Introduction
Lung cancer is one of the most common cancers
leading to patient death (1).
Non-small cell lung cancer (NSCLC) is the most frequent subtype of
lung cancer, and the predicted 5-year survival rate remains poor
despite recent therapeutic advances (2,3).
Although molecular-targeted drugs have been developed against
specific molecules in NSCLC, including epidermal growth factor
receptor (EGFR) and anaplastic lymphoma kinase, there are many
NSCLC patients who do not receive treatment with these drugs
(4–6). Therefore, it is necessary to clarify
further the molecular mechanisms involved in NSCLC tumourigenesis
and progression for improving the therapeutic strategies for
NSCLC.
Solute carrier organic anion transporter family
member 1B3 (SLCO1B3) is a member of the organic anion transporting
polypeptide (OATP/SLCO) superfamily. SLCO1B3 plays a role in the
uptake of a variety of endogenous compounds (e.g., bile acids,
cholecystokinin, conjugated steroids, and thyroid hormones) as well
as clinically relevant drugs (7–11).
SLCO1B3 was initially considered as a liver-specific transporter;
however, recent research has demonstrated that this gene is
transcribed at low levels in several extrahepatic tissues (10). Previous studies have revealed that
some cancerous tissues and cells (e.g. colon, lung, and pancreatic
cancers) express an alternative splicing form of SLCO1B3 [cancer
type-SLCO1B3 (Ct-SLCO1B3)] (12–14).
Since Ct-SLCO1B3 mRNA is transcribed from within intron 2 of the
SLCO1B3 gene, it lacks a part of the N-terminal coding
region of full-length SLCO1B3. The function of Ct-SLCO1B3 remains
largely unknown in cancer.
In the present study, we detected significantly
upregulated Ct-SLCO1B3 in NSCLC tissues compared to that noted in
normal lung tissues. Ct-SLCO1B3 contributed to
anchorage-independent cell growth, cell migration, and tumour
growth in NSCLC cells, whereas Lt-SLCO1B3 did not show these
phenotypes. Moreover, we found that Ct-SLCO1B3 is involved in
maintaining the expression of epithelial-mesenchymal transition
(EMT)-related genes. Reductions in anchorage-independent cell
growth and migration by Ct-SLCO1B3 knockdown were mediated by the
upregulation of E-cadherin. To the best of our knowledge, this is
the first study to show that Ct-SLCO1B3 promotes NSCLC progression
via regulation of EMT.
Materials and methods
NSCLC clinical samples and NSCLC cell
lines
Gene expression analysis in clinical
specimens
Specimens of NSCLC tissues and adjacent noncancerous
tissues were obtained from 101 patients who had undergone primary
curative resection of lung tumours at Kagoshima University Hospital
(Japan). Information concerning the clinical specimens from the
NSCLC patients is shown in Table
SI. This study was approved by the Ethics Committee of
Kagoshima University Hospital. All patients signed informed consent
prior to their inclusion in the study. The samples were stored
immediately at −80°C by immersing in RNAlater (Qiagen, Germany) 24
h prior to RNA extraction. mRNA was purified using an miRNeasy Mini
kit (Qiagen). Real-time polymerase chain reaction (real-time PCR)
analysis was conducted to validate Ct-SLCO1B3 expression in NSCLC
using 101 matched paired specimens of NSCLC.
NSCLC cell lines
Human lung adenocarcinoma cell lines (A549, HLC-1,
LC-2/ad, and RERF-LC-KJ) and human lung squamous cancer cell lines
(EBC-1, LC-1F, LK-2, RERF-LC-AI, and Sq-1) were obtained from RIKEN
Cell Bank (Japan). Human lung adenocarcinoma cell lines (NCI-H1650
NCI-H1975, NCI-H2228, II-18, PC-14, Calu-3, NCI-H1755, NCI-H1792,
NCI-H1838, NCI-H23, and NCI-H1437) and human lung squamous cancer
cell lines (NCI-H226 and NCI-H520) were obtained from the American
Type Culture Collection.
Exon array analysis
Total RNA (100 ng) from 4 matched paired samples of
NSCLC were subjected to cDNA synthesis by Ambion WT Expression kit
(Thermo Fisher Scientific, Inc.). The obtained cDNA was fragmented
and biotinylated by fragmentation using the GeneChip WT Terminal
Labeling Kit (Affymetrix; Thermo Fisher Scientific, Inc.) and was
hybridised to the GeneChip Human Exon 1.0 ST Array (Affymetrix;
Thermo Fisher Scientific, Inc.) for 17 h. After hybridization,
GeneChips were washed and stained with the GeneChip Hybridization
Wash and Stain Kit (Affymetrix; Thermo Fisher Scientific, Inc.)
using the Fluidics Station (Affymetrix; Thermo Fisher Scientific,
Inc.), and the chips were scanned using the 3000 GeneChip Scanner.
Gene Spring 12.1 (Agilent Technologies, Inc.) was used for data
analysis.
Cell culture
RPMI-1640 medium (Wako Japan, for NCI-H23, HLC-1,
LC-2/ad, RERF-LC-KJ, EBC-1, LC-1F, LK-2, RERF-LC-AI, and Sq-1) and
DMEM (Wako, for A549, NCI-H1650 NCI-H1975, NCI-H2228, II-18, PC-14,
Calu-3, NCI-H1755, NCI-H1792, NCI-H1838, NCI-H23, NCI-H1437,
NCI-H226, and NCI-H520) supplemented with 10% fetal calf serum
(FCS) and 100 mg/l kanamycin were used as cell culture media at
37°C under a 5% CO2 atmosphere.
