Scientific Research Foundation of Guangdong Province of
China (A2017070 and A2017122), and the Project of Administration of
Traditional Chinese Medicine of Guangdong Province of China
(20191009).
Availability of data and materials
The datasets generated and analyzed during the
present study are available from the corresponding author on
reasonable request.
Authors' contributions
JM and ZSL conceived and designed the research
study, and had primary responsibility for the final content. LLX
and JM collected the data and conducted the research. XQY, LSX and
WHS analyzed and interpreted the data. JM wrote the initial
manuscript, and WHS revised the manuscript. SMZ and SL performed
additional cell experiments according to the reviewers'
suggestions. All authors read and approved the manuscript and agree
to be accountable for all aspects of the research in ensuring that
the accuracy or integrity of any part of the work are appropriately
investigated and resolved.
Ethics approval and consent to
participate
This study was approved by Guangdong General
Hospital Ethics Committee. All procedures performed in studies
involving animals were in accordance with the ethical standards of
the institution or practice at which the studies were
conducted.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2016. CA Cancer J Clin. 66:7–30. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
He S, Del Viso F, Chen CY, Ikmi A, Kroesen
AE and Gibson MC: An axial Hox code controls tissue segmentation
and body patterning in Nematostella vectensis. Science.
361:1377–1380. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Holland PW: Evolution of homeobox genes.
Wiley Interdiscip Rev Dev Biol. 2:31–45. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Stoffers DA, Heller RS, Miller CP and
Habener JF: Developmental expression of the homeodomain protein
IDX-1 in mice transgenic for an IDX-1 promoter/lacZ transcriptional
reporter. Endocrinology. 140:5374–5381. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ma J, Chen M, Wang J, Xia HH, Zhu S, Liang
Y, Gu Q, Qiao L, Dai Y, Zou B, et al: Pancreatic duodenal
homeobox-1 (PDX1) functions as a tumor suppressor in gastric
cancer. Carcinogenesis. 29:1327–1333. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Guz Y, Montminy MR, Stein R, Leonard J,
Gamer LW, Wright CV and Teitelman G: Expression of murine STF-1, a
putative insulin gene transcription factor, in beta cells of
pancreas, duodenal epithelium and pancreatic exocrine and endocrine
progenitors during ontogeny. Development. 121:11–18.
1995.PubMed/NCBI
|
|
9
|
Liu T, Gou SM, Wang CY, Wu HS, Xiong JX
and Zhou F: Pancreas duodenal homeobox-1 expression and
significance in pancreatic cancer. World J Gastroenterol.
13:2615–2618. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Herring BP, Kriegel AM and Hoggatt AM:
Identification of Barx2b, a serum response factor-associated
homeodomain protein. J Biol Chem. 276:14482–14489. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Naka T and Yokose S: Immunohistochemical
localization of barx2 in the developing fetal mouse submandibular
glands. Acta Histochem Cytochem. 42:47–53. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Meech R, Edelman DB, Jones FS and
Makarenkova HP: The homeobox transcription factor Barx2 regulates
chondrogenesis during limb development. Development. 132:2135–2146.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Olson LE, Zhang J, Taylor H, Rose DW and
Rosenfeld MG: Barx2 functions through distinct corepressor classes
to regulate hair follicle remodeling. Proc Natl Acad Sci USA.
102:3708–3713. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Jones FS, Kioussi C, Copertino DW,
Kallunki P, Holst BD and Edelman GM: Barx2, a new homeobox gene of
the Bar class, is expressed in neural and craniofacial structures
during development. Proc Natl Acad Sci USA. 94:2632–2637. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Stevens TA, Iacovoni JS, Edelman DB and
Meech R: Identification of novel binding elements and gene targets
for the homeodomain protein BARX2. J Biol Chem. 279:14520–14530.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Mi Y, Zhao S, Zhou C, Weng J, Li J, Wang
Z, Sun H, Tang H, Zhang X, Sun X, et al: Downregulation of homeobox
gene Barx2 increases gastric cancer proliferation and metastasis
and predicts poor patient outcomes. Oncotarget. 7:60593–60608.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Sellar GC, Li L, Watt KP, Nelkin BD,
Rabiasz GJ, Stronach EA, Miller EP, Porteous DJ, Smyth JF and Gabra
H: BARX2 induces cadherin 6 expression and is a functional
suppressor of ovarian cancer progression. Cancer Res. 61:6977–6981.
