Introduction
Renal cell carcinoma (RCC) accounts for 3–5% of all
cancer diagnoses worldwide (1). The
incidence rate of RCC has been increasing annually, and at present,
~20% of newly diagnosed RCC cases result in death. RCC is one of
the leading causes of cancer-associated death worldwide, according
to the World Health Organization (WHO) (2) and 20–30% of patients with RCC present
with metastases at the initial diagnosis (2). Due to the limitations of current
treatment methods, which may result in the development of
resistance to chemoradiotherapy, as well as the difficulty of
radical resection, novel therapeutic targets are required to
improve outcomes of patients with RCC. Wei et al (3) demonstrated that the expression levels
of certain hub genes [CDKN3, TPX2, BUB1B, CDCA8, UBE2C, NDC80,
RRM2, NCAPG, NCAPH, PTTG1, FAM64A, ANLN, KIF4A, CEP55, centromere
protein (CENP)-F, KIF20A, ASPM and Holliday junction recognition
protein (HJURP)] were significantly associated with overall
survival and recurrence-free survival in RCC (3). In recent years, HJURP has been
demonstrated to involve in the cell viability and cell cycle in
numerous types of human tumor (4–7).
However, the mechanism of HJURP in RCC has remained elusive.
Therefore, in the present study, HJURP was selected as the research
subject to investigate its role in the development of RCC.
HJURP, also known as hFLEG1, is a histone chaperone
in the nucleosome, which recruits CENP-C and CENP-A (5,6). HJURP
has been indicated to regulate chromosomal stability and
amplification of the centromere (7,8). HJURP
is also involved in the accurate segregation of chromosomes during
mitosis. HJURP binds directly to soluble CENP-A, stabilizing it and
regulating its binding to centromeres during the G1 phase of the
cell cycle (9). HJURP is
upregulated by ataxia telangiectasia-mutated signaling during a DNA
double-strand break response and participates in the homologous
recombination pathways during repair of double-stranded breaks
(4). Downregulation of HJURP levels
results in a significant reduction of CENP-A levels at centromeres,
which may affect centromere assembly and microtubule attachment,
and may thus underlie defects in chromosome segregation during
mitosis (10–12).
Valente et al (13) demonstrated that the upregulation of
HJURP expression may serve an important role in the survival of
cells with a high degree of proliferative activity in high-grade
malignancy gliomas. Furthermore, HJURP has been demonstrated to be
involved in cell viability and the cell cycle in lung carcinoma
(4), breast carcinoma (14), bladder cancer (15) and hepatocellular carcinoma (16), and may be a novel and important
prognostic indicator involved in the development and progression of
several types of cancer (17–19).
Peroxisome-proliferator-activated receptors (PPARs)
are nuclear receptors that regulate tumor growth (20,21).
Activation/deactivation of PPARs may affect the expression of genes
associated with cellular metabolism, proliferation, lipid
peroxidation and stress responses, including reactive oxygen
species (ROS) (22). The nuclear
receptor PPARγ, a key member of the PPAR family, is involved in
cell cycle regulation (23,24), where it binds to the promoter region
of sirtuin (SIRT)1 to regulate its transcription (25). SIRT1 functions as a key regulator of
genes regulating apoptosis and cell survival, including PPARγ
(26) and p53 (27). The association between PPARγ and
SIRT1 and the presence of a negative feedback loop between
PPARγ/SIRT1 has been previously reported (25).
Tumor cells are characterized by an accumulation of
mutations that drive tumor development and progression, including
changes in the number of copies of chromosomes, chromosomal
rearrangements, point mutations and small deletions and insertions
(28–30). Although the association between RCC
and HJURP has not been previously demonstrated, to the best of our
knowledge, preliminary results by our group indicated that HJURP
participates in the regulation of the cell cycle and cell apoptosis
in RCC. Thus, it was hypothesized that changes in HJURP expression
levels may serve an important role in the regulation of cell
viability and the cell cycle of RCC cells. In the present study,
HJURP expression levels were determined and the expression of
ROS-associated genes was detected in RCC tissues; furthermore, the
roles of HJURP were assessed in vitro using RCC cell
lines.
Materials and methods
Patients
RCC tissue samples (n=15) and adjacent paracancerous
renal tissue samples (n=15) were obtained from patients who
underwent radical resection at the Affiliated Hospital of Qingdao
University (Qingdao, China). The age of the subjects ranged from 35
to 76 years with a median age of 56 years, including 10 males and 5
females. These samples were immediately stored in liquid nitrogen
following surgical removal for further analysis. All patients
provided written informed consent and the diagnosis of RCC was
determined according to the WHO criteria (31). The Medical Ethics Committee at the
Affiliated Hospital of Qingdao University (Qingdao, China) approved
the use of human RCC tissue samples for protein analysis and RNA
extraction (approval no. 201801), and all experiments involving the
RCC tissue samples were performed in accordance with the criteria
approved by the Ethics Committee.
Cell culture
The RCC cell lines A498 (KG414) and Caki-1 (KG211),
as well as the renal tubular epithelial cell line HK2 (KG350), were
purchased from Nanjing KeyGen Biotech Co., Ltd. The A498 and Caki-1
cells were cultured in RPMI-1640 medium (Gibco; Thermo Fisher
Scientific, Inc.), while HK2 cells were cultured in Dulbecco's
modified Eagle's medium (Gibco; Thermo Fisher Scientific, Inc.)
supplemented with 100 U/ml penicillin, 0.1 mg/ml streptomycin and
10% fetal bovine serum (Gibco; Thermo Fisher Scientific, Inc.).
Cells were maintained in a humidified incubator with 5%
CO2 at 37°C.
Reverse transcription-quantitative
(RT-qPCR) analysis
RNA was extracted from A498 cells, Caki-1 cells and
RCC tissues using TRIzol® (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol.
Complementary DNA synthesis was performed using oligo(dT), MLV
Reverse Transcriptase, Reverse Transcription 5X Buffer and dNTP
Mixture (10 mM each dNTP) according to the manufacturer's protocol
(Takara, Bio, Inc.) after RNA was treated with DNAse I (Sangon
Biotech, Co., Ltd.) and RNAse inhibitor (Invitrogen; Thermo Fisher
Scientific, Inc.). The SYBR Green I Real-Time PCR kit (Takara Bio,
Inc.) was used to perform qPCR according to the manufacturer's
protocol and the amplification was performed in an ABI Prism 7500
(Perkin-Elmer, Inc.). The following thermocycling conditions were
used for the qPCR: 40 cycles of 95°C for 15 sec, 60°C for 20 sec
and 72°C for 40 sec. The expression was normalized to GAPDH. The
sequences of the primers used were as follows: HJURP forward,
5′-CCGCAGCAGACATCTGACCTTC-3′ and reverse,
5′-TCCGTGGCCTGGCACTTCTT-3′; and GAPDH forward,
5′-AGATCATCAGCAATGCCTCCT-3′ and reverse,
5′-TGAGTCCTTCCACGATACCAA-3′. The mRNA expression levels were
calculated using the 2−∆∆Cq method (32).
