Introduction
The microRNA (miR)-29 family consists of the
miR-29a, miR-29b and miR-29c members, whose aberrant expression is
involved in tumorigenesis, as their specific target genes include
oncogenes and members of the DNA methyltransferase family (1,2). miR-29b
dysregulation has been reported in breast cancer (3–5), in which
low miR-29b expression has been found to be positively correlated
with large tumor size and advanced cancer stage (6,7).
Accordingly, miR-29b has been reported as a sensitive marker for
predicting patient outcome in all breast tumor subtypes (7), including triple-negative breast cancer
(TNBC) (8). In TNBC cell lines,
miR-29b has been shown to suppress viability and migration and
increase sensitivity to chemotherapeutic agents (2,9),
suggesting that the restoration of miR-29b levels may be crucial
for this unfavorable breast cancer subset. More recently, the
predominantly expressed mature miR-29b-3p and the less expressed
miR-29b1-5p have been reported to have opposite effects on tumor
cells with a triple-negative phenotype (9–13);
therefore, the role of miR-29b in TNBC remains elusive.
A number of positive or negative transcriptional
modulators of miR-29b expression have been identified (14), but little is known concerning the
regulation of miR-29b in breast cancer (15). The multidomain protein Vav1 has been
found to play a peculiar role in miR-29b expression in acute
myeloid leukemia (AML) cells, in which the PU.1-mediated expression
of miR-29b is almost completely dependent on adequate levels of
Vav1 inside the nuclear compartment (16). Vav1 is ectopically expressed in the
majority of breast carcinomas, in which it displays a prevalent
localization inside the nucleus, which is positively correlated
with a low incidence of relapse (17). In TNBC cells, Vav1 negatively
regulates invasiveness in vitro and metastatic efficiency
in vivo by affecting the expression of genes involved in the
invasion and/or metastasis of breast tumors (17,18).
The aim of the present study was to assess the role
of Vav1 in the regulation of miR-29b levels in breast cancer cells.
This study focused on cells with a triple-negative phenotype, for
which target-based therapies are not currently available, and
sought to determine whether Vav1 can promote the miR-29b
transcriptional process.
Materials and methods
All reagents were purchased from Merck KGaA unless
otherwise specified.
Cells and treatments
The non-transformed MCF10A cells (RRID: CVCL_0598)
and the malignant MCF7 (RRID: CVCL_0031), MDA-MB-453 (RRID:
CVCL_0418), MDA-MB-468 (RRID: CVCL_0419) and MDA-MB-231 (RRID:
CVCL_0062) cell lines were purchased from the American Type Culture
Collection. BT-474 cells (RRID: CVCL_0179) were obtained from
Interlab Cell Line Collection.
MCF10A cells were cultured in DMEM-F12 (Thermo
Fisher Scientific, Inc.) containing 10 µg/ml bovine insulin, 100
ng/ml cholera toxin, 0.5 µg/ml hydrocortisone, 20 ng/ml recombinant
human epidermal growth factor, and 10% horse serum. The BT-474 cell
line was cultured in RPMI-1640 medium (Thermo Fisher Scientific,
Inc.) supplemented with 10% fetal bovine serum (FBS), 1 mM Na
pyruvate, and 0.01 mg/ml bovine insulin. MCF7, MDA-MB-453,
MDA-MB-468 and MDA-MB-231 cell lines were maintained in DMEM
(Thermo Fisher Scientific, Inc.) supplemented with 10% FBS
(17–19).
Established cell lines from breast tumor
patient-derived xenografts (PDXs) with a triple-negative phenotype
(HBCx-2, HBCx-9, HBCx-17, HBCx-39 and T174) were provided by
Xentech and cultured in Gibco™ Advanced DMEM-F12 (Thermo Fisher
Scientific, Inc.) supplemented with 8% FBS (Thermo Fisher
Scientific, Inc.), 1% penicillin- streptomycin solution and 20 µM
Rho-associated kinase inhibitor Y-27632 (DBA ITALIA SRL).
All cell lines were cultured at 37°C in a humidified
atmosphere of 5% CO2 in air, and were tested for
mycoplasma and other contaminations monthly.
In all cell lines, the upregulation and
downregulation of Vav1 were performed as previously described
(18).
Immunochemical analysis
Total lysates (50 µg protein) were separated on
polyacrylamide denaturing gels, blotted to nitrocellulose membranes
(GE Healthcare), and treated with primary antibodies against Vav1
(diluted 1:500, cat. no. sc-8039), CEBPα (diluted 1:250, cat. no.
sc-61) (both from Santa Cruz Biotechnology, Inc.), GATA3 (diluted
1:500, cat. no. ab199428; Abcam), and β-tubulin (diluted 1:1,000,
cat. no. T4026; Merck KGaA), as previously described (18). The membranes were then incubated with
appropriate peroxidase-conjugated secondary antibodies (goat
anti-mouse, diluted 1:2,000, cat. no A4416; goat anti-rabbit,
diluted 1:2,000, cat. no. A6154; Merck KGaA) and visualized using
the ECL system (PerkinElmer, Inc.). Chemiluminescence images of the
bands were captured by ImageQuant™ LAS 4000 biomolecular imager (GE
Healthcare), and densitometric analysis was performed using
ImageQuant TL version 7.0 software (RRID:SCR_018374; GE
Healthcare).
