Introduction
Osteosarcoma (OS) is a common primary malignant
tumor, that accounts for 1 out of 5 of all diagnosed primary
malignant tumors. Children and adolescents are high-risk OS
populations (1). OS originates from
primitive mesenchymal bone cells and often occurs in long bones,
such as the proximal humerus and distal femur (2). For the past 20 years, epidemiological
studies report that the global annual incidence of OS is ~3
million, with a male to female incidence ratio of 5:1, and OS is
notorious for its high degree of malignancy, rapid proliferation,
high recurrence rate and low long-term survival rates (3). Current treatments have evolved from
traditional amputation surgery to local radiotherapy and adjuvant
chemotherapy (4). Although the cure
rate of OS has increased, due to its high degree of malignancy and
metastasis, recurrence and low 5-year survival rates (≤20%) persist
(5,6).
Clinically, OS patients show multiple metastases in
the body. Although treatment with neoadjuvant chemotherapy combined
with surgery improves the survival rates of OS patients, hearing
impairment, liver and kidney damage, and hematopoietic dysfunction
frequently occur (7). Cadmium is a
common industrial heavy metal that is widely used in manufacturing.
Cadmium has toxic effects on various organs and systems, causing
bone damage, osteoporosis and osteomalacia (8,9). Since
Coogan and colleagues reported the antitumor effects of cadmium, it
has become the focus of intensive research efforts (10). Studies have shown that cadmium
inhibits cell proliferation and induces DNA damage and apoptosis
(11). Toxicology studies show that
bone is its major site of accumulation in the human body and
represents the main target organ (12).
In the present study, we explored the potential of
cadmium chloride to reduce the proliferation, migration and
apoptosis of OS cells. Using gene chip technology and in
vivo and in vitro assessments, cadmium chloride was
found to reduce the growth of OS cells and enhance their
sensitivity to cisplatin (DDP) through the regulation of forkhead
box protein M1 (FOXM1). Cadmium chloride was also found to enhance
cisplatin sensitivity in OS nude-mouse models.
Materials and methods
Reagents and antibodies
Cadmium chloride (CdCl2), Cisplatin (DDP), and
2,7-dichlorofluorescin diacetate were obtained from
Sigma-Aldrich/Merck KGaA. Dulbecco's modified Eagle's medium (DMEM)
with high glucose, penicillin, streptomycin and fetal bovine serum
(FBS) were obtained from Thermo Fisher Scientific, Inc. The MTT
Cell Proliferation and Cytotoxicity Assay Kit was purchased from
Beyotime Institute of Biotechnology. The following antibodies were
used: Cleaved caspase-3 antibody [dilution, 1:1,000 for Western
blot analysis (WB); cat. #9664; Cell Signaling Technology, Inc. USA
(CST)], Bcl-2 (dilution 1:1,000 for WB; cat. #15071; CST), BAX
(dilution 1:1,000 for WB; cat. #5023; CST), MMP-2 (dilution 1:1,000
for WB; cat. #4022; CST), MMP-9 (dilution 1:1,000 for WB; cat.
#3852; CST), E-cadherin (dilution 1:2,000 for WB; cat. #3195; CST),
FOXM1 (dilution 1:80 for IHC, 1:1,000 for WB; cat. no. ab232649;
Abcam) and glyceraldehyde-3-phosphate dehydrogenase (GAPDH)
(dilution 1:1,000 for WB).
Tumor cell lines and culture
The human embryo immortalized osteoblast cell line
Hfob1.19 and OS cell lines MG63, U2OS, 143B and SaoS2 were
purchased from Yong Jin Biotech and cultured in DMEM containing 10%
FBS, penicillin 100 U/ml and streptomycin 100 pg/ml, in 5%
CO2 at 37°C. Cells were assessed when in the logarithmic
growth phase.
Projection electron microscopy
Cells in the logarithmic growth phase were plated
into 6-well plates at a density of 5×105 cells/well.
After incubation for 12–24 h, the cells were treated with 20 µM
cadmium chloride (CdCl2) for 24 h and fixed in 2.5%
glutaraldehyde solution overnight at 4°C. The cells were washed in
PBS, fixed in 1% citric acid for 1–2 h, and dehydrated with
ethanol. Cells were mounted using embedding agent, and the
ultrastructural changes of the cells were observed under an
electron microscope (magnification, ×1,000 and ×5,000).
Drug toxicity
Cells (1×105 cells/ml) were seeded in
96-well plates at 200 µl per well. After the cells had grown to a
confluent state, the culture medium was discarded and 200 µl of
serum-free medium containing different final concentrations of
CdCl2 (0, 10, 20, 30, 40, 50 µM) or DDP (0, 5, 10, 15,
20, 25 µM) was added to each well. Three replicates were plated for
each group. After 24 h of incubation at room temperature (RT), the
culture medium was discarded. Then, 200 µl thiazole blue (0.5
mg/ml) was added to each well. After incubation for 4 h at RT, the
waste solution was discarded and dimethyl sulfoxide (150 µl/well)
was added and mixed thoroughly for 10 min; the absorbance A
(wavelength: 570 nm) of each well was detected with a microplate
reader. The cell inhibition rate and half maximal inhibitory
concentration (IC50) were calculated.
Cell proliferation
Cells were seeded into 96-well plates at
1×105 cells per well, and cultured for 24 h at RT.
Different concentrations of CdCl2 were then added to the
culture medium for different times. Control groups were treated
with an equal volume of dimethyl sulfoxide (DMSO). MTT reagent (20
µl) was added to each well, and supernatants were discarded after 4
h. DMSO (150 µl) was added to each well to dissolve the MTT reagent
and absorbances were measured at 490 nm. Inhibition rate formula:
Inhibition rate (%) = (Control group value-Treatment group
value)/Control group value ×100%.
Transwell assay
A total of 1×106 cells in serum-free
medium were seeded into the upper chamber, while the lower chamber
was maintained in 10% FBS medium. After incubation for 24 h at RT,
migratory cells at the bottom of the upper chamber were fixed with
4% paraformaldehyde for 30 min, stained with 0.5% crystal violet
for 15 min and then counted under an inverted microscope
(magnification, ×400). Data were analyzed using ImageJ V1.8.0
software (National Institutes of Health).
Lentivirus infection
Human FOXM1-specific silenced and overexpressing
lentivirus and corresponding negative control lentivirus were
purchased from Genechem. The sh-FOXM1 group was transfected with
the specific silenced sequence, the FOXM1 group was transfected
with the specific overexpressing sequence. The sh-NC and the vector
group were transfected with corresponding negative control
lentivirus. MG63 cells were seeded into 6-well plates and
transfected with the indicated lentiviruses, and the multiplicity
of infection (MOI) was 20. At ~30% confluency, the cells were
infected for 10 h and the culture media was replaced. Fluorescence
intensities were measured at 72 h post infection to confirm
lentivirus transduction, which reached an efficiency of ~70-80%.
Forty-eight hours after observing fluorescence, cells were
collected and used for follow-up studies.
Apoptosis and cell counting
Adherent MG63 cells were fixed in 4%
paraformaldehyde at 4°C for 15 min. Cells were treated with 0.1%
Triton-X for 10 min and terminal deoxynucleotidyl
transferase-mediated dUTP-biotin nick end labeling (TUNEL) was
performed for 1 h at 37°C to allow diaminobenzidine (DAB)
development. Imaged sections showing brown granules in the nucleus
were deemed TUNEL-positive cells. Five fields (×400) were selected
for each slice and the nucleus and TUNEL-positive cells were then
observed under a fluorescence microscope (Nikon), and the apoptosis
rate (TUNEL-positive cells/nucleus) of five random fields were
calculated.
