Introduction
Over the last 30 years, systemic therapeutic (drug
therapy) guidelines for patients with recurrent or metastatic (R/M)
head and neck squamous cell carcinoma (HNSCC) have not changed
(1). In 2008, the Extreme trial
(2) found that the addition of
cetuximab (trade name Erbitux™), a molecular targeted agent against
epidermal growth factor receptor (EGFR), to the standard platinum
and 5-fluorouracil (5-FU) regimen improved median overall survival
(OS) and progression-free survival (PFS) in HNSCC. The National
Comprehensive Cancer Network (NCCN) Clinical Practice Guidelines
have also suggested that systemic therapy, including cetuximab,
should be listed as the recommended category 2B for its
effectiveness in treating very advanced HNSCC (3). Based on this evidence, cetuximab
therapy was selected for R/M oral squamous cell carcinoma (OSCC),
and the tumor growth was controlled for unresectable R/M OSCC
(4,5). However, some OSCC tumors acquired
resistance with long-term cetuximab administration, and new lesions
appeared in the brain following 2 years of treatment (6); thus management of these lesions is a
future priority.
Although cetuximab is reported to have significant
therapeutic efficacy against HNSCC (2,7,8), the
phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit α
isoform (PIK3CA) gene is a candidate gene involved in the acquired
resistance to long-term cetuximab treatment due to point mutations
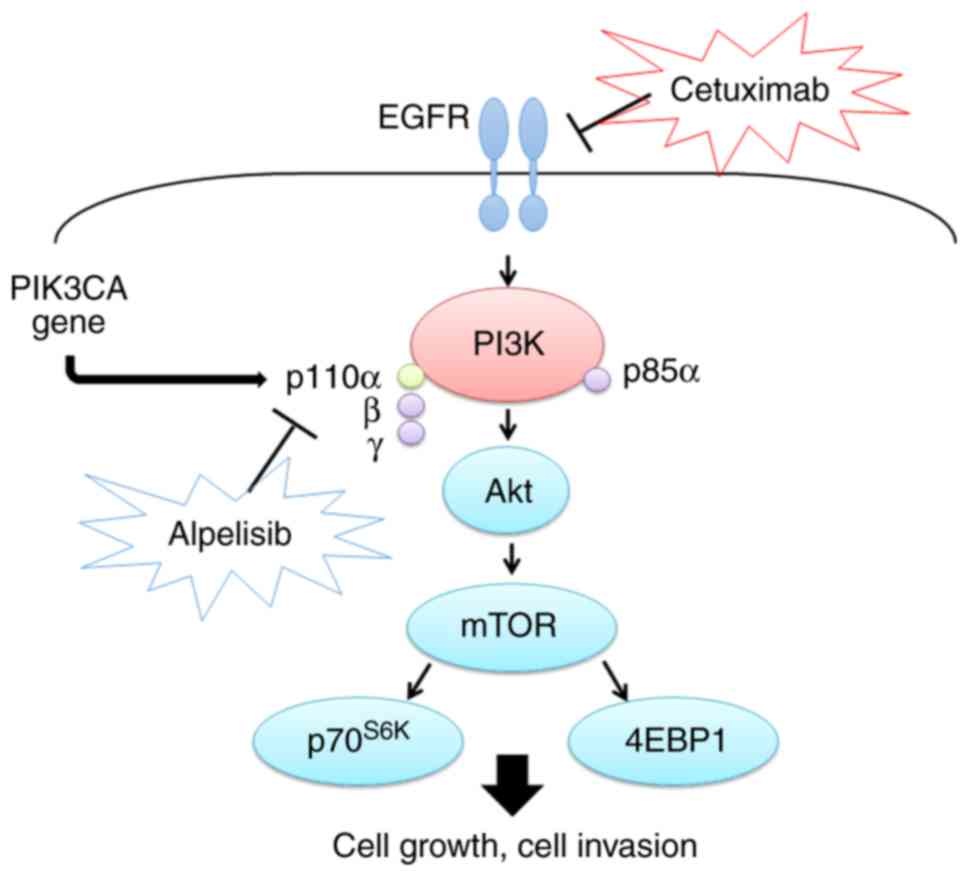
within the gene (9–11). PIK3CA, a key element of the PI3K/AKT
pathway (Fig. 1), is located on
chromosomal 3q26.3, encoding the 110 kDa catalytic subunit of class
IA PI3K (PI3Kp110α) (12,13). Somatic PIK3CA gene point mutations
have been identified as cancer-specific or heterozygous, and these
mutations have been proven to activate the PI3K/Akt-mTOR signaling
pathway in human cancers, including HNSCC (12,13).
Most somatic PIK3CA gene mutations are clustered in the helical
domain encoded by exon 9 and the kinase domain encoded by exon 20
in OSCC (12,13). In addition, three hotspot mutations
in these exons, E542K, E545K and H1047R, were proven to activate
the PI3K/AKT pathway through the phosphorylation of AKT (12,13).
Overexpression of the mutant PIK3CA gene markedly increased
Akt/mTOR pathway activation compared with that in overexpressed
wild-type PIK3CA in HNSCC (12).
Consequently, the PIK3CA point mutation may promote cetuximab
resistance in HNSCC (8,9). Copy number amplifications and gene
point mutations of PIK3CA have also been reported in OSCC (12,14)
and observed in advanced stages of OSCC (12). To the best of our knowledge,
regarding clinical outcomes of PIK3CA mutations and cetuximab
therapy, PIK3CA point mutations in colorectal cancer are associated
with clinical resistance to EGFR-targeted monoclonal antibodies
(cetuximab or panitumumab) (15,16)
however, the clinical outcome of PIK3CA mutants against OSCC has
not been investigated.
Therefore, the present study aimed to determine the
clinicopathological significance of the tumor response to cetuximab
therapy in R/M OSCC. Furthermore, the antitumor effects of PI3Kp110
inhibitor, alone or in combination with cetuximab, were examined to
clarify the additional benefit of combination therapy in the
preclinical model of OSCC.
Materials and methods
Patients
The present study was approved by the independent
Ethics Committee of Nagasaki University Hospital (approval no.
19081915). The medical records of 25 patients who received
cetuximab therapy for R/M OSCC between December 2012 and March 2017
were retrospectively reviewed at Nagasaki University Hospital
(Table I). The Eastern Cooperative
Oncology Group (ECOG) (17)
performance status score was used as follows: 0, Fully active, able
to perform all pre-disease performance without restriction; 1,
restricted in physically strenuous activity but ambulatory and able
to perform light or sedentary work, e.g., light house work and
office work, 2, ambulatory and capable of all selfcare but unable
to perform any work activities for example walking about >50% of
waking hours. Survival data was reviewed in March 2019.
Furthermore, 4-µm paraffin-embedded sections (fixed in buffered 10%
formalin for 24 h at room temperature) from biopsy or resected
specimens obtained immediately prior to cetuximab therapy were
obtained. Tumor response was assessed every 4–8 weeks with repeated
clinical and enhanced computed tomography assessments. in
accordance with the RECIST guidelines (18). Normal oral mucosal specimens were
obtained from 10 healthy individuals undergoing routine surgical
removal of their third molars during the aforementioned study
period at Nagasaki University Hospital. The tumor stage was
classified according to the TNM classification of the Union for
International Cancer Control (19).
Tumor histological differentiation was defined according to the
World Health Organization classification (20). The pattern of invasion was assessed
according to Bryne's classification (score 1, well-defined margins;
score 2, solid strings and/or inlets; score 3, small cell groups
n<15; score 4, cellular diffuse dissociation and characterized
in small and/or isolated cell groups) (21).
 | Table I.Clinicopathological characteristics
of the 25 patients with oral squamous cell carcinoma. |
Table I.