Reverse transcription quantitative PCR
(RT-qPCR)
High-quality RNA, including small RNAs, was
extracted from all cell lines using miRNeasy Micro Kit (Qiagen SpA
Italia), according to the manufacturer's instructions.
miR-29b and CEBPα expression were evaluated by
RT-qPCR using TaqMan Assays (ID 000413; ID Hs05650633_s1: Thermo
Fisher Scientific, Inc), as previously described (16,19,20).
miR-29b and CEBPα expression levels were normalized to U6 snRNA (ID
001973, Thermo Fisher Scientific, Inc.) and to RPL13A (ID
Hs03043885_g1, Thermo Fisher Scientific, Inc.), respectively.
To measure Vav1 mRNA in PDX-derived cell lines, and
pri-miR-29b1 and pri-miR-29b2 in all cell lines, RT-qPCR was
performed using the iTaq Universal SYBR-Green SuperMix on a CFX96™
Real-time detection system (RRID:SCR_018064; Bio-Rad Laboratories
Inc.). The following primers were used: Vav1,
5′-ACGTCGAGGTCAAGCACATT-3′ forward and 5′-GGCCTGCTGATGGTTCTCTT-3′
reverse; pri-miR-29b1, 5′-AAATGGCAGTCAGGTCTCTG-3′ forward and
5′-GCAATGCAAATGTATGCAAAT-3′ reverse; pri-miR-29b2,
5′-TTGAGTGTGGCGATTGTCAT-3′ forward and 5′-ATCAACGCCGAATACTCCAG-3′
reverse.
Levels of Vav1, pri-miR-29b1 and pri-miR-29b2 were
normalized to RPL32 content (5′-CATCTCCTTCTCGGCATCA-3′ forward and
5′-AACCCTGTTGTCAATGCCTC-3′ reverse).
All reactions were performed in triplicate, and the
experiments were repeated 3 times.
Quantitative chromatin
immunoprecipitation (Q-ChIP) assay
Q-ChIP assays were performed as previously described
(16). Samples were
immunoprecipitated with antibodies against CEBPα or with a
non-specific IgG antibody (cat. no. sc-53344; Santa Cruz
Biotechnology, Inc). qPCR of a 131-bp DNA fragment (primers:
5′-GCAGGTTTTCAGTTGGTGGTTT-3′ forward and 5′-GCCGTGACAGTTCAGTAGGA-3′
reverse), encompassing the putative CEBPα binding site at −89/+42
bp from the transcriptional start in the pri-miR29a/b1 promoter on
Chr 7q32.3 was performed using an iTaq Universal SYBR-Green
SuperMix. PCR products were separated on Tris-acetate 1% agarose
gels, stained with ethidium bromide and visualized by a UV light
apparatus.
Statistical analysis
The association between Vav1 mRNA, pre-miR-29b1 and
pre-miR-29b2 levels was evaluated within each of the 918 invasive
ductal carcinomas of ‘The Cancer Genome Atlas’ (TCGA;
cancergenome.nih.gov). Pearson's correlation coefficient (r) and
Spearman's rank correlation coefficient (ρ) were calculated for all
values (https://jasp-stats.org/). All miR-29b
values in cell lines are expressed as mean ± standard deviation and
analyzed by one-way ANOVA followed by Dunnett's multiple comparison
test for more than two groups, using GraphPad Prism 6.0 statistical
package (RRID: SCR_002798; GraphPad Software, Inc.). P<0.05 was
considered to indicate a statistically significant difference.
Results and Discussion
The involvement of the multidomain protein Vav1 in
gene transcription has been demonstrated in both myeloid leukemia
and breast cancer cells. In acute promyelocytic leukemia cells, it
has been shown that Vav1 promotes the access of the transcription
factor PU.1 to its consensus regions on DNA (16,21), and
that adequate levels of Vav1 are essential for the PU.1-mediated
expression of miRNAs, including that of miR-29b (16).