Western blot analysis
RCC tissue samples and adjacent paracancerous renal
tissue samples were homogenized in 150 U/ml DNase I buffer in 5
mmol/l MgCl2, 1 mmol/l CaCl2 and 20 mmol/l
Tris-HCl (pH 6.8; Takara Bio, Inc.). Cells were sonicated and lysed
in radioimmunoprecipitation assay lysis buffer supplemented with
protease and phosphatase inhibitors (Nanjing KeyGen Biotech Co.,
Ltd.) for 15 min at 4°C. The solution was centrifuged at 14,000 × g
for 15 min at 4°C and the supernatant was collected. The
concentrations of proteins were determined using a bicinchoninic
acid assay kit (Nanjing KeyGen Biotech Co., Ltd.), with bovine
serum albumin (BSA) used as the standard (Nanjing KeyGen Biotech
Co., Ltd.). Proteins (10 µg) were loaded on a 10% SDS gel, resolved
using SDS-PAGE and transferred to polyvinylidene difluoride
membranes (Nanjing KeyGen Biotech Co., Ltd.). Membranes were
blocked in 5% BSA for 2 h at room temperature. Subsequently,
membranes were incubated with primary antibodies, including those
to HJURP (1:2,000; cat. no. ab100800; Abcam), catalase (1:10,000;
cat. no. ab76024; Abcam), superoxide dismutase (SOD)2 (1:1,000;
cat. no. ab68155; Abcam), PPARγ (1:1,000; cat. no. ab59256; Abcam),
phosphorylated (p)-SIRT1 (1:1,000; cat. no. ab76039; Abcam), SIRT1
(1:1,000; cat. no. ab110304; Abcam), forkhead box (FOX)O3a (1:100;
cat. no. BM4734; Wuhan Boster Biological Technology, Ltd.),
p-FOXO3a (1:1,000; cat. no. ab154786; Abcam) and cyclin D1 (CCND1;
1:1,000; cat. no. ab40754; Abcam) overnight at 4°C, followed by
horseradish peroxidase-labeled secondary antibody (1:10,000; cat.
no. KGAA35; Nanjing KeyGen Biotech Co., Ltd.) for 1 h at room
temperature. GAPDH (1:3,000; cat. no. K200057M; Solarbio Life
Science) was selected as the loading control. As GAPDH and CCND1
have a similar molecular weight, the same specimen was added to two
different gels, electrophoresis was performed on the same apparatus
and labeling with the antibodies was performed separately. Signals
were visualized using enhanced chemiluminescence reagent (Bio-Rad
Laboratories, Inc.) and bands were imaged using a G:BOX chemiXR5
system (Syngene Europe).
Overexpression of HJURP in RCC
cells
A pcDNA3.1(+)-HJURP vector was synthesized by
Nanjing KeyGen Biotech Co., Ltd. The A498 RCC cells (2,000
cells/well) were plated in 96-well plates and pcDNA3.1(+)-HJURP
vector were introduced into cells using Oligofectamine (Invitrogen;
Thermo Fisher Scientific, Inc.). Cells were fixed and processed for
the subsequent experiment 72 h after transfection. The A498 RCC
cells were transfected with the pcDNA3.1(+)-HJURP vector to
increase the mRNA expression levels of HJURP (A498 HJURP OE). The
A498 cells were used as the control group and the A498 cells
transfected with pcDNA3.1(+) were used as a negative control (A498
NC).
Immunocytochemistry
Cells were washed three times with cold PBS and
fixed in 4% paraformaldehyde for 30 min at room temperature. RCC
cells were placed in 3% H2O2-methanol
solution (Nanjing KeyGen Biotech Co., Ltd.) for 10 min at room
temperature, washed in PBS and blocked in standard goat serum
(Nanjing KeyGen Biotech Co., Ltd.) at room temperature for 20 min.
Following blocking, cells were incubated with the HJURP primary
antibody (1:100; cat. no. ab100800, Abcam) for 2 h at 37°C, washed
using PBS and subsequently incubated with FITC-labeled secondary
antibody (1:100; cat. no. BA1105; BosterBiological Technology) for
1 h at 37°C. To visualize the nuclei, cells were stained with DAPI
(2 µg/ml) for 5 min at room temperature. Immunofluorescence
staining was observed using a fluorescence microscope (×200
magnification).
Cell Counting Kit-8 (CCK-8) assay
RCC cells were collected (5×104 cells/ml)
following transfection for 48 h and seeded in 96-well plates and
cultured for 72 h. Subsequently, 10 µl CCK-8 solution (5 mg/ml) was
added to each well and cells were incubated at 37°C for 2 h. The
absorbance was measured using a microplate reader at 450 nm (BioTek
ELx800; BioTek Instruments, Inc.).
Colony formation assay
Following transfection for 48 h, RCC cells were
cultured in 6-well plates (5×104 cells/ml) at 37°C for
12 days. Subsequently, cells were washed twice with PBS, fixed in
4% paraformaldehyde for 20 min at room temperature, stained using
Giemsa stain for 10 min at room temperature, and subsequently, the
colonies (>50 cells/colony) in the wells were counted and imaged
using a fluorescence microscope (Olympus Corp).
Cell cycle and apoptosis analysis
A total of 5×105 cells were collected and
centrifuged (300 × g for 5 min at 4°C). Cell pellets were washed
and resuspended in a solution consisting 400 µl propidium iodide
(PI), 100 µl RNase A and permeabilization solution (Nanjing KeyGen
Biotech Co., Ltd.) for 30 min at 4°C in the dark. Cell cycle
analysis was performed using flow cytometry (FACSCalibur™; BD
Biosciences). Cell apoptosis was measured using an Annexin
V-FITC/PI Apoptosis Detection kit I (Nanjing KeyGen Biotech Co.,
Ltd.) according to the manufacturer's protocol and assessment was
performed using flow cytometry (FACSCalibur™; BD Biosciences).
