Introduction
The design and development of novel therapeutic
small molecules is a major research field and a main focus of this
research is the identification of more effective anticancer
solutions (1). A number of powerful
and diverse array of methods have been developed to improve the
success of drug design based on the three-dimensional structure of
the pharmacological target (2–5);
however, numerous researchers are still successfully using the
ligand-based strategies. These methods are based on the simple
principle that similar structures will produce the same biological
effect and have the advantage of a lower risk of failure (6,7). A
closer analysis of the drug design process has revealed that
medicinal chemistry specialists rely on their intuition when
deciding the direction of the research, and often, they are
affected by cognitive biases (8,9). One
of the most important is the so-called confirmation bias. This
represents the tendency of researchers to consider the data that
support their hypotheses to a greater extent and not to actively
search for evidence that would contradict their hypothesis
(9).
The confirmation bias can be easily observed by
reading most of the medicinal chemistry articles in which
researchers are supporting their focus on a particular type of
structure by presenting various similar compounds sharing the
targeted biological effect. For example, Sharma et al
considered the thiazole ring as an essential core scaffold for
anticancer drugs by presenting clinically proved drugs, such as
dasatinib, dabrafenib, or ixabepilone (10). Indole is another heterocycle that is
regarded as useful in the target-based design of anticancer agents
based on similar reasoning (11).
Natural scaffolds, such as the flavonoid core structure, are
extensively used in the design of novel cancer drugs (12,13).
Ismail et al argued that pyrazolo[3,4-d]pyrimidine
derivatives have a good chance to target protein kinases and to be
developed as anticancer agents (14). Our own research group has focused on
a particular chemophore, the pyrazole ring, as a scaffold for the
design of anticancer agents (15–18).
The objective of the present study was to identify
an unbiased quantitative method which may be used to measure the
usefulness of the most important heterocyclic structures in the
development of anti-proliferative drugs. The scope of the present
study was to help chemists focus on the chemical structure with
better potential, as well as to understand their bias towards a
particular scaffold.
Cancer cell lines serve as a major model for
antineoplastic drug discovery and development. The National Cancer
Institute (NCI) established a systematic screening program by
assembling a panel of 60 cancer cell lines (NCI-60 panel) from
multiple tumor types (brain, blood and bone marrow, breast, colon,
kidney, lung, ovary, prostate and skin) (19,20).
Over the past decades, a large number of compounds have been tested
on the NCI-60 panel and several cytotoxic agents have emerged as
first-line treatment options for a number of tumor types (21).
A compound is first tested at a single concentration
and then, if found active, it is tested at five different
concentrations with 48-h drug exposure and 50% growth inhibition
(GI50), total growth inhibition (TGI), and 50% lethal
concentration (LC50) are computed (22). Data analysis tools, such as COMPARE
use these outputs on all 60 cancer cell lines to create a
fingerprint profile that allows classification and can predict the
mechanisms of action (23,24). The fingerprint of cellular response
in the NCI-60 assay can be used to determine similar prototype
compounds, the usefulness of this data mining approach being
demonstrated in various studies (25,26).
Materials and methods
Creation and preparation of
datasets
Two sets of data were collected freely from the DTP
website (https://dtp.cancer.gov/databases_tools/default.htm)
representing one-dose screening values and five-dose screening
data. The number reported in the one-dose data set is each cell
line growth percentage (GI%) following 48 h of exposure to
10−5 M solution of drug, relative to the no-drug
control, for a specific cell line. A GI% value <100 and >0
indicates a growth inhibition and a value <0 represents a lethal
effect of the tested compound. The 5-dose screening data set
contains the negative log10 of the 50% growth inhibitory
concentration expressed as molar concentration (pGI50)
for each tested compound.