Vav1 has been found to be ectopically expressed in
breast cancer, where it has been reported to accumulate inside the
nuclear compartment of tumor cells and to be involved in gene
expression (17); these findings were
similar to those observed in leukemia cells (16,21). The
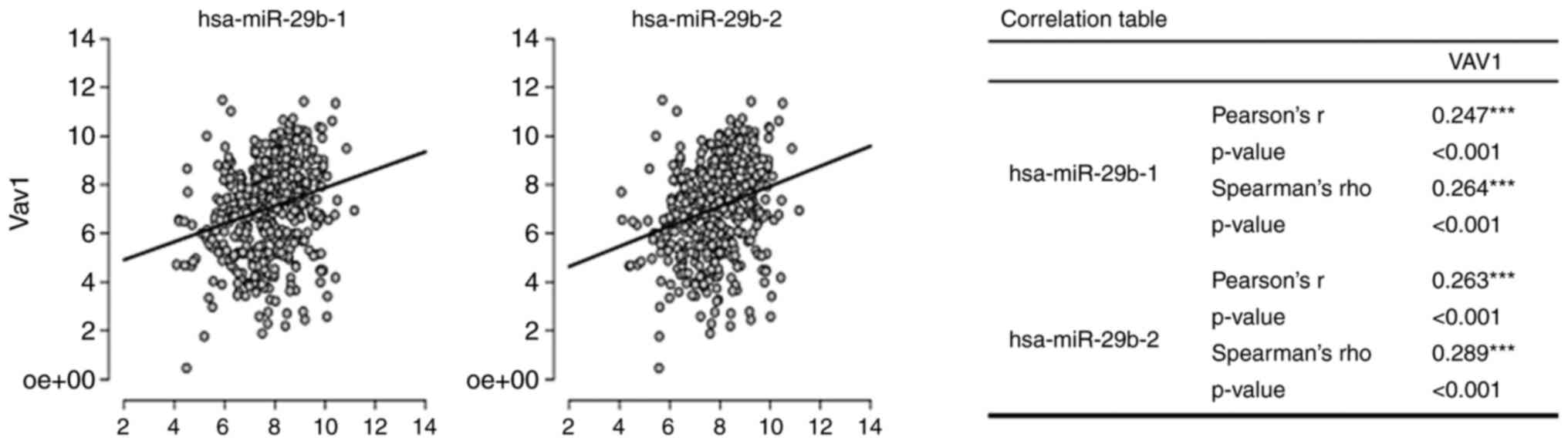
positive correlation (P<0.001) between Vav1 mRNA and the levels
of both pre-miR-29b1 and pre-miR-29b2 contributors to mature
miR-29b that was observed in the well-characterized TCGA cohort of
invasive breast tumors (Fig. 1)
suggest a potential regulatory role of Vav1 in the expression of
miR-29b in breast cancer cells, on which this miRNA has been shown
to have controversial effects (9–13, 22).
The association between Vav1 and miR-29b was first
investigated in the BT-474, MCF7, MDA-MB-453, MDA-MB-468, and
MDA-MB-231 breast cancer cell lines, with different phenotypes,
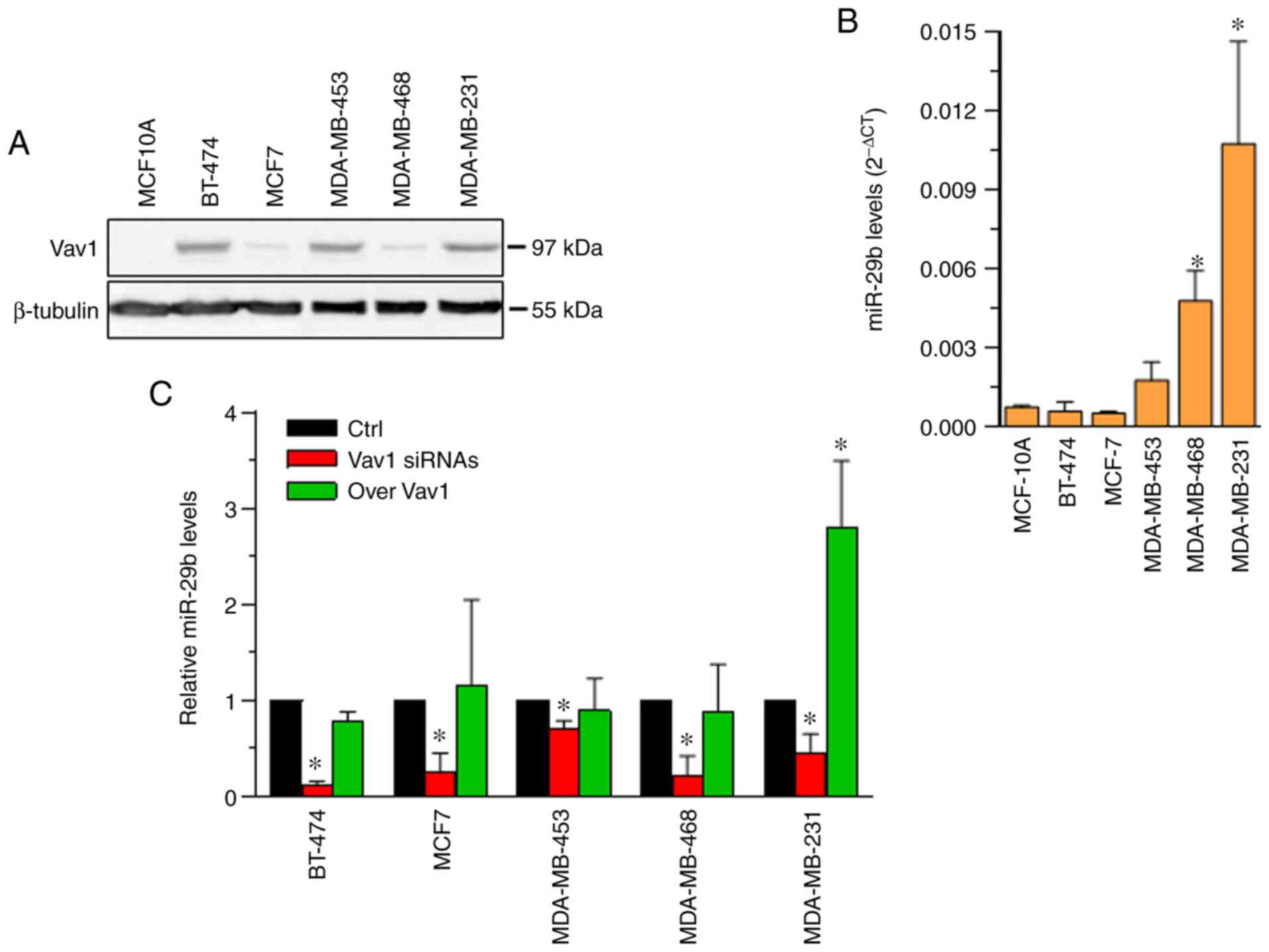
representative of the most common breast tumors (23). As expected, based on our previous
results (13,17), the examined cell lines exhibited
various levels of Vav1, which were not correlated with the tumor
phenotype, and the protein expression was absent in the
non-malignant MCF10A cell line (Fig.