Terminal deoxynucleotidyl transferase
deoxyuridine triphosphate nick end labelling (TUNEL) assay
Following transfection for 72 h, RCC cells were
fixed using 4% paraformaldehyde for 30 min at room temperature and
subsequently washed using cold PBS. Cells were permeabilized using
1% Triton X-100 for 15 min and washed three times in PBS. Analysis
of apoptosis was performed using a TUNEL assay (cat. no. KGA7061;
Nanjing KeyGen Biotech Co., Ltd.) according to the manufacturer's
protocol. Nuclear staining was performed using 100 µl DAPI for 5
min at room temperature, followed by washing with PBS. Cells were
imaged using a fluorescence microscope (magnification, ×200) and
counted using a manual cell counter.
Statistical analysis
Values are expressed as the mean ± standard
deviation of three experimental repeats. Expression levels between
RCC and adjacent paracancerous renal tissue samples were analyzed
by a parametric paired t-test. One-way analysis of variance
followed by the least significant difference test was used to
analyze the values for each condition/group. Analysis was performed
using SPSS version 17.0 (SPSS, Inc.). P<0.05 was considered to
indicate a statistically significant difference.
Results
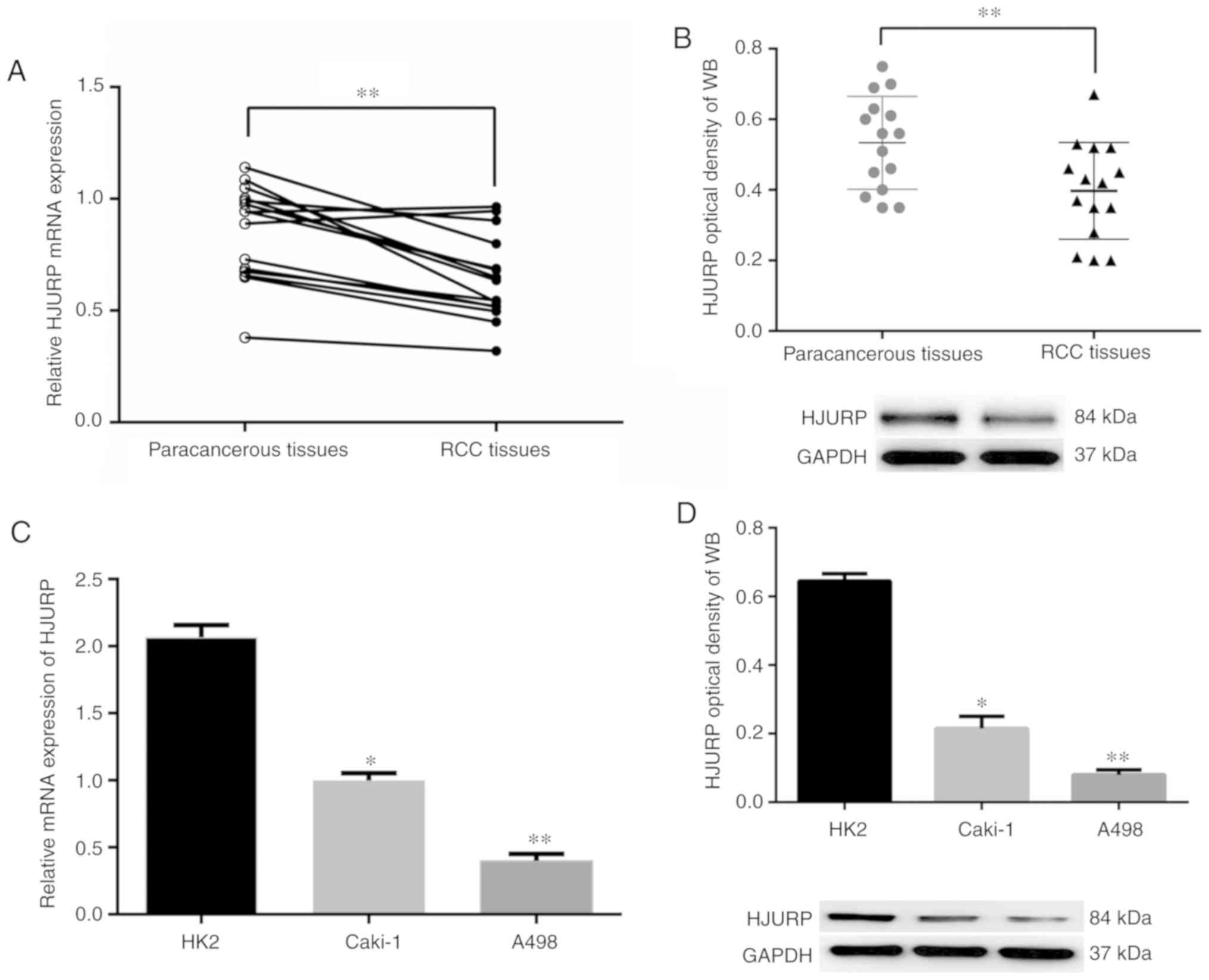
HJURP expression is downregulated in
RCC vs. paracancerous tissues
mRNA expression levels of HJURP in 15 pairs of RCC
samples and the adjacent matching tissues were analyzed using
RT-qPCR. The results indicated that HJURP expression was
downregulated in RCC tissues compared with that in adjacent normal
tissue samples (Fig. 1A). Similar
results were obtained at the protein expression level, where HJURP
was indicated to be downregulated in the RCC tissues compared with
the matched normal tissues (Fig.
1B). Furthermore, HJURP protein and mRNA expression levels in
the RCC cell lines were downregulated compared with those in renal
tubular epithelial cells (Fig. 1C and
D).