Immunohistochemical staining
For immunohistochemical analysis, mouse cancer
tissues were fixed in 10% formalin at RT for 24 h and
paraffin-embedded. Sections (5-µm thick) were dewaxed and
dehydrated in a gradient ethanol series. Endogenous peroxidase
activity was quenched by incubating sections in methanol containing
3% H2O2 for 10 min and heating sections in
decanoic acid at 65°C for 10 h for antigen retrieval. Paraffin
sections were added with anti-human FOXM1, followed by incubation
overnight at 4°C. Then the sections were reacted with goat
anti-rabbit lgG-HRP polymer (Beyotime Institute of Biotechnology)
at 37°C for 20 min, followed by color exposure using DAB.
Western blot analysis
Total proteins were extracted from the tissues or
cells. Protein concentrations were measured via bicinchoninic acid
assays (Nanjing Keygen Biotechnology Co., Ltd.). Protein samples
(50 µg) were separated on 10% polyacrylamide gels and transferred
to Hybond® polyvinylidene difluoride membranes
(Millipore). Membranes were blocked in 5% fat-free milk for 2 h at
room temperature and probed with primary antibodies (cleaved
caspase-3, Bcl-2, Bax, MMP-2, MMP-9, E-cadherin, FOXM1 and GAPDH)
overnight at 4°C. Membranes were labeled with the appropriate
secondary antibody for 2 h, and protein bands were visualized using
electrochemiluminescence hypersensitive luminescent solution. Data
were analyzed using ImageJ software (V1.8.0; National Institutes of
Health).
Quantitative real-time polymerase
chain reaction (RT-qPCR) analysis
Cells or tissues were lysed in TRIzol reagent
(Sigma-Aldrich/Merck KGaA) and air dried following the addition of
chloroform, isopropanol, and ethanol. FOXM1 primers were:
5′-GGAGGAAATGCCACACTTAGCG-3′ and 5′-TAGGACTTCTTGGGTCTTGGGGTG-3.
Samples were reverse transcribed into cDNA and RT-qPCR was
performed using reverse transcription reagent on a LightCycler 96
(Roche Life Sciences) using Real-Time PCR Mix (Vazyme Biotech Co.
Ltd.). The reaction conditions were: 95°C pre-heated for 3 min,
95°C denaturation for 30 sec, 60°C annealing for 30 sec and
involving 40 cycles. Gene expression was assessed using the
2−ΔΔCq method relative to GAPDH expression (13). Two independent experiments were
performed in triplicate for each RT-qPCR.
RNA sequencing and functional
enrichment analysis
Total RNA was isolated from MG63 cells using TRIzol
(Invitrogen/Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol. RNA purity was assessed using the ND-1000
(Nanodrop; Thermo Fisher Scientific, Inc.). Each RNA sample had an
A260:A280 ratio above 1.8 and an A260:A230 ratio above 2.0. RNA
integrity was evaluated using the Agilent 2200 TapeStation (Agilent
Technologies). Briefly, rRNAs were removed from total RNA using the
EpicenterRibo-Zero rRNA Removal Kit (Illumina, Inc.) and fragmented
to approximately 200 bp. Subsequently, the purified RNAs were
subjected to first-strand and second-strand cDNA synthesis followed
by adaptor ligation and enrichment with a low-cycle according to
the instructions of the NEBNext® Ultra™ RNA Library Prep
Kit for Illumina [New England BioLabs, Inc. (NEB)]. The purified
library products were evaluated using the Agilent 2200 TapeStation
and Qubit® 2.0 (Thermo Fisher Scientific, Inc.) and then
diluted to 10 pM for cluster generation in situ on the
pair-end flow cell followed by sequencing (2×150 bp) HiSeq 3000
system (Illumina, Inc.). Clean reads were obtained after removal of
reads containing adapter, poly-N and at low quality from raw data.
Differential expression was assessed by DESeq using read counts as
input. The Benjamini-Hochberg multiple test correction method was
enabled. Differentially expressed genes were chosen according to
the criteria of fold change >2 and adjusted P-value <0.05.
All of the differentially expressed genes were used for heat map
analysis.
Grouping of nude mice
Sixteen BALB/c nude mice (6 weeks, ~18 g, random
sex) were obtained from the Experimental Animal Center of Southern
China Medical University and used for tumor formation experiments.
After MG63 cells were digested, they were washed twice with PBS to
remove the residual medium. A total of 1×106 cells were
injected into the right forelegs of the nude mice subcutaneously.
The status of nude mice and tumor growth were observed every week.
When the transplanted tumor reached 60–80 mm3, the nude
mice were randomly divided into 4 groups: i) negative controls
[intraperitoneal (i.p.) injection of normal saline, once every
day); ii) DDP group (peritoneal injection of cisplatin 5 mg/kg,
once every other day); iii) cadmium chloride (CdCl2)
(i.p. injection of cadmium chloride 1 mg/kg), once every other
day); and iv) DDP+ CdCl2 (cisplatin 5 mg/kg + cadmium
chloride 1 mg/kg, once every other day).
Treatment of nude mice
Drug administration was performed according to the
above grouping. Every 2–3 nude mice were kept in a cage. The
feeding conditions were as follows: room temperature was maintained
at 26–28°C, relative humidity was maintained at 40–60%, and 10 h of
light and 14 h of dark were kept daily. The drinking water,
defecation and mental state of the nude mice were observed every
day, and a vernier caliper was used to measure the longest and
shortest diameters of the transplanted tumor every two days. The
volume (V) of the subcutaneous transplanted tumor was calculated (V
= L × W2 × 0.5), and the tumor growth curve was drawn.
Forty-eight hours after the last dose (after 2 weeks, about 23 g),
mice were anesthetized by using carbon dioxide. The specific method
is as follows: The nude mouse was placed in a closed chamber
(50×40×40 cm), and the air in the chamber was gradually replaced by
a mixture of carbon dioxide and oxygen. The flow rate of gas was
16,000 cm3/min and the ratio of carbon dioxide and
oxygen was 6:4. After 20 min when the nude mice lost consciousness,
the CO2 concentration was then raised to 100% for 20 min
until the nude mice died. The criteria for judging the death of the
nude mice were: Stopped heartbeat, dilated pupils, and halted
breathing. Tumor tissue was then removed, weighed and imaged. At
the same time, blood was collected from the posterior orbital
venous plexus of the nude mice, and blood routine and liver and
kidney functions were measured. Automatic blood cell analyzer
(ProCyte Dx; IDEXX, Inc.) was used to detect the following
indicators: white blood cells (WBCs), red blood cells (RBCs),
platelets (PLTs) and lymphocytes (LYMs). An automatic biochemical
analyzer (Catalyst One; IDEXX, Inc.) was used to detect the
following indicators: Aspartate aminotransferase (AST), alanine
aminotransferase (ALT), alkaline phosphatase (ALP), uric acid (UA),
urea nitrogen (BUN), and creatinine (CR). The animal experiment was
performed in strict accordance with the National Institutes of
Health Guide for the Care and Use of Laboratory Animals. The animal
study complied with the ARRIVE guidelines and the AVMA euthanasia
guidelines from 2013 (14,15). All protocols described in this study
were reviewed and approved by the Ethics Committee of the
Affiliated Yuebei People's Hospital of Shantou University Medical
College (Shaoguan, Guangdong, China).
Statistical analysis
All data were analyzed using SPSS 17.0 software
(SPSS, Inc.). Qualitative variables are expressed as counts (%),
and quantitative variables are expressed as the mean ± standard
deviation. Measurement data that conformed to the normal
distribution were analyzed using independent sample t-test and the
Mann-Whitney rank sum tests were used to analyze those that did not
conform to a normal distribution, while for multiple specimens,
statistical analysis was performed using one-way ANOVA and a post
hoc test was performed after pairwise comparison test. On the
premise of homogeneity of variance, the post hoc test was performed
by Scheffe and Tukey method. Differences were statistically
significant at the P<0.05 level.
