Introduction
Breast cancer (BRCA) is the most common type of
malignancy in females worldwide (1,2). Over
the past decades, patients with BRCA have been usually treated with
radiotherapy, chemotherapy or their combination according to the
tumor type, histological stage, tumor size, lymph node metastasis
and metastatic site (3). However,
the prognosis of patients with BRCA remains poor (4). Furthermore, the incidence of BRCA is
increasing (5). In 2018, ~2.1
million newly diagnosed cases of BRCA were reported worldwide,
accounting for almost a quarter of the total number of diagnosed
cancers in female subjects (5).
Therefore, understanding the molecular mechanisms underlying BRCA
progression can be used to identify novel biomarkers for the early
detection of this disease.
An increasing number of studies have demonstrated
that the abnormal regulation of metabolism-associated pathways is
associated with tumorigenesis and the development of human cancer
(6–9). Lipid metabolism disorders have been
reported to be associated with a variety of diseases, such as
cancer (10). Lipid metabolism is
involved in regulating multiple cellular biological activities in
cancer cells, such as cell proliferation and apoptosis. The
steroidogenic acute regulatory protein-related lipid transfer
(STARD) domain family serves a crucial role in the transportation
of cholesterol (11,12). STARD family proteins serve a role in
non-vesicular lipid transport and cell signaling via their
interaction with cholesterol, phospholipids, sphingolipids and bile
acids (13). The STARD4 protein was
initially identified by Soccio et al (14) in 2002. STARD4 is located in both the
nuclear and cytoplasmic regions (15). STARD4 is involved in the regulatory
mechanism of intracellular cholesterol homeostasis (16). Silencing of STARD4 results in
elevated levels of intracellular free cholesterol (17). Furthermore, overexpression of STARD4
promotes the formation of cholesterol esters, suggesting that
STARD4 can facilitate the transport of cholesterol to the
endoplasmic reticulum and mitochondria (18). A previous study has indicated that
knockdown of STARD4 in the liver cancer HepG2 cell line disrupts
cholesterol trafficking, suggesting that STARD4 may be associated
with cancer progression (19).
However, the roles of STARD4 in the development of BRCA require
further investigation.
The present study initially investigated the role of
STARD4 in BRCA and elucidated the effects of STARD4 on the
proliferation of BRCA cells. The data provide important information
for understanding BRCA pathogenesis.
Materials and methods
Tissue samples
A total of 121 BRCA samples and 49 matched normal
tissues were collected from the Department of Breast Surgery,
Xiangya Hospital, Central South University (Changsha, China).
Patients with BRCA were aged 31–76 years, with a median age of 63
years. The samples were collected from female patients with BRCA
who were diagnosed according to the postoperative pathological
report between January 2016 and December 2018 after surgical
resection. Cases of invasive ductal carcinoma, intraductal
carcinoma and mucinous adenocarcinoma were included in the present
study. Cases with multiple primary cancers and a postoperative
positive margin were excluded. The postoperative
immunohistochemical (IHC) staining results of the patients for the
markers estrogen receptor (ER), progesterone receptor (PR), HER2
and Ki67 were recorded in detail. Clinical and pathological data,
including age, histopathological grade, number of axillary lymph
node dissection, and metastasis and site of metastasis, were
collected (Table I). The patients
were all female and the median age was 63 years. The present study
was approved by the Ethics Committee of Xiangya Hospital (Changsha,
China). Written informed consent was obtained from all
patients.
 | Table I.Clinicopathological characteristics
of patients and STARD4 expression in breast cancer. |
Table I.
Clinicopathological characteristics
of patients and STARD4 expression in breast cancer.
|
Characteristics | Number of cases
(%) |
|---|
| Age, years |
|
|
£40 | 21 (17.4) |
|
>40 | 100 (82.6) |
| T stage |
|
|
T1-T2 | 101 (83.5) |
|
T3-T4 | 20 (16.5) |
| M stage |
|
| M0 | 117 (96.7) |
| M1 | 4 (3.3) |
| N stage |
|
| N0 | 55 (45.5) |
|
N1-N3 | 66 (54.5) |
| TNM stage |
|
|
I–II | 82 (67.8) |
|
III–IV | 39 (32.2) |
| Histological
stage |
|
|
Intraductal carcinoma | 10 (8.3) |
| I | 5 (4.1) |
| II | 78 (64.5) |
|
III | 27 (22.3) |
|
Mucinous adenocarcinoma | 1 (0.8) |
| Metastasis |
|
| No | 68 (56.2) |
|
Yes | 53 (43.8) |
| Lymph node
metastasis, % |
|
|
<20 | 84 (69.4) |
|
≥20 | 37 (30.6) |
| Molecular type |
|
|
Luminal | 69 (57.0) |
|
HER2+ | 43 (35.5) |
|
TNBC | 9 (7.4) |
| ER |
|
|
Negative | 37 (30.6) |
|
Positive | 84 (69.4) |
| PR |
|
|
Negative | 51 (42.1) |
|
Positive | 70 (57.9) |
| HER |
|
|
Negative | 78 (64.5) |
|
Positive | 43 (35.5) |
| STARD4
expression |
|
|
Low | 59 (48.8) |
|
High | 62 (51.2) |
The present study first evaluated the expression
levels of STARD4 in 57 BRCA cell lines by analyzing data in the
Cancer Cell Line Encyclopedia (CCLE) database (https://portals.broadinstitute.org/ccle)
(20,21). The expression levels of STARD4 in
T1, T2, T3 and T4 stage BRCA samples were analyzed using the
Betastasis database (http://www.betastasis.com). The expression levels of
STARD4 in different types of BRCA were analyzed using the Tumor
Immune Estimation Resource (TIMER) database (https://cistrome.shinyapps.io/timer/), which is based
on The Cancer Genome Atlas RNA-sequence data (https://portal.gdc.cancer.gov/). TIMER provides
six major analytic modules that allow users to interactively
explore the associations between immune infiltrates and a wide
spectrum of factors, including gene expression, clinical outcomes,
somatic mutations and somatic copy number alterations (22). The prognostic value of STARD4 was
evaluated using Kaplan-Meier Plotter (www.kmplot.com). To analyze the association between
STARD4 levels and distant metastasis-free survival (DMFS) time,
patient samples were divided into two groups (low and high
expression) based on median mRNA levels with a hazard ratio with
95% confidence intervals and log-rank P-values (23). Log-rank P-values <0.05 were
considered statistically significant.
IHC staining and evaluation
Tissue samples were fixed in 10% (v/v) formaldehyde
at room temperature for 24 h, embedded in paraffin and cut into
5-µm sections. All IHC staining assays used in the present study
were conducted as previously described (24). The biotin-labeled IgG (cat. no.
Sp-9001; 1:1,000; OriGene Technologies, Inc.) was used as secondary
antibody for 30 min at room temperature. Dehydration was
subsequently performed and sample sections were sealed using cover
glasses. Five fields were randomly selected and imaged under an
optical microscope (BX51T-PHD-J11; Olympus Corporation). The images
were analyzed using Image-Pro Plus 6.0 (Media Cybernetics, Inc.).
The staining was assessed independently by two pathologists in a
blinded manner and all discrepancies were resolved by
consensus.
STARD4 immunohistochemical staining was performed
using a polyclonal antibody against STARD4 (dilution, 1:200; cat.
no. ab202060; Abcam). The staining intensity was divided into the
following categories: 0 points, negative (−); 1–2 points, weakly
positive (+); 3–4 points, positive (++); 6–9 points, strongly
positive (+++). The percentage of positively-stained cells was
scored as follows: 0, 0–5%; 1, 5–25%; 2, 25–50%; 3, 50–100%.
ER and PR immunohistochemical staining was performed
using a mouse monoclonal anti-human ER antibody (clone, 1D5; Dako;
Agilent Technologies, Inc.) and a mouse monoclonal anti-human PR
antibody (clone, 636; Dako; Agilent Technologies, Inc.) at a
dilution of 1:100. The cut-off value for a positive result was
positive staining for ER and PR in ≥1% of tumor cells in 10
selected tumor sub-regions (25).
The results were recorded as the percentage of positively-stained
nuclei, and the intensity was graded between 0 and 3+ as follows: 0
(negative result), positive staining in <1% of the tumor cells;
1+, mildly distinct, positive staining in ≤25% of the tumor cells;
2+, moderately distinct, positive staining in 25–50% of the tumor
cells; and 3+, strong, positive staining in >50% of the tumor
cells.
HER2 immunohistochemical staining was performed
using the HercepTest™ assay (Dako; Agilent Technologies, Inc.)
according to manufacturer's protocols. The expression levels of
HER2 were initially determined by immunohistochemistry and graded
between 0 and 3+ as follows: 0 (negative result), absence or
presence of HER2 in <10% of the tumor cells; 1+ (negative
result), membranous, weak and discontinuous staining in >10% of
the tumor cells; 2+ (questionable result), membranous, low/moderate
and continuous staining in >10% of the tumor cells, or
membranous, intense and continuous staining in ≤30% of the tumor
cells; and 3+ (positive result), membranous, intense and continuous
staining in >30% of the tumor cells. Samples with HER2 scores of
2+ were confirmed to be HER2-negative or HER2-positive using
fluorescence in situ hybridization analysis as previously
described (26).
Ki67 immunohistochemical staining was performed
using a mouse monoclonal anti-human Ki67 antibody (dilution, 1:100;
clone, MIB-1; Dako; Agilent Technologies, Inc.). At least three
fields in particular staining ‘hot-spots’ were selected in order to
represent the spectrum of staining observed upon the initial
overview of the entire section. The cancer cells in the three
micrographs were manually counted (500–1,000 cells were counted),
and the percentage of positively-stained cancer cells was
considered to be the Ki67 score (27).
Cell culture, and apoptosis,
proliferation and migration assays
HCC1937 and MDA-MB-231 cells were obtained from The
Cell Bank of Type Culture Collection of the Chinese Academy of
Sciences and cultured in low-glucose DMEM (Gibco; Thermo Fisher
Scientific, Inc) with 10% FBS (Gibco; Thermo Fisher Scientific,
Inc.) and 1% penicillin-streptomycin (Gibco; Thermo Fisher
Scientific, Inc.) in a 37°C incubator with 5% CO2.
Briefly, recombinant lentiviral vectors were obtained Shanghai
Genechem Co., Ltd. A total of 6 µg shRNA plasmids and shRNA control
were transfected to knockdown target expression levels using
Lipofectamine 3000 reagent (Invitrogen; Thermo Fisher Scientific,
Inc.), according to standard molecular techniques. At 48 h after
transfection, the transfection efficiency was determined using
reverse transcription-quantitative (RT-q)PCR and western blotting.
The short hairpin RNA (sh)STARD4 sequence was
5′-CCGGTCCTATACTGTGGGCTATAAACTCGAGTTTATAGCCCACAGTATAGGATTTTTG-3′.
