Introduction
Airborne carcinogens, including tobacco smoke, are strongly implicated in the etiology of both squamous cell carcinomas of the head and neck and squamous cell carcinoma of the lung (1). Similar to the case for all head and neck cancers (HNC), squamous cell carcinoma is the most common histologic subtype of oral cancer (OSCC), accounting for nearly 90% of all cases of oral cancer (2). OSCC ranks sixth in cancer incidence worldwide, with approximately 300,400 newly diagnosed cases and 145,400 deaths each year, and the cancer is often diagnosed at an advanced stage (3,4). The two most important risk factors for OSCC, accounting for over 80% of all patients, are tobacco and alcohol (5). Smoking is also a strong risk factor for lung squamous cell carcinoma (LSCC), which accounts for 15 to 30% of all cases of lung cancer; a trend towards decrease in the incidence rates associated with a decrease in smoking rates has been reported in developed countries (6). Due to the occurrence of lung cancer in epidemic proportions worldwide, however, urgent development of countermeasures is still required for LSCC.
Recent advances in molecular-targeted therapies have brought about significant improvements in the treatment outcomes of some advanced cancers (7). Especially, tyrosine-kinase inhibitors (TKIs) have proven to be significantly beneficial for patients with advanced adenocarcinomas of the lung harboring driver oncogene mutations, including activating epidermal growth factor receptor (EGFR) mutations and rearrangements of the anaplastic lymphoma kinase gene (8). On the other hand, in the case of LSCC, no driver mutations or effective TKIs have been identified yet, thus no effective molecular-targeted therapies against this cancer are available to date. Although cetuximab, an anti-EGFR antibody, administered in combination with radiation (9) or cytotoxic chemotherapy (10) has been revealed to be effective for advanced HNC, including OSCC, and another anti-EGFR antibody, necitumumab, administered in combination with cytotoxic chemotherapy (11) has been revealed to be effective for advanced LSCC, most patients with advanced OSCC or LSCC eventually exhibit disease progression. Therefore, novel molecular targets must be sought for developing novel molecular-targeted therapies for OSCC and LSCC.
The epithelial stromal interaction 1 gene (EPSTI1) is a gene mapped to human chromosome 13q13.3, that has received increasing attention in recent years. It has been revealed to be expressed in some normal tissues, including the spleen, small intestine, salivary glands, testes, germinal centers of lymph nodes, and placenta (12–15). In addition, overexpression of the gene has been reported in primary cancers of the breast, colorectal and pancreas (12,16,17). In breast cancers, high expression levels of EPSTI1 have been revealed to be strongly associated with enhanced tumor cell migration ability and tumor invasiveness, a high propensity for metastasis, and escape from apoptosis, suggesting that EPSTI1 expression may be an independent prognostic marker in patients with breast cancer (13,18,19). Furthermore, accumulated evidence has revealed multiple roles of EPSTI1 in the immune response; it has been demonstrated to be associated with the establishment of immune privilege, development of autoimmune diseases, control of viral infections, and activation of macrophages. The gene has been revealed to have a role in the immune privilege of the testes (20), and a genome-wide association study has linked the gene to male fertility (21). Overexpression of EPSTI1 has been reported in the peripheral blood cells of patients with systemic lupus erythematosus (22), and such overexpression was revealed to be associated with abnormal B-cell activation via the NF-κB signaling pathway in primary Sjogren syndrome (23); expression of the gene induced by HCV infection has been revealed to effectively inhibit viral replication and exert antiviral effects through upregulation of IL-28A (24). Finally, the gene has also been revealed to play a role in modulating macrophage activation and polarization (15). However, despite these already known functions of EPSTI1, its roles in human OSCC and LSCC remain unclear.
Materials and methods
Cell lines
A human normal oral keratinocyte cell line, HNOKs, derived from a mixture of healthy gingival specimens of 22- to 35-year-old patients, was established and maintained as previously described (25). Human OSCC cell lines HSC2, HSC3, and HSC4 were purchased from RIKEN Bioresource Center, and HSC3-M3 was purchased from the Human Science Resources Bank. The cells were cultured in DMEM (Sigma-Aldrich; Merck KGaA). A normal human lung cell line, BEAS-2B, and the LSCC cell lines LK-2, EBC-1 and H226 were purchased from JCRB cell bank. BEAS-2B, LK-2 and H226 were cultured in PRMI-1640 medium, and EBC-1 was cultured in MEM (both from Sigma-Aldrich; Merck KGaA). All the culture media were supplemented with 10% fetal bovine serum and 1% antibiotics, including penicillin and streptomycin (all from Life Technologies; Thermo Fisher Scientific, Inc.). All the cell lines were cultured in a humidified incubator at 37°C in a 5% CO2 atmosphere.
Expression of mRNA and protein
The cells were washed three times with phosphate-buffered saline (PBS), followed by extraction of total RNA and protein. The mRNA expression levels were quantified by real-time reverse transcription-quantitative polymerase chain reaction (RT-qPCR) with SYBR green expression assays (Roche Diagnostics), in accordance with the manufacturer's instructions. In brief, total RNA was extracted using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.) when the cells reached 80% confluence in the 10-cm culture plates. Reverse transcription was performed using the ReverTra Ace qPCR RT Master Mix (Toyobo Life Science). Gene expression was measured using LightCycler 480 Instrument (Roche Diagnostics). The thermocycling conditions for the PCR were as follows: initial denaturation at 95°C for 10 min, followed by 45 cycles of amplification at 95°C (10 sec) for denaturation, 60°C (10 sec) for annealing and 72°C for extension, followed by a cooling step at 40°C for 30 sec. GAPDH was used as the internal control. Transcript amounts were estimated from respective standard curves and normalized to GAPDH. Primers were designed using the Universal Probe Library Assay Design Center (http://lifescience.roche.com/). The primers for EPSTI1 were 5′-CCGGAGAAATGAGATACAAAGAAT-3′ (forward) and 5′-GGTGAACCGGTTTAGCTCTG−3′ (reverse), and the primers for GAPDH were 5′-AACATCATCCCTGCCTCTACTGG-3′ (forward) and 5′-TTGAAGTCAGAGGAGACCACTG-3′ (reverse).