Extraction of total RNAs and real-time
PCR analysis
Total RNA was isolated using Trizol reagent
(Invitrogen; Thermo Fisher Scientific, Inc.), and cDNA was
synthesised using the Primescript RT reagent Kit (Takara Bio,
Japan) according to the manufacturer's protocol. A light cycler
(Roche Diagnostics, Switzerland) was used for real-time PCR
analysis. The thermal cycling conditions were as follows: An
initial step at 95°C for 30 sec and 40 cycles at 95°C for 15 sec,
58°C for 30 sec (GAPDH) or 40 cycles at 95°C for 15 sec, and 62°C
for 30 sec (Ct-SLCO1B3 exon 1*-3, SLCO1B3 exon 9–10). The primer
sequences used for the gene amplification were as follows:
Ct-SLCO1B3 exon 1*-3 forward primer, 5′-TTCCCTGGGGAGAGGGACATACA-3′
and reverse primer, 5′-CCAGCAAGAGAAGAGGATATGTCA-3′; Ct-SLCO1B3 exon
9–10 forward primer, 5′-GTCCAGTCATTGGCTTTGCA-3′ and reverse primer,
5′-CAACCCAACGAGAGTCCTTAGG-3′; GAPDH forward primer,
5′-CCATCACCATCTTCCAGGAG-3′ and reverse primer
5′-AATGAGCCCCAGCCTTCTCC-3′. Extraction of total RNAs and real-time
PCR analysis for Snail and Slug are outlined in Data S1.
RT-PCR analysis of full-length
Ct-SLCO1B3 and Lt-SLCO1B3
Total RNA was isolated using the Trizol reagent
(Invitrogen; Thermo Fisher Scientific, Inc.). The PrimeScript RT
reagent Kit (Takara Bio) was used for the preparation of cDNA from
500 ng total RNA samples. The thermal cycling conditions were as
follows: An initial step at 94°C for 120 sec, 40 cycles at 94°C for
30 sec, and 68°C for 150 sec using Ct-SLCO1B3 or Lt-SLCO1B3
full-length primer as follows: Ct-SLCO1B3 forward primer,
5′-ATGGGATGGCTTGGCTTGG-3′ and reverse primer,
5′-TTAGTTGGCAGCAGCATTGTCTTG-3′; Lt-SLCO1B3 forward primer,
5′-ATGGACCAACATCAACATTTGAATAAAAC-3′ and reverse primer
5′-ATGGGATGGCTTGGCTTGG-3′.
Small interfering RNA (siRNA)
transfection
siRNA duplexes used to downregulate Ct-SLCO1B3 mRNA
(Ct-SLCO1B3 siRNA: ACGUUACUGAAUCUACAUGTT) and the negative control
siRNA duplex (Control siRNA: AUCCGCGCGAUAGUACGUATT) were purchased
from Gene Design Inc. (Japan). Stealth RNAi siRNAs used to
downregulate E-cadherin mRNA (E-cadherin siRNA #1,
ACACUGCCAACUGGCUGGAGAUUAA and E-cadherin siRNA #2,
GAGCACGUGAAGAACAGCACGUAUA) and the negative control Stealth RNAi
siRNA duplex (the sequence has not been published) were purchased
from Life Technologies. Ct-siRNAs of SLCO1B3 and E-cadherin were
transfected at concentrations of 10 nM and the indicated
concentrations, respectively, using Lipofectamine RNAiMAX
(Invitrogen; Thermo Fisher Scientific, Inc.) in accordance with the
manufacturer's protocol.
Western blotting
The whole-cell lysates were separated by sodium
dodecyl sulphate (SDS)-polyacrylamide gel (10%) electrophoresis
(PAGE) and then transferred to a polyvinylidene difluoride (Merck
Millipore) membrane using XCell SureLock Mini-Cell and XCell II
Blot Module (Invitrogen; Thermo Fisher Scientific, Inc.). The
membranes were probed with specific antibodies and then incubated
with horseradish peroxidase-conjugated antibodies against mouse or
rabbit immunoglobulin (Cell Signaling Technology, Inc.).
Immunoreactive proteins were detected by treatment with a detection
reagent (ECL Prime Western Blotting Detection Reagent, GE
Healthcare). The ImageQuant LAS4000 Mini System (GE Healthcare) was
used for chemiluminescence detection. The following antibodies were
used for immunological analysis in this study: Polyclonal
anti-β-actin antibodies (dilution 1:1,000; cat. no. sc-69879; Santa
Cruz Biotechnology, Inc. USA), anti-E-cadherin (dilution 1:1,000;
cat. no. 3195; Cell Signaling Technology, Inc.), Snail (dilution
1:1,000; cat. no. 3879; Cell Signaling Technology, Inc.), Slug
(dilution 1:1,000; cat. no. 9585; Cell Signaling Technology, Inc.),
TCF8/ZEB1 (dilution 1:1,000; cat. no. 3396; Cell Signaling
Technology, Inc.), and vimentin antibodies (dilution 1:1,000; cat.
no. 5741P; Cell Signaling Technology, Inc.).
Construction of Ct-SLCO1B3 and
Lt-SLCO1B3 expression plasmids
cDNAs of Ct-SLCO1B3 and Lt-SLCO1B3 were amplified
via PCR using total RNAs of A549 cells and normal liver cells,
respectively. The primer sequences for gene amplification were as
follows: Ct-SLCO1B3 forward primer,
5′-CCCTGAGATGGGATGGCTTGGCTTGG-3′ and reverse primer,
5′-CCGGATCCGTTGGCAGCAGCATTGTCTTG-3′; Lt-SLCO1B3 forward primer,
5′-CCCTGAGAATGGACCAACATCAACATTTGAATAAAAC-3′ and reverse primer
5′-CCGGATCCGTTGGCAGCAGCATTGTCTTG-3′. The gel-purified PCR products
ligated into the pT7-blue vector (Merck Millipore) using the
Perfectly Blunt Cloning Kit (Merck Millipore) were transformed into
E. coli DH5α (Takara Bio). The plasmid was purified from
DH5α and treated with a restriction enzyme for insertion into the
pcDNA3.0 vector (Invitrogen; Thermo Fisher Scientific, Inc.) using
Ligation High (Toyobo).