The sh-cAMP responsive element binding protein 1 (CREB1) sequence
was
5′-CCGGTCCCAACAAATGACAGTTCAACTCGAGTTGAACTGTCATTTGTTGGGATTTTTG-3′.
The shcontrol sequence was
5′-CCGGTCTTCTCCGAACGTGTCACGTCTCGAGACGTGACACGTTCGGAGAAGATTTTTG-3′.
Lentivirus construction and infection were performed as previously
described (28).
For the cell proliferation assay, 2,000 HCC1937 or
MDA-MB-231 cells/well were seeded into 96-well plates. The optical
density was detected at 450 nm using a microplate reader after
adding 10 µl Cell Counting Kit-8 (CCK-8; Dojindo Molecular
Technologies, Inc.) reagent to each well at each time point (day 1,
2, 3, 4 and 5) according to the manufacturer's protocol.
The cell counting assay was performed using the
Celigo image cytometer (Nexcelom Bioscience LLC). For cell
cytometry system Celigo® analysis, HCC1937 and
MDA-MB-231 cells transfected with shSTARD4 or negative control were
seeded into 96-well plates (1,500 cells/well) and cultured for 5
days. The cell images were captured and analyzed using the using a
fluorescence microscope system (Celigo®; Nexcelom
Bioscience LLC). Each experiment was performed three times.
The apoptosis of HCC1937 and MDA-MB-231 cells was
analyzed using a FACS Calibur flow cytometer (BD Biosciences) with
a FITC Annexin V Apoptosis Detection kit (BD Biosciences. Cell
apoptosis data were analyzed using FCS Express software (version
3.0; De Novo Software). Each experiment was performed three
times.
The cell migration assay was performed as previously
described using a Transwell assay (29). For the wound healing assay, HCC1937
and MDA-MB-231 cells were seeded into 6-well plates in duplicate.
Once the confluency of the cells reached ~80%, the monolayer of
cells was wounded using a 10-µl plastic pipette tip and then the
scratched cells were rinsed with PBS and cultured with serum-free
DMEM for 24 h. The wound was imaged at 0, 12 and 24 h using a
fluorescence microscope (CK40-F200; Olympus Corporation) and then
analyzed using ImageJ software (version 1.62; National Institutes
of Health). The following formula was used: Wound closure rate
(%)=migrated cell surface area/total surface area ×100. Each
experiment was performed three times.
RT-qPCR
Total RNA was extracted from BRCA cell lines using
the TRIzol reagent (Thermo Fisher Scientific, Inc.) and was
transcribed to cDNA using the RevertAid First Strand cDNA Synthesis
kit (Thermo Fisher Scientific, Inc.). Reverse transcription was
conducted at 37°C for 1 h, followed by 85°C for 5 sec according to
the manufacturer's protocol. RT-qPCR was performed using the
QuantiTect SYBR-Green PCR kit (Roche Diagnostics) as previously
described (30). The primer
sequences are listed in Table SI.
The 2−ΔΔCq method was used to calculate the relative
expression levels of the target genes (31). GAPDH was used for normalization. The
reaction conditions were as follows: 95°C for 10 min, followed by
95°C for 30 sec, 60°C for 30 sec and 72°C for 30 sec (39 cycles),
and finally 72°C for 10 min.
In vivo tumor assays
A total of 3×106 STARD4-knockdown or
control HCC1937 cells were suspended in PBS and injected
subcutaneously into the dorsal skin of 6-week-old female BALB/c
nude mice (10 mice/group) with a median weight of 20 g (Shanghai
SLAC Laboratory Animal Co., Ltd.). The protocol for the animal
experiments was approved by the Shanghai Medical Experimental
Animal Care Commission (approval no. ShCI-14-008). Mice were housed
under specific pathogen-free conditions at 25°C with a 12-h
light/dark cycle, and free access to food and water. The
maintenance conditions for the mice were as follows: Humidity,
40–60%; ventilation, 15 times/h. STARD4 knockdown and control
luciferase-expressing HCC1937 cells were injected into the mice and
monitored using an in vivo Imaging System (IVIS;
PerkinElmer, Inc.). The animals were sacrificed when the tumors
reached 1 cm in diameter. The mice were sacrificed 7 weeks after
injection using CO2 (with a flow rate of CO2
for euthanasia displacing ≤30% of the chamber volume/min). The
volume and weight of all nude mice were measured after 4 weeks.
Tumor growth was calculated using the following formula: Volume =
0.5 × length × width2.
Western blotting
Total protein was extracted from cells with RIPA
buffer (Beyotime Institute of Biotechnology). The BCA assay kit
(Boster Biological Technology) was used to measure the total
protein concentration. Protein (30 µg/lane) was subsequently
separated by 12% SDS-PAGE, followed by transfer to PVDF membranes.
Subsequently, the membranes were blocked with 3% BSA (Sangon
Biotech Co., Ltd.)/TBS with 0.05% Tween-20 (TBST; Sangon Biotech
Co., Ltd.) for 2 h at room temperature and incubated with primary
antibodies at 4°C overnight. The PVDF membranes were rinsed three
times for 5 min each with TBST and incubated with HRP-conjugated
secondary antibodies for 1 h at room temperature. The antibodies
are listed in Table SII. The total
protein levels were assessed using enhanced chemiluminescence
reagents (Thermo Fisher Scientific, Inc.). ImageJ 6.0 software
(National Institutes of Health) was used to analyze the images.
Microarray and expression
analyses
Total RNA was extracted using TRIzol reagent
(Invitrogen; Thermo Fisher Scientific, Inc.) and quantified by the
NanoDrop ND-2000 (Thermo Fisher Scientific, Inc.). The RNA
integrity was assessed using the Agilent Bioanalyzer 2100 (Agilent
Technologies, Inc). Whole-genome gene expression levels were
examined using the Gene Chip Prime View Human Gene Expression Array
(Thermo Fisher Scientific, Inc.). Sample labeling, microarray
hybridization, washing and gene normalization were performed
according to the manufacturer's protocols. RNA integrity was also
examined using an Agilent Bioanalyzer 2100 (cat. no. G2938A;
Agilent Technologies, Inc.). To obtain biotin-tagged cDNA, total
RNA was subsequently amplified, labeled and purified using a WT
PLUS Reagent kit (cat. no. 902280; Affymetrix; Thermo Fisher
Scientific, Inc.). Array hybridization was performed using an
Affymetrix GeneChip Human Gene 2.0 ST Array (Affymetrix; Thermo
Fisher Scientific, Inc.) and Hybridization Oven 645 (cat. no.
00-0331-220V; Affymetrix; Thermo Fisher Scientific, Inc.). The Gene
Chip was subsequently washed using a Hybridization, Wash and Stain
kit (cat. no. 900720; Affymetrix; Thermo Fisher Scientific, Inc.)
in a Fluidics Station 450 (cat. no. 00-0079, Affymetrix; Thermo
Fisher Scientific, Inc.). A GeneChip Scanner 3000 (cat. no.
00-00213; Affymetrix; Thermo Fisher Scientific, Inc.) was used to
scan the results, which were analyzed by Command Console Software
4.0 (Affymetrix; Thermo Fisher Scientific, Inc.) to summarize probe
cell intensity data, namely, the CEL files with default settings.
Following this, CEL files were normalized according to gene and
exon levels using Expression Console Software 4.0 (Affymetrix;
Thermo Fisher Scientific, Inc.). All procedures, including array
hybridization and scanning, were independently performed according
to a standard protocol (32) for
microarray experiments (n=3).
Differentially expressed mRNAs were subsequently
identified using fold change values. The threshold for upregulated
and downregulated genes was ≥1.5. Genes that were differentially
expressed were identified based on the following criteria:
P<0.05 and absolute fold change >1.5.
Statistical analysis
All statistical analyses were performed using
GraphPad Prism 6.0 (GraphPad Software, Inc.). All data are
presented as the mean ± SEM. All assays were conducted at least
three times. The unpaired Student's t-test, Mann-Whitney U test and
Wilcoxon matched-pairs signed rank test were used to analyze the
differences between two groups according to the test condition.
One-way ANOVA with Tukey's post hoc test was used to analyze the
differences among multiple groups. Survival curves were estimated
using the Kaplan-Meier method and the log-rank test. P<0.05 was
considered to indicate a statistically significant difference.
Results
STARD4 is upregulated in breast
carcinoma
In order to evaluate the expression levels of STARD4
in BRCA, 121 BRCA samples and 49 adjacent normal tissue samples
were collected from patients who underwent surgical resection. The
clinicopathological characteristics and the expression levels of
STARD4 in BRCA are shown in Table
I. STARD4 was expressed in the nuclear regions in both BRCA and
normal breast tissues (Fig. 1A-D).
The comparison of the STARD4 levels between BRCA and normal samples
demonstrated that STARD4 protein levels were significantly
upregulated in 121 BRCA samples compared with in 49 adjacent normal
tissues (Fig. 1E). The BRCA levels
were upregulated in 49 paired BRCA samples (Fig. 1F).