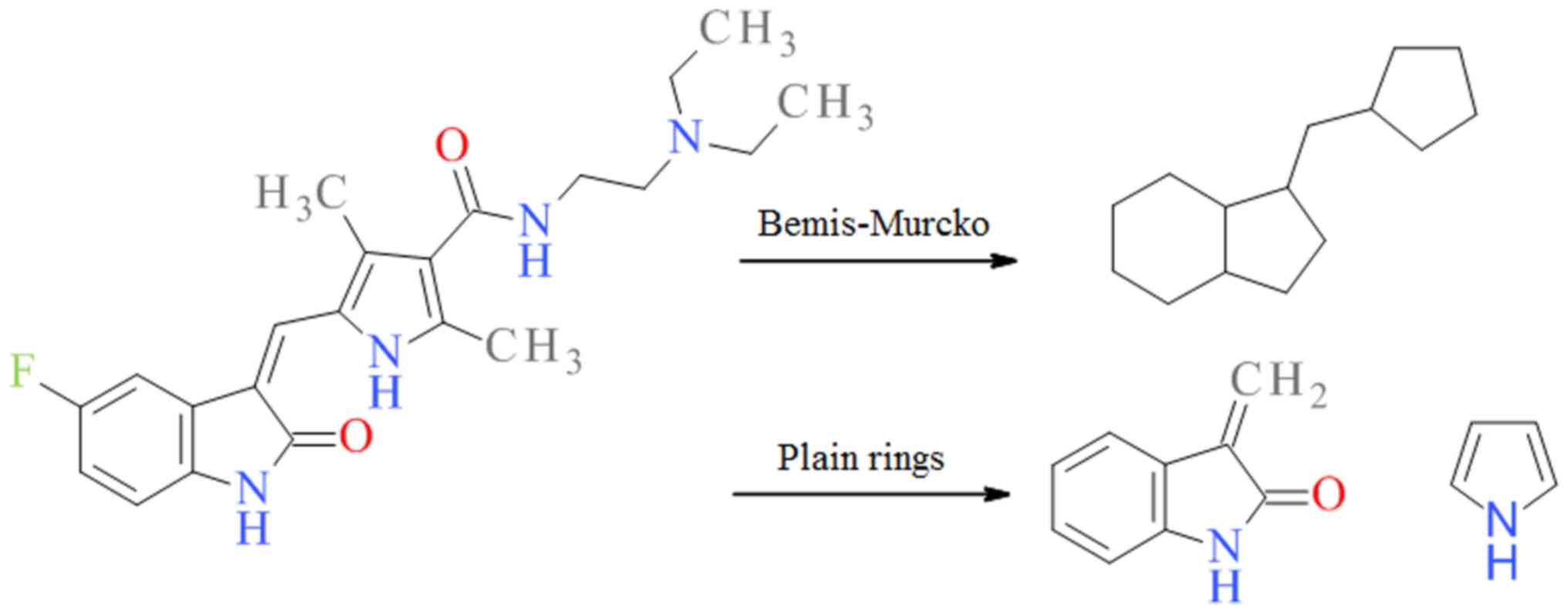
Bemis-Murcko scaffold analysis
Bemis-Murcko skeletons represent the molecular
frameworks resulting after the removal of the side-chain atoms and
atom labels. All bond types are transformed into single ones
(10). The analysis was performed
using DataWarrior 5.2.0 (http://www.openmolecules.org/datawarrior/) to generate
the Bemis-Murcko scaffolds representing the cyclic frameworks
incorporating only the rings and the chains connecting them.
Bemis-Murcko scaffolds underline the importance of molecular
topology and have been proven to be useful in several drug design
studies (11–15).
Plain ring analysis
DataWarrior 5.2.0 software was used to identify all
ring systems in each compound without substituents. The double
bonded heteroatoms connected directly to the ring system were taken
into consideration. Both procedures for Bemis-Murcko scaffold and
plain ring using sunitinib the Cancer Chemotherapy National Service
Center number [(NSC Number) 750690] as an example are illustrated
in Fig. 1.
Scoring methods
The one-dose data set matrix was analyzed in order
to assess the performance of each chemical feature on generating
potent anti-proliferative candidates. The average (A1D)
of all GI% values registered for a specific scaffold was calculated
as an indicator of the anti-proliferative potency of that chemical
feature as follows:
A1D=∑GI%No.of cell lines
For each scaffold, a performance score
(P1D) was defined as the number of GI% values under the
50% value threshold reported to the total number of GI% values
recorded for that scaffold. The corresponding mathematical formula
is as follows:
P1D=100×No of cell lines GI%≤50No. of
cell lines
The scoring formula is similar to the measure of
incidence in epidemiology (27)
considering an observed event an anti-proliferative effect
>50%.
A third score was implemented to assess the
selectivity of the anti-proliferative effect based on the number of
outliers. Considering that each compound is tested on a panel of 60
cancer cell lines, a vector of data results. The lower outliers
were identified as any GI% data below the Tukey's lower boundary, a
threshold (TLi) based on the interquartile range (IQR) and the
first quartile (Q1) using the following formula:
IQR=Q3−Q1TLi=Q1−1.5×IQR
where ‘I’ represents the compound number, Q1 and Q3
represent first and third quartiles in the data vector of each
compound, while IQR represents the difference between Q3 and Q1.
The presence of a low outlier indicates that the corresponding cell
is specifically sensitive to the tested compound.
The selectivity score (O1D) was
calculated as the number of GI% values below the TLi value reported
to the total number of GI% values recorded for each scaffold. The
corresponding mathematical formula is as follows:
O1D=100×No. of cell lines GI%≤TLiNo. of
cell lines
The same formulas were adjusted to calculate similar
scores using the pGI data. In the case of the performance score
(PpGI), the counted values where those >5 and in the
case of the selectivity score, the counted values were those above
the Ti fence.
ApGI=∑pGINo. of cell linesPpGI=100×No. of
cell lines pGI>5No. of cell linesOpGI=100×No. cell lines
pGI≥TUiNo. of cell lines
where TUi represents the upper boundary for outlier
identification and it is calculated using the following
formula:
TUi=Q3+1.5×IQR
In order to reduce false high-positive values caused
by a low number of tested cells, all the scores were analyzed only
if the total number of data points registered for a specific
structure was >500.
Results
Creation of data sets
A total of 284,176 structures were downloaded from
the DTP website and were manually curated by eliminating 119
erroneous cases. From this set, a total of 192,619 structures were
eliminated as no GI% or pGI50 value were associated with
the corresponding NSC code. The set of compounds with GI% values
resulted after the analysis of the one-dose set and represents
45,020 unique substances. The compounds with no associated
structure were eliminated, yielding a final set of 44,960 compounds
(set GI1D). The pGI50 set contains 52,769
compounds. The compounds with pGI50 values expressed as
µg/ml were eliminated, resulting in a final set of 51,968 compounds
(set PGI). The third set (AL) consists of the union of the 2
sets.