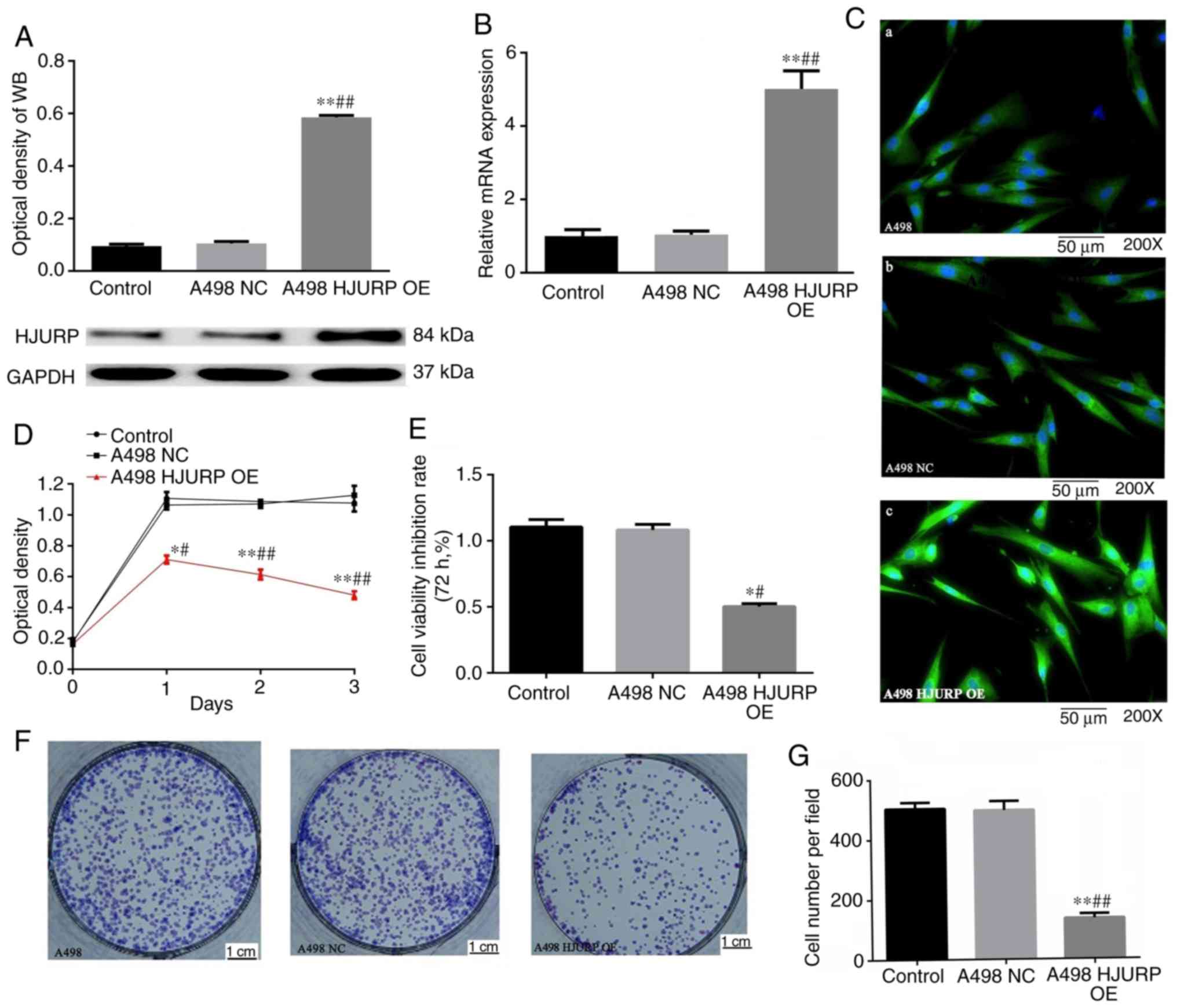
Reduced HJURP expression increases
proliferation and viability of RCC cells
For subsequent in vitro experiments, the A498
cell line was used, as the endogenous expression levels of HJURP
were lower than those of the other cell lines. The A498 cells were
transfected with a pcDNA3.1(+) vector containing an HJURP
transcript and successful upregulation was confirmed using RT-qPCR
and western blot analysis at 72 h after transfection (Fig. 2A and B). Immunofluorescence staining
suggested that the protein expression levels of HJURP were also
significantly increased in the HJURP-transfected cells (Fig. 2C), further confirming the successful
establishment of an HJURP overexpression cell line. A CCK-8 assay
and colony formation assay indicated that A498 cells overexpressing
HJURP exhibited reduced viability and colony formation compared
with the empty vector-transfected cells (Fig. 2D-G). Taken together, these results
suggest that HJURP inhibits RCC in vitro.
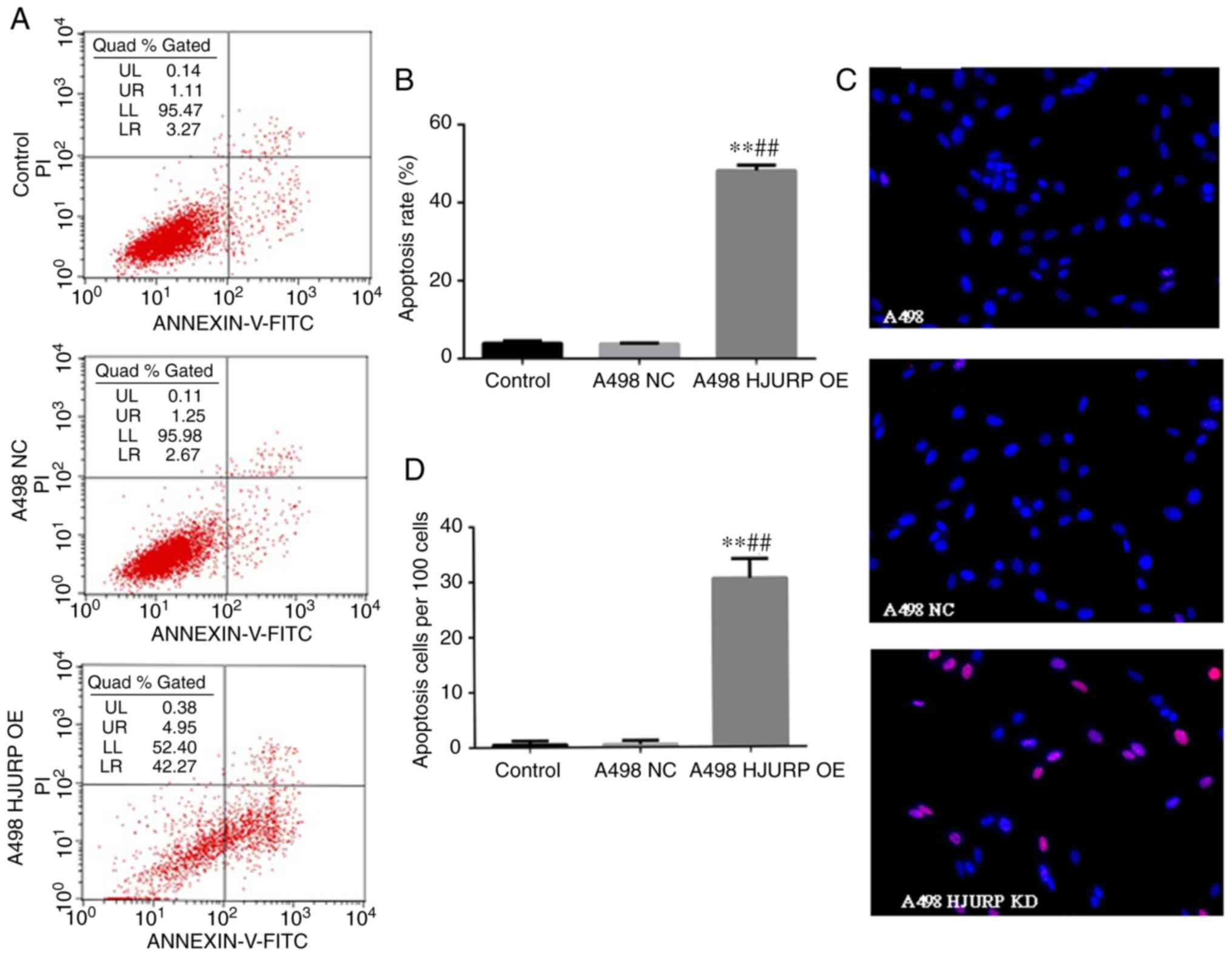
HJURP overexpression in A498 cells
increases apoptosis
As the growth of HJURP-overexpressing A498 cells was
significantly reduced, the apoptotic status of the RCC cells was
also assessed. The ratio of the cells in early apoptosis in the
HJURP-overexpressing A498 cells was significantly increased
compared with that in the empty vector-transfected cells (Fig. 3A and B). Furthermore, a TUNEL assay
confirmed total apoptosis results (Fig.