Anchorage-independent cell growth
assay
siRNA and Stealth RNAi siRNA transfected A549 cells
were suspended in FCeM-D (Nissan Chemical) with 10% FCS and were
seeded in a HydroCell 96-well plate (CellSeed) (1,000 cells/well)
48 h after transfection. NCI-H23 cells were seeded in a 96-well
plate (4,000 cells/well). Two hours after incubation with the WST-1
reagent (Dojindo) at 37°C in 5% CO2, the optical density
was read at a wavelength of 450/630 nm (Ex/Em).
Wound healing assay
Cell migration was examined by the wound healing
assay. A549 (6×104 cells/well) and NCI-H23
(1×105 cells/well) cells were seeded in a 48-well plate
48 h after transfection. A wound was created in a monolayer of
about 80–90% confluent A549 or NCI-H23 cells using a sterile 1-ml
pipette tip. Cell migration images were recorded at 0, 6, 12, 24,
36, and 48 (only A549) h with an Olympus IX71 fluorescence
microscope (Olympus) at a ×100 magnification.
siRNA administration in an in vivo
xenograft model
A549 cells were suspended in an equal volume of
Matrigel Matrix High Concentration (Corning, Inc.) and injected
subcutaneously into 7-week-old male BALB/c nu-nu mice. Eleven days
after the injection, 14 tumour-bearing mice were divided into two
groups. Tumour size was measured on days 11, 15, 18, 22, 25, 29 and
32 after tumour cell injection, and siRNA was administered on days
12, 19 and 26 after the injection. The siRNA solution was prepared
with AteloGene Local Use ‘Quick Gelation’ (Koken) as a carrier, and
4 nmol/mouse/150 µl was injected. On day 32 after tumour cell
injection, mice were sacrificed, tumours were enucleated from each
mouse, and the tumour weight was measured.
Statistics
Results are expressed as the mean ± standard
deviation of the mean (SD). Differences between values were
statistically analysed using a Student's t-test or one-way ANOVA
with Bonferroni post-hoc test (GraphPad Prism 5.0, GraphPad
Software). A P-value <0.05 was considered to indicate
statistical significance.
Results
Ct-SLCO1B3 is expressed in NSCLC
To explore the alternative splicing signature in
NSCLC, we performed an exon array analysis using four matched pairs
of postoperative specimens of NSCLC patients. We focused on SLCO1B3
since the exon usage for this gene is different between NSCLC
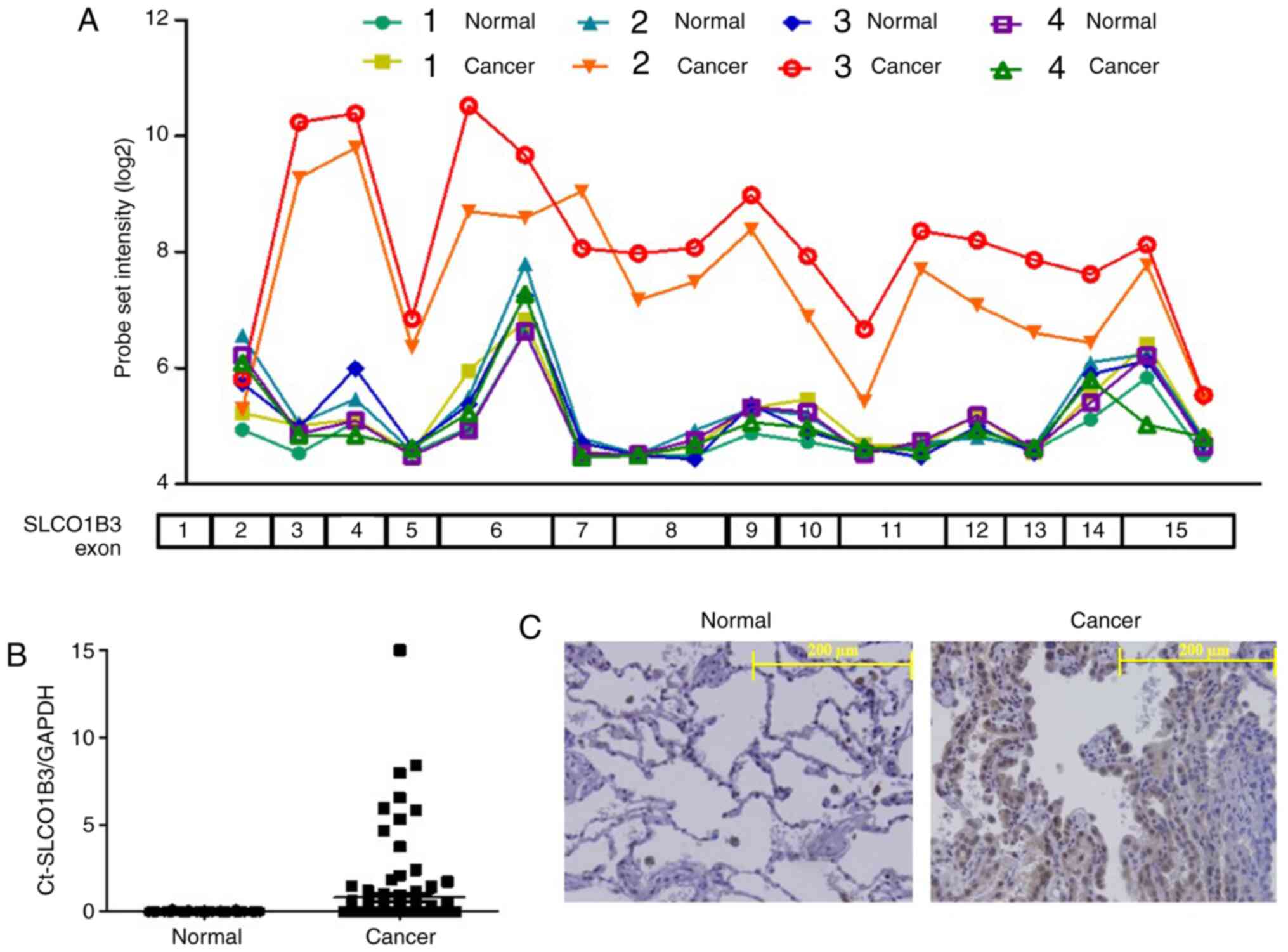
tissues and the adjacent normal tissues. As shown in Fig. 1A, we found a high probe set
intensity of SLCO1B3 exons 3–15 in only 2 samples from cancerous
tissues of NSCLC patients (cancers 2 and 3). Interestingly, the
probe intensity of SLCO1B exon 2 in these two samples was as low as
that in all the other samples, including the adjacent normal lung
tissues. This result suggested that SLCO1B3 in the two samples was
transcribed with its 5′-end missing. The expression of SLCO1B3
lacking exon 2 (Ct-SLCO1B3) has been detected in some cancers
(Fig. S1A). Therefore, we
amplified the gene fragment in the two samples using primers
designed around SLCO1B3 exons 1 and 3 and confirmed the deletion of
SLCO1B3 exon 2 by sequencing. Then, we determined Ct-SLCO1B3
expression in 101-matched paired NSCLC specimens by real-time PCR
using a specific primer set as described above. As shown in
Fig. 1B, Ct-SLCO1B3 was detected in
26 specimens of NSCLC tissues but not in adjacent normal tissue
specimens. Ct-SLCO1B3 expression in NSCLC was independent of EGFR
mutation, patient sex, and NSCLC subtype and was also detected in
specimens with stage IA LSCLC cancer (Fig. S2). Immunohistochemical examination
showed that Ct-SLCO1B3 was localised to the cytoplasm of NSCLC
cells, but no signal was found in normal lung tissue (Fig. 1C). Lt-SLCO1B was localised in the
cell membrane of hepatocytes, as described previously (Fig. S1B) (13).