Clinicopathological characteristics
of the 25 patients with oral squamous cell carcinoma.
|
Characteristics | Value |
|---|
| Sex, n (%) |
|
|
Male | 11 (44.0) |
|
Female | 14 (56.0) |
| Median age,
years | 68 |
| Age, range,
years | 50-90 |
| Primary site, n
(%) |
|
|
Tongue | 8
(32.0) |
|
Gingiva | 14 (56.0) |
| Oral
floor | 1 (4.0) |
|
Intraosseous | 1 (4.0) |
|
Buccal | 1 (4.0) |
| Differentiation, n
(%) |
|
|
Well | 22 (88.0) |
|
Moderate | 1 (4.0) |
|
Poor | 1 (4.0) |
| Not
specified | 1 (4.0) |
| ECOG Performance
status, n (%) |
|
| 0 | 13 (52.0) |
| 1 | 9
(36.0) |
| 2 | 3
(12.0) |
| Initial treatment,
n (%) |
|
| Surgery
alone | 15 (60.0) |
| Surgery
+ adjuvant RT | 3
(12.0) |
| Surgery
+ adjuvant CCRT | 7
(28.0) |
| Cetuximab regimen,
n (%) |
|
| Cet +
RT | 15
(60.0) |
| Cet +
PTX | 8
(32.0) |
| Cet +
FP | 1
(4.0) |
| Cet
alone | 1
(4.0) |
| Median number of
treatment cycles, n | 10 |
| Number of treatment
cycles, range, n | 2-70 |
Immunohistochemistry staining and
evaluation
Sections were deparaffinized in xylene, rehydrated
in a descending alcohol series (70-100%), incubated in 10 mM
citrate buffer (pH 6.0), and heated at 121°C for 5 min for antigen
retrieval. Endogenous peroxidases were blocked by incubation with
0.3% H2O2 in methanol for 30 min at room
temperature. Immunohistochemistry staining was performed using the
EnVision kit (EnVision+; Dako; Agilent Technologies, Inc.). The
PI3Kp110α (cat. no. 4249S; dilution 1:400; Cell Signaling
Technology, Inc.) rabbit polyclonal primary antibody was used.
Tissue sections were washed in phosphate-buffered saline, followed
by incubation with the primary antibody overnight at 4°C, then with
a secondary antibody (cat. no. K4003; pre-diluted; Dako; Agilent
Technologies, Inc.) for 30 min. Reaction products were visualized
by immersing the sections in diaminobenzidine solution. The samples
were counterstained with Meyer's hematoxylin for 10 min at room
temperature and subsequently mounted.
The expression of PI3Kp110α protein was evaluated by
calculating the total immunostaining score as the product of the
number of positive cells (proportional score) and intensity scores
at the invasion front (inside surface) of the tumor in two fields
of view. The proportional scores were based on the calculated
fraction of positively stained tumor cells (0, none; 1, <10%; 2,
10–50%; 3, 50–80%; 4, >80%). The intensity score represented the
calculated staining intensity (0, no staining; 1, weak; 2,
moderate; 3, strong). Total immunostaining scores ranged from 0 to
12, with positive cases defined as total scores >4. Patient
samples showed a bimodal distribution of immunohistochemistry
protein expression, with the most common score of 3–4, therefore
the cut-off for positive expression was set to >4. All
immunohistochemical assessments were performed by both an oral
cancer surgeon and an oral cancer pathologist, who were blinded to
the samples.
Cell lines and reagents
A total of three human OSCC cell lines, SAS
(wild-type PIK3CA gene), HSC-2 and HSC-3 (both with mutant PIK3CA
gene) were used in the present study, as found in a previous study
(12) and were cultured in a 1:1
mixture of Ham's F-12/Dulbecco's modified Eagle's medium
supplemented with 10% fetal bovine serum (all Trace Scientific,
Ltd; Thermo Fisher Scientific, Inc.). All cells were maintained at
37°C in a humidified incubator with 5% CO2 and 19%
O2. Alpelisib, a selective PI3Kα inhibitor, was
purchased from Selleck Chemicals (cat. no. BS119QD), dissolved in
dimethyl sulfoxide (DMSO), and adjusted to a range of
concentrations (0.1–100 µM) with culture medium (1:1 mixture of
Ham's F-12 and DMEM supplemented with 10% fetal bovine serum) (all
Trace Scientific, Ltd; Thermo Fisher Scientific, Inc.). Cetuximab
(cat. no. C225) was purchased from Merck KGaA. Working solutions
were freshly prepared from the stock solution by diluting with cell
culture medium on the day of the experiment.
Cell cytotoxicity assay
Cells were seeded in 96-well plates at a
concentration of 1.5×103 per well and incubated for 24
h. Cells were exposed to alpelisib or cetuximab at concentrations
ranging from 0.1 to 100 µM. At the end of the 72-h treatment, cells
were incubated with 0.5 mg/ml MTT (Sigma-Aldrich; Merck KGaA). The
medium was replaced with 100 µl DMSO and vortexed for 10 min, 4 h
later. Absorbance was recorded at 570 nm, using a microplate auto
reader (Multiskan FC; Thermo Fisher Scientific, Inc.). Cell
viability was calculated as the percentage of cells from that in
the control group, which received medium only. IC50 is a widely
used indicator of the effectiveness of inhibitors in pharmaceutical
research (22) and was subsequently
calculated from the results. IC50 was calculated from
the results of MTT assay, using the following equation,
IC50=10^ [Log(A/B) × (50-C)/(D-C) + Log(B)] was used. A
indicates high concentration across 50%; B, indicates low
concentration across 50%; C, indicates inhibition rate at B and D
indicates inhibition rate at A. Therefore, the appropriate
concentration was used to compare the efficacy of alpelisib and
cetuximab with that in the control group.
Western blot analysis
Cells were harvested using trypsinization, washed
with PBS, and precipitated by centrifugation (4°C; 22,140 × g, for
5 min). The Mammalian Cell Extraction kit (BioVision, Inc.) was
used to extract the total proteins. All subsequent manipulations
were performed on ice. Cells were incubated in Extraction Buffer
Mix. The protein concentration of each sample was measured using
the micro-bicinchoninic acid protein assay (Pierce; Thermo Fisher
Scientific, Inc.). After the samples were denatured in SDS sample
buffer, they were incubated at 70°C for 10 min and then 10 µg was
loaded onto a 4–12% NuPAGE NOVEX bis-tris polyacrylamide gel or a
3–8% NuPAGE NOVEX tris-acetate polyacrylamide gel. After
electrophoresis, the separated proteins were transferred to iBlot
polyvinylidene difluoride membranes, using the iBlot Dry Blotting
System (Thermo Fisher Scientific, Inc.). The membrane was blocked
for 30 min using blocking reagent (part A and B) from the Western
Breeze Immunodetection kit (Thermo Fisher Scientific, Inc.) at room
temperature. Primary antibodies against PI3Kp110α (cat. no. 4249S;
Cell Signaling Technology, Inc.), Akt (cat. no. 9272S; Cell
Signaling Technology, Inc.), phosphorylated (p)-Akt (cat. no.
4060S; Cell Signaling Technology, Inc.), mTOR (cat. no. 2972S; Cell
Signaling Technology, Inc.), p-mTORSer2448 (cat. no.
2971S; Cell Signaling Technology, Inc.), p70S6K (cat. no. 9202S;
Cell Signaling Technology, Inc.), p-p70S6KTyr389 (cat.
no. 9205S; Cell Signaling Technology, Inc.), 4EBP-1 (cat. no.
9452S; Cell Signaling Technology, Inc.), p-4EBP1 (cat. no. 9459S;
Cell Signaling Technology, Inc.), E-cadherin, and N-cadherin (DAKO;
Agilent Technologies, Inc.) were used at 1:1,000 dilution and the
membrane was incubated for one h at room temperature. β-actin
(4970S; Cell Signaling Technology, Inc.) was used as the loading
control. The secondary antibody (Western Breeze Immunodetection
kit; Thermo Fisher Scientific, Inc.) was used at a dilution of 1:10
at room temperature for 30 min. Signals were detected using the
Western Breeze Immunodetection kit (Thermo Fisher Scientific,
Inc.). PI3Kp110α/β-actin ratio was determined using Image J
software v1.51 (National Institutes of Health).