3C and D).
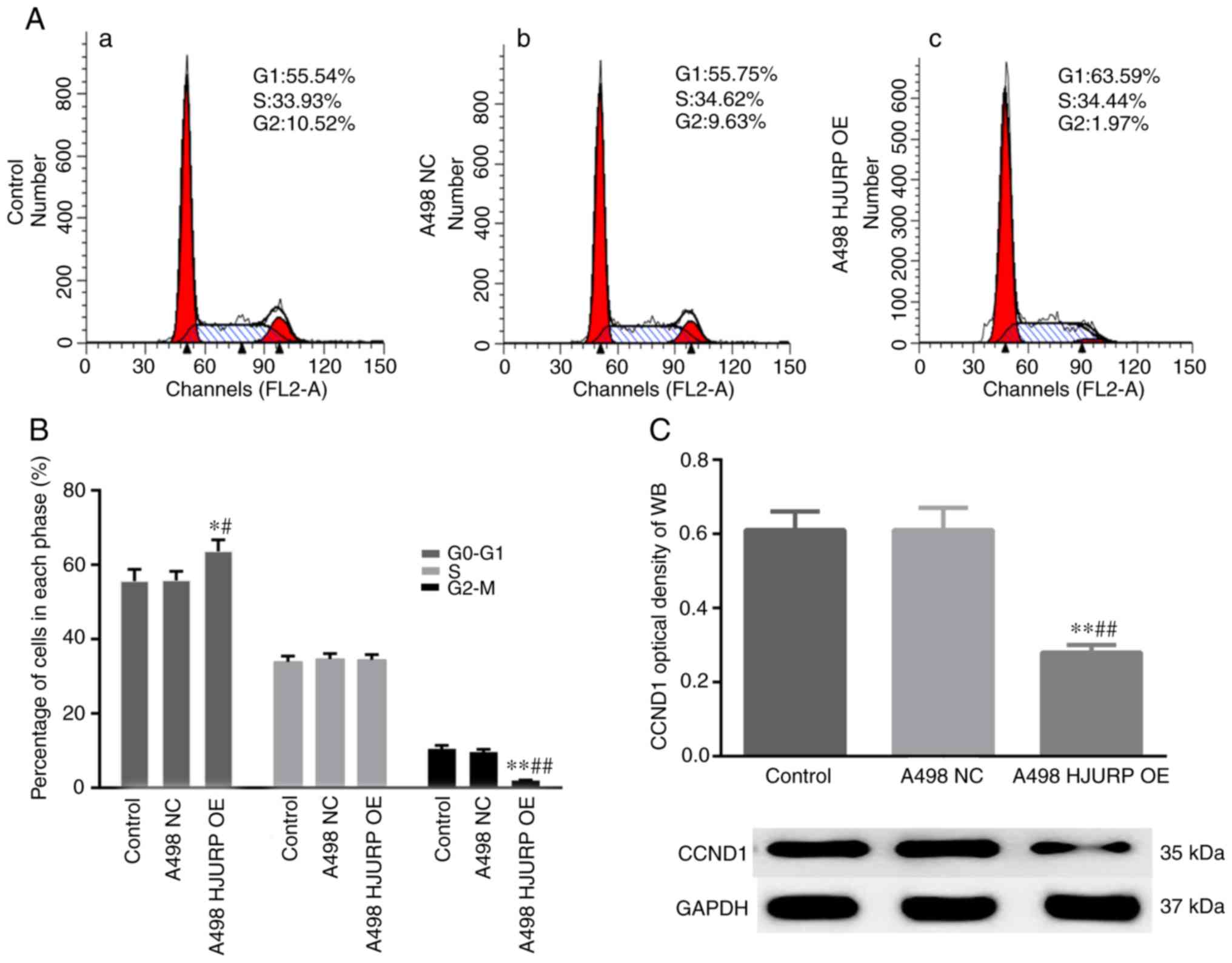
HJURP overexpression results in cell
cycle arrest at the G0/G1 phase
Changes in the cell cycle were assessed using flow
cytometry. The results suggested that the proportion of cells in
G0/G1 phase was significantly increased and the ratio of cells at
the G2/M phase was significantly decreased in the
HJURP-overexpressing cells (Fig. 4A and
B). Furthermore, CCND1 expression was decreased in the
HJURP-overexpressing cells compared with the empty
vector-transfected control cells (Fig.
4C).
Upregulation of HJURP increases ROS
stress in A498 cells
Catalase and SOD2 protein expression levels, both of
which are associated with the antioxidant response and ROS
metabolism, were significantly decreased in the
HJURP-overexpressing cells compared with the control cells
(Fig. 5A). In addition, the levels
of p-SIRT1/total (t)-SIRT1 were decreased in the
HJURP-overexpressing cells compared with the control cells.
Furthermore, the p-FOXO3a/t-FOXO3a ratio was decreased in the
HJURP-overexpressing cells (Fig.
