Introduction
Pituitary adenoma (PA) occurs in anterior and
posterior pituitary and residual cells of the craniopharynageal
duct epithelium and accounts for ~10% of intracranial tumors
(1). The disease is more common in
men than in women (1.32:1) in China (2), often affecting the patient's growth and
development, as well as reproductive, learning and working ability
(3). The clinical manifestation of
pituitary tumors includes headaches, vision loss, visual field
defects and abnormal hormone secretion; the primary manifestation
in women also includes menstrual disorders, amenorrhea and
galactorrhea (4). The current
treatment of pituitary tumors generally includes surgery,
medication and radiation therapy (5).
The goal of these three treatment methods is to decrease or
eliminate the impact of tumor-occupying lesions, correct excessive
hormone secretion by the tumor and retain normal pituitary function
(6). However, the recurrence rate
following pituitary tumor surgery is high at ~7–35% (7). In addition, pituitary tumors may lead to
complications following surgery, such as diabetes insipidus,
sphenoid sinusitis, cerebrospinal fluid leakage, worsening visual
impairment, cerebral palsy and meningitis (8). It is therefore important to develop
novel effective treatments.
Long non-coding (lnc)RNA is a class of RNA
molecules, a number of which have been identified as biomarkers of
cancer, such as exosome H19 and Clarin 1 antisense RNA 1 (9–13). Iyer
et al (10) performed data
analysis on 7,256 tumor and normal tissue samples and cell lines,
and screened 7,942 lncRNAs associated with cancer; these lncRNAs
may be biomarkers for specific types of cancer tissue. Numerous
lncRNAs have been confirmed to be associated with the development
of pituitary tumor (11). For
example, exosome H19 inhibits the growth of distal pituitary tumors
and the expression levels of exosome H19 in plasma can predict the
prognosis and treatment effectiveness (12). D'Angelo et al (13) demonstrated that expression levels of
ribosomal protein SA pseudogene 52 are significantly higher in
pituitary tumor cells than in controls and accelerate pituitary
tumor development via regulating high mobility group A. Clarin 1
antisense RNA 1 has also been confirmed to participate in the
Wnt/β-actin signaling pathway, inhibit autophagy progress and
suppress pathogenesis of pituitary tumors (14). There has been research on differential
expression of lncRNAs in PA and the functional mechanisms involved
in PA occurrence, development and prognosis (15,16).
However, the expression levels, function and regulatory mechanisms
of lncRNAs in PA need further investigation.
lncRNA maternally expressed 3 (MEG3) has been
reported to be expressed at significantly lower levels in a number
of tumors (17,18). In the development of normal anterior
pituitary tissue to PA, MEG3 expression levels gradually decrease,
whereas those of proliferating cell nuclear antigen (PCNA)
significantly increase in non-functional (NF)PA (19). PCNA is negatively associated with MEG3
(20). Therefore, the expression
levels of MEG3 may be associated with the development and invasive
ability of NFPA. Previous studies have also found that MEG3 levels
are decreased in NF and corticotrophic cell PA, which may be
associated with the pathogenesis of this disease (15,21). To
the best of our knowledge, however, the detailed molecular
mechanism underlying MEG3 in PA has not been identified. The
present study aimed to investigate this mechanism, which may
provide a theoretical basis for gene therapy of pituitary
tumor.
Materials and methods
Analysis of GSE77517 profile
GSE77517 (ncbi.nlm.nih.gov/geo/query/acc.cgi) was downloaded
from the Gene Expression Omnibus (GEO) database, which comprises 5
patients with NFPA. The relative MEG3 expression level compared
with that in normal tissue was calculated.
Patients and tissues
A total of 34 patients who underwent surgical
treatment of pituitary tumors at First Affiliated Hospital of
Soochow University (Nantong, China) from January to December 2019
were enrolled in the present study, including 19 males and 15
females. Patients had not been treated with radiotherapy or
chemotherapy before surgery. The age range of the patients 25–67
years, with an average age of 46.5 years. The present study was
approved by the ethics committee of First Affiliated Hospital of
Soochow University and all patients provided written informed
consent for the use of their samples in scientific research.
Patient information, including age, sex and tumor size, stage and
invasiveness were recorded. Pituitary tumor samples were collected
during surgery. After the tissue was obtained, it was placed in
liquid nitrogen to freeze and stored in a refrigerator at
−80°C.
Cell culture and transfection
Pituitary tumor cell lines (GH3 and MMQ) were
purchased from the American Type Culture Collection (ATCC). GH3 and
MMQ cells were maintained in RPMI-1640 medium containing 10% fetal
bovine serum (HyClone; GE Healthcare Life Sciences) at 37°C and 5%
CO2 for routine culture and passage. Cells in the
logarithmic growth phase were used for experiments. Cells were
transfected using Lipofectamine® 2000 (Invitrogen;
Thermo Fisher Scientific, Inc.) according to the manufacturer's
instructions. When confluence reached 80%, the cells were digested
by trypsin digestion solution at room temperature for 1 min and
resuspended in RPMI-1640 medium. Cells were seeded in a 6-well
plate at a density of 2×105/l and divided into
pcDNA3.1(+)-MEG3 and pcDNA3.1(+)-negative control (NC) transfection
groups, with 3 replicate wells in each group. The concentration of
pcDNA3.1(+)-MEG3 and pcDNA3.1(+)-negative control was 20 pmol/l.
The sequence of lncRNA MEG3 was obtained from ddbj database
(getentry.ddbj.nig.ac.jp/getentry/na/AB032607/?filetype=html).
pcDNA3.1(+)-MEG3 and pcDNA3.1(+)-negative control (NC) were
constructed by Sangon Biotech ltd. When confluence reached 70–80%,
cells were washed twice with PBS, then serum-free RPMI-1640 was
added and DNA-liposome complex was made at a ratio of 1 µg:3 µl.
Following incubation at 37°C and 5% CO2 for 5 h, the
culture was replaced with RPMI-1640 containing 10% fetal bovine
serum. After culturing at 37°C for 48 h, cells were collected for
subsequent experiments. MicroRNA (miRNA or miR)-23b-3p mimic and
controls, as well as short hairpin (sh)MEG3, miR-23b-3p inhibitor
or their controls, were also transfected as aforementioned.
Reverse transcription-quantitative
(RT-q)PCR assay
RNA was extracted from tissue samples and pituitary
tumor cells (~100 mg) using TRIzol® (Invitrogen; Thermo
Fisher Scientific, Inc.). Purity and concentration of total RNA was
detected by NanoDrop 1000 (Thermo Fisher Scientific, Inc.) and
A260/A280 should be ~2.0. The Platinum SYBR Green qPCR super Mix-UD
kit (Invitrogen; Thermo Fisher Scientific, Inc.) was used to
establish a 25 µl reaction system according to the manufacturer's
instructions. All primers were designed and purchased from Nanjing
Kingsray Biotechnology Co., Ltd.. Primer sequences were as follows:
MEG3, forward, 5′-CTGCCCATCTACACCTCACG-3′ and reverse,
5′-CTCTCCGCCGTCTTGCGCTAGGGGCT-3′; miR-23b-3p, forward,
5′-CACAAAGGCATCTCTACGCCCATC-3′ and reverse,
5′-TGTAGCTGCCTCGCCAGAGGC-3′; FOXO4, forward,
5′-CTTTCTGAAGACTGGCAGGAATGTG-3′ and reverse,
5′-GATCTAGGTCTATGATCGCGGCAG-3′; GAPDH, forward,
5′-AGAGGCAGGGATGATGTTCTG-3′ and reverse,
5′-GACTCATGACCACAGTCCATGC-3′; and U6, forward,
5′-TGCGGGTGCTCGCTTCGGCAGC-3′ and reverse,
5′-CCAGTGCAGGGTCCGAGGT-3′. The amplification conditions were as
follows: Holding stage, 50°C for 2 min, 95°C for 2 min; cycling
stage, 95°C for 15 sec, 60°C for 30 sec, 40 cycles; melt curve
stage, 95°C for 15 sec, 60°C for 1 min. The relative expression was
calculated as 2−ΔΔCq (22). For miR-23b-3p expression, U6 was
chosen as the internal reference control.
