Introduction
Pancreatic cancer has a 5-year survival rate less
than 8% and is one of the deadliest types of cancer worldwide.
Pancreatic ductal adenocarcinoma (PDAC) is the most common
pathological type of pancreatic cancer (1–4). For
patients with advanced stage PDAC, therapeutic options are limited,
and their prognosis is extremely poor (1–4). Thus,
there is an urgent need to improve the current understanding of the
mechanisms underlying progression of PDAC to identify novel
therapeutic targets.
Long non-coding RNAs (lncRNAs) are a type of RNA
molecule of >200 nucleotides in length, which have limited or no
protein-coding capabilities (5).
Previously, it was hypothesized that lncRNAs were transcriptional
noise, and that they did not possess any biological function
(5). However, in the last decade, a
growing number of studies have demonstrated that lncRNAs
participate in a range of cellular biological processes, including
cell proliferation, migration, differentiation and apoptosis
(6–8).
lncRNAs also exhibit crucial roles in the development and/or
progression of cancers. For example, knockdown of lncRNA actin
filament associated protein 1-antisense RNA 1 was found to impede
the proliferation and cell cycle progression of colon cancer cells
(9). It has been reported that lncRNA
TMPO antisense RNA 1 (TMPO-AS1) expression is upregulated in
bladder cancer tissues and cells, where it promotes cell growth,
migration and invasion (10). In
non-small lung cancer, knockdown of lncRNA colon cancer associated
transcript 1 suppressed cancer cell proliferation and sensitized
cancer cells to gefitinib (11).
lncRNA PC-esterase domain containing 1B antisense RNA 1
(PCED1B-AS1) was found to be involved in the regulation of
macrophage apoptosis and autophagy in active tuberculosis (12,13);
additionally, it has been demonstrated that PCED1B-AS1 is
abnormally expressed in gliomas and breast cancer tissues, where it
functions as an oncogenic lncRNA (14,15).
However, its biological function, mechanistic partners and clinical
value in PDAC have not been assessed.
lncRNAs can function as competitive endogenous RNAs
(ceRNAs), competitively interacting with microRNAs (miRNAs/miRs)
and indirectly regulating the expression of target genes (16,17). For
example, lncRNA Pvt1 oncogene acts as a molecular sponge, absorbing
miR-448 and upregulating SERPINE1 mRNA binding protein 1, thus
promoting pancreatic cancer cell proliferation and migration
(18). It has also been reported that
lncRNA X inactive specific transcript can facilitate the migration,
invasion and epithelial-mesenchymal transition (EMT) of pancreatic
cancer cells by repressing miR-429, indirectly resulting in
upregulation of zinc finger E-box binding homeobox 1 expression
(19). Moreover, highly upregulated
in liver cancer (HULC) was found to promote the proliferation,
migration and invasion of pancreatic cancer cells by downregulating
miR-15a and activating the PI3K/AKT pathway (20). These studies suggest that lncRNAs act
as ceRNAs and participate in the progression of PDAC.
Hypoxia-inducible factor-1α (HIF-1α), a dominant
regulator of a tumor cell's response to hypoxia (21), is closely associated with the
progression and metastasis of several types of cancer, including
PDAC (22–24). In the present study, it was shown that
PCED1B-AS1 expression was significantly upregulated in PDAC tissues
and cell lines. PCED1B-AS1 overexpression facilitated the malignant
biological behaviors of cancer cells. Mechanistically, it acted as
a ceRNA of miR-411-3p, resulting in upregulation of HIF-1α. The
results of the present study clarify the mechanism by which HIF-1α
expression is dysregulated in PDAC, and identified PCED1B-AS1 as a
novel oncogenic lncRNA in PDAC.
Materials and methods
Tissue sample collection
A total of 47 pairs of PDAC tissue samples and the
corresponding adjacent normal tissues were surgically removed from
patients between January 2017 and January 2019 from The People's
Hospital of Three Gorges University and collected. The patients had
a mean age of 45 years (range, 28–77 years; 22 male and 25 female)
and did not receive any radiotherapy or chemotherapy prior to
surgery. Written informed consent was provided by each patient and
the collection of human samples was approved by the Ethics
Committee of the People's Hospital of Three Gorges University. All
tissues were stored in liquid nitrogen (−196°C).
Cell culture and transfection
Five PDAC cell lines (AsPC-1, PANC-1, CFPAC-1,
SW1990 and BxPC-3), normal human pancreatic ductal epithelial cell
line HPDE6-C7, and human embryonic kidney cell line, 293T, were all
purchased from The Cell Bank Type Culture Collection of the Chinese
Academy of Sciences. Cells were cultured in DMEM (Gibco; Thermo
Fisher Scientific, Inc.) supplemented with 10% FBS (Invitrogen;
Thermo Fisher Scientific, Inc.) in a humidified incubator at 37°C
with 5% CO2.
Small interfering (si)RNAs targeting PCED1B-AS1
(si-PCED1B-AS1), negative control siRNAs (si-NC), miR-411-3p mimic,
mimic negative control (mimic NC), miR-411-3p inhibitor, inhibitor
negative control (NC), HIF-1α overexpression plasmid, and the
negative control plasmid were purchased from Shanghai GenePharma
Co., Ltd.. According to the manufacturer's protocols, PDAC cells
were transfected using Lipofectamine® 2000 transfection
reagent (Invitrogen; Thermo Fisher Scientific, Inc.). Cells were
collected for subsequent analysis 48 h after the transfection.
Reverse transcription-quantitative
(RT-q) PCR
TRIzol® (Invitrogen; Thermo Fisher
Scientific, Inc.) was used to obtain total RNA from PDAC tissues
and cells. Total RNA was reverse transcribed into cDNA using a
PrimeScript RT kit (Takara Bio, Inc.). qPCR was performed using a
SYBR-Green PCR MasterMix kit (Takara Bio, Inc.) on an ABI 7500
real-time PCR system (Applied Biosystems; Thermo Fisher Scientific,
Inc.). Expression of PCED1B-AS1 and HIF-1α was normalized to GAPDH.
Expression of miR-411-3p was normalized to U6. The relative
expression level of each gene was quantified using the
2−ΔΔCq method (25). The
sequences of the primers are listed in Table I.
 | Table I.Sequences of the primers used for
RT-qPCR. |
Table I.
Sequences of the primers used for
RT-qPCR.
| Gene | Sequence,
5′-3′ |
|---|
| PCED1B-AS1 |
|
|
Forward |
TTTGATGTTGGCCAATGCCG |
|
Reverse |
GGGCAGGGAGTCTTCATAGC |
| HIF-1α |
|
|
Forward |
AGTAATCGGACTACCGGACGTG |
|
Reverse |
TGGGCATTACATCGCATGCATC |
| GAPDH |
|
|
Forward |
GTCAGGATCCACTCATCACG |
|
Reverse |
GATCGGACTTACGGACTCACATC |
|
microRNA-411-3p |
|
|
Forward |
TAGTAGACCGTATAGCGTACG |
| U6 |
|
|
Forward |
AAAGACCTGTACGCCAACAC |
|
Reverse |
GTCATACTCCTGCTTGCTGAT |
Dual luciferase reporter assay
A dual luciferase reporter assay was performed using
the 293T cell line. First, the target sites of miR-411-3p on
PCED1B-AS1 or HIF-1α 3′ untranslated region (3′UTR) were predicted
using bioinformatics analysis. The wild-type (WT) and mutant (MUT)
PCED1B-AS1 and HIF-1α 3′UTR regions were amplified and inserted
into a pmir-GLO luciferase reporter vector (Promega Corp.). The
recombinant plasmids PCED1B-AS1-WT, PCED1B-AS1-MUT, HIF-1α-WT and
HIF-1α-MUT were subsequently co-transfected into 293T cells with
the miR-411-3p mimic or NC mimic, respectively. After 48 h, the
luciferase activities were measured using a Dual-Luciferase
Reporter Assay system (Promega Corp.) according to the
manufacturer's protocol.