Cell migration and invasion assay
The method used for migration and invasion was the
same, in that a Biocoat Matrigel invasion chamber containing an
internal chamber with an 8-µm porous membrane bottom was used for
both assays; however, for the invasion assay the chamber was coated
with Matrigel for 2 h at 37°C (Becton, Dickinson and Company). A
total of 12 cell culture inserts and a 24-well multiwall companion
plate were used for the experiment. The membranes were rehydrated
with warm serum-free 1:1 mixture of Ham's F-12 and DMEM for 2 h.
Cells were collected using trypsinization, followed by seeding, at
a density of 1.25×105, in the internal chamber with
serum-free DMEM. The lower chamber was filled with medium
containing 10% FBS as a chemoattractant. Cells were incubated for
72 h, and non-invading, and non-migrating cells were removed from
the top of the wells with a cotton swab, and cells that transferred
to the inverse surface of the membrane were subjected to Diff-Quick
staining for 15 sec at room temperature. Cells were counted using a
light microscope at ×100 magnification. Cells that passed through a
control chamber without Matrigel were used for calculating the
migration index. The number of cells that passed through the
Matrigel chamber were divided by the control cell count to
calculate the percentage of invasion. All experiments were
performed in triplicate, and cell numbers were counted in at least
two fields of view/well.
Xenograft model assay
The animal protocol was approved by the Animal Care
and Use Committee at Nagasaki University Graduate School of
Biomedical Sciences (approval no. 1901041500). Briefly, a total of
16, female BALB/c-nu/nu mice (6 weeks old) weighing 18–20 g were
purchased from Japan SLC Inc. and maintained in a barrier unit,
under standard conditions (temperature, 20–26°C; humidity, 40–70°C;
12-h light/dark cycle; with ad libitum access to food and
water). HSC-3 cells (1×107) were suspended in 100 µl PBS
and injected subcutaneously into the mice using a 21-gauge needle.
After growing to 10–15 mm in diameter, the mice were sacrificed
using carbon dioxide. When the flow rate had displaced >30% of
the volume of air in the chamber (infusion of carbon dioxide was 1
min), the mice were checked for >5 min and death was confirmed
by observing lack of respiration and cardiac output. The HSC-3
tumor was extracted, cut into 1-mm3 sections, and
transplanted subcutaneously into the back of 12 different
BALB/c-nu/nu mice. These mice were divided into four treatment
groups: Control, cetuximab, alpelisib, and cetuximab plus
alpelisib. The tumors were allowed to grow to 5–10 mm in diameter,
following which the tumor-bearing mice were treated for 4 weeks
with cetuximab (20 mg/kg, three times/week) and/or alpelisib (20
mg/kg, three times/week). Cetuximab was diluted 1:4 in saline,
while alpelisib was dissolved in 64.5% saline, and the
concentration was adjusted using 30% polyethylene glycol 400, 0.5%
polyoxyethylene sorbitan monooleate 80, and 5% propylene glycol.
The control group received saline only. All treatments were
delivered by intraperitoneal injection. The mice were treated for 4
weeks, after which time the xenografted tumors were excised, fixed
in buffered 10% formalin for 24 h at room temperature, and embedded
in paraffin for histological examination using hematoxylin and
eosin staining, and immunohistochemical examination using PI3Kp110α
(cat. no. 4249S; dilution 1:400), EGFR (cat. no. 4267S; dilution
1:100), and p-mTORSer2448 (cat. no. 2976S; dilution
1:100) antibodies (all Cell Signaling Technology, Inc.), and
incubated overnight at 4°C. Sections (4-µm thick) were
deparaffinized in xylene, rehydrated in a descending alcohol series
(70-100%), then incubated with Meyer's hematoxylin stain for 4 min
and Eosin stain for 1 min at room temperature.
Statistical analysis
All statistical analyses were performed using Excel
software v3.0 (Microsoft Corporation). Associations between the
expression level of proteins of interest and clinicopathological
characteristics were analyzed using Fisher's exact test. Continuous
data are presented as mean ± standard deviation. Survival analyses
were calculated using the Kaplan-Meier method and compared using
the log-rank test. The correlations between protein expression
levels of PI3Kp110α and cell migration and invasion, and with
cetuximab sensitivity, in the OSCC cell lines, were analyzed using
the Spearman's rank correlation test. A multiple comparison test
between two groups in MTT assay, migration and invasion assays was
performed using the Scheffe's method. P<0.05 were considered to
indicate a statistically significant result.
Results
Expression of PI3Kp110α in OSCC
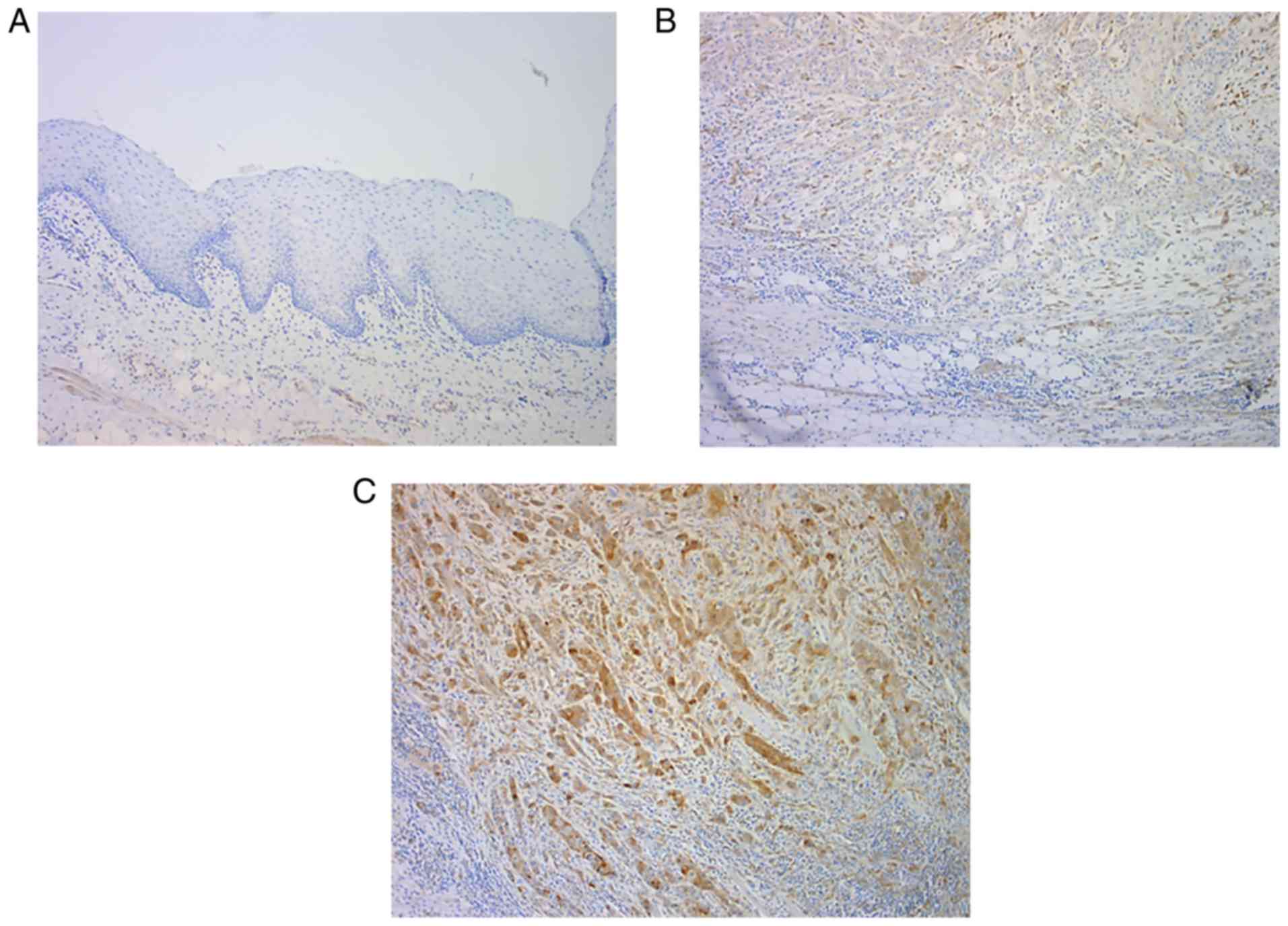
In normal oral epithelium, the expression of
PI3Kp110α was negative (Fig. 2A).