Cell Counting Kit (CCK)8 assay
The CCK8 method was used to determine cell
proliferation according to the manufacturer's instructions (APeXBIO
Technology LLC). Cells grown to the logarithmic phase were
digested, passaged, and seeded into 96-well plates at
2×103 cells/well. The volume of culture medium per well
was 100 µl. After 24 h of incubation, 10 µl CCK8 solution was added
to each well. After 1 h incubation, the absorbance of each well was
measured with a microplate reader at a wavelength of 450 nm. The
experiment was repeated 3 times. A proliferation curve was plotted
with time as the abscissa and average absorbance value as the
ordinate.
Western blot assay
Pre-chilled PBS was used to wash adherent cells (GH3
and MMQ) and centrifugation was performed at 300 × g at 4°C for
5–10 min. The cell lysate (obtained using RIPA lysis buffer;
Beyotime Institute of Biotechnology) was evenly added to the entire
surface of the vessel and pipetted several times with a pipette.
The lysed cells were transferred to pre-cooled centrifuge tubes and
the total protein was precipitated. The cells were centrifuged at
400 × g for 15 min in a 4°C centrifuge (pre-cooled) and the
supernatant was collected in a 0.5-ml eppendorf (EP) tube. Total
protein samples (100 µg) were separated by 10% SDS-PAGE and
transferred to a PVDF membrane. After the protein was blocked with
5% BSA (BioVision, Inc.) at room temperature for 4 h, membranes
were incubated with 1:1,000 antibodies from Abcam (cleaved
Caspase-3, cat. no. ab32042; Survivin, cat. no. ab76424; Bcl-2,
cat. no. ab182858; Bax, cat. no. ab32503; E-cadherin, cat. no.
ab233611; Twist, cat. no. ab50887; Slug, cat. no. ab27568; MMP-7,
cat. no. ab207299; FOXO4, cat. no. ab128908 and GAPDH, cat. no.
ab8245) overnight at 4°C. The membrane was washed and incubated
with horseradish peroxidase-conjugated goat anti-mouse secondary
antibody (1:10,000, cat. no. ab150113; Abcam) at room temperature
for 4 h. The protein was developed via ECL (Merck KGaA) scanned by
gel imaging system and quantified by densitometry (Quantity One
Version 4.6.3; both Bio-Rad Laboratories, Inc.). The expression
level of each protein was expressed as the ratio of the protein
optical density value of each group to that of GAPDH.
Flow cytometry assay
A total of 4×105 cells (GH3 and MMQ cell
lines) were seeded in 6-well plates for 24 h, then digested with
trypsin without EDTA at room temperature for 1 min for collection.
Following centrifugation at 400 × g at 4°C for 5 min, the
supernatant was discarded. The cells were washed twice with
pre-chilled PBS. A total of 3 µl Annexin V-FITC and 3 µl PI
(Sigma-Aldrich; Merck KGaA) were added and mixed gently, then 300
µl combined buffer solution was added and the reaction was allowed
to progress for 15 min at room temperature in the dark. Flow
cytometry (FACSCalibur; BD Biosciences) was used to measure cell
apoptosis. Software used for analysis was MultiSET V3.0.1 (BD
Biosciences).
Transwell assay
Transwell experiments were used to verify cell
invasion and migration ability. Cells from each group were as
aforementioned collected and resuspended in serum-free RPMI-1640
medium (Gibco; Thermo Fisher Scientific, Inc.) (cell density,
5×104/ml). The upper Transwell chamber was pre-coated
with Matrigel at 37°C for 1 h, then cell suspension (100 µl/well)
was added. A total of 600 µl medium containing 10% fetal bovine
serum (Gibco; Thermo Fisher Scientific, Inc.) was added to the
lower chamber and incubated at 37°C in a 5% CO2
incubator for 24 h. The Transwell chamber was obtained, washed with
PBS, fixed with 5% glutaraldehyde at room temperature for 2 h and
stained with 0.01% crystal violet at room temperature for 20 min.
The number of transmembrane cells in 5 random fields of view was
counted under a light microscope (magnification, ×200). A total of
three parallel wells was set up for each group of cells and the
experiment was repeated 3 times. For migration experiments, the
same procedure was followed but Matrigel was not used.
Luciferase reporter experiment
Starbase (starbase.sysu.edu.cn/) was used to predict the binding
sites between miR-23b-3p and MEG3 or FOXO4. In addition,
MEG3-mutant (Mut) (5′-GGGACUGACU-ACUUACACAA-3′) and FOXO4-Mut were
designed (5′-AGCAAUGCUGUUGGAAUACACAA-3′). In order to confirm that
MEG3 can target miR-23b-3p, the 3′ untranslated region (UTR) of
MEG3 was inserted into a luciferase reporter gene vector and
co-transfected with miR-23b-3p mimic into 293T cells. pMIR-REPORT
Luciferase plasmid was purchased from Promega Corporation.
miR-23b-5p mimics and control were synthesized by Shanghai Jima
Pharmaceutical Technology Co., Ltd.. The sequences were as follows:
miR-23b-3p mimics, AUCACAUUGCCAGGGAUUACC and miR-NC,
CAGUACUUUUGUGUAGUACAA. The carrier contained two luciferase
reporter genes (Renilla and firefly luciferase reporter
gene). The 3′UTR of the lncRNA MEG3 was inserted directly
downstream of the Renilla luciferase reporter gene. If the
3′UTR was recognized by miR-23b-3p, MEG3 expression would be
suppressed. The 293T cells were seeded in 24-well plates and
transfected with reporter gene vector and miR-23b-3p mimic by
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) at 37°C. After 24 h, the luciferase report was
performed using a dual luciferase assay system (Promega
Corporation) according to the manufacturer's instructions. The
binding of miR-23b-3p and FOXO4 was detected in the same
manner.
Pull-down assay
Pull-down assay was performed according to the
manufacturer's instructions (cat. no. KT103-01; Guangzhou Sai Cheng
Biotechnology Co., Ltd.). Cells were collected into an RNase-free
EP tube and were centrifuged with 300 × g at 4°C for 5 min. The
supernatant was discarded, and cells were washed 1–2 times with PBS
solution. Then, 1 ml Cell lysis buffer containing protease
inhibitor was added. The cells were centrifuged at 4°C at 14,000 ×
g for 15 min and the supernatant was transferred to a new 1.5-ml EP
tube. The magnetic beads were suspended upside down slightly, 50 µl
suspended magnetic bead suspension was placed in an RNase-free
1.5-ml EP tube and the supernatant was removed. Binding Buffer (500
µl) was added to each tube, gently inverted and mixed to wash the
magnetic beads twice. Centrifugation was performed at 5,000 × g at
4°C for 1 min and the supernatant was removed. Binding Buffer (500
µl) was used to resuspend the magnetic beads and 2 µg probe was
added to the EP tube. Samples were placed in a refrigerator at 4°C
and incubated for 8 h. The magnetic bead-probe mixture was
centrifuged at 5,000 × g at 4°C for 2 min and the supernatant was
removed. Magnetic beads were washed once with 500 µl binding
buffer, and then 1 ml binding buffer, 5 µl RNase Inhibitor and 300
µl cell lysate were added in sequence, mixed slightly, sealed,
placed in a refrigerator at 4°C and incubated overnight. The
mixture was centrifuged at 5,000 × g at 4°C for 2 min, the
supernatant was removed and 1 ml washing buffer was added. The
magnetic beads were washed 5 times, centrifuged at 5,000 × g at 4°C
for 1 min, and the supernatant was removed. Finally, 50 µl 2X
Loading buffer was added to each group, boiled at 100°C for 10 min,
centrifuged at 5,000 × g at 4°C for 1 min and the supernatant was
transferred to a new EP tube, then a 20 µl sample was used for
western blotting, as aforementioned. The primary antibody FOXO4 was
purchased from Abcam (1:1,000; cat. no ab154520).
Statistical analysis
SPSS19.0 software (IBM Corp.) was used for
statistical analysis. The experimental data are expressed as the
mean ± SD (n=3). The two-tailed paired t-test was used to compare
differences between two groups being compared once. For comparisons
in datasets containing multiple groups or single groups being
compared more than once, one-way ANOVA with Tukey's post hoc test
was applied to analyze the difference. Differences in patient
information, including age, sex, tumor size, stage and invasiveness
between two groups were analyzed by χ2 test. Pearson
correlation analysis was used to analyze the correlation between
expression levels of MEG3, miR-23b-3p and FOXO4. P<0.05 was
considered to indicate a statistically significant difference.
Results
MEG3 expression levels are lower in PA
tumor tissue
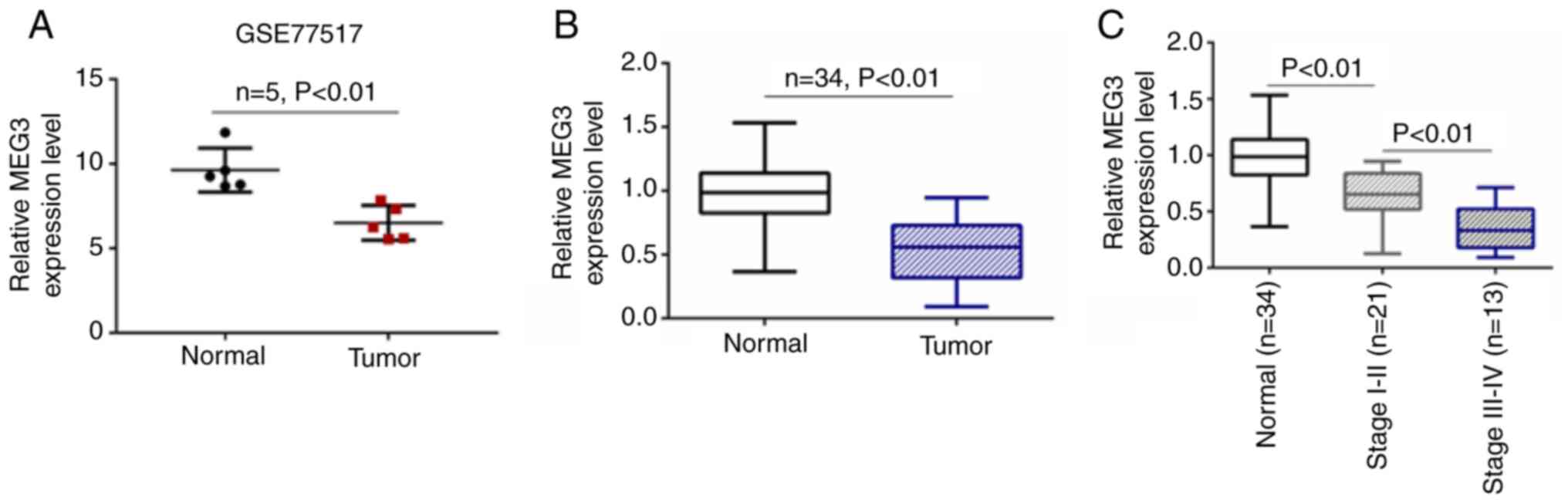
GSE77517 was downloaded from the GEO database and 5
patients with NFPA were included. Based on the profile of GSE77517,
expression levels of lncRNA MEG3 were lower compared with those in
normal tissue (Fig. 1A). Tumor and
adjacent tissue samples (n=34) were collected from patients with PA
and the expression levels of MEG3 were detected by RT-qPCR. MEG3
expression was significantly lower in PA tumor tissue compared with
normal samples (Fig. 1B). MEG
expression levels were assessed in samples from 34 patients with
NFPA (21 patients with stage I–II and 13 with stage III–IV). MEG3
expression levels were lower in patients with stage III–IV than
those with stage I–II NFPA (Fig. 1C).
Based on the median value of MEG2, these patients were divided into
lncRNA MEG3 high and low expression groups. Tumor size,
invasiveness and clinical stage were significantly different but
there was no significant difference between sex and age in these
two groups (Table I).
 | Table I.Correlation between MEG3 expression
levels and clinical pathology in patients with pituitary
adenoma. |
Table I.
Correlation between MEG3 expression
levels and clinical pathology in patients with pituitary
adenoma.
|
|
| MEG3 expression
levels |
|
|---|
|
|
|
|
|
|---|
| Characteristic | Total patients
(n=34) | High (≥median,
n=15) | Low (<median,
n=19) | P-value |
|---|
| Age, years |
|
|
| 0.620 |
| <50 | 16 | 7 | 9 |
|
| ≥50 | 18 | 8 | 10 |
|
| Sex |
|
|
| 0.468 |
| Female | 15 | 6 | 9 |
|
| Male | 19 | 9 | 10 |
|
| Tumor size, cm |
|
|
| 0.045 |
| ≤3 | 16 | 10 | 6 |
|
| >3 | 18 | 5 | 13 |
|
| Invasiveness |
|
|
| 0.018 |
| Non-IPA | 17 | 11 | 6 |
|
| IPA | 17 | 4 | 13 |
|
| Clinical stage |
|
|
| 0.009 |
| I–II | 21 | 13 | 8 |
|
| III–IV | 13 | 2 | 11 |
|
MEG3 inhibits proliferation and
improves apoptosis of PA tumor cells
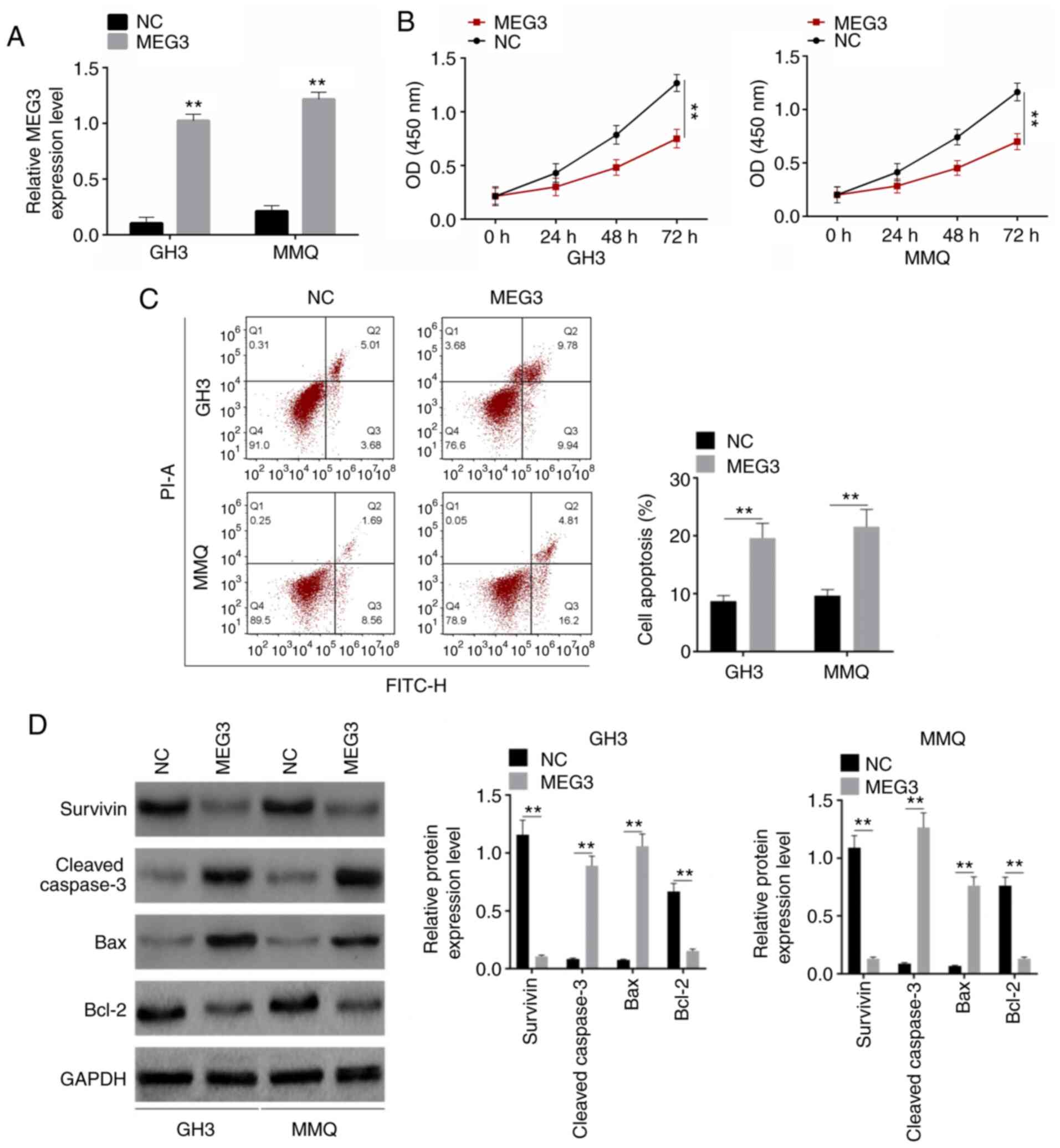
PA cell lines GH3 and MMQ (purchased from ATCC) were
transfected with pcDNA3.1(+)-NC, pcDNA3.1(+)-MEG3. RT-qPCR was used
to detect the transfection efficiency. MEG3 was overexpressed,
indicating that the transfection was successful in both GH3 and MMQ
cells (Fig. 2A). Following
transfection, proliferation significantly decreased, whereas the
rate of cell apoptosis significantly increased (Fig. 2B and C). Survivin, cleaved-Caspase-3,
Bax and Bcl-2 are proliferation- and apoptosis-associated proteins
(23). Western blot analysis was
performed to detect expression levels of these proteins. The
expression levels of Survivin and Bcl-2 significantly decreased,
whereas those of cleaved-Caspase-3 and Bax were increased following
transfection (Fig. 2D). These results
suggested that MEG3 inhibited proliferation and improved apoptosis
of PA tumor cells.
MEG3 inhibits invasion, migration and
the EMT process
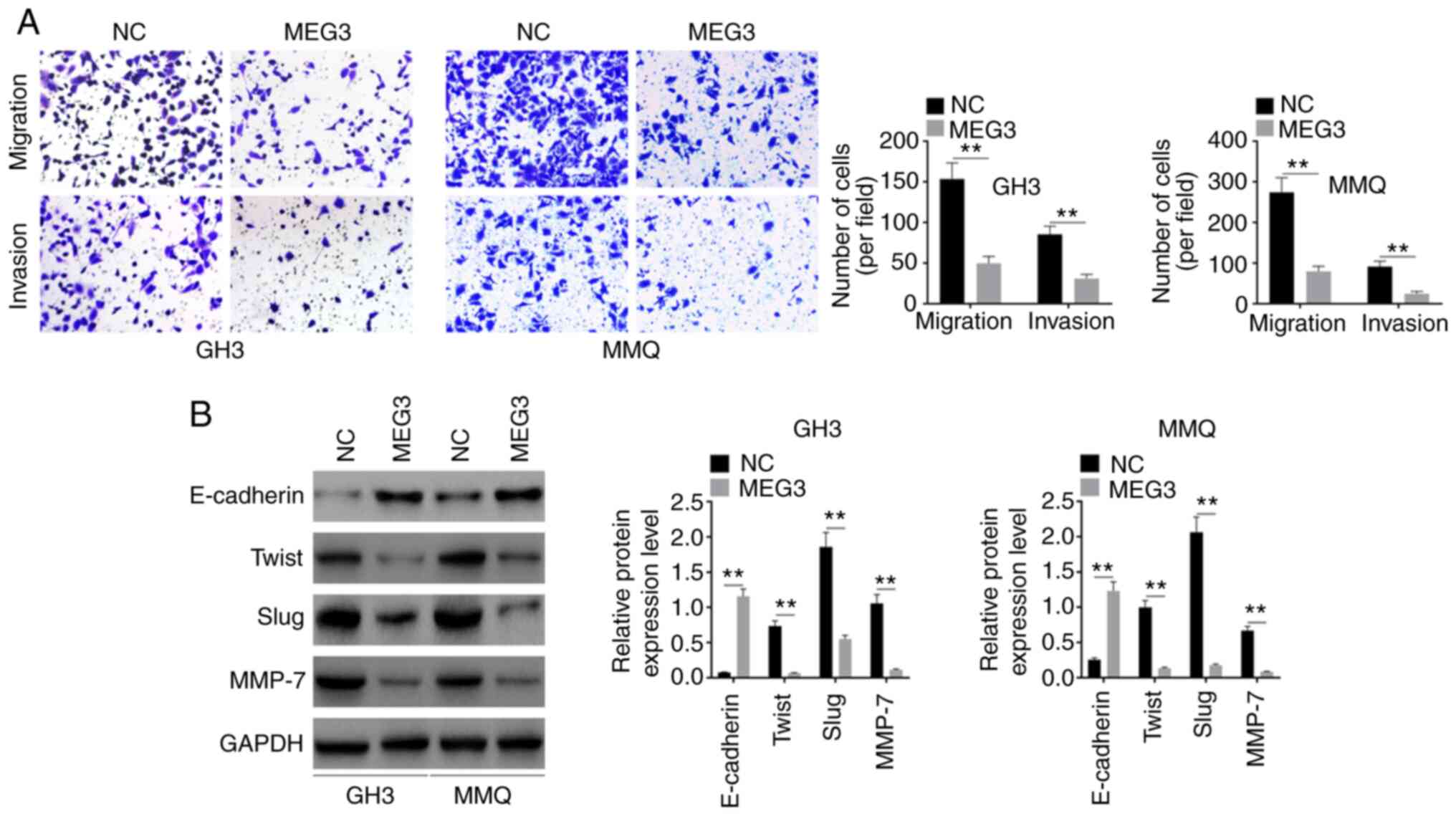
Following transfection, migration and invasion was
detected by Transwell assay. Cell migration and invasion
significantly decreased in both GH3 and MMQ cell lines (Fig. 3A). Expression levels of E-cadherin,
Twist, Slug and MMP-7 were also detected. The levels of these EMT
markers were regulated by overexpressed MEG3: E-cadherin was
upregulated, while Twist, Slug and MMP-7 were downregulated by
overexpression of MEG3 (Fig. 3B).
This suggested that MEG3 inhibited pituitary tumor development by
participating in the EMT process.
MEG3 targets and downregulates
miR-23b-3p
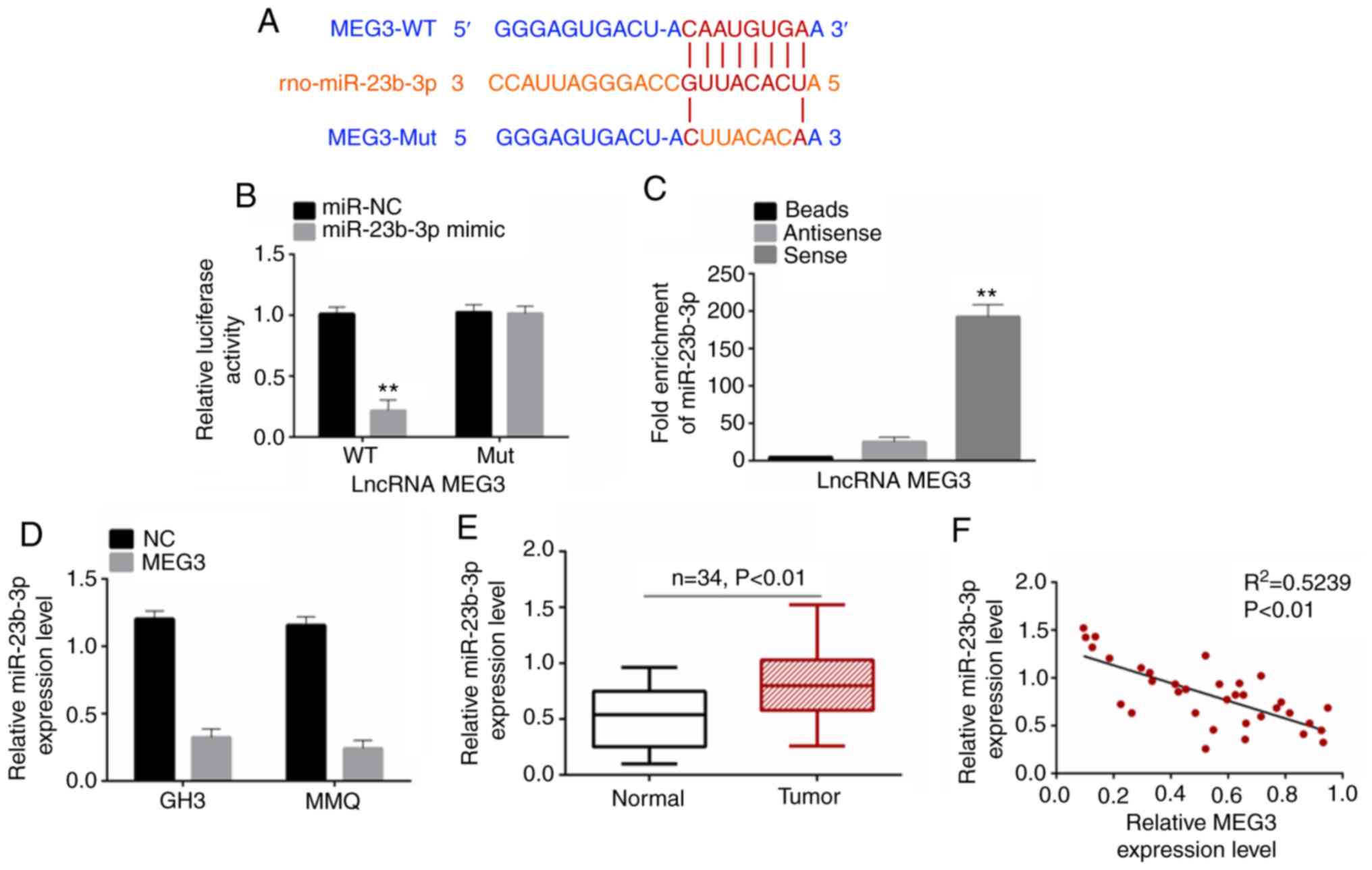
Based on starbase (starbase.sysu.edu.cn/), binding sites were predicted
between MEG3 and miR-23b-3p. The binding was shown in Fig. 4A. Dual luciferase reporter gene assay
showed that, when co-transfected miR-23b-3p entered 293T cells, the
relative luciferase activity of wild-type (WT) MEG3 reporter gene
was significantly inhibited. By contrast, the luciferase activity
of the Mut MEG3 reporter gene was not affected by miR-23b-3p
co-transfection (Fig. 4B). RNA
pull-down assay also confirmed the binding interaction between MEG3
and miR-23b-3p (Fig. 4C). RT-qPCR was
performed to detect the expression levels of miR-23b-3p in cells
following transfection and in tissue. miR-23b-3p was significantly
decreased in cells following MEG3 overexpression (Fig. 4D). miR-23b-3p was more highly
expressed in PA tumor than normal tissue (Fig. 4E). Pearson's correlation analysis was
used to analyze the correlation between expression levels of MEG3
and miR-23b-3p; the results showed that MEG3 and miR-23b-3p were
negatively correlated (Fig. 4F). This
indicated that MEG3 targeted miR-23b-3p.
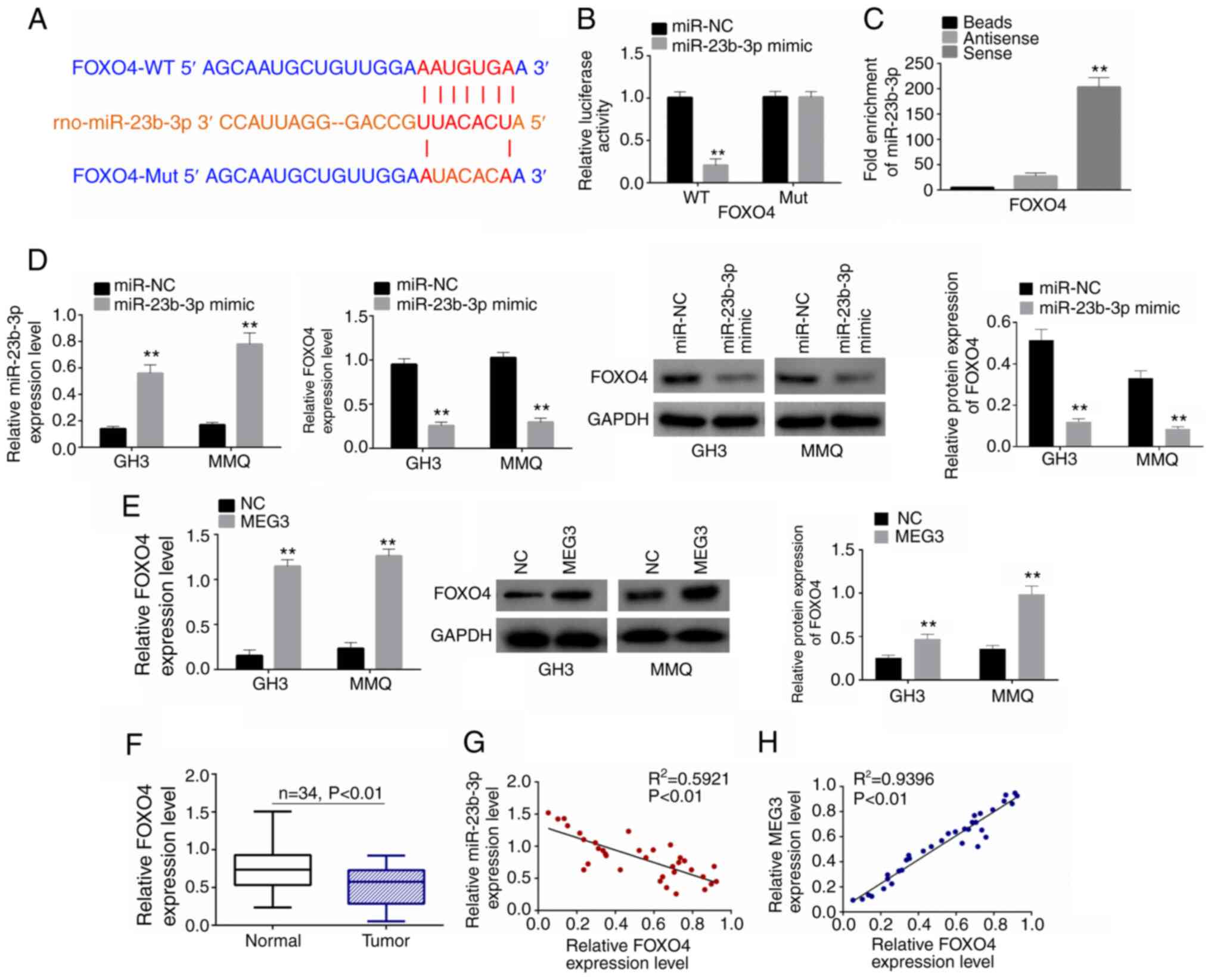
miR-23b-3p targets FOXO4
Based on the starbase, binding sites between FOXO4
and miR-23b-3p were predicted. The binding was shown in Fig. 5A). When co-transfected with miR-23b-3p
mimic, the relative luciferase activity in the FOXO4-WT group was
significantly inhibited (Fig. 5B).
RNA pull-down assay also confirmed binding between miR-23b-3p and
FOXO4 (Fig. 5C). RT-qPCR and western
blot were used to detect the expression levels of FOXO4 in cells
following miR-23b-3p mimic or MEG3 transfection. Following
miR-23b-3p mimic transfection, miR-23b-3p expression was
significantly increased in both GH3 and MMQ cells, indicating that
transfection was successful (Fig.
5D). FOXO4 was downregulated following miR-23b-3p mimic
transfection (Fig. 5D) but
upregulated following MEG3 transfection (Fig. 5E). Moreover, relative FOXO4 expression
levels were lower in pituitary tumor than in normal tissue samples
(Fig. 5F). Pearson's correlation
analysis showed that FOXO4 was negatively correlated with
miR-23b-3p but positively correlated with MEG3 (Fig. 5G and H).
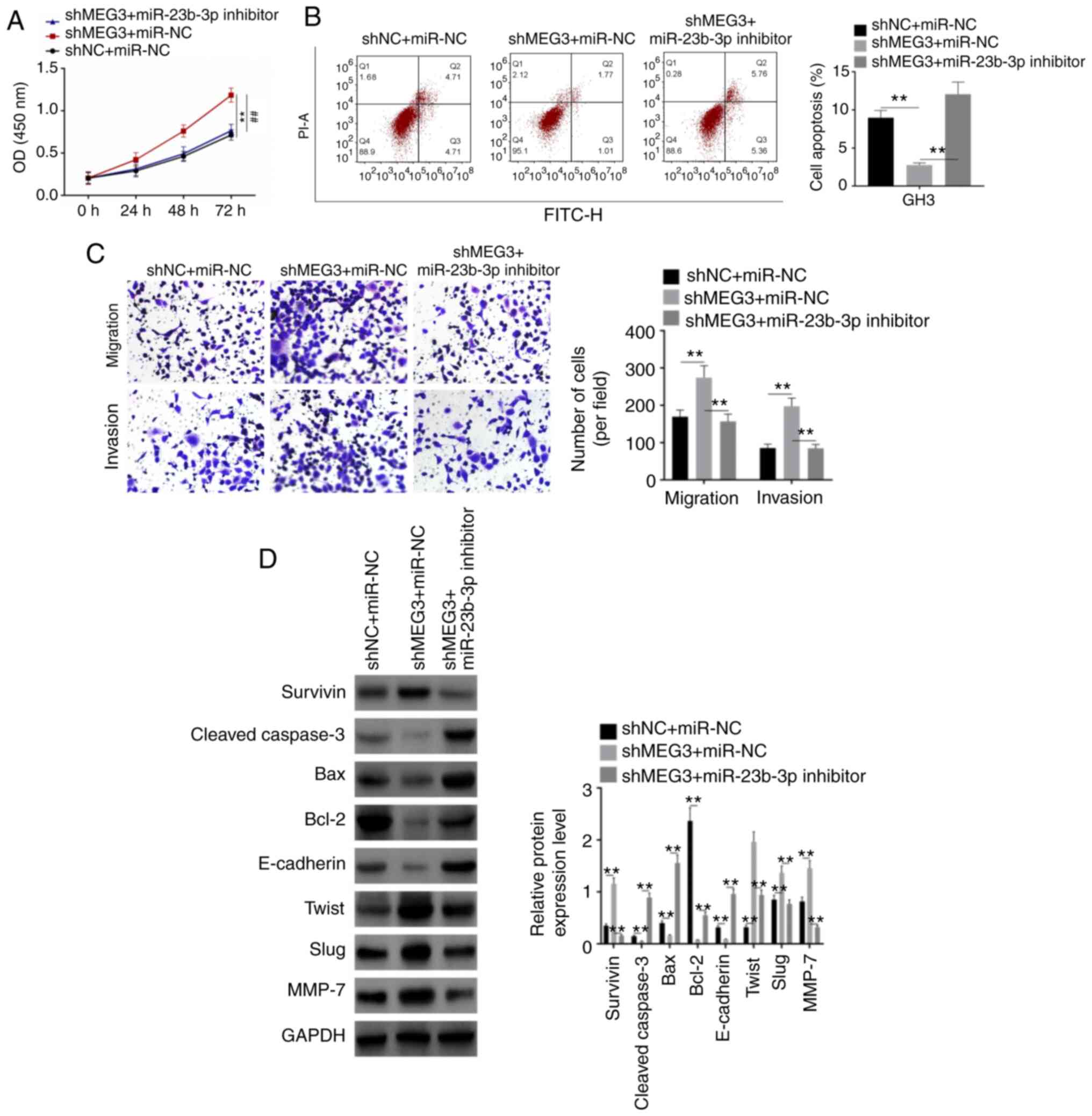
Rescue assay
For the rescue assay, GH3 cells were divided into 3
groups (shNC + miR-NC, shMEG3 + miR-NC and shMEG3 + miR-23b-3p
inhibitor). shMEG3 significantly increased proliferation, migration
and invasion of GH3 cells and inhibited apoptosis, whereas
miR-23b-3p inhibitor rescued the effects of shMEG3 on these
processes (Fig. 6A-C). Then, western
blot analysis was performed to detect expression levels of
apoptosis and EMT-associated proteins, including Survivin,
Cleaved-Caspase-3, Bax, Bcl-2, E-cadherin, Twist, Slug, MMP-7 and
GAPDH. Expression levels of Survivin, Twist, Slug and MMP-7 were
upregulated by shMEG3, whereas those of Cleaved-Caspase-3, Bax,
Bcl-2 and E-cadherin were downregulated (Fig. 6D). When miR-23-3p was co-transfected,
the expression levels of these proteins were rescued. These results
indicated that shMEG3 facilitated pituitary tumor cell
proliferation, migration and invasion by participating in the EMT
process, whereas miR-23b-3p inhibitor rescued the effect of
shMEG3.
Discussion
Pituitary tumor is a common type of intracranial
tumor (24). Due to its complicated
pathogenesis, the treatment of pituitary tumor presents a clinical
problem (25). Therefore, seeking
effective therapeutic targets is key for the treatment of pituitary
tumors. In the present study, MEG3 expression levels were lower in
pituitary tumor tissues and cells than in normal samples.
Overexpression of MEG3 inhibited proliferation, invasion and
migration of pituitary tumor cells and accelerated apoptosis.
MEG3, a maternally expressed imprinted gene, encodes
an lncRNA with a length of ~1,600 nt (26). Previous studies have demonstrated that
MEG3 stimulates transcriptional activity mediated by the tumor
suppressor p53, causes p53 protein to aggregate in the cell,
decreases expression levels of MDM2 protein and selectively
activates growth differentiation factor 15 (27,28). The
expression of MEG3 is controlled by epigenetics and abnormal CpG
methylation of MEG3 occurs in various types of tumor, such as
cervical cancer and pituitary adenoma (23,29,30). In a
mouse model, MEG3 has been shown to regulate cerebral angiogenesis
via the VEGF pathway and inactivation of MEG3 expression levels in
NFPA has been demonstrated clinically (31,32). In
the present study, patients with stage III–IV pituitary tumor
exhibited lower MEG3 mRNA expression levels than those with I–II
pituitary tumor. Therefore, lncRNA MEG3 was considered to be a
critical inhibitor of pituitary tumor development.
Here, MEG3 negatively regulated miR-23b-3p
expression levels. The role of miR-23b-3p has been confirmed in
multiple types of cancer. For example, miR-23b-3p is a carcinogenic
miRNA that inhibits PTEN expression levels in renal cell carcinoma
(33). PTEN inactivation has been
demonstrated in invasive pituitary tumor tissue; decreased
expression levels of PTEN protein result in a decreased tumor
suppressive effect of PTEN protein and enhance tumor invasion
(34). Additionally, miR-23b-3p is
upregulated in radiation-induced thymic lymphoma (35). Downregulation of miR-23b-3p increases
autophagy, which promotes resistance to radiation in pancreatic
cancer cells (36). The present study
verified that miR-23-3p inhibitor rescued the effect of shMEG3 on
proliferation, invasion, migration and apoptosis of pituitary tumor
cells. Based on the aforementioned evidence, it was concluded that
MEG3 inhibits pituitary tumor development by downregulating
miR-23b-3p expression levels.
miR-23b-3p was verified to negatively regulate FOXO4
expression levels in the present study. Previous studies have found
that FOXO4 is a target of NAD+-dependent deacetylase
silencing information regulator 1 (Sir1) to regulate cell growth
under normal and non-stress conditions (37,38). FOXO4
regulates apoptosis by inhibiting expression levels of Bcl-xl
(39). FOXO4 specifically binds to
the Bcl-6 promoter to increase Bcl-6 transcription, while Bcl-6
directly binds to the Bcl-xl promoter to inhibit its expression.
Under stress, FOXO4 promotes Bcl-2 transcription by binding to the
Bim promoter and further promotes endogenous apoptosis (40). In addition to inducing apoptosis,
FOXO4 overexpression blocks the cell cycle at the G1
phase, which is associated with p27 (41). Therefore, changes in apoptosis induced
by abnormal expression of FOXO4 may be an underlying mechanism of
tumor pathogenesis.
In the present study, overexpression of MEG3
inhibited the EMT process in pituitary tumor. EMT refers to the
biological process in which epithelial cells are transformed into
cells with mesenchymal phenotype via a specific procedure (42). In this process, the adhesion
structure, polarity and cytoskeleton between epithelial cells
change, thereby increasing the ability of epithelial cells to
deform, migrate and invade, as well as enhancing anti-apoptotic
ability (43). EMT is an important
biological process by which malignant tumors derived from
epithelial cells acquire migration and invasion capabilities and
serves a key role in tumor metastasis.
There were certain limitations in the present study.
Firstly, the number of clinical samples included was relatively
small; the role of MEG3 must be verified using a larger patient
cohort. Secondly, the regulatory mechanism of MEG3 is not
unidirectional-it may simultaneously regulate multiple miRNAs,
genes and pathways. Therefore, it is necessary to identify a
regulatory network to further study the molecular mechanism
underlying the development of pituitary tumors. Finally, the
effects of MEG3 in vivo need be confirmed using animal
models.
In conclusion, MEG3 inhibited pituitary tumor
development by participating in cell proliferation, apoptosis and
the EMT process, which may be a novel target for treatment of
pituitary tumors.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
XW contributed to the conception of the study. XL
contributed significantly to analysis and manuscript preparation;
ZW performed the data analysis. XW, XW and ZW confirmed the
authenticity of all the raw data. All authors read and approved the
final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of First Affiliated Hospital of Soochow University
(approval no. 2018013). All patients provided written informed
consent for the use of their samples in scientific research.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Haddad N, Tedoro AJ and Soares NP:
Carotenoids inhibits cell proliferation, arrest cell cycle and
induces apoptosis in pituitary tumor cells. 13th European Congress
of Endocrinology. BioScientifica; 2011
|
|
2
|
Hui L, Shengwu C and Mingwei Z: Surgical
treatment of pituitary adenoma: Analysis of 508 cases at a single
institution. Chin J Oncol Surg. 5:42013.
|
|
3
|
Sheary CB: Possible influence of
orthodontics on pituitary gland function and learning ability. J
Clin Orthod. 19:889–890. 1985.PubMed/NCBI
|
|
4
|
Bürgi U and Seiler R: Hypophyseal
dysfunction and tumors. Ther Umsche. 49:136–141. 1992.(In
German).
|
|
5
|
Rao G and Apfelbaum RI: Symptomatic
pneumocephalus occurring years after transphenoidal surgery and
radiation therapy for an invasive pituitary tumor: A case report
and review of the literature. Pituitary. 6:49–52. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bolognani F, Albariño CG, Romanowski V,
Carri NG and Goya RG: In vitro and in vivo herpetic vector-mediated
gene transfer in the pituitary gland: Impact on hormone secretion.
Eur J Endocrinol. 145:497–503. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Santoro A, Minniti G, Ruggeri A, Esposito
V, Jaffrain-Rea ML and Delfini R: Biochemical remission and
recurrence rate of secreting pituitary adenomas after
transsphenoidal adenomectomy: Long-term endocrinologic follow-up
results. Surg Neurol. 68:513–518. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhong A, Pu J, Ruan L, Jin J, Tan S, Wang
F, Mou J and Yang G: The complications of endoscopic
transsphenoidal surgery for pituitary neoplasms. Int J Clin Exp
Med. 9:20026–20031. 2016.
|
|
9
|
Prensner JR and Chinnaiyan AM: The
emergence of lncRNAs in cancer biology. Cancer Discov. 1:391–407.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Iyer MK, Niknafs YS, Malik R, Singhal U,
Sahu A, Hosono Y, Barrette TR, Prensner JR, Evans JR, Zhao S, et
al: The landscape of long noncoding RNAs in the human
transcriptome. Nat Genet. 47:1992015. View
Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhou K, Li S, Du G, Fan Y, Wu P, Sun H and
Zhang T: LncRNA XIST depletion prevents cancer progression in
invasive pituitary neuroendocrine tumor by inhibiting bFGF via
upregulation of microRNA-424-5p. Onco Targets Ther. 12:7095–7109.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhang Y, Liu YT, Tang H, Xie WQ, Yao H, Gu
WT, Zheng YZ, Shang HB, Wang Y, Wei YX, et al: Exosome-transmitted
lncRNA H19 inhibits the growth of pituitary adenoma. J Clin
Endocrinol Metab. 104:6345–6356. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
D'Angelo D, Mussnich P, Sepe R, Raia M,
Del Vecchio L, Cappabianca P, Pellecchia S, Petrosino S, Saggio S,
Solari D, et al: RPSAP52 lncRNA is overexpressed in pituitary
tumors and promotes cell proliferation by acting as miRNA sponge
for HMGA proteins. J Mol Med (Berl). 97:1019–1032. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wang C, Tan C, Wen Y, Zhang D, Li G, Chang
L, Su J and Wang X: FOXP1-induced lncRNA CLRN1-AS1 acts as a tumor
suppressor in pituitary prolactinoma by repressing the autophagy
via inactivating Wnt/β-catenin signaling pathway. Cell Death Dis.
10:4992019. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhu H, Jing G, Shen Y, Dong W, Gao H, Miao
Y, Li C and Zhang Y: Functions and mechanisms of tumor necrosis
factor-α and noncoding RNAs in bone-invasive pituitary adenomas.
Clin Cancer Res. 24:5757–5766. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yue X, Dong C, Ye Z, Zhu L, Zhang X, Eang
X, Mo F, Li Z and Pan B: LncRNA SNHG7 sponges miR-449a to promote
pituitary adenomas progression. Metab Brain Dis. 36:123–132. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Modali SD, Parekh VI, Kebebew E and
Agarwal SK: Epigenetic regulation of the lncRNA MEG3 and its target
c-MET in pancreatic neuroendocrine tumors. Mol Endocrinol.
29:224–237. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Liu LX, Deng W, Zhou XT, Chen RP, Xiang
MQ, Guo YT, Pu ZJ, Li R, Wang GF and Wu LF: The mechanism of
adenosine- mediated activation of lncRNA MEG3 and its antitumor
effects in human hepatoma cells. Int J Oncol. 48:421–429. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Modali SD, Desai SS, Parekh VI, Kebebew E,
Emmert-Buck M and Agarwal SK: Abstract LB-249: Reduced expression
of the long non-coding RNA MEG3 in sporadic and MEN1-associated
tumors. Cancer Res. 79 (Suppl 8):LB–249. 2013.
|
|
20
|
Fan FY, Deng R, Yi H, Sun HP, Zeng Y, He
GC and Su Y: The inhibitory effect of MEG3/miR-214/AIFM2 axis on
the growth of T-cell lymphoblastic lymphoma. Int J Oncol.
51:316–326. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Mezzomo LC, Gonzales PH, Pesce FG,
Kretzmann Filho N, Ferreira NP, Oliveira MC and Kohek MB:
Expression of cell growth negative regulators MEG3 and GADD45γ is
lost in most sporadic human pituitary adenomas. Pituitary.
15:420–427. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Method. 25:402–408. 2001.
View Article : Google Scholar
|
|
23
|
Zhang J, Yao T, Lin Z and Gao Y: Aberrant
methylation of MEG3 functions as a potential plasma-based biomarker
for cervical cancer. Sci Rep. 7:62712017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kouhara H, Tatekawa T, Koga M, Hiraga S,
Arita N, Mori H and Sato B: Intracranial and intraspinal
dissemination of an ACTH-secreting pituitary tumor. Endocrinol Jpn.
39:177–184. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Shimon I and Melmed S: Pituitary tumor
pathogenesis. J Clin Endocrinol Metab. 82:1675–1681. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Tian ZZ, Guo XJ, Zhao YM and Fang Y:
Decreased expression of long non-coding RNA MEG3 acts as a
potential predictor biomarker in progression and poor prognosis of
osteosarcoma. Int J Clin Exp Pathol. 8:15138–15142. 2015.PubMed/NCBI
|
|
27
|
Hu D, Su C, Jiang M, Shen Y, Shi A, Zhao
F, Chen R, Shen Z, Bao J and Tang W: Fenofibrate inhibited
pancreatic cancer cells proliferation via activation of p53
mediated by upregulation of LncRNA MEG3. Biochem Biophys Res
Commun. 471:290–295. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kim Y, Noren Hooten N and Evans MK: CRP
stimulates GDF15 expression in endothelial cells through p53.
Mediators Inflamm. 2018:82780392018. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
House JS, Hall J, Park SS, Planchart A,
Money E, Maguire RL, Huang Z, Mattingly CJ, Skaar D, Tzeng JY, et
al: Cadmium exposure and MEG3 methylation differences between
Whites and African Americans in the NEST cohort. Environ Epigenet.
5:dvz0142019. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Gejman R, Batista DL, Zhong Y, Zhou Y,
Zhang X, Swearingen B, Stratakis CA, Hedley-Whyte ET and Klibanski
A: Selective loss of MEG3 expression and intergenic differentially
methylated region hypermethylation in the MEG3/DLK1 locus in human
clinically nonfunctioning pituitary adenomas. J Clin Endocrinol
Metab. 93:4119–4125. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Cheunsuchon P, Zhou Y, Zhang X, Lee H,
Chen W, Nakayama Y, Rice KA, Tessa Hedley-Whyte E, Swearingen B and
Klibanski A: Silencing of the imprinted DLK1-MEG3 locus in human
clinically nonfunctioning pituitary adenomas. Am J Pathol.
179:2120–2130. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Su W, Xie W, Shang Q and Su B: The long
noncoding RNA MEG3 is downregulated and inversely associated with
VEGF levels in osteoarthritis. Biomed Res Int. 2015:3568932015.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zaman MS, Thamminana S, Shahryari V,
Chiyomaru T, Deng G, Saini S, Majid S, Fukuhara S, Chang I, Arora
S, et al: Inhibition of PTEN gene expression by oncogenic
miR-23b-3p in renal cancer. PLoS One. 7:e502032012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhou K, Zhang T, Fan Y, Du G, Wu P and
Geng D: MicroRNA-106b promotes pituitary tumor cell proliferation
and invasion through PI3K/AKT signaling pathway by targeting PTEN.
Tumour Biol. 37:13469–13477. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Li B, Sun M, Gao F, Liu W, Yang Y, Liu H,
Cheng Y, Liu C and Cai J: Up-regulated expression of miR-23a/b
targeted the pro-apoptotic Fas in radiation-induced thymic
lymphoma. Cell Physiol Biochem. 32:1729–1740. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wang P, Zhang J, Zhang L, Zhu Z, Fan J,
Chen L, Zhuang L, Luo J, Chen H, Liu L, et al: MicroRNA 23b
regulates autophagy associated with radioresistance of pancreatic
cancer cells. Gastroenterology. 145:1133–1143.e12. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Folmer F, Orlikova B, Schnekenburger M,
Dicato M and Diederich M: Naturally occurring regulators of histone
acetylation/deacetylation. Curr Nutr Food Sci. 6:78–99. 2010.
View Article : Google Scholar
|
|
38
|
Chuang PY, Dai Y, Liu R, He H, Kretzler M,
Jim B, Cohen CD and He JC: Alteration of forkhead box O (foxo4)
acetylation mediates apoptosis of podocytes in diabetes mellitus.
PLoS One. 6:e235662011. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Lam EF, Francis R and Petkovic M: FOXO
transcription factors: Key regulators of cell fate. Biochem Soc
Trans. 34:722–726. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Urbich C, Knau A, Fichtlscherer S, Walter
DH, Brühl T, Potente M, Hofmann WK, de Vos S, Zeiher AM and
Dimmeler S: FOXO-dependent expression of the proapoptotic protein
Bim: Pivotal role for apoptosis signaling in endothelial progenitor
cells. FASEB J. 19:974–976. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Brenkman AB, van den Broek NJ, de Keizer
PL, van Gent DC and Burgering BM: The DNA damage repair protein
Ku70 interacts with FOXO4 to coordinate a conserved cellular stress
response. FASEB J. 24:4271–4280. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Zuo JH, Zhu W, Li MY, Li XH, Yi H, Zeng
GQ, Wan XX, He QY, Li JH, Qu JQ, et al: Activation of EGFR promotes
squamous carcinoma SCC10A cell migration and invasion via inducing
EMT-like phenotype change and MMP-9-mediated degradation of
E-cadherin. J Cell Biochem. 112:2508–2517. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Klymkowsky MW and Savagner P:
Epithelial-mesenchymal transition: A cancer researcher's conceptual
friend and foe. Am J Pathol. 174:1588–1593. 2009. View Article : Google Scholar : PubMed/NCBI
|