Introduction
Acute myeloid leukaemia (AML) is one of the most
common malignant clonal diseases of the circulatory system; it
carries a high mortality rate (including a high treatment-related
mortality rate of approximately 1.57/100,000 individuals per year
in China) and a high recurrence rate (1). The prognosis of most AML patients is
poor. The cure rate of AML patients (except for that of acute
promyelocytic leukaemia) under 60 years of age is 35–40%, but this
rate is only 5–15% in patients over 60 years of age in China.
Strategies designed to improve prognosis and to improve these high
rates are the foci of investigation among researchers.
Histone modification is an epigenetic event related
to the prognosis of malignant haematologic diseases; histone
deacetylase was recently found to be associated with the prognosis
in lymphoma (2). A study reported
that loss of function and deletions in zeste homolog 2 (EZH2) [a
histone methyltransferase that is responsible for transcriptional
repression of target genes by trimethylation of lysine 27 on
histone H3 (H3K27me3)] are frequent in myeloid malignancies such as
myelodysplastic syndromes (MDS), atypical chronic myelogenous
leukaemia (CML), T cell acute lymphoblastic leukaemia (T-ALL) and
myelofibrosis; these mutations are generally associated with poorer
patient prognosis with reduced overall survival (OS) and event-free
survival (3–9). Mixed-lineage leukaemia
(MLL)-rearranged leukaemias have distinct clinical features and
poor prognosis. The majority of MLL translocations result in
oncogenic fusion proteins in which the native methyltransferase
domain is replaced with sequences that interact with disruptor of
telomeric silencing 1-like (DOT1L) directly or indirectly.
MLL-rearranged leukaemia depends on aberrant histone H3 lysine 79
(H3K79) methylation by DOT1L (10,11).
Another study showed that H3K9me3 deregulation in AML occurred
preferentially as a decrease in histone H3 lysine 9 trimethylation
(H3K9me3) levels at core promoter regions. When the H3K9me3
signature was combined with established clinical prognostic
markers, it outperformed prognosis predictions based on clinical
parameters alone (12). Taken
together, these studies suggest that histone methylation is
important for prognosis; nevertheless, there have been no studies
of the pathways or regulatory mechanisms of H3K9me3 in AML cell
lines.
Previous research from our team found that levels of
H3K9me3 could be affected by chidamide, a novel benzamide chemical
class of histone deacetylase inhibitor (HDACi), an agent that
alters expression levels of sirtuin 1 (SIRT1) (a histone
deacetylase), and enhances the cytotoxicity of drugs in AML cells
(13). Therefore, we performed the
present study to explore the pathway of H3K9me3 as well as the
regulatory mechanisms of SIRT1 on H3K9me3. This study may also
provide effective treatment strategies consequently improving the
prognosis of AML patients.
Materials and methods
Samples and databases
A total of 108 primary AML samples and 36 control
samples were selected from the Gene Expression Omnibus (GEO)
database [GEO accession no. GSE20452 (12), last update, March 22, 2012]. In
GSE20452, blasts from patients with AML were obtained at the time
of diagnosis. Two batches of experiments were performed and
analysed separately. One group of specimens contained AML samples
(n=38) and the other contained AML samples (n=70), CD34+
progenitor cells (n=21) and white blood cells (n=15) as controls.
We used GEO2R (https://www.ncbi.nlm.nih.gov/geo/geo2r/) to compare
two groups of samples so as to identify genes that are
differentially expressed across experimental conditions. GEO2R
terms with corrected P-values <0.05 were considered significant,
as were logFC of GEO2R terms >1.0.
Different peak analysis
Different peak analysis was based on the
fold-enrichment of peaks in various experiments. A peak was defined
as different when the odds ratio (OR) between two groups was more
than 2. Using the same method, genes associated with different
peaks were identified and subjected to Gene Ontology (GO)
enrichment analysis. We used KEGG Orthology-Based Annotation System
(KOBAS) 3.0 online (14) to test
the statistical enrichment of peak-related genes in Kyoto
Encyclopedia of Genes and Genomes (KEGG) (15–17)
pathways. GO terms with corrected P-values <0.05 were considered
significantly enriched by peak-related genes.
Analysis of protein interactions
The STRING (https://string-db.org/) database provides
protein-protein interaction (PPI) information, including direct
(physical) and indirect (functional) associations (18). Pathways from KEGG and the extended
network were constructed for m1A regulators and related
protein-coding genes signatures using Cytoscape 3.5.1.
Cell line
The AML cell line THP-1 was kindly donated by
Professor Ravi Bhatia (City of Hope National Medical Center,
Duarte, CA, USA). The THP-1 cell line was cultured in Iscoves
modified Dulbeccos medium or RPMI-1640 medium (Invitrogen; Thermo
Fisher Scientific, Inc.) with 10% foetal bovine serum (FBS) at 37°C
in a humidified incubator containing 5% CO2.
Drug treatment
Chidamide was kindly donated by Shenzhen Chipscreen
Co. AML cells were treated for 24 h. For THP-1 cells, the dose of
chidamide was 0.5 µM (20).
Chromatin immunoprecipitation
sequencing (ChIP-seq)
Chromatin immunoprecipitation (ChIP) experiments
were performed as previously described (13,21),
and ChIP-seq was based on the Illumina Technology Sequencing
platform (Illumina, Inc., USA). The single/paired-end method
(13,21) was used to complete the ChIP-seq
sequencing analysis of the THP-1 cell line. The antibody against
histone was H3K9me3 (cat. #4260, RRID: AB_10828006; Cell Signaling
Technology, Inc.).
Statistical analysis
The significances of differences were calculated
using the moderated t-statistic (only available when two groups of
samples were defined). P-values were adjusted for multiple testing.
Genes with the smallest P-values were considered the most reliable.
P<0.05 after adjustment was considered statistically
significant. Log2-fold change between two experimental conditions
(only available when two groups of samples were defined) was
carried out. Moderated F-statistic combined the t-statistics was
conducted for all pair-wise comparisons into an overall test of
significance for that gene (only available when more than two
groups of samples are defined).
Results
Differential expression of genes
across experimental conditions between AML and control samples
When samples were collected from the GEO database
(accession no. GSE20452), the differential expression of genes
between AML samples and control samples was analysed using GEO2R.
There were more than 2,000 genes that showed significant
differential expression. The definition of the value in log(FC) for
upregulation was >1.0, and the definition of the value in
log(FC) for downregulation was <-1.0. According to the
definition of upregulation or downregulation together with
P<0.05, there were 147 genes related to alterations in H3K9me3
showing downregulation and 170 genes related to the change in
H3K9me3 showing upregulation (data not shown).
