Introduction
Chemotherapy, as the first-line of treatment or
adjuvant treatment for tumors, often delivers mixed outcomes
(1). The primary reason for this is
drug resistance encountered during chemotherapy. Overcoming drug
resistance by tumors is a basic and challenging initiative to
resolve the insensitivity of tumors toward chemotherapy. Recent
studies have focused on microRNAs (miRNAs) as a tool to suppress
tumorigenesis, which is expected to assist in the development of
effective chemotherapy drugs (2).
miRNAs are endogenous non-coding RNAs with an
approximate length of 23 nucleotides, which can regulate the
expression of target genes via interaction with its 3′-untranslated
regions (3). miRNAs are involved in
diverse biological and pathological processes, including the
generation and development of cancer. They also act on target genes
to inhibit tumor resistance to chemotherapy drugs (4). Various known mechanisms of miRNA
regulation include the overexpression of multidrug resistance (MDR)
transporters, defects in cell-cycle and the apoptotic machinery,
induction of autophagy, alternation of anticancer drug metabolism,
alteration in drug targets, DNA repair, and disruption of redox
homeostasis. Among these, autophagy regulation is one of the most
critical mechanisms (5) that
demonstrates an important mechanism to overcome chemotherapy
resistance of various tumors (6).
In 2016, Yoshinori Ohsumi won the Nobel Prize in
physiology or medicine in recognition of his outstanding
achievements in emphasizing the importance of autophagy as the key
player in understanding human diseases. Autophagy is the
quality-control mechanism of a cell to regulate its cellular
homeostasis (7). It plays a dual
role in tumorigenesis, either as a promoter or suppressor of tumors
(8). Recent studies, however, have
focused on the protective effect of autophagy on tumor cells and
have further strengthened the belief that autophagy can promote
apoptosis of tumor cells and inhibit their proliferation (6). A large number of molecules are
involved in each step of the autophagic pathway (6,9–11)
(Fig. 1), and, during this process,
miRNAs target some of these molecules and thereby participate in
the generation of tumor chemotherapy resistance.
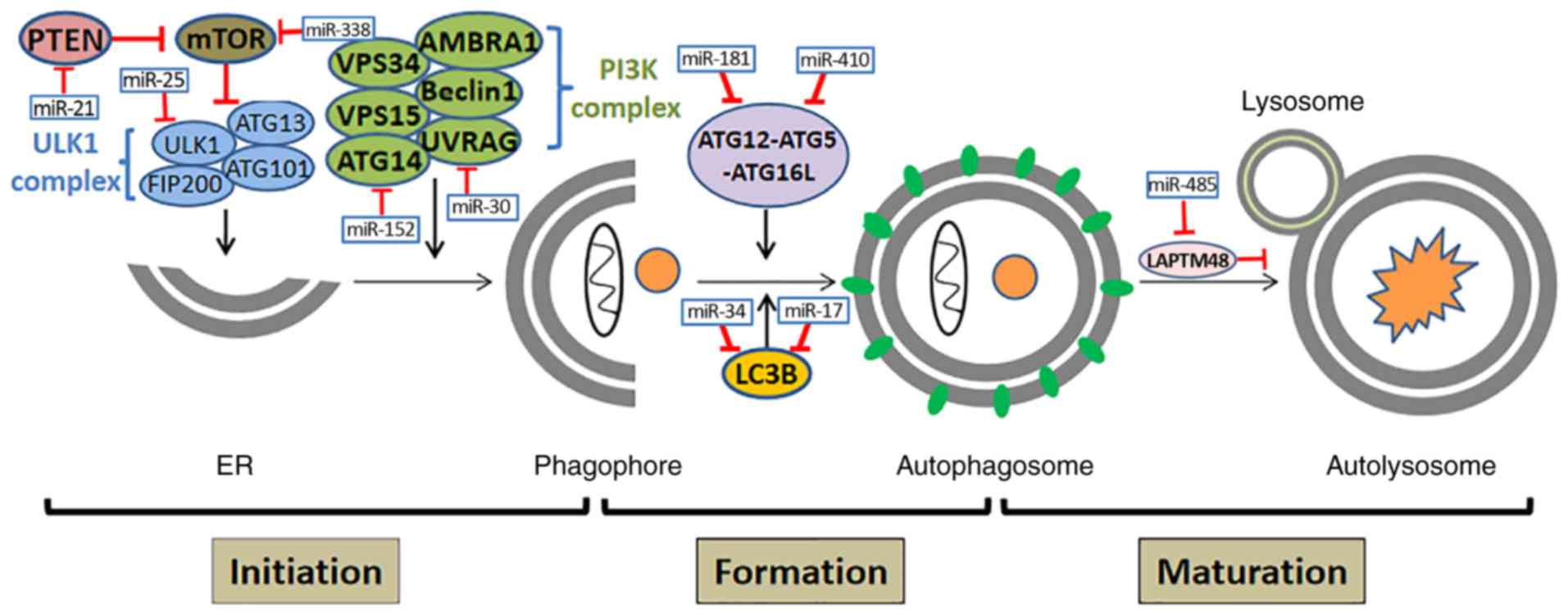 | Figure 1.The molecules involved in each step
of the autophagy process. Autophagy initiation depends on two vital
complexes: The ULK1 complex (ULK1, FIP200, ATG13, ATG101) and PI3K
complex (VPS34, VPS15, ATG14, AMBRA1, Beclin1, UVRAG). mTOR plays a
suppressive role in the initiation of autophagy; autophagosome
formation involves the formation of a complex of ATG12-ATG5-ATG16L
and LC3B; other molecules participate in the maturation of the
autolysosome. Some key miRNAs regulate their target genes to
participate in every stage of autophagy. ULK1, UNC-51-like
autophagy-activating kinase 1; PI3K, the class III PI 3-kinase;
FIP20, focal adhesion kinase family interacting protein of 200 kDa;
ATG, autophagy-related gene; ABCG2, ATP-binding cassette sub-family
G member 2; VPS, vacuolar protein sorting; AMBRA1, autophagy and
Beclin-1 regulator 1; UVRAG, UV resistance-associated gene; LC3B,
microtubule-associated protein light chain 3; LAPTM4B,
lysosome-associated protein transmembrane 4 β; EGR1, early growth
response 1; HMGB1, high-mobility group box 1; PTEN, phosphatase and
tension homolog. |
The present review summarizes some of the relevant
works in recent years and their perspective on the role of
miRNA-mediated autophagy in tumor chemotherapy resistance.