5B). All of these results suggest that upregulation of HJURP
induced oxidative stress in RCC cells via deactivation of
PPARγ/SIRT1 and downstream FOXO3a signaling.
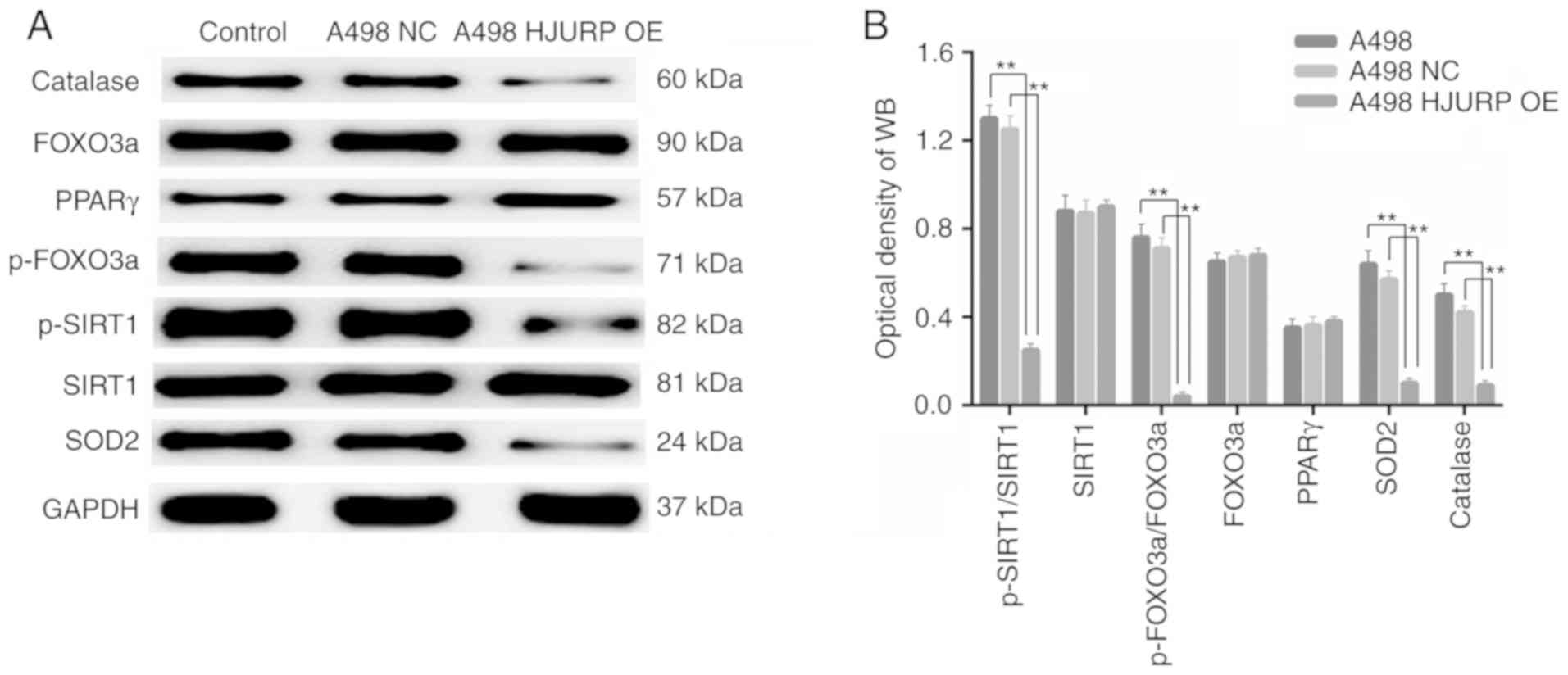 | Figure 5.Overexpression of HJURP increases ROS
stress in renal cell carcinoma cells. (A) Expression of proteins
associated with ROS status in the (a) A498 control cells, (b) NC
group and (c) HJURP-OE group. (B) Densitometry analysis indicated
that the expression of ROS-associated proteins was significantly
increased in the HJURP-OE group. *P<0.05, **P<0.01 as
indicated. WB, western blot; HJURP, Holliday junction recognition
protein; ROS, reactive oxygen species; OE, overexpression; NC,
negative control; PPAR, peroxisome proliferator-activated receptor;
p-FOXO3, phosphorylated forkhead box O3; SIRT1, sirtuin 1; SOD,
superoxide dismutase. |
Discussion
RCC is a life-threatening disease with a high rate
of morbidity and mortality. There are several high-risk factors,
including hypertension, obesity, smoking and chronic kidney
disease, associated with the development of RCC (33,34).
Patients with RCC are frequently diagnosed with distant metastases,
at which point surgical removal is not a viable option. Thus, there
is an urgency for novel accurate diagnostic and therapeutic
biomarkers to improve individualized treatments and increase
survival. HJURP has been demonstrated to serve as a key chaperone
of CENP-A, which is involved in nucleosome assembly, and has been
reported to serve a key role in different types of cancer, which
contributes to promoting hepatocellular carcinoma proliferation and
dysregulating the cell cycle and ROS metabolism in bladder cancer
(15,16,35).
In the present study, the role of HJURP in RCC was
determined. The expression of HJURP was downregulated at the mRNA
and protein expression levels in RCC cell lines and tissues. This
result is in contrast with studies on other types of cancer
(4,14–16),
suggesting that the specific role of HJURP varies and is
tissue-dependent, and may be associated with organ specificity, or
highlights the possibility that HJURP may exert its effects on
different target genes in different tissues (36–38).
In the present study, overexpression of HJURP in RCC cells reduced
colony formation and cell growth. Furthermore, the apoptotic rate
of HJURP-overexpressing RCC cells was significantly higher compared
with that in the respective controls. Thus, it was hypothesized
that HJURP may regulate the cell cycle in RCC cells.
To further investigate the mechanism underlying the
increased apoptosis induced by overexpression of HJURP, the
expression of ROS-associated proteins was assessed and it was
demonstrated that their expression was downregulated when HJURP
expression was increased, and this may have been associated with
apoptosis, cell cycle regulation and oxidative stress (22,39–41).
These results suggest that downregulation of HJURP in RCC cells
disrupted homeostasis of ROS metabolism, resulting in oxidative
modification of lipids, proteins or DNA, followed by apoptosis
induced by oxidative stress (23).
Thus, the increased apoptotic rate in RCC cells induced by the
increase in HJURP expression may be associated with oxidative
stress and ROS-associated pathways.