Ct-SLCO1B3 plays roles in
anchorage-independent cell growth and migration of A549 cells
To investigate Ct-SLCO1B3 function in NSCLC cells,
we first evaluated Ct-SLCO1B3 mRNA expression levels in 22 lung
cancer cell lines. Among the lung cancer cell lines, Ct-SLCO1B3
mRNA expression was predominant in the adenocarcinoma cell lines
(Fig. S3). We selected A549 and
NCI-H23 cells based on expression levels of Ct-SLCO1B3 and
transfection efficiency for subsequent experiments. To assess the
knockdown efficiency of Ct-SLCO1B3 siRNAs, real-time PCR was
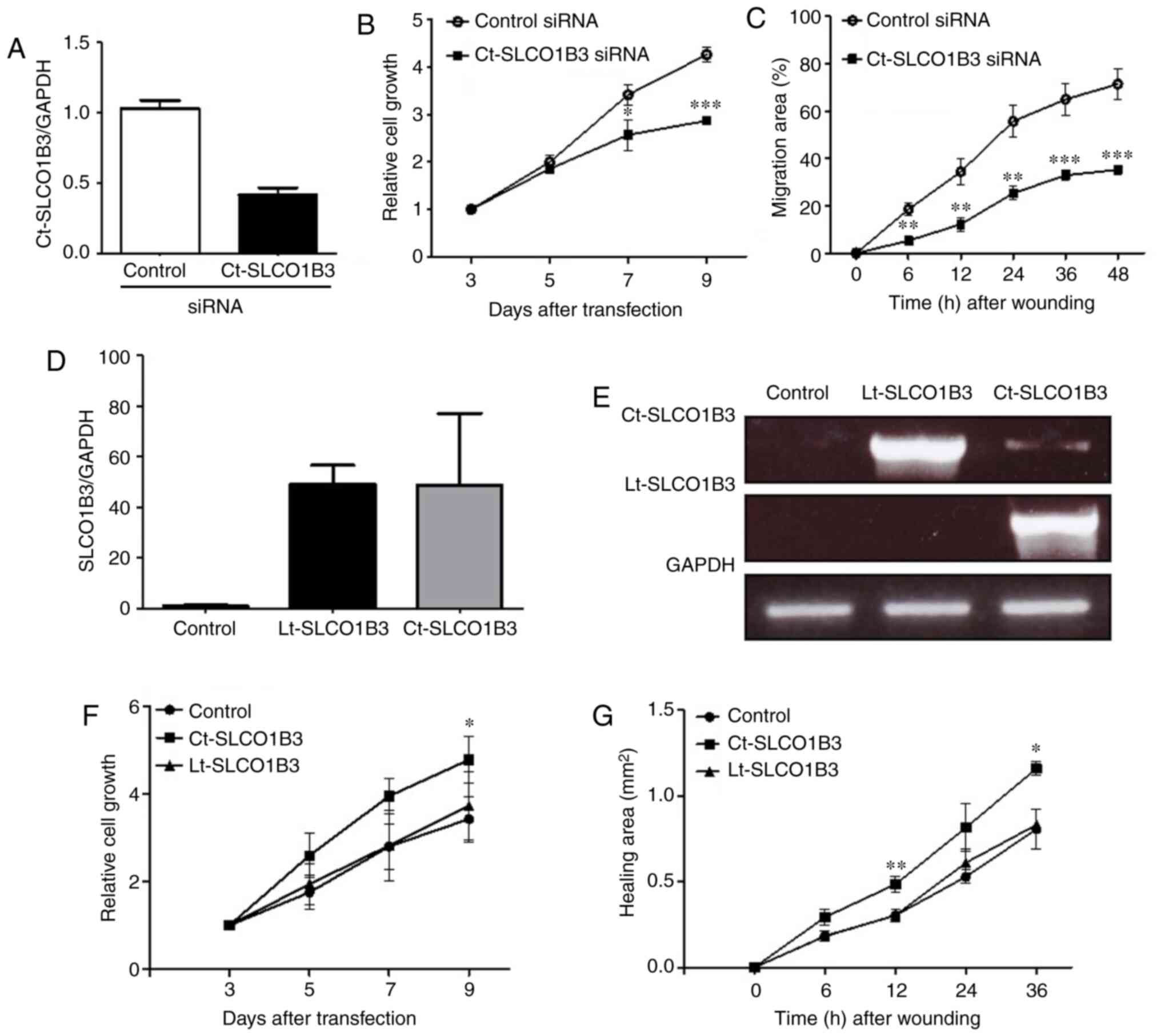
conducted. As shown in Fig. 2A, the
expression level of Ct-SLCO1B3 mRNA was sufficiently reduced by
Ct-SLCO1B3 siRNA in the A549 cells. To investigate the potential
function of Ct-SLCO1B3, we examined the effect of Ct-SLCO1B3 siRNA
on anchorage-independent cell growth and migration in A549 cells.