2A). At variance with Vav1 expression, the level of miR-29b was
significantly higher in cell lines with a triple-negative phenotype
(MDA-MB-468 and MDA-MB-231), as compared with the non-malignant
MCF10A cell line; while the expression of the miRNA remained low in
the luminal BT-474 (luminal B) and MCF7 (luminal A) cell lines
(Fig. 2B).
Regardless of the apparent lack of a correlation
between the basal levels of the two molecules, Vav1 was found to be
essential for the expression of miR-29b in all examined cell lines
(Fig. 2C), which suggest a crucial
role of this multidomain protein in the mechanism(s) leading to the
production of mature miRNA in breast cancer cells, independent of
their phenotype. However, the forced expression of Vav1 induced
miR-29b only in MDA-MB-231 cells (Fig.
2C), suggesting a phenotype-related mechanism involving Vav1 in
the expression of this miRNA in breast cancer. Despite the fact
that no experimental evidence or predictive analysis has suggested
Vav1 as a direct target of miR-29b, mimic and inhibitors were used
in the present study to exclude any effects of the miRNA on Vav1
levels (data not shown).
Since Vav1 is involved in miRNA expression as a
facilitator of transcription factors in AML cells (16,21), the
role of Vav1 in regulating miR-29b in breast cancer cells was
explored at the transcriptional level. The investigation excluded
the transcription factor PU.1, with which Vav1 acts synergistically
in AML cells (16,21,24), as it
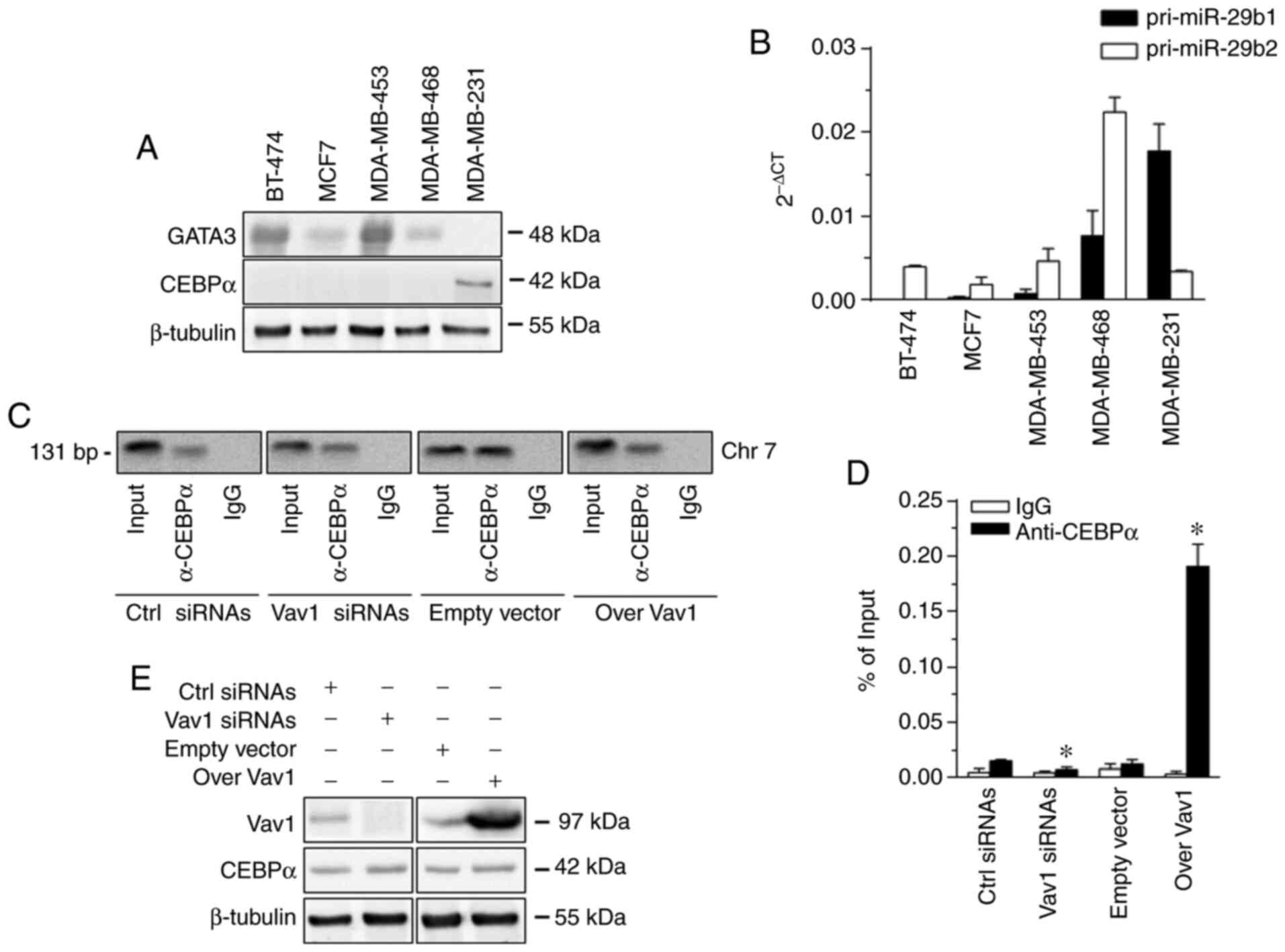
was not expressed in the examined cell lines (25). GATA binding protein 3 (GATA3), the
only transcription factor known to regulate miR-29b in breast tumor
cells (15), was also excluded from
the investigation, since it was found to be expressed in the cell
lines examined except in MDA-MB-231 (Fig.