miRNA-mediated autophagy changes with
respect to drug resistance in tumor treatment
miRNA-mediated autophagy initiation
changes the pattern of drug resistance in tumors
Autophagy initiation involves 2 major complexes: The
UNC-51–like autophagy-activating kinase (ULK)1 complex and the
class III PI 3-kinase (PI3K) complex (12). Some miRNAs target any one or more of
the above-mentioned 2 compounds or their inhibitors and trigger
autophagy alteration, thereby changing the tumor drug resistance
pattern (Table I).
 | Table I.miRNA-targets related to autophagy
initiation. |
Table I.
miRNA-targets related to autophagy
initiation.
| A, ULK1
complex |
|---|
|
|---|
| miRNAs | miRNA
expression | Targets | Target
expression | Autophagy
level | Cancer | Chemotherapy
resistance |
|---|
| miR-26a/b | Up | ULK1 | Down | Down | HCC | Down |
| miR-106a | Up | ULK1 | Down | Down | Lung
adenocarcinoma | Down |
| miR-25 | Down | ULK1 | Up | Up | Breast cancer | Up |
| miR-409-3p | Up | Fip200 | Down | Down | Ovarian cancer | Down |
|
| B, PI3K
complex |
|
| miRNAs | miRNA
expression | Targets | Target
expression |
Autophagylevel | Cancer | Chemotherapy
resistance |
|
| miR-140-5p | Up | ATG14 | Down | Down | Myeloma | Down |
| miR-152 | Up | ATG14 | Down | Down | Ovarian cancer | Down |
| miR-30a | Up | Beclin-1 | Down | Down | Osteosarcoma | Down |
| miR-30d | Down | Beclin-1 | Up | Up | UTC | Up |
| miR-216b | Up | Beclin-1, UVRAG and
ATG5 | Down | Down | Melanoma | Down |
| miR-409-3p | Up | Beclin-1 | Down | Down | Colon cancer | Down |
| miR-216b | Up | Beclin-1 | Down | Down | NSCLC | Down |
|
| C, mTOR and mTOR
pathway |
|
| miRNAs | miRNA
expression | Targets | Targets
expression | Autophagy
level | Cancer | Chemotherapy
resistance |
|
| miR-338-3p | Up | mTOR | Down | Up | Colon cancer | Up |
| miR-193-5p | Up | mTOR | Down | Up | Glioma | Up |
| miR-155 | Up | PTEN | Down | Down | Osteosarcoma | Down |
| miR-21 | Down | PTEN | Up | Up | Breast cancer | Up |
| miR-21 | Up | PTEN | Down | Down | HCC | Down |
| miR-142-3p | Up | HMGB1 | Down | Up | NSCLC | Up |
| miR-410-3p | Up | HMGB1 | Down | Up | PDAC | Up |
| miR-218 | Up | HMGB1 | Down | Up | Endometrial
cancer | Up |
| miR-22 | Up | HMGB1 | Down | Up | Osteosarcoma | Up |
| miR-34a | Up | HMGB1 | Down | Up | Retinoblastoma | Up |
The ULK1 complex includes ULK1, autophagy-related
gene (ATG)13 protein, focal adhesion kinase family interacting
protein of 200 kDa (FIP200) and ATG101 in humans. ULK1 is a part of
a family of ULK1-4 kinases that play an important role in autophagy
(12). ULK1 is targeted by
miRNA-26a/b, and the in vitro overexpression of ULK1
promotes apoptosis by inhibiting autophagy; moreover, the
overexpression of miRNA-26a/b has been shown to enhance the
sensitivity of hepatocellular carcinoma cells (HCC) to doxorubicin
in vivo in xenograft models of nude mice (13). In another report, ULK1-mediated
autophagy protected lung adenocarcinoma cells from the toxic
effects of tyrosine kinase inhibitors (TKIs), and high expression
of miR-106a was found to inhibit ULK1 expression, reduce
autophagy, and increase the cytotoxic effects of TKIs (14). Another study showed that
isoliquiritugenin (ISL), a natural flavonoid, possesses anticancer
properties and that it could inhibit the expression of miR-25,
leading to the upregulation of its target gene ULK1 and an
increase in the extent of autophagy. These events ultimately led to
the accelerated degradation of ATP-binding cassette sub-family G
member 2 (ABCG2) through the autophagy-lysosomal pathway, resulting
in improved toxicity of chemotherapy drugs against breast cancer
cells (15). FIP200, a large
ULK1-interacting protein, has a predicted coiled-coil
protein involved in scaffolding (16). Cheng et al reported that the
expression of miR-409-3p was enhanced in cisplatin-sensitive cells
when compared with cisplatin-resistant cells, and the
overexpression of miR-409-3p decreased the expression of
FIP200 and the related autophagy, thereby increasing the
cisplatin sensitivity of ovarian cancer cells (17).