Ct-SLCO1B3 siRNA significantly suppressed anchorage-independent
cell growth (Fig. 2B) and decreased
the migration ability (Figs. 2C and
S4) of the A549 cells.
Next, we carried out overexpression experiments of
Ct-SLCO1B3 and Lt-SLCO1B3 using NCI-H23 cells with a deficient
expression of Ct-SLCO1B3. Plasmid vector pcDNA3.0 containing either
Lt-SLCO1B3 or Ct-SLCO1B3 cDNA was transfected into NCI-H23 cells,
and similar expression levels of each SLCO1B3 mRNA were confirmed
by real-time PCR using specific primer sets of Lt-SLCO1B3 and
Ct-SLCO1B3 (Fig. 2D and E). Then,
we investigated the effect of Lt-SLCO1B3 or Ct-SLCO1B3
overexpression on anchorage-independent cell growth and migration.
Overexpression of Ct-SLCO1B3 but not Lt-SLCO1B3 significantly
increased anchorage-independent cell growth and migration (Fig. 2F and G). These results indicated
that Ct-SLCO1B3 and Lt-SLCO1B3 exhibit different biological
functions, and only Ct-SLCO1B3 may contribute to NSCLC
development.
Ct-SLCO1B3 regulates
anchorage-independent cell growth and cell migration via
EMT-related gene expression
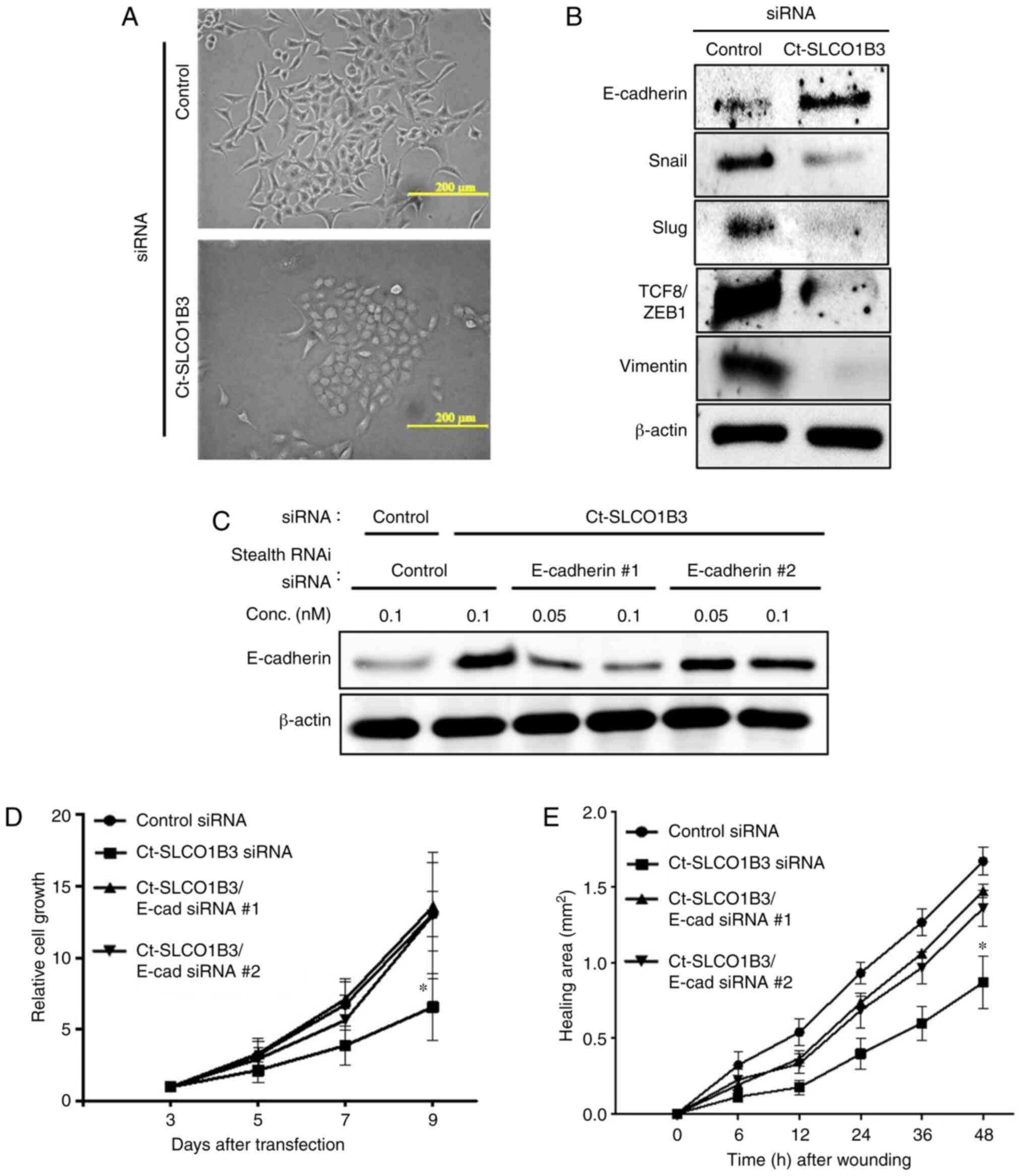
In siRNA transfection experiments, we observed
morphological changes in Ct-SLCO1B3-knockdown A549 cells, which
resembled epithelial cells with tight cell-cell interaction
(Fig. 3A). From this observation,
we assumed that Ct-SLCO1B3 regulates EMT. Since EMT is a crucial
step in cancer progression, that facilitates cancer cells to lose
cell polarity and cell-to-cell adhesion to gain migratory and
invasive properties (15–17), we investigated the mRNA (Fig. S5A) and protein (Fig. 3B) expression of EMT-related factors.