Results
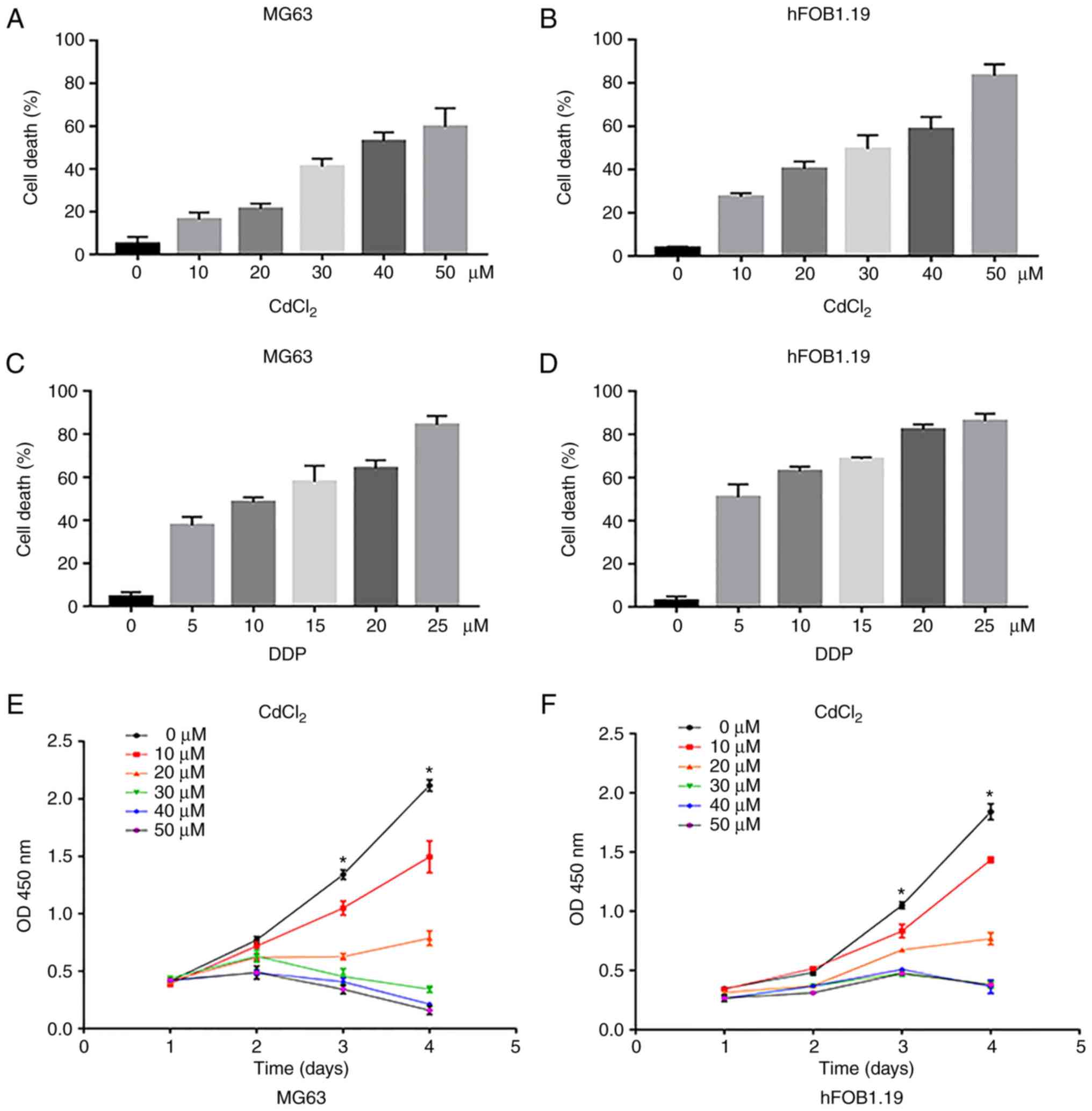
Assessment of the cytotoxicity of both
cadmium chloride and DDP in osteosarcoma and normal cells
To evaluate the cytotoxicity of both cadmium
chloride (CdCl2) and DDP in OS and normal cells, we
first investigated the effects of CdCl2 and DDP on MG63
human OS cells and hFOB1.19 human osteoblasts. MTT assays revealed
that the cell death increased in a concentration-dependent manner
(Fig. 1A-D). Compared to
CdCl2, DDP was more cytotoxic. Normal cells were more
sensitive to the drugs than tumor cells. The half maximal
inhibitory concentration (IC50) values of
CdCl2 and DDP on MG63 cells were calculated to be
39.95±1.99 and 12.79±0.07 µM, respectively. To compare the effects
of different concentrations of CdCl2 on cell
proliferation, MTT assays were performed. The results demonstrated
that CdCl2 at a concentration of 10 µM slightly
inhibited cell proliferation, and when the concentration was
greater than 20 µM, cell proliferation was significantly inhibited
at day 3 and 4 compared with the untreated cells (Fig. 1E and F). The concentration of 20 µM
of CdCl2 was selected for subsequent experiments.
Investigation of the mechanism(s) of
CdCl2 cytotoxicity
OS MG63 cells were treated with 20 µM
CdCl2 for 24 h, and apoptosis was assessed by electron
microscopy. After treatment with CdCl2, the
characteristic morphological changes of apoptosis were observed
which included: Shrinkage of cell membranes, concentrated chromatin
and organelles, folding of endoplasmic reticulum and appearance of
apoptotic bodies outside the membrane (Fig. 2A). MTT assay results showed that
compared with the control (DMSO) group, cell proliferation was
significantly decreased after treatment with CdCl2 for 7
days (Fig. 2B). Meanwhile, cell
apoptosis was significantly increased after treatment with DDP or
CdCl2 compared with the control group, and the combined
use of DDP and CdCl2 further promoted cell apoptosis
(Fig. 2C-E). Western blot analysis
confirmed these effects, as DDP+CdCl2 treatment led to a
loss of Bcl-2 expression and increased Bax and cleaved caspase-3
levels (Fig. 2F). Then, Transwell
assays were performed to investigate the cell migration level. The
results revealed that DDP+CdCl2 treatment significantly
inhibited MG63 cell migration ability compared with the use of DDP
or CdCl2 only, or the control group (Fig. 2G and H). Western blot results
confirmed that the expression of MMP2 and MMP9 was significantly
inhibited while E-cadherin was increased in the
DDP+CdCl2 treatment group compared with the use of DDP
or CdCl2 only, or the control group (Fig. 2I). This implies that DDP and
CdCl2 may downregulate the migration of MG63 through the
Wnt pathway.
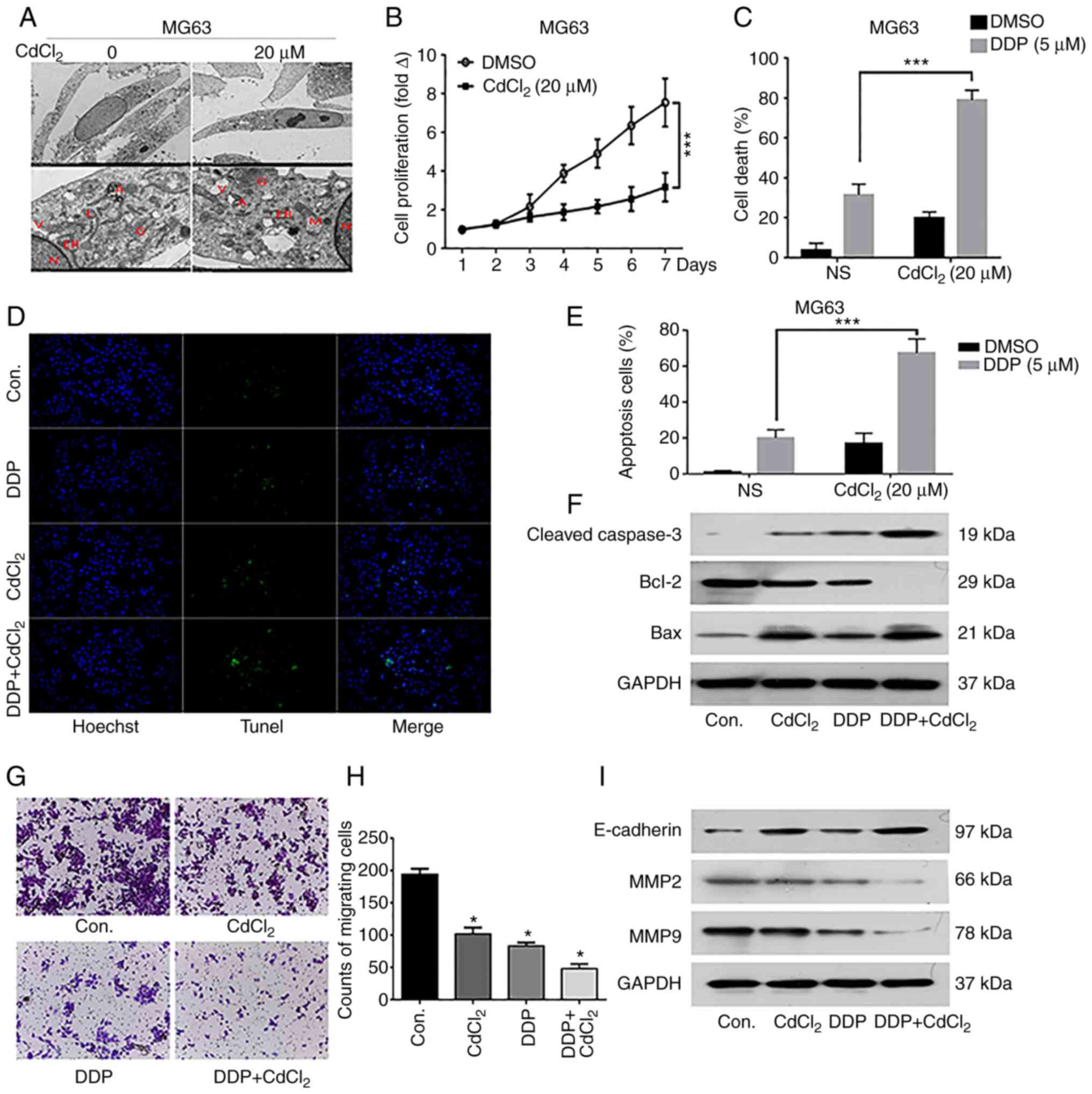 | Figure 2.Effects of cadmium chloride
(CdCl2) on the survival of osteosarcoma (OS) cells. (A)
Electron microscopy images of OS MG63 cells treated with
CdCl2. N, M, V, A, L, ER and G indicate the nucleus,
mitochondria, vacuole, autophagosome, lysosome, endoplasmic
reticulum and Golgi respectively. (B) MTT results showing the cell
proliferation level over 4 consecutive days in each group. (C-E)
TUNEL results show the cell death and apoptosis of MG63 cells
treated with DDP and/or CdCl2. ***P<0.001. (F)
Western blot results indicate that CdCl2 promotes
apoptosis in MG63 cells. (G and H) The migration capacity of MG63
cells treated with DDP and/or CdCl2 was analyzed by
Transwell assays. (I) Western blot results indicate that
CdCl2 inhibits migration in MG63 cells. *P<0.05,
compared with the control (cont.) group. |
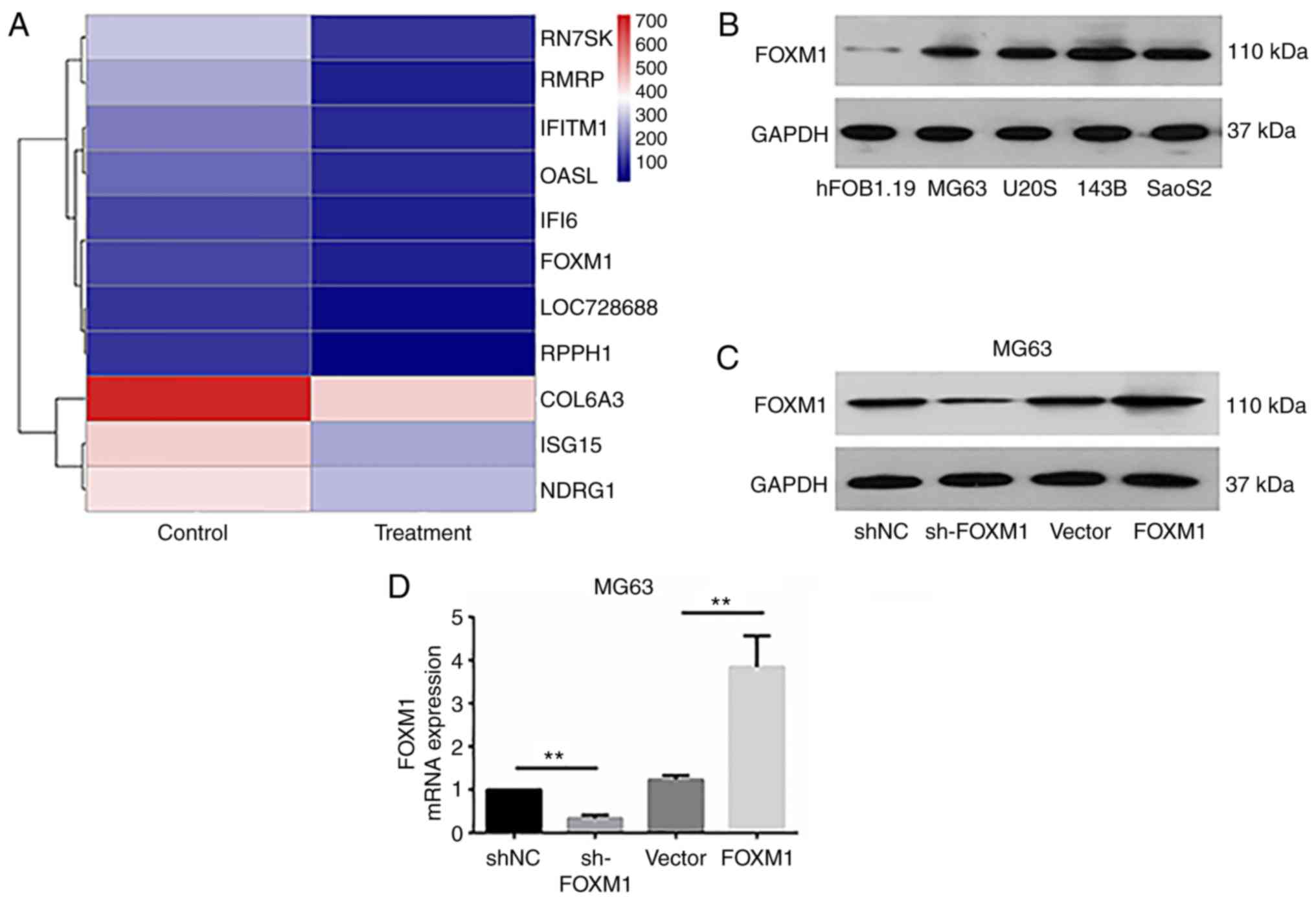
Investigation of the mechanism(s) by
which cadmium chloride inhibits OS
To investigate the mechanism(s) by which
CdCl2 inhibits OS, we performed gene chip screening of
CdCl2-treated MG63 cells and the untreated cells.
Heatmaps were constructed based on the differentially expressed
genes between the treatment groups (Fig. 3A, Table
I), of which FOXM1 was identified as being significantly
downregulated in response to treatment with CdCl2. To
investigate the effects of FOXM1 on OS cells, Western blotting was
adopted to explore the expression of FOXM1 in OS cell lines. The
results demonstrated that the expression of FOXM1 in MG63, U2OS,
143B and SaoS2 OS cells was higher than that in the hFOB.19 cells
(Fig. 3B). Then,
FOXM1-overexpressing and -silenced lentiviruses were constructed.
Results of the Western blot analysis and qRT-PCR showed that the
expression of FOXM1 in the sh-FOXM1 and the FOXM1 group were
significantly lower and higher, respectively, than that in the
control (shNC and Vector) groups (Fig.
3C and D). In the FOXM1-silenced (sh-FOXM1) cells, low
concentrations of DDP significantly reduced OS cell viability
(Fig. 3E). Conversely, high DPP
concentrations were required to reduce the viability of
FOXM1-overexpressing (FOXM1) cells when compared to the Vector
group (Fig. 3F), thus confirming a
role for FOXM1 in DPP-mediated cell death. DPP treatment of
FOXM1-silenced MG63 cells induced cell apoptosis, while DPP
treatment of FOXM1-overexpressing MG63 cells inhibited cell
apoptosis (Fig. 3G and H). These
results were verified by Western blot analysis. Compared with the
control group, silencing of FOXM1 upregulated the expression of
cleaved caspase-3 and Bax levels, and downregulated the expression
of Bcl-2 levels. Addiitonally, overexpression of FOXM1 led to the
opposite trend in regards to the expression of cleaved caspase-3,
Bax and Bcl-2 levels (Fig. 3I).
 | Table I.Differential gene expression levels
between MG63 cells and MG63-CdCl2 cells in the gene
chips. |
Table I.
Differential gene expression levels
between MG63 cells and MG63-CdCl2 cells in the gene
chips.
| Gene | Gene_type | MG63-ceM |
MG63-CdCl2-ceM | log2(FC) | P-value | q-value |
|---|
| ISG15 | Protein_coding | 454.746 | 250.452 | −0.861 | <0.001 | <0.001 |
| IFITM1 | Protein_coding | 196.566 | 87.550 | −1.167 | <0.001 | <0.001 |
| OASL | Protein_coding | 164.334 | 88.149 | −0.899 | <0.001 | <0.001 |
| IFI6 | Protein_coding | 122.105 | 68.162 | −0.841 | <0.001 | 0.007 |
|
LOC728688 | Pseudogene | 96.470 | 38.221 | −1.336 | <0.001 | <0.001 |
| COL6A3 | Protein_coding | 726.174 | 453.297 | −0.680 | <0.001 | <0.001 |
| NDRG1 | Protein_coding | 428.946 | 281.864 | −0.606 | <0.001 | <0.001 |
| RPPH1 | RNase_P_RNA | 92.824 | 22.347 | −2.054 | <0.001 | <0.001 |
| RN7SK | snRNA | 298.014 | 100.370 | −1.570 | <0.001 | <0.001 |
| RMRP | RNase_MRP_RNA | 253.020 | 74.464 | −1.765 | <0.001 | <0.001 |
| FOXM1 | Protein_coding | 121.922 | 66.312 | −0.879 | <0.001 | 0.004 |
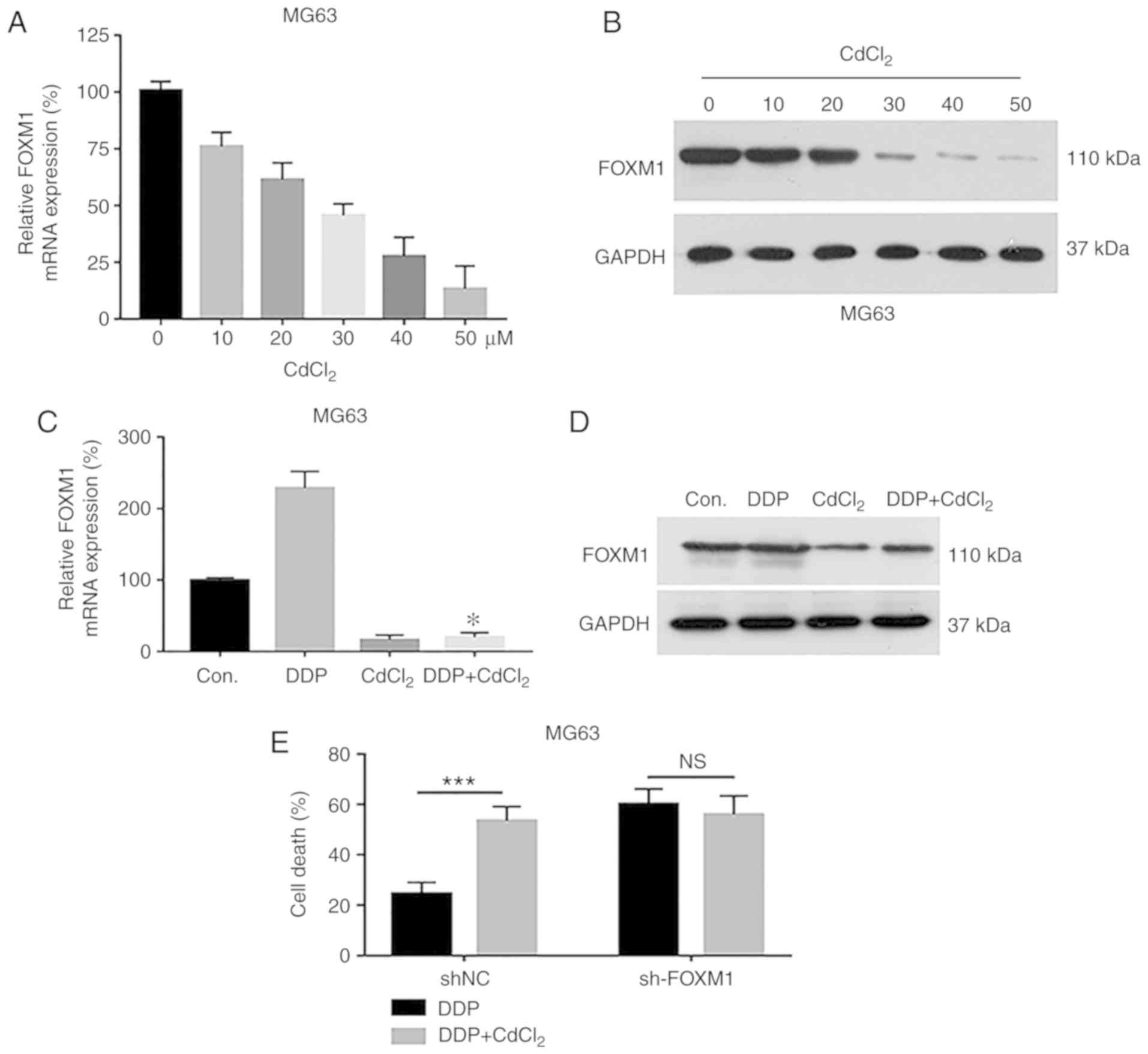
Assessment of the regulatory
relationship between cadmium chloride and FOXM1 in OS cells
To verify the regulatory relationship between
CdCl2 and FOXM1 in OS cells, MG63 cells were treated
with various concentrations of CdCl2, and the expression
of FOXM1 was verified by qPCR and Western blot analysis. FOXM1
levels were decreased with increasing CdCl2
concentrations at both the mRNA and protein levels (Fig. 4A and B). We next investigated the
effects of FOXM1 expression on the response of OS cells to
DDP+CdCl2 treatment. qPCR and Western blotting results
demonstrated that after MG63 cells were treated with DPP, FOXM1
expression was upregulated, and the combined use of
DDP+CdCl2 significantly suppressed the expression of
FOXM1 in MG63 cells compared to the control cells (Fig. 4C and D). In FOXM1-silenced MG63
cells, MTT results showed that DDP led to an increase in the MG63
cell death rate compared to the shNC cells. DDP+CdCl2
treatment of the sh-FOXM1 cells yielded no significant change in
cell death. Meanwhile, DDP+CdCl2 treatment of the shNC
cells resulted in higher apoptosis compared with the DDP alone
treated cells (Fig. 4E).
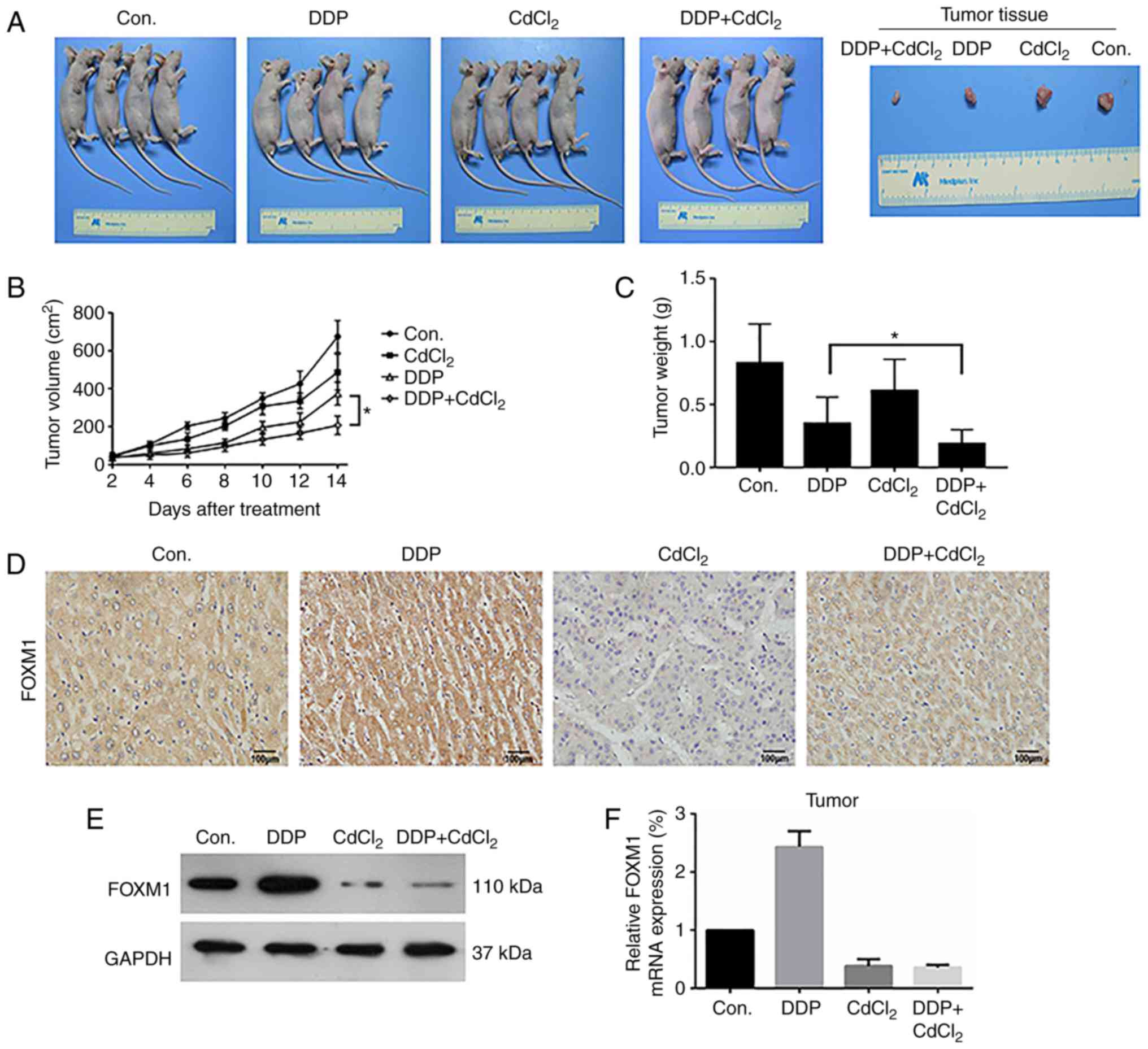
In vivo nude mouse OS model
To confirm these findings in vivo, nude mice
were implanted with OS MG63 cells (Fig.
5A), and the effects of DDP+CdCl2 combination
therapy were assessed after 14 days of treatment in the i) control
(Con.), ii) DDP, iii CdCl2 and iv) DDP+CdCl2
groups. We first tested the safety of the DDP+CdCl2
combination. DDP+CdCl2 caused changes in blood routine
and liver and kidney function in nude mice (Tables II and III). The results showed that
DDP+CdCl2 increased WBCs, PLTs, LYMs, ALT, ALP, UA, BUN,
and CR levels compared with control group in venous blood. There
was no significant effect on red RBCs or AST levels. Then, we
measured the tumor volumes and weight (Fig. 5B and C). Compared with the control
group and the use of DDP or CdCl2 group, treatment with
DDP+CdCl2 reduced tumor volume and weight, and
DDP+CdCl2 had the most significant effect compared with
the use of DDP or CdCl2 alone. Tumor tissues were
isolated, and FOXM1 expression was assessed by immunohistochemical
(IHC) staining (Fig. 5D), Western
blot analysis (Fig. 5E) and qPCR
(Fig. 5F). Similar to the results
of the in vitro experiments, DDP upregulated FOXM1
expression while CdCl2 inhibited FOXM1 expression.
 | Table II.Analysis of routine blood indicators
of the mice in each group. |
Table II.
Analysis of routine blood indicators
of the mice in each group.
|
| Treatment
group |
|---|
|
|
|
|---|
| Index | Con. |
CdCl2 | DDP |
DDP+CdCl2 | P-value |
|---|
| WBCsa | 2.528±0.057 | 2.618±0.120 | 2.743±0.185 | 2.848±0.174 | 0.005 |
| RBCsb | 8.918±0.036 | 8.833±0.079 | 8.893±0.073 | 8.858±0.059 | 0.116 |
| PLTsa | 693.29±2.614 | 725.72±53.35 | 772.38±64.68 | 840.57±90.46 | 0.003 |
| LYMsa | 2.123±0.082 | 2.153±0.067 | 2.440±0.346 | 2.660±0.102 | 0.001 |
 | Table III.Analysis of the biochemical blood
indices of the mice in each group. |
Table III.
Analysis of the biochemical blood
indices of the mice in each group.
|
| Treatment
group |
|---|
|
|
|
|---|
| Index | Con. |
CdCl2 | DDP |
DDP+CdCl2 | P-value |
|---|
| ASTa | 127.13±2.09 | 127.23±3.20 | 128.00±3.36 | 124.70±2.51 | 0.209 |
| ALTa | 44.35±0.48 | 44.80±0.71 | 49.33±0.97 | 56.18±2.16 | 0.001 |
| ALPa | 363.8±11.65 | 367.9±14.76 | 343.0±17.2 | 366.8±17.6 | 0.031 |
| UAb | 143.80±0.91 | 148.4±2.30 | 159.63±1.63 | 162.20±2.51 | 0.001 |
| BUNc | 7.49±0.07 | 7.66±0.05 | 8.24±0.25 | 8.91±0.21 | 0.001 |
| CRb | 105.20±1.54 | 104.32±2.06 | 127.03±1.02 | 108.03±1.38 | 0.002 |
Discussion
Osteosarcoma (OS) is a common malignant tumor in
childhood and adolescence, with an global annual incidence of ~3.1
million for the past 20 years (16,17).
OS occurs in the metaphyseal end of adolescent long tubular bones
and shows early blood metastasis and rapid progression. Current
therapies include neoadjuvant chemotherapy combined with limb
salvage, but specific treatments are lacking. Although this
approach preserves joint function in the limbs, metastasis and OS
recurrence are frequent. Surgery is also challenging and requires
tumor resection and limb reconstruction (3). Chemotherapy resistance presents an
additional challenge. Although next-generation sequencing to
explore drug resistance genes in patients with OS has been proposed
with the ultimate aim of precision therapy, this approach is
limited by the high mutational rates associated with OS (18–20).
Further studies to explore OS pathogenesis and new therapeutic
targets are urgently required.
Commonly used chemotherapy drugs include adriamycin,
cisplatin and methotrexate. Other less frequently used drugs
include ifosfamide and vincristine. DDP is commonly used for OS but
its therapeutic effect has not improved over the last 30 years,
primarily due to drug-resistance (2,21). As
a classic chemotherapy drug, cisplatin binds DNA after entering
tumor cells, forming intrachain and interchain adducts destroying
the normal DNA structure, promoting DNA damage, inhibiting DNA
replication and transcription, and enhancing tumor cell death. In
resistant cells, cisplatin accumulation is reduced (22) and the DNA damage responses of tumor
cells are enhanced (23). These
cells are also resistant to autophagy (24), an important cause of OS tolerance to
cisplatin. Overcoming the tolerance of OS to cisplatin can increase
its sensitivity during clinical treatment (25). We therefore explored whether
short-term cadmium chloride treatment can enhance the sensitivity
of OS to cisplatin to improve its efficacy.
Previous studies have shown that forkhead box
protein M1 (FOXM1) regulates cell proliferation, and its expression
varies according to the cell cycle status. FOXM1 levels are low in
G0, gradually increase in G1, peak in late G1 and early S, and
remain high during M phase. When cells enter the M phase, FOXM1 is
rapidly degraded, and its expression sharply declines. FOXM1
regulates cell cycle progression through its role in DNA repair,
chromatin assembly, and maintenance of chromosomal stability to
ensure that mitosis proceeds and that cell proliferation occurs
(26,27). FOXM1 is highly expressed in a
variety of solid tumor tissues, and its inhibition prevents tumor
proliferation and apoptosis (28,29).
In addition, FOXM1 is highly expressed in proliferating cells,
where it regulates DNA damage repair, tissue homeostasis and
chemoresistance (30). FOXM1
promotes tumor cell survival in hypoxic environments, and the loss
of FOXM1 expression impairs the proliferation of hypoxic tumor
cells (23). FOXM1 also induces
breast cancer cell resistance to epirubicin by upregulating MRN
complexes (31,32).
In the present study, cadmium chloride was found to
induce MG63 cell apoptosis, inhibit MG63 cell proliferation and
migration, and enhance the regulatory effect of cisplatin (DDP) in
OS MG63 cells. Wnt signaling pathway is widely involved in the
regulation of cell proliferation, survival, apoptosis, migration
and metastasis (33,34). We further found that chromium
chloride can activate the Wnt signaling pathway and the Bcl-2
protein family. Therefore, the regulation of chromium chloride on
MG63 cells may be related to the activation of Wnt signaling
pathway. Cadmium chloride was also found to increase the
sensitivity of OS cells to cisplatin in vitro and in
vivo. OS cells treated with cadmium chloride were screened
through gene chip assays in which the expression of FOXM1 was found
to be significantly decreased. FOXM1 silencing enhanced the effects
of DDP and cadmium chloride on growth inhibition and apoptosis,
while FOXM1 overexpression increased cell survival. These findings
were confirmed in in vivo models, in which mice treated with
DDP and/or cadmium chloride showed reduced OS tumor growth. IHC
staining confirmed altered FOXM1 expression in tumor cells treated
with cadmium chloride alone or in combination with DPP, confirming
its role in DPP resistance. This highlighted the ability of cadmium
chloride to improve the therapeutic effects of DPP.
The use of cadmium chloride in combination with DPP
as a treatment for OS poses challenges. First, cadmium chloride
enhances DPP sensitization, but its optimal concentrations for use
are unknown. Second, the effects of cadmium chloride on tissue
toxicity and immune responses must be assessed. In addition, the
pharmacokinetics of cadmium chloride in vivo require
assessment to confirm its absorption, distribution and metabolic
elimination.
Taken together, this study demonstrated that
exogenous cadmium chloride reduces the expression of the resistance
gene FOXM1 in OS cells and enhances the therapeutic effects of DPP
on OS. This highlights cadmium chloride as a therapeutic adjuvant
to reduce cisplatin-induced drug resistance.
Acknowledgements
Not applicable.
Funding
The present study was supported in part by a grant
from the Guangdong Provincial Science and Technology Project
(2017A020215080).
Availability of data and materials
The datasets used during the present study are
available from the corresponding author upon reasonable
request.
Authors' contributions
YD and KH conceived and designed the study. KH, WX
and WQ performed the experiments. KH, SN, SY and GT analyzed the
data and wrote the manuscript. KH, WQ and YD reviewed the data and
edited the manuscript. All authors read and approved the manuscript
and agree to be accountable for all aspects of the research in
ensuring that the accuracy or integrity of any part of the work are
appropriately investigated and resolved.
Ethics approval and consent to
participate
The animal experiment was performed in strict
accordance with the National Institutes of Health Guide for the
Care and Use of Laboratory Animals. The animal study complied with
the ARRIVE guidelines and the AVMA euthanasia guidelines from 2013.
All protocols described in this study were reviewed and approved by
the Ethics Committee of the Affiliated Yuebei People's Hospital of
Shantou University Medical College (Shaoguan, Guangdong,
China).
Patient consent for publication
Not applicable.
Competing interests
The authors state that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
CdCl2
|
cadmium chloride
|
|
DAB
|
diaminobenzidine
|
|
DMEM
|
Dulbecco's modified Eagle's medium
|
|
DMSO
|
dimethyl sulfoxide
|
|
FBS
|
fetal bovine serum
|
|
FOXM1
|
forkhead box protein M1
|
|
GAPDH
|
glyceraldehyde-3-phosphate
dehydrogenase
|
|
OS
|
osteosarcoma
|
|
RT-qPCR
|
quantitative real-time polymerase
chain reaction
|
|
TUNEL
|
terminal deoxynucleotidyl
transferase-mediated dUTP-biotin nick end labeling
|
References
|
1
|
Anninga JK, Gelderblom H, Fiocco M, Kroep
JR, Taminiau AH, Hogendoorn PC and Egeler RM: Chemotherapeutic
adjuvant treatment for osteosarcoma: Where do we stand? Eur J
Cancer. 47:2431–2445. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kansara M, Teng MW, Smyth MJ and Thomas
DM: Translational biology of osteosarcoma. Nat Rev Cancer.
14:722–735. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ritter J and Bielack SS: Osteosarcoma. Ann
Oncol. 21 (Suppl 7):vii320–325. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ferrari S and Serra M: An update on
chemotherapy for osteosarcoma. Expert Opin Pharmacother.
16:2727–2736. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Moriarity BS, Otto GM, Rahrmann EP, Rathe
SK, Wolf NK, Weg MT, Manlove LA, LaRue RS, Temiz NA, Molyneux SD,
et al: A Sleeping Beauty forward genetic screen identifies new
genes and pathways driving osteosarcoma development and metastasis.
Na Genet. 47:615–624. 2015. View
Article : Google Scholar
|
|
6
|
Baranski Z, Booij TH, Cleton-Jansen AM,
Price LS, van de Water B, Bovée JV, Hogendoorn PC and Danen EH:
Aven-mediated checkpoint kinase control regulates proliferation and
resistance to chemotherapy in conventional osteosarcoma. J Pathol.
236:348–359. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Holmboe L, Andersen AM, Mørkrid L, Slørdal
L and Hall KS: High dose methotrexate chemotherapy:
Pharmacokinetics, folate and toxicity in osteosarcoma patients. Br
J Clin Pharmacol. 73:106–114. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wallin M, Barregard L, Sallsten G, Lundh
T, Karlsson MK, Lorentzon M, Ohlsson C and Mellström D: Low-level
cadmium exposure is associated with decreased bone mineral density
and increased risk of incident fractures in elderly men: The MrOS
Sweden Study. J Bone Miner Res. 31:732–741. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Akesson A, Bjellerup P, Lundh T, Lidfeldt
J, Nerbrand C, Samsioe G, Skerfving S and Vahter M: Cadmium-induced
effects on bone in a population-based study of women. Environ
Health Perspect. 114:830–834. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
10
|
Coogan TP, Achanzar WE and Waalkes MP:
Spontaneous transformation of cultured rat liver (TRL 1215) cells
is associated with down-regulation of metallothionein: Implications
for sensitivity to cadmium cytotoxicity and genotoxicity. J Environ
Pathol Toxicol Oncol. 19:261–273. 2000.PubMed/NCBI
|
|
11
|
Benbrahim-Tallaa L, Waterland RA, Dill AL,
Webber MM and Waalkes MP: Tumor suppressor gene inactivation during
cadmium-induced malignant transformation of human prostate cells
correlates with overexpression of de novo DNA methyltransferase.
Environ Health Perspect. 115:1454–1459. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Xie D and Sheng Z: Low-level cadmium
exposure and bone health. J Bone Miner Res. 32:4192017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kahler SC: Integrating cultural
competence: Animal health, food safety ultimately benefit as AVMA
incorporates cultural competence in CE. J Am Vet Med Assoc.
242:1186–1187. 2013.PubMed/NCBI
|
|
15
|
Rice ASC, Morland R, Huang W, Currie GL,
Sena ES and Macleod MR: Transparency in the reporting of in vivo
pre-clinical pain research: The relevance and implications of the
ARRIVE (Animal research: Reporting in vivo experiments) guidelines.
Scand J Pain. 4:58–62. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Picci P: Osteosarcoma (osteogenic
sarcoma). Orphanet J Rare Dis. 2:62007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Jaffe N: Osteosarcoma: Review of the past,
impact on the future. The American experience. Cancer Treat Res.
152:239–262. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Do SI, Jung WW, Kim HS and Park YK: The
expression of epidermal growth factor receptor and its downstream
signaling molecules in osteosarcoma. Int J Oncol. 34:797–803.
2009.PubMed/NCBI
|
|
19
|
Man TK, Lu XY, Jaeweon K, Perlaky L,
Harris CP, Shah S, Ladanyi M, Gorlick R, Lau CC and Rao PH:
Genome-wide array comparative genomic hybridization analysis
reveals distinct amplifications in osteosarcoma. BMC Cancer.
4:452004. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lau CC, Harris CP, Lu XY, Perlaky L,
Gogineni S, Chintagumpala M, Hicks J, Johnson ME, Davino NA, Huvos
AG, et al: Frequent amplification and rearrangement of chromosomal
bands 6p12-p21 and 17p11.2 in osteosarcoma. Genes, Chromos Cancer.
39:11–21. 2004. View Article : Google Scholar
|
|
21
|
Luetke A, Meyers PA, Lewis I and Juergens
H: Osteosarcoma treatment-Where do we stand? A state of the art
review. Cancer Treat Rev. 40:523–532. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Martelli L, Di Mario F, Botti P, Ragazzi
E, Martelli M and Kelland L: Accumulation, platinum-DNA adduct
formation and cytotoxicity of cisplatin, oxaliplatin and
satraplatin in sensitive and resistant human osteosarcoma cell
lines, characterized by p53 wild-type status. Biochem Pharmacol.
74:20–27. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Liang Q, Dexheimer TS, Zhang P, Rosenthal
AS, Villamil MA, You C, Zhang Q, Chen J, Ott CA, Sun H, et al: A
selective USP1-UAF1 inhibitor links deubiquitination to DNA damage
responses. Nat Chem Biol. 10:298–304. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Levy JM, Towers CG and Thorburn A:
Targeting autophagy in cancer. Nat Rev Cancer. 17:528–542. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Shibata A, Moiani D, Arvai AS, Perry J,
Harding SM, Genois MM, Maity R, van Rossum-Fikkert S, Kertokalio A,
Romoli F, et al: DNA double-strand break repair pathway choice is
directed by distinct MRE11 nuclease activities. Mol Cell. 53:7–18.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Koo CY, Muir KW and Lam EW: FOXM1: From
cancer initiation to progression and treatment. Biochim Biophys
Acta. 1819:28–37. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Uddin S, Hussain AR, Ahmed M, Siddiqui K,
Al-Dayel F, Bavi P and Al-Kuraya KS: Overexpression of FoxM1 offers
a promising therapeutic target in diffuse large B-cell lymphoma.
Haematologica. 97:1092–1100. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wang M and Gartel AL: The suppression of
FOXM1 and its targets in breast cancer xenograft tumors by siRNA.
Oncotarget. 2:1218–1226. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Pandit B and Gartel AL: FoxM1 knockdown
sensitizes human cancer cells to proteasome inhibitor-induced
apoptosis but not to autophagy. Cell Cycle. 10:3269–3273. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Nestal de Moraes G, Bella L, Zona S,
Burton MJ and Lam EW: Insights into a critical role of the
FOXO3a-FOXM1 axis in DNA damage response and genotoxic drug
resistance. Curr Drug Targets. 17:164–177. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Peake BF, Eze SM, Yang L, Castellino RC
and Nahta R: Growth differentiation factor 15 mediates epithelial
mesenchymal transition and invasion of breast cancers through
IGF-1R-FoxM1 signaling. Oncotarget. 8:94393–94406. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Khongkow P, Karunarathna U, Khongkow M,
Gong C, Gomes AR, Yagüe E, Monteiro LJ, Kongsema M, Zona S, Man EP,
et al: FOXM1 targets NBS1 to regulate DNA damage-induced senescence
and epirubicin resistance. Oncogene. 33:4144–4155. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zerlin M, Julius MA and Kitajewski J:
Wnt/Frizzled signaling in angiogenesis. Angiogenesis. 11:63–69.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Oltvai ZN, Milliman CL and Korsmeyer SJ:
Bcl-2 heterodimerizes in vivo with a conserved homolog, Bax, that
accelerates programmed cell death. Cell. 74:609–619. 1993.
View Article : Google Scholar : PubMed/NCBI
|