3A), confirming previous data in MCF7 and MDA-MB-231 cell lines
(26). Therefore, CCAAT enhancer
binding protein α (CEBPα), the main regulator of miR-29b in
hematopoietic cells, which interacts specifically with its promoter
on chromosome 7 and is responsible for the transcription of the
miR-29a/b1 locus (27), was taken
into consideration. It was revealed that, of the examined cell
lines, only MDA-MB-231 exhibited a low expression of CEBPα
(Fig. 3A), which, substantiating the
high expression of the pri-miR-29b1 observed in this cell line
(Fig. 3B), was actually recruited by
the miR-29b promoter on Chr 7 (Fig.
3C). The overexpression of Vav1 in MDA-MB-231 cells induced an
association between CEBPα and DNA, which was significantly reduced
by the silencing of the protein (Fig.
3D), justifying the effects of the upregulation and
downregulation of Vav1 on miR-29b levels in this cell line. The
evaluation of CEBPα expression in MDA-MB-231 cells, in which Vav1
was forcedly regulated, excluded any effects of Vav1 on this
transcription factor (Fig. 3E).
These data suggest that, in breast cancer cells,
similar to AML cells, Vav1 can regulate the interaction of
transcription factors with their DNA consensus sequences. In
particular, they highlight the specific role of Vav1 in regulating
the CEBPα-dependent expression of miR-29b in MDA-MB-231 cells,
while clearly suggesting the existence of other mechanisms through
which Vav1 supports the production of miR-29b in breast tumor
cells. Furthermore, the lack of effects of Vav1 overexpression on
the miR-29b levels in MDA-MB-453 and MDA-MB-468 cells, which
exhibited a triple-negative phenotype (28) but a lack of CEBPα (Fig. 3A), support the Vav1/CEBPα cooperation
but highlight the need to examine this phenomenon in depth in TNBC,
which is highly heterogeneous (29)
and characterized by controversial roles of miR-29b (9–13).