An epithelial cell marker, E-cadherin, was upregulated in
Ct-SLCO1B3-knockdown A549 cells. On the other hand, suppression of
Ct-SLCO1B3 expression reduced the expression of mesenchymal related
genes, Snail, Slug, TCF8/ZEB1, and vimentin. Moreover, prominent
E-cadherin localization in the cell-cell contact region was
observed in Ct-SLCO1B3 siRNA-transfected A549 cells (Fig. S5B). These results suggest that
Ct-SLCO1B3 facilitates EMT in NSCLC cells. To demonstrate that
phenotype induction by Ct-SLCO1B3 knockdown occurred via the
upregulation of E-cadherin, we performed Ct-SLCO1B3 and E-cadherin
double knockdown experiments. Upregulation of E-cadherin by
Ct-SLCO1B3 siRNA was diminished following transfection with two
types of E-cadherin stealth siRNAs (Fig. 3C). Suppression of
anchorage-independent cell growth and migration by Ct-SLCO1B3
siRNAs was reversed by transfection of both E-cadherin stealth
siRNAs (Fig. 3D and E). These
experiments indicated that Ct-SLCO1B3 is related to EMT in NSCLC
cells, and E-cadherin plays an important role in
anchorage-independent cell growth and migration in
Ct-SLCO1B3-knockdown A549 cells.
Ct-SLCO1B3 facilitates tumourigenesis
in NSCLC xenograft model mice
Since anchorage-independent cancer cell growth plays
an important role in tumourigenesis, we supposed that Ct-SLCO1B3
contributes to tumour growth in vivo. To elucidate that
Ct-SLCO1B3 regulates tumour formation in vivo, we
established a xenograft model by subcutaneous injection of A549
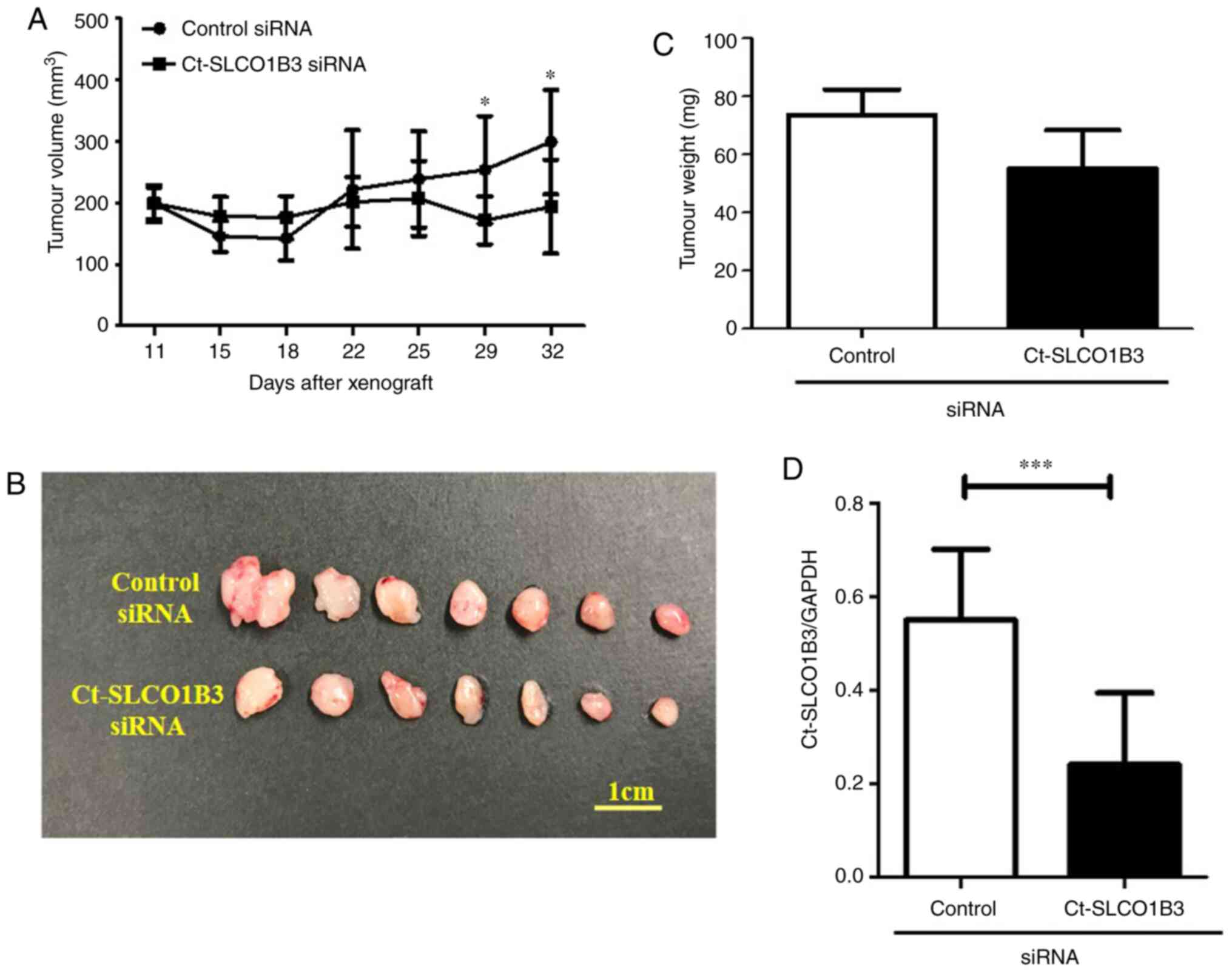
cells in nude mice. After tumour formation was confirmed, we
administered control or Ct-SLCO1B3 siRNAs subcutaneously for 7
days. Tumour volume measurement was performed after tumour volumes
reached approximately 200 mm3. As shown in Fig. 4A, Ct-SLCO1B3 siRNA significantly
decreased tumour volume after 29 days of its administration. On day
32, formed tumours were resected and imaged (Fig. 4B). There was a reduction in tumour
weight (Fig. 4C) as well as
Ct-SLCO1B3 levels (Fig. 4D) in the
group of mice treated with Ct-SLCO1B3 siRNA in comparison to the
control siRNA group. These results demonstrated that Ct-SLCO1B3 is
involved in tumour growth in vivo.