The PPARγ/SIRT1 feedback loop is closely associated
with the oxidative stress response, cell cycle regulation and
apoptosis (25,42). These two proteins may be associated
with the regulation of cell apoptosis, cell cycle progression and
ROS metabolism. In the present study, HJURP overexpression in RCC
cells decreased the ratio of p-SIRT1/t-SIRT1 significantly. Cell
cycle arrest at the G0/G1 phase was observed, accompanied with a
decrease in the expression of CCND1 protein. CCND1 is associated
with cell cycle progression from the G0/G1 to the Sphase (43–45).
In conclusion, HJURP downregulation increased the
proliferation of RCC cells and overexpression of HJURP inhibited
RCC cell proliferation and promoted apoptosis. HJURP may be
involved in the regulation of ROS metabolism and thus contributes
to the progression of RCC. Taken together, these results suggest
that HJURP may be a novel potential molecular therapeutic target
for the treatment of RCC and further investigation into its
suitability as a biomarker is warranted.
Acknowledgements
Not applicable.
Funding
The present study was supported by National Natural
Science Foundation of China (grant no. 31800680).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
KW and ZLZ designed the study; ZLZ, JSY and ZSC
performed the experiments and analyzed the data; KW and ZLZ
participated in the analysis of clinical specimens. JSY and ZLZ
wrote the manuscript. KW and ZLZ revised the manuscript. All
authors have read the manuscript and approved its submission.
Ethics approval and consent to
participate
All patients provided written informed consent. The
Medical Ethics Committee at the Affiliated Hospital of Qingdao
University (Qingdao, China) approved the use of human RCC tissue
samples for protein analysis and RNA extraction (approval no.
201801), and all experiments involving the RCC tissue samples were
performed in accordance with the criteria approved by the Ethics
Committee.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2018. CA Cancer J Clin. 68:7–30. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Capitanio U and Montorsi F: Renal cancer.
Lancet. 387:894–906. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wei W, Lv Y, Gan Z, Zhang Y, Han X and Xu
Z: Identification of key genes involved in the metastasis of clear
cell renal cell carcinoma. Oncol Lett. 17:4321–4328.
2019.PubMed/NCBI
|
|
4
|
Kato T, Sato N, Hayama S, Yamabuki T, Ito
T, Miyamoto M, Kondo S, Nakamura Y and Daigo Y: Activation of
holliday junction recognizing protein involved in the chromosomal
stability and immortality of cancer cells. Cancer Res.
67:8544–8553. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Liu C and Mao Y: Diaphanous formin mDia2
regulates CENP-A levels at centromeres. J Cell Biol. 213:415–424.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Tachiwana H, Muller S, Blumer J, Klare K,
Musacchio A and Almouzni G: HJURP involvement in de novo CenH3
(CENP-A) and CENP-C recruitment. Cell Rep. 11:22–32. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Perpelescu M, Hori T, Toyoda A, Misu S,
Monma N, Ikeo K, Obuse C, Fujiyama A and Fukagawa T: HJURP is
involved in the expansion of centromeric chromatin. Mol Biol Cell.
26:2742–2754. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhao H, Winogradoff D, Bui M, Dalal Y and
Papoian GA: Promiscuous histone mis-assembly is actively prevented
by chaperones. J Am Chem Soc. 138:13207–13218. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Mellone BG, Zhang W and Karpen GH: Frodos
found: Behold the CENP-A ‘Ring’ bearers. Cell. 137:409–412. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Dunleavy EM, Roche D, Tagami H, Lacoste N,
Ray-Gallet D, Nakamura Y, Daigo Y, Nakatani Y and
Almouzni-Pettinotti G: HJURP is a cell-cycle-dependent maintenance
and deposition factor of CENP-A at centromeres. Cell. 137:485–497.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Foltz DR, Jansen LE, Bailey AO, Yates JR
III, Bassett EA, Wood S, Black BE and Cleveland DW:
Centromere-specific assembly of CENP-a nucleosomes is mediated by
HJURP. Cell. 137:472–484. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Shuaib M, Ouararhni K, Dimitrov S and
Hamiche A: HJURP binds CENP-A via a highly conserved N-terminal
domain and mediates its deposition at centromeres. Proc Natl Acad
Sci USA. 107:1349–1354. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Valente V, Serafim RB, de Oliveira LC,
Adorni FS, Torrieri R, Tirapelli DP, Espreafico EM, Oba-Shinjo SM,
Marie SK, Paçó-Larson ML, et al: Modulation of HJURP (Holliday
Junction-Recognizing Protein) levels is correlated with
glioblastoma cells survival. PLoS One. 8:e622002013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Montes de Oca R, Gurard-Levin ZA, Berger
F, Rehman H, Martel E, Corpet A, de Koning L, Vassias I, Wilson LO,
Meseure D, et al: The histone chaperone HJURP is a new independent
prognostic marker for luminal A breast carcinoma. Mol Oncol.
9:657–674. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Cao R, Wang G, Qian K, Chen L, Qian G, Xie
C, Dan HC, Jiang W, Wu M, Wu CL, et al: Silencing of HJURP induces
dysregulation of cell cycle and ROS metabolism in bladder cancer
cells via PPARγ-SIRT1 feedback loop. J Cancer. 8:2282–2295. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chen T, Huang H, Zhou Y, Geng L, Shen T,
Yin S, Zhou L and Zheng S: HJURP promotes hepatocellular carcinoma
proliferation by destabilizing p21via the MAPK/ERK1/2 and AKT/GSK3β
signaling pathways. J Exp Clin Cancer Res. 37:1932018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Bravaccini S, Tumedei MM, Scarpi E, Zoli
W, Rengucci C, Serra L, Curcio A, Buggi F, Folli S, Rocca A, et al:
New biomarkers to predict the evolution of in situ breast cancers.