For the evaluation of the protein expression, the cells were washed twice with cold phosphate-buffered saline (PBS) and collected with lysis buffer [7 M urea, 2M thiourea, 4% (w/v) CHAPS, and 10 mM Tris] with a proteinase inhibitor cocktail (Roche Diagnostics). The protein concentration was first assessed by the Bradford method, followed by western blot analysis. Protein extracts (20 µg/lane) were separated by a 4–12% Bis-Tris gel (Thermo Fisher Scientific, Inc.), and transferred to nitrocellulose membranes. Subsequently, the membrane was blocked for 1 h at room temperature in 5% skim milk, and then incubated with primary antibodies (1:1,000) overnight at 4°C. The membranes were washed with 0.1% Tween-20 in Tris-buffered saline, three times (15 min for each wash), followed by incubation with a secondary antibody with anti-mouse (cat. no. S3721) or anti-rabbit IgG (cat. no. S3731) (1:2,000; Promega Corporation) for 1.5 h at room temperature. Finally, the bands were detected using clarity Western ECL Substrate (Bio-Rad Laboratories, Inc.), and immunoblotting images were visualized by exposing the membranes to the BioRad ChemiDoc™ XRS System (BioRad Laboratories, Inc.). The densitometric analysis of the protein expression was performed using Image Lab software 6.0.1 (Bio Rad Laboratories, Inc.).
The antibodies used were mouse anti-EPSTI1 monoclonal antibody (cat. no. AT1934a; Abcepta; Abgent, Inc.), rabbit anti-E-cadherin (product no. 3195), anti-N-cadherin (product no. 13116), anti-p21 (product no. 2947), anti-cyclin D1 (product no. 55506), and anti-CDK2 (product no. 18048) monoclonal antibodies (all from Cell Signaling Technology, Inc.), and mouse anti-fibronectin (product no. SAB4200760), anti-β-actin (product no. SAB1305567) (both from Sigma Aldrich; Merck KGaA), and anti-α-tubulin monoclonal antibodies (cat. no. sc-5286; Santa Cruz Biotechnology, Inc.).
Transfection of plasmids
OSCC-derived cells (HSC3-M3 and HSC4) were transfected with EPSTI1-suppressing shRNA or the control plasmid, shMock (cat. no. sc-105335-SH and cat. no. sc-108060, respectively, Santa Cruz Biotechnology, Inc.), at a concentration of 2.5 µg/well, using Lipofectamine 3000 (Thermo Fisher Scientific, Inc.), and then incubated overnight at 37°C. After the transfection, the cells exhibiting stable transfection were selected by taking advantage of the selection marker and its inhibitor, puromycin (Santa Cruz Biotechnology, Inc.). LSCC-derived cells (LK-2 and EBC-1) were transfected with EPSTI1-overexpressing plasmid (OriGene Technologies, Inc.) or the control plasmid (OriGene Technologies, Inc.) at a concentration of 2.5 µg/well, using Lipofectamine 3000 and incubated overnight at 37°C. Following the transfection, the cells exhibiting stable transfection were selected by utilizing the selection marker and its inhibitor G-418 Solution (Roche Diagnostics). Then, incubation of the selected cells with the inhibitors for 2 to 3 weeks yielded individual clones from each of the EPSTI1-suppressed OSCC cell lines and EPSTI1-overexpressing LSCC cell lines; these clones and the control cell lines were used for further experiments.
Proliferation assay
To evaluate the cell proliferation ability, shEPSTI1 and shMock cells (HSC3-M3 and HSC4), oeEPSTI1 and OeMock cells (5×104 cells/plate) were seeded on to a 6-cm plate, and the number of cells were counted every 24 h in triplicate using a hemocytometer to measure the chronological changes.
Wound healing assay
For evaluating the cell migration ability, 2×105 cells/well were seeded on to a 6-well plate overnight until they reached 95% confluence; the surface of the plate was then scratched with the tip of a 1,000-µl micropipette and the culture medium was immediately washed off with PBS; the cells were then cultured in serum-free medium for a further 24 h. The scratch scars were viewed under an inverted microscope (×10 magnification of objective) (ECLIPSE TS100; Nikon Corporation), and the scratched areas were quantified using the Lenaraf220b free software (available at http://www.vector.co.jp/soft/dl/win95/art/se312811.html). The wound closure rates were determined as a percentage of the total repaired area per hour and normalized to the control.
Cell cycle analysis
Cells were harvested with trypsin and centrifuged for 5 min at 300 × g at room temperature, and then the concentration was adjusted to 1.0×106 cells/ml. Cell cycle analyses were performed using the Cycletest™ Plus DNA reagent kit (BD Biosciences) and the BD Accuri C6 Flow Cytometer (BD Biosciences), in accordance with the manufacturers' instructions. For data analysis FCS Express 4 (De Novo Software) was used.
PCR array
Differences in the mRNA expression of the EPSTI1-related genes in the two EPSTI1-downregulated OSCC cell lines, HSC3-M3 and HSC4, and two EPSTI1-overexpressing LSCC cell lines, LK2 and EBC-1, as compared to the expression levels in the corresponding parental cells, were detected using the RT2 Profiler PCR Array Human Cancer Pathway Finder (cat. no. PAHS-033ZF-6; Qiagen, Inc.). In brief, cDNA was synthesized from the total RNA extracted from each cell line using the RT2 First Strand Kit (Qiagen, Inc.). RT2 SYBR Green qPCR Master Mix (Qiagen, Inc.) was used for the PCR arrays, in accordance with the manufacturer's instructions.
Bioinformatics analysis
The gene expression in clinical samples was obtained from the Oncomine Platform (http://www.oncomine.org).
Statistical analysis
The unpaired Student's t-test was used to analyze the statistical significance of differences between two groups. One-way ANOVA with Dunnett's post hoc test calculated by GraphPad Prism software (version 8; GraphPad software, Inc.) were used to evaluate the differences of multiple comparisons. All experiments were performed in triplicate and repeated 3 times, unless otherwise specified. P<0.05 was considered to indicate statistically significant differences. The data are expressed as the means ± standard error (SE).
Results
EPSTI1 expression in the OSCC and LSCC cell lines
The series of RT-qPCR and western blot analyses in the four OSCC (HSC2, HSC3, HSC3-M3 and HSC4) and three LSCC (EBC-1, LK-2 and H226) cell lines revealed significant overexpression of EPSTI1 in all of the OSCC cell lines (Fig. 1A and C), except for protein expression in HSC2, and significantly suppressed expression of the gene in all of the LSCC cell lines (with the exception of EPST11 mRNA expression in the LK-2 cell line) (Fig. 1B and D), as compared to the expression levels in the corresponding control cell lines, HNOKs and BEAS-2B, respectively (Fig. 1B).