RNA-binding protein
immunoprecipitation (RIP) assay
PDAC cells transfected with miR-411-3p mimics or NC
mimics were collected, and according to the manufacturer's
protocols, RIP was performed using an anti-Ago2 antibody (EMD
Millipore) and an RIP assay kit (EMD Millipore). Mouse anti-human
immunoglobulin G (IgG) antibody was used as the control.
Subsequently, RNA was extracted using TRIzol, and the expression of
PCED1B-AS1 was assessed using RT-qPCR.
Cell Counting Kit-8 (CCK-8) assay
The viability of PDAC cells was detected using a
CCK-8 assay (Beyotime Institute of Biotechnology). CFPAC-1 and
SW1990 cells were plated into a 96-well plate. After 24, 48, 72 and
96 h, 10 µl of CCK-8 solution was added to each well, and the cells
were further incubated at 37°C for 2 h. Subsequently, the
absorbance of each well was assessed at an optical density of 450
nm using a microplate reader. A proliferation curve was plotted
with time as the abscissa and the value of absorbance as the
ordinate.
EdU staining assay
Cell proliferation was also evaluated using an EdU
assay. Transfected cells were plated in a 96-well plate
(5×103 cells/well) and cultured for 24 h. Then, 100 µl
of EdU solution (50 µM; Guangzhou RiboBio Co., Ltd.) was added to
each well, and the cells were subsequently incubated at 37°C for 2
h. Cells were washed 3 times with PBS and then fixed using
paraformaldehyde/glycine for 30 min. Cells were permeabilized using
0.5% Triton X-100, then stained with Apollo fluorescent staining
reaction solution for 30 min in the dark and washed twice with
methanol and PBS. Cells were subsequently counterstained with DAPI
staining solution for 30 min, and washed with PBS 3 times.
Fluorescence was observed using a fluorescence microscope, and the
percentage of EdU-positive cells was counted and calculated. Cell
proliferation rate=number of EdU-positive cells/number of
DAPI-positive cells ×100%.
Transwell assay
Transwell chambers (8-µm pore size; BD Biosciences)
were used to assess the invasive ability of PDAC cells. CFPAC-1 and
SW1990 cells were suspended in serum-free DMEM and added to the
upper chamber, which had been pre-coated with Matrigel. The lower
chamber was filled with 600 µl of medium supplemented with 10% FBS.
A total of 24 h after incubation at 37°C, the chambers were
removed, and the residual cells remaining on the upper surface of
the membrane were wiped off using a cotton swab. The cells which
had invaded to the lower surface of the membrane were fixed using
4% paraformaldehyde, and stained using 0.1% crystal violet for 10
min. Membranes were washed using tap water and dried, and the cells
were observed using an inverted microscope and counted. The number
of cells from five fields in each well were counted, and the
experiments were performed in triplicate.
Western blotting
PDAC cells were lysed using RIPA lysis buffer
(Beyotime Institute of Biotechnology). An equivalent amount of
protein was loaded per lane on SDS-gel (stacking gel 4%, separation
gel 10%), resolved using SDS-PAGE, transferred to PVDF membranes
(EMD Millipore) and blocked using 5% skimmed milk. Subsequently,
the membranes were incubated with the primary antibodies overnight
at 4°C. The primary antibodies used were: Anti-HIF-1α antibody
(cat. no. ab51608; 1:1,000; Abcam), anti-N-cadherin antibody (cat.
no. ab202030; 1:1,000; Abcam), anti-E-cadherin antibody (cat. no.
ab40772; 1:1,000; Abcam), anti-Vimentin antibody (cat. no. ab92547;
1:1,000; Abcam), anti-Snail antibody (cat. no. ab53519; 1:1,000;
Abcam) or anti-β-actin antibody (cat. no. ab179467; 1:2,000;
Abcam). Subsequently, the membranes were incubated with an
HRP-conjugated secondary antibody (cat. no. ab205718; 1:2,000;
Abcam) at room temperature for 1 h. Signals were visualized using
an ECL kit (Beyotime Institute of Biotechnology). Densitometry
analysis was performed using ImageJ_v1.8.0 (National Institutes of
Health).
Bioinformatics analysis
The expression pattern of PCED1B-AS1 in PAAD and
normal tissues was predicted using the Gene Expression Profiling
Interactive Analysis (GEPIA) database (http://gepia.cancer-pku.cn/) (26). The potential target miRNAs of
PCED1B-AS1 was predicted using the LncBase Predicted version 2
database (http://carolina.imis.athena-innovation.gr/diana_tools/web/index.php?r=lncbasev2%2Findex)
(27). The interaction between HIF-1α
and miR-411-3p was predicted using the TargetScan database
(http://www.targetscan.org/vert_72/)
(28).
Statistical analysis
Statistical analysis was performed using SPSS
version 21.0 (IBM Corp.). Data are presented as the mean ± standard
deviation of at least three independent experiments. Distribution
of the data was examined using a Kolmogorov-Smirnov test. A
two-tailed student's t-test was used to determine the differences
between two groups. A one-way ANOVA with Tukey's post-hoc test was
used to determine the differences among ≥3 groups. For data that
were not normally distributed, comparison of expression in PDAC
tissue samples and the corresponding adjacent normal tissues was
performed using a paired sample Wilcoxon signed-rank test. A
χ2 test was used to analyze the association between the
expression of PCED1B-AS1 and the clinicopathological
characteristics of patients with PDAC. Pearson's correlation
coefficient analysis was utilized to determine the correlation
between PCED1B-AS1 expression and miR-411-3p expression or HIF-1α
expression.
Results
PCED1B-AS1 expression is upregulated
in pancreatic cancer tissues and cells
Using the Gene Expression Profiling Interactive
Analysis (GEPIA) database, 171 cases of normal tissues and 179
cases of cancerous tissues were compared. PCED1B-AS1 expression was
significantly higher in the 179 PDAC tissues (Fig. 1A). To confirm the upregulation of
PCED1B-AS1 in PDAC tissues, the expression of PCED1B-AS1 in PDAC
tissues and corresponding non-tumor tissues was further examined
using RT-qPCR. Compared with the corresponding non-tumor tissues,
the expression of PCED1B-AS1 was upregulated in PDAC tissues
(Fig. 1B). To assess the association
between the expression of PCED1B-AS1 and the clinicopathological
characteristics of 47 patients with PDAC, patients were divided
into a high expression group (n=25) and low expression group
(n=22), based on the median expression level of PCED1B-AS1. The
results demonstrated that increased expression of PCED1B-AS1 was
positively correlated with advanced TNM stage (stage III–IV) and
lymph node metastasis (Table II).
Additionally, the expression levels of PCED1B-AS1 in PDAC cell
lines (AsPC-1, PANC-1, CFPAC-1, SW1990 and BxPC-3) was
significantly higher compared with the normal pancreatic ductal
epithelial cell line HPDE6-C7 (Fig.
1C). Among the five PDAC cell lines, PCED1B-AS1 expression was
highest in CFPAC-1 and SW1990 cells, thus these two cell lines were
used for subsequent experiments.