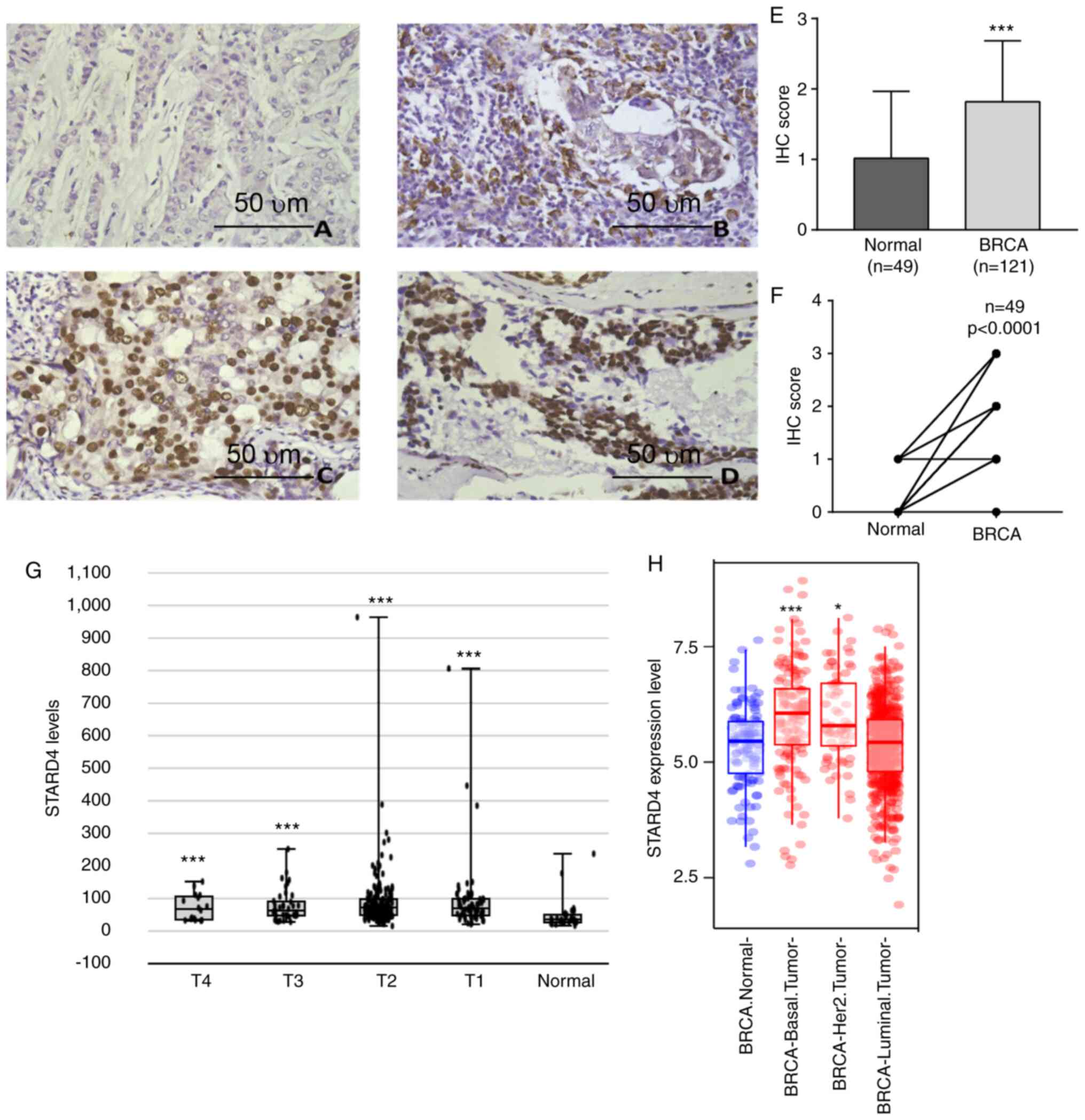 | Figure 1.STARD4 expression is upregulated in
BRCA samples. STARD4 expression was (A) negative, (B) weakly
positive, (C) positive and (D) strongly positive in clinical BRCA
samples as detected by IHC. Scale bar, 50 µm. (E) STARD4 protein
levels were significantly upregulated in 121 BRCA samples compared
with 49 adjacent normal tissue, and (F) upregulated in 49 paired
BRCA samples. (G) STARD4 was upregulated in stage T1, T2, T3 and T4
BRCA samples compared with normal breast samples. (H) STARD4
expression was upregulated in basal and Her2-positive, but not in
luminal BRCA samples compared with in normal breast tissues. For
(E) statistical comparisons between groups of normalized data were
performed using Mann-Whitney U test according to the test
condition. For (F) statistical comparisons between groups of
normalized data were performed using the Wilcoxon matched-pairs
signed rank test according to the test condition. *P<0.05,
***P<0.001 vs. normal group. BRCA, breast cancer; IHC,
immunohistochemistry; STARD4, steroidogenic acute regulatory
protein-related lipid transfer 4. |
The associations between STARD4 protein expression
and the clinicopathological features of patients with BRCA are
summarized in Table II. Positive
protein expression of STARD4 was associated with advanced
histological stage, metastatic status and ER expression. However,
it was not observed to be significantly associated with age, T
stage, M stage, N stage, PR and HER-2 expression. These results
demonstrated that STARD4 was significantly associated with ER
expression, but not PR expression. When analyzing the association
between ER and STARD4 using the TIMER database, a negative
correlation was observed between the two genes in all types of BRCA
(Fig. S1A), HER2-positive BRCA
(Fig. S1B) and luminal BRCA
(Fig. S1C). However, STARD4
expression was positively correlated with ER expression in basal
BRCA (Fig. S1D).
 | Table II.Association between STARD4 expression
and clinicopathologic characteristics of patients with breast
cancer. |
Table II.
Association between STARD4 expression
and clinicopathologic characteristics of patients with breast
cancer.
|
| STARD4 |
|
|
|---|
|
|
|
|
|
|---|
|
Characteristics | Low, n | High, n | χ2 | P-value |
|---|
| Age, years |
|
|
|
|
|
≤40 | 11 | 10 | 0.133 | 0.812 |
|
>40 | 48 | 52 |
|
|
| T stage |
|
|
|
|
|
T1-T2 | 53 | 48 | 3.375 | 0.087 |
|
T3-T4 | 6 | 14 |
|
|
| M stage |
|
|
|
|
| M0 | 58 | 59 | 0.935 | 0.619 |
| M1 | 1 | 3 |
|
|
| N stage |
|
|
|
|
| N0 | 29 | 26 | 0.635 | 0.468 |
|
N1-N3 | 30 | 36 |
|
|
| TNM stage |
|
|
|
|
|
I–II | 45 | 37 | 3.811 | 0.055 |
|
III–IV | 14 | 25 |
|
|
| Histological
stage |
|
|
|
|
|
Intraductal carcinoma | 8 | 2 | 9.537 | 0.049 |
| I | 4 | 1 |
|
|
| II | 37 | 41 |
|
|
|
III | 9 | 18 |
|
|
| Mucinous
adenocarcinoma | 1 | 0 |
|
|
| Metastasis |
|
|
|
|
| No | 41 | 27 | 8.266 | 0.006 |
|
Yes | 18 | 35 |
|
|
| Lymph node
metastasis, % |
|
|
|
|
|
<20 | 44 | 40 | 1.441 | 0.244 |
|
≥20 | 15 | 22 |
|
|
| Molecular type |
|
|
|
|
|
Luminal | 32 | 37 | 1.87 | 0.395 |
|
HER2+ | 24 | 19 |
|
|
|
TNBC | 3 | 6 |
|
|
| ER |
|
|
|
|
|
Negative | 11 | 26 | 7.726 | 0.006 |
|
Positive | 48 | 36 |
|
|
| PR |
|
|
|
|
|
Negative | 25 | 26 | 0.002 | >0.999 |
|
Positive | 34 | 36 |
|
|
| HER |
|
|
|
|
|
Negative | 36 | 42 | 0.597 | 0.454 |
|
Positive | 23 | 20 |
|
|
Furthermore, online public datasets were analyzed.
When analyzing the Betastasis database, the data indicated that
STARD4 expression was upregulated in T1, T2, T3 and T4 stage BRCA
samples compared with in normal breast samples (Fig. 1G). However, STARD4 expression was
not significantly different among the different stages.
Furthermore, the TCGA dataset was analyzed to detect STARD4 levels
in different types of BRCA. STARD4 expression was upregulated in
basal and Her2-positive BRCA tissues, whereas it was not
upregulated in luminal BRCA samples compared with in normal breast
tissues (Fig. 1H). The analysis
demonstrated that STARD4 expression may be associated with BRCA
progression and may serve as an early diagnostic biomarker.
However, the dysregulation of STARD4 could not be used for the
prediction of BRCA stage.
Upregulation of STARD4 expression is
associated with poor BRCA prognosis
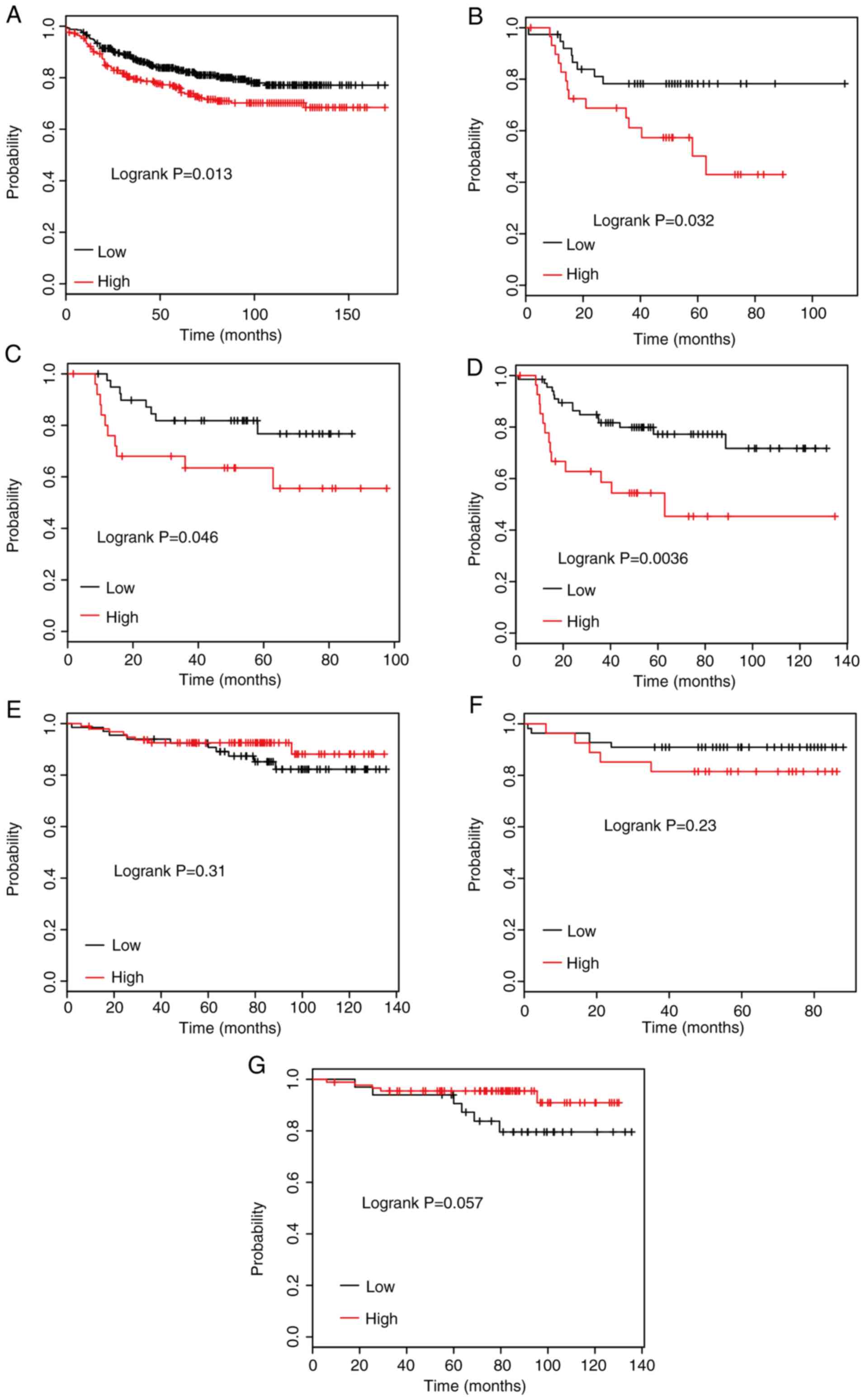
The association between STARD4 levels and DMFS time
was determined using the Kaplan-Meier survival Plotter database.
The median expression of STARD4 in all BRCA cases was selected as a
cut-off to divide the BRCA samples into STARD4-high and -low
groups. Kaplan-Meier survival curve analysis indicated that higher
expression levels of STARD4 were significantly associated with
shorter DMFS time in patients with BRCA (Fig. 2A).
Furthermore, the association between STARD4
expression and DMFS time was examined in patients with BRCA
according to their clinicopathological characteristics, including
ER, PR and HER2 status. The results indicated that high STARD4
expression was associated with poor DMFS rates compared with low
STARD4 expression in patients with ER-negative (Fig. 2B), PR-negative (Fig. 2C) and HER2-positive (Fig. 2D) BRCA. However, dysregulation of
STARD4 was not significantly associated with DMFS time in patients
with ER-positive (Fig. 2E),
PR-positive (Fig. 2F) and
HER2-negative (Fig. 2G) BRCA.
Furthermore, the present study evaluated the prognostic utility of
STARD4 based on subtype (luminal and HER2). As presented in
Fig. S2, although a potential
tendency that higher expression levels of STARD4 were associated
with shorter DMFS time in luminal A, luminal B and HER2-positive
BRCA was observed, no significant association between STARD4
expression and DMFS time was observed in groups of patients based
on subtype (luminal and HER2).
Knockdown of STARD4 suppresses cell
proliferation in BRCA
Upregulation of STARD4 in BRCA suggested that STARD4
may serve as an oncogene in BRCA. Therefore, loss-of-functions
assays were used to detect the effects of STARD4 on cell
proliferation. The present study first evaluated the expression
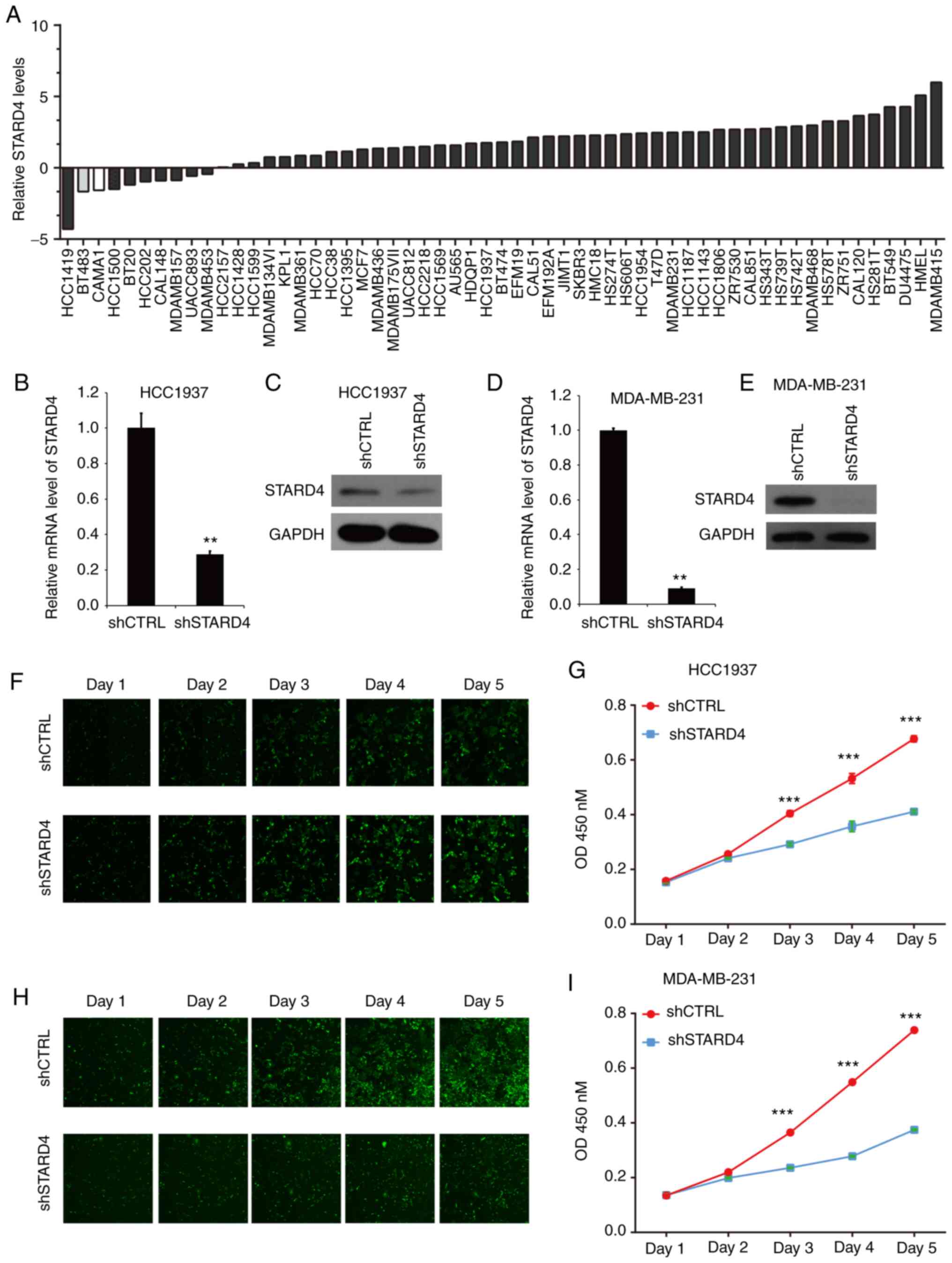
levels of STARD4 in 57 BRCA cell lines by analyzing the CCLE
database (20,21). The results demonstrated that STARD4
was highly expressed in multiple BRCA cell lines, including
MDA-MB-231 and HCC1937 cells, which do not have the highest STARD4
levels; however, they are widely used in triple-negative BRCA
research (Fig. 3A). Following
transfection of HCC1937 (Fig. 3B and
C) and MDA-MB-231 (Fig. 3D and
E) cells with shSTARD4 lentivirus, the mRNA and protein levels
of STARD4 in BRCA cells were reduced compared with those of the
control group.
Subsequently, the effects of STARD4 on BRCA
proliferation were assessed using a Celigo cell counting assay for
5 days. Knockdown of STARD4 reduced cell proliferation in HCC1937
(Fig. 3F) and MDA-MB-231 (Fig. 3H) cells compared with in control
cells. In addition, a CCK-8 assay revealed that knockdown of STARD4
suppressed the HCC1937 (Fig. 3G)
and MDA-MB-231 (Fig. 3I) cell
proliferation rate at day 3, 4 and 5.
Silencing of STARD4 suppresses
G0/G1 phase transition in BRCA cells
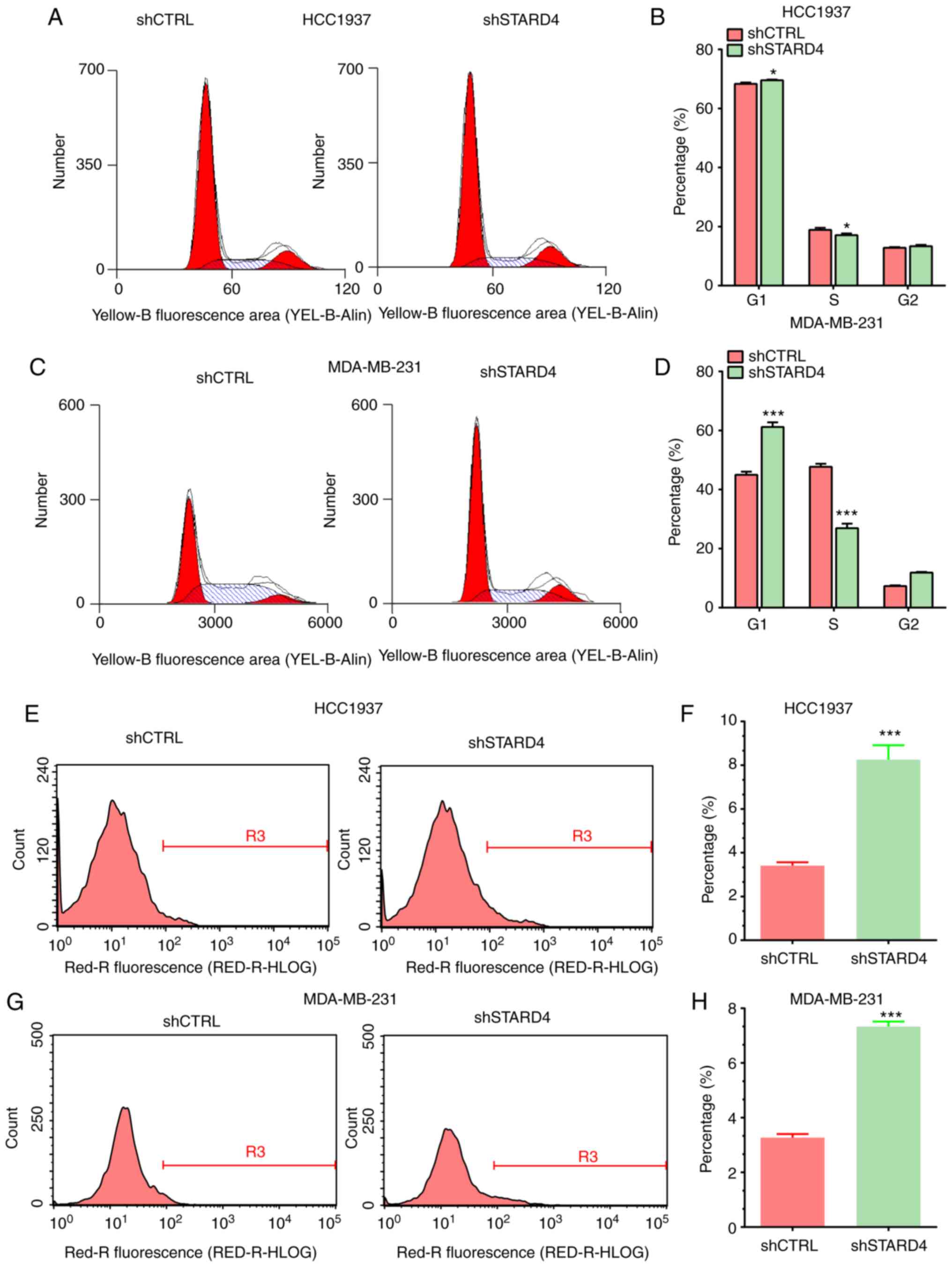
The effect of STARD4 on the cell cycle progression
of BRCA cells was investigated. Flow cytometry demonstrated that
knockdown of STARD4 expression in BRCA cells inhibited the cell
cycle progression at the G0/G1 phase
(Fig. 4A-D). The proportion of
cells arrested at the G1 phase was significantly higher
in shSTARD4-transfected cells compared with that noted in the
control cells. However, the percentage of cells arrested at the S
phase was significantly lower in the shSTARD4-transfected cells
compared with that in the control cells.
Silencing of STARD4 induces apoptosis
of BRCA cells
The effects of STARD4 on cell apoptosis were also
investigated using flow cytometry. STARD4 knockdown induced
apoptosis in BRCA cells compared with the control group. The
apoptotic rates of STARD4-silenced HCC1937 and MDA-MB-231 cells
were higher than those of the corresponding control groups
(Fig. 4E-H). These results
indicated that STARD4 may promote cell proliferation by reducing
cell apoptosis.
Knockdown of STARD4 inhibits migration
of BRCA cells
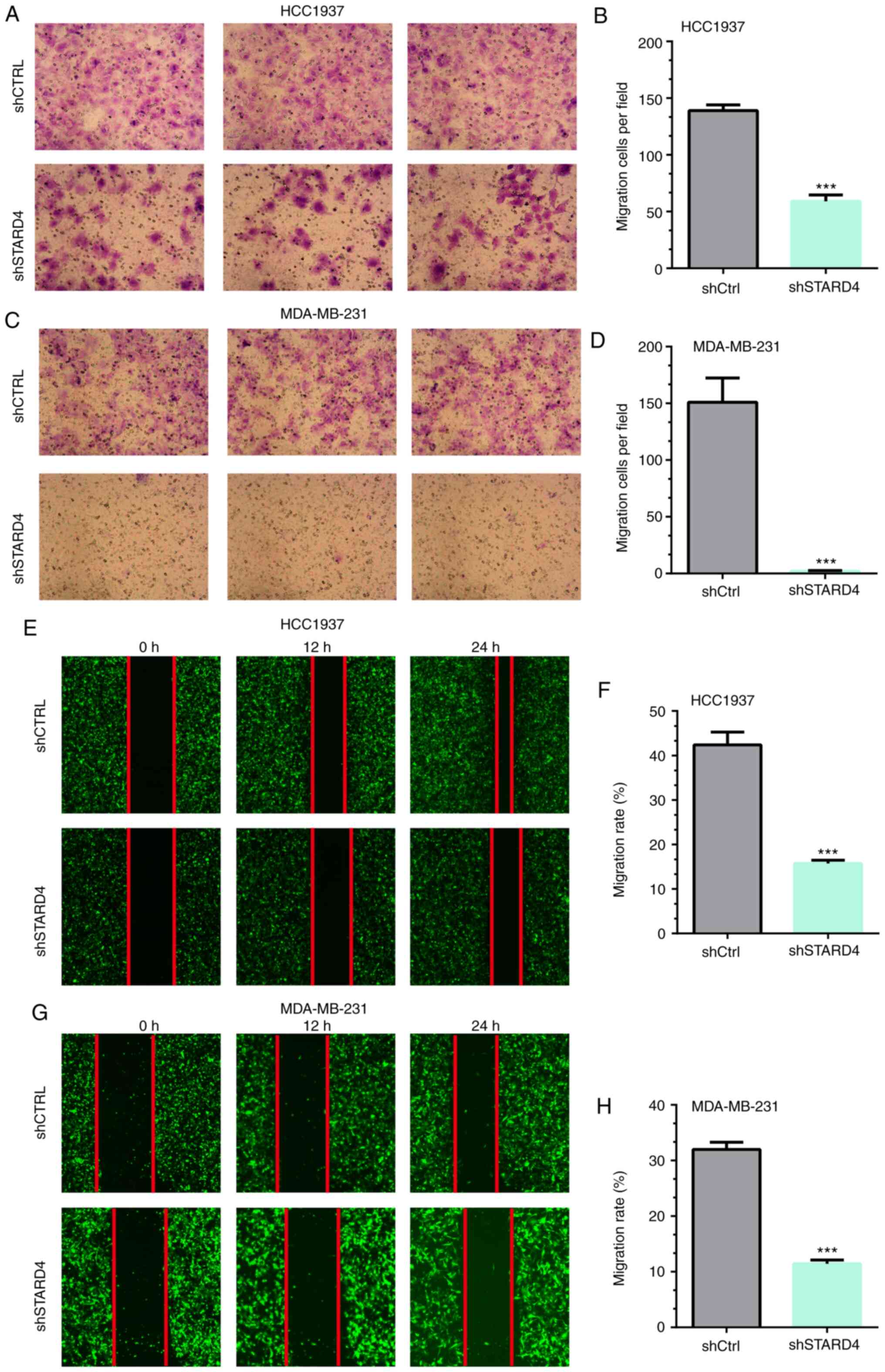
The ability of STARD4 to affect cell migration was
examined using Transwell and wound healing assays. The results of
the Transwell assay demonstrated that the migratory activity of
HCC1937 and MDA-MB-231 cells was significantly attenuated following
transfection with STARD4 shRNA compared with that of the negative
control cells (by 59 and 99%, respectively; Fig. 5A-D). Similar results were observed
in the wound healing assay (Fig.
5E-H).
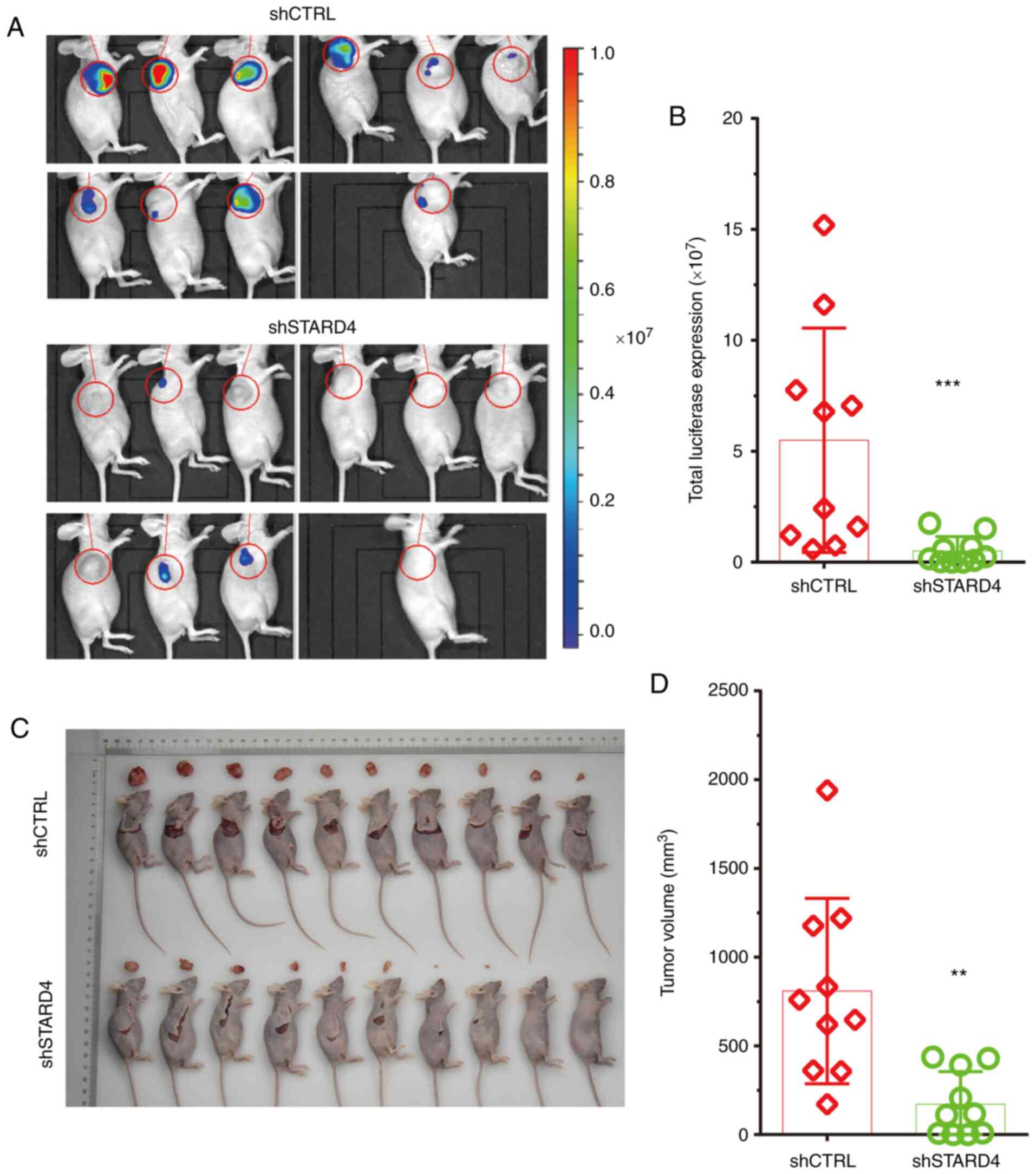
Knockdown of STARD4 suppresses BRCA
growth in vivo
The effects of STARD4 knockdown on tumor growth were
analyzed in vivo using a nude mouse model. The
luciferase-expressing MDA-MB-231 cells were constructed to implant
tumors in live animals. Using an IVIS system, the data indicated
that luciferase signaling in the STARD4 knockdown group was
decreased compared with that in the control group (Fig. 6A and B). It was revealed that the
tumor volume in the STARD4 knockdown group was lower than that in
the negative control group (Fig. 6C and
D).
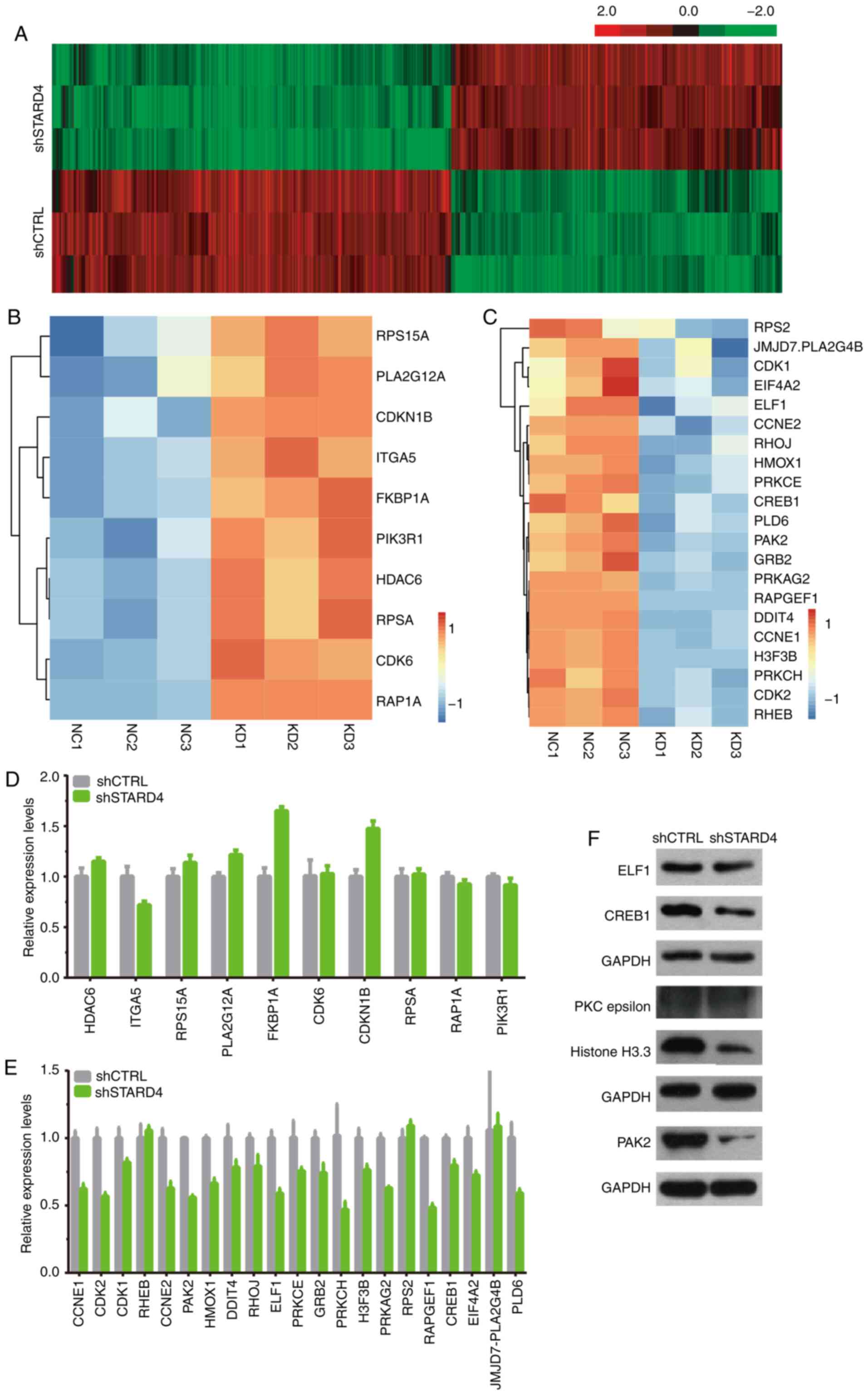
Identification of STARD4 downstream
targets in BRCA using microarray analysis
To understand the mechanisms of STARD4 in regulating
BRCA progression, a gene expression microarray was used to identify
targets of STARD4 in MDA-MB-231 cells. A total of 772 genes,
including 350 upregulated and 422 downregulated genes, were
identified as downstream targets of STARD4 in BRCA (Fig. 7A; Table
SIII).
In order to validate the downstream targets of
STARD4 identified by microarray analysis, the expression levels of
the potential targets were detected following silencing of STARD4.
A total of 10 upregulated (RPS15A, PLA2G12A, CDKN1B, ITGA5, FKBP1A,
PIK3R1, HDAC6, RPSA, CDK6 and RAP1A; Fig. 7B) and 21 downregulated genes (RPS2,
JMJD7.PLA2G4B, CDK1, EIF4A2, ELF1, CCNE2, RHOJ, HMOX1, PRKCE,
CREB1, PLD6, PAK2, GRB2, PRKAG2, RAPGEF1, DDIT4, CCNE1, H3F3B,
PRKCH, CDK2 and RHEB; Fig. 7C) were
selected for further validation. The results indicated that
knockdown of STARD4 increased the expression levels of CDKN1B and
FKBP1A (Fig. 7D), but decreased the
expression levels of PRKCH, RAPGEF1, p21 (RAC1) activated kinase 2
(PAK2), CDK2, CREB1, E74 like ETS transcription factor 1 (ELF1),
PLD6, CCNE1, CCNE2, PRKAG2 and HMOX1 (Fig. 7E). Furthermore, western blot
analysis indicated that the protein expression levels of ELF1,
CREB1 and PAK2 were reduced in the STARD4 knockdown group compared
with those in the control group (Fig.
7F). However, the protein levels of PKC epsilon, and Histone
H3.3 were not changed following STARD4 knockdown (Fig. 7F).
Knockdown of CREB1 suppresses cell
proliferation and migration in BRCA
The present study focused on CREB1, which showed the
most significant decreased expression following STARD4 knockdown.
When analyzing the correlation between CREB1 and STARD4 using the
TIMER database, a significant positive correlation between the two
genes was observed in all types of BRCA (Fig. 8A), HER2 positive BRCA (Fig. 8B), luminal BRCA (Fig. 8C) and basal BRCA (Fig. 8D).
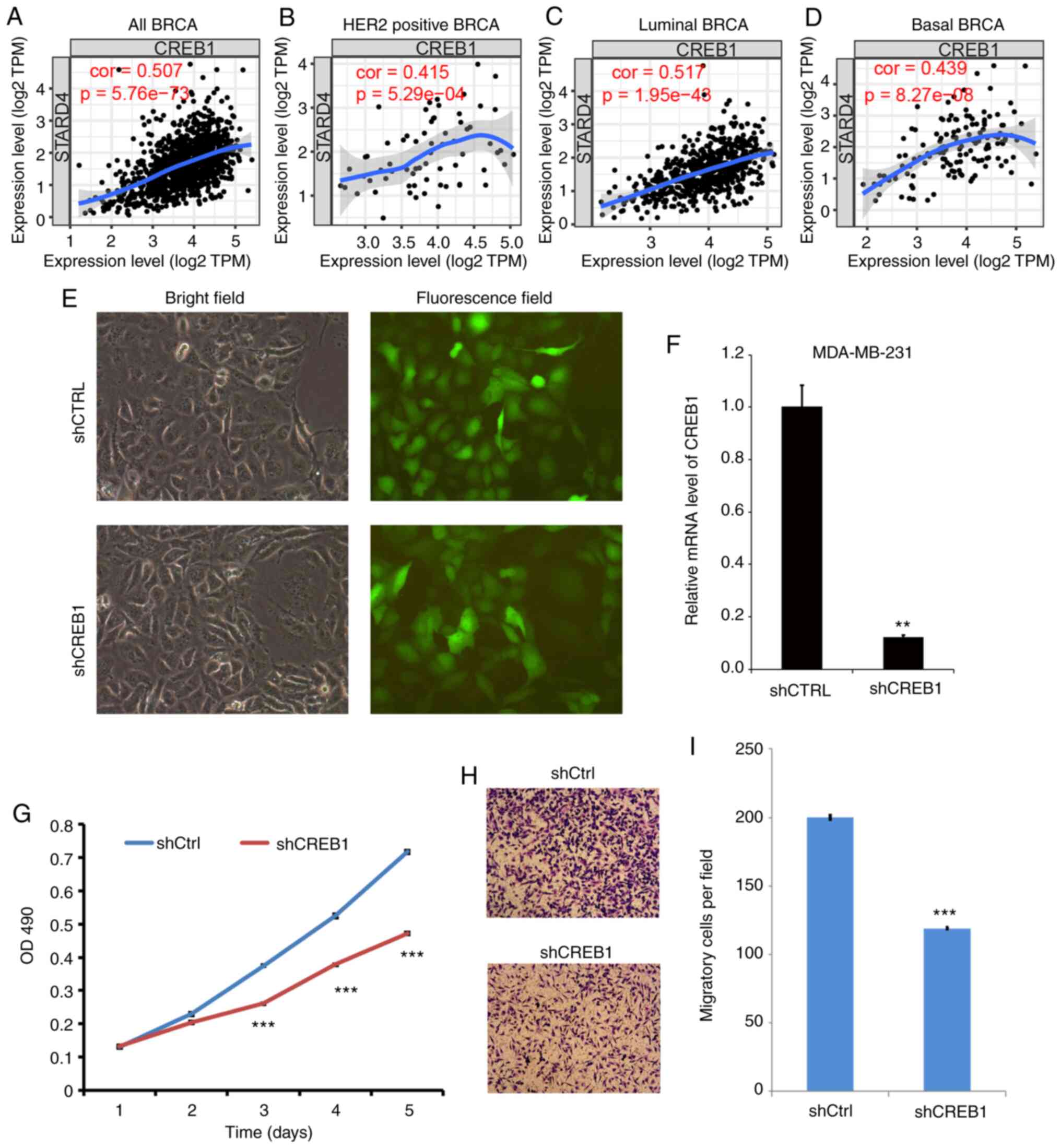 | Figure 8.Knockdown of CREB1 suppresses cell
proliferation and migration in BRCA. (A-D) Correlations between
CREB1 and STARD4 in (A) all types of BRCA, (B) HER2-positive BRCA,
(C) luminal BRCA and (D) basal BRCA were analyzed using the Tumor
Immune Estimation Resource database. (E) Transfection of MDA-MB-231
cells with control vector and shCREB1. Magnification ×200. (F)
CREB1 expression was detected in different groups. (G) Cell
Counting Kit-8 assay demonstrating that knockdown of CREB1
suppressed MDA-MB-231 cell proliferation. (H and I) Transwell assay
revealing that knockdown of CREB1 suppressed MDA-MB-231 cell
migration. Magnification ×100. At least three independent
experiments were performed. **P<0.01, ***P<0.001 vs. shCTRL.
BRCA, breast cancer; CREB1, cAMP responsive element binding protein
1; CTRL, control; OD, optical density; sh, short hairpin RNA;
STARD4, steroidogenic acute regulatory protein-related lipid
transfer 4; shCtrl, shcontrol. |
Loss-of-functions assays were used to detect the
effects of CREB1 on cell proliferation and migration. The infection
efficiency and knockdown efficiency are shown in Fig. 8E and F. As shown in Fig. 8G, knockdown of CREB1 significantly
reduced cell proliferation in MDA-MB-231 cells compared with in
control cells at day 3, 4 and 5. The results of the Transwell assay
demonstrated that the migratory activity of MDA-MB-231 cells
transfected with CREB1 shRNA was significantly attenuated compared
with that of the negative control cells (Fig. 8H and I).
Discussion
The mechanisms underlying the progression of BRCA
remain largely unclear (33,34).
In addition, the molecular functions of STARD4 in human cancers
have not been fully examined. The present study indicated that the
expression levels of STARD4 were upregulated in BRCA samples.
Higher expression levels of STARD4 were significantly associated
with shorter DMFS time in patients with ER-negative, HER2-positive
and PR-negative BRCA. The results indicated that STARD4 may be
associated with the regulation of BRCA progression. Additional
validation demonstrated that STARD4 promoted BRCA progression by
inducing cell proliferation, cell cycle progression and migration,
and by suppressing apoptosis in BRCA cells. To the best of our
knowledge, the present study was the first to indicate that STARD4
may function as an oncogene and promote the tumorigenesis of
BRCA.
By detecting the protein levels of STARD4 in BRCA,
the present study further validated the results of the public
dataset analysis which suggested that STARD4 was upregulated in
BRCA samples. Upregulation of this protein was noted in BRCA
samples. The present study demonstrated that positive protein
expression of STARD4 was associated with ER expression, whereas it
was not significantly associated with PR status, HER-2 status or
TNM stage. These results suggested that STARD4 may be involved in
regulating the imbalance of cholesterol in patients with
ER-negative BRCA, which requires further validation in future
studies. ERα and ERβ are highly expressed in >75% of patients
with BRCA and serve a crucial role in mammary gland development
(35). ER is involved in the
regulation of energy metabolism, including insulin sensitivity,
glucose metabolism balance and fat synthesis (36). For example, ER interacts with
serine/threonine kinase 11, which negatively interferes with the
phosphorylation of protein kinase AMP-activated catalytic subunit
α2, thereby inhibiting TSC2/mTOR/p70S6K signaling (37). Notably, the present study revealed a
significant negative correlation between ER expression and STARD4
levels using public databases and clinical samples. STARD4 is
involved in the regulatory mechanism of intracellular cholesterol
homeostasis and serves a key role in transporting
7α-hydroperoxycholesterol (7α-OOH) from liposomes to isolated
mitochondria (38). 7α-OOH exposure
contributes to a significant reduction in 27-hydroxycholesterol
(27-HC) (39). The potential roles
of 27-HC in affecting ER expression and activity have been revealed
in multiple studies (40–43). Exogenous 27-HC increases ER-mediated
transcription and expression of the endogenous estrogen-regulated
gene trefoil factor 1 in ER+ long-term estrogen
deprivation (LTED) cells but not in the ER− LTED cells
(40). Additionally, a previous
study has demonstrated that 27-HC exerts agonist activity on both
the estrogen and liver X receptors (41–43).
Despite this, the detail mechanisms remain to be investigated
further. To the best of our knowledge, the present study was the
first to reveal the correlation between STARD4 and ER expression in
BRCA, which may be modulated by cholesterol homeostasis.
In human cells, STARD4 promotes the transport of
cholesterol to the mitochondria via the bile acid synthesis pathway
and leads to an increase in cholesterol metabolites (38). In addition, STARD4 can transport
cholesterol to the reticular vesicles, which exhibit high activity
levels of acetyl-CoAzyme A (cholesterol acyltransferases; ACAT) for
esterification and storage of cholesterol (45). STARD4 is co-localized with ACAT1 in
the endoplasmic (44) reticulum and
serves a crucial role in the formation of lipid droplets (45). A previous study further demonstrated
that only STARD4 from the STARD family of proteins could increase
the activity of ACAT (14).
Previous studies have reported that ACAT1 is upregulated in a
series of human cancers and that it is involved in regulating
cancer proliferation, invasion and glycolysis (46–48).
Antalis et al (48) reported
that ACAT1 could promote BRCA proliferation and metastasis.
Considering the interaction between ACAT1 and STARD4, these reports
indicated the potential regulatory roles of STARD4 in BRCA. The
present study explored the potential functions of STARD4 in BRCA
using loss-of-function assays. The data indicated that knockdown of
STARD4 significantly suppressed MDA-MB-231 and HCC1937 cell
proliferation, cell cycle progression and migration, whereas it
promoted cell apoptosis, suggesting that this gene acted as an
oncogene in BRCA.
In order to investigate the mechanisms of STARD4
underlying BRCA progression, microarray and bioinformatics analyses
were performed. Several crucial regulators implicated in cancer
progression were identified as STARD4-regulated genes, including
ELF1, CREB1 and PAK2, which suggested that STARD4 acted as an
oncogene. ELF1 is an important ETS family member and can act both
as an activator and a repressor in the regulation of the
transcription of various genes (49). ELF1 regulates several important
factors, such as SCL, in hematopoietic stem cells (49). A limited number of studies have
demonstrated that ELF1 is involved in regulating the progression of
human cancer (50,51). For example, ELF1 is overexpressed in
acute myeloid leukemia (52).
However, in prostate cancer, ELF1 has been reported as a tumor
suppressor, which suppresses cancer metastasis and cellular
senescence (51). CREB1 is a
nuclear transcription factor which is overexpressed in multiple
human cancers, including lung and colorectal cancer, glioma and
BRCA (53). CREB1 can affect a
series of key regulators in cancer, such as the
apoptosis-associated gene Bcl-2, the invasion-associated gene MMP9
and the cell cycle-associated genes cyclin A1, cyclinB1 and cyclin
D1 (54). PAK2 is a member of the
group I P21 activated kinases family of serine/threonine kinases,
which are involved in modulating apoptosis (55,56).
Previous studies have demonstrated that PAK2 is overexpressed in
different human cancer types, including ovarian (57), gastric (58) and head and neck cancer (59). The present study revealed that CREB1
was a potential target of STARD4 in patients with BRCA. Knockdown
of CREB1 significantly suppressed BRCA cell proliferation and
migration.
The present study had several limitations. First,
the roles of STARD4 in HER2-positive BRCA require further
investigation. More in vivo and in vitro assays
should be performed to explore the potential roles of STARD4 in
HER2-positive cells. Second, the mechanisms of STARD4 in the
regulation of CREB1 expression should be explored further. Third,
STARD4 was only significantly associated with the survival time in
patients with BRCA in ER negative, HER2 positive and PR negative
groups. These results suggested that STARD4 may have different
roles in different types of BRCA. The present study mainly explored
the potential roles of STARD4 in TNBC. In a future study, more
clinical samples will be collected to confirm this finding and
explore the roles in all type of BRCA.
In conclusion, the results of the present study
indicated that the expression levels of STARD4 were increased in
BRCA, and that STARD4 expression was associated with BRCA
malignancy. Furthermore, the present study demonstrated that STARD4
acted as an oncogene in BRCA. These results suggest that STARD4 can
be used as a potential promising biomarker and provide a novel
therapeutic target for BRCA treatment.
Supplementary Material
Supporting Data
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by grants from The
National Natural Science Foundation of China (grant no. 81572612),
The National Key Clinical Specialist Construction Programs of China
(grant no.2014kll), and the Research Innovation Program for
Graduate Students of Central South University (grant nos.
2018zzts912;502211901).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
MZ and WBZ conceived the experiments and drafted the
manuscript. MZ, ZX, FW and RS conducted the experiments. LL, JC,
BAL, JH and LQS analyzed and interpreted the data. All authors
agree to be accountable for all aspects of the research in ensuring
that the accuracy or integrity of any part of the work are
appropriately investigated and resolved. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committees of Central South University. Informed consent was
obtained from all individual participants in the study. All animal
handling and experimental procedures were approved by the Animal
Ethics Committees of Central South University and were in
accordance with the guidelines of the China Council of Animal
Care.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Wagner J, Rapsomaniki MA, Chevrier S,
Anzeneder T, Langwieder C, Dykgers A, Rees M, Ramaswamy A, Muenst
S, Soysal SD, et al: A single-cell atlas of the tumor and immune
ecosystem of human breast cancer. Cell. 177:1330–1345 e1318. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ciriello G, Gatza ML, Beck AH, Wilkerson
MD, Rhie SK, Pastore A, Zhang H, McLellan M, Yau C, Kandoth C, et
al: Comprehensive molecular portraits of invasive lobular breast
cancer. Cell. 163:506–519. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Jung SY, Rosenzweig M, Sereika SM, Linkov
F, Brufsky A and Weissfeld JL: Factors associated with mortality
after breast cancer metastasis. Cancer Causes Control. 23:103–112.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Li XH, Song JW, Liu JL, Wu S, Wang Ls,
Gong Ly and Lin X: Serine-Arginine protein kinase 1 is associated
with breast cancer progression and poor patient survival. Med
Oncol. 31:832014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhu J and Thompson CB: Metabolic
regulation of cell growth and proliferation. Nat Rev Mol Cell Biol.
20:436–450. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kamphorst JJ and Gottlieb E: Cancer
metabolism: Friendly neighbours feed tumour cells. Nature.
536:401–402. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wagner EF and Petruzzelli M: Cancer
metabolism: A waste of insulin interference. Nature. 521:430–431.
2015. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Tasselli L and Chua KF: Cancer: Metabolism
in ‘the driver's seat’. Nature. 492:362–363. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Cao Y: Adipocyte and lipid metabolism in
cancer drug resistance. J Clin Invest. 129:3006–3017. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chang IY, Ohn T, Ko GS, Yoon Y, Kim JW and
Yoon SP: Immunolocalization of steroidogenic acute regulatory
protein-related lipid transfer (START) domain-containing proteins
in the developing cerebellum of normal and hypothyroid rats. J Chem
Neuroanat. 43:28–33. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ishikawa T, Hwang K, Lazzarino D and
Morris PL: Sertoli cell expression of steroidogenic acute
regulatory protein-related lipid transfer 1 and 5 domain-containing
proteins and sterol regulatory element binding protein-1 are
interleukin-1beta regulated by activation of c-jun n-terminal
kinase and cyclooxygenase-2 and cytokine induction. Endocrinology.
146:5100–5111. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Clark BJ: The mammalian START domain
protein family in lipid transport in health and disease. J
Endocrinol. 212:257–275. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Soccio RE, Adams RM, Romanowski MJ,
Sehayek E, Burley SK and Breslow JL: The cholesterol-regulated
starD4 gene encodes a StAR-related lipid transfer protein with two
closely related homologues, starD5 and starD6. Proc Natl Acad Sci
USA. 99:6943–6948. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Alpy F and Tomasetto C: Give lipids a
START: The StAR-related lipid transfer (START) domain in mammals. J
Cell Sci. 118:2791–2801. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Calderon-Dominguez M, Gil G, Medina MA,
Pandak WM and Rodríguez-Agudo D: The starD4 subfamily of
steroidogenic acute regulatory-related lipid transfer (START)
domain proteins: New players in cholesterol metabolism. Int J
Biochem Cell Biol. 49:64–68. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Riegelhaupt JJ, Waase MP, Garbarino J,
Cruz DE and Breslow JL: Targeted disruption of steroidogenic acute
regulatory protein D4 leads to modest weight reduction and minor
alterations in lipid metabolism. J Lipid Res. 51:1134–1143. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Rodriguez-Agudo D, Ren S, Wong E, Marques
D, Redford K, Gil G, Hylemon P and Pandak WM: Intracellular
cholesterol transporter starD4 binds free cholesterol and increases
cholesteryl ester formation. J Lipid Res. 49:1409–1419. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Garbarino J, Pan M, Chin HF, Lund FW,
Maxfield FR and Breslow JL: STARD4 knockdown in hepG2 cells
disrupts cholesterol trafficking associated with the plasma
membrane, ER, and ERC. J Lipid Res. 53:2716–2725. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Bouhaddou M, DiStefano MS, Riesel EA,
Carrasco E, Holzapfel HY, Jones DC, Smith GR, Stern AD, Somani SS,
Thompson TV and Birtwistle MR: Drug response consistency in CCLE
and CGP. Nature. 540:E9–E10. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Dey-Rao R, Smith JR, Chow S and Sinha AA:
Differential gene expression analysis in CCLE lesions provides new
insights regarding the genetics basis of skin vs. systemic disease.
Genomics. 104:144–155. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Li T, Fan J, Wang B, Traugh N, Chen Q, Liu
JS, Li B and Liu XS: TIMER: A web server for comprehensive analysis
of tumor-infiltrating immune cells. Cancer Res. 77:e108–e110. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Gyorffy B, Lánczky A and Szállási Z:
Implementing an online tool for genome-wide validation of
survival-associated biomarkers in ovarian-cancer using microarray
data from 1287 patients. Endocr Relat Cancer. 19:197–208. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Li K, Ma YB, Zhang Z, Tian YH, Xu XL, He
YQ, Xu L, Gao Y, Pan WT and Song WJ: Upregulated IQUB promotes cell
proliferation and migration via activating
Akt/GSK3beta/beta-catenin signaling pathway in breast cancer.
Cancer Med. 7:3875–3888. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Hammond ME, Hayes DF, Dowsett M, Allred
DC, Hagerty KL, Badve S, FitzgibbonsP L, Francis G, Goldstein NS,
Hayes M, et al: American society of clinical oncology/college of
american pathologists guideline recommendations for
immunohistochemical testing of estrogen and progesterone receptors
in breast cancer. J Clin Oncol. 28:2784–2795. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wolff AC, Hammond ME, Hicks DG, Dowsett M,
McShane LM, Allison KH, Allred DC, Bartlett JM, Bilous M,
Fitzgibbons P, et al: Recommendations for human epidermal growth
factor receptor 2 testing in breast cancer: American society of
clinical oncology/college of American pathologists clinical
practice guideline update. J Clin Oncol. 31:3997–4013. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Dowsett M, Nielsen TO, A'Hern R, Bartlett
J, Coombes RC, Cuzick J, Ellis M, Henry NL, Hugh JC, Lively T, et
al: Assessment of Ki67 in breast cancer: Recommendations from the
international Ki67 in breast cancer working group. J Natl Cancer
Inst. 103:1656–1664. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Yang F, Liu H, Zhao J, Ma X and Qi W:
POLR1B is upregulated and promotes cell proliferation in non-small
cell lung cancer. Oncol Lett. 19:671–680. 2020.PubMed/NCBI
|
|
29
|
Wan X, Kong Z, Chu K, Yi C, Hu J, Qin R,
Zhao C, Fu F, Wu H, Li Y and Huang Y: Co-Expression analysis
revealed PTCH1-3′UTR promoted cell migration and invasion by
activating miR-101-3p/SLC39A6 axis in non-small cell lung cancer:
Implicating the novel function of PTCH1. Oncotarget. 9:4798–4813.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Cai Z, Sanchez A, Shi Z, Zhang T, Liu M
and Zhang D: Activation of toll-like receptor 5 on breast cancer
cells by flagellin suppresses cell proliferation and tumor growth.
Cancer Res. 71:2466–2475. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wu L, Wei Y, Zhou WB, Zhang YS, Chen QH,
Liu MX, Zhu ZP, Zhou J, Yang LH, Wang HM, et al: Gene expression
alterations of human liver cancer cells following borax exposure.
Oncol Rep. 42:115–130. 2019.PubMed/NCBI
|
|
33
|
Wang Z, Chen J, Zhong MZ, Huang J, Hu YP,
Feng DY, Zhou ZJ, Luo X, Liu ZQ, Jiang WZ and Zhou WB:
Overexpression of ANLN contributed to poor prognosis of
anthracycline-based chemotherapy in breast cancer patients. Cancer
Chemother Pharmacol. 79:535–543. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhou W, Wang Z, Shen N, Pi W, Jiang W,
Huang J, Hu Y, Li X and Sun L: Knockdown of ANLN by lentivirus
inhibits cell growth and migration in human breast cancer. Mol Cell
Biochem. 398:11–19. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zhou W, Guan X, Wang L, Liao Y and Huang
J: P12(CDK2-AP1) inhibits breast cancer cell proliferation and in
vivo tumor growth. J Cancer Res Clin Oncol. 138:2085–2093. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Gandhi N and Das GM: Metabolic
reprogramming in breast cancer and its therapeutic implications.
Cells. 26:89019.
|
|
37
|
Davis KE, Neinast MD, Sun K, Skiles WM,
Bills JD, Zehr JA, Zeve D, Hahner LD, Cox DW, Gent LM, et al: The
sexually dimorphic role of adipose and adipocyte estrogen receptors
in modulating adipose tissue expansion, inflammation, and fibrosis.
Mol Metab. 2:227–242. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Korytowski W, Rodriguez-Agudo D, Pilat A
and Girotti AW: StarD4-Mediated translocation of
7-hydroperoxycholesterol to isolated mitochondria: Deleterious
effects and implications for steroidogenesis under oxidative stress
conditions. Biochem Biophys Res Commun. 392:58–62. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Korytowski W, Wawak K, Pabisz P, Schmitt
JC, Chadwick AC, Sahoo D and Girotti AW: Impairment of macrophage
cholesterol efflux by cholesterol hydroperoxide trafficking:
Implications for atherogenesis under oxidative stress. Arterioscler
Thromb Vasc Biol. 35:2104–2113. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Simigdala N, Gao Q, Pancholi S,
Roberg-Larsen H, Zvelebil M, Ribas R, Folkerd E, Thompson A, Bhamra
A, Dowsett M and Martin LA: Cholesterol biosynthesis pathway as a
novel mechanism of resistance to estrogen deprivation in estrogen
receptor-positive breast cancer. Breast Cancer Res. 18:582016.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Munir MT, Ponce C, Powell CA, Tarafdar K,
Yanagita T, Choudhury M, Gollahon LS and Rahman SM: The
contribution of cholesterol and epigenetic changes to the
pathophysiology of breast cancer. J Steroid Biochem Mol Biol.
183:1–9. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
He S and Nelson ER: 27-Hydroxycholesterol,
an endogenous selective estrogen receptor modulator. Maturitas.
104:29–35. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
McDonnell DP, Chang CY and Nelson ER: The
estrogen receptor as a mediator of the pathological actions of
cholesterol in breast cancer. Climacteric. 17:60–65. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Iaea DB, Dikiy I, Kiburu I, Eliezer D and
Maxfield FR: STARD4 membrane interactions and sterol binding.
Biochemistry. 54:4623–4636. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Rodriguez-Agudo D, Calderon-Dominguez M,
Ren S, Marques D, Redford K, Medina-Torres MA, Hylemon P, Gil G and
Pandak WM: Subcellular localization and regulation of starD4
protein in macrophages and fibroblasts. Biochim Biophys Acta.
1811:597–606. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Garcia-Bermudez J and Birsoy K: Drugging
ACAT1 for cancer therapy. Mol Cell. 64:856–857. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Saraon P, Trudel D, Kron K,
Dmitromanolakis A, Trachtenberg J, Bapat B, van der Kwast T, Jarvi
KA and Diamandis EP: Evaluation and prognostic significance of
ACAT1 as a marker of prostate cancer progression. Prostate.
74:372–380. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Antalis CJ, Arnold T, Rasool T, Lee B,
Buhman KK and Siddiqui RA: High ACAT1 expression in estrogen
receptor negative basal-like breast cancer cells is associated with
LDL-induced proliferation. Breast Cancer Res Treat. 122:661–670.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Oikawa T and Yamada T: Molecular biology
of the ets family of transcription factors. Gene. 303:11–34. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Xiao XH and He SY: ELF1 activated long
non-coding RNA CASC2 inhibits cisplatin resistance of non-small
cell lung cancer via the miR-18a/IRF-2 signaling pathway. Eur Rev
Med Pharmacol Sci. 24:3130–3142. 2020.PubMed/NCBI
|
|
51
|
Budka JA, Ferris MW, Capone MJ and
Hollenhorst PC: Common ELF1 deletion in prostate cancer bolsters
oncogenic ETS function, inhibits senescence and promotes docetaxel
resistance. Genes Cancer. 9:198–214. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Fukushima T, Miyazaki Y, Tsushima H,
Tsutsumi C, Taguchi J, Yoshida S, Kuriyama K, Scadden D, Nimer S
and Tomonaga M: The level of MEF but not ELF-1 correlates with FAB
subtype of acute myeloid leukemia and is low in good prognosis
cases. Leukemia Res. 27:387–392. 2003. View Article : Google Scholar
|
|
53
|
Rao M, Zhu Y, Cong X and Li Q: Knockdown
of CREB1 inhibits tumor growth of human gastric cancer in
vitro and in vivo. Oncol Rep. 37:3361–3368. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Sakamoto KM and Frank DA: CREB in the
pathophysiology of cancer: Implications for targeting transcription
factors for cancer therapy. Clin Cancer Res. 15:2583–2587. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Shao Y, Qi Y, Huang Y, Liu Z, Ma Y, Guo X,
Jiang S, Sun Z and Ruan Q: Human cytomegalovirus miR-US4-5p
promotes apoptosis via downregulation of p21-activated kinase 2 in
cultured cells. Mol Med Rep. 16:4171–4178. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Eron SJ, Raghupathi K and Hardy JA: Dual
site phosphorylation of caspase-7 by PAK2 blocks apoptotic activity
by two distinct mechanisms. Structure. 25:27–39. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Flate E and Stalvey JR: Motility of select
ovarian cancer cell lines: Effect of extra-cellular matrix proteins
and the involvement of PAK2. Int J Oncol. 45:1401–1411. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Gao C, Ma T, Pang L and Xie R: Activation
of P21-activated protein kinase 2 is an independent prognostic
predictor for patients with gastric cancer. Diagn Pathol. 9:552014.
View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Park J, Kim JM, Park JK, Huang S, Kwak SY,
Ryu KA, Kong G, Park J and Koo BS: Association of p21-activated
kinase-1 activity with aggressive tumor behavior and poor prognosis
of head and neck cancer. Head Neck. 37:953–963. 2015. View Article : Google Scholar : PubMed/NCBI
|