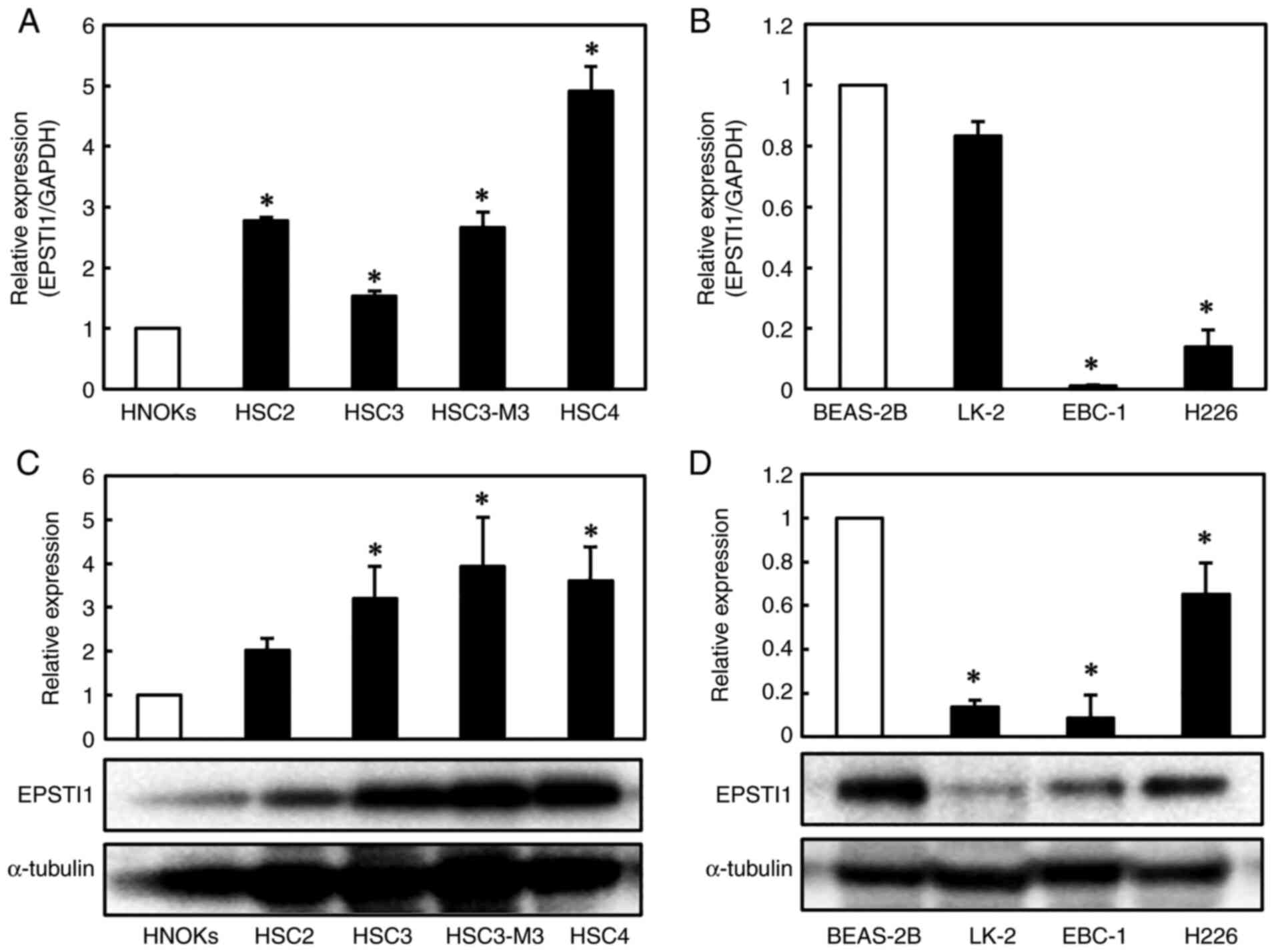 |
Figure 1.
Expression of EPSTI1 in human OSCC and LSCC cell lines. Quantification of EPSTI1 mRNA expression levels by RT-qPCR revealed (A) significantly increased expression levels in the four human OSCC cell lines (HSC2, HSC3, HSC3-M3 and HSC4) as compared to the levels in the control cell line, HNOKs, and (B) significantly reduced expression levels in two of the three human LSCC cell lines (EBC-1 and H226, but not LK-2) as compared to the levels in the normal control cell line, BEAS-2B. Western blotting, using a-tubulin as the internal control, confirmed (C) significant upregulation of the gene in the OSCC cell lines and (D) significant downregulation of the gene in the LSCC cell lines, including LK-2. The values on the y-axes represent the ratios to the expression levels in the normal control cell lines. Each experiment was performed in triplicate and repeated three times, and data are presented as the means ± SE. *P<0.05. EPSTI, epithelial-stromal interaction 1; OSCC, oral squamous cell carcinoma; LSCC, lung squamous cell carcinoma; SE, standard error.
|
Establishment of cells with EPSTI1 knockdown and EPST11 overexpression
Based on the results of the experiments aforementioned (Fig. 1C and D), the OSCC cell lines, HSC3-M3 and HSC4, and LSCC cell lines, LK-2 and EBC-1, were selected for further experiments because the former exhibited the strongest expression, among all the OSCC cell lines, of the EPSTI1 protein, and the latter exhibited the weakest expression, among all the LSCC cell lines, of the EPSTI1 protein. HSC3-M3 and HSC4, were transfected with EPSTI1 shRNA plasmid to suppress the gene expression, which yielded two clones for each cell line, whereas LK-2 and EBC-1 were transfected with the EPSTI1 overexpression plasmid, which yielded a single clone per cell line, to further investigate the roles of the gene. As transfection of the EPSTI1 overexpression plasmid into the LSCC cell lines was somewhat difficult, only a single clone was obtained for each cell line. The alterations in the expression levels of the gene were verified by RT-qPCR and western blot analysis. The results revealed significant downregulation of EPSTI1 at the mRNA and/or protein level in the shRNA-transfected OSCC cell lines as compared to the shMock-transfected cells (Fig. 2A and C); similarly, significant upregulation of the gene at the mRNA and/or protein level was confirmed in the LSCC cells transfected with the overexpression vector as compared to the oeMock-transfected cells (Fig. 2B and D).