Among the 25 patients with OSCC, PI3Kp110α expression was detected
in 56% of the patients using immunohistochemical staining.
PI3Kp110α was expressed primarily in the cytoplasm and nucleus of
the tumor cells, and the staining intensity ranged from weak to
strong (Fig. 2B and C).
Association of PI3Kp110α expression
with clinicopathological characteristics and survival
The expression levels of PI3Kp110α in OSCC were
examined as a function of the clinical response to cetuximab
therapy. Positive expression of PI3Kp110α was significantly
associated with stable disease (SD)/progressive disease (PD) in the
clinical response (P<0.05). The overall response rate was 68.0%,
with 9 patients achieving CR and 8 achieving PR. The disease
control rate was 84.0%, which included 4 patients with SD (Table II). The cases of positive PI3Kp110α
expression originated from the following tissues: Tongue (n=6),
gingiva (n=6), buccal mucosa (n=1), and one primary intraosseous
(n=1), and no significant difference was found with respect to
origin.
 | Table II.Tumor response of cetuximab therapy
and association between expression of PI3Kp110α and tumor response.
(n=25). |
Table II.
Tumor response of cetuximab therapy
and association between expression of PI3Kp110α and tumor response.
(n=25).
| Factor | Value | P-value |
|---|
| Best response, n
(%) |
|
|
| CR | 9 (36.0) |
|
| PR | 8 (32.0) |
|
| SD | 4 (16.0) |
|
| PD | 4 (16.0) |
|
| Overall response
ratea, % | 68.0 |
|
| Disease control
rateb, % | 84.0 |
|
| PI3Kp110α |
|
|
| CR/PR,
n |
| 0.042 |
| − | 10 |
|
| + | 7 |
|
| SD/PD, n |
|
|
| − | 1 |
|
| + | 7 |
|
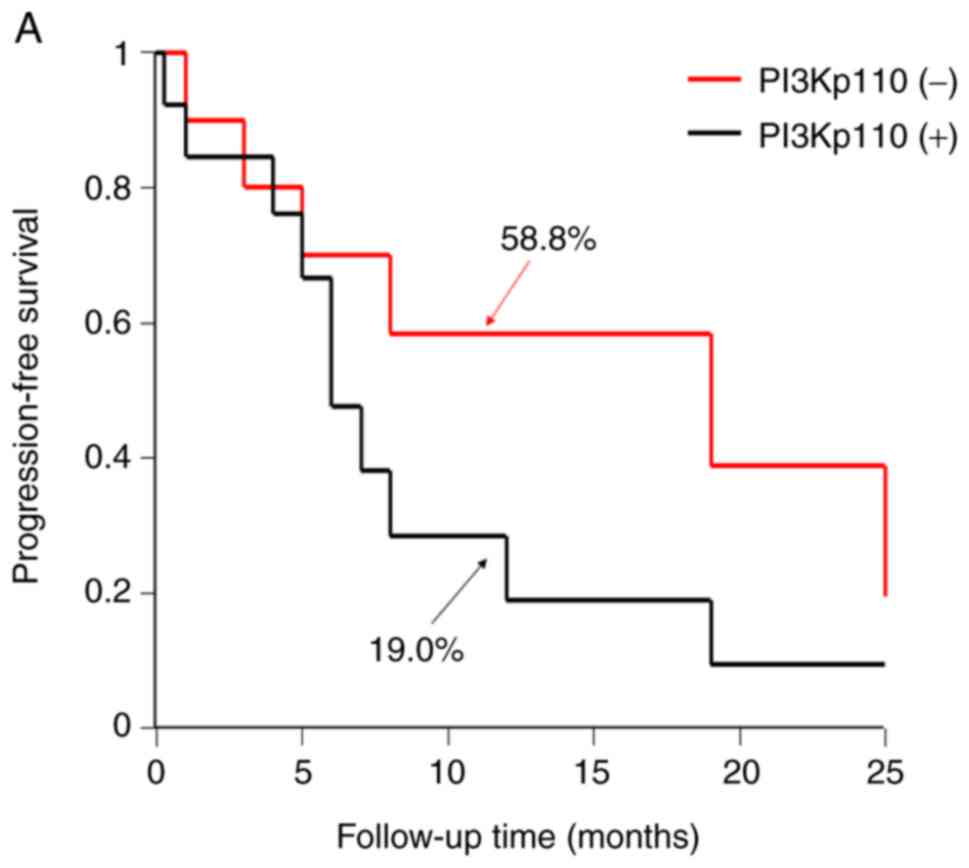
The 1-year progression-free survival (PFS) and
overall survival (OS) rates, according to the expression level of
PI3Kp110α, were determined. The Kaplan-Meier method revealed that
the PI3Kp110α high expression group had a lower PFS and OS compared
with that in the low expression group. However, the 2-year PFS and
OS between PI3Kp110α high and low expression groups were equal
(Fig. 3A and B).
Effect of PI3Kp110α expression on the
cell migration and invasion potential of OSCC cell lines
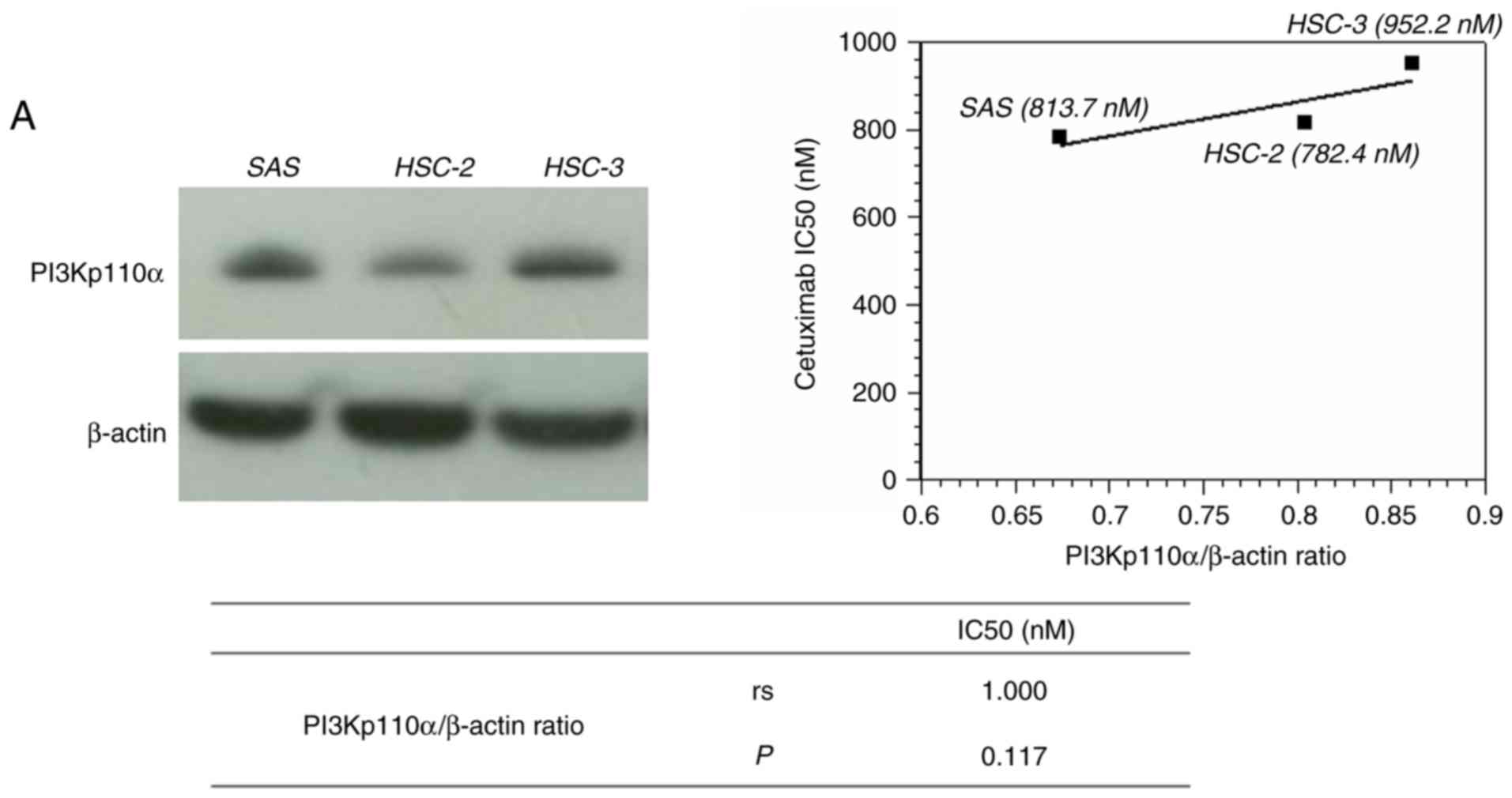
At the protein level, the expression of PI3Kp110α
was evaluated using western blot analysis in three OSCC cell lines,
following treatment with cetuximab (SAS, 813.7 nM; HSC-2, 82.4 nM;
HSC-3, 952.2 nM). There was no significant difference between
cetuximab sensitivity and PI3Kp110α protein level; however, a high
correlation was observed (Fig. 4A).