The study was then extended to triple-negative cell
lines selected on the basis of their molecular subtype, according
to the Lehmann classification (30)
which, considering age at diagnosis and local and distant disease
progression, may prove useful for identifying appropriate targeted
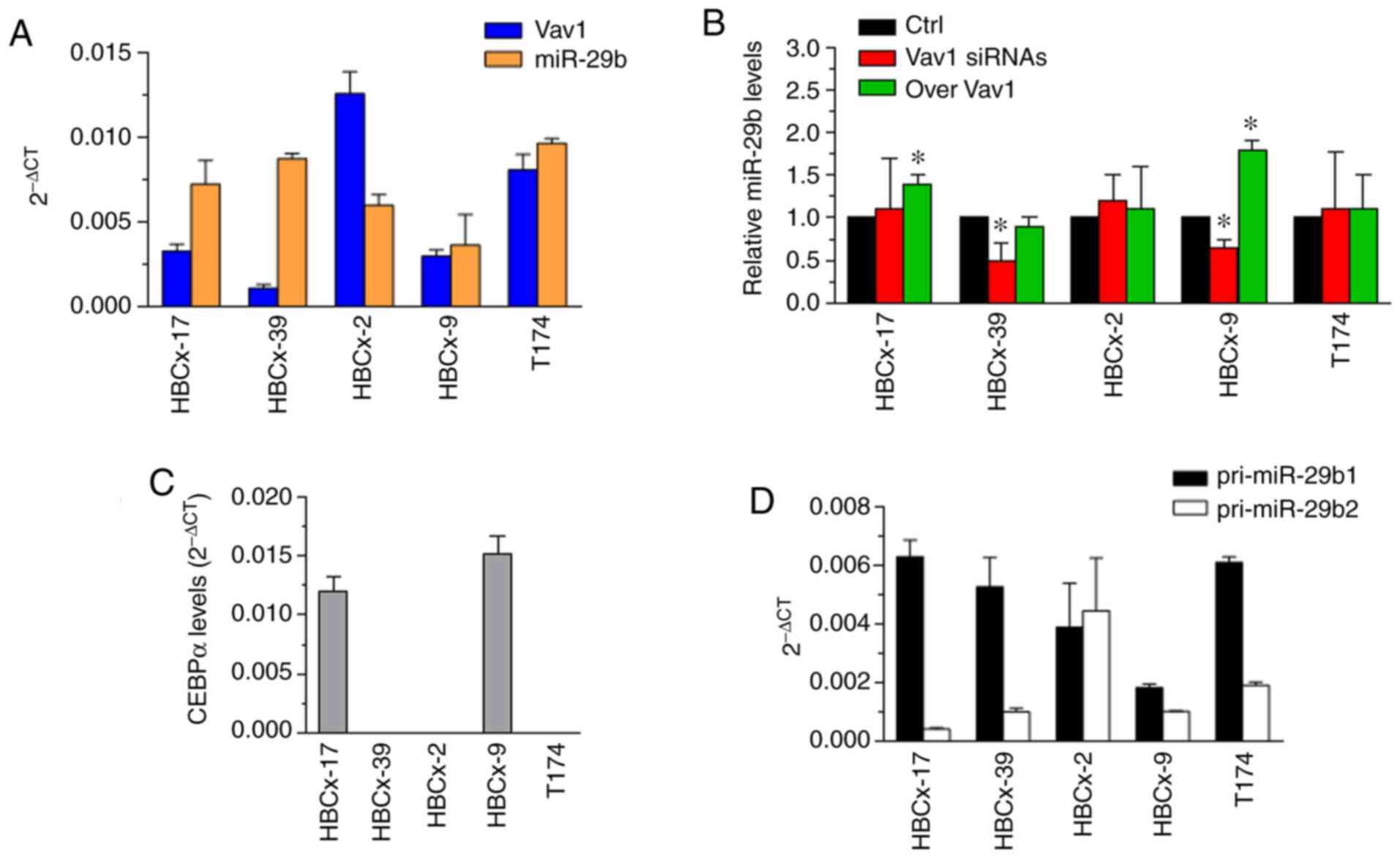
therapies for patients with TNBC (30). The HBCx-2, HBCx-9, HBCx-17, HBCx-39
and T174 PDX-derived TNBC cell lines of the mesenchymal (M),
basal-like 1 (BL1), basal-like 2 (BL2) and luminal androgen
receptor (LAR) subtypes, which expressed various and clearly not
correlated basal levels of Vav1 and miR-29b (Fig. 4A), were investigated to determine the
association between Vav1 and miR-29b. The overexpression of Vav1
(Fig. S1A) was found to be
sufficient to increase miR-29b levels only in the HBCx-9 and
HBCx-17 cell lines (Fig. 4B), which
were characterized by a mesenchymal phenotype and expressed CEBPα
(Fig. 4C), similar to the MDA-MB-231
cell line, which also had a mesenchymal molecular phenotype
(2,28). Possibly due to the low amount of Vav1
in these cell lines, its silencing (Fig.
S1B) allowed to reveal a significant decrease in miR-29b only
in the HBCx-9 cells (Fig. 4B).
The overexpression of Vav1 was ineffective in the
other examined PDX-derived cell lines that did not express the
transcription factor (Fig. 4B and C),
supporting the existence of a Vav1/CEBPα cooperation in TNBC cells.
On the other hand, the silencing of Vav1 reduced the levels of
miR-29b (Fig. 4B) in PDX-derived
cells with a BL1 phenotype (HBCx-39) that did not express CEBPα
(Fig. 4C); this finding was similar
to that in the MDA-MB-468 cell line, which is also classified as
BL1 (28). No effects of Vav1 on
miR-29b were observed in PDX-derived cell lines of the less
frequent BL2 and LAR molecular subtypes (Fig. 4B), which expressed relatively high
basal levels of the two molecules (Fig.
4A) and were negative for CEBPα (Fig.