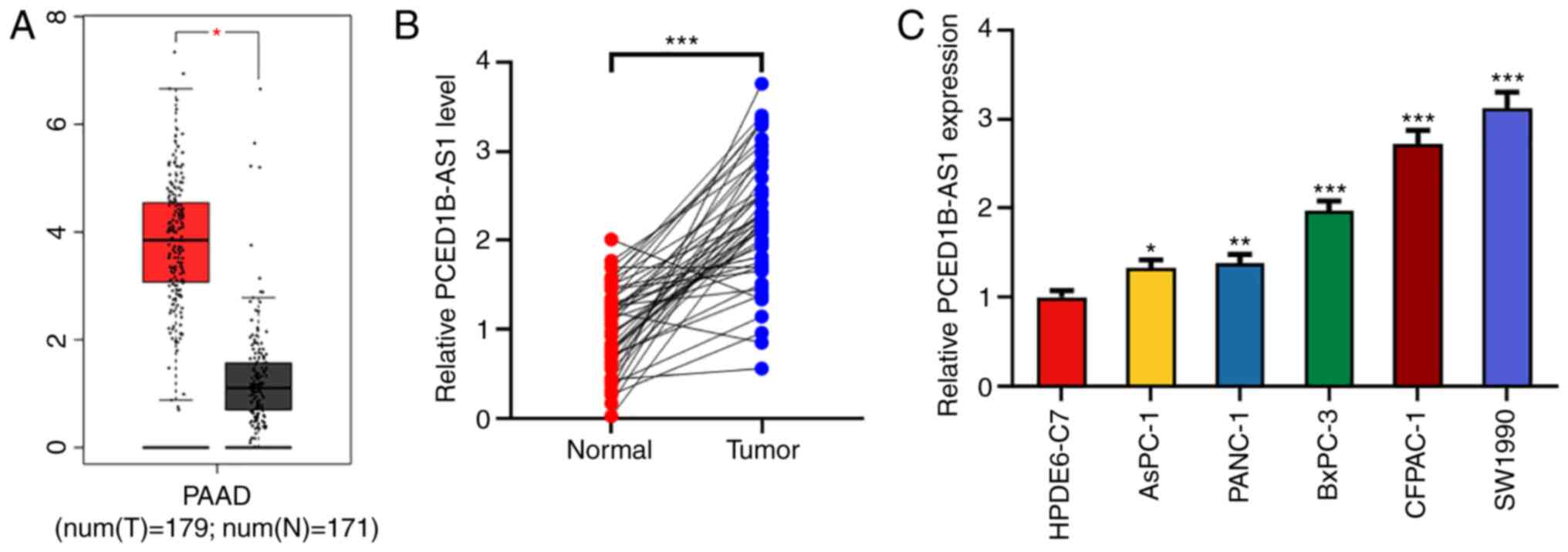 | Figure 1.PCED1B-AS1 is upregulated in PDAC
tissues and cells. (A) Expression levels of PCED1B-AS1 in the PDAC
tissues based on data obtained from GEPIA. (B) Relative expression
levels of PCED1B-AS1 in 47 cases of PDAC tissues and the
corresponding non-tumor tissues were detected using RT-qPCR. (C)
RT-qPCR was used to detect the expression levels of PCED1B-AS1 in
the normal human pancreatic ductal epithelial cell line HPDE6-C7
and the five PDAC cell lines, AsPC-1, PANC-1, CFPAC-1, SW1990 and
BxPC-3. *P<0.05, **P<0.01, ***P<0.001 vs. normal tissues
or the HPDE6-C7 cell line. GEPIA, Gene Expression Profiling
Interactive Analysis; PDAC, pancreatic ductal adenocarcinoma;
PCED1B-AS1, PC-esterase domain containing 1B-antisense RNA 1;
RT-qPCR, reverse transcription-quantitative PCR. |
 | Table II.Correlation between PCED1B-AS1
expression levels and the clinicopathologic characteristics of the
47 patients with PDAC. |
Table II.
Correlation between PCED1B-AS1
expression levels and the clinicopathologic characteristics of the
47 patients with PDAC.
|
|
| PCED1B-AS1
expression |
|
|---|
|
|
|
|
|
|---|
| Clinicopathological
characteristic | n | High, n=25 | Low, n=22 | P-value |
|---|
| Age, years |
|
|
| 0.282 |
|
≥60 | 21 | 13 | 8 |
|
|
<60 | 26 | 12 | 14 |
|
| Sex |
|
|
| 0.106 |
|
Male | 22 | 15 | 8 |
|
|
Female | 25 | 10 | 14 |
|
| Tumor size, cm |
|
|
| 0.119 |
|
>2 | 27 | 17 | 10 |
|
| ≤2 | 20 | 8 | 12 |
|
|
Differentiation |
|
|
| 0.191 |
|
Poor | 24 | 15 | 9 |
|
|
Moderate and well | 23 | 10 | 13 |
|
|
Tumor-Node-Metastasis stage |
|
|
| 0.028a |
|
I+II | 14 | 4 | 10 |
|
|
III+IV | 33 | 21 | 12 |
|
| Distant
metastasis |
|
|
| 0.118 |
|
Negative | 33 | 20 | 13 |
|
|
Positive | 14 | 5 | 9 |
|
| Lymph node
metastasis |
|
|
| 0.0141a |
|
Absent | 21 | 7 | 14 |
|
|
Present | 26 | 18 | 8 |
|
PCED1B-AS1 knockdown reduces
proliferation, invasion and EMT of pancreatic cancer cells
To further study the biological function of
PCED1B-AS1 on the progression of PDAC, si-PCED1B-AS1 was
transfected into CFPAC-1 and SW1990 cells to knockdown its
expression. Compared with the si-NC group, transfection of
si-PCED1B-AS1 significantly reduced the expression of PCED1B-AS1 in
CFPAC-1 and SW1990 cells, suggesting that PCED1B-AS1 was
successfully knocked down in CFPAC-1 and SW1990 cells (Fig. 2A). The results of the CCK-8 and EdU
assay revealed that the PCED1B-AS1 knockdown significantly reduced
PDAC cell proliferation (Fig. 2B and
C). Transwell invasion assays and western blotting were used to
determine the effect of PCED1B-AS1 knockdown on cell invasion and
EMT. Compared with cells transfected with si-NC, CFPAC-1 and SW1990
cells transfected with si-PCED1B-AS1 exhibited significantly
reduced invasion (Fig. 2D).
Furthermore, knockdown of PCED1B-AS1 significantly increased the
expression of E-cadherin, and significantly reduced the expression
of N-cadherin, Vimentin and Snail in both PDAC cell lines (Fig. 2E).