Discussion
To develop a novel drug for non-small cell lung
cancer (NSCLC) therapy, it is crucial not only to identify a
molecule with characteristic expression in cancer cells but also to
understand how the molecule is associated with the development and
progression of NSCLC. Sun et al reported that cancer
type-solute carrier organic anion transporter family member 1B3
(Ct-SLCO1B3) expression is detected in fewer lung cancer specimens,
including NSCLC and small cell carcinoma (14). In the present study, we examined
Ct-SLCO1B3 expression in 101-matched paired NSCLC specimens,
including its correlation with EGFR mutation as well as
pathological stages. Moreover, we clarified a molecular mechanism
involving Ct-SLCO1B3 in the development and progression of
NSCLC.
Loss of E-cadherin and upregulation of
mesenchymal-related molecules such as vimentin, matrix
metalloproteinase (MMP)-9, integrin-αvβ6, and N-cadherin are
associated with poor clinical outcome in NSCLC (18,19).
Moreover, it is well established that the state of
epithelial-to-mesenchymal transition (EMT) affects the tumour
response to various drugs, including epidermal growth factor
receptor (EGFR) kinase inhibitors (20,21),
indicating a crucial role of EMT in the progression and treatment
of NSCLC. In this study, we showed that Ct-SLCO1B3 siRNA
downregulated Snail, Slug, Zinc finger E-box binding homeobox 1
(ZEB1), and vimentin, and upregulated E-cadherin. Snail, Slug, and
ZEB1 are known to be transcriptional regulators of E-cadherin.
Therefore, Ct-SLCO1B3 knockdown was proposed to induce the
expression of E-cadherin via the downregulation of these
transcriptional regulators. Ct-SLCO1B3 and E-cadherin double
knockdown experiments strongly supported that Ct-SLCO1B3 regulates
E-cadherin expression in NSCLC. Ct-SLCO1B3 but not liver type
(Lt)-SLCO1B3 facilitated the upregulation of anchorage-independent
cell growth and cell migration in NSCLC cells by regulatory
mechanisms. Research focusing on structural differences between
Lt-SLCO1B3 and Ct-SLCO1B3 will lead to the elucidation of the
detailed EMT-related molecular mechanisms in NSCLC. Furthermore,
Ct-SLCO1B3 siRNA significantly decreased tumour growth in an in
vivo xenograft mouse model experiment; however, whether
Ct-SLCO1B3 regulates EMT via Snail, Slug, or ZEB1 in vivo
remains unclear.
Lt-SLCO1B3 functions as a liver-specific organic
anion transporter. However, it is not clear whether Ct-SLCO1B3 also
functions as a transporter similar to Lt-SLCO1B3 in NSCLC.
Additionally, Lt-SLCO1B3 is expressed on the cell membrane
(22), while Ct-SLCO1B3 is
localised in the cytoplasm of NSCLC cells. Based on this, we
speculate that Ct-SLCO1B3, with its abnormal structure, may not
exhibit similar transporter activity.
In conclusion, our present study showed that a
subset of NSCLC patients express Ct-SLCO1B3, and Ct-SLCO1B3
significantly promoted anchorage-independent cell growth and cell
migration through EMT induction in NSCLC cells. Moreover, in
vivo tumour formation of NSCLC cells was suppressed by
Ct-SLCO1B3 knockdown. These findings indicate that Ct-SLCO1B3 may
be involved in the development and/or progression of NSCLC. Drugs
specifically targeting Ct-SLCO1B3 could be a promising NSCLC
therapy.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
This study was partially supported by Japan Agency
for Medical Research and Development (AMED) (grant no.
JP19lm0203007).
Availability of data and materials
The data sets generated in the present study are
available from the corresponding author on reasonable request.
Authors' contributions
HH conceived and designed the study and participated
in analysing the data. KMa and SN carried out the experiments,
analysed the data, and drafted the manuscript. MA, TN, AT, KU, MS,
KMi, MY and TF participated in providing and analysing the patient
samples and supported the study. KK, KJ and YU participated in
administrative, technical, or material support. KT developed a plan
for whole this study, reviewed the draft manuscript and edited the
manuscript. All authors read and approved the manuscript and agree
to be accountable for all aspects of the research in ensuring that
the accuracy or integrity of any part of the work are appropriately
investigated and resolved.
Ethics approval and consent to
participate
The study was approved by the Ethics Committee of
Kagoshima University Hospital (Kogoshima, Japan). Written informed
consent was obtained from each participant involved regarding the
use of their tissues for research purposes.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2016. CA Cancer J Clin. 66:7–30. 2016. View Article : Google Scholar
|
|
2
|
Ettinger DS, Wood DE, Aggarwal C, Aisner
DL, Akerley W, Bauman JR, Bharat A, Bruno DS, Chang JY, Chirieac
LR, et al: NCCN guidelines insights: Non-small cell lung cancer,
version 1.2020. J Natl Compr Canc Netw. 17:1464–1472. 2019.