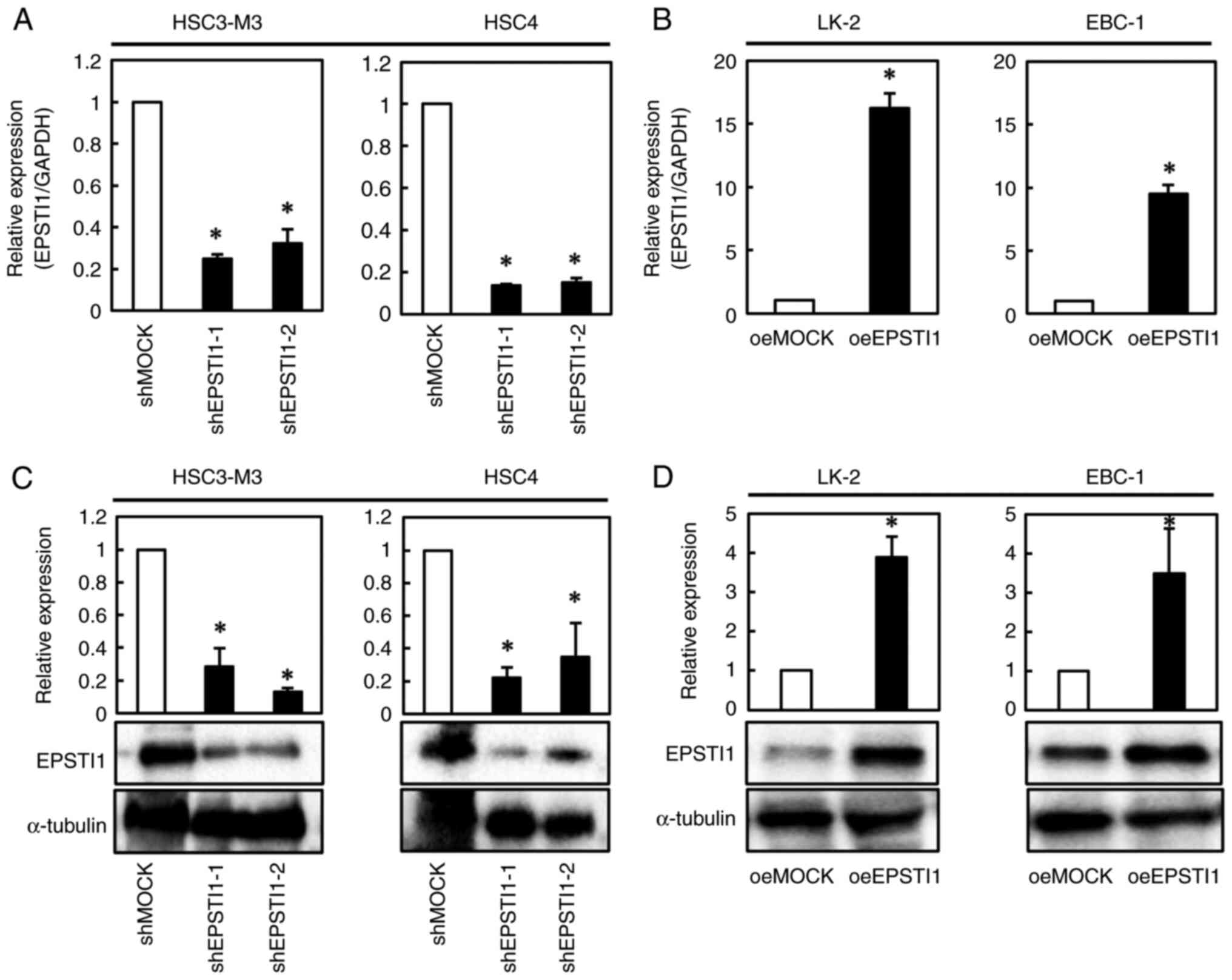 |
Figure 2.
Confirmation of the modified gene expression of EPSTI1 in the OSCC and LSCC cell lines. For further elucidation of the functions of EPSTI1, expression of the gene was knocked down in two OSCC cell lines, HSC3-M3 and HSC4, by transfection of shRNA, and overexpressed in two LSCC cell lines, LK-2 and EBC-1, using overexpressing vectors. Consequently, EPST11 mRNA expression was (A) significantly suppressed in the two OSCC cells as compared to that in the mock transfectants, and (B) significantly overexpressed in the two LSCC cells as compared to that in the mock-transfected cells. In addition, western blotting confirmed (C) reduced EPSTI1 protein expression in the shRNA-transfected OSCC cell lines, and (D) overexpression of EPST11 protein in the overexpressing vector-transfected LSCC cell lines. Each experiment was performed in triplicate and repeated three times, and data are presented as the means ± SE. *P<0.05. EPSTI, epithelial-stromal interaction 1; OSCC, oral squamous cell carcinoma; LSCC, lung squamous cell carcinoma; SE, standard error.
|
Altered expression of EPSTI1 induces alterations in the cell proliferation ability and cell cycle progression
Cell propagation was significantly suppressed by downregulation of EPSTI1 in the two OSCC cell lines (Fig. 3A), and by upregulation of the gene in the two LSCC cell lines (Fig. 3B). Cell cycle analysis demonstrated accumulation of cells in the G1 phase in every OSCC cell line with downregulated EPSTI1 expression and every LSCC cell line with upregulated EPSTI1 expression with statistical significance except for HSC4 which revealed a trend of G1 accumulation (P=0.053) (Fig. 3C-F). To confirm this finding, the expression levels of the cell cycle-related genes, p21, CDK2, and cyclin D1, were investigated. As anticipated, upregulation of p21 and downregulation of CDK2 and cyclin D1 were observed in every OSCC cell line with downregulated EPSTI1 expression (Fig. 4A and C) and in every LSCC cell line with upregulated EPSTI1 expression (Fig. 4B and D), although statistical significance was marginal for p21 (P=0.08) and cyclin D1 (P=0.06) in HSC3-M3 cells.