4C). This bulk of data clearly indicate that, in TNBC cells,
Vav1 plays a phenotype-specific role, which is mainly correlated
with the expression of CEBPα but also suggests that Vav1 may
sustain the activity of other transcription factors regulating
miR-29b. This latter hypothesis was supported by the presence of
both miR-29b primary transcripts in all examined TNBC cell lines
(Figs. 3B and 4D), indicating that transcription factors
other than CEBPα, which is only responsible for miR-29b1
expression, are involved in positive or negative transcriptional
regulation of this miRNA in breast cancers. Given the multiple
roles that Vav1 can play in cells, both cytoplasmic and nuclear,
and the controversial role of miR-29b in breast cancer that takes
into account the 3p and 5p variants, our results were unable to
establish the functional meaning of the Vav1/miR-29b axis in the
different subtypes of breast cancer.
Considering that miR-29b affects multiple oncogenic
characteristics of breast tumors, a better knowledge of the
machinery that regulates its expression could constitute an
important contribution to the management of this type of cancer.
Although the complex positive and negative regulation of miR-29b
precursors requires further study to understand the mechanisms
involved, the present results indicated that, in breast cancer
cells, Vav1 is involved in the regulation of miR-29b levels at the
transcriptional level. Of note, the forced modulation of Vav1
expression affects the levels of mature miR-29b in specific
molecular subtypes of TNBC, suggesting that approaches targeting
Vav1 may be useful in tumor subsets for which there is lack of
targeted therapies, and the patient prognosis is generally
unfavorable despite the high chemosensitivity.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
This research study was supported by grants from the
University of Ferrara (Italy) to VB.
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
VB was responsible for the study concept, and
supervised all the experiments. VB, SC, MDM and JGJ integrated the
results. SG, FV and FB performed experiments and prepared the
figures. SV performed the statistical analysis. VB and MDM drafted
the manuscript. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
DNMT
|
DNA methyltransferase
|
|
TNBC
|
triple-negative breast cancer
|
|
AML
|
acute myeloid leukemia
|
|
RT-qPCR
|
reverse transcription quantitative
PCR
|
|
Q-ChIP
|
quantitative chromatin
immunoprecipitation
|
References
|
1
|
Qi Y, Huang Y, Pang L, Gu W, Wang N, Hu J,
Cui X, Zhang J, Zhao J, Liu C, et al: Prognostic value of the
microRNA-29 family in multiple human cancers: A meta- analysis and
systematic review. Clin Exp Pharmacol Physiol. 44:441–454. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kwon JJ, Factora TD, Dey S and Kota J: A
systematic review of miR-29 in cancer. Mol Ther Oncolytics.
12:173–194. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sandhu R, Rivenbark AG, Mackler RM, Livasy
CA and Coleman WB: Dysregulation of microRNA expression drives
aberrant DNA hypermethylation in basal-like breast cancer. Int J
Oncol. 44:563–572. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Muluhngwi P, Alizadeh-Rad N, Vittitow SL,
Kalbfleisch TS and Klinge CM: The miR-29 transcriptome in
endocrine- sensitive and resistant breast cancer cells. Sci Rep.
7:52052017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Darbeheshti F, Rezaei N, Amoli MM,
Mansoori Y and Tavakkoly Bazzaz J: Integrative analyses of triple
negative dysregulated transcripts compared with non-triple negative
tumors and their functional and molecular interactions. J Cell
Physiol. 234:22386–22399. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Shinden Y, Iguchi T, Akiyoshi S, Ueo H,
Ueda M, Hirata H, Sakimura S, Uchi R, Takano Y, Eguchi H, et al:
MiR-29b is an indicator of prognosis in breast cancer patients. Mol
Clin Oncol. 3:919–923. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Papachristopoulou G, Papadopoulos EI,
Nonni A, Rassidakis GZ and Scorilas A: Expression analysis of
miR-29b in malignant and benign breast tumors: A promising
prognostic biomarker for invasive ductal carcinoma with a possible
histotype-related expression status. Clin Breast Cancer.
18:305–312.e3. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Milevskiy MJ, Sandhu GK, Wronski A, Korbie
D, Brewster BL, Shewan A, Edwards SL, French JD and Brown MA:
MiR-29b-1-5p is altered in BRCA1 mutant tumours and is a biomarker
in basal-like breast cancer. Oncotarget. 9:33577–33588. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Drago-Ferrante R, Pentimalli F, Carlisi D,
De Blasio A, Saliba C, Baldacchino S, Degaetano J, Debono J,
Caruana-Dingli G, Grech G, et al: Suppressive role exerted by
microRNA-29b-1-5p in triple negative breast cancer through SPIN1
regulation. Oncotarget. 8:28939–28958. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
De Blasio A, Di Fiore R, Pratelli G,
Drago-Ferrante R, Saliba C, Baldacchino S, Grech G, Scerri C, Vento
R and Tesoriere G: A loop involving NRF2, miR-29b-1-5p and AKT,
regulates cell fate of MDA-MB-231 triple-negative breast cancer
cells. J Cell Physiol. 235:629–637. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wang C, Bian Z, Wei D and Zhang JG:
miR-29b regulates migration of human breast cancer cells. Mol Cell
Biochem. 352:197–207. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wang H, An X, Yu H, Zhang S, Tang B, Zhang
X and Li Z: MiR-29b/TET1/ZEB2 signaling axis regulates metastatic
properties and epithelial-mesenchymal transition in breast cancer
cells. Oncotarget. 8:102119–102133. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang B, Shetti D, Fan C and Wei K:
miR-29b-3p promotes progression of MDA-MB-231 triple-negative
breast cancer cells through downregulating TRAF3. Biol Res.