The PI3K complex occurs downstream of ULK1.
The PI3K complex consists of autophagy and Beclin-1 regulator 1
(AMBRA1), Beclin-1, vacuolar protein sorting 34 (VPS34)
(phosphatidylinositol), VPS15, UV resistance-associated gene
(UVRAG) and ATG14L. ATG14L is a Beclin-1-associated autophagy key
regulator (18). lncRNAs are
non-coding RNA with a length of more than 200 nucleotides. lncRNA
lacks the coding ability, but it is a functional molecule (19) with multiple effects on miRNAs
(20). An lncRNA can be used as an
miRNA sponge as it competes with miRNA to bind to the target mRNA,
and some lncRNAs can also be converted into miRNAs. It was found
that knockdown of Lnc0515 induced the overexpression of miR-140-5p,
which targets ATG14, one of the components of the PI3K
complex. This event eventually caused inhibition of autophagy,
thereby alleviating the chemoresistance of myeloma cells (18). ATG14 is also known to be
targeted by miR-152, while the transcription factor early growth
response 1 (EGR1) regulates miR-152 upstream, the
overexpression of either EGR1 or miR-152 led to the inhibition of
ATG14 expression as well as the inhibition of autophagy,
eventually increasing the cisplatin toxicity to ovarian cancer
cells and thereby weakening drug resistance (21). Luciferase reporter assay identified
that several miRNAs, such as miR-30a, which directly binds to the
3′-UTR of Beclin-1, inhibited Beclin-1 expression. When
miR-30a was overexpressed, Beclin-1-mediated autophagy was
inhibited, which not only promoted the chemotherapy-induced
apoptosis of osteosarcoma cells but also enhanced the toxicity of
sorafenib to renal cell carcinoma (RCC) cells (22,23).
The sensitivity of undifferentiated thyroid carcinoma (UTC) to
cisplatin is heterogeneous, which may be related to the
downregulation of miR-30d; both in vivo and in vitro
experiments confirmed that the downregulation of miR-30d could
target and promote Beclin-1 expression, leading to an
increase in autophagy and in the insensitivity of UTC cells to
cisplatin (24). miR-216b was found
to target Beclin-1, UVRAG, and ATG5, 3 essential autophagy
genes, directly to weaken autophagy and enhance the antitumor
activity of the drug vemurafenib in BRAF (V600E) melanoma cells
(25). In addition, Tan et
al reported that the overexpression of miR-409-3p inhibited
Beclin-1 expression and autophagy, and thus enhanced the
chemosensitivity of colon cancer cells to the drug oxaliplatin
in vivo and in vitro (26). Other research also noted a reduction
in the expression level of miR-216b in paclitaxel-treated non-small
cell lung carcinoma (NSCLC) cells. As the expression level of
miR-216b increased, the Beclin-1 mRNA translation level decreased,
autophagy was inhibited, and the toxicity of paclitaxel to NSCLC
was enhanced (27). High-mobility
group box 1 (HMGB1) is a type of non-histone protein that has been
widely reported to play a key role in the induction of autophagy.
Several studies have suggested that HMGB1 binds to the PI3K complex
to facilitate autophagic progression (28–33).
miR-142-3p was found to directly downregulate HMGB1, inhibit
autophagy induced by anticancer drugs, and increase the
chemosensitivity of NSCLC in both in vitro and in
vivo experiments (29). In
vivo and in vitro experiments showed that the expression
of miR-410-3p was downregulated in tumor cells and in transplanted
tumor tissues with chemotherapy-resistance to gemcitabine; after
intervention, high expression of miR-410-3p was targeted to inhibit
HMGB1 and its induced autophagy, which resulted in enhanced
sensitivity of chemotherapy (30).
