Introduction
Cervical cancer (CC) is a common malignant
gynaecological tumour. In 2015, worldwide data showed that CC was
the second most common type of cancer (second only to breast
cancer) among women in developing countries (1). In 2018, 8,622,539 cases of cancer were
newly diagnosed in women worldwide, and cancer-associated mortality
among women reached 4,169,387 cases, where CC (7.5%) was the fourth
leading cause of cancer mortality after breast cancer (15.0%), lung
cancer (13.8%), and colorectal cancer (9.5%) (2). In China, the prevalence of CC is 28.2
per 1,000 women aged 30–44 years (3). Given the prevalence of CC, identifying
improved treatment methods for patients at different stages is
important. Early CC (stage IA) is usually treated with surgery, and
advanced CC (stage IIB-IV) is usually treated with non-surgical
options. For intermediate-stage CC (stages IB and IIA) (4), no consensus on the optimal treatment
has been established, and the majority of patients undergo surgery
or radiotherapy; however, currently available tests cannot reliably
predict risk factors in these patients. If such patients undergo
surgery and risk factors are confirmed during the operation,
radiotherapy will be required after surgery. Numerous studies have
demonstrated a high incidence of complications following radical
surgery combined with radiotherapy (5–9). Every
effort should be made to avoid radical surgery combined with pelvic
radiotherapy. These patients are not recommended to receive
surgery, and at-risk patients may benefit more from
chemoradiotherapy. Proper preoperative selection of non-surgical
treatment over unnecessary surgery will greatly improve the
prognosis (6,8–10).
Hence, high-risk predictors must be identified to enable stratified
management. Filamin A (FLNa) can be used as a predictor of
chemotherapy sensitivity and as a biomarker of prognosis (11), and uncoupling protein 2 (UCP2) can
be used as an indicator of chemotherapy sensitivity in patients
with advanced CC (12). The present
study focused on FLNa and UCP2 and investigated their expression
levels in CC, their roles in the development and progression of CC,
and their value for early diagnosis and prognosis prediction of
CC.
As a non-muscular actin-binding protein and an
important scaffolding protein, FLNa interacts with a number of
proteins involved in cell signalling and cytoskeletal
reorganization, and is also regulated by phosphorylation (13). FLNa is widely distributed in the
cytoplasm and is involved in the adhesion between cells. FLNa plays
roles in signal transduction and protein sorting (14,15).
Increased or decreased FLNa expression contributes to conditions
such as inflammation and cancer (14,15).
FLNa is aberrantly expressed or mutated in numerous different types
of cancer, such as breast cancer, colon cancer, melanoma and
prostate cancer (16–21).
UCP2 is a member of the mitochondrial UCP family,
which is widely distributed throughout the body (22). As a transmembrane transporter of the
inner mitochondrial membrane, UCP2 decreases the production of ATP
and reactive oxygen species (ROS) by decoupling mitochondrial
oxidation and phosphorylation, thereby protecting tissues and
organs from oxidative stress (23).
Previous studies have demonstrated increased UCP2 expression in
numerous malignant tumours, including colon cancer,
cholangiocarcinoma, breast cancer and leukaemia (24–27).
To the best of our knowledge, few studies have
investigated the roles of FLNa and UCP2 in CC. To detect the
mechanisms of FLNa and UCP2 in the development and progression of
CC, the present study used paraffin sections of cervical tissues,
including normal cervix (NC), low-grade intraepithelial neoplasia
(LSIL), high-grade intraepithelial neoplasia (HSIL) and CC tissues,
and performed immunohistochemical staining to determine the
expression levels of FLNa and UCP2. The expression of FLNa and UCP2
was knocked down in CC cell lines in order to investigate the
effects on cancer cell proliferation, cell cycle arrest, apoptosis,
migration and invasion. In addition, the present study investigated
the effects of interfering with FLNa and UCP2 expression on
apoptosis-associated proteins [extracellular signal-regulated
kinase (ERK), phosphorylated (p) ERK, protein kinase B (AKT), p-AKT
and B-cell lymphoma-2 (Bcl-2)] and the mRNA levels of Ras, matrix
metalloproteinase (MMP)-2 and MMP-9 in CC cell lines (SiHa and
HeLa) with the aim of determining the mechanisms of action
underlying FLNa and UCP2 in the development and progression of CC,
and their roles in the early diagnosis and prognosis prediction of
CC.
Materials and methods
Tissue samples from patients
In the present study, paraffin sections were
collected from 33 patients with NC, 33 patients with LSIL, 40
patients with HSIL and 45 patients with CC among patients
(47.51±11.41 years) treated at Feicheng People's Hospital, (Tai'an,
China), between January 2010 and December 2019. The tissues were
cut into 4-µm sections and 0% neutral buffer formalin solution was
used to fix the samples at room temperature for 24–48 h. The
clinical stage was determined according to the 2018 International
Federation of Gynecology and Obstetrics (FIGO) staging guidelines
(4). Patient age, previous human
papilloma virus (HPV) test results, thyrocalcitonin levels,
colposcopy findings, and pathology findings were recorded. For each
tissue collection, a written consent from the subject was obtained.
The present study was approved by the Ethics Committee of Qilu
Hospital (approval no. KYLL-2017-560).
Cell lines and cell culture
Human CC cell lines (C33A, SiHa, HeLa, CaSKi, ME-180
and HH-8) and HUCEC cells (normal human cervical epithelial cells),
which were used as a control, were provided by Shanghai Meixuan
Biotechnology Co., Ltd. Cells were cultured in 90% Dulbecco's
Modified Eagle's Medium (DMEM) (high glucose; H) + 10% foetal
bovine serum (FBS) (Hyclone; Cytiva). Cells were transferred under
sterile conditions, seeded into 8–10 ml of complete medium (90%
DMEM [H] + 10% FBS), and cultured at 37°C and 5% CO2 in
an incubator. The SiHa and HeLa cell lines, which have high FLNa
and UCP2 expression levels, were selected for the subsequent
experiments.
Antibodies and reagents
The following antibodies were used for
immunofluorescence staining: Anti-GAPDH (1:5,000; cat. no. Ab8245;
Abcam); anti-UCP2 (1:100; cat. no. 11081-1-AP; ProteinTech Group,
Inc.) and anti-FLNa (1:100; cat. no. ab76289; Abcam). The
antibodies were used to detect UCP2 and FLNa expression in
different CC cell lines.
The following antibodies were used for
immunohistochemical staining: Anti-UCP2 (1:200; cat. no.
11081-1-AP; ProteinTech Group, Inc.), anti-FLNa (1:200; cat. no.
ab76289; Abcam), anti-P16 (1:200; cat. no. 10883-1-AP; ProteinTech
Group, Inc.), and anti-Ki67 (1:1,000; cat. no. ab92742; Abcam).
The following primary antibodies were used for
western blotting of relevant proteins: Anti-UCP2 (1:1,000; cat. no.
11081-1-AP; ProteinTech Group, Inc.), anti-FLNa (1:1,000; cat. no.
ab76289; Abcam), anti-GAPDH (1:5,000; cat. no. ab8245; Abcam),
anti-ERK1/2 (1:1,000; cat. no. 4695; Cell Signaling Technology,
Inc.), anti-P-ERK1/2 (1:1,000; cat. no. 4370; Cell Signaling
Technology, Inc.), anti-AKT1 (1:1,000; cat. no. ab227100; Abcam),
anti-P-AKT1 (1:10,000; cat. no. ab81283; Abcam), and anti-BCL-2
(1:1,000 cat. no. ab32124; Abcam).
Western blotting
After the cell samples were fully lysed using Lysis
Buffer (Beyotime Institute of Biotechnology), they were centrifuged
at 12,000 × g, 4°C for 5 min, and part of the supernatant was
collected for protein determination using the bicinchoninic acid
(BCA) method.
For the western blotting, the protein was separated
via SDS-PAGE (10 and 12% gels), and the concentration was adjusted
to 1 mg/ml, the protein quality was 20 µg, and the volume was 20 µl
per lane. Before separating the proteins on a gel, a standard curve
was drawn according to the optical density (OD) value and the
standard protein concentration to calculate the total protein
concentration in the sample. After transferring the sample
solution, the membrane was blocked at room temperature for 2 h.,
using 1% BSA (Roche Diagnostics). The membrane was removed and then
washed with tris-buffered saline Tween-20 (TBST) on a shaking table
for 5 min, 3 times. In an incubation bag, the primary antibodies
(FLNa and UCP2) were diluted with sealing solution and incubated
overnight at 4°C. The film was washed with TBST for 5 min, 3 times,
and then the sheep anti-rabbit secondary antibody (1:5,000; cat.
no. SA00001-2 Jackson ImmunoResearch Laboratories, Inc.)
horseradish peroxidase-conjugated was added and BCA Protein
Quantitation kit (Beyotime Institute of Biotechnology) was
used.