View Article : Google Scholar
|
|
3
|
Chen Z, Fillmore CM, Hammerman PS, Kim CF
and Wong KK: Non-small-cell lung cancers: A heterogeneous set of
diseases. Nat Rev Cancer. 14:535–546. 2014. View Article : Google Scholar
|
|
4
|
Besse B, Adjei A, Baas P, Meldgaard P,
Nicolson M, Paz-Ares L, Reck M, Smit EF, Syrigos K, Stahel R, et
al: 2nd ESMO consensus conference on lung cancer: Non-small-cell
lung cancer first-line/second and further lines of treatment in
advanced disease. Ann Oncol. 25:1475–1484. 2014. View Article : Google Scholar
|
|
5
|
Pillai RN and Ramalingam SS: Advances in
the diagnosis and treatment of non-small cell lung cancer. Mol
Cancer Ther. 13:557–564. 2014. View Article : Google Scholar
|
|
6
|
Rolfo C, Giovannetti E, Hong DS, Bivona T,
Raez LE, Bronte G, Buffoni L, Reguart N, Santos ES, Germonpre P, et
al: Novel therapeutic strategies for patients with NSCLC that do
not respond to treatment with EGFR inhibitors. Cancer Treat Rev.
40:990–1004. 2014. View Article : Google Scholar
|
|
7
|
Hagenbuch B and Meier PJ: The superfamily
of organic anion transporting polypeptides. Biochim Biophys Acta.
1609:1–18. 2003. View Article : Google Scholar
|
|
8
|
Abe T, Unno M, Onogawa T, Tokui T, Kondo
TN, Nakagomi R, Adachi H, Fujiwara K, Okabe M, Suzuki T, et al:
LST-2, a human liver-specific organic anion transporter, determines
methotrexate sensitivity in gastrointestinal cancers.
Gastroenterology. 120:1689–1699. 2001. View Article : Google Scholar
|
|
9
|
Hagenbuch B and Meier PJ: Organic anion
transporting polypeptides of the OATP/SLC21 family: Phylogenetic
classification as OATP/SLCO superfamily, new nomenclature and
molecular/functional properties. Pflugers Arch. 447:653–665. 2004.
View Article : Google Scholar
|
|
10
|
Obaidat A, Roth M and Hagenbuch B: The
expression and function of organic anion transporting polypeptides
in normal tissues and in cancer. Annu Rev Pharmacol Toxicol.
52:135–151. 2012. View Article : Google Scholar
|
|
11
|
Yamaguchi H, Okada M, Akitaya S, Ohara H,
Mikkaichi T, Ishikawa H, Sato M, Matsuura M, Saga T, Unno M, et al:
Transport of fluorescent chenodeoxycholic acid via the human
organic anion transporters OATP1B1 and OATP1B3. J Lipid Res.
47:1196–1202. 2006. View Article : Google Scholar
|
|
12
|
Nagai M, Furihata T, Matsumoto S, Ishii S,
Motohashi S, Yoshino I, Ugajin M, Miyajima A, Matsumoto S and Chiba
K: Identification of a new organic anion transporting polypeptide
1B3 mRNA isoform primarily expressed in human cancerous tissues and
cells. Biochem Biophys Res Commun. 418:818–823. 2012. View Article : Google Scholar
|
|
13
|
Thakkar N, Kim K, Jang ER, Han S, Kim K,
Kim D, Merchant N, Lockhart AC and Lee W: A cancer-specific variant
of the SLCO1B3 gene encodes a novel human organic anion
transporting polypeptide 1B3 (OATP1B3) localized mainly in the
cytoplasm of colon and pancreatic cancer cells. Mol Pharm.
10:406–416. 2013. View Article : Google Scholar
|
|
14
|
Sun Y, Furihata T, Ishii S, Nagai M,
Harada M, Shimozato O, Kamijo T, Motohashi S, Yoshino I, Kamiichi
A, et al: Unique expression features of cancer-type organic anion
transporting polypeptide 1B3 mRNA expression in human colon and
lung cancers. Clin Transl Med. 3:372014. View Article : Google Scholar
|
|
15
|
Thiery JP: Epithelial-mesenchymal
transitions in tumour progression. Nat Rev Cancer. 2:442–454. 2002.
View Article : Google Scholar
|
|
16
|
De Craene B and Berx G: Regulatory
networks defining EMT during cancer initiation and progression. Nat
Rev Cancer. 13:97–110. 2013. View
Article : Google Scholar
|
|
17
|
Lamouille S, Xu J and Derynck R: Molecular
mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell
Biol. 15:178–196. 2014. View
Article : Google Scholar
|
|
18
|
Prudkin L, Liu DD, Ozburn NC, Sun M,
Behrens C, Tang X, Brown KC, Bekele BN, Moran C, Moran C and
Wistuba II: Epithelial-to-mesenchymal transition in the development
and progression of adenocarcinoma and squamous cell carcinoma of
the lung. Mod Pathol. 22:668–678. 2009. View Article : Google Scholar
|
|
19
|
Liu D, Huang C, Kameyama K, Hayashi E,
Yamauchi A, Kobayashi S and Yokomise H: E-Cadherin expression
associated with differentiation and prognosis in patients with
non-small cell lung cancer. Ann Thorac Surg. 71:949–955. 2001.
View Article : Google Scholar
|
|
20
|
Thomson S, Buck E, Petti F, Griffin G,
Brown E, Ramnarine N, Iwata KK, Gibson N and Haley JD: Epithelial
to mesenchymal transition is a determinant of sensitivity of
non-small-cell lung carcinoma cell lines and xenografts to
epidermal growth factor receptor inhibition. Cancer Res.
65:9455–9462. 2005. View Article : Google Scholar
|
|
21
|
Rho JK, Choi YJ, Lee JK, Ryoo BY, Na II,
Yang SH, Kim CH and Lee JC: Epithelial to mesenchymal transition
derived from repeated exposure to gefitinib determines the
sensitivity to EGFR inhibitors in A549, a non-small cell lung
cancer cell line. Lung Cancer. 63:219–226. 2009. View Article : Google Scholar
|
|
22
|
Chun SE, Thakkar N, Oh Y, Park JE, Han S,
Ryoo G, Hahn H, Maeng SH, Lim YR, Han BW and Lee W: The N-terminal
region of organic anion transporting polypeptide 1B3 (OATP1B3)
plays an essential role in regulating its plasma membrane
trafficking. Biochem Pharmacol. 131:98–105. 2017. View Article : Google Scholar
|