AL=GI1D∪PGI
The AL set consists of 91,438 unique compounds
characterized by an NSC code, a chemical structure, and at least
one biological endpoint (GI%, pGI, or both).
Bemis-Murcko scaffolds
The structures of the compounds from all 3 data sets
were transformed into their Bemis-Murcko skeleton using the
procedure described in the methods section. The transformation of
all the 91,438 structures in the AL set resulted in 11,763 distinct
Bemis-Murcko scaffolds. For a number of 4,106 compounds
representing 4.49% of the set, the results were blank as they did
not contain any ring structures.
Despite the high diversity of scaffolds, only 86 of
these appear with a frequency above the 0.1% threshold. The
distribution of the scaffolds in all 91,438 structures follows the
power law (R2 = 0.961) with the frequency of occurrence
for the i-ranked scaffold obeying the following formula:
F(i)=7.066×i−0.972
The same type of distribution was observed in a
study on the distribution of Bemis-Murcko scaffolds in the organic
subset Chemical Abstracts Service Registry (CAS Registry) (28). There are two main reasons for such a
distribution: Popularity or performance; however, the distribution
alone is not sufficient to understand which of these factors causes
it (29). In order to provide an
answer to this question, the performance and selectivity scores
were implemented.
The top-ranking 10 scaffolds are presented in
Table I along with their frequency
and corresponding scores A1D, ApGI,
P1D, PpGI, O1D and
OpGI.
 | Table I.Most commonly used Bemis-Murcko
scaffolds and their anti-proliferative performance scores. |
Table I.
Most commonly used Bemis-Murcko
scaffolds and their anti-proliferative performance scores.
The results clearly demonstrate that the most
popular Bemis-Murcko scaffolds provide little guarantee that the
compounds containing them will determine sufficient
anti-proliferative effects. The BM10 scaffold presents the optimal
scores amongst the top 10 used ones. The scores registered for the
non-cyclic compounds are better than those of some BM scaffolds,
indicating that the presence of a cyclic structure is not
essential.
For all the compounds in the PGI set, the
Bemis-Murcko analysis returned 9,540 scaffolds. For each scaffold,
the aforementioned scores were calculated. The values were analyzed
only if the total number of data points used was >500. A total
of 662 scaffolds met this criterion. Analyzing the ApGI
values, only 142 scaffolds (21.45%) had values >5, and only 28
scaffolds (4.23%) presented ApGI values >6. Of these
28 scaffolds, apart from one two-ringed skeleton, all had between 3
to 7 rings, and at least 14 carbon atoms. Similarly, based on the
PpGI values, only 121 of the scaffolds produced at least
a 50% growth inhibition in at least 50% of the tested cells
(PpGI >50). Taking both scores into account, the
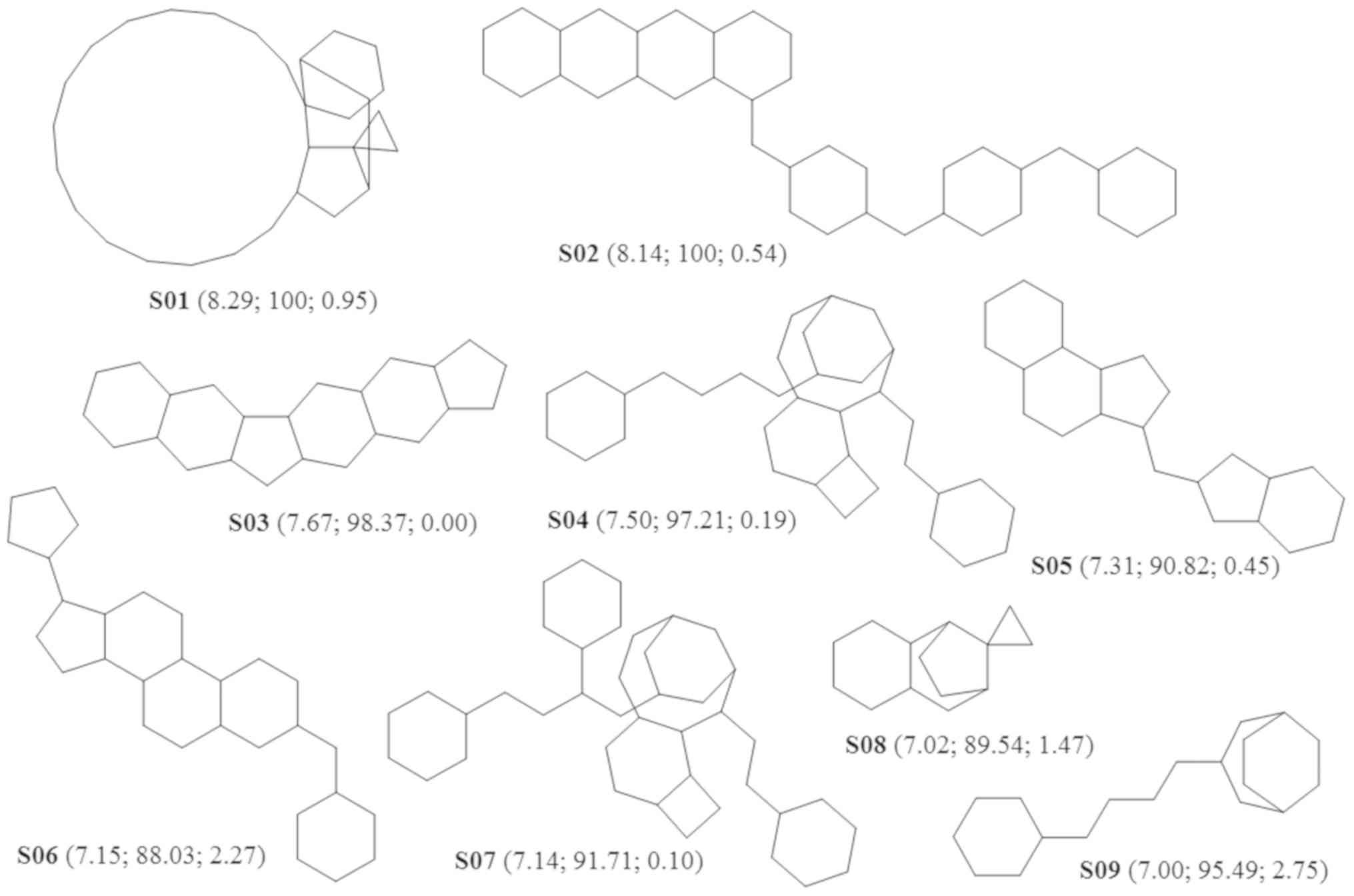
optimal Bemis-Murcko skeletons are presented in Fig. 2.
The results indicated that the most successful
Bemis-Murcko scaffolds had a very large complexity, which explains
their limited use in drug design. It seems that the simple chemical
structures are preferred, even if their potential to generate good
leads is weak.
Plain ring global analysis
The analysis of all of the 91,438 structures in the
AL set resulted in 10,074 distinct ring scaffolds. A total of
30,577 compounds (33.44%) had 1 ring, 31,705 (34.67%) contained 2
plain rings in their structure and 18.47% contained 3 rings.
Considering the frequency of appearance, the first ranked ring was
benzene, which had a very high recurrence being present in 45.50%
of the whole AL set, while the second-ranked ring (pyridine) had a
frequency of 3.18%.
Only 73 ring scaffolds registered a frequency
>0.1%. Based on the number of ring closures in each scaffold,
the 1-ring scaffolds appear in 70.33% of compounds, followed by the
2-ring scaffolds (16.82%) and the 3-ring ones (5.94%).
The analysis of the anti-proliferative potential of
each plain ring was performed firstly at a global level, taking
into account all occurrences of a certain ring in the analyzed data
set. This method ignores the synergistic or antagonistic effect
produced by the presence in the same molecule of other rings. The
top most frequently used scaffolds (A01-A20) are presented in
Table II along with their
frequency (in descending order) and corresponding global scores
A1D, ApGI, P1D, PpGI,
O1D, and OpGI.
 | Table II.Most commonly used scaffolds and
their corresponding global scores. |
Table II.
Most commonly used scaffolds and
their corresponding global scores.
The analysis of the A1D and
ApGI scores indicated that of these top favorite rings,
the optimal anti-proliferative effects were produced by quinoline
(A08), tetrahydropyran (A05), benzimidazole (A11) and pyrazole
(A14). The same 4 rings exhibited the optimal P1D and
PpGI scores, even if the ranking order differed
slightly. The outlier type scores, O1D and
OpGI, exhibited similar values for all 20 rings. The
importance of these rings can be also observed by comparing their
scores with the corresponding Bemis-Murcko structures, such as BM05
in the case of pyrazole, BM02 for quinoline, BM04 for benzimidazole
and BM01 for tetrahydropyran, highlighting the weight of the
heteroatoms nature over the general topology.
The present study subsequently focused on the PGI
set, as the compounds in this set have a higher potency. The
analysis returned 8,725 distinct rings. The scores were analyzed if
the total number of data points used was >500, yielding 685
rings.
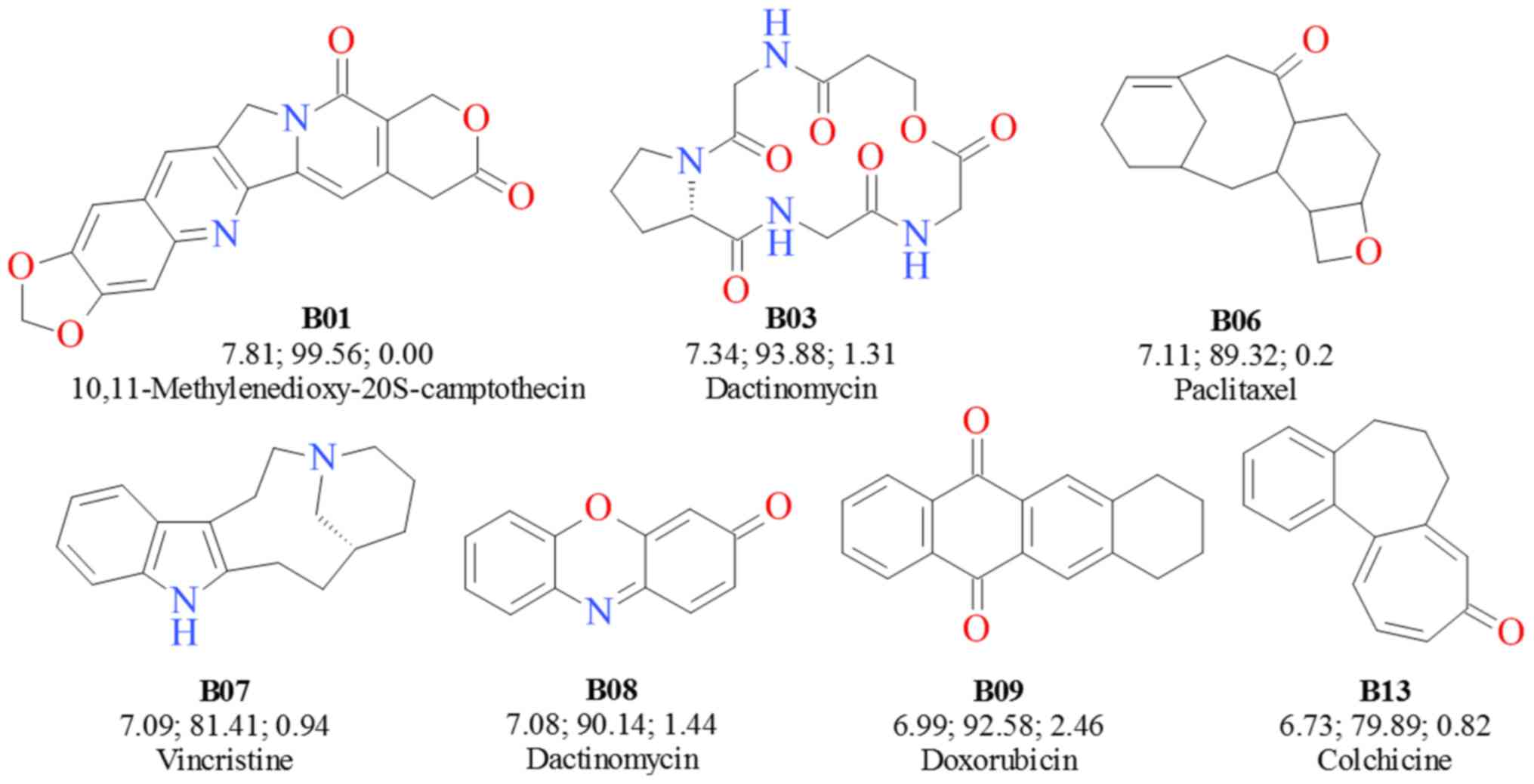
The structural analysis focused on the 40 rings
(B01-B40) that had both ApGI values >6 and
PpGI >60. The majority of these rings have complex
structures and originate from natural compounds with
well-established anticancer properties. Some representative ring
structures are presented in Fig. 3
together with their ApGI, PpGI, and
OpGI scores and with examples of well-known drugs that
feature them.
Plain ring independent analysis. The analysis of the
global effect of each ring structure ignored that the
anti-proliferative effect can be influenced by the presence in the
molecular structure of other rings. In order to better evaluate the
contribution of each ring structure, the analysis was performed
only on the 30,577 compounds of the AL set that contain only one
ring in their structure. The majority of the A01-A20 rings
presented lower performance scores in the independent analysis when
compared with the global approach. The most significant difference
was observed in the case of the quinoxaline structure (A18), which
has a PpGI value of 39.94 and an ApGI value
of 4.89 in compounds that have only one ring vs. a PpGI
value of 15.90 and an ApGI value of 4.48 in all the
compounds. These results indicate that the association of the
quinoxaline ring with other rings lowers the anticancer
potential.
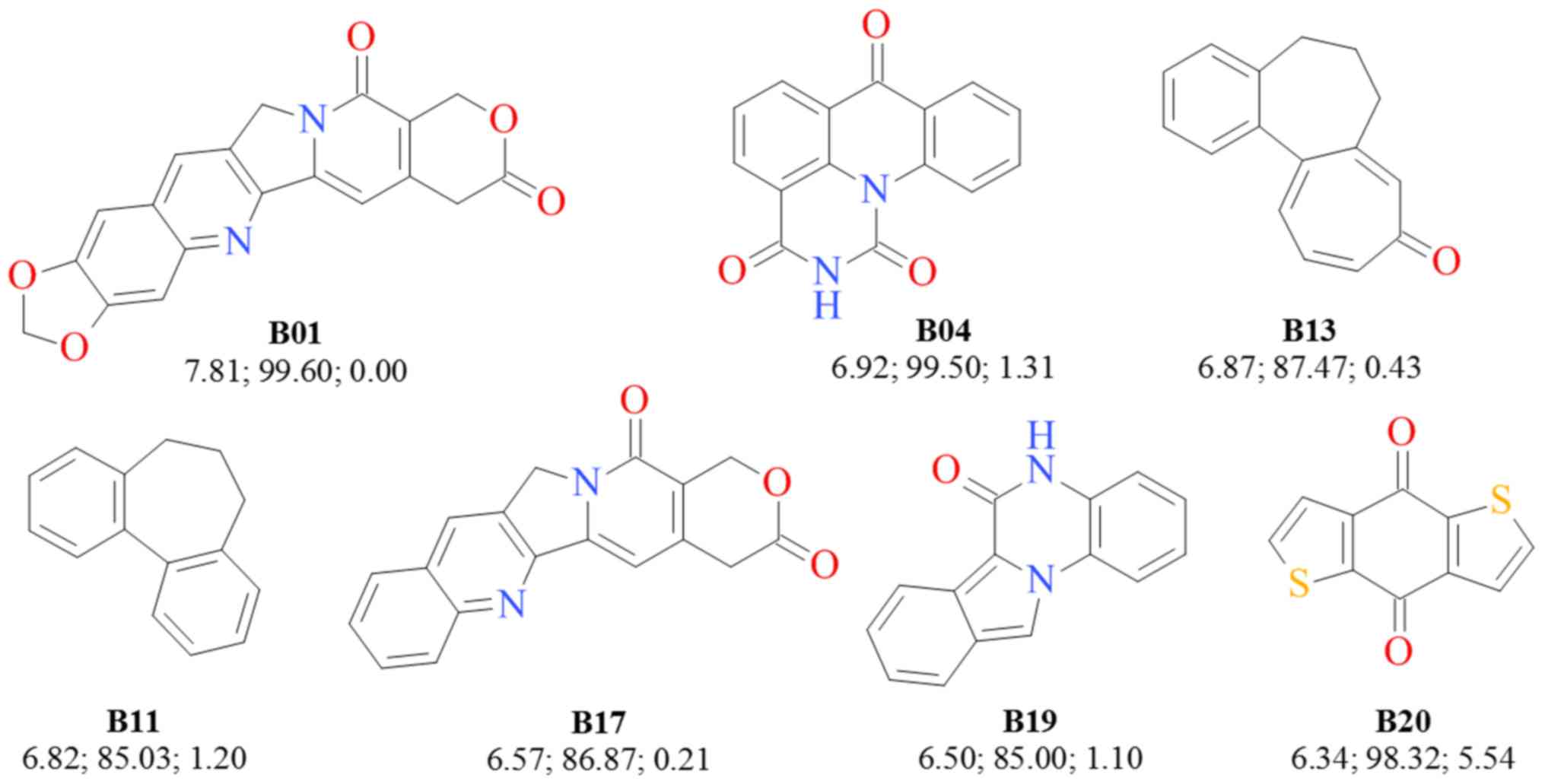
Analyzing the optimal PpGI and
ApGI scores for the independent effect of each ring for
which the 500 data point rule was respected, yielded as the optimal
results, the structures presented in Fig. 3. It can be observed that these 7
rings presented in Fig. 4 belong to
the B01-B40 set, but not all B01-B40 rings exhibited significant
score values in the independent analysis.
The most interesting rings appear to be B11, B13 and
B20. The ring B13 is the skeleton of colchicine, a tricyclic
alkaloid extracted from the plant Colchicum autumnale that
strongly inhibits cellular mitosis by binding tubulin (30), while B11 is a bioisosteric
derivative of B13 found in the tubulin-binding allocolchicine
(31). Based on the OpGI
score, the 4,8-dihydrobenzodithiophene-4,8-dione structure (B20)
may be used to develop novel potent, yet selective,
anti-proliferative drugs.
Discussion
The scope of present study was to perform an
objective analysis of the impact of ring structures on the
anti-proliferative effects in drug design. The results indicate
that medicinal chemistry specialists focus their research on simple
scaffolds, even if most of these have no significant pay-off
guarantees. The reason is probably that the optimal chemical
structures are complex and difficult to synthesize, and are
therefore associated with high research costs. The use of a simple
scaffold is based on the low-risk approach and is very similar to
the repurposing strategy (32).
However, the present study has some limitations, considering the
use of only one chemical repository. Anticancer research is far
more complex and the NCI data represent just one sample.
Drug repurposing or drug repositioning is the
identification of novel therapeutic uses for known drugs as an
alternative to the long and expensive drug development programs
beginning from scratch (33–35).
It is considered that the same thinking paradigm is used for
heterocyclic rings. The repurposing of chemical scaffolds is based
on the privileged structure concept. These types of structures are
chemically accessible, have flexible and potent binding affinities
towards a large type of biological targets, and possess good
drug-like properties (36).
If the privileged structure concept is focused on
the assessment of the potential for a particular ring structure to
interact with a number of targets, the present study was centered
on the objective evaluation of the anti-proliferative potential of
cyclic structures as scaffolds for anticancer drug design,
regardless of their mechanisms of action. The scoring method in the
present study provides an important new tool that could be
successfully combined with the privileged structure analysis
methods to identify better scaffolds. For example, the quinoline
ring (A08) emerged in the present study as an effective ring and
has been demonstrated as a privileged scaffold in cancer drug
development (37,38). The pyrazole ring (A14) is another
privileged structure, particularly when targeting protein kinases
(39,40), which can generate interesting leads
providing that it is joined by other heterocyclic structures.
The present study used a dual strategy, a
topological method using Bemis-Murcko scaffolds to assess the
importance of rings interconnection networks (39), and a plain ring analysis to reveal
the importance of specific heteroatoms and their relative position.
In some cases, the results of both methods were similar. One
important example is in the case of taxene derivatives. Paclitaxel
is represented by the Bemis-Murcko scaffold S07 and by the plain
ring B06, both with ApGI scores >7. Docetaxel, a
congener of paclitaxel, shares the ring B06, but is represented by
the more potent Bemis-Murcko scaffold S04. Another important
observation is the practice of structural simplification, a method
of molecular truncation used to avoid large and low drug-like
scaffolds (41).
In conclusion, the scoring method developed and
implemented herein can easily be used in any other similar research
based on the NSC data. The method could focus on a particular
cyclic structure and it can be used to find other ring structures
with a similar inhibition profile. Even if the method used in the
present study ignored the effects of substituents on the cyclic
structures, the results clearly indicate that the nature of the
ring structure can have a significant impact on the anticancer
potential. Likewise, this method can be used in future research to
quantify the impact of the ring structure compared to that of the
substituents nature or position.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
Data sharing is not applicable to this article, as
no datasets were generated or analyzed during the current
study.
Authors' contributions
GMN was involved in the conceptualization of the
study. All authors (GNDI, OTO, GN, IIO, AT, TIB, DAS and GMD) were
involved in the study methodology. All authors have read and
approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
DAS is the Editor-in-Chief for the journal, but had
no personal involvement in the reviewing process, or any influence
in terms of adjudicating on the final decision, for this article.
All the other authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
CAS Registry
|
Chemical Abstracts Service
Registry
|
|
GI50
|
half maximal growth inhibition
|
|
IQR
|
interquartile range
|
|
LC50
|
half maximal lethal concentration
|
|
NCI
|
National Cancer Institute
|
|
NSC Number
|
Cancer Chemotherapy National Service
Center number
|
|
TGI
|
total growth inhibition
|
References
|
1
|
Schirrmacher V: From chemotherapy to
biological therapy: A review of novel concepts to reduce the side
effects of systemic cancer treatment (Review). Int J Oncol.
54:407–419. 2019.PubMed/NCBI
|
|
2
|
Ferreira LG, Dos Santos RN, Oliva G and
Andricopulo AD: Molecular docking and structure-based drug design
strategies. Molecules. 20:13384–13421. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Śledź P and Caflisch A: Protein
structure-based drug design: From docking to molecular dynamics.
Curr Opin Struct Biol. 48:93–102. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lionta E, Spyrou G, Vassilatis DK and
Cournia Z: Structure-based virtual screening for drug discovery:
Principles, applications and recent advances. Curr Top Med Chem.
14:1923–1938. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chen KC, Juang SH and Lien JC:
Identification of antiproliferative emodin analogues as inhibitors
of epidermal growth factor receptor in cancer. Int J Mol Med.
43:1281–1288. 2019.PubMed/NCBI
|
|
6
|
Martin YC, Kofron JL and Traphagen LM: Do
structurally similar molecules have similar biological activity? J
Med Chem. 45:4350–4358. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sun H, Tawa G and Wallqvist A:
Classification of scaffold-hopping approaches. Drug Discov Today.
17:310–324. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Gomez L: Decision Making in Medicinal
Chemistry: The Power of Our Intuition. ACS Med Chem Lett.
9:956–958. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Segall M and Chadwick A: The risks of
subconscious biases in drug-discovery decision making. Future Med
Chem. 3:771–774. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Sharma PC, Bansal KK, Sharma A, Sharma D
and Deep A: Thiazole-containing compounds as therapeutic targets
for cancer therapy. Eur J Med Chem. 188:1120162020. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Dadashpour S and Emami S: Indole in the
target-based design of anticancer agents: A versatile scaffold with
diverse mechanisms. Eur J Med Chem. 150:9–29. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Mazumder A, Cerella C and Diederich M:
Natural scaffolds in anticancer therapy and precision medicine.
Biotechnol Adv. 36:1563–1585. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Munteanu AC, Badea M, Olar R, Silvestro L,
Dulea C, Negut CD and Uivarosi V: Synthesis and Structural
Investigation of New Bio-Relevant Complexes of Lanthanides with
5-Hydroxyflavone: DNA Binding and Protein Interaction Studies.
Molecules. 21:17372016. View Article : Google Scholar
|
|
14
|
Ismail NSM, Ali EMH, Ibrahim DA, Serya RAT
and Abou El Ella DA: Pyrazolo[3,4-d]pyrimidine based scaffold
derivatives targeting kinases as anticancer agents. Futur J Pharm
Sci. 2:20–30. 2016. View Article : Google Scholar
|
|
15
|
Nitulescu GM, Matei L, Aldea IM, Draghici
C, Olaru OT and Bleotu C: Ultrasound-assisted synthesis and
anticancer evaluation of new pyrazole derivatives as cell cycle
inhibitors. Arab J Chem. 12:816–824. 2019. View Article : Google Scholar
|
|
16
|
Nitulescu GM, Draghici C, Olaru OT, Matei
L, Ioana A, Dragu LD and Bleotu C: Synthesis and apoptotic activity
of new pyrazole derivatives in cancer cell lines. Bioorg Med Chem.
23:5799–5808. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Nitulescu GM, Draghici C and Olaru OT: New
potential antitumor pyrazole derivatives: Synthesis and cytotoxic
evaluation. Int J Mol Sci. 14:21805–21818. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Nitulescu GM, Paunescu H, Draghici C,
Missir AV, Coman OA and Fulga I: Synthesis and pharmacological
evaluation of some new pyrazole derivatives. Farmacia. 58:190–197.
2010.
|
|
19
|
Takimoto CH: Anticancer drug development
at the US National Cancer Institute. Cancer Chemother Pharmacol. 52
(Suppl 1):S29–S33. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
DeVita VT Jr and Chu E: A history of
cancer chemotherapy. Cancer Res. 68:8643–8653. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kim HS, Sung YJ and Paik S: Cancer Cell
Line Panels Empower Genomics-Based Discovery of Precision Cancer
Medicine. Yonsei Med J. 56:1186–1198. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Shoemaker RH: The NCI60 human tumour cell
line anticancer drug screen. Nat Rev Cancer. 6:813–823. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zaharevitz DW, Holbeck SL, Bowerman C and
Svetlik PA: COMPARE: A web accessible tool for investigating
mechanisms of cell growth inhibition. J Mol Graph Model.
20:297–303. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Fang X, Shao L, Zhang H and Wang S:
Web-based tools for mining the NCI databases for anticancer drug
discovery. J Chem Inf Comput Sci. 44:249–257. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Nitulescu GM, Soriga SG, Socea LI, Olaru
OT and Plesu V: Structure-activity relationships and
chemoinformatic analysis of the anticancer profile of an
aminopyrazole derivative. Rev Chim. 67:162–165. 2016.
|
|
26
|
Nitulescu GM, Iancu G, Nitulescu G, Iancu
RC, Bogdanici C and Vasile D: Brave new hope for breast cancer:
Aminopyrazole derivates between rational design and clinical
efficacy. Rev Chim. 68:754–757. 2017. View Article : Google Scholar
|
|
27
|
Herbowski L: Skeletal muscle metastases
from papillary and follicular thyroid carcinomas: An extensive
review of the literature. Oncol Lett. 15:7083–7089. 2018.PubMed/NCBI
|
|
28
|
Lipkus AH, Yuan Q, Lucas KA, Funk SA,
Bartelt WF III, Schenck RJ and Trippe AJ; CAS Registry, :
Structural diversity of organic chemistry. A scaffold analysis of
the CAS Registry. J Org Chem. 73:4443–4451. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Néda Z, Varga L and Biró TS: Science and
Facebook: The same popularity law! PLoS One. 12:e01796562017.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ahmed K, Zaidi SF, Cui Z-G, Zhou D, Saeed
SA and Inadera H: Potential proapoptotic phytochemical agents for
the treatment and prevention of colorectal cancer. Oncol Lett.
18:487–498. 2019.PubMed/NCBI
|
|
31
|
Paymode DJ and Ramana CV: Total Synthesis
of (±)-Allocolchicine and Its Analogues Using Co-Catalyzed Alkyne
[2 + 2 + 2]-Cyclotrimerization. ACS Omega. 2:5591–5600. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Armando RG, Mengual Gómez DL and Gomez DE:
New drugs are not enough drug repositioning in oncology: An update.
Int J Oncol. 56:651–684. 2020.PubMed/NCBI
|
|
33
|
Pushpakom S, Iorio F, Eyers PA, Escott KJ,
Hopper S, Wells A, Doig A, Guilliams T, Latimer J, McNamee C, et
al: Drug repurposing: Progress, challenges and recommendations. Nat
Rev Drug Discov. 18:41–58. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Ion GND, Mihai DP, Lupascu G and Nitulescu
GM: Application of molecular framework-based data-mining method in
the search for beta-secretase 1 inhibitors through drug
repurposing. J Biomol Struct Dyn. 37:3674–3685. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Yang X, Huang W-T, Wu H-Y, He R-Q, Ma J,
Liu A-G and Chen G: Novel drug candidate for the treatment of
several soft tissue sarcoma histologic subtypes: A computational
method using survival associated gene signatures for drug
repurposing. Oncol Rep. 41:2241–2253. 2019.PubMed/NCBI
|
|
36
|
Schneider P and Schneider G: Privileged
Structures Revisited. Angew Chem Int Ed Engl. 56:7971–7974. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Musiol R: An overview of quinoline as a
privileged scaffold in cancer drug discovery. Expert Opin Drug
Discov. 12:583–597. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Afzal O, Kumar S, Haider MR, Ali MR, Kumar
R, Jaggi M and Bawa S: A review on anticancer potential of
bioactive heterocycle quinoline. Eur J Med Chem. 97:871–910. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Cui YJ, Tang LQ, Zhang CM and Liu ZP:
Synthesis of Novel Pyrazole Derivatives and Their Tumor Cell Growth
Inhibitory Activity. Molecules. 24:2792019. View Article : Google Scholar
|
|
40
|
Nitulescu GM, Nedelcu G, Buzescu A and
Olaru OT: Aminopyrazoles as privileged structures in anticancer
drug design - an in silico study. Izv Him. 48:55–60. 2016.
|
|
41
|
Wang S, Dong G and Sheng C: Structural
simplification: An efficient strategy in lead optimization. Acta
Pharm Sin B. 9:880–901. 2019. View Article : Google Scholar : PubMed/NCBI
|