Immunofluorescence
The SiHa and HeLa cells were blocked with 3% BSA-PBS
(Roche 735094) for 30 min at room temperature. The present study
used anti-UCP2 and anti-FLNa as primary antibodies and FITC
[Fluorescein (FITC)-conjugated Affinipure Goat Anti-Rabbit IgG;
1:50; cat. no. SA00003-2; ProteinTech Group, Inc.] as a secondary
antibody. The cell samples were fixed in 4% precooled
paraformaldehyde at room temperature for 15 min, washed with
phosphate-buffered saline (PBS) 3 times (3 min each time), and
incubated in 0.5% Triton X-100 for 20 min at room temperature.
Next, diluted antibody solution (in PBS containing 5% bovine serum
albumin) (Hyclone; Cytiva) was added (PBS 0.01 M, pH 7.4 was used
in the blank control group), and the samples were incubated at 4°C
overnight. On the next morning, the samples were removed from the
refrigerator, placed at room temperature for 15 min, and then
washed with PBS (0.01 M, pH 7.4) 5 times (5 min each). After
removal of any extra PBS, FITC was added, and then DAPI was added,
followed by incubation in the dark for 2 min. The nucleus was
identified by blue fluorescence microscope (Olympus Corporation;
×10 magnification).
Immunohistochemical staining and
evaluation
Antigen retrieval was performed using TE buffer pH
9.0, heating in the microwave on band 3 for 10–15 min (700 W oven),
the retrieval solution was allowed to cool at room temperature.
Endogenous peroxidase activity was blocked by incubating sections
in 3% H2O2 solution in ddH2O for
10 min at room temperature. Tissue sections (4 mm-thick) were
stained with FLNa antibody (diluted in PBS, 1:200) and UCP2
antibody (diluted in PBS, 1:200). The expression of FLNa and UCP2
was evaluated in cancer cells. (DAB Detection Kit, Ventana Medical
Systems, Inc)The expression scores were grouped as follows: 0,
<10% positive cells; 1, 10–25% positive cells; 2, 26–50%
positive cells; 3, 51–75% positive cells; and 4, >75% positive
cells. The cells were stained with DAB for 40 sec and haematoxylin
and eosin for 8 min, both at room temperature, and the staining
intensity was rated as 0, none; 1, weak; 2, mild; and 3, strong.
The final score was the product of the quantitative expression
score and staining intensity and was denoted as negative (0–1) or
positive (≥2). All immunohistochemical results were reported by the
pathologists at Feicheng People's Hospital (Tai'an, China) and
reviewed by another experienced pathologist. A confocal microscope
was used to view the images (Olympus Corporation; ×40 and ×200
magnification).
Reverse transcription-quantitative PCR
(RT-qPCR)
The primers (Table
I) were designed and synthesized by Shanghai Meixuan
Biotechnology Co., Ltd. TRIzol® reagent (Invitrogen;
Thermo Fisher Scientific, Inc.) was used to extract the total RNA
from SiHa and HeLa cells, which was stored at −80°C. The total RNA
(2 µg) was used for the synthesis of cDNA according to a
PrimeScript RT Reagent kit with g DNA Eraser. Reverse transcription
was performed as 50°C for 10 min). The fluorophore was provided by
Shanghai Meixuan Biotechnology Co., Ltd. (SYBR Premix Ex Taq). The
PCR conditions were predenaturation at 95°C for 30 sec, then 45
cycles of 95°C for 5 sec and 60°C for 30 sec. The data were
analysed using the 2−ΔΔCq relative-expression method
(28).
 | Table I.Primers for mRNA detection of
relevant genes. |
Table I.
Primers for mRNA detection of
relevant genes.
| Gene | Species | Forward, 5′-3′ | Reverse, 5′-3′ |
|---|
| hGAPDH | h |
GCACCGTCAAGGCTGAGAAC |
TGGTGAAGACGCCAGTGGA |
| RAS | h |
GAGTAGCGCAGTCGCCAAAG |
GCTCGGGGTCCAATAGTAGC |
| MMP2 | h |
CGCATCTGGGGCTTTAAACA |
GCACTGCCAACTCTTTGTCC |
| MMP9 | h |
TCTATGGTCCTCGCCCTGAA |
CATCGTCCACCGGACTCAAA |
Silencing of the FLNa and UCP2 genes
in SiHa and HeLa cells
Small interfering RNA (siRNA) sequences were
designed and synthesized by Shanghai Meixuan Biotechnology Co.,
Ltd. Once the cells reached 40–50% confluency, the cells were
seeded into 24-well plates and transfected with
Lipofectamine® 2000 according to the manufacturer's
instructions (Thermo Fisher Scientific, Inc.). First, 20 pmol siRNA
was dissolved in 50 µl Opti-MEM culture medium with free FBS. Then,
1 µl Lipofectamine® 2000 was dissolved in 50 µl Opti-MEM
culture medium for 5 min at room temperature. Finally, the
aforementioned two solutions were mixed while resting for 20 min at
room temperature, then 400 µl mixed solution was added to each well
in the 24-well plates, and the transfection medium was changed to
cultured medium (Hyclone; Cytiva) containing 90% DMEM high sugar
and 10% FBS) after 4–6 h. The cells were extracted for the PCR
experiment after 48 h. siRNAs were transfected into cells with
Lipofectamine® 2000 for 48 h, and then cells were
collected for qPCR to analyse the mRNA levels of UCP2 and FLNa.
After 48 h, a sequence with good interference was selected for
later experiments; a third siRNA was selected to knockdown UCP2,
and a second siRNA was selected to knockdown FLNa in HeLa and SiHa
cells. Each in vitro experiment was performed three
independent times and in three replicates.
Cell proliferation assay
Cell Counting Kit-8 (CCK-8) (Beyotime Institute of
Biotechnology) was used to detect cell survival. The absorbance at
450 nm was measured with a microplate reader (Multiskan 3; Thermo
Fisher Scientific, Inc.). The measurement was 5×103/100
µl for each sample. All experiments were repeated at least three
times. The equation used for the calculation of the cell
proliferation inhibition rate was as follows: Inhibition ratio=(OD
value of control group-OD value of experimental group)/(OD value of
control group-OD value of blank hole) ×100%.
Migration and invasion assays
Scratch test
The transfected cells were digested and seeded into
6-well plates. Once the cells reached 100% confluency, the medium
was replaced with serum-free medium to starve the cells overnight.
Different siRNAs were transfected into different groups of cells,
and the medium was replaced with fresh serum-free medium after 4–6
h. A 100-µl pipette tip was used to scratch the plates, an image of
which was captured and recorded as 0 h. After 48 h, another image
was captured to analyse cell confluency relative to that at 0 h.
The present study used Image Pro Plus software (version 6.0; Media
Cybernetics, Inc.) to test and calculate the distance between the
cells. The formula used for the calculation of the cell migration
distance was as follows: Cell migration distance=0 h intercellular
distance-48 h intercellular distance. A confocal microscope was
used to detect the cells (Olympus Corporation; ×100
magnification).
Transwell assay
A total of 5×104 cells were inoculated
into a Transwell chamber (BD Biosciences), and 500 µl of fresh
medium containing 20% FBS was added to the lower chamber. The
chamber was incubated at 37°C and 5% CO2 for 48 h. Next,
the cells were fixed in 4% paraformaldehyde for 10 min, stained
with crystal violet solution for 5 min at room temperature and
washed with water until the base membrane was transparent. Lastly,
images of the cells were captured using a confocal microscope
(Olympus Corporation; ×100 magnification), then the cells were
counted and analysed.
Cell cycle and apoptosis assays
The transfected cells were washed twice using
precooled PBS and then digested in 70% alcohol at 4°C overnight.
For cell cycle detection, a cell cycle detection kit was obtained
from Beyotime Institute of Biotechnology. A FACSCalibur flow
cytometer (BD Biosciences) was used to detect red fluorescence at
an excitation wavelength of 488 nm. Appropriate analysis software
(FlowJo; version 10; FlowJo LLC) was used for DNA content and light
scattering analyses. For apoptosis detection, an Annexin V-FITC
apoptosis assay kit was obtained from Beyotime Institute of
Biotechnology. Cells were collected and treated as described in the
kits and analysed by flow cytometry.
Statistical analysis
SPSS software (version 21.0; IBM Corp.) was used for
immunohistochemical staining statistical analysis. The
χ2 and Fisher's exact test were performed. For the cell
line experiments, all assays were performed in at least triplicate.
The results are expressed as the mean ± SD and analysed by unpaired
t-test or one-way ANOVA followed by Tukey's post hoc test, with
SPSS software (version 21.0). P<0.05 was considered to indicate
a statistically significant difference.
Results
Expression of FLNa/UCP2 in human
cervical cancer tissues and cell lines
To detect the expression levels of FLNa and UCP2 in
CC, the present study performed immunohistochemical staining on 33
NC, 33 LSIL, 40 HSIL and 45 CC tissues. Representative images are
presented in Fig. 1A. The
expression levels of FLNa and UCP2 in CC were rated as the product
of the quantitative score and staining intensity (n=45). The scores
ranged from 0 to 12, and scores of 0–1 were rated as negative,
whereas scores ≥2 were rated as positive. The median score of each
protein in the tissues was used as the threshold to separate low
expression from high expression (Table
II). The results show that FLNa and UCP2 were highly expressed
in the CC tissues (Table III).