52:382019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kollinerova S, Vassanelli S and Modriansky
M: The role of miR-29 family members in malignant hematopoiesis.
Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub. 158:489–501.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chou J, Lin JH, Brenot A, Kim JW, Provot S
and Werb Z: GATA3 suppresses metastasis and modulates the tumour
microenvironment by regulating microRNA-29b expression. Nat Cell
Biol. 15:201–213. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
16
|
Vezzali F, Grassilli S, Lambertini E,
Brugnoli F, Patergnani S, Nika E, Piva R, Pinton P, Capitani S and
Bertagnolo V: Vav1 is necessary for PU.1 mediated upmodulation of
miR-29b in acute myeloid leukaemia-derived cells. J Cell Mol Med.
22:3149–3158. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Grassilli S, Brugnoli F, Lattanzio R,
Rossi C, Perracchio L, Mottolese M, Marchisio M, Palomba M, Nika E,
Natali PG, et al: High nuclear level of Vav1 is a positive
prognostic factor in early invasive breast tumors: A role in
modulating genes related to the efficiency of metastatic process.
Oncotarget. 5:4320–4336. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Grassilli S, Brugnoli F, Lattanzio R,
Marchisio M, Perracchio L, Piantelli M, Bavelloni A, Capitani S and
Bertagnolo V: Vav1 downmodulates Akt in different breast cancer
subtypes: A new promising chance to improve breast cancer outcome.
Mol Oncol. 12:1012–1025. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Bertagnolo V, Grassilli S, Volinia S,
Al-Qassab Y, Brugnoli F, Vezzali F, Lambertini E, Palomba M,
Piubello Q, Orvieto E, et al: Ectopic expression of PLC-β2 in
non-invasive breast tumor cells plays a protective role against
malignant progression and is correlated with the deregulation of
miR-146a. Mol Carcinog. 58:708–721. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Grassilli S, Nika E, Lambertini E,
Brugnoli F, Piva R, Capitani S and Bertagnolo V: A network
including PU.1, Vav1 and miR-142-3p sustains ATRA-induced
differentiation of acute promyelocytic leukemia cells-a short
report. Cell Oncol (Dordr). 39:483–489. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yan B, Guo Q, Fu FJ, Wang Z, Yin Z, Wei YB
and Yang JR: The role of miR-29b in cancer: Regulation, function,
and signaling. Onco Targets Ther. 8:539–548. 2015.PubMed/NCBI
|
|
23
|
Dai X, Cheng H, Bai Z and Li J: Breast
cancer cell line classification and its relevance with breast tumor
subtyping. J Cancer. 8:3131–3141. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Brugnoli F, Lambertini E, Varin-Blank N,
Piva R, Marchisio M, Grassilli S, Miscia S, Capitani S and
Bertagnolo V: Vav1 and PU.1 are recruited to the CD11b promoter in
APL-derived promyelocytes: Role of Vav1 in modulating
PU.1-containing complexes during ATRA-induced differentiation. Exp
Cell Res. 316:38–47. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
He J, Pan Y, Hu J, Albarracin C, Wu Y and
Dai JL: Profile of Ets gene expression in human breast carcinoma.
Cancer Biol Ther. 6:76–82. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yan W, Cao QJ, Arenas RB, Bentley B and
Shao R: GATA3 inhibits breast cancer metastasis through the
reversal of epithelial-mesenchymal transition. J Biol Chem.
285:14042–14051. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Eyholzer M, Schmid S, Wilkens L, Mueller
BU and Pabst T: The tumour-suppressive miR-29a/b1 cluster is
regulated by CEBPA and blocked in human AML. Br J Cancer.
103:275–284. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Lehmann BD, Bauer JA, Chen X, Sanders ME,
Chakravarthy AB, Shyr Y and Pietenpol JA: Identification of human
triple-negative breast cancer subtypes and preclinical models for
selection of targeted therapies. J Clin Invest. 121:2750–2767.
2011. View
Article : Google Scholar : PubMed/NCBI
|
|
29
|
Garrido-Castro AC, Lin NU and Polyak K:
Insights into molecular classifications of triple-negative breast
cancer: Improving patient selection for treatment. Cancer Discov.
9:176–198. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Lehmann BD, Jovanovic B, Chen X, Estrada
MV, Johnson KN, Shyr Y, Moses HL, Sanders ME and Pietenpol JA:
Refinement of triple-negative breast cancer molecular subtypes:
Implications for neoadjuvant chemotherapy selection. PLoS One.
11:e01573682016. View Article : Google Scholar : PubMed/NCBI
|