For the migration and invasion indices, the correlation between
protein expression level of PI3Kp110α and migration and invasion
potential were determined. There was a significant difference in
the invasion index (P=0.042; Fig.
4B), but not with migration index. As the HSC-3 cell line,
which is a PIK3CA mutant cell line, exhibited high expression of
PI3Kp110α amongst the three cell lines, it was used for subsequent
experiments.
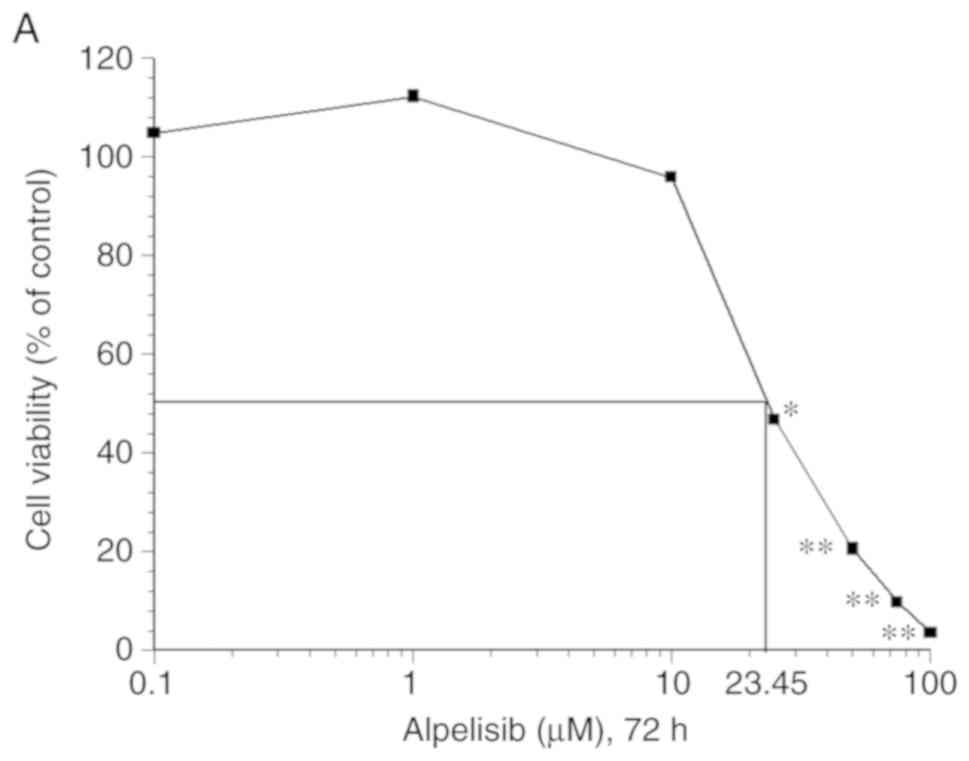
The MTT assay revealed that alpelisib significantly
inhibited cell proliferation in a dose-dependent manner in the
HSC-3 cell line (P<0.05) (Fig.
5A). Moreover, western blot analysis revealed that alpelisib
decreased the levels of PI3Kp110α, p-Akt, p-mTOR, p-p70S6K, and
p-4EBP1 at the IC50 concentration (23.54 µM), compared
with that in the controls (Fig.
5B). In addition, the expression of E-cadherin was increased,
while that of N-cadherin was decreased.
Alpelisib and cetuximab alone or in
combination decreases HSC-3 cell migration and invasion
potential
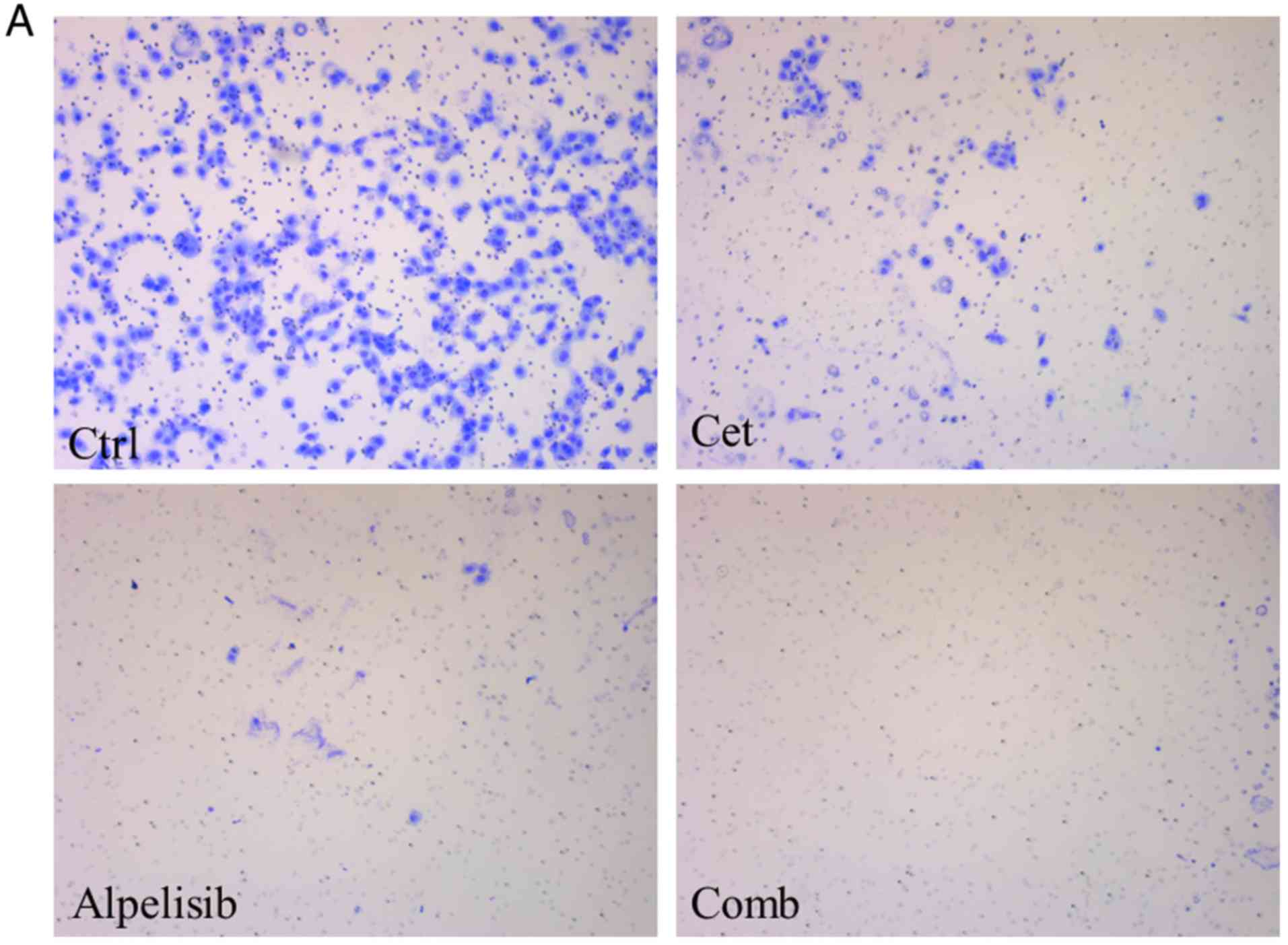
Following treatment of HSC-3 cells with alpelisib or
cetuximab alone or in combination there was a decrease in cell
migration (Fig. 6A), which was
found to be significantly different; the cell migration was
significantly lower in the alpelisib or cetuximab only treatment
groups and in combination compared with that in the control group
(Fig. 6B; P<0.01). In addition,
there was a significant decrease in migration with the combined
treatment group compared with that in cetuximab alone. No
significant differences were found between the alpelisib or
cetuximab only treatment groups.