The FLNa and UCP2 expression levels were detected by PCR and
western blotting in six human CC cell lines (C33A, SiHa, HeLa,
CaSKi, ME-180 and HH-8), and HUCEC cells (normal human cervical
epithelial cells) as control cells. All experiments were repeated
at least three times (Fig. 2A-C).
The SiHa and HeLa cell lines, which had the highest FLNa and UCP2
expression levels, were selected for the subsequent experiments. To
investigate the expression of FLNa/UCP2 in cells, the present study
performed immunofluorescence staining in SiHa and HeLa cells. The
immunofluorescence quantitative analysis is presented in (Fig. 2D). It was revealed that FLNa and
UCP2 were mainly located in the cytoplasm of CC cells, as
determined by immunofluorescence staining (Fig. 2D). According to the results of the
present study, FLNa and UCP2 were highly expressed in CC cells. The
immunohistochemical staining experiment showed the same results,
and thus, SiHa and HeLa cells were selected for the subsequent
experiments in order to determine the function of FLNa/UCP2 in CC
cell lines.
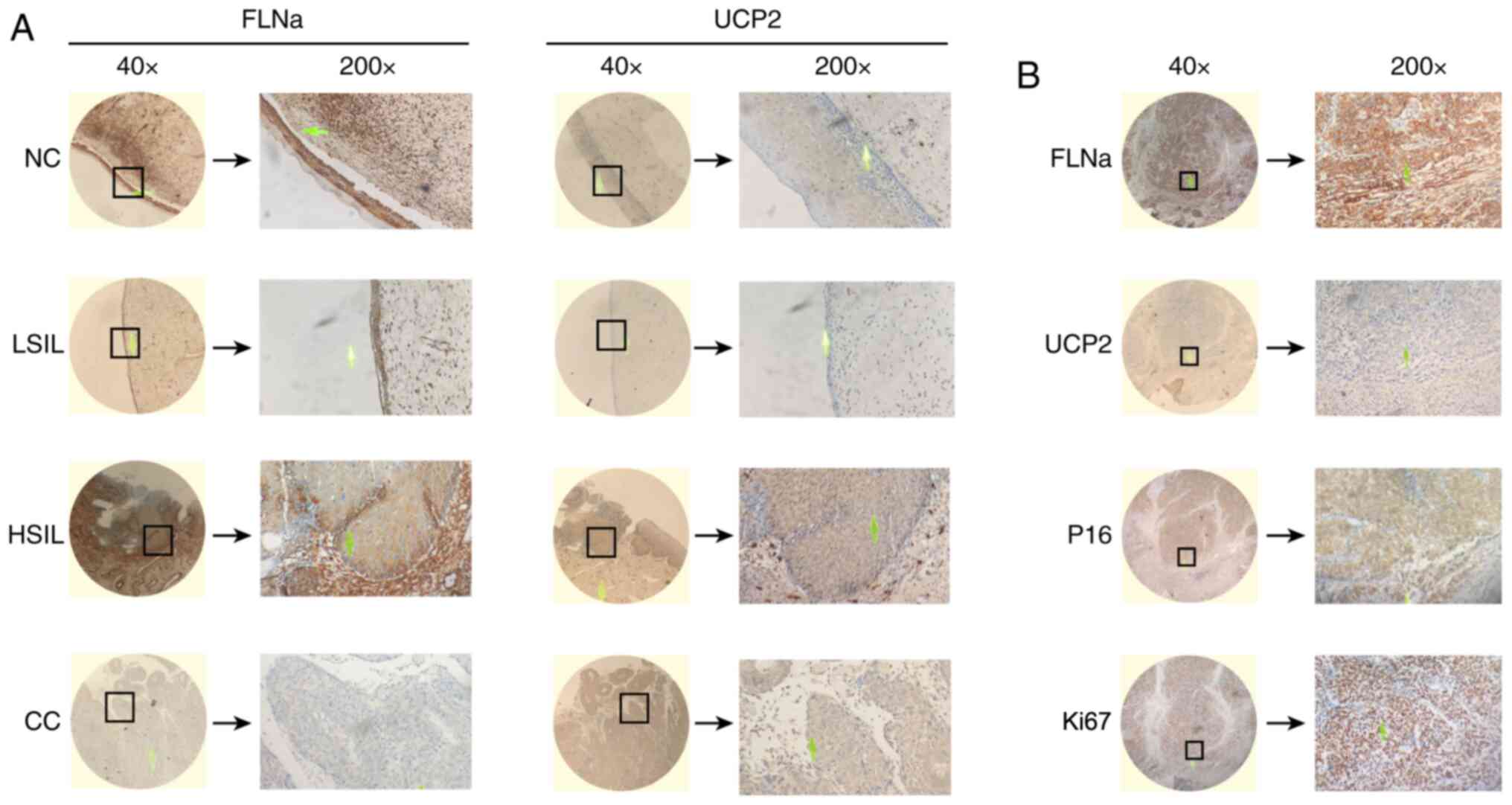 | Figure 1.Immunohistochemical staining of FLNa,
UCP2, p16 and Ki67 in different groups of cervical tissues. (A)
Representative images for the immunohistochemical staining of FLNa
and UCP2 in NC, LSIL, HSIL and CC tissues (magnification ×40 and
×200). (B) Representative images for the immunohistochemical
staining of FLNa, UCP2, p16 and Ki67 in CC tissues (magnification
×40 and ×200). FLNa, filamin A; UCP2, uncoupling protein 2; NC,
normal cervix; LSIL, low-grade squamous intraepithelial lesion;
HSIL, high-grade intraepithelial neoplasia; CC, cervical
cancer. |
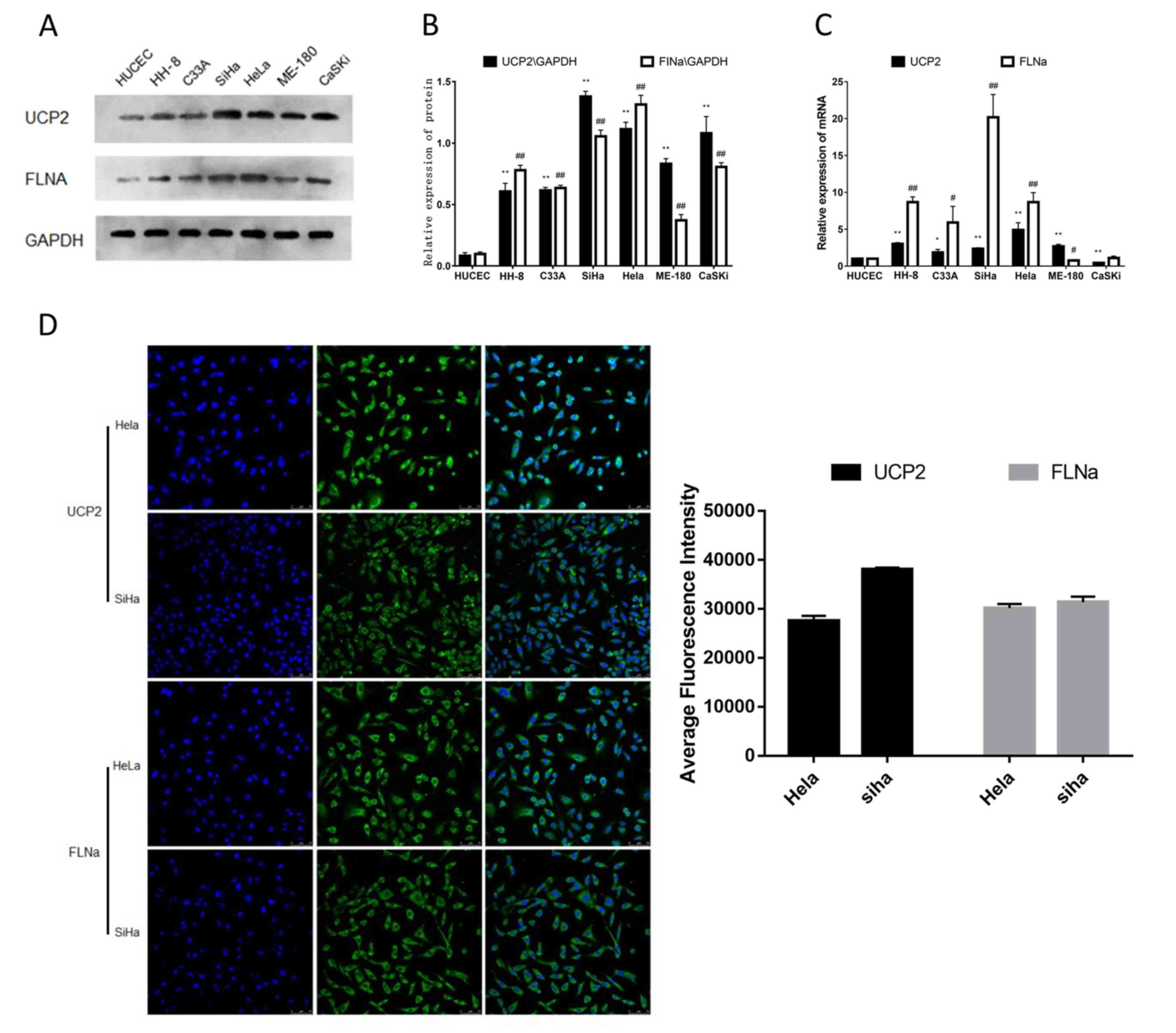 | Figure 2.FLNa and UCP2 expression in different
CC cell lines. (A) Relative expression of FLNa and UCP2 detected by
western blotting in six human CC cell lines (C33A, SiHa, HeLa,
CaSKi, ME-180 and HH-8) and HUCEC cells (normal human cervical
epithelial cells) as control cells. (B) Quantification of the
protein expression levels in (A) compared with the HUCEC cells. (C)
Relative expression of FLNa and UCP2 detected by reverse
transcription-quantitative PCR in six human CC cell lines (C33A,
SiHa, HeLa, CaSKi, ME-180 and HH-8) and in HUCEC cells (normal
human cervical epithelial cells) as control cells, compared with
the HUCEC cells. (D) Immunofluorescence staining showing that FLNa
and UCP2 were mainly located in the cytoplasm of HeLa and SiHa
cells. Quantification of the immunofluorescence is shown.