Function and network analysis
The function of genes collected from GEO were
further analysed using geometric mean of semantic similarities in
biological processes (BPs), molecular functions (MFs), and cellular
components (CCs), that were assessed using the GOSemSim package
(22) by considering the GO
topological structure in a more precise and unbiased manner. In
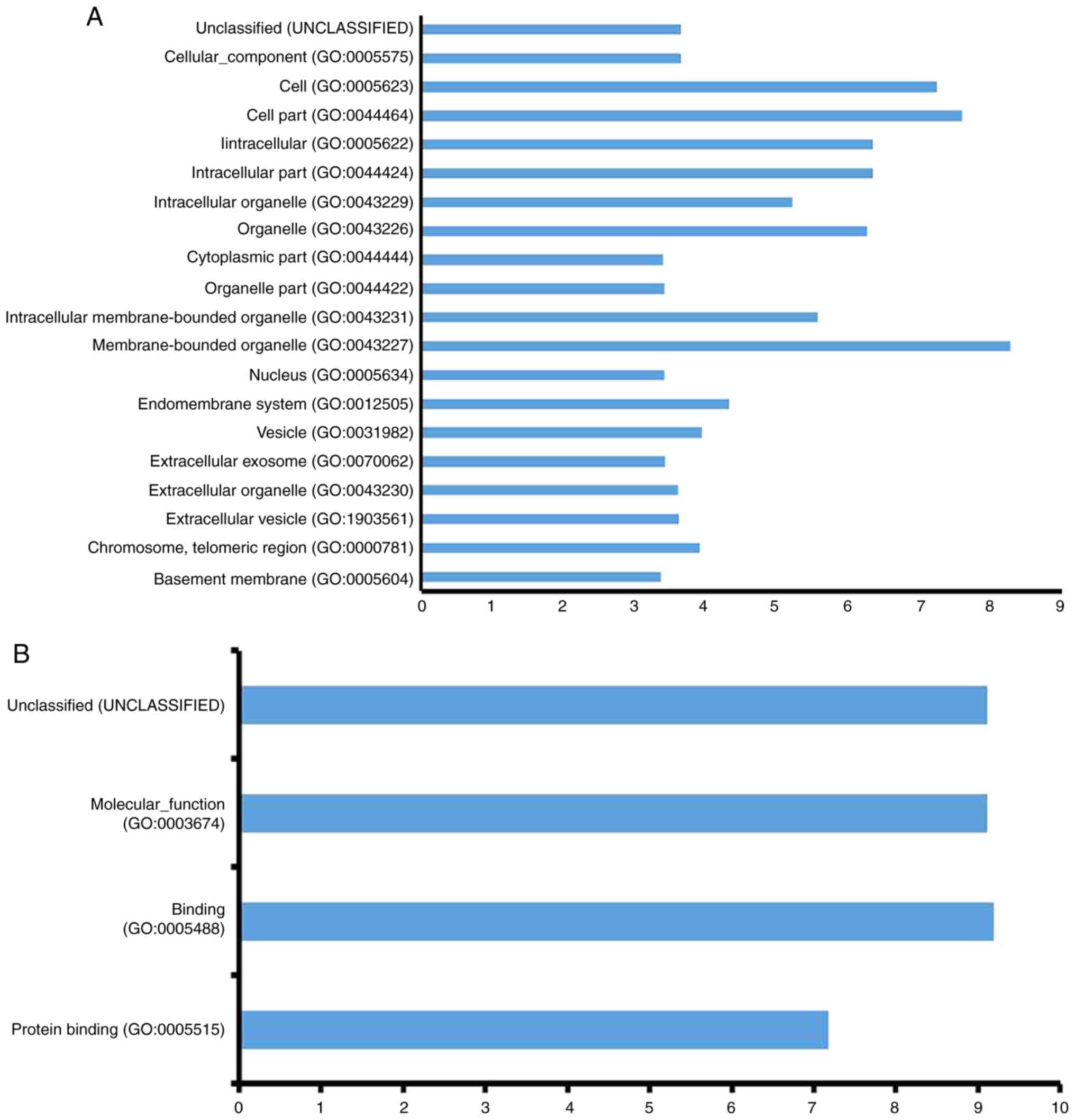
BPs, there were 20 processes that showed significant changes
(Fig. 1A). The main processes were
related to metabolism, including cofactor metabolic process,
cellular metabolic processes, and others. In MFs, there were only
three significantly altered processes: Protein binding, binding,
and molecular-function (Fig. 1B).
In CCs, there were 20 processes that showed significant
alterations, including intracellular and extracellular processes
(Fig. 1C). The relationship of GO
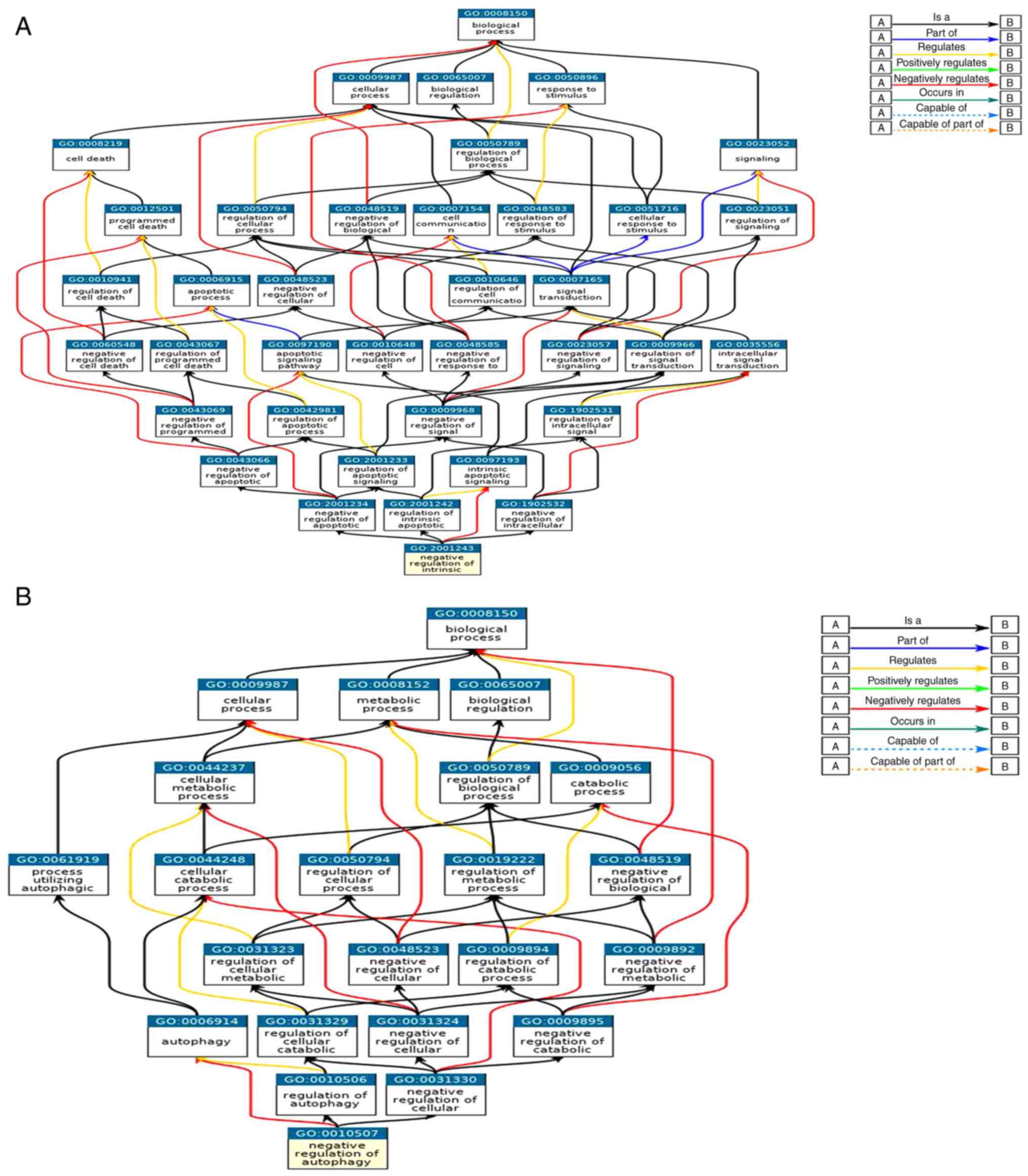
items in the regulation of apoptosis and autophagy are shown in
Fig. 2A and B. These GO items were
included in BPs, MFs, and CCs.