Furthermore, there was also a decrease in cell
invasion (Fig. 6C), which was also
significant; cell invasion was significantly lower in the
combination treatment group (P<0.01) and in the alpelisib or
cetuximab only treatment groups (P<0.05) compared with that in
the control group (Fig. 6D).
Effects of alpelisib and cetuximab
alone or in combination in vivo
As alpelisib and cetuximab revealed antitumor
effects in vitro, the antitumor potential was subsequently
investigated in vivo effects. The weight of the mice in the
control, cetuximab and alpelisib only treatment groups and in the
combined treatment groups, at the end of the experiment, were 24.2,
22.6, 23.3 and 22.8 g, respectively, while the weight of the
resected tumors were 1.8, 0.14, 0.34 and 0.09 g, respectively. The
tumors in mice treated with cetuximab only grew markedly slower
compared with that in the control group, and the tumors treated
with alpelisib and cetuximab in combination also grew markedly
slower compared with that in the control, alpelisib- and
cetuximab-only treatment groups (Fig.
7A). However, there was no significant association between the
type of treatment and tumor size. The protein expression levels of
PI3Kp110α, EGFR, and p-mTOR was evaluated using
immunohistochemistry in the excised tumors, and were highly
expressed in the tumors from the control group. However, the
expression levels of both EGFR and PI3Kp110α were negative in the
majority of the tumor cells in the tumors treated with
cetuximab-only. The expression of EGFR was strongly observed in the
tumors treated with alpelisib only, whereas both PI3Kp110α and
p-mTOR were negative in the majority of the tumor cells. No
expression of PI3Kp110α, EGFR, or p-mTOR expression was observed in
the tumors treated with a combination of cetuximab and alpelisib
(Fig. 7B).
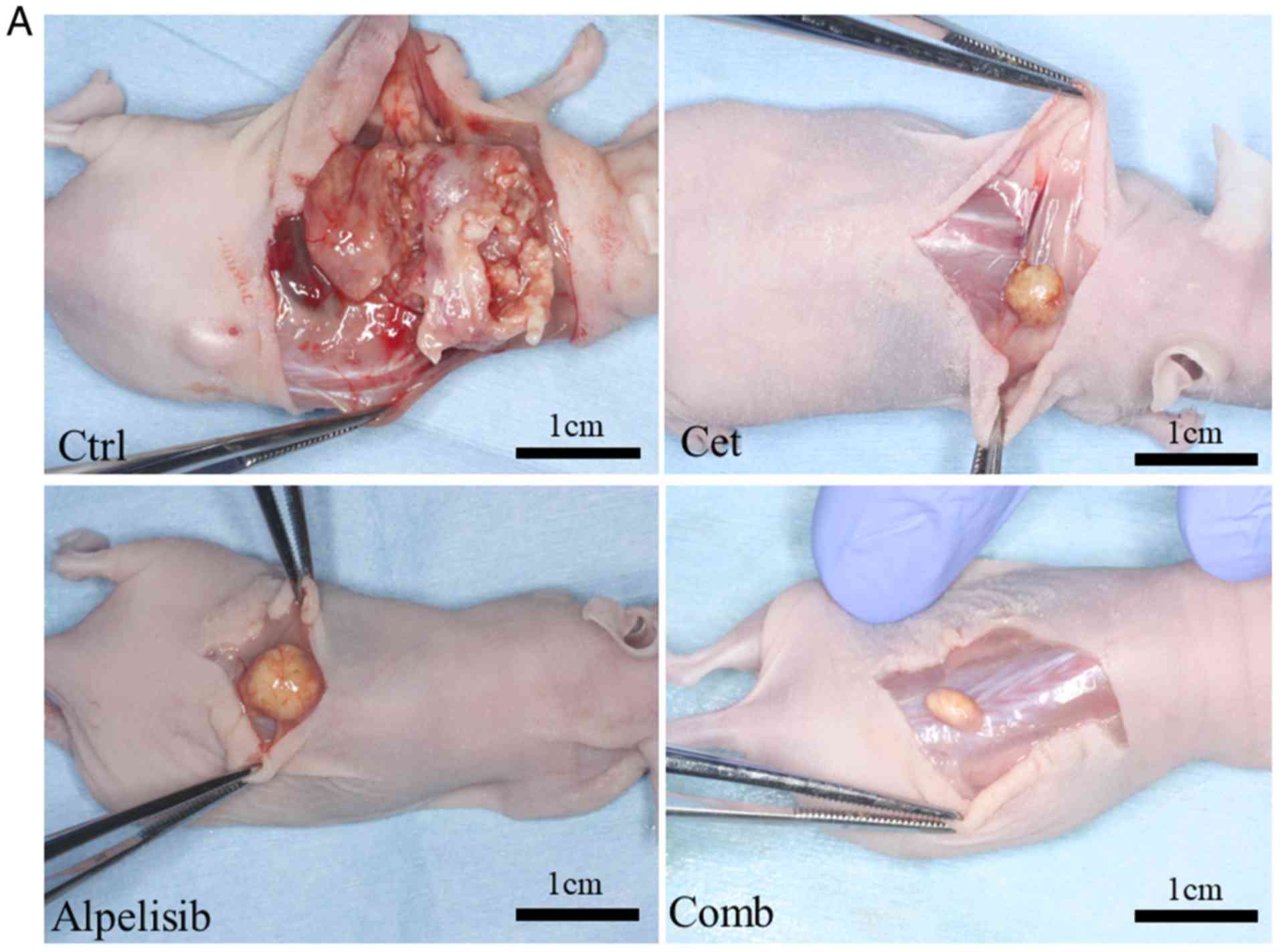 | Figure 7.Effects of alpelisib and cetuximab
alone or in combination in vivo. (A) Representative images
of the HSC-3 ×enograft tumors in mice treated with either control
(saline-treated), cetuximab-only, alpelisib-only, or in
combination. (B) Representative images of paraffin-embedded
sections of HSC-3 tumors treated with either control,
cetuximab-only, alpelisib-only, and in combination in vivo
stained with H&E or EGFR) PI3Kp110α, and mTOR Ser2448
Magnification, ×100. H&E, hematoxylin and eosin; cont, control;
cet, cetuximab; comb, combination. |
Discussion
The NCCN Clinical Practice Guideline has suggested
that systemic therapy (drug therapy), including cetuximab is the
standard treatment for very advanced HNSCC; however, in some cases
resistance to long-term administration of cetuximab develops, and
new lesions can appear, including the brain (3,6). The
present study demonstrated that high protein expression of
PI3Kp110α, which encodes the mutant PIK3CA gene, is associated with
cetuximab resistance and survival. Furthermore, administration of
alpelisib in combination with cetuximab, inhibits the Akt-mTOR
pathway, providing additional antitumor effects, such as reduced
growth in OSCC.