*P<0.05, **P<0.01, #P<0.05,
##P<0.01 vs. FLNa, filamin A; UCP2, uncoupling
protein 2; CC, cervical cancer. |
 | Table II.Expression of FLNa and UCP2 in
cervical cancer. |
Table II.
Expression of FLNa and UCP2 in
cervical cancer.
|
|
| Low expression | High
expression |
|---|
|
|
|
|
|
|---|
| Protein | Total | Score | n | Score | n |
|---|
| UCP2 | 45 | 0-1 | 35 | 2-12 | 10 |
| FLNa | 45 | 0-6 | 17 | 9-12 | 28 |
 | Table III.Positive expression of FLNa and UCP2
in NC, LSIL, HSIL and CC. |
Table III.
Positive expression of FLNa and UCP2
in NC, LSIL, HSIL and CC.
|
|
| FLNa
expression | UCP2
expression |
|---|
|
|
|
|
|
|---|
|
|
| Positive | P-value | Positive | P-value |
|---|
|
|
|
|
|
|
|
|---|
| Group | n | n | % | vs. NC | vs. LSIL | vs. HSIL | n | % | vs. NC | vs. LSIL | vs. HSIL |
|---|
| NC | 33 | 8 | 23.3 | N/A | N/A | N/A | 0 | 0 | N/A | N/A | N/A |
| LSIL | 33 | 19 | 57.6 | 0.006 | N/A | N/A | 3 | 9.1 | 0.237 | N/A | N/A |
| HSIL | 40 | 24 | 72.5 | 0.002 | 0.834 | N/A | 2 | 5.0 | 0.498 | 0.823 | N/A |
| CC | 45 | 39 | 86.7 | 0.000 | 0.004 | 0.005 | 10 | 22.2 | 0.011 | 0.124 | 0.023 |
Associations between the expression of
FLNa and UCP2 in CC tissues and clinicopathological factors
To investigate the associations between FLNa/UCP2
expression in CC tissues and clinicopathological factors, the
present study analysed age, tumour size, tumour differentiation,
clinical stage, lymph node metastasis and histological type of the
patients. No significant association was observed between the
expression levels of FLNa in CC and age (P=0.714), tumour size
(P=0.064), tumour differentiation (P=0.317), clinical stage
(P=0.828), lymph node metastasis (P=0.737) or histology (P=0.060)
(all P>0.05). No significant association was observed between
the expression level of UCP2 in CC and age (P=0.276), tumour size
(P=0.194), tumour differentiation (P=0.064) or histology
(P>0.999). However, significant associations were observed for
clinical stage (P=0.009) and lymph node metastasis (P=0.022) (both
P<0.05) (Table IV).
 | Table IV.Association between the expression
levels of FLNa and UCP2 and the clinicopathological features of
cervical cancer. |
Table IV.
Association between the expression
levels of FLNa and UCP2 and the clinicopathological features of
cervical cancer.
|
|
| FLNa, n | UCP2, n |
|---|
|
|
|
|
|
|---|
| Group | Total | Low expression
(n=17) | High expression
(n=28) | P-value | Low expression
(n=35) | High expression
(n=10) | P-value |
|---|
| Age, years |
|
|
| 0.714 |
|
| 0.276 |
|
≤45 | 17 | 7 | 10 |
| 15 | 2 |
|
|
>45 | 28 | 10 | 18 |
| 20 | 8 |
|
| Diameter of tumour,
cm |
|
|
| 0.064 |
|
| 0.194 |
| ≤4 | 35 | 16 | 19 |
| 29 | 6 |
|
|
>4 | 10 | 1 | 9 |
| 6 | 4 |
|
| Degree of
differentiation |
|
|
| 0.317 |
|
| 0.064 |
|
Low | 28 | 9 | 19 |
| 19 | 9 |
|
|
Moderate or high | 17 | 8 | 9 |
| 16 | 1 |
|
| Stage |
|
|
| 0.828 |
|
| 0.009 |
| I | 30 | 11 | 19 |
| 27 | 3 |
|
|
II–III | 15 | 6 | 9 |
| 8 | 7 |
|
| Lymph node
metastasis |
|
|
| 0.737 |
|
| 0.022 |
|
Yes | 13 | 4 | 9 |
| 7 | 6 |
|
| No | 32 | 13 | 19 |
| 28 | 4 |
|
| Pathological
pattern |
|
|
| 0.060 |
|
| >0.999 |
|
SCC | 40 | 13 | 27 |
| 31 | 9 |
|
|
Adenocarcinoma | 5 | 4 | 1 |
| 4 | 1 |
|
Expression of four immunohistochemical
indicators, FLNa, UCP2, p16 and Ki67, in CC tissues
To investigate whether the predictive function of
FLNa/UCP2 was better than that of p16 and Ki-67 in cervical HSIL
progression to CC, the present study stained for FLNa, UCP2, P16
and Ki-67 as four CC markers in immunohistochemical staining. The
expression levels of these four markers were observed under ×200
and ×40 magnification under a confocal microscope (Olympus
Corporation). FLNa and UCP2 were primarily stained in the cytoplasm
of CC cells as brown or dark-brown particles. The positive rate of
FLNa was 86.7% (39/45), and the positive rate of UCP2 was 22.2%
(10/45). p16 was expressed in the nucleus as brown or dark-brown
particles, and the positive rate was 97.8% (44/45). Ki67 was
expressed in the nucleus as brown or dark-brown particles, and the
positive rate was 100% (45/45). Representative images are presented
in Fig. 1B. The present study
compared the positive rates of FLNa, UCP2, p16 and Ki67 between the
CC and HSIL tissues (Table V).
Significant differences in FLNa (P=0.005) and UCP2 (P=0.023) were
observed between HSIL and CC tissues. No significant difference was
observed in p16 (P=0.917) or Ki67 (P=0.471). FLNa and UCP2 were
superior to p16 and Ki67 for early prediction of progression from
HSIL to CC. The present study also analysed the associations
between the expression levels of FLNa, UCP2 and p16 in CC and HSIL
tissues (Table VI). It was
revealed that in the CC tissues, both FLNa and p16 were positively
expressed in 39 cases, and neither was expressed in one case
(P<0.01). Both UCP2 and p16 were positively expressed in 10
cases, and neither was expressed in one case, indicating a positive
association, though this result was not statistically significant
(P>0.05). In HSIL tissues, both FLNa and p16 were positively
expressed in 29 cases, and neither were expressed in two cases, but
this was not statistically significant (P>0.01). Both UCP2 and
p16 were positively expressed in two cases, and neither was
expressed in two cases, but this result was not statistically
significant (P>0.05). The aforementioned analysis indicated that
there is an association between the expression levels of FLNa and
p16 (P<0.01).
 | Table V.Positive expression of FLNa, UCP2,
p16 and Ki67 in CC and HSIL. |
Table V.
Positive expression of FLNa, UCP2,
p16 and Ki67 in CC and HSIL.
|
|
| FLNa | UCP2 | p16 | Ki67 |
|---|
|
|
|
|
|
|
|
|---|
|
|
| Positive |
| Positive |
| Positive |
| Positive |
|
|---|
|
|
|
|
|
|
|
|
|
|
|
|---|
| Group | Total, n | n | % | P-value | n | % | P-value | n | % | P-value | n | % | P-value |
|---|
| HSIL | 40 | 24 | 72.5 | 0.005 | 2 | 5 | 0.023 | 38 | 95 | 0.917 | 39 | 97.5 | 0.471 |
| CC | 45 | 39 | 86.7 |
| 10 | 22.5 |
| 44 | 97.8 |
| 45 | 100 |
|
 | Table VI.Co-expression of FLNa, UCP2 and p16
in CC and HSIL. |
Table VI.