HMGB1 is also the target gene of miR-218, which binds to the
3′-UTR of this gene, inhibits the expression of the target gene,
inhibits autophagy, and was found to improve the sensitivity of
endometrial cancer cells to paclitaxel chemotherapy (31). Similarly, miR-22 and miR-34a were
also found to target HMGB1 and inhibit its expression,
inhibit autophagy, and restore the sensitivity of osteosarcoma and
retinoblastoma cancer cells to chemotherapy drugs (32,33).
mTOR, a serine/threonine kinase, is a master
regulator of cellular metabolism and the key regulator of
autophagy. It forms 2 complexes: mTORC1 and mTORC2; the former
inhibits autophagy-initiation by preventing the formation of ULK1
and PI3K complexes (34).
miR-338-3p was found to increase autophagy by downregulating the
expression of its target mTOR, leading to the resistance of
colon cancer cells containing P53 mutants to 5-fluorouracil
(35). Downregulation of
lncRNA-CASC2 in glioma resulted in an increase in the miR-193a-5p
expression level and a decrease in the mTOR expression, which
further induced protective autophagy, resulting in temozolomide
resistance (36). The
PI3K/PTEN/Akt/mTOR signaling pathway has been implicated in
the resistance to chemoradiotherapy (37). Phosphatase and tension homolog
(PTEN), which encodes a phosphatase protein, is an essential
tumor-suppressor gene. It activates the mTOR pathway and inhibits
autophagy (38). In MG-63/ADM
osteosarcoma cells, miR-155, p-AKT, and p-mTOR were found to be
overexpressed. PTEN, the target gene of miR-155, is a
positive regulator of autophagy, and the overexpression of miR-155
was found to inhibit PTEN and reduce autophagy induction,
simultaneously. In addition, expression of AKT and
mTOR was found to activate the PI3K/AKT/mTOR
signaling pathway and inhibit autophagy, which in turn led to
increased adriamycin resistance in tumor cells (39). The knockdown of miR-21 expression in
breast cancer cells was found to lead to an increase in the
expression of PTEN and the inhibition of the activation of
AKT. This event blocked the PI3K/AKT/mTOR signaling
pathway, increased autophagic cell death, and partially overcame
the endocrine resistance of tumor cells to tamoxifen and
fulvestrant (40). Another study
reported that miR-21 expression was higher in HCC cells resistant
to sorafenib, which resulted in decreased expression of
PTEN, increased activation of the AKT/mTOR pathway,
and significant inhibition of autophagy (41).
miRNA-mediated autophagosome formation
changes the pattern of drug resistance in tumors
The two essential complexes of autophagosomes are
ATG12-ATG5-ATG16L and LC3B (42,43).
Other miRNAs target any one or more of the above-mentioned two
compounds, trigger autophagy alteration, and change the response of
the tumor to chemotherapy drugs (Table
II). ATG5-ATG12 conjugation depends on ATG7 (an E1-like
ubiquitin-activating enzyme) and ATG10 (an E2-like ubiquitin
carrier protein), which pairs with ATG16L dimers to form the
ATG5-ATG12-ATG16L complex; this complex associates with the outer
membrane of the extending phagophore (10). By competitively inhibiting
miR-23b-3p, lncRNA MALAT1 was found to increase the expression of
ATG12, which in turn led to increased autophagy and drug
resistance in gastric cancer cells (44). miR-410 was found to reduce
expression of ATG16L1 by directly targeting its 3′-UTR,
subsequently inhibiting autophagy and improving the response of
osteosarcoma cells to chemotherapy drugs (rapamycin, adriamycin,
and cisplatin) (45). ATG5
is an miR-181a target, and overexpression of miR-181a was found to
inhibit ATG5 and increase the sensitivity of gastric cells
SGC7901/CDDP to cisplatin in a mouse model, in which tumor
xenografts were downsized (46).
PU.1-miR-142-3p targets ATG5/ATG16L1. Upregulation of
heterotopic miR-142-3p, was found to reduce sorafenib-induced
autophagy, enhance sorafenib-induced apoptosis, and inhibit cell
growth, enhancing HCC cell sensitivity to sorafenib (47). ATG12 and HMGB2, target
genes of miR-23b-3p, are essential elements of autophagy. Following
miR-23b-3p overexpression, the expression and ATG12 and
HMGB2-mediated autophagy are inhibited, and as a result, the
sensitivity of gastric cancer cells to chemotherapy drugs is
increased (48).
 | Table II.miRNA-targets related to
autophagosome formation. |
Table II.
miRNA-targets related to
autophagosome formation.