The clinicopathological results revealed that the
protein expression of PI3Kp110α was associated with response to
cetuximab (P<0.05). In addition, patients with high expression
of PI3Kp110α had poor 1-year PFS and OS compared with that in
patients with low expression. To the best of our knowledge, this is
the first study to report the association of high PI3Kp110α
expression with cetuximab response. The point mutant PIK3CA gene
has been associated with PFS, OS, and the clinical response to
anti-EGFR antibodies, including cetuximab, in colorectal cancer
(15). Moreover, somatic point
mutations in exon 20 of PIK3CA are significantly associated with a
low response to cetuximab in metastatic colorectal cancer (16). In HNSCC, treatment using cetuximab
was not effective in seven patients with R/M HNSCC who also
harbored the point mutant PIK3CA gene, as recurrence or metastasis
was found (23). However, there was
no association between patients with R/M OSCC, who also harbored
the mutant PIK3CA gene and the cetuximab response. It is unclear
whether it is appropriate to evaluate the same clinical response of
cetuximab with the mutant PIK3CA gene and the protein expression of
PI3Kp110α; although, overexpression of mutant PIK3CA gene notably
increased the activation of the PI3K/Akt/mTOR pathway (13). Therefore, this suggests that high
expression of PI3Kp110α is a novel biomarker for the clinical
response to cetuximab. With respect to prognosis, it was previously
reported by our laboratory that cetuximab therapy provided
additional benefits (such as improved 1-year OS) in patients with
distant metastasis compared with patients receiving non-cetuximab
therapy in 1-year, but not 2-year OS (5). It is difficult to maintain the
therapeutic effect of cetuximab for >2 years; however, PI3Kp110α
could be a useful marker of one-year PFS and OS.
Biological evidence from the present study supports
our hypothesis that the protein expression of PI3Kp110α extracted
from OSCC cell lines is associated with the cetuximab response and
is significantly associated with invasive potential. Alpelisib was
selected to inhibit the PI3Kp110α protein, as it is a single agent
with proven effectiveness by inhibiting proliferation, apoptosis
and inducing cell cycle arrest in HNSCC in a preclinical study
(24). In the preclinical study of
HNSCC, alpelisib was shown to overcome cetuximab resistance in
HNSCC (25). In addition, alpelisib
treatment reduced Akt activation and suppressed tumor growth in
HNSCC, in vitro and in vivo (25,26).
In the present study the HSC-3 OSCC cell line were found to be
sensitive to the anti-proliferative effects of alpelisib. Moreover,
there was also a decrease in phosphorylation of downstream markers
of the PI3K-Akt pathway in a dose-dependent manner. In particular,
alpelisib-mediated suppression of the epithelial-to-mesenchymal
transition (EMT), which was confirmed using western blot analysis.
Kimura et al (27) reported
that cetuximab-resistant cell lines (including HOC313) increased
the mRNA and protein expression levels of EMT-associated genes
(such as E-cadherin, N-cadherin, vitmentin and Snail). Schmitz
et al (28) also determined
the association between the increase in protein expression of EMT
markers (ZEB1, TWIST1, TWIST2, LEF1, VIM, SNAI1 and SNAI2) and
cetuximab resistance using 20 pre- and post-cetuximab treatment
HNSCC biopsy specimens (P<0.05). Moreover, in our previous study
there was a case of distant metastasis and the appearance of new
lesions following long-term cetuximab therapy was observed
(6). In addition, based on the
immunohistochemical staining of recurrent tumors in this case
before and after long-term cetuximab administration, cetuximab
resistance may be associated with EMT (6). These reports suggest that
cetuximab-resistant cells possess high invasive and migration
potential (associated with EMT) and that inhibition of PI3Kp110α
might inhibit the potential of cetuximab-resistant cells to undergo
EMT in OSCC.
We hypothesized that cetuximab and alpelisib would
have additive anti-cancer effects in vitro, through
selective blocking of the extracellular ligand-binding domain of
EGFR by cetuximab, while alpelisib competed for the intracellular
catalytic site of PI3K. It has been reported that alpelisib in
combination with cetuximab had an overall response rate of 11% and
a disease control rate of 54% in cetuximab-resistant HNSCC
(25); however, OSCC was not
included in this study. The EXTREME trial revealed that the benefit
of including cetuximab to platinum-based chemotherapy for patients
with HNSCC was more effective in patients with OSCC. However, the
results should be interpreted with caution, as this does not
suggest that OSCCs would be the only HNSCC tumor types to respond
to this type of therapy (2).
Therefore, studies with OSCC alone are required. In the present
study, both cetuximab and alpelisib inhibited cell invasion and
migration potential, and their combined treatment resulted in
additive inhibition of OSCC migration and invasion potential
compared with that in either agent alone. In our previous study the
inhibition of the PI3K downstream pathway decreased the migration
and invasion ability in the HSC-3 cell line (29). Therefore, combined treatment with
alpelisib and cetuximab was effective in OSCC invasion in
vitro.
The combined treatment of alpelisib and cetuximab
markedly inhibited OSCC progression in vivo compared with
that in either agent alone; however, there was no significant
association with tumor size. This could be due to the antitumor
effects of alpelisib or cetuximab alone in the xenograft tumor,
which was evident from the decreased protein expression levels of
PI3Kp110α from immunohistochemistry analysis.
The immunohistochemical study is retrospective
nature and includes a small number of cases from a single
institution, which is a limitation to the present study. Therefore,
an intergroup study with an increased number of cases is
required.
In conclusion, the results of the present study
suggest that the PI3K pathway plays an important role in
cetuximab-resistant OSCC, and that combined treatment with
alpelisib and cetuximab may overcome cetuximab resistance. Previous
studies have revealed that, nivolmab, an anti-programmed cell death
protein 1 (PD-1) monoclonal antibody, has been recommended as
another treatment option for R/M HNSCC (3,30).
However, the overall response rate is not high, thus additional
treatment options are required. Therefore, novel systemic drug
treatment regimens that include PI3K inhibitors are expected to
further improve the survival of patients with cetuximab-resistant
OSCC in the future.
Acknowledgements
Not applicable.
Funding
This study was partially supported by the Ministry
of Education, Culture, Sports, Science and Technology, Japan (grant
no 17K17267).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
TN, SY and MU designed the study. HT, TN, KOk, KF
and KOm performed the experiments. HT, TN and KOk analyzed the
data. HT, TN and MU wrote the manuscript. All authors read and
approved the final manuscript.
Ethical approval and consent to
participate
This study was approved by the independent ethics
committee of Nagasaki University Hospital (approval no.
19081915).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Ho AS, Kraus DH, Ganly I, Lee NY, Shah JP
and Morris LG: Decision making in the management of recurrent head
and neck cancer. Head Neck. 36:144–151. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Vermorken JB, Mesia R, Rivera F, Remenar
E, Kawecki A, Rottey S, Erfan J, Zabolotnyy D, Kienzer HR, Cupissol
D, et al: Platinum-based chemotherapy plus cetuximab in head and
neck cancer. N Engl J Med. 359:1116–1127. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
National Comprehensive Cancer Network
(NCCN), . NCCN Guidelines®. Head and Neck Cancers.
Version 2. NCCN, Plymouth Meeting. (PA). 2019.
|
|
4
|
Yanamoto S, Umeda M, Kioi M, Kirita T,
Yamashita T, Hiratsuka H, Yokoo S, Tanzawa H, Uzawa N, Shibahara T,
et al: Multicenter retrospective study of cetuximab plus
platinum-based chemotherapy for recurrent or metastatic oral
squamous cell carcinoma. Cancer Chemother Pharmacol. 81:549–554.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Naruse T, Yanamoto S, Matsushita Y,
Sakamoto Y, Morishita K, Ohba S, Shiraishi T, Yamada SI, Asahina I
and Umeda M: Cetuximab for the treatment of locally advanced and
recurrent/metastatic oral cancer: An investigation of distant
metastasis. Mol Clin Oncol. 5:246–252. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Naruse T, Yanamoto S, Matsushita Y,
Sakamoto Y, Morishita K, Ohba S, Shiraishi T, Yamada SI, Asahina I
and Umeda M: Lower gingival squamous cell carcinoma with brain
metastasis during long-term cetuximab treatment: A case report.