Biomed Res Int. 2014:1597652014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Coates P, Dewar J and Thompson AM: At
last, a predictive and prognostic marker for radiotherapy? Breast
Cancer Res. 12:1062010. View
Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hu Z, Huang G, Sadanandam A, Gu S, Lenburg
ME, Pai M, Bayani N, Blakely EA, Gray JW and Mao JH: The expression
level of HJURP has an independent prognostic impact and predicts
the sensitivity to radiotherapy in breast cancer. Breast Cancer
Res. 12:R182010. View
Article : Google Scholar : PubMed/NCBI
|
|
20
|
Belfiore A, Genua M and Malaguarnera R:
PPAR-g agonists and their effects on IGF-I receptor signaling:
Implications for cancer. PPAR Res. 2009:8305012009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Berger J and Moller DE: The mechanisms of
action of PPARs. Annu Rev Med. 53:409–435. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Vecchione G, Grasselli E, Voci A, Baldini
F, Grattagliano I, Wang DQ, Portincasa P and Vergani L: Silybin
counteracts lipid excess and oxidative stress in cultured steatotic
hepatic cells. World J Gastroenterol. 22:6016–6026. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Li S, Zhou Q, He H, Zhao Y and Liu Z:
Peroxisome proliferator-activated receptor g agonists induce cell
cycle arrest through transcriptional regulation of Kruppel-like
factor 4 (KLF4). J Biol Chem. 288:4076–4084. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yamamoto Y, Ono T, Dhar DK, Yamanoi A,
Tachibana M, Tanaka T and Nagasue N: Role of peroxisome
proliferator-activated receptor-gamma (PPARgamma) during liver
regeneration in rats. J Gastroenterol Hepatol. 23:930–937. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Han L, Zhou R, Niu J, McNutt MA, Wang P
and Tong T: SIRT1 is regulated by a PPAR{g}-SIRT1 negative feedback
loop associated with senescence. Nucleic Acids Res. 38:7458–7471.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Picard F, Kurtev M, Chung N, Topark-Ngarm
A, Senawong T, Machado De Oliveira R, Leid M, McBurney MW and
Guarente L: Sirt1 promotes fat mobilization in white adipocytes by
repressing PPAR-gamma. Nature. 429:771–776. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Vaziri H, Dessain SK, Ng Eaton E, Imai SI,
Frye RA, Pandita TK, Guarente L and Weinberg RA: hSIR2 (SIRT1)
functions as an NAD-dependent p53 deacetylase. Cell. 107:149–159.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Clark MJ, Homer N, O'Connor BD, Chen Z,
Eskin A, Lee H, Merriman B and Nelson SF: U87MG decoded: The
genomic sequence of a cytogenetically aberrant human cancer cell
line. PLoS Genet. 6:e10008322010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Pleasance ED, Cheetham RK, Stephens PJ,
McBride DJ, Humphray SJ, Greenman CD, Varela I, Lin ML, Ordóñez GR,
Bignell GR, et al: A comprehensive catalogue of somatic mutations
from a human cancer genome. Nature. 463:191–196. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Stephens PJ, McBride DJ, Lin ML, Varela I,
Pleasance ED, Simpson JT, Stebbings LA, Leroy C, Edkins S, Mudie
LJ, et al: Complex landscapes of somatic rearrangement in human
breast cancer genomes. Nature. 462:1005–1010. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Eble JN, Sauter G, Epstein JI and
Sesterhenn IA: Pathology and genetics of tumours of the urinary
system and male genital organs. (1st). IARC Press. (Lyon). 12–43.
2004.
|
|
32
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Capitanio U, Bensalah K, Bex A, Boorjian
SA, Bray F, Coleman J, Gore JL, Sun M, Wood C and Russo P:
Epidemiology of renal cell carcinoma. Eur Urol. 75:74–84. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Chow WH, Dong LM and Devesa SS:
Epidemiology and risk factors for kidney cancer. Nat Rev Urol.
7:245–257. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Hu B, Wang Q, Wang Y, Chen J, Li P and Han
M: Holliday junction-recognizing protein promotes cell
proliferation and correlates with unfavorable clinical outcome of
hepatocellular carcinoma. Onco Targets Ther. 10:2601–2607. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wei Y, Ouyang GL, Yao WX, Zhu YJ, Li X,
Huang LX, Yang XW and Jiang WJ: Knockdown of HJURP inhibits
non-small cell lung cancer cell proliferation, migration, and
invasion by repressing Wnt/β-catenin signaling. Eur Rev Med
Pharmacol Sci. 23:3847–3856. 2019.PubMed/NCBI
|
|
37
|
Chen YF, Liang YX, Yang JA, Yuan DZ, Li J,
Zheng SS, Wan YP, Wang B, Han ZD and Zhong WD: Upregulation of
holliday junction recognition protein predicts poor prognosis and
biochemical recurrence in patients with prostate cancer. Oncol
Lett. 18:6697–6703. 2019.PubMed/NCBI
|
|
38
|
Serafim RB, Cardoso C, Di Cristofaro LFM,
Pienna Soares C, Araújo Silva W Jr, Espreafico EM, Paçó-Larson ML,
Price BD and Valente V: HJURP knockdown disrupts clonogenic
capacity and increases radiation-induced cell death of glioblastoma
cells. Cancer Gene Ther. 27:319–329. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Liu J, Zhou J, Wu Z, Wang X, Liu L and Yao
C: Cyanidin 3-O-beta-glucoside ameliorates ethanol-induced acute
liver injury by attenuating oxidative stress and apoptosis: The
role of SIRT1/FOXO1 signaling. Alcohol Clin Exp Res. 40:457–466.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Ruan Y, Dong C, Patel J, Duan C, Wang X,
Wu X, Cao Y, Pu L, Lu D, Shen T and Li JL: SIRT1 suppresses
doxorubicin-induced cardiotoxicity by regulating the oxidative
stress and p38MAPK pathways. Cell Physiol Biochem. 35:1116–1124.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Circu ML and Aw TY: Reactive oxygen
species, cellular redox systems, and apoptosis. Free Radic Biol
Med. 48:749–762. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Wang Y, Liang X, Chen Y and Zhao X:
Screening SIRT1 activators from medicinal plants as bioactive
compounds against oxidative damage in mitochondrial function. Oxid
Med Cell Longev. 2016:42063922016. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Wang Q, He G, Hou M, Chen L, Chen S, Xu A
and Fu Y: Cell cycle regulation by alternative polyadenylation of
CCND1. Sci Rep. 8:68242018. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Guardavaccaro D, Corrente G, Covone F,
Micheli L, D'Agnano I, Starace G, Caruso M and Tirone F: Arrest of
G(1)-S progression by the p53-inducible gene PC3 is Rb dependent
and relies on the inhibition of cyclin D1 transcription. Mol Cell
Biol. 20:1797–1815. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Musgrove EA, Lee CS, Buckley MF and
Sutherland RL: Cyclin D1 induction in breast cancer cells shortens
G1 and is sufficient for cells arrested in G1 to complete the cell
cycle. Proc Natl Acad Sci USA. 91:8022–8026. 1994. View Article : Google Scholar : PubMed/NCBI
|