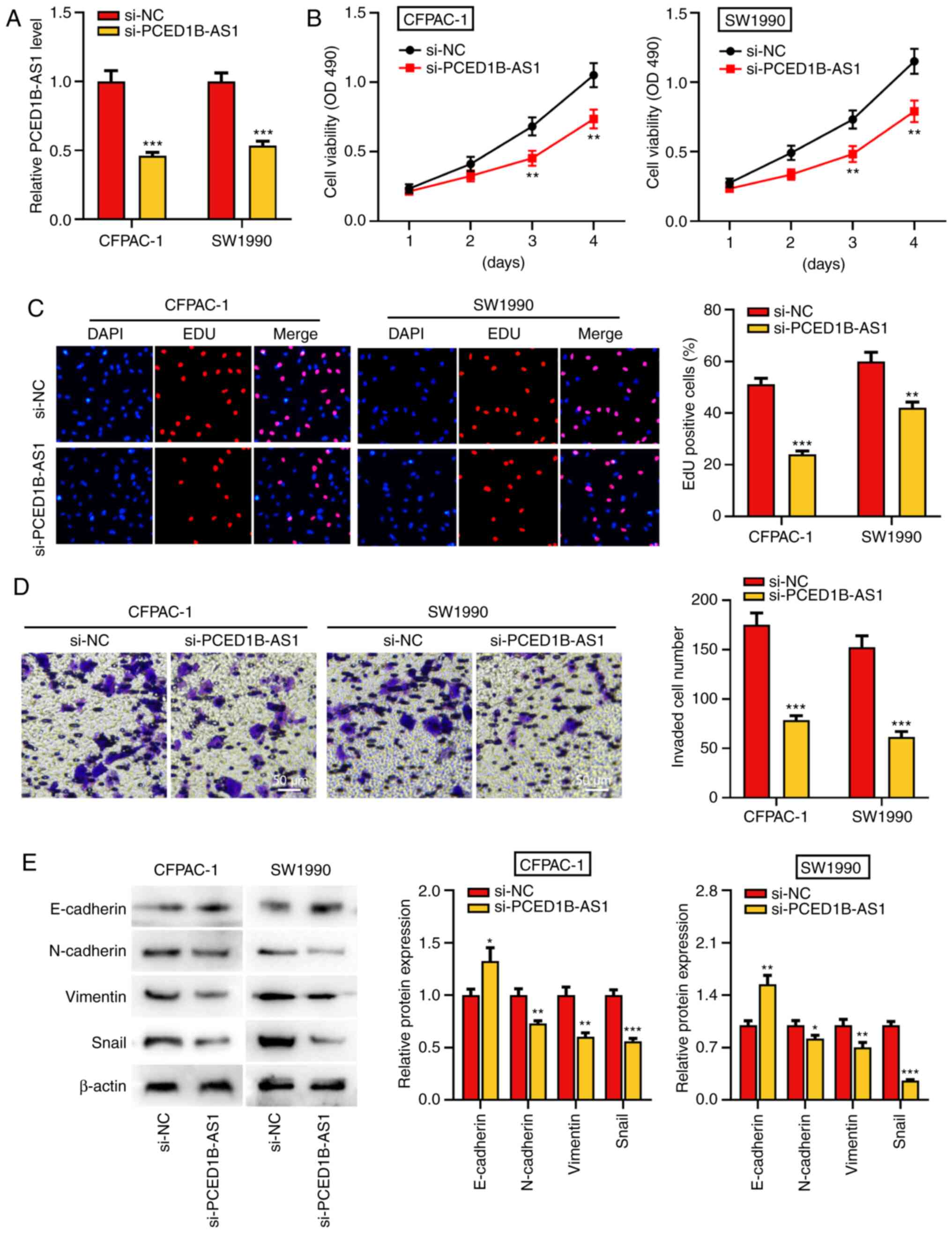 | Figure 2.PCED1B-AS1 knockdown represses PDAC
cell proliferation, invasion, and EMT. (A) RT-qPCR was utilized to
investigate the expression of PCED1B-AS1 in CFPAC-1 and SW1990
cells transfected with si-NC or si-PCED1B-AS1. (B and C) CCK-8 and
EdU staining assay were used to assess the effect of PCED1B-AS1
knockdown on proliferation of CFPAC-1 and SW1990 cells. (D)
Transwell invasion assays were used to assess the effects of
PCED1B-AS1 knockdown on the invasion of CFPAC-1 and SW1990 cells.
(E) Western blotting was used to assess the expression of the EMT
markers, E-cadherin, N-cadherin, Vimentin and Snail, following
transfection. *P<0.05, **P<0.01, ***P<0.001 vs. si-NC.
siRNA, small interfering RNA; si-NC, si-negative control; PDAC,
pancreatic ductal adenocarcinoma; PCED1B-AS1, PC-esterase domain
containing 1B-antisense RNA 1; RT-qPCR, reverse
transcription-quantitative PCR; EMT, epithelial-mesenchymal
transition; CCK-8, Cell Counting Kit-8. |
PCED1B-AS1 negatively regulates
miR-411-3p expression
To elucidate whether PCED1B-AS1 acts as a ceRNA in
the progression of PDAC, the online bioinformatics tool LncBase
Predicted version 2 was used to predict the potential target miRNAs
of PCED1B-AS1 (Table SI). The
results indicated that miR-411-3p was a potential target of
PCED1B-AS1 (Fig. 3A). The subcellular
localization of PCED1B-AS1 in CFPAC-1 and SW1990 cells was then
determined. RT-qPCR results showed that PCED1B-AS1 was primarily
expressed in the cytoplasm of CFPAC-1 and SW1990 cells (Fig. 3B). Subsequently, dual luciferase
reporter assays and RIP experiments were used to confirm the
targeted binding. The results showed that miR-411-3p mimics could
reduce the luciferase activity of cells in the PCED1B-AS1-WT group,
but had no significant effect on the PCED1B-AS1-MUT group (Fig. 3C). RIP analysis further confirmed
increased enrichment of miR-411-3p and PCED1B-AS1 in the
Ago2-immunoprecipitation complex (Figs.
3D and S1). Additionally,
compared with the non-tumor tissues, miR-411-3p was significantly
downregulated in PDAC tissues (Fig.
3E). The expression levels of PCED1B-AS1 in PDAC tissues was
negatively correlated with the expression levels of miR-411-3p
(Fig. 3F). Compared with the si-NC
transfected control group, PCED1B-AS1 knockdown significantly
increased the expression of miR-411-3p in PDAC cell lines (Fig. 3G). These results suggest that
PCED1B-AS1 can effectively reduce the expression of miR-411-3p.
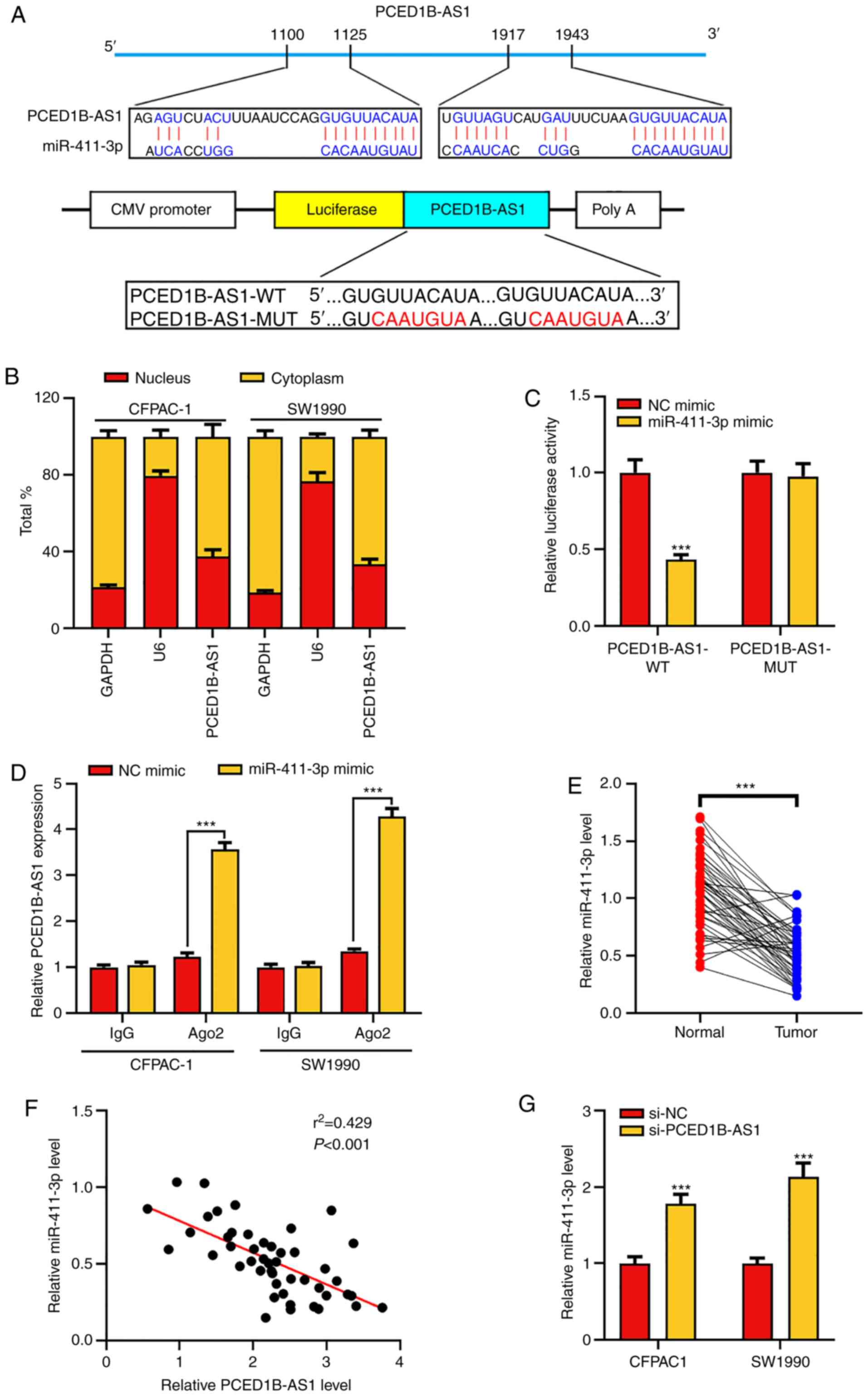 | Figure 3.PCED1B-AS1 sponges miR-411-3p. (A)
Predicted binding sites between miR-411-3p and PCED1B-AS1, and the
WT and MUT sequences. (B) Expression of PCED1B-AS1 in the nuclei
and cytoplasm of CFPAC-1 and SW1990 cells was evaluated using
RT-qPCR. (C) 293T cells were co-transfected with miR-411-3p or NC
mimic and luciferase reporter vectors containing PCED1B-AS1 WT or
MUT. The relative luciferase activity of cells was measured. (D)
Direct binding between miR-411-3p and PCED1B-AS1 in CFPAC-1 and
SW1990 cells was examined using RIP experiments. (E) Relative
expression levels of miR-411-3p in the 47 PDAC tissues and the
corresponding non-tumor tissues were detected using RT-qPCR. (F)
Correlation analysis of miR-411-3p and PCED1B-AS1 expression in the
47 PDAC patients was analyzed using Pearson's correlation analysis.
(G) RT-qPCR was used to investigate the expression of miR-411-3p in
CFPAC-1 and SW1990 cells transfected with si-NC or si-PCED1B-AS1.
***P<0.001. siRNA, small interfering RNA; si-NC, si-negative
control; PDAC, pancreatic ductal adenocarcinoma; PCED1B-AS1,
PC-esterase domain containing 1B-antisense RNA 1; RT-qPCR, reverse
transcription-quantitative PCR; WT, wild-type; MUT, mutant; RIP,
RNA immunoprecipitation; miR, microRNA. |
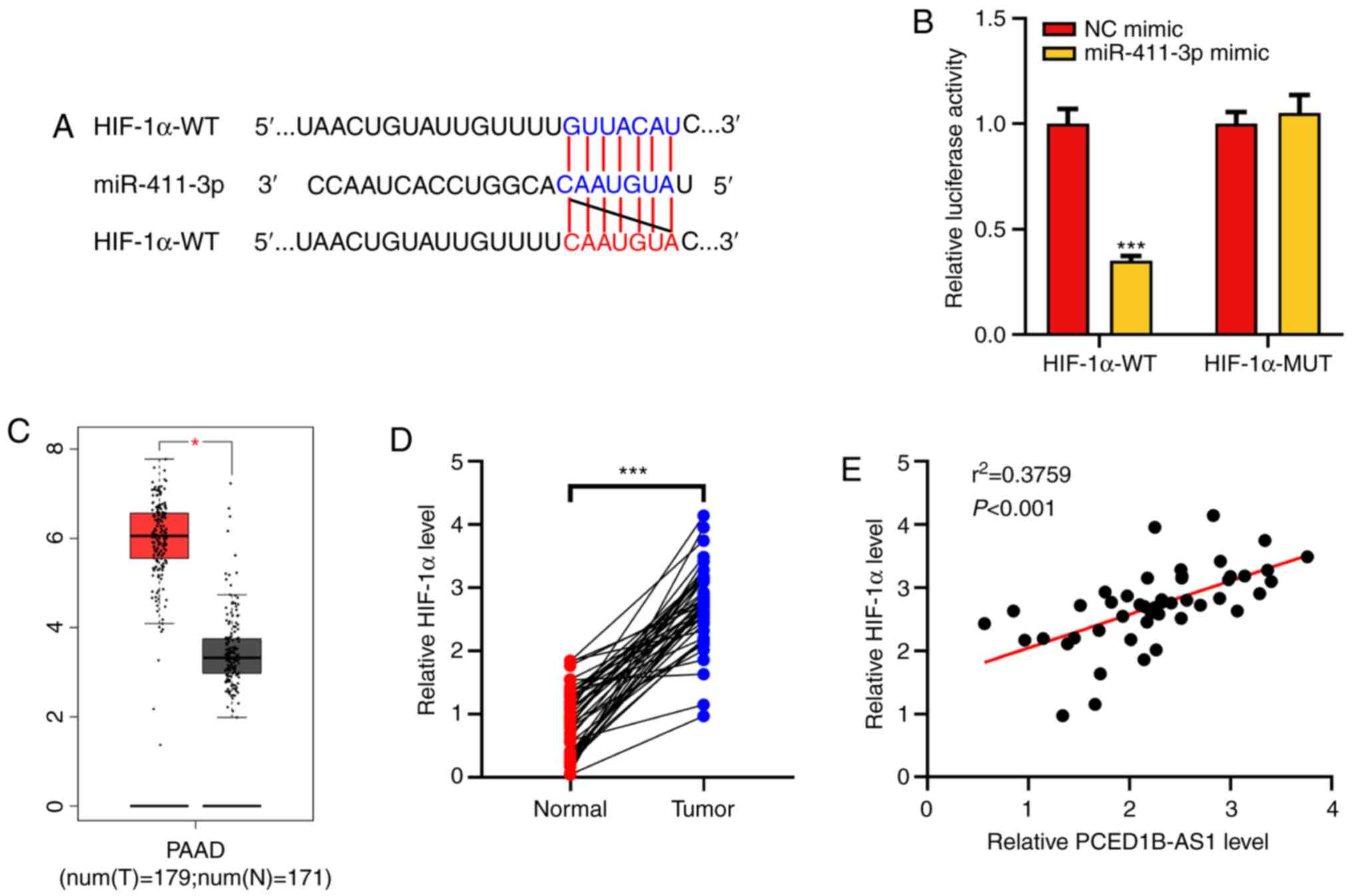
HIF-1α is a target gene of
miR-411-3p
HIF-1α was predicted as a potential target for
miR-411-3p using the TargetScan database (Fig. 4A). Dual-luciferase reporter assays
were then performed to confirm this prediction. It was demonstrated
that following co-transfection with the miR-411-3p mimics, the
luciferase activity of HIF-1α-WT reporter was significantly
reduced, whereas the luciferase activity of HIF-1α-MUT reporter was
not altered (Fig. 4B). Analysis of
data obtained from the GEPIA database showed that HIF-1α expression
was upregulated in PDAC tissues (Fig.
4C). The expression of HIF-1α mRNA in the clinical PDAC tissues
was then determined using RT-qPCR. Its expression was significantly
higher in tumor tissues of patients with PDAC and was positively
correlated with the expression of PCED1B-AS1 (Fig. 4D and E). These results suggest that
miR-411-3p can target HIF-1α expression, and PCED1B-AS1 may exert
its biological functions via a miR-411-3p/HIF-1α axis.
PCED1B-AS1 regulates the
miR-411-3p/HIF-1α axis to reduce PDAC cell proliferation, invasion
and EMT
To further investigate the effect of PCED1B-AS1 and
miR-411-3p on the biological behaviors of PDAC cells,
si-PCED1B-AS1, miR-411-3p inhibitor or HIF-1α overexpression
plasmids were co-transfected into CFPAC-1 and SW1990 cells
(Figs. S2 and S3). Western blotting was used to
investigate the expression of HIF-1α; PCED1B-AS1 knockdown
significantly reduced the expression of HIF-1α in CFPAC-1 and
SW1990 cells, whereas the transfection of miR-411-3p inhibitor and
HIF-1α overexpression plasmid restored the expression of HIF-1α
(Fig. 5A). Cell proliferation,
invasion and EMT in each group were then evaluated. CCK-8 and EdU
staining assays demonstrated that compared with the control group,
the proliferation of CFPAC-1 and SW1990 cells in the si-PCED1B-AS1
group was reduced; however, this reduction was reversed by the
transfection of miR-411-3p inhibitor or HIF-1α overexpression
plasmid (Fig. 5B and C). Transwell
assays and western blotting were used to assess invasion and EMT.
In the si-PCED1B-AS1 transfected cells, invasion and EMT were
reduced, and co-transfection with the miR-411-3p inhibitor or
HIF-1α attenuated this inhibitory effect (Fig. 5D and E). These results show that
PCED1B-AS1 modulates PDAC cell proliferation, invasion and EMT via
regulation of a miR-411-3p/HIF-1α axis.
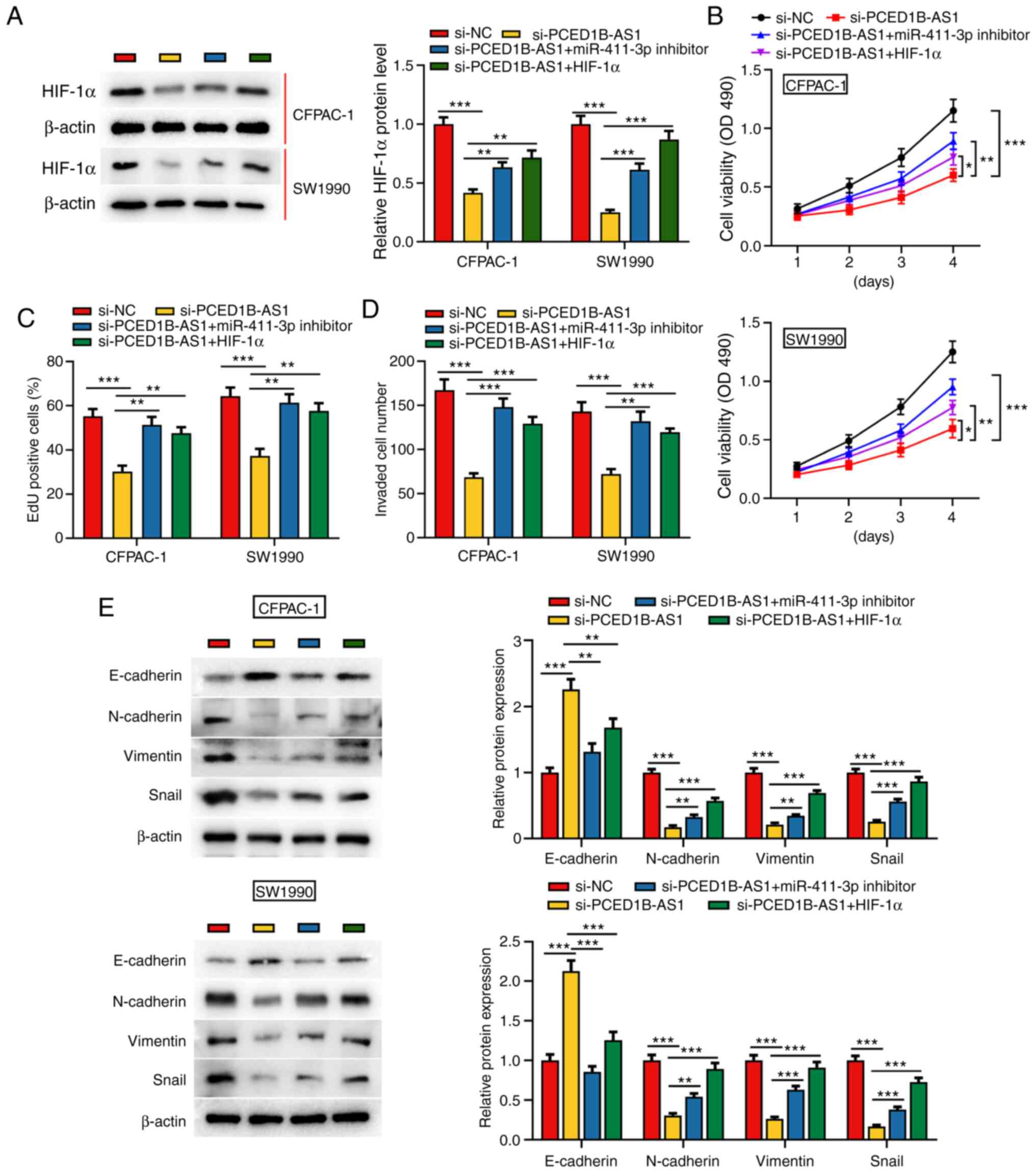 | Figure 5.PCED1B-AS1 modulates the biological
behaviors of PDAC cells via regulation of a miR-411-3p/HIF-1α axis.
(A) Western blotting was used to detect the expression of HIF-1α
expression in CFPAC-1 and SW1990 cells transfected with si-NC,
si-PCED1B-AS1, si-PCED1B-AS1 + miR-411-3p inhibitor or
si-PCED1B-AS1 + HIF-1α overexpression plasmid. (B-E) Proliferation,
invasion and expression of EMT markers in CFPAC-1 and SW1990 cells
were detected using a CCK-8 assay, EdU assay, Transwell invasion
assay and western blotting, respectively. *P<0.05, **P<0.01,
***P<0.001. siRNA, small interfering RNA; si-NC, si-negative
control; PDAC, pancreatic ductal adenocarcinoma; PCED1B-AS1,
PC-esterase domain containing 1B-antisense RNA 1; miR, microRNA;
HIF-1α, hypoxia inducible factor-1α; CCK-8, Cell Counting
Kit-8. |
Discussion
Pancreatic cancer is one of the most aggressive and
fatal types of cancer (1–4). An increasing number of studies have
shown that non-coding RNAs, such as long non-coding RNAs (lncRNAs)
and microRNAs (miRNAs), serve prominent roles in regulating the
occurrence and progression of pancreatic ductal adenocarcinoma
(PDAC). The present study showed that lncRNA PC-esterase domain
containing 1B-antisense RNA 1 (PCED1B-AS1) expression is
upregulated in PDAC tissues and cell lines, and it is closely
associated with TNM stage and lymph node metastasis of the
patients. Additionally, PCED1B-AS1 knockdown impaired
proliferation, invasion and EMT of PDAC cells. These results show
that PCED1B-AS1 functions as an oncogenic lncRNA in PDAC.
lncRNAs can function as competitive endogenous RNA
(ceRNAs), regulating the expression and function of miRNAs
(29). For example, as a ceRNA for
miR-520a-3p, lncRNA non-coding RNA activated by DNA damage (NORAD)
was found to modulate the PI3K/AKT/mTOR signaling pathway to
promote the occurrence and progression of non-small cell lung
cancer (30). It has been reported
that lncRNA ADPGK-AS1 upregulates orthodenticle homeobox 1
expression, promotes breast cancer cell proliferation and
migration, induces EMT, and impedes apoptosis by sponging miR-3196
(31). PCED1B-AS1 is upregulated in
several types of cancer (14,15). In gliomas, PCED1B-AS1 promotes cancer
cell proliferation and reduces apoptosis by modulating a
miR-194-5p/PCED1B axis (14). In the
present study, bioinformatics analysis, luciferase reporter gene
experiments and RIP experiments confirmed that PCED1B-AS1 directly
interacted with miR-411-3p in PDAC. Furthermore, PCED1B-AS1
knockdown reduced PDAC cell proliferation, invasion and EMT;
conversely, co-transfection with miR-411-3p inhibitors reversed
these effects. These data suggest that PCED1B-AS1 regulates PDAC
proliferation, invasion and EMT by sponging miR-411-3p.
MiR-411-3p is a tumor-suppressive miRNA. Low
expression of miR-411-3p is significantly correlated with reduced
overall survival in patients with non-small cell lung cancer
(32). It has also been reported that
CDKN2B-AS1 interacts with miR-411-3p and regulates ovarian cancer
progression via a HIF-1α/VEGF/p38 pathway (33). In the present study, it was confirmed
through bioinformatics analysis and luciferase reporter gene assays
that HIF-1α was a direct target of miR-411-3p in PDAC, and that
hypoxia inducible factor-1α (HIF-1α) was positively regulated by
PCED1B-AS1. Previous studies report that HIF-1α is involved in
regulating the malignant biological behaviors of cancer cells, such
as cell proliferation, migration and angiogenesis, in several types
of cancer (34–38). For example, HIF-1α is upregulated in
colorectal cancer cell lines and contributes to angiogenesis by
modulating the expression of EMT-related molecules claudin-4,
E-cadherin and Vimentin (38). In
pancreatic cancer, HIF-1α expression has been reported to be
upregulated, and it is involved in the regulation of the Warburg
effect, cancer metastasis and chemoresistance; upregulated
expression of HIF-1α is associated with unfavorable prognosis of
the patients (39–41). A recent study showed that ascorbate
inhibits tumor growth of PDAC by reducing the expression of HIF-1α
at the protein level under hypoxic condition via post-translational
regulation (42). HIF-1α regulates
granulocyte-macrophage colony-stimulating factor (GM-CSF)
expression via direct binding to the hypoxia response element in
the promoter region of GM-CSF gene, and participates in tumor-nerve
interaction in PDAC (43). In
addition, HIF-1α can directly bind to the hypoxia response element
in the promoter region of cyclophilin A, regulating cyclophilin A
expression and thus promoting PDAC cell proliferation and invasion,
and suppressing apoptosis in vitro (44). In the present study, it was found that
HIF-1α was significantly upregulated in PDAC tissues, consistent
with previous reports (39–41). The upregulation of HIF-1α was
primarily due to the presence of a hypoxic tumor microenvironment
(21). Importantly, in the present
study, it was also demonstrated that the expression of HIF-1α was
regulated by a PCED1B-AS1/miR-411-3p axis. Functional experiments
showed that HIF-1α overexpression partially reversed the inhibition
of PCED1B-AS1 knockdown on PDAC cell proliferation, invasion and
EMT. These results may explain the mechanism by which HIF-1α
expression is dysregulated in PDAC.
In summary, it was demonstrated that PCED1B-AS1 was
significantly upregulated in PDAC tissues and PDAC cell lines, and
it was associated with a less favorable outcome in patients with
PDAC. PCED1B-AS1 knockdown impeded PDAC cell proliferation,
invasion and EMT. PCED1B-AS1 was shown to directly target
miR-411-3p, acting as a ceRNA, indirectly increasing HIF-1α
expression, thereby promoting PDAC progression. Collectively, these
results provide an improved understanding of the characteristics of
the PCED1B-AS1/miR-411-3p/HIF-1α axis in PDAC progression, which
may provide novel directions for improvement of PDAC diagnosis and
treatment. In future studies, in vivo models will be used to
further validate the findings of the present study. Additionally, a
larger cohort will be enrolled and survival analysis will be
performed to evaluate the potential of PCED1B-AS1 as a biomarker in
PDAC.
Supplementary Material
Supporting Data
Supporting Data
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used during the present study are
available from the corresponding author upon reasonable
request.
Authors' contributions
YZ and CC conceived and designed the study. ZY and
HM performed the experiments. YZ, HM and CC wrote the paper. YZ, HM
and CC reviewed and edited the manuscript. All authors read and
approved the manuscript and agree to be accountable for all aspects
of the research in ensuring that the accuracy or integrity of any
part of the work are appropriately investigated and resolved.
Ethics approval and consent to
participate
All experimental protocols were approved by the
Ethics Committee of the People's Hospital of Three Gorges
University.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2017. CA Cancer J Clin. 67:7–30. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Steeg PS: Targeting metastasis. Nat Rev
Cancer. 16:201–218. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Heestand GM and Kurzrock R: Molecular
landscape of pancreatic cancer: Implications for current clinical
trials. Oncotarget. 6:4553–4561. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Melisi D, Calvetti L, Frizziero M and
Tortora G: Pancreatic cancer: Systemic combination therapies for a
heterogeneous disease. Curr Pharm Des. 20:6660–6669. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ponting CP, Oliver PL and Reik W:
Evolution and functions of long noncoding RNAs. Cell. 136:629–641.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wu H, Yang L and Chen LL: The diversity of
long noncoding RNAs and Their Generation. Trends Genet. 33:540–552.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhou X, Liu S, Cai G, Kong L, Zhang T, Ren
Y, Wu Y, Mei M, Zhang L and Wang X: Long non coding RNA MALAT1
promotes tumor growth and metastasis by inducing
epithelial-mesenchymal transition in oral squamous cell carcinoma.
Sci Rep. 5:159722015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Khaitan D, Dinger ME, Mazar J, Crawford J,
Smith MA, Mattick JS and Perera RJ: The melanoma-upregulated long
noncoding RNA SPRY4-IT1 modulates apoptosis and invasion. Cancer
Res. 71:3852–3862. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Tang J, Zhong G, Wu J, Chen H and Jia Y:
Long noncoding RNA AFAP1-AS1 facilitates tumor growth through
enhancer of zeste homolog 2 in colorectal cancer. Am J Cancer Res.
8:892–902. 2018.PubMed/NCBI
|
|
10
|
Luo H, Yang L, Liu C, Wang X, Dong Q, Liu
L and Wei Q: TMPO-AS1/miR-98-5p/EBF1 feedback loop contributes to
the progression of bladder cancer. Int J Biochem Cell Biol.
122:1057022020. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Jin X, Liu X, Zhang Z and Guan Y: lncRNA
CCAT1 Acts as a MicroRNA-218 sponge to increase gefitinib
resistance in NSCLC by targeting HOXA1. Mol Ther Nucleic Acids.
19:1266–1275. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Li M, Cui J, Niu W, Huang J, Feng T, Sun B
and Yao H: Long non-coding PCED1B-AS1 regulates macrophage
apoptosis and autophagy by sponging miR-155 in active tuberculosis.
Biochem Biophys Res Commun. 509:803–809. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhao Y, Wang Z, Zhang W and Zhang L:
MicroRNAs play an essential role in autophagy regulation in various
disease phenotypes. Biofactors. 45:844–856. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yang J, Yu D, Liu X, Changyong E and Yu S:
LncRNA PCED1B-AS1 activates the proliferation and restricts the
apoptosis of glioma through cooperating with miR-194-5p/PCED1B
axis. J Cell Biochem. 121:1823–1833. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yuan CL, Jiang XM, Yi Y, E JF, Zhang ND,
Luo X, Zou N, Wei W and Liu YY: Identification of differentially
expressed lncRNAs and mRNAs in luminal-B breast cancer by
RNA-sequencing. BMC Cancer. 19:11712019. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ye Y, Li SL and Wang SY: Construction and
analysis of mRNA, miRNA, lncRNA, and TF regulatory networks reveal
the key genes associated with prostate cancer. PLoS One.
13:e01980552018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Duguang L, Jin H, Xiaowei Q, Peng X,
Xiaodong W, Zhennan L, Jianjun Q and Jie Y: The involvement of
lncRNAs in the development and progression of pancreatic cancer.
Cancer Biol Ther. 18:927–936. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhao L, Kong H, Sun H, Chen Z, Chen B and
Zhou M: LncRNA-PVT1 promotes pancreatic cancer cells proliferation
and migration through acting as a molecular sponge to regulate
miR-448. J Cell Physiol. 233:4044–4055. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Shen J, Hong L, Yu D, Cao T, Zhou Z and He
S: LncRNA XIST promotes pancreatic cancer migration, invasion and
EMT by sponging miR-429 to modulate ZEB1 expression. Int J Biochem
Cell Biol. 113:17–26. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Feng H, Wei B and Zhang Y: Long non-coding
RNA HULC promotes proliferation, migration and invasion of
pancreatic cancer cells by down-regulating microRNA-15a. Int J Biol
Macromol. 126:891–898. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Puppo M, Battaglia F, Ottaviano C, Delfino
S, Ribatti D, Varesio L and Bosco MC: Topotecan inhibits vascular
endothelial growth factor production and angiogenic activity
induced by hypoxia in human neuroblastoma by targeting
hypoxia-inducible factor-1alpha and −2alpha. Mol Cancer Ther.
7:1974–1984. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ren W, Mi D, Yang K, Cao N, Tian J, Li Z
and Ma B: The expression of hypoxia-inducible factor-1α and its
clinical significance in lung cancer: A systematic review and
meta-analysis. Swiss Med Wkly. 143:w138552013.PubMed/NCBI
|
|
23
|
Hung JJ, Yang MH, Hsu HS, Hsu WH, Liu JS
and Wu KJ: Prognostic significance of hypoxia-inducible
factor-1alpha, TWIST1 and Snail expression in resectable non-small
cell lung cancer. Thorax. 64:1082–1089. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wu YL, Hu LN, Zheng CD, Sun RC, Zhang SX,
Yan Q and Li YX: Expression of hypoxia-inducible factor 1α in
gastric cancer and its clinical signficance. Zhonghua Yi Xue Za
Zhi. 96:1418–1423. 2016.(In Chinese). PubMed/NCBI
|
|
25
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Tang Z, Li C, Kang B, Gao G, Li C and
Zhang Z: GEPIA: A web server for cancer and normal gene expression
profiling and interactive analyses. Nucleic Acids Res. 45:W98–W102.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Paraskevopoulou MD, Vlachos IS, Karagkouni
D, Georgakilas G, Kanellos I, Vergoulis T, Zagganas K, Tsanakas P,
Floros E, Dalamagas T and Hatzigeorgiou AG: DIANA-LncBase v2:
Indexing microRNA targets on non-coding transcripts. Nucleic Acids
Res. 44:D231–D238. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Agarwal V, Bell GW, Nam JW and Bartel DP:
Predicting effective microRNA target sites in mammalian mRNAs.
Elife. 4:e050052015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Abdollahzadeh R, Daraei A, Mansoori Y,
Sepahvand M, Amoli MM and Tavakkoly-Bazzaz J: Competing endogenous
RNA (ceRNA) cross talk and language in ceRNA regulatory networks: A
new look at hallmarks of breast cancer. J Cell Physiol.
234:10080–10100. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wan Y, Yao Z, Chen W and Li D: The lncRNA
NORAD/miR-520a-3p facilitates malignancy in non-small cell lung
cancer via PI3k/Akt/mTOR signaling pathway. Onco Targets Ther.
13:1533–1544. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Yang J, Wu W, Wu M and Ding J: Long
noncoding RNA ADPGK-AS1 promotes cell proliferation, migration, and
EMT process through regulating miR-3196/OTX1 axis in breast cancer.
In VitroCell Dev Biol Anim. 55:522–532. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Halvorsen AR, Sandhu V, Sprauten M, Flote
VG, Kure EH, Brustugun OT and Helland Å: Circulating microRNAs
associated with prolonged overall survival in lung cancer patients
treated with nivolumab. Acta Oncol. 57:1225–1231. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wang Y, Huang Y, Liu H, Su D, Luo F and
Zhou F: Long noncoding RNA CDKN2B-AS1 interacts with miR-411-3p to
regulate ovarian cancer in vitro and in vivo through
HIF-1a/VEGF/P38 pathway. Biochem Biophys Res Commun. 514:44–50.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Lu Y, Li Y, Wang Z, Xie S, Wang Q, Lei X,
Ruan Y and Li J: Downregulation of RGMA by HIF-1A/miR-210-3p axis
promotes cell proliferation in oral squamous cell carcinoma. Biomed
Pharmacother. 112:1086082019. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Sohn SH, Kim B, Sul HJ, Choi BY, Kim HS
and Zang DY: Foretinib inhibits cancer stemness and gastric cancer
cell proliferation by decreasing CD44 and c-MET signaling. Onco
Targets Ther. 13:1027–1035. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Ma D, Fan SB, Hua N, Li GH, Chang Q and
Liu X: Hypermethylation of single CpG dinucleotides at the promoter
of CXCL13 gene promoting cell migration in cervical cancer. Curr
Cancer Drug Targets. 20:355–363. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Yang QC, Zeng BF, Shi ZM, Dong Y, Jiang
ZM, Huang J, Lv YM, Yang CX and Liu YW: Inhibition of
hypoxia-induced angiogenesis by trichostatin A via suppression of
HIF-1a activity in human osteosarcoma. J Exp Clin Cancer Res.
25:593–599. 2006.PubMed/NCBI
|
|
38
|
Li W, Zong S, Shi Q, Li H, Xu J and Hou F:
Hypoxia-induced vasculogenic mimicry formation in human colorectal
cancer cells: Involvement of HIF-1a, claudin-4, and E-cadherin and
Vimentin. Sci Rep. 6:375342016. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Shen Y, Chen G, Zhuang L, Xu L, Lin J and
Liu L: ARHGAP4 mediates the Warburg effect in pancreatic cancer
through the mTOR and HIF-1α signaling pathways. Onco Targets Ther.
12:5003–5012. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Shukla SK, Purohit V, Mehla K, Gunda V,
Chaika NV, Vernucci E, King RJ, Abrego J, Goode GD, Dasgupta A, et
al: MUC1 and HIF-1alpha signaling crosstalk induces anabolic
glucose metabolism to impart gemcitabine resistance to pancreatic
cancer. Cancer Cell. 32:71–87.e7. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Colbert LE, Fisher SB, Balci S, Saka B,
Chen Z, Kim S, El-Rayes BF, Adsay NV, Maithel SK, Landry JC and
Curran WJ Jr: High nuclear hypoxia-inducible factor 1 alpha
expression is a predictor of distant recurrence in patients with
resected pancreatic adenocarcinoma. Int J Radiat Oncol Biol Phys.
91:631–639. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Wilkes JG, O'Leary BR, Du J, Klinger AR,
Sibenaller ZA, Doskey CM, Gibson-Corley KN, Alexander MS, Tsai S,
Buettner GR and Cullen JJ: Pharmacologic ascorbate (P-AscH-)
suppresses hypoxia-inducible Factor-1α (HIF-1α) in pancreatic
adenocarcinoma. Clin Exp Metastasis. 35:37–51. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Wang H, Jia R, Zhao T, Li X, Lang M, Lan
C, Wang H, Li Z, Zhou B, Wu L, et al: HIF-1α mediates tumor-nerve
interactions through the up-regulation of GM-CSF in pancreatic
ductal adenocarcinoma. Cancer Lett. 453:10–20. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Zhang H, Chen J, Liu F, Gao C, Wang X,
Zhao T, Liu J, Gao S, Zhao X, Ren H and Hao J: CypA, a gene
downstream of HIF-1α, promotes the development of PDAC. PLoS One.
9:e928242014. View Article : Google Scholar : PubMed/NCBI
|