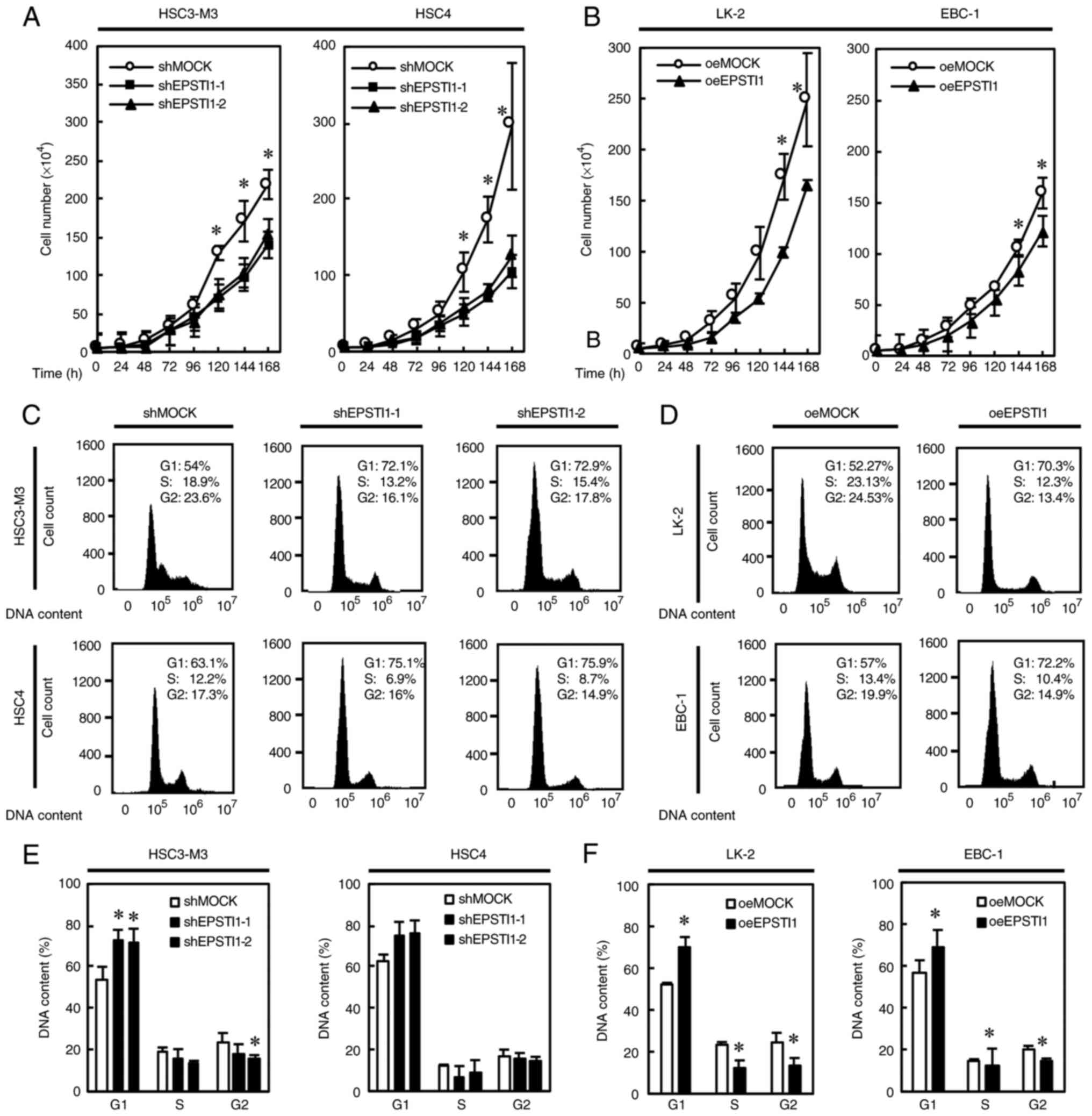 |
Figure 3.
Inhibition of cell proliferation and induction of cell cycle arrest in the G1 phase is associated with modified EPSTI1 expression. Genetically modified expression of EPSTI1; (A) downregulation of the gene in two OSCC cell lines was associated with significantly suppressed cell proliferation and (C and E) increase in the G1 population, although statistical significance was marginal in HSC4 (P=0.0533), suggesting cell cycle arrest. Similarly, (B) overexpression of the gene in two LSCC cell lines significantly suppressed cell proliferation and (D and F) induced cell cycle arrest in the G1 phase, with a significant decrease in the population of cells in the S and G2 phases in the LK-2 cell line, and in the population of cells in the G2 phase in the EBC-1 cell line. Each experiment was performed in triplicate and repeated six times for A and B, and three times for C-F. Data are presented as the means ± SE. *P<0.05. EPSTI, epithelial-stromal interaction 1; OSCC, oral squamous cell carcinoma; LSCC, lung squamous cell carcinoma; SE, standard error.
|
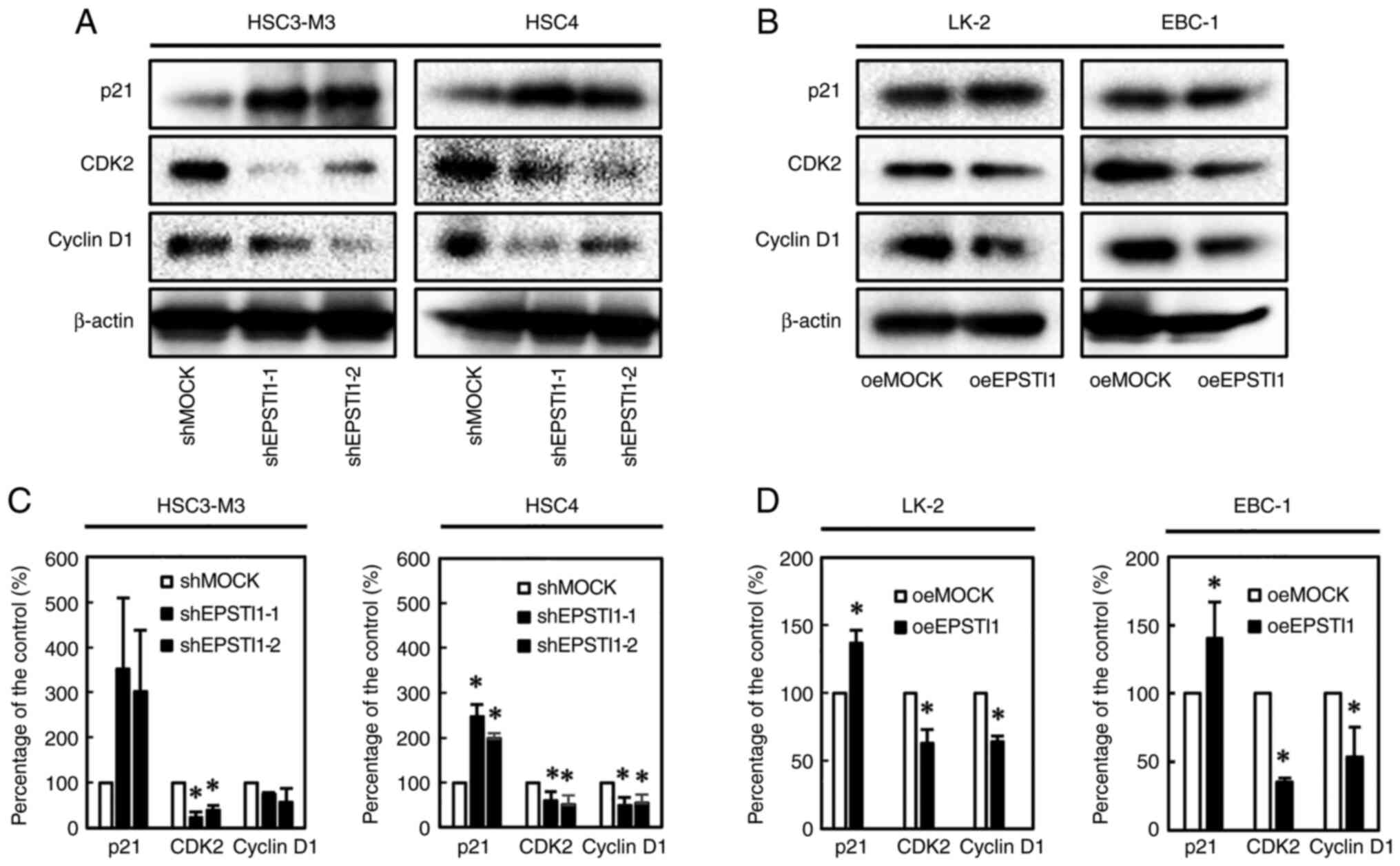 |
Figure 4.