| A,
ATG12-ATG5-ATG16L complex |
|---|
|
|---|
| miRNAs | miRNA
expression | Targets | Target
expression | Autophagy
level | Cancer | Chemotherapy
resistance |
|---|
| miR-23b-3p | Down | ATG12 | Up | Up | Gastric cancer | Up |
| miR-410 | Up | ATG16L | Down | Down | Osteosarcoma | Down |
| miR-181a | Up | ATG5 | Down | Down | Gastric cancer | Down |
| miR-142-3p | Up | ATG5/ATG16L | Down | Down | HCC | Down |
| miR-23b-3p | Up | ATG12 | Down | Down | Gastric cancer | Down |
|
| B, LC3B
complex |
|
| miRNAs | miRNA
expression | Targets | Target
expression | Autophagy
level | Cancer | Chemotherapy
resistance |
|
| miR-34a | Up | ATG4B | Down | Down | Prostate
cancer | Down |
| miR-34c-5p | Up | ATG4B | Down | Down | Cervical
cancer | Down |
| miR-375 | Up | ATG7 | Down | Down | Breast cancer | Down |
| miR-520b | Up | ATG7 | Down | Down | HCC | Down |
| miR-17 | Up | ATG7 | Down | Down | NSCLC | Down |
| miR-423-5p | Up | ATG7 | Down | Down | HCC | Down |
| miR-17 | Down | ATG7 | Up | Up | Glioblastoma | Up |
| miR-119a-5p | Up | ATG7 | Down | Down | HCC | Down |
Microtubule-associated protein light chain 3 (LC3B)
is another ubiquitin-like system involved in autophagosome
formation. LC3B is encoded by ATG8 and it is a product of the
cleavage of ProLC3 by ATG4 and ATG7. One previous study showed that
hypermethylation causes downregulation of miR-34a, which results in
the upregulation of miR-34. Demethylated miR-34a targets
ATG4B, thereby downregulating ATG4B-induced autophagy
by activating the AMPK/mTOR pathway, with an increased
sensitivity to chemotherapy in prostate cancer (PCa) cells
(49). THP (pirarubicin) treatment
significantly reduced the size of the transplanted tumor. In
vivo experiments revealed that the overexpression of miR-34c-5p
in cells led to a decrease in the inhibition of autophagy by
ATG4B, which increased the sensitivity to THP (50). Several studies have shown that
multiple miRNAs target ATG7 to mediate autophagy in order to alter
the pattern of drug resistance in tumors. For example, MTT assay
demonstrated that dual treatment of inhibitors of abelson
non-receptor tyrosine kinase (c-ABL) (imatinib) and EGFR
(lapatinib) suppressed breast cancer cell growth and upregulated
miRNA-375; the overexpression of miR-375 decreased the expression
of its target ATG7, which in turn mediated autophagy, which
is beneficial in preventing fulvestrant resistance (51–56).
Apigenin was found to significantly increase adriamycin
sensitivity, induce the overexpression of miR-520b, and inhibit
ATG7-related autophagy in BEL-7402/ADM cells; these results
were verified in a nude mouse xenograft as well as in vitro
(52). The expression level of
lncRNA-XIST (an oncogene in colorectal cancer) was found to be
significantly increased in cisplatin-resistant A549 cells. Hence,
lncRNA-turbulence/miR-17/autophagy may be a target for improving
chemotherapy resistance in NSCLC patients by the knockdown of
lncRNA-XIST to regulate miR-7 expression and inhibit ATG7
expression so as to restore the sensitivity of cisplatin-resistant
A549 cells (53). miR-423-5p is
inactivated by ceramide. A previous study reported that miR-423-5p
downregulates the expression of its target ATG7, leading to
autophagy inhibition. In addition, miR-423-5p was found to
downregulate the expression of ERK to inhibit the
proliferation of cells. Altogether, these results in a previous
study implied that miR-423-5p induced the inhibition of autophagy
and cell proliferation increased the apoptosis of cells and
improved the sensitivity of HCC cells to sorafenib (54). miR-17 was demonstrated to negatively
regulate ATG7, and the downregulation of miR-17 was found to
increase the expression of ATG7. miR-17 also enhanced
autophagy, drug sensitivity of temozolomide, and the sensitivity of
glioblastoma to low-dose ionizing radiation (55). Moreover, miR-199a-5p was found to
specifically inhibit the expression of ATG7, reduce
autophagy, and enhance the inhibitory effect of cisplatin in
regards to the proliferation of HCC cells (56).
miRNA-mediated autolysosome maturation
changes the pattern of drug resistance in tumors
The development from autophagosome to autolysosome
involves a complex system of new lysosomes and several large
molecules that affect this process by altering the lysosomal
activity. Lysosome-associated protein transmembrane 4 β (LAPTM4B)
is one such molecule whose knockdown not only increases the
lysosomal pH and inhibits lysosome and autophagosome fusion
(57) but also causes lysosomal
membrane permeabilization (58).
LAPTM4B was reported as the target of miR-489 and was
downregulated by miR-489. Overexpression of miR-489 was found to
induce LAPTM4B-related autophagy and inhibit drug resistance
in breast cancer cells (59).
LAMP-1 and LAMP-2 are proteins that play a vital role in lysosomal
biogenesis (60). miR-487-5p
targets LAMP2, and knockdown of miR-487-5p was found to markedly
increase levels of LAMP-2, enhance autophagy, and decrease cellular
proliferation, whereas, miR-487b-5p suppression promoted
temozolomide (TMZ) resistance in lung cancer cells (61).
Conclusion and future directions
The regulation of autophagy by non-coding miRNAs has
been extensively researched in the last couple of years. Although a
change in chemotherapy resistance for a single tumor may need the
involvement of several different types of miRNAs, a single miRNA is
capable of imparting chemotherapy resistance in a wide variety of
tumors. Thus, resistance to a tumor chemotherapy drug can develop
through various regulatory mechanisms of miRNAs via different
target genes at every stage of autophagy. However, there are
limitations to the studies on miRNAs and the involvement of
autophagy in drug resistance. On one hand, some studies have
focused on the relationship between an miRNA and its target genes
related to autophagy at the molecular level through in vitro
experiments. In addition, other studies have performed both in
vitro experiments as well as animal models. In some studies,
the experimental verification was confined to tumor
transplantation, but not to human tissues. On the other hand, when
multiple miRNAs are involved in the autophagy process of
tumorigenesis, it is important to explore the specificity of the
miRNA(s) or the specific steps(s) involved by using a large group
of studies. miRNAs are thus regulated by lncRNAs or other upstream
factors, making the regulation process extremely complex, which
necessitates further research on the elaborate mechanisms involved
in this process. Further insight into the exciting and complex
mechanisms of miRNA-regulated autophagy is expected to be valuable
in a variety of therapeutic applications, including the
chemotherapy resistance of various types of tumors.
Acknowledgements
We thank all the individuals who assisted in this
research.
Funding
No funding was received.
Availability of data and materials
All information included in this Review was
supported by relevant references.
Authors' contributions
YH reviewed and edited the manuscript. TH prepared
the original draft and completed the manuscript. ZX and LC designed
the figures and YL completed the document retrieval. All authors
read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Konieczkowski DJ, Johannessen CM and
Garraway LA: A convergence-based framework for cancer drug
resistance. Cancer Cell. 33:801–815. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
An X, Sarmiento C, Tan T and Zhu H:
Regulation of multidrug resistance by microRNAs in anti-cancer
therapy. Acta Pharm Sin B. 7:38–51. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Baek D, Villén J, Shin C, Camargo FD, Gygi
SP and Bartel DP: The impact of microRNAs on protein output.
Nature. 455:64–71. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Rupaimoole R and Slack FJ: MicroRNA
therapeutics: Towards a new era for the management of cancer and
other diseases. Nat Rev Drug Discov. 16:203–222. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Pan ST, Li ZL, He ZX, Qiu JX and Zhou SF:
Molecular mechanisms for tumour resistance to chemotherapy. Clin
Exp Pharmacol Physiol. 43:723–737. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Levy JMM, Towers CG and Thorburn A:
Targeting autophagy in cancer. Nat Rev Cancer. 17:528–542. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Tooze SA and Dikic I: Autophagy captures
the nobel prize. Cell. 167:1433–1435. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kimmelman AC and White E: Autophagy and
tumor metabolism. Cell Metab. 25:1037–1043. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Mizushima N and Komatsu M: Autophagy:
Renovation of cells and tissues. Cell. 147:728–741. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Glick D, Barth S and Macleod KF:
Autophagy: Cellular and molecular mechanisms. J Pathol. 221:3–12.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yu L, Chen Y and Tooze SA: Autophagy
pathway: Cellular and molecular mechanisms. Autophagy. 14:207–215.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Hurley JH and Young LN: Mechanisms of
autophagy initiation. Annu Rev Biochem. 86:225–244. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Jin F, Wang Y, Li M, Zhu Y, Liang H, Wang
C, Wang F, Zhang CY, Zen K and Li L: miR-26 enhances
chemosensitivity and promotes apoptosis of hepatocellular carcinoma
cells through inhibiting autophagy. Cell Death Dis. 8:e25402017.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Rothschild SI, Gautschi O, Batliner J,
Gugger M, Fey MF and Tschan MP: MicroRNA-106a targets autophagy and
enhances sensitivity of lung cancer cells to Src inhibitors. Lung
Cancer. 107:73–83. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wang Z, Wang N, Liu P, Chen Q, Situ H, Xie
T, Zhang J, Peng C, Lin Y and Chen J: MicroRNA-25 regulates
chemoresistance-associated autophagy in breast cancer cells, a
process modulated by the natural autophagy inducer
isoliquiritigenin. Oncotarget. 5:7013–7026. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hara T, Takamura A, Kishi C, Iemura S,
Natsume T, Guan JL and Mizushima N: FIP200, a ULK-interacting
protein, is required for autophagosome formation in mammalian
cells. J Cell Biol. 181:497–510. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Cheng Y, Ban R, Liu W, Wang H, Li S, Yue
Z, Zhu G, Zhuan Y and Wang C: miRNA-409-3p enhances
cisplatin-sensitivity of ovarian cancer cells by blocking the
autophagy mediated by Fip200. Oncol Res. Jan 2–2018.(Epub ahead of
print). View Article : Google Scholar
|
|
18
|
Lu D, Yang C, Zhang Z, Cong Y and Xiao M:
Knockdown of Linc00515 inhibits multiple myeloma autophagy and
chemoresistance by upregulating miR-140-5p and downregulating
ATG14. Cell Physiol Biochem. 48:2517–2527. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Marchese FP, Raimondi I and Huarte M: The
multidimensional mechanisms of long noncoding RNA function. Genome
Biol. 18:2062017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yoon JH, Abdelmohsen K and Gorospe M:
Functional interactions among microRNAs and long noncoding RNAs.
Semin Cell Dev Biol. 34:9–14. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
He J, Yu JJ, Xu Q, Wang L, Zheng JZ, Liu
LZ and Jiang BH: Downregulation of ATG14 by EGR1-MIR152 sensitizes
ovarian cancer cells to cisplatin-induced apoptosis by inhibiting
cyto-protective autophagy. Autophagy. 11:373–384. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Xu R, Liu S, Chen H and Lao L:
MicroRNA-30a downregulation contributes to chemoresistance of
osteosarcoma cells through activating Beclin-1-mediated autophagy.
Oncol Rep. 35:1757–1763. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zheng B, Zhu H, Gu D, Pan X, Qian L, Xue
B, Yang D, Zhou J and Shan Y: MiRNA-30a-mediated autophagy
inhibition sensitizes renal cell carcinoma cells to sorafenib.
Biochem Biophys Res Commun. 459:234–239. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhang Y, Yang WQ, Zhu H, Qian YY, Zhou L,
Ren YJ, Ren XC, Zhang L, Liu XP, Liu CG, et al: Regulation of
autophagy by miR-30d impacts sensitivity of anaplastic thyroid
carcinoma to cisplatin. Biochem Pharmacol. 87:562–570. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Luo M, Wu L, Zhang K, Wang H, Wu S,
O'Connell D, Gao T, Zhong H and Yang Y: miR-216b enhances the
efficacy of vemurafenib by targeting Beclin-1, UVRAG and ATG5 in
melanoma. Cell Signal. 42:30–43. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Tan S, Shi H, Ba M, Lin S, Tang H, Zeng X
and Zhang X: miR-409-3p sensitizes colon cancer cells to
oxaliplatin by inhibiting Beclin-1-mediated autophagy. Int J Mol
Med. 37:1030–1038. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Chen K and Shi W: Autophagy regulates
resistance of non-small cell lung cancer cells to paclitaxel.
Tumour Biol. 37:10539–10544. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Huang J, Ni J, Liu K, Yu Y, Xie M, Kang R,
Vernon P, Cao L and Tang D: HMGB1 promotes drug resistance in
osteosarcoma. Cancer Res. 72:230–238. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Chen Y, Zhou X, Qiao J and Bao A:
MiR-142-3p overexpression increases chemo-sensitivity of NSCLC by
inhibiting HMGB1-mediated autophagy. Cell Physiol Biochem.
41:1370–1382. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Xiong J, Wang D, Wei A, Ke N, Wang Y, Tang
J, He S, Hu W and Liu X: MicroRNA-410-3p attenuates gemcitabine
resistance in pancreatic ductal adenocarcinoma by inhibiting
HMGB1-mediated autophagy. Oncotarget. 8:107500–107512. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ran X, Yang J, Liu C, Zhou P, Xiao L and
Zhang K: miR-218 inhibits HMGB1-mediated autophagy in endometrial
carcinoma cells during chemotherapy. Int J Clin Exp Pathol.
8:6617–6626. 2015.PubMed/NCBI
|
|
32
|
Guo S, Bai R, Liu W, Zhao A, Zhao Z, Wang
Y, Wang Y, Zhao W and Wang W: miR-22 inhibits osteosarcoma cell
proliferation and migration by targeting HMGB1 and inhibiting
HMGB1-mediated autophagy. Tumour Biol. 35:7025–7034. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Liu K, Huang J, Xie M, Yu Y, Zhu S, Kang
R, Cao L, Tang D and Duan X: MIR34A regulates autophagy and
apoptosis by targeting HMGB1 in the retinoblastoma cell. Autophagy.
10:442–452. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Kim YC and Guan KL: mTOR: A pharmacologic
target for autophagy regulation. J Clin Invest. 125:25–32. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Han J, Li J, Tang K, Zhang H, Guo B, Hou N
and Huang C: miR-338-3p confers 5-fluorouracil resistance in p53
mutant colon cancer cells by targeting the mammalian target of
rapamycin. Exp Cell Res. 360:328–336. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Jiang C, Shen F, Du J, Fang X, Li X, Su J,
Wang X, Huang X and Liu Z: Upregulation of CASC2 sensitized glioma
to temozolomide cytotoxicity through autophagy inhibition by
sponging miR-193a-5p and regulating mTOR expression. Biomed
Pharmacother. 97:844–850. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Hildebrandt MA, Yang H, Hung MC, Izzo JG,
Huang M, Lin J, Ajani JA and Wu X: Genetic variations in the
PI3K/PTEN/AKT/mTOR pathway are associated with clinical outcomes in
esophageal cancer patients treated with chemoradiotherapy. J Clin
Oncol. 27:857–871. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Lim HJ, Crowe P and Yang JL: Current
clinical regulation of PI3K/PTEN/Akt/mTOR signalling in treatment
of human cancer. J Cancer Res Clin Oncol. 141:671–689. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Wang L, Tang B, Han H, Mao D, Chen J, Zeng
Y and Xiong M: miR-155 affects osteosarcoma MG-63 cell autophagy
induced by adriamycin through regulating PTEN-PI3K/AKT/mTOR
signaling pathway. Cancer Biother Radiopharm. 33:32–38. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Yu X, Li R, Shi W, Jiang T, Wang Y, Li C
and Qu X: Silencing of MicroRNA-21 confers the sensitivity to
tamoxifen and fulvestrant by enhancing autophagic cell death
through inhibition of the PI3K-AKT-mTOR pathway in breast cancer
cells. Biomed Pharmacother. 77:37–44. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
He C, Dong X, Zhai B, Jiang X, Dong D, Li
B, Jiang H, Xu S and Sun X: MiR-21 mediates sorafenib resistance of
hepatocellular carcinoma cells by inhibiting autophagy via the
PTEN/Akt pathway. Oncotarget. 6:28867–28881. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Mizushima N: Autophagy: Process and
function. Genes Dev. 21:2861–2873. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Kirkin V, McEwan DG, Novak I and Dikic I:
A role for ubiquitin in selective autophagy. Mol Cell. 34:259–269.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
YiRen H, YingCong Y, Sunwu Y, Keqin L,
Xiaochun T, Senrui C, Ende C, XiZhou L and Yanfan C: Long noncoding
RNA MALAT1 regulates autophagy associated chemoresistance via
miR-23b-3p sequestration in gastric cancer. Mol Cancer. 16:1742017.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Chen R, Li X, He B and Hu W: MicroRNA-410
regulates autophagy-related gene ATG16L1 expression and enhances
chemosensitivity via autophagy inhibition in osteosarcoma. Mol Med
Rep. 15:1326–1334. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Zhao J, Nie Y, Wang H and Lin Y: MiR-181a
suppresses autophagy and sensitizes gastric cancer cells to
cisplatin. Gene. 576:828–833. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Zhang K, Chen J, Zhou H, Chen Y, Zhi Y,
Zhang B, Chen L, Chu X, Wang R and Zhang C: PU.1/microRNA-142-3p
targets ATG5/ATG16L1 to inactivate autophagy and sensitize
hepatocellular carcinoma cells to sorafenib. Cell Death Dis.
9:3122018. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
An Y, Zhang Z, Shang Y, Jiang X, Dong J,
Yu P, Nie Y and Zhao Q: miR-23b-3p regulates the chemoresistance of
gastric cancer cells by targeting ATG12 and HMGB2. Cell Death Dis.
6:e17662015. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Liao H, Xiao Y, Hu Y, Xiao Y, Yin Z, Liu
L, Kang X and Chen Y: Methylation-induced silencing of miR-34a
enhances chemoresistance by directly upregulating ATG4B-induced
autophagy through AMPK/mTOR pathway in prostate cancer. Oncol Rep.
35:64–72. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Wu Y, Ni Z, Yan X, Dai X, Hu C, Zheng Y,
He F and Lian J: Targeting the MIR34C-5p-ATG4B-autophagy axis
enhances the sensitivity of cervical cancer cells to pirarubicin.
Autophagy. 12:1105–1117. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Liu L, Shen W, Zhu Z, Lin J, Fang Q, Ruan
Y and Zhao H: Combined inhibition of EGFR and c-ABL suppresses the
growth of fulvestrant-resistant breast cancer cells through
miR-375-autophagy axis. Biochem Biophys Res Commun. 498:559–565.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Gao AM, Zhang XY, Hu JN and Ke ZP:
Apigenin sensitizes hepatocellular carcinoma cells to doxorubic
through regulating miR-520b/ATG7 axis. Chem Biol Interact.
280:45–50. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Sun W, Zu Y, Fu X and Deng Y: Knockdown of
lncRNA-XIST enhances the chemosensitivity of NSCLC cells via
suppression of autophagy. Oncol Rep. 38:3347–3354. 2017.PubMed/NCBI
|
|
54
|
Stiuso P, Potenza N, Lombardi A,
Ferrandino I, Monaco A, Zappavigna S, Vanacore D, Mosca N,
Castiello F, Porto S, et al: MicroRNA-423-5p promotes autophagy in
cancer cells and is increased in serum from hepatocarcinoma
patients treated with sorafenib. Mol Ther Nucleic Acids.
4:e2332015. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Comincini S, Allavena G, Palumbo S, Morini
M, Durando F, Angeletti F, Pirtoli L and Miracco C: MicroRNA-17
regulates the expression of ATG7 and modulates the autophagy
process, improving the sensitivity to temozolomide and low-dose
ionizing radiation treatments in human glioblastoma cells. Cancer
Biol Ther. 14:574–586. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Xu N, Zhang J, Shen C, Luo Y, Xia L, Xue F
and Xia Q: Cisplatin-induced downregulation of miR-199a-5p
increases drug resistance by activating autophagy in HCC cell.
Biochem Biophys Res Commun. 423:826–831. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Li Y, Iglehart JD, Richardson AL and Wang
ZC: The amplified cancer gene LAPTM4B promotes tumor growth and
tolerance to stress through the induction of autophagy. Autophagy.
8:273–274. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Blom T, Li S, Dichlberger A, Bäck N, Kim
YA, Loizides-Mangold U, Riezman H, Bittman R and Ikonen E: LAPTM4B
facilitates late endosomal ceramide export to control cell death
pathways. Nat Chem Biol. 11:799–806. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Soni M, Patel Y, Markoutsa E, Jie C, Liu
S, Xu P and Chen H: Autophagy, cell viability, and chemoresistance
are regulated by miR-489 in breast cancer. Mol Cancer Res.
16:1348–1360. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Eskelinen EL: Roles of LAMP-1 and LAMP-2
in lysosome biogenesis and autophagy. Mol Aspects Med. 27:495–502.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Bao L, Lv L, Feng J, Chen Y, Wang X, Han S
and Zhao H: miR-487b-5p regulates temozolomide resistance of lung
cancer cells through LAMP2-medicated autophagy. DNA Cell Biol.
35:385–392. 2016. View Article : Google Scholar : PubMed/NCBI
|