Co-expression of FLNa, UCP2 and p16
in CC and HSIL.
|
|
|
|
| FLNa | UCP2 |
|---|
|
|
|
|
|
|
|
|---|
| Protein | Type | n | Group | Negative | Positive | P-value | Negative | Positive | P-value |
|---|
| p16 | HSIL | 40 | Negative (n=2) | 2 | 0 | 0.071 | 2 | 0 | 1 |
|
|
|
| Positive
(n=38) | 9 | 29 |
| 36 | 2 |
|
|
| CC | 45 | Negative (n=1) | 1 | 0 | 0.009 | 1 | 0 | 0.599 |
|
|
|
| Positive
(n=44) | 5 | 39 |
| 34 | 10 |
|
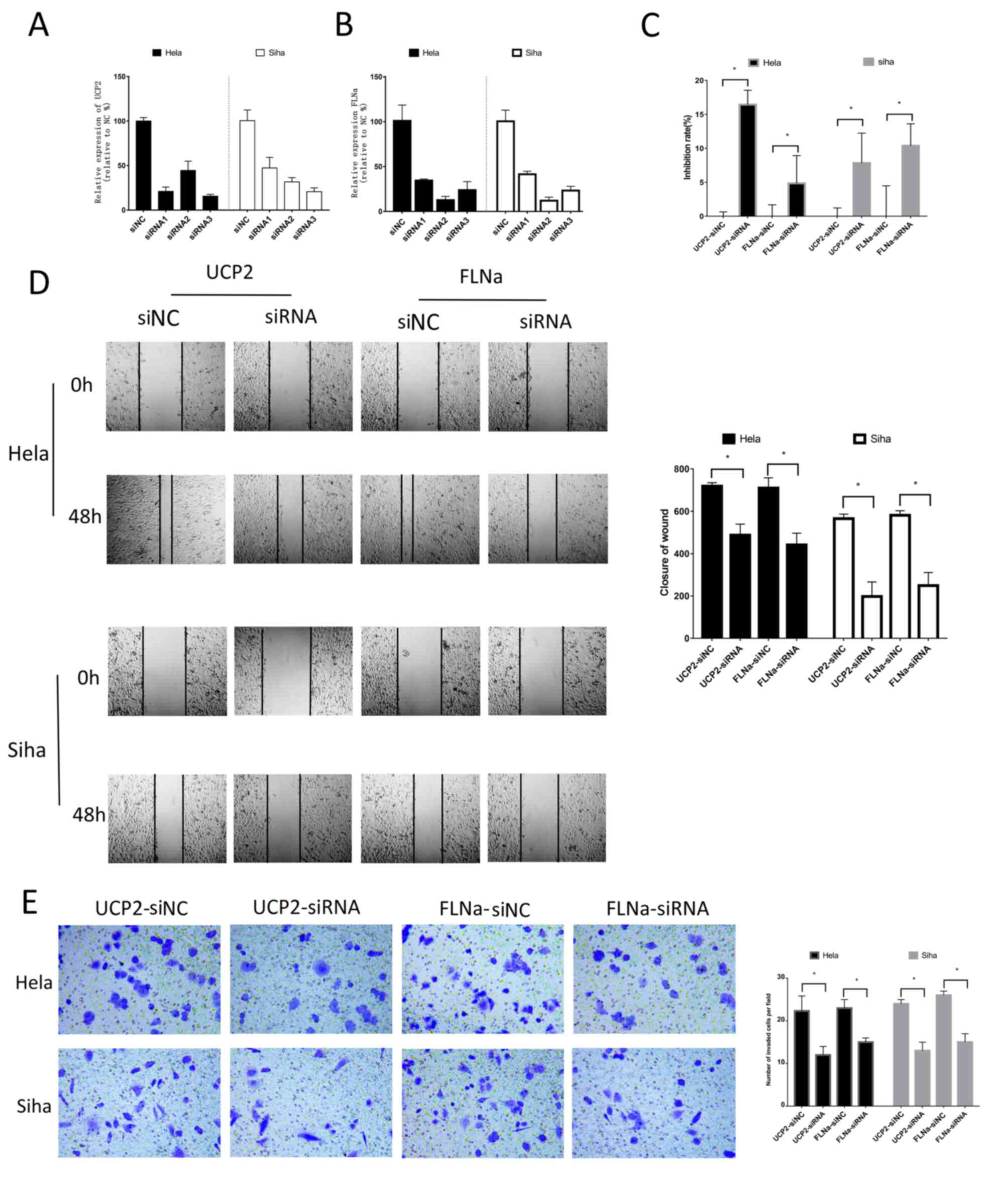
Knockdown of FLNa/UCP2 influences cell
proliferation, apoptosis and the cell cycle
To investigate the functions and signalling pathways
of FLNa and UCP2 in the development and progression of CC, FLNa and
UCP2 siRNAs (Table VII) were
designed and synthesized to downregulate the expression of FLNa and
UCP2 via plasmid transfection, and the knockdown effect was
confirmed via western blotting HeLa and SiHa cells with stable,
high expression levels of FLNa and UCP2 were used for subsequent
experiments. All experiments were repeated at least three times.
Based on the qPCR, a third siRNA was selected to knockdown UCP2,
and a second siRNA was selected to knockdown FLNa in HeLa and SiHa
cells (Fig. 3A and B). To
investigate the functions and mechanisms of action underlying UCP2
and FLNa, UCP2 and FLNa expression was knocked down in HeLa and
SiHa cells, and the CCK-8 assay was used to analyse cell
proliferation. The present study used the cellular proliferation
inhibition rate to present the results of the CCK-8 assays, which
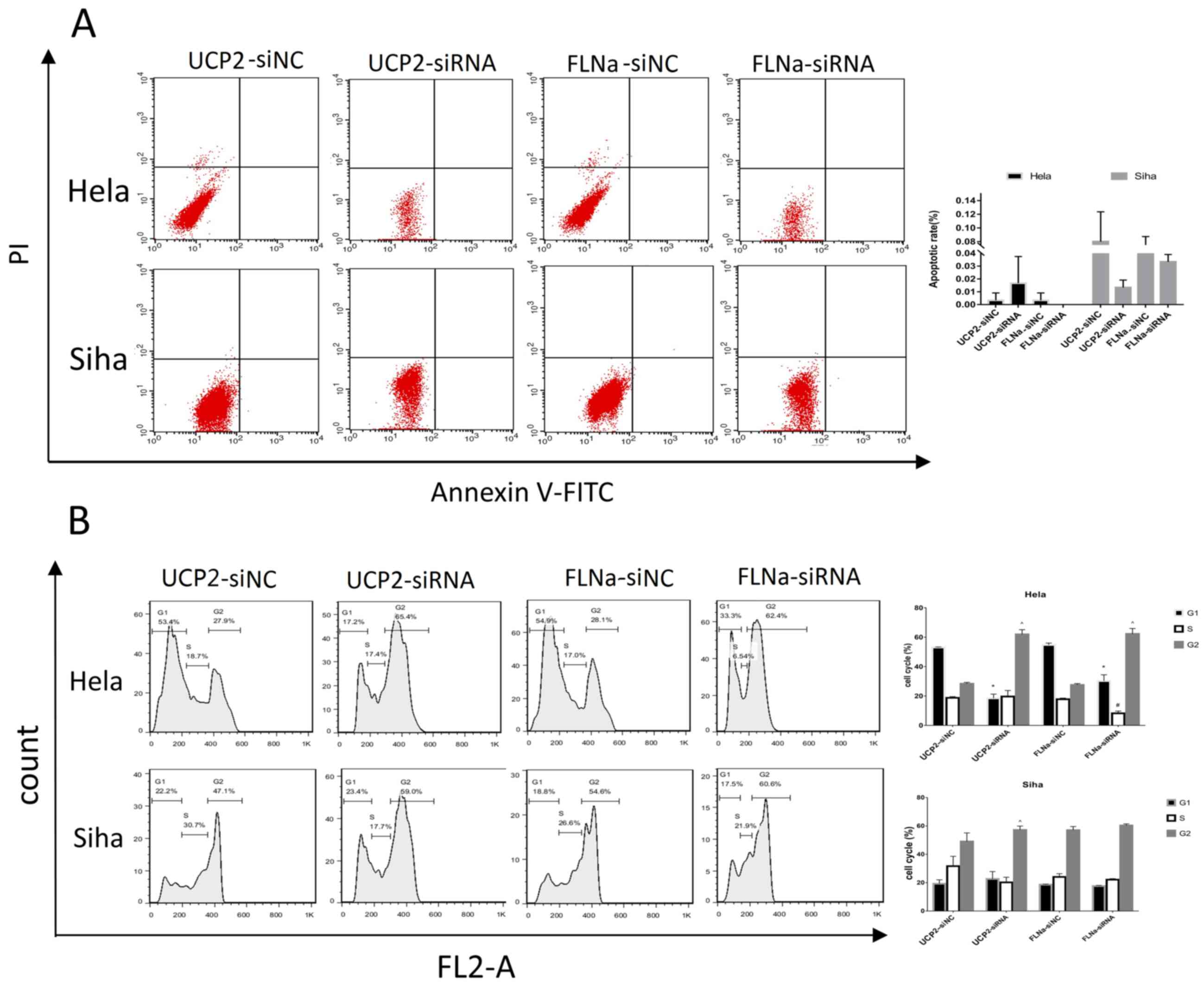
demonstrated that cell proliferation was inhibited (Fig. 3C). The present study also used flow
cytometry to analyse cell apoptosis, which revealed that following
UCP2 and FLNa knockdown in HeLa and SiHa cells, there was no
significant difference in apoptosis compared with the control
groups (Fig. 4A). Following UCP2
and FLNa knockdown in HeLa cells and UCP2 knockdown in SiHa cells,
more cells were arrested at the G2 phase. Following UCP2
knockdown in SiHa cells G2 cells content increased. Following FLNa
knockdown in SiHa cells, no significant cell cycle arrest was
observed (Fig. 4B).
 | Table VII.FLNa and UCP2 interference
sequences. |
Table VII.
FLNa and UCP2 interference
sequences.
|
| Sequence,
5′-3′ |
|---|
| UCP2 |
|
| 1 |
GCCUGUAUGAUUCUGUCAATT |
|
|
UUGACAGAAUCAUACAGGCTT |
| 2 |
CCUGUAUGAUUCUGUCAAATT |
|
|
UUUGACAGAAUCAUACAGGTT |
| 3 |
GGUAAAGGUCCGAUUCCAATT |
|
|
UUGGAAUCGGACCUUUACCTT |
| Control
1 |
ACGGGCUCUUAAAGGAUCATT |
|
|
UGAUCCUUUAAGAGCCCGUTT |
| FLNa |
| 1 |
GCACUUACAGCUGCUCCUATT |
|
|
UAGGAGCAGCUGUAAGUGCTT |
| 2 |
GCUGGCAGCUACACCAUUATT |
|
|
UAAUGGUGUAGCUGCCAGCTT |
| 3 |
GCACAUGUUCCGUGUCCUATT |
|
|
UAGGACACGGAACAUGUGCTT |
| Control
2 |
AGUAGUCGUAUCGGACAACTT |
|
|
GUUGUCCGAUACGACUACTTT |
Knockdown of FLNa/UCP2 influences cell
invasion and migration
The scratch test and Transwell assay were used to
detect cell migration and invasion. The results revealed that cell
migration (Fig. 3D) and cell
invasion were significantly decreased (Fig. 3E) with knockdown of these two
proteins.
Knockdown of FLNa/UCP2 influences cell
signalling pathway proteins
Western blotting was performed in order to detect
the expression levels of relevant proteins (Fig. 5A), including ERK, p-ERK, AKT, p-AKT
and Bcl-2. The results showed no significant change in ERK/p-ERK,
AKT1 or Bcl-2 levels (Fig. 5D), but
p-ERK1/2 (Fig. 5B) and p-AKT1
(Fig. 5C) were downregulated. PCR
showed that the mRNA levels of Ras (Fig. 5E), MMP-2 (Fig. 5F) and MMP-9 (Fig. 5G) were decreased.
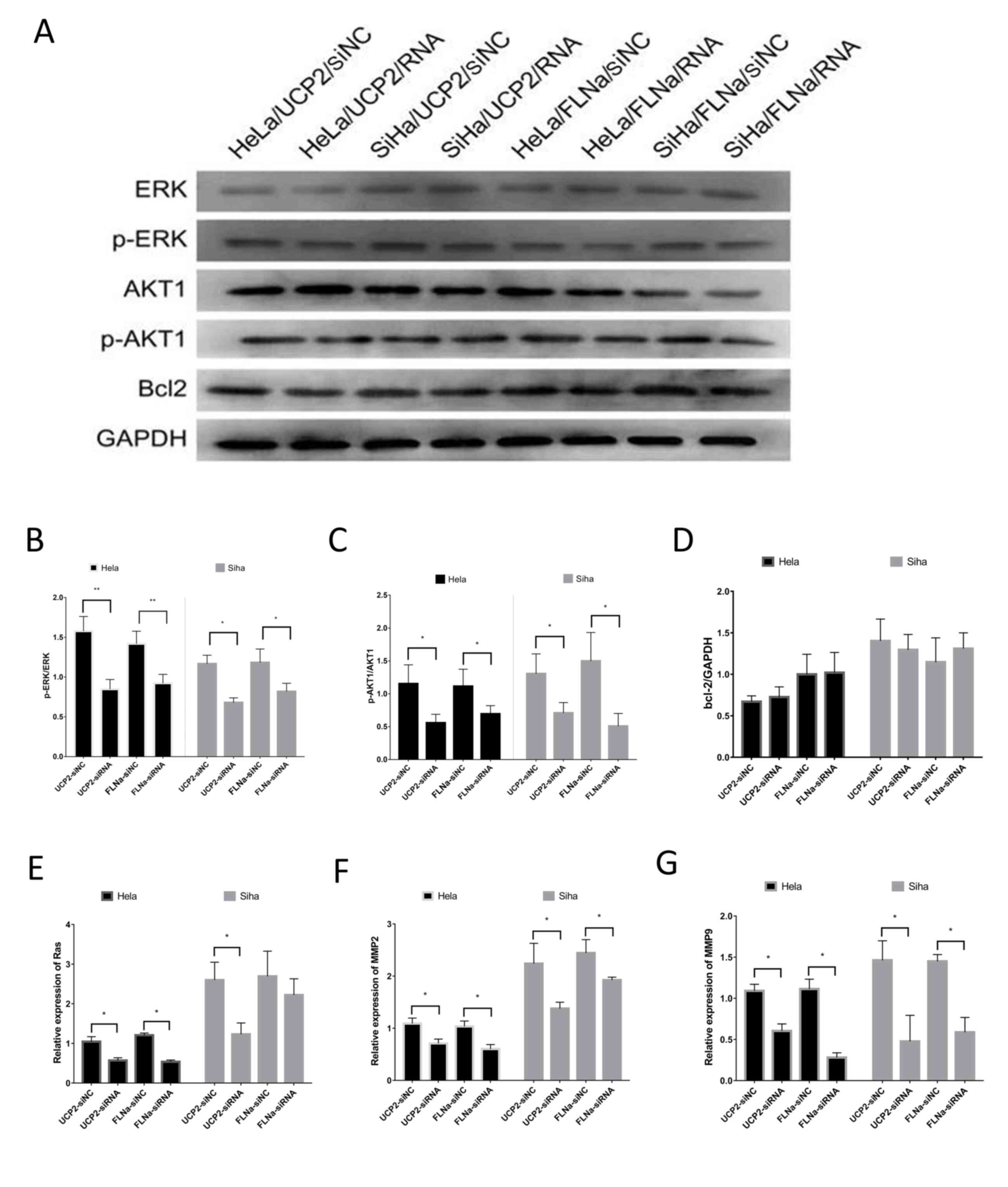 | Figure 5.Investigation of the signalling
pathways of UCP2 and FLNa. (A) Protein levels detected by western
blotting, including ERK, p-ERK, AKT, p-AKT and Bcl-2. (B) After
UCP2 and FLNa knockdown in HeLa and SiHa cells, p-ERK1/2 was
downregulated; (C) p-AKT1 was downregulated; (D) there was no
significant change in Bcl-2 expression; (E) the RAS mRNA levels
were decreased in HeLa cells, but there was no significant change
in SiHa cells; (F) MMP-2 mRNA levels were decreased; and (G) MMP-9
mRNA levels were decreased. *P<0.05; **P<0.01. UCP2,
uncoupling protein 2; FLNa, filamin A; ERK, extracellular
signal-regulated kinase; p, phosphorylated; AKT, protein kinase B;
Bcl-2, B-cell lymphoma-2. |
Discussion
For decades, population-wide cytology screening for
CC in Europe, North America, Australia and New Zealand has
contributed to a rapid decline in the prevalence of CC (29,30).
In Eastern Europe and Central Asia, however, CC-associated
premature mortality continues to rise due to the lack of effective
screening and treatment strategies (31). Given the scale of the patient
population, prognostic indicators of CC must be identified to
enable stratified management of patients.
The present study showed that the positive rate of
FLNa was significantly higher in CC tissues than in NC tissues,
contradicting the results of a previous study (32) showing that the positive rate of FLNa
was higher in normal prostate tissue than in prostate cancer
tissue. This discrepancy may be associated with the different roles
of FLNa in cancer cells at different sites and to the different
pathological types, and further research is required in order to
investigate the mechanism underlying increasing or decreasing FLNa
expression levels. The results of the present study showed that the
positive rate of UCP2 was significantly higher in CC tissues than
in NC tissues, which is consistent with a study by Horimoto et
al (24). Furthermore, the
present study revealed that the positive rate of FLNa was 86.7%
(39/45) in CC tissues (positive, n=39; negative, n=6). Among the
six tissue samples with negative FLNa expression, two had positive
UCP2 expression, indicating that UCP2 was a useful complementary
indicator to FLNa and may help to decreased missed diagnoses during
screening. Immunohistochemical staining showed that FLNa may be
expressed in both CC and HSIL tissues. Thus, a positive screening
result suggests high-grade cervical lesions, and that there is an
association between the expression levels of FLNa and p16, which is
very important for precancer screening, although the absence of
negative individuals in the present study was a limitation, and so
more data are required. The positive rate of UCP2 was significantly
higher in CC tissues than in NC tissues, with no significant
difference between NC, LSIL and HSIL tissues. UCP2 is more specific
with regard to clinical stage and lymph node metastasis, and
positive screening may help predict at-risk patients or advanced
clinical stages.
The necessity for surgical treatment should be
evaluated in each patient. Non-surgical options (such as
radiotherapy) may be more advantageous than surgery. In clinical
practice, p16 and Ki67 have been widely used for CC screening.
Among high-risk HPV-positive subjects, p16/Ki67 dual staining is
more sensitive and specific than thyrocalcitonin for cervical
lesion screening (33). In the
present study, the positive immunohistochemical staining rates of
FLNa, UCP2, p16 and Ki67 were analysed in both CC and HSIL tissues.
The results demonstrated significant differential expression of
FLNa and UCP2, but not p16 or Ki67, between CC and HSIL.
Furthermore, FLNa and UCP2 were superior to p16 and Ki67 for early
prediction of progression from HSIL to CC. The positive rates of
p16 and Ki67 were extremely high in CC tissues [p16, 97.8% (44/45);
Ki67, 100% (45/45)], with no significant differential expression
between patients with different clinical stages or between those
with and without risk factors, potentially due to p16/Ki67 dual
staining being advantageous for precancerous screening; however, it
cannot be used to predict the prognosis. The present study showed
that UCP2 was more advantageous for clinical staging and prediction
of lymph node metastasis in patients with CC compared with p16/Ki67
dual staining.
Tumour development and progression depend on the
activation state of signalling pathways that regulate cell
proliferation and differentiation (34). Among the numerous signalling
pathways associated with tumour development and progression,
mitogen-activated protein kinase (MAPK) signalling is an important
pathway. As the most important MAPK family member in mammalian
cells, ERK is widely expressed in various tissues and is involved
in the regulation of cell proliferation and differentiation.
Abnormal activation of this signalling pathway leads to abnormal
cell proliferation and differentiation, which are important steps
in tumour development and progression (35,36).
Loss of integrity of the extracellular matrix and
basal membrane (mainly collagen, laminin and fibronectin; lytic
enzymes can destroy these components) is an important step in
tumour metastasis (37). MMPs
consist of a large family of proteolytic enzymes, and their
expression is increased in a number of malignant tumour types,
cultured tumour cells and oncogene-transformed cells. In
vitro experiments have demonstrated that a high invasion
capacity of tumour cells is associated with increased expression of
MMP-9 (38,39). MMP-2 and MMP-9 are upregulated in CC
and precancerous lesions, and studies have investigated the value
of MMP-2 and MMP-9 overexpression in predicting cervical diseases
(40).
In the present study, the effects and underlying
mechanisms of FLN and UCP2 on the biological behaviour of cultured
CC cell lines in vitro were investigated. Previous studies
on FLNa, such as that by Zhu et al (41), demonstrated that FLNa regulates the
Ras/ERK and Ras/guanine nucleotide exchange factor-1 pathways to
decrease intracellular MMP-9, inhibit degradation of the
extracellular matrix, and thereby prevent tumour cell migration,
which is consistent with the findings of the present study. With
regard to the mechanism underlying UCP2 in the development of colon
cancer, it has been suggested that UCP2 may be involved in the
development and progression of colon cancer by regulating the
production of ROS, which then activate ERK1/2 and JNK1/2, resulting
in hyperproliferation of intestinal epithelial cells (42). These studies suggest that FLNa and
UCP2 play roles in tumour development and progression via the
Ras/MAPK/ERK signalling pathway. Our results from cultured CC cell
lines in vitro show that FLNa and UCP2 were mainly
distributed in the cytoplasm of CC cells and that knockdown of FLNa
or UCP2 slowed cancer cell proliferation, arrested the cells at the
G2 phase, with no significant apoptosis, and decreased
cell migration and invasion, suggesting that FLNa or UCP2 knockdown
affected the biological behaviour of CC cells by decreasing the
expression levels of cell-associated proteins (such as p-ERK1/2 and
p-AKT1) and the mRNA levels of Ras, MMP-2 and MMP-9. Taken
together, these data suggest that FLNa and UCP2 play a role in
tumour development and progression via the Ras/MAPK/ERK signalling
pathway, which is consistent with findings of previous studies
(41,42).
Due to limitations of the experimental conditions,
the present study did not analyse the tumourigenicity of FLNa or
UCP2 in vitro. Several studies have demonstrated the roles
of FLNa and UCP2 in the development of colon and prostate cancer.
In the present study, in vitro experiments were performed
with cultured CC cells and showed that FLNa and UCP2 play roles in
the development and progression of CC via the Ras/MAPK/ERK
signalling pathway. FLNa and UCP2 are upregulated in CC cells and
show relatively low expression levels in NC cells, reflecting the
specificity of their high expression in CC. FLNa and UCP2 are
superior to p16 and Ki67 for early prediction of the progression of
HSIL to CC. FLNa expression levels were associated with and p16
expression levels, and may be used as indicators for this purpose.
UCP2 is specific for clinical stage and lymph node metastasis and
may be used as a prognostic indicator. Combined FLNa and UCP2
screening facilitates stratified management of patients.
Particularly for patients who are UCP2-positive, the necessity for
surgery should be re-evaluated.
The present study had a relatively small sample
size; therefore, experimental data from studies with larger sample
sizes are required to further validate the application of FLNa and
UCP2 as clinical diagnostic strategies.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National Key
Research and Development Program of China (grant nos.
2016YFC1302900 and 2016YFC090290), the National Natural Science
Foundation of China (grant no. 81572559), and the Key Research and
Development Program of Shandong Province, China (grant nos.
2017CXGC1210 and 2015GSF118097).
Availability of data and materials
The datasets used and/or analysed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
AW and YZ conceived and designed the experiments. AW
performed the experiments, analysed the data and wrote the paper.
LL, MY, SH and XY participated in the cell experiments. HZ and FL
participated in the clinical research. All authors read and
approved the final version of this manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of Qilu Hospital (approval no. KYLL-2017-560). All
patients provided written informed consent prior to the study
start.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
FLNa
|
filamin A
|
|
UCP2
|
uncoupling protein 2
|
|
CC
|
cervical cancer
|
|
NC
|
normal cervix
|
|
LSIL
|
low-grade squamous intraepithelial
lesion
|
|
HSIL
|
high-grade intraepithelial
neoplasia
|
|
MAPK
|
mitogen-activated protein kinase
|
|
ERK
|
extracellular signal-regulated
kinase
|
|
AKT
|
protein kinase B
|
|
MMP
|
matrix metalloproteinase
|
|
Bcl-2
|
B-cell lymphoma-2
|
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Pecorelli S, Zigliani L and Odicino F:
Revised FIGO staging for carcinoma of the cervix. Int J Gynaecol
Obstet. 105:107–108. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Sedlis A, Bundy BN, Rotman MZ, Lentz SS,
Muderspach LI and Zaino RJ: A randomized trial of pelvic radiation
therapy versus no further therapy in selected patients with stage
IB carcinoma of the cervix after radical hysterectomy and pelvic
lymphadenectomy: A Gynecologic Oncology Group Study. Gynecol Oncol.
73:177–183. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Keys HM, Bundy BN, Stehman FB, Muderspach
LI, Chafe WE, Suggs CL III, Walker JL and Gersell D: Cisplatin
radiation and adjuvant hysterectomy compared with radiation and
adjuvant hysterectomy for bulky stage IB cervical carcinoma. N Engl
J Med. 340:1154–1161. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Nakamura K, Kitahara Y, Satoh T, Takei Y,
Takano M, Nagao S, Sekiguchi I and Suzuki M: Analysis of the effect
of adjuvant radiotherapy on outcomes and complications after
radical hysterectomy in FIGO stage IB1 cervical cancer patients
with intermediate risk factors (GOTIC Study). World J Surg Oncol.
14:1732016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kim H, Park W, Kim YS and Kim YJ:
Chemoradiotherapy is not superior to radiotherapy alone after
radical surgery for cervical cancer patients with intermediate-risk
factor. J Gynecol Oncol. 31:e352020. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Song S, Song C, Kim HJ, Wu HG, Kim JH,
Park NH, Song YS, Kim JW, Kang SB and Ha SW: 20 year experience of
postoperative radiotherapy in IB-IIA cervical cancer patients with
intermediate risk factors: Impact of treatment period and
concurrent chemotherapy. Gynecol Oncol. 124:63–67. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ryu SY, Park SI, Nam BH, Cho CK, Kim K,
Kim BJ, Kim MH, Choi SC, Lee ED and Lee KH: Is adjuvant
chemoradiotherapy overtreatment in cervical cancer patients with
intermediate risk factors? Int J Radiat Oncol Biol Phys.
79:794–799. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Jin YZ, Pei CZ and Wen LY: FLNA is a
predictor of chemoresistance and poor survival in cervical cancer.
Biomark Med. 10:711–719. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Imai K, Fukuda T, Wada T, Kawanishi M,
Tasaka R, Yasui T and Sumi T: UCP2 expression may represent a
predictive marker of neoadjuvant chemotherapy effectiveness for
locally advanced uterine cervical cancer. Oncol Lett. 14:951–957.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Stossel TP, Condeelis J, Cooley L, Hartwig
JH, Noegel A, Schleicher M and Shapiro SS: Filamins as integrators
of cell mechanics and signalling. Nat Rev Mol Cell Biol. 2:138–145.
2001. View
Article : Google Scholar : PubMed/NCBI
|
|
14
|
Robertson SP, Twigg SR, Sutherland-Smith
AJ, Biancalana V, Gorlin RJ, Horn D, Kenwrick SJ, Kim CA, Morava E,
Newbury-Ecob R, et al: Localized mutations in the gene encoding the
cytoskeletal protein filamin A cause diverse malformations in
humans. Nat Genet. 33:487–491. 2003. View
Article : Google Scholar : PubMed/NCBI
|
|
15
|
Keshamouni VG, Michailidis G, Grasso CS,
Anthwal S, Strahler JR, Walker A, Arenberg DA, Reddy RC, Akulapalli
S, Thannickal VJ, et al: Differential protein expression profiling
by iTRAQ-2DLC-MS/MS of lung cancer cells undergoing
epithelial-mesenchymal transition reveals a migratory/invasive
phenotype. J Proteome Res. 5:1143–1154. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ai J, Huang H, Lv X, Tang Z, Chen M, Chen
T, Duan W, Sun H, Li Q, Tan R, et al: FLNA and PGK1 are two
potential markers for progression in hepatocellular carcinoma. Cell
Physiol Biochem. 27:207–216. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lin JF, Xu J, Tian HY, Gao X, Chen QX, Gu
Q, Xu GJ, Song JD and Zhao FK: Identification of candidate prostate
cancer biomarkers in prostate needle biopsy specimens using
proteomic analysis. Int J Cancer. 121:2596–2605. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Larriba MJ, Martín-Villar E, Garcia JM,
Pereira F, Peña C, de Herreros AG, Bonilla F and Muñoz A: Snail2
cooperates with snail1 in the repression of vitamin D receptor in
colon cancer. Carcinogenesis. 30:1459–1468. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Flanagan LA, Chou J, Falet H, Neujahr R,
Hartwig JH and Stossel TP: Filamin A, the Arp2/3 complex, and the
morphology and function of cortical actin filaments in human
melanoma cells. J Cell Biol. 155:511–517. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhang L, Komurov K, Wright WE and Shay JW:
Identification of novel driver tumor suppressors through functional
interrogation of putative passenger mutations in colorectal cancer.
Int J Cancer. 132:732–737. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhang L, Kim S, Jia G, Buhmeida A, Dallol
A, Wright WE, Fornace AJ, Al-Qahtani M and Shay JW: Exome
sequencing of normal and isogenic transformed human colonic
epithelial cells (HCECs) reveals novel genes potentially involved
in the early stages of colorectal tumorigenesis. BMC Genomics. 16
(Suppl 1):S82015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Pecqueur C, Alves-Guerra MC, Gelly C,
Levi-Meyrueis C, Couplan E, Collins S, Ricquier D, Bouillaud F and
Miroux B: Uncoupling protein 2, in vivo distribution, induction
upon oxidative stress, and evidence for translational regulation. J
Biol Chem. 276:8705–8712. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Bo J, Xie S, Guo Y, Zhang C, Guan Y, Li C,
Lu J and Meng QH: Methylglyoxal impairs insulin secretion of
pancreatic beta-cells through increased production of ROS and
mitochondrial dysfunction mediated by upregulation of UCP2 and
MAPKs. J Diabetes Res. 2016:20298542016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Horimoto M, Resnick MB, Konkin TA,
Routhier J, Wands JR and Baffy G: Expression of uncoupling
protein-2 in human colon cancer. Clin Cancer Res. 10:6203–6207.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Berthiaume E, Derdak Z, Konkin TA, Resnick
MB, Wands JR and Baffy G: Increased expression of uncoupling
protein-2 in cholangiocarcinoma cells may confer resistance to
apoptosis. Hepatology. 40:372A–373A. 2004.
|
|
26
|
Sayeed A, Meng Z, Luciani G, Chen LC,
Bennington JL and Dairkee SH: Negative regulation of UCP2 by TGFβ
signaling characterizes low and intermediate-grade primary breast
cancer. Cell Death Dis. 1:e532010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Harper ME, Antoniou A, Villalobos-Menuey
E, Russo A, Trauger R, Vendemelio M, George A, Bartholomew R, Carlo
D, Shaikh A, et al: Characterization of a novel metabolic strategy
used by drug-resistant tumor cells. FASEB J. 16:1550–1557. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Bray F, Carstensen B, Møller H, Zappa M,
Zakelj MP, Lawrence G, Hakama M and Weiderpass E: Incidence trends
of adenocarcinoma of the cervix in 13 European countries. Cancer
Epidemiol Biomarkers Prev. 14:2191–2199. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Bray F, Loos AH, McCarron P, Weiderpass E,
Arbyn M, Møller H, Hakama M and Parkin DM: Trends in cervical
squamous cell carcinoma incidence in 13 European countries:
Changing risk and the effects of screening. Cancer Epidemiol
Biomarkers Prev. 14:677–686. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Bray F, Lortet-Tieulent J, Znaor A,
Brotons M, Poljak M and Arbyn M: Patterns and trends in human
papillomavirus-related diseases in Central and Eastern Europe and
Central Asia. Vaccine. 31:H32–H45. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Bedolla RG, Wang Y, Asuncion A, Chamie K,
Siddiqui S, Mudryj MM, Prihoda TJ, Siddiqui J, Chinnaiyan AM, Mehra
R, et al: Nuclear versus cytoplasmic localization of filamin A in
prostate cancer: Immunohistochemical correlation with metastases.
Clin Cancer Res. 15:788–796. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yu LL, Chen W, Lei XQ, Qin Y, Wu ZN, Pan
QJ, Zhang X, Chang BF, Zhang SK, Guo HQ and Qiao YL: Evaluation of
p16/Ki-67 dual staining in detection of cervical precancer and
cancers: A multicenter study in China. Oncotarget. 7:21181–21189.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Cairns RA, Harris IS and Mak TW:
Regulation of cancer cell metabolism. Nat Rev Cancer. 11:85–95.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Niault TS and Baccarini M: Targets of raf
in tumorigenesis. Carcinogenesis. 31:1165–1174. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wong KK: Recent developments in
anti-cancer agents targeting the Ras/Raf/MEK/ERK pathway. Recent
Pat Anticancer Drug Discov. 4:28–35. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Watanabe H: Extracellular
matrix-regulation of cancer invasion and metastasis. Gan To Kagaku
Ryoho. 37:2058–2061. 2010.(In Chinese). PubMed/NCBI
|
|
38
|
Ogata Y, Matono K, Nakajima M, Sasatomi T,
Mizobe T, Nagase H and Shirouzu K: Efficacy of the MMP inhibitor
MMI270 against lung metastasis following removal of orthotopically
transplanted human colon cancer in rat. Int J Cancer. 118:215–221.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Lai WC, Zhou M, Shankavaram U, Peng G and
Wahl LM: Differential regulation of lipopolysaccharide-induced
monocyte matrix metalloproteinase (MMP)-1 and MMP-9 by p38 and
extracellular signal-regulated kinase 1/2 mitogen-activated protein
kinases. J Immunol. 170:6244–6249. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Rauvala M, Aglund K, Puistola U,
Turpeenniemi-Hujanen T, Horvath G, Willén R and Stendahl U: Matrix
metalloproteinases-2 and −9 in cervical cancer: Different roles in
tumor progression. Int J Gynecol Cancer. 16:1297–1302. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Zhu TN, He HJ, Kole S, D'Souza T, Agarwal
R, Morin PJ and Bernier M: Filamin A-mediated down-regulation of
the exchange factor Ras-GRF1 correlates with decreased matrix
metalloproteinase-9 expression in human melanoma cells. J Biol
Chem. 282:14816–14826. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Derdak Z, Mark NM, Beldi G, Robson SC,
Wands JR and Baffy G: The mitochondrial uncoupling protein-2
promotes chemoresistance in cancer cells. Cancer Res. 68:2813–2819.
2008. View Article : Google Scholar : PubMed/NCBI
|