Expression of the cell-cycle-related genes, p21, CDK2, and Cyclin D1. Western blotting revealed enhanced expression of p21, and suppressed expression of CDK2 and Cyclin D1 associated with modified EPSTI1 expression (upregulation and downregulation, respectively) in the (A and C) OSCC and (B and D) LSCC cell lines, although statistical significance was marginal for p21 (P=0.08) and cyclin D1 (P=0.06) in HSC3-M3, which appears consistent with cell cycle arrest in each of the cell lines. Each experiment was performed in triplicate and repeated three times, and data are presented as the means ± SE. *P<0.05. EPSTI, epithelial-stromal interaction 1; OSCC, oral squamous cell carcinoma; LSCC, lung squamous cell carcinoma; SE, standard error.
|
Altered expression of EPSTI1 induces altered cell migration ability and epithelial-mesenchymal transition
The wound healing assay revealed significantly decreased cell migration ability in OSCC cell lines with downregulated EPSTI1 expression, HSC3-M3 and HSC4 (Fig. 5A) and in the LSCC cell line EBC-1 with upregulated EPSTI1 expression (LK-2 was not used since the cells were scattered once the cells reached confluence) (Fig. 5B). The expression levels of the epithelial-mesenchymal transition (EMT)-related proteins were evaluated by western blot analysis. Significant upregulation of E-cadherin and downregulation of N-cadherin and fibronectin, suggestive of a suppressed EMT phenotype, were observed in OSCC cell line with downregulated EPSTI1 expression and LSCC cell line with upregulated EPSTI1 expression with exception in fibronectin in HSC4 which exhibited marginal statistical significance (P=0.08) (Fig. 5C-F).
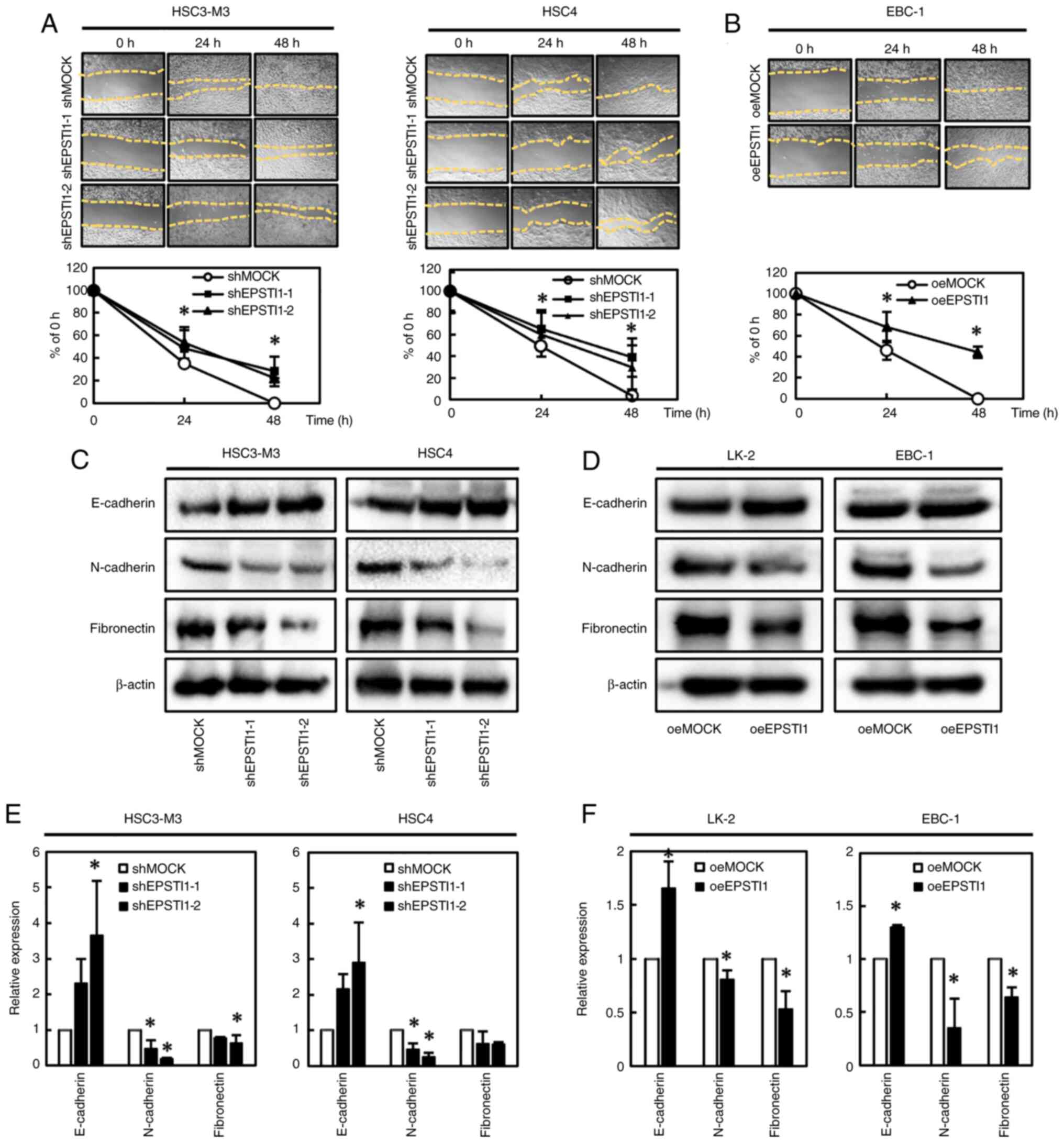 |
Figure 5.
Decreased cell migration capability and reversal of the EMT phenotype in tumor cells with modified EPSTI1 expression. (A) Downregulation of EPSTI1 in OSCC and (B) upregulation of the gene in LSCC were associated with significant suppression of the cell migration ability, suggesting reversal of the EMT phenotype. Accordingly, (C and E) downregulation of EPSTI1 in OSCC and (D and F) upregulation of the gene in LSCC were also associated with significantly increased expression levels of E-cadherin and significantly suppressed expression levels of N-cadherin and fibronectin with exception in fibronectin in HSC4 which exhibited marginal statistical significance (P=0.08), again suggesting reversal of the EMT phenotype. The cell migration assay in LSCC was performed using only EBC-1, but not LK-2. Each experiment was performed in triplicate and repeated three times, and data are presented as the means ± SE. *P<0.05. EMT, epithelial-mesenchymal transition; EPSTI, epithelial-stromal interaction 1; OSCC, oral squamous cell carcinoma; LSCC, lung squamous cell carcinoma; SE, standard error.
|
EPSTI1-related genes as shown by PCR arrays
PCR arrays identified some upstream and downstream genes in relation to the altered EPSTI1 expression in the two OSCC and two LSCC cell lines, as summarized in Tables I and II. Overexpression of carbonic anhydrase 9 (CA9) and downregulation of C-C motif chemokine ligand 2 (CCL2) were identified in both the EPSTI1-suppressed OSCC cell lines, as compared to the corresponding control cell line (Table I), whereas downregulation of cyclin D2 (CCND2), baculoviral IAP repeat containing 3 (BIRC3) and insulin-like growth factor binding protein 7 (IGFBP7) were identified in all the EPSTI1-overexpressing LSCC cell lines, as compared to the corresponding control cell line (Table II).
 |
Table I.