2001.PubMed/NCBI
|
|
18
|
Stevens TA and Meech R: BARX2 and estrogen
receptor-alpha (ESR1) coordinately regulate the production of
alternatively spliced ESR1 isoforms and control breast cancer cell
growth and invasion. Oncogene. 25:5426–5435. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhang Y, Zhang JX, Huang LL, He LJ, Liao
YJ, Lai YR, Deng HX, Tian XP, Kung HF, Xie D and Zhu SL: Low
expression of BARX2 in human primary hepatocellular carcinoma
correlates with metastasis and predicts poor prognosis. Hepatol
Res. 45:228–237. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Mi Y, Zhao S, Zhang W, Zhang D, Weng J,
Huang K, Sun H, Tang H, Zhang X, Sun X, et al: Down-regulation of
Barx2 predicts poor survival in colorectal cancer. Biochem Biophys
Res Commun. 478:67–73. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chen H, Zhang M, Zhang W, Li Y, Zhu J,
Zhang X, Zhao L, Zhu S and Chen B: Downregulation of BarH-like
homeobox 2 promotes cell proliferation, migration and aerobic
glycolysis through Wnt/β-catenin signaling, and predicts a poor
prognosis in non-small cell lung carcinoma. Thorac Cancer.
9:390–399. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wong NA, Britton MP, Choi GS, Stanton TK,
Bicknell DC, Wilding JL and Bodmer WF: Loss of CDX1 expression in
colorectal carcinoma: Promoter methylation, mutation, and loss of
heterozygosity analyses of 37 cell lines. Proc Natl Acad Sci USA.
101:574–579. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Guo M, House MG, Suzuki H, Ye Y, Brock MV,
Lu F, Liu Z, Rustgi AK and Herman JG: Epigenetic silencing of CDX2
is a feature of squamous esophageal cancer. Int J Cancer.
121:1219–1226. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ma J, Wang JD, Zhang WJ, Zou B, Chen WJ,
Lam CS, Chen MH, Pang R, Tan VP, Hung IF, et al: Promoter
hypermethylation and histone hypoacetylation contribute to
pancreatic-duodenal homeobox 1 silencing in gastric cancer.
Carcinogenesis. 31:1552–1560. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ushijima T: Detection and interpretation
of altered methylation patterns in cancer cells. Nat Rev Cancer.
5:223–231. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Esteller M: Cancer epigenomics: DNA
methylomes and histone-modification maps. Nat Rev Genet. 8:286–298.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2 (-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhao F, Xuan Z, Liu L and Zhang MQ: TRED:
A transcriptional regulatory element database and a platform for in
silico gene regulation studies. Nucleic Acids Res. 33
(Suppl):D103–D107. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Li LC and Dahiya R: MethPrimer: Designing
primers for methylation PCRs. Bioinformatics. 18:1427–1431. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Sander GR and Powell BC: Expression of the
homeobox gene barx2 in the gut. J Histochem Cytochem. 52:541–544.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Verbrugge I, Johnstone RW and Smyth MJ:
SnapShot: Extrinsic apoptosis pathways. Cell. 143:1192–1192.e2,
1192.e1-1192.e2. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Green DR and Llambi F: Cell death
signaling. Cold Spring Harb Perspect Biol. 7:72015. View Article : Google Scholar
|
|
33
|
Chiurillo MA: Role of the Wnt/β-catenin
pathway in gastric cancer: An in-depth literature review. World J
Exp Med. 5:84–102. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhuang L, Hulin JA, Gromova A, Tran Nguyen
TD, Yu RT, Liddle C, Downes M, Evans RM, Makarenkova HP and Meech
R: Barx2 and Pax7 have antagonistic functions in regulation of wnt
signaling and satellite cell differentiation. Stem Cells.
32:1661–1673. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wang G, Liu J, Cai Y, Chen J, Xie W, Kong
X, Huang W, Guo H, Zhao X, Lu Y, et al: Loss of Barx1 promotes
hepatocellular carcinoma metastasis through up-regulating MGAT5 and
MMP9 expression and indicates poor prognosis. Oncotarget.
8:71867–71880. 2017.PubMed/NCBI
|
|
36
|
Chen C, Lu Y, Liu J, Li L, Zhao N and Lin
B: Genome-wide ChIP-seq analysis of TCF4 binding regions in
colorectal cancer cells. Int J Clin Exp Med. 7:4253–4259.
2014.PubMed/NCBI
|
|
37
|
Debebe A, Medina V, Chen CY, Mahajan IM,
Jia C, Fu D, He L, Zeng N, Stiles BW, Chen CL, et al: Wnt/β-catenin
activation and macrophage induction during liver cancer development
following steatosis. Oncogene. 36:6020–6029. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Sarkar A, Huebner AJ, Sulahian R, Anselmo
A, Xu X, Flattery K, Desai N, Sebastian C, Yram MA, Arnold K, et
al: Sox2 suppresses gastric tumorigenesis in mice. Cell Rep.
16:1929–1941. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Laurent A, Calabrese M, Warnatz HJ, Yaspo
ML, Tkachuk V, Torres M, Blasi F and Penkov D: ChIP-Seq and RNA-Seq
analyses identify components of the Wnt and Fgf signaling pathways
as Prep1 target genes in mouse embryonic stem cells. PLoS One.
10:e01225182015. View Article : Google Scholar : PubMed/NCBI
|