Oncol Lett. 15:7158–7162. 2018.PubMed/NCBI
|
|
7
|
Bonner JA, Harari PM, Giralt J, Azarnia N,
Shin DM, Cohen RB, Jones CU, Sur R, Raben D, Jassem J, et al:
Radiotherapy plus cetuximab for squamous-cell carcinoma of the head
and neck. N Engl J Med. 354:567–578. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hitt R, Irigoyen A, Cortes-Funes H, Grau
JJ, García-Sáenz JA and Cruz-Hernandez JJ; Spanish Head and Neck
Cancer Cooperative Group (TTCC), : Phase II study of the
combination of cetuximab and weekly paclitaxel in the first-line
treatment of patients with recurrent and/or metastatic squamous
cell carcinoma of head and neck. Ann Oncol. 23:1016–1022. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wang Z, Martin D, Molinolo AA, Patel V,
Iglesias-Bartolome R, Degese MS, Vitale-Cross L, Chen Q and Gutkind
JS: mTOR co-targeting in cetuximab resistance in head and neck
cancers harboring PIK3CA and RAS mutations. J Natl Cancer Inst.
106:dju2152014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Swick AD, Prabakaran PJ, Miller MC, Javaid
AM, Fisher MM, Sampene E, Ong IM, Hu R, Iida M, Nickel KP, et al:
Co-targeting mTORC and EGFR signaling as a therapeutic strategy in
HNSCC. Mol Cancer Ther. 16:1257–1268. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Rampias T, Giagini A, Siolos S, Matsuzaki
H, Sasaki C, Scorilas A and Psyrri A: RAS/PI3K crosstalk and
cetuximab resistance in head and neck squamous cell carcinoma. Clin
Cancer Res. 20:2933–2946. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kozaki K, Imoto I, Pimkhaokham A, Hasegawa
S, Tsuda H, Omura K and Inazawa J: PIK3CA mutation is an oncogenic
aberration at advanced stages of oral squamous cell carcinoma.
Cancer Sci. 97:1351–1358. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ligresti G, Militello L, Steelman LS,
Cavallaro A, Basile F, Nicoletti F, Stivala F, McCubrey JA and
Libra M: PIK3CA mutations in human solid tumors: Role in
sensitivity to various therapeutic approaches. Cell Cycle.
8:1352–1358. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Chang YS, Hsu HT, Ko YC, Yeh KT, Chang SJ,
Lin CY and Chang JG: Combined mutational analysis of RAS, BRAF,
PIK3CA, and TP53 genes in Taiwanese patients with oral squamous
cell carcinoma. Oral Surg Oral Med Oral Pathol Oral Radiol.
118:110–116.e1. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Sartore-Bianchi A, Martini M, Molinari F,
Veronese S, Nichelatti M, Artale S, Di Nicolantonio F, Saletti P,
De Dosso S and Mazzucchelli L: PIK3CA mutations in colorectal
cancer are associated with clinical resistance to EGFR-targeted
monoclonal antibodies. Cancer Res. 69:1851–1857. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
De Roock W, Claes B, Bernasconi D, De
Schutter J, Biesmans B, Fountzilas G, Kalogeras KT, Kotoula V,
Papamichael D, Laurent-Puig P, et al: Effects of KRAS, BRAF, NRAS,
and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy
in chemotherapy-refractory metastatic colorectal cancer: A
retrospective consortium analysis. Lancet Oncol. 11:753–762. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Sok MZ, Zavrl M, Greif B and Srpčič M:
Objective assessment of WHO/ECOG performance status. Support Care
Cancer. 27:3793–3798. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Nishino M, Jackman DM, Hatabu H, Yeap BY,
Cioffredi LA, Yap JT, Jänne PA, Johnson BE and Van den Abbeele AD:
New response evaluation criteria in solid tumors: Revised RECIST
guideline version 1.1. Eur J Cancer. (Suppl 6):S132008.
|
|
19
|
Pinborg JJ, Reichart PA, Smith CJ and van
der Waal I: Histological typing of cancer and precancer of the oral
mucosa. (2nd). (Berlin, Germany). Springer. 1997.
|
|
20
|
Wright JM and Vered M: Update from the 4th
edition of the world health organization classification of head and
neck tumours: Odontogenic and maxillofacial bone tumors. Head Neck
Pathol. 11:68–77. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Bryne M, Boysen M, Alfsen CG, Abeler VM,
Sudbø J, Nesland JM, Kristensen GB, Piffko J and Bankfalvi A: The
invasive front of carcinomas. The most important area for tumour
prognosis? Anticancer Res. 18:4757–4764. 1998.PubMed/NCBI
|
|
22
|
Vis DJ, Bombardelli L, Lightfoot H, Iorio
F, Garnett MJ and Wessels LF: Multilevel models improve precision
and speed of IC50 estimates. Pharmacogenomics. 17:691–700. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Jimeno A, Shirai K, Choi M, Laskin J,
Kochenderfer M, Spira A, Cline-Burkhardt V, Winquist E, Hausman D,
Walker L and Cohen RB: A randomized, phase II trial of cetuximab
with or without PX-866, an irreversible oral phosphatidylinositol
3-kinase inhibitor, in patients with relapsed or metastatic head
and neck squamous cell cancer. Ann Oncol. 26:556–561. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Keam B, Kim S, Ahn YO, Kim TM, Lee SH, Kim
DW and Heo DS: In vitro anticancer activity of PI3K alpha selective
inhibitor BYL719 in head and neck cancer. Anticancer Res.
35:175–182. 2015.PubMed/NCBI
|
|
25
|
Munster P, Elkabets M, Gilbert J, Razak
ARA, Ahn MJ, Yen CJ, Lee SH, Wang HM, Herpen C and Lim WT: Abstract
A46: Inhibition of PIK3CA with BYL719 can overcome resistance to
cetuximab in squamous cell carcinoma of the head and neck (SCCHN).
Mol Cancer Ther. 14 (Suppl 7):A46. 2015.
|
|
26
|
Meister KS, Godse NR, Khan NI, Hedberg ML,
Kemp C, Kulkarni S, Alvarado D, LaVallee T, Kim S, Grandis JR and
Duvvuri U: HER3 targeting potentiates growth suppressive effects of
the PI3K inhibitor BYL719 in pre-clinical models of head and neck
squamous cell carcinoma. Sci Rep. 9:91302019. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kimura I, Kitahara H, Ooi K, Kato K,
Noguchi N, Yoshizawa K, Nakamura H and Kawashiri S: Loss of
epidermal growth factor receptor expression in oral squamous cell
carcinoma is associated with invasiveness and
epithelial-mesenchymal transition. Oncol Lett. 11:201–207. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Schmitz S, Bindea G, Albu RI, Mlecnik B
and Machiels JP: Cetuximab promotes epithelial to mesenchymal
transition and cancer associated fibroblasts in patients with head
and neck cancer. Oncotarget. 6:34288–34299. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Naruse T, Yanamoto S, Okuyama K, Yamashita
K, Omori K, Nakao Y, Yamada SI and Umeda M: Therapeutic implication
of mTORC2 in oral squamous cell carcinoma. Oral Oncol. 65:23–32.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ferris RL, Blumenschein G Jr, Fayette J,
Guigay J, Colevas AD, Licitra L, Harrington K, Kasper S, Vokes EE,
Even C, et al: Nivolumab for recurrent squamous-cell carcinoma of
the head and neck. N Engl J Med. 375:1856–1867. 2016. View Article : Google Scholar : PubMed/NCBI
|