The related pathways were predicted using KOBAS 3.0
online. One of these pathways, the longevity pathway (Fig. 3), may related to the survival of
leukaemia cells, including the PI3K/AKT pathway, the AMPK pathway,
the FOXO pathway, the P53 pathway, and others. During analysis,
SIRT1 was also found to be related to the AMPK pathway and
autophagy pathway, consistent with findings from our previous study
(23).
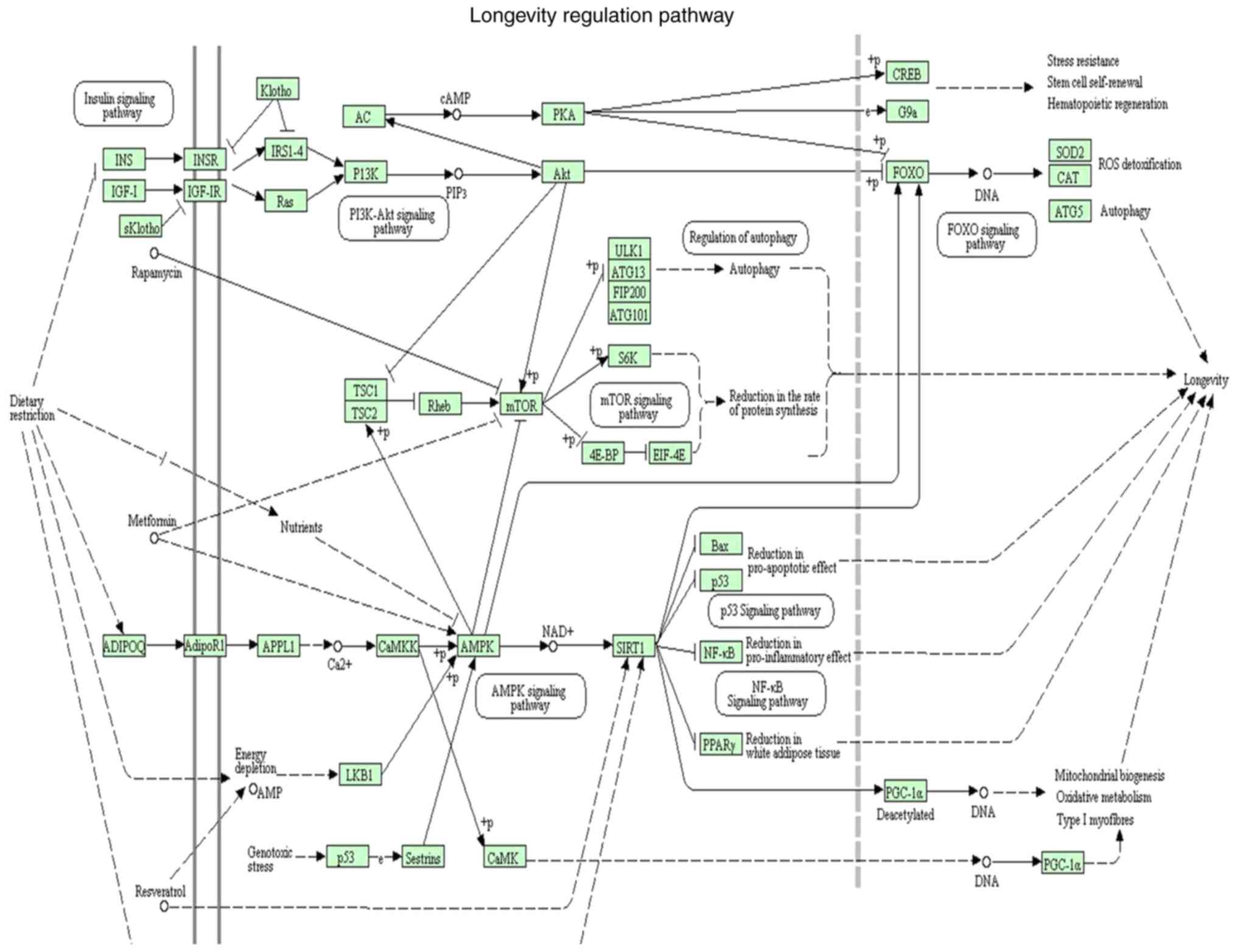 | Figure 3.Related pathways were predicted using
KEGG Orthology-Based Annotation System (KOBAS) 3.0 online. This
pathway was called the longevity pathway. The pathway contained
regular pathways related to the survival of leukaemia cells,
including the PI3K/AKT pathway, the AMPK pathway, the FOXO pathway,
the P53 pathway, mTOR pathway and NF-κB pathway. During analysis,
SIRT1 was found to be related to the AMPK pathway or the autophagy
pathway, consistent with our previous studies. PI3K,
phosphatidylinositol 3-kinase; AKT, serine/threonine kinase; AMPK,
protein kinase AMP-activated catalytic subunit α1; FOXO, forkhead
box; mTOR, mechanistic target of rapamycin kinase; SIRT1, sirtuin
1. |
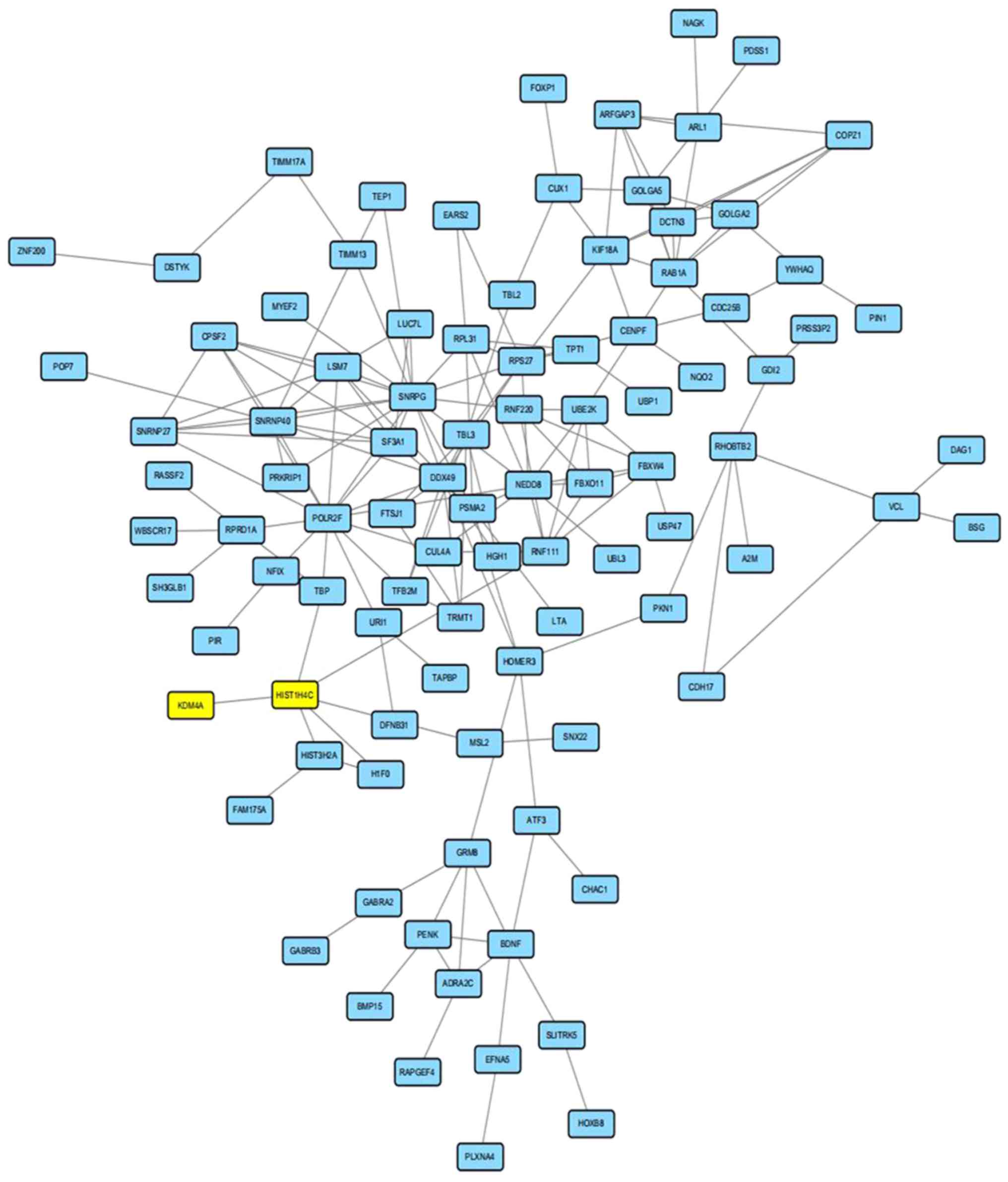
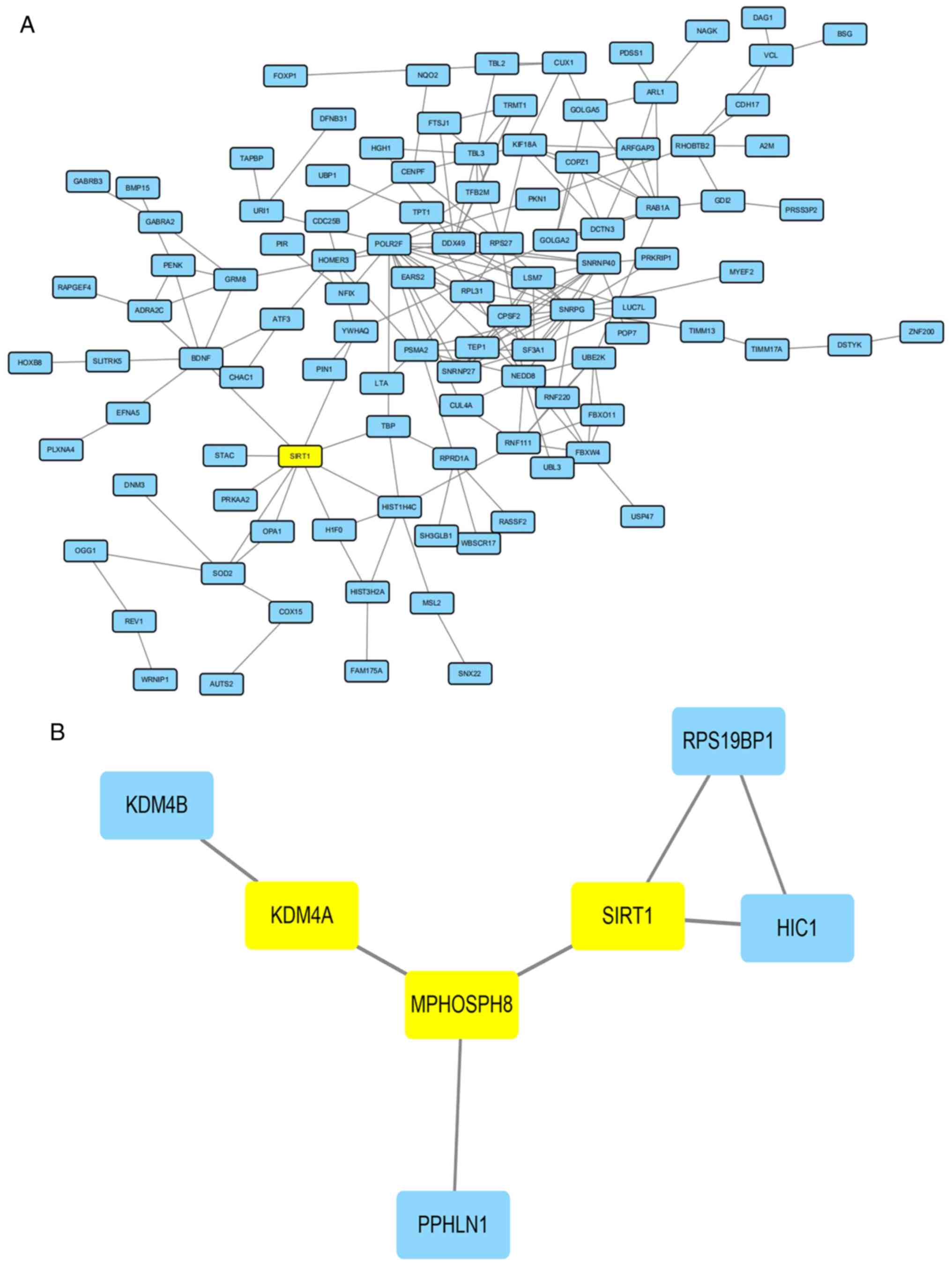
A total of 317 regulator genes, together with
lysine-specific demethylase 4A (KDM4A), a regulation protein for
the H3K9me3 network containing 220 nodes and 202 edges, were
obtained from the STRING online database and Cytoscape software.
The PPI network showed detailed protein interactions; however, the
PPI enrichment P-value was above 0.05, suggesting that the network
did not have significantly more interactions than expected.
Nevertheless, the modulation enzyme lysine-specific demethylase 4A
(KDM4A) for H3K9me3 could have an interaction with other proteins
in the network by HISTIH4C (Histone cluster 1, H4c, a modulation
enzyme for histone H4). There were also many protein interactions
that could help identify the mechanism of H3K9me3 modulation
(Fig. 4).
Distribution of peaks for gene
function and the analysis of GO using KEGG network in AML cells
treated with chidamide
The ChIP-seq of H3K9me3 was performed in AML cells
in which levels of SIRT1 were inhibited by chidamide; this was
conducted to identify genes that may be involved in the mechanisms
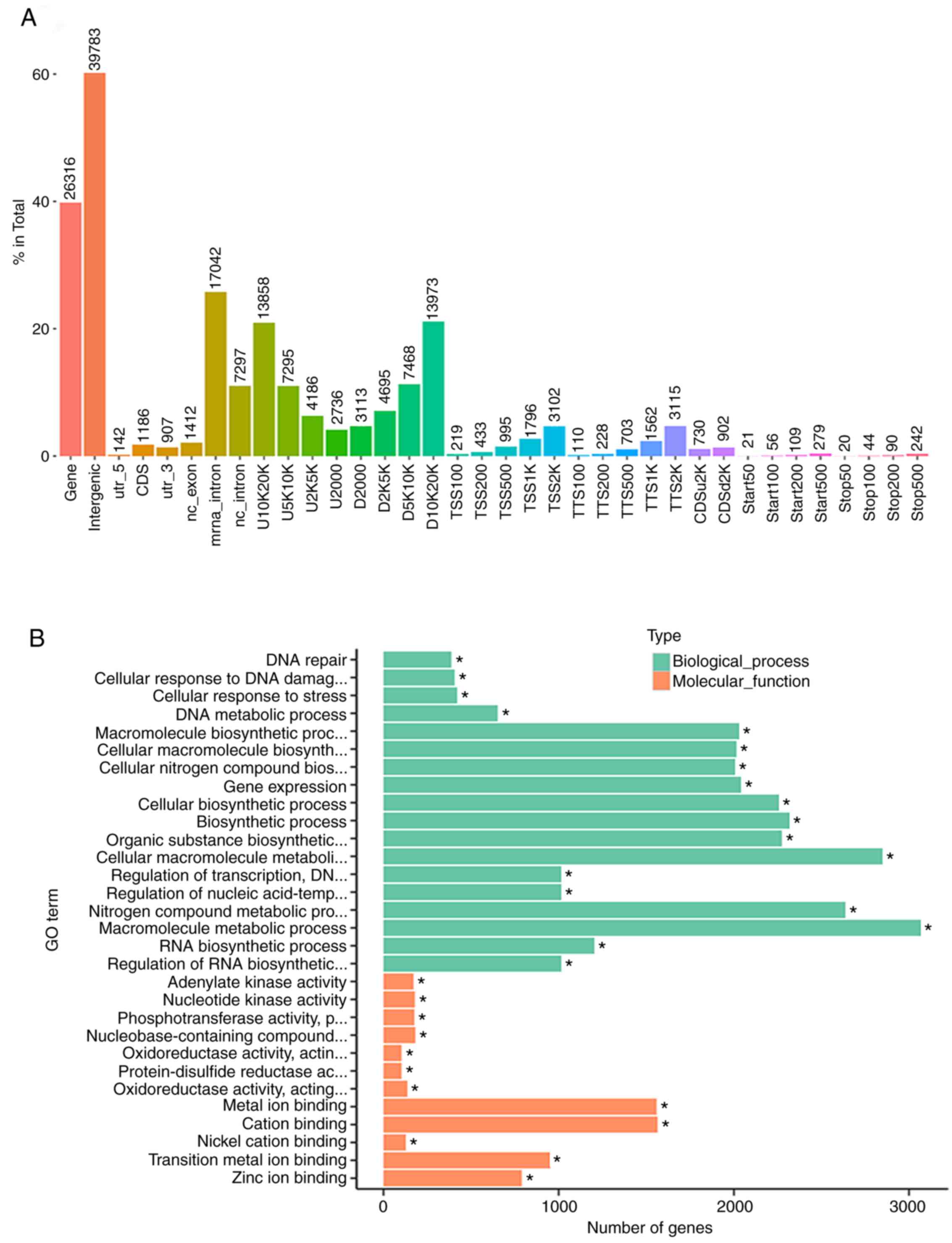
of cell death. The peaks almost clustered in the gene and
intergenic areas (nearly 99%); others clustered in areas such as
CDS and D10K20K (Fig. 5A).
GO analysis showed that the peak of processes that
had significant changes for AML cells treated by chidamide were BPs
and MFs. In the BPs, the most significant peaks involved metabolic
processes such as the macromolecule metabolic process and cellular
macromolecule metabolic processes. In the MFs, the most significant
peaks involved metal ion binding or cation binding. KEGG analysis
also revealed that the richest peak involved metabolic processes
(Fig. 5B).
Network analysis of GO enrichment in AML cells
treated by chidamide were in BPs. Units responsive to stress in
directed acyclic graph (DAG) had the most significant changes,
especially for the DNA repair and cells responsive to stress. In
the CCs, the ribosome unit showed the most significant change;
other units also showed significant changes, including cytoplasm
and intracellular ribonucleoprotein. In the MFs,
phosphotransferase, nucleotide kinase, and adenylate kinase showed
the most significant changes.
Our previous research (13) suggested that the level of H3K9me3
could be affected by a low dose of chidamide, thus we aimed to
ascertain in the present study whether the network of GO enrichment
in AML cells similar to AML patients could be affected by
chidamide. This result may be evidence for further research
concerning the use of chidamide in AML patients. An interesting
result was that the network analysis of GO enrichment in AML cells
treated with vhidamide were similar in AML patients. In GO0050789,
GO0050794, GO0050896, and GO0080090, all of which are related to
BPs, GO enrichment showed significant changes in AML patients
compared with control groups; the same also appeared in AML cells
treated with chidamide (Fig. 5E),
which was different from THP-1 cells without chidamide treatment
(Fig. 5C). In the CCs, GO
enrichment such as GO005623, GO005622, and GO0044444 showed
significant changes in AML patients as well as in the AML cells
treated with chidamide. However, in MFs, only GO005488 showed
significant changes in AML patients and in AML cells treated with
chidamide (Fig. 5F), which was
different from THP-1 cells without chidamide treatment (Fig. 5D).
Interactions between SIRT1 and H3K9me3
regulatory proteins that showed significant changes in AML
patients
The network of analysis for AML cells treated with
chidamide showed several results of GO enrichment that were the
same as the results of GO enrichment obtained from the patients in
GSE20452. Our previous study showed that SIRT1 (located on
chromosome 10) was in the domain of H3K9me3 on chromosome 10 that
was downregulated by chidamide, suggesting there may be interaction
between SIRT1 and the regulation of H3K9me3 (17). A total of 317 regulator genes
together with SIRT1, containing 219 nodes and 209 edges, were
obtained from the STRING online database and Cytoscape software.
The PPI network showed detailed protein interactions; however, the
PPI enrichment P-value was above 0.05, suggesting that the network
did not have significantly more interactions than expected.
Nevertheless, SIRT1 had a direct interaction with several proteins
in the network compared with KDM4A; however, there were also many
other protein interactions that were identified so as to uncover
the relationship between SIRT1 and regulatory genes acting on
H3K9me3 (Fig. 6A).
Interactions between SIRT1 and
KDM4A
Our previous study (13) showed that the location of SIRT1 on
chromosome 10 was in the domain of H3K9me3, suggesting there also
may exist interactions between SIRT1 and KDM4A. The gene SIRT1,
together with KDM4A, contained eight nodes and seven edges,
obtained from STRING online database and Cytoscape software. The
PPI network showed the detailed protein interactions, and the PPI
enrichment P-value was below 0.05, suggesting that the network had
significantly more interactions than expected. Nevertheless, there
were no direct interactions between SIRT1 and KDM4A. The protein
M-phase phosphoprotein 8 (MPHOSPH8) may be the bridge for SIRT1 and
KDM4A, and interactions between MPHOSPH8 and KDM4A need to be
demonstrated (Fig. 6B).
Discussion
Histone H3 lysine 9 trimethylation (H3K9me3) has a
role not only in malignancies but also in normal cellular
development. It acts as a repressor of lineage-inappropriate genes
and it maintains early cell integrity and genomic stability. In the
early 2000s (24), a number of
groups provided evidence of its important interactions with
evolutionarily conserved amino terminal chromodomain of
heterochromatin protein 1 (HP1), a hallmark of heterochromatin,
thereby recruiting it to specific chromatin loci. To date, roles
for H3K9me3 have been revealed in regulating apoptosis (25,26),
autophagy (27), development
(28,29), DNA repair (30–33),
splicing (34–38), self-renewal (39,40),
transcriptional elongation (36),
viral latency (41–43), imprinting (44), aging (45), and cell identity (46). In acute myeloid leukaemia (AML),
alterations in H3K9 methylation at promoter regions were found to
be associated with inactivation of tumour-suppressor genes and
blockade of differentiation and deregulated proliferation (47,48).
Given the reversible nature of H3K9 trimethylation, this represents
an attractive therapeutic target in AML.
Correct identification of the signalling pathways in
AML is the foundation of the discovery of therapeutic targets for
H3K9me3. In the present study, data from AML patients from the GEO
dataset were analysed using GEOR2. Compared with the control group
(CD34+ white cells), there were several genes related to
changes in H3K9me3 that were significantly differentially
upregulated or downregulated. These genes were related to
biological processes (BPs), molecular functions (MFs), and cellular
components (CCs). Some genes were related to the development of AML
and some took part in drug resistance in AML. These changes showed
that the change in histone methylation also may be an important
factor for the development of AML or drug resistance. In the
present study, various changes in THP-1 cells treated with
chidamide were the same as that in AML patients. The network
analysis of GO enrichment showed that alterations in H3k9me3 may
cause changes in cells in regards to apoptosis, autophag. These
databases became a potential foundation for our subsequent analysis
of chidamide on AML cells in vitro. THP-1 is a cell line
that is resistant to cytarabine, and chidamide could enhance the
cytotoxicity of cytarabine in THP-1 cells by modulating H3K9me3
(13). H3K9me3 was also reported to
be related to poor patient prognosis, thus it was thought that
H3K9me3 may be related to drug-resistance which is a main factor
for poor prognosis (12). There are
no studies concerning similar changes in gene expression in other
AML cell lines that have been carried out. Changes in the THP-1
cell line in this research may give us a direction for further
research in regards to other drug-resistant AML cell lines. The
functions of gene expression changes in other drug-resistant AML
cells will be carried out in next stage in future research.
The interaction analysis for related proteins of
H3K9me3 showed only a few proteins with interactions that had been
previously demonstrated in other studies (49–51).
The modulation protein lysine-specific demethylase 4A (KDM4A) for
H3K9me3 in this analysis was found only with an interaction with
HISTIH4C that was a modulation enzyme for another histone (H4).
These findings suggest that there are many interactions between
KDM4A and other proteins that warrant further investigation.
Meanwhile further research concerning the function of KDM4A on AML
cells will be carried out in subsequent research.
A recent review highlighted the emerging theme that
histone modifications can influence one another, such that one
modification recruits or activates chromatin-modifying complexes to
generate additional histone modifications (12). Our previous study also showed that
the drug chidamide (a histone deacetylation inhibitor (HDACi)
developed in China) was the first oral subtype-selective HDACi in
the world that could enhance the cytotoxicity of drugs in AML cells
(17). One of the potential
mechanisms might be related to an effect on H3K9me3. The data
suggest that there may be the same changes in vitro as those
in patients. The ChIP-seq test for THP-1 cells treated with
chidamide showed that significant peaks of GO analysis were BPs and
MFs, which lack the course of CCs compared with the results in
patients. However, the network analysis of GO enrichment in
vitro found that some changes were related to apoptosis and
autophagy. As previously suspected, some forecast changes in
vitro were the same as the forecast changes in patients,
including GO0050789, GO0050794, GO0050896, and GO0080090, related
to the BPs or the GO enrichment such as GO005623, GO005622, and
GO0044444 in the CCS. These results may be a theoretical basis for
further usage of chidamide in AML patients. We believe that these
results may suggest target treatments involving H3K9me3.
Changes in H3K9me3 in patients or in vitro
may cause autophagy. Our previous study regarding the potential
mechanisms of action of chidamide in enhancing the cytotoxicity of
drugs in AML cells suggested that chidamide inhibits autophagy by
inhibiting sirtuin 1 (SIRT1), a histone deacetylation enzyme
(17). SIRT1 may also have an
interaction with changes in H3k9me3 or the modulation enzyme KDM4A.
The pathway of KEGG analysis of H3k9me3 in patients showed several
pathways related to the survival of leukaemia cells. These included
some related to the survival of leukaemia cells, including the
PI3K/AKT pathway, the AMPK pathway, the FOXO pathway, the P53
pathway, and others. SIRT1 had an effect on the FOXO pathway that
was downstream of the PI3K/AKT pathway, related to drug resistance.
This result may be evidence to support the mechanism of chidamide
in reversing drug resistance in AML cells via the SIRT1
gene. This research is currently being conducted by our research
group. In the STRING database analysis, the interaction of SIRT1
with proteins related to a change in H3K9me3 were more evident even
more than KDM4A; however, the network did not have significantly
more interactions than expected, suggesting that further research
needs to be conducted. A compelling result for the interaction of
SIRT1 with KDM4A may be a relationship between SIRT1 and KDM4A,
although there was not a direct interaction, and there may be
involvement of a bridge called the MPHOSPH8 gene. Research
for further verification of the relationship between KDM4A and
MPHOSPH8 or SIRT1 must be carried out. This result also suggests a
link between histone deacetylation and methylation, as reported in
other studies about histone modifications influencing one another
(52–55).
In conclusion, bioinformatics analysis of H3k9me3 in
patients and in AML cells in vitro showed that H3K9me3 may
be a target for the treatment for AML; it also suggested that
chidamide may be a target drug for AML patients. Finally, our data
suggest several directions for the further study of drug resistance
in AML.
Acknowledgements
Not applicable.
Funding
This research study was supported by Zhejiang
Natural Science Foundation Program (LY19H080002), and the Breeding
Program of the Second Affiliated Hospital and Yumiao Childrens
Hospital of Wenzhou Medical University (Wenzhou, Zhejiang,
China).
Availability of data and materials
We declared that materials described in the
manuscript, including all relevant raw data, will be freely
available to any scientist wishing to use them for non-commercial
purposes, without breaching participant confidentiality.
Authors contributions
AD, WY and WG performed the analysis of the genes
for the GEO database peak analysis, as well as analysis of protein
interactions. HH, ZH and XZ performed the ChIP-seq analysis. RY
conducted the statistical analysis. WG wrote the paper. All authors
read and approved the manuscript and agree to be accountable for
all aspects of the research in ensuring that the accuracy or
integrity of any part of the work are appropriately investigated
and resolved.
Ethics approval and consent to
participate
This article does not contain any studies with human
participants or animals performed by any of the authors.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Goyama S and Kitamura T: Epigenetics in
normal and malignant hematopoiesis: An overview and update 2017.
Cancer Sci. 108:553–562. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Cao R, Wang L, Wang H, Xia L,
Erdjument-Bromage H, Tempst P, Jones RS and Zhang Y: Role of
histone H3 lysine 27 methylation in Polycomb-group silencing.
Science. 298:1039–1043. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Morin RD, Johnson NA, Severson TM, Mungall
AJ, An J, Goya R, Paul JE, Boyle M, Woolcock BW, Kuchenbauer F, et
al: Somatic mutations altering EZH2 (Tyr641) in follicular and
diffuse large B-cell lymphomas of germinal-center origin. Nat
Genet. 42:181–185. 2010. View
Article : Google Scholar : PubMed/NCBI
|
|
5
|
Abd Al Kader L, Oka T, Takata K, Sun X,
Sato H, Murakami I, Toji T, Manabe A, Kimura H and Yoshino T: In
aggressive variants of non-Hodgkin lymphomas, Ezh2 is strongly
expressed and polycomb repressive complex PRC1.4 dominates over
PRC1.2. Virchows Arch. 463:697–711. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Asangani IA, Ateeq B, Cao Q, Dodson L,
Pandhi M, Kunju LP, Mehra R, Lonigro RJ, Siddiqui J, Palanisamy N,
et al: Characterization of the EZH2-MMSET histone methyltransferase
regulatory axis in cancer. Mol Cell. 49:80–93. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ernst T, Chase AJ, Score J, Hidalgo-Curtis
CE, Bryant C, Jones AV, Waghorn K, Zoi K, Ross FM, Reiter A, et al:
Inactivating mutations of the histone methyltransferase gene EZH2
in myeloid disorders. Nat Genet. 42:722–726. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Nikoloski G, Langemeijer SM, Kuiper RP,
Knops R, Massop M, Tönnissen ER, van der Heijden A, Scheele TN,
Vandenberghe P, de Witte T, et al: Somatic mutations of the histone
methyltransferase gene EZH2 in myelodysplastic syndromes. Nat
Genet. 42:665–667. 2010. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ntziachristos P, Tsirigos A, Van
Vlierberghe P, Nedjic J, Trimarchi T, Flaherty MS, Ferres-Marco D,
da Ros V, Tang Z, Siegle J, et al: Genetic inactivation of the
polycomb repressive complex 2 in T cell acute lymphoblastic
leukemia. Nat Med. 18:298–301. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
10
|
Daigle SR, Olhava EJ, Therkelsen CA,
Basavapathruni A, Jin L, Boriack-Sjodin PA, Allain CJ, Klaus CR,
Raimondi A, Scott MP, et al: Potent inhibition of DOT1L as
treatment of MLL-fusion leukemia. Blood. 122:1017–1025. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Muntean AG and Hess JL: The pathogenesis
of mixed-lineage leukemia. Annu Rev Pathol. 7:283–301. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Müller-Tidow C, Klein HU, Hascher A, Isken
F, Tickenbrock L, Thoennissen N, Agrawal-Singh S, Tschanter P,
Disselhoff C, Wang Y, et al Study Alliance Leukemia, : Profiling of
histone H3 lysine 9 trimethylation levels predicts transcription
factor activity and survival in acute myeloid leukemia. Blood.
116:3564–3571. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Huang H, Wenbing Y, Dong A, He Z, Yao R
and Guo W: Chidamide enhances the cytotoxicity of cytarabine and
sorafenib in acute myeloid leukemia cells by modulating H3K9me3 and
autophagy levels. Front Oncol. 9:12762019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Xie C, Mao X, Huang J, Ding Y, Wu J, Dong
S, Kong L, Gao G, Li CY and Wei L: KOBAS 3.0: A web server for
annotation and identification of enriched pathways and diseases.
Nucleic Acids Res. 39:W316–W322. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kanehisa M and Goto S: KEGG: Kyoto
encyclopedia of genes and genomes. Nucleic Acids Res. 28:27–30.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kanehisa M, Sato Y, Furumichi M, Morishima
K and Tanabe M: New approach for understanding genome variations in
KEGG. Nucleic Acids Res. 47:D590–D595. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kanehisa M: Toward understanding the
origin and evolution of cellular organisms. Protein Sci.
28:1947–1951. 2019. View
Article : Google Scholar : PubMed/NCBI
|
|
18
|
Szklarczyk D, Franceschini A, Wyder S,
Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos
A, Tsafou KP, et al: STRING v10: Protein-protein interaction
networks, integrated over the tree of life. Nucleic Acids Res.
43:D447–D452. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Li X, Yan X, Guo W, Huang X, Huang J, Yu
M, Ma Z, Xu Y, Huang S, Li C, et al: Chidamide in FLT3-ITD positive
acute myeloid leukemia and the synergistic effect in combination
with cytarabine. Biomed Pharmacother. 90:699–704. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kleine-Kohlbrecher D, Christensen J,
Vandamme J, Abarrategui I, Bak M, Tommerup N, Shi X, Gozani O,
Rappsilber J, Salcini AE and Helin K: A functional link between the
histone demethylase PHF8 and the transcription factor ZNF711 in
X-linked mental retardation. Mol Cell. 38:165–178. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yu G, Li F, Qin Y, Bo X, Wu Y and Wang S:
GOSemSim: an R package for measuring semantic similarity among GO
terms and gene products. Bioinformatics. 26:976–978. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Guo W, Jin J, Pan J, Yao R, Li X, Huang X,
Ma Z, Huang S, Yan X, Jin J and Dong A: The change of nuclear LC3
distribution in acute myeloid leukemia cells. Exp Cell Res.
369:69–79. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Mandrioli M and Borsatti F: Analysis of
heterochromatic epigenetic markers in the holocentric chromosomes
of the aphid Acyrthosiphon pisum. Chromosome Res.
15:1015–1022. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Olcina MM, Leszczynska KB, Senra JM, Isa
NF, Harada H and Hammond EM: H3K9me3 facilitates hypoxia-induced
p53-dependent apoptosis through repression of APAK. Oncogene.
35:793–799. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lu C, Yang D, Sabbatini ME, Colby AH,
Grinstaff MW, Oberlies NH, Pearce C and Liu K: Contrasting roles of
H3K4me3 and H3K9me3 in regulation of apoptosis and gemcitabine
resistance in human pancreatic cancer cells. BMC Cancer.
18:1492018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Biga PR, Latimer MN, Froehlich JM,
Gabillard JC and Seiliez I: Distribution of H3K27me3, H3K9me3, and
H3K4me3 along autophagy-related genes highly expressed in starved
zebrafish myotubes. Biol Open. 6:1720–1725. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Fujiwara K, Fujita Y, Kasai A, Onaka Y,
Hashimoto H, Okada H and Yamashita T: Deletion of JMJD2B in neurons
leads to defective spine maturation, hyperactive behavior and
memory deficits in mouse. Transl Psychiatry. 6:e7662016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Magaraki A, van der Heijden G,
Sleddens-Linkels E, Magarakis L, van Cappellen WA, Peters AHFM,
Gribnau J, Baarends WM and Eijpe M: Silencing markers are retained
on pericentric heterochromatin during murine primordial germ cell
development. Epigenetics Chromatin. 10:112017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Sun Y, Jiang X, Xu Y, Ayrapetov MK, Moreau
LA, Whetstine JR and Price BD: Histone H3 methylation links DNA
damage detection to activation of the tumour suppressor Tip60. Nat
Cell Biol. 11:1376–1382. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ayrapetov MK, Gursoy-Yuzugullu O, Xu C, Xu
Y and Price BD: DNA double-strand breaks promote methylation of
histone H3 on lysine 9 and transient formation of repressive
chromatin. Proc Natl Acad Sci USA. 111:9169–9174. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Khoury-Haddad H, Nadar-Ponniah PT, Awwad S
and Ayoub N: The emerging role of lysine demethylases in DNA damage
response: Dissecting the recruitment mode of KDM4D/JMJD2D to DNA
damage sites. Cell Cycle. 14:950–958. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wu R, Wang Z, Zhang H, Gan H and Zhang Z:
H3K9me3 demethylase Kdm4d facilitates the formation of
pre-initiative complex and regulates DNA replication. Nucleic Acids
Res. 45:169–180. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Hausmann M, Wagner E, Lee JH, Schrock G,
Schaufler W, Krufczik M, Papenfuß F, Port M, Bestvater F and
Scherthan H: Super-resolution localization microscopy of
radiation-induced histone H2AX-phosphorylation in relation to
H3K9-trimethylation in HeLa cells. Nanoscale. 10:4320–4331. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Saint-André V, Batsché E, Rachez C and
Muchardt C: Histone H3 lysine 9 trimethylation and HP1γ favor
inclusion of alternative exons. Nat Struct Mol Biol. 18:337–344.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Bieberstein NI, Kozáková E, Huranová M,
Thakur PK, Krchňáková Z, Krausová M, Carrillo Oesterreich F and
Staněk D: TALE-directed local modulation of H3K9 methylation shapes
exon recognition. Sci Rep. 6:299612016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Barrand S, Andersen IS and Collas P:
Promoter-exon relationship of H3 lysine 9, 27, 30 and 79
methylation on pluripotency-associated genes. Biochem Biophys Res
Commun. 401:611–617. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Pedersen MT, Kooistra SM, Radzisheuskaya
A, Laugesen A, Johansen JV, Hayward DG, Nilsson J, Agger K and
Helin K: Continual removal of H3K9 promoter methylation by Jmjd2
demethylases is vital for ESC self-renewal and early development.
EMBO J. 35:1550–1564. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Loh YH, Zhang W, Chen X, George J and Ng
HH: Jmjd1a and Jmjd2c histone H3 Lys 9 demethylases regulate
self-renewal in embryonic stem cells. Genes Dev. 21:2545–2557.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Vakoc CR, Mandat SA, Olenchock BA and
Blobel GA: Histone H3 lysine 9 methylation and HP1gamma are
associated with transcription elongation through mammalian
chromatin. Mol Cell. 19:381–391. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Maksakova IA, Goyal P, Bullwinkel J, Brown
JP, Bilenky M, Mager DL, Singh PB and Lorincz MC: H3K9me3-binding
proteins are dispensable for SETDB1/H3K9me3-dependent retroviral
silencing. Epigenetics Chromatin. 4:122011. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Imai K, Kamio N, Cueno ME, Saito Y, Inoue
H, Saito I and Ochiai K: Role of the histone H3 lysine 9
methyltransferase Suv39 h1 in maintaining Epsteinn-Barr virus
latency in B95-8 cells. FEBS J. 281:2148–2158. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Lang F, Li X, Vladimirova O, Hu B, Chen G,
Xiao Y, Singh V, Lu D, Li L, Han H, et al: CTCF interacts with the
lytic HSV-1 genome to promote viral transcription. Sci Rep.
7:398612017. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Fukuda A, Tomikawa J, Miura T, Hata K,
Nakabayashi K, Eggan K, Akutsu H and Umezawa A: The role of
maternal-specific H3K9me3 modification in establishing imprinted
X-chromosome inactivation and embryogenesis in mice. Nat Commun.
5:54642014. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Mendelsohn AR and Larrick JW: Stem cell
depletion by global disorganization of the H3K9me3 epigenetic
marker in aging. Rejuvenation Res. 18:371–375. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Koide S, Oshima M, Takubo K, Yamazaki S,
Nitta E, Saraya A, Aoyama K, Kato Y, Miyagi S, Nakajima-Takagi Y,
et al: Setdb1 maintains hematopoietic stem and progenitor cells by
restricting the ectopic activation of nonhematopoietic genes.
Blood. 128:638–649. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
McDonald OG, Li X, Saunders T,
Tryggvadottir R, Mentch SJ, Warmoes MO, Word AE, Carrer A, Salz TH,
Natsume S, et al: Epigenomic reprogramming during pancreatic cancer
progression links anabolic glucose metabolism to distant
metastasis. Nat Genet. 49:367–376. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Chung YR, Schatoff E and Abdel-Wahab O:
Epigenetic alterations in hematopoietic malignancies. Int J
Hematol. 96:413–427. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Lehmann U, Brakensiek K and Kreipe H: Role
of epigenetic changes in hematological malignancies. Ann Hematol.
83:137–152. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Becker JS, Nicetto D and Zaret KS:
H3K9me3-dependent heterochromatin: Barrier to cell fate changes.
Trends Genet. 32:29–41. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Wang C, Liu X, Gao Y, Yang L, Li C, Liu W,
Chen C, Kou X, Zhao Y, Chen J, et al: Reprogramming of
H3K9me3-dependent heterochromatin during mammalian embryo
development. Nat Cell Biol. 20:620–631. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Nicetto D and Zaret KS: Role of H3K9me3
heterochromatin in cell identity establishment and maintenance.
Curr Opin Genet Dev. 55:1–10. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Adamkova K, Yi YJ, Petr J, Zalmanova T,
Hoskova K, Jelinkova P, Moravec J, Kralickova M, Sutovsky M,
Sutovsky P and Nevoral J: SIRT1-dependent modulation of methylation
and acetylation of histone H3 on lysine 9 (H3K9) in the zygotic
pronuclei improves porcine embryo development. J Anim Sci
Biotechnol. 8:832017. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Lee MG, Norman J, Shilatifard A and
Shiekhattar R: Physical and functional association of a trimethyl
H3K4 demethylase and Ring6a/MBLR, a polycomb-like protein. Cell.
128:877–887. 2017. View Article : Google Scholar
|
|
55
|
Li L and Wang Y: Cross-talk between the
H3K36me3 and H4K16ac histone epigenetic marks in DNA double-strand
break repair. J Biol Chem. 292:11951–11959. 2017. View Article : Google Scholar : PubMed/NCBI
|