Altered gene expression in OSCC with EPSTI1 knockdown as assessed by PCR array.
|
Table I.
Altered gene expression in OSCC with EPSTI1 knockdown as assessed by PCR array.
| HSC3-M3 |
HSC4 |
|
|
| Gene symbola (ratiob) |
Gene symbol (ratio) |
|
|
| Upregulated |
Downregulated |
Upregulated |
Downregulated |
| CA9 (7.2)c |
CCL2 (−3.4) |
LDHA (79.9) |
XIAP (−224.9) |
| SERPINF1 (4.2) |
BIRC3 (−2.5) |
MKI67 (15.0) |
IGFBP3 (−38.5) |
| FGF2 (3.5) |
SERPINB2 (−2.4) |
SERPINB2 (14.7) |
HMOX1 (−35.7) |
| TBX2 (3.0) |
|
BMI1 (7.9) |
CCL2 (−26.5) |
| PGF (2.9) |
|
PINX1 (5.8) |
TBX2 (−8.3) |
| DDIT3 (2.1) |
|
DKC1 (4.3) |
DDIT3 (−7.3) |
| |
|
CA9 (3.9) |
FLT1 (−5.8) |
| |
|
WEE1 (2.5) |
SOX10 (−5.8) |
| |
|
LIG4 (2.4) |
TEK (−5.8) |
| |
|
AURKA (2.4) |
EPO (−5.4) |
| |
|
APAF1 (2.3) |
IGFBP5 (−5.4) |
| |
|
VEGFC (2.3) |
LPL (−5.2) |
| |
|
MAPK14 (2.1) |
PPP1R15A (−4.6) |
| |
|
CDH2 (2.1) |
GADD45G (−3.4) |
| |
|
CDC20 (2.0) |
COX5A (−3.3) |
| |
|
|
CASP7 (−3.1) |
| |
|
|
GSC (−2.9) |
| |
|
|
G6PD (−2.3) |
| |
|
|
ANGPT2 (−2.2) |
| |
|
|
KDR (−2.2) |
| |
|
|
CASP9 (−2.2) |
| |
|
|
TINF2 (−2.1) |
| |
|
|
SNAI1 (−2.1) |
 |
Table II.
Altered gene expression in LSCC with EPSTI1 overexpression, as assessed by PCR array.
|
Table II.
Altered gene expression in LSCC with EPSTI1 overexpression, as assessed by PCR array.
| LK-2 |
EBC-1 |
|
|
| Gene symbola (ratiob) |
Gene symbol (ratio) |
|
|
| Upregulated |
Downregulated |
Upregulated |
Downregulated |
| TBXT2 (28.0) |
CCND2 (−17.3)c |
HMOX1 (9.5) |
CDC20 (−14.5) |
| LPL (16.2) |
SNAI2 (−11.3) |
DDIT3 (9.5) |
MKI67 (−11.1) |
| IGFBP5 (5.3) |
KRT14 (−6.2) |
XIAP (3.4) |
IGFBP7 (−10.7) |
| CDH2 (3.9) |
SERPINB2 (−4.1) |
LIG4 (3.4) |
LPL (−4.7) |
| KDR (3.7) |
SLC2A1 (−2.4) |
CASP9 (3.0) |
STMN1 (−4.6) |
| HGDC (3.1) |
BIRC3 (−2.3) |
ARNT (2.9) |
TEK (−4.0) |
| SERPINF1 (3.1) |
IGFBP7 (−2.2) |
ERCC5 (2.7) |
CCL2 (−3.7) |
| GSC (2.6) |
ACSL4 (−2.1) |
IGFBP3 (2.7) |
AURKA (−3.6) |
| GADD45G (2.4) |
|
PPP1R15A (2.6) |
CCND3 (−2.9) |
| SOD1 (2.2) |
|
SOX10 (2.2) |
LDHA (−2.8) |
| SNAL1 (2.1) |
|
TNKS (2.2) |
CCND2 (−2.7) |
| ANGPT1 (2.1) |
|
PINX1 (2.1) |
PGF (−2.6) |
| |
|
OCLN (2.0) |
BIRC3 (−2.3) |
| |
|
|
SKP2 (−2.1) |
Discussion
The present study revealed significantly higher expression levels of ESPTI1 in all the four OSCC cell lines examined in the present study as compared to the corresponding control cell line, and significantly lower expression levels of the EPSTI1 gene in all the three LSCC cell lines examined as compared to the corresponding control cell line. Both OSCC cell lines with forced downregulation of the gene and LSCC cell lines with overexpression of the gene exhibited significantly suppressed tumor cell proliferation ability. Analyses of the cell cycle and of genes related to the cell cycle, i.e., p21, CDK2 and cyclin D1, generally supported the finding of cell cycle arrest in the G1 phase in both the aforementioned OSCC and LSCC cell lines. OSCC cell lines with downregulation of EPSTI1 and LSCC cell lines with upregulation of EPSTI1 similarly revealed suppressed EMT, as assessed by the cell migration activity and cellular expression levels of E-cadherin, N-cadherin and fibronectin. Therefore, EPSTI1 was demonstrated to play roles in enhancing the malignant phenotypes, including the cell proliferation ability and EMT in OSCC, whereas it played the opposite roles in LSCC.
Overexpression of EPSTI1 was first identified in breast cancer, and overexpression of the gene is considered to drive the malignant phenotype, including the cell proliferation and migration abilities, invasiveness, EMT, and escape from apoptosis (12–14,18,19). It has also been identified as being upregulated in cell lines derived from peritoneal disseminations of colorectal cancer as compared to the cell lines derived from primary colorectal cancers (16). In pancreatic cancer, gene expression was associated with resistance to therapy with an oncolytic virus (17).
In the present study, EPSTI1 was demonstrated to enhance the malignant phenotype in OSCC, similar to breast (13,14,18,19) and colorectal cancers (16); notably, the gene played the opposite role in the LCSS cell lines. The gene expression in clinical samples obtained from the Oncomine database revealed higher expression levels of the gene in cancer tissues as compared to normal tissues (26). Unfortunately, relevant data could not be obtained in the Oncomine database regarding EPSTI1 expression in LSCC (27). Although the contrasting roles of the same gene in different cancers, i.e., OSCC and LSCC, appear somewhat unusual, a few similar examples have been reported previously. Specifically, a T-box family member, TBX2, was reported to be amplified and overexpressed in breast cancer (28) and its overexpression has been revealed to be associated with a high pathological grade in prostatic cancer (29); conversely, the gene has been revealed to be downregulated in non-small cell lung cancer (30); although further details have not yet been elucidated. Therefore, it is considered that the present study findings, provide further insight into understanding the multi-functional roles of EPSTI1.
To elucidate the mechanisms underlying these phenomena, a series of PCR array assays were performed. Upregulation of the CA9 gene, observed in both the OSCC cell lines with forced downregulation of EPSTI1 expression, has been revealed to augment the cell proliferation and transformation ability under hypoxic conditions, under the regulation of the transcription factor HIF-1α (31). Downregulation of the CCL2 gene, a member of the chemokine superfamily, observed in the same cell lines, has been demonstrated to promote cancer progression through recruiting inflammatory cells in multiple cancers (32–34), and to promote EMT-associated cell migration ability in OSCC (35). Conversely, downregulation of CCND2, BIRC3, and IGFBP7, observed in both the EPSTI1-overexpressing LSCC cell lines, has been revealed to promote G1/S phase transition (CCND2) (36), suppress apoptosis by inhibiting caspase-3 (BIRC3) (37), and to promote TGF-b-induced EMT in human renal proximal tubular epithelial cells (IGFBP7) (38). These studies, except for the information concerning CA9, are all consistent with the phenomena observed in the OSCC and LSCC cell lines in the present study. There were other inconsistencies in the PCR array results, in addition to the case of CA9. Although western blotting revealed downregulation of N-cadherin in the HSC4 cell line with EPSTI1 knockdown, the PCR array disclosed upregulation of CDH2 in the same cell line. Since the results of western blot analysis are likely to be more reliable, comprehensive gene analysis by PCR array may be susceptible to error. MKI67, as well as CA9, revealed to be upregulated in the HSC4 cells with EPSTI1 knockdown, reportedly augment cell proliferation, whereas knockdown of EPSTI1 in the HSC4 suppressed cell proliferation in the present research. The finding that there were no commonly altered gene expression between the OSCC and LSCC cell lines may indicate that EPSTI1 acts differently in the two cancers, which is also consistent with the findings of the present study.
The present study had some limitations. The results were obtained solely from studies using cell lines. The findings remain to be validated in tumor samples collected from patients with OSCC and LSCC. Expression of EPSTI1 in normal tissue and cancer tissue according to the patient characteristics, including the clinical stage of the cancer, tumor doubling time, and sensitivity to chemotherapy, are expected to further clarify the clinical relevance of EPSTI1 in these diseases. Although the comprehensive analyses with a PCR array were solely exploratory, the findings failed to explain mechanisms involved in the contrasting functions of the gene. New studies to elucidate the clinical relevance of the gene and to explore the mechanisms underlying the diverse functions of the gene according to the cancer type are warranted.
In conclusion, the present research revealed, for the first time, the opposite functions of EPSTI1, namely, of promoting and suppressing cancer progression, according to the type of cancer; i.e., its overexpression promoted the malignant phenotype in OSCC, whereas its downregulation promoted the malignant phenotype in LSCC.
Acknowledgements
Not applicable.
Funding
The present study was financially supported by grants from the Ministry of Education, Culture, Sports, Science and Technology in Japan (Kiban-C grant nos. 17K09647 and 20K08561), and the Kanto Academic Alliance for Fostering Cancer Professionals. The abstract was presented at the 111rd Annual Meeting of the American Association for Cancer research; June 22-24 2020 in a virtual meeting, and published as abstract no. 2580 in Cancer Res 80 (Suppl 16): 2580.
Availability of data and materials
Data supporting the findings of the present study are available upon reasonable request from the corresponding author.
Authors' contributions
MF, MA, KU, MS, NK, HT and YT contributed to the design of the study. Cell culture and data acquisition were performed by MF, TC and RF. MF, MA, and AT performed statistical analyses, and interpretation of data was performed by all the authors. MF, MA and YT contributed to drafting the manuscript. All authors contributed to revising the manuscript critically for important intellectual content, as well as reading and approving the final version of the manuscript to be published.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
|
1
|
Dotto GP and Rustgi AK: Squamous cell cancers: A unified perspective on biology and genetics. Cancer Cell. 29:622–637. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chi AC, Day TA and Neville BW: Oral cavity and oropharyngeal squamous cell carcinoma-an update. CA Cancer J Clin. 65:401–421. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Silverman S Jr: Demographics and occurrence of oral and pharyngeal cancers. The outcomes, the trends, the challenge. J Am Dent Assoc. 132 (Suppl):S7–S11. 2001. View Article : Google Scholar
|
|
5
|
Shield KD, Ferlay J, Jemal A, Sankaranarayanan R, Chaturvedi AK, Bray F and Soerjomataram I: The global incidence of lip, oral cavity, and pharyngeal cancers by subsite in 2012. CA Cancer J Clin. 67:51–64. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Li Y, Gu J, Xu F, Zhu Q, Ge D and Lu C: Transcriptomic and functional network features of lung squamous cell carcinoma through integrative analysis of GEO and TCGA data. Sci Rep. 8:158342018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sharma B and Kanwar SS: Phosphatidylserine: A cancer cell targeting biomarker. Semin Cancer Biol. 52:17–25. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Loong HH, Kwan SS, Mok TS and Lau YM: Therapeutic strategies in EGFR mutant non-small cell lung cancer. Curr Treat Options Oncol. 19:582018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bonner JA, Harari PM, Giralt J, Azarnia N, Shin DM, Cohen RB, Jones CU, Sur R, Raben D, Jassem J, et al: Radiotherapy plus cetuximab for squamous-cell carcinoma of the head and neck. New Engl J Med. 354:567–578. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Vermorken JB, Mesia R, Rivera F, Remenar E, Kawecki A, Rottey S, Erfan J, Zabolotnyy D, Kienzer HR, Cupissol D, et al: Platinum-based chemotherapy plus cetuximab in head and neck cancer. N Engl J Med. 359:1116–1127. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Thatcher N, Hirsch FR, Luft AV, Szczesna A, Ciuleanu TE, Dediu M, Ramlau R, Galiulin RK, Bálint B, Losonczy G, et al: Necitumumab plus gemcitabine and cisplatin versus gemcitabine and cisplatin alone as first-line therapy in patients with stage IV squamous non-small-cell lung cancer (SQUIRE): An open-label, randomised, controlled phase 3 trial. Lancet Oncol. 16:763–774. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Nielsen HL, Ronnov-Jessen L, Villadsen R and Petersen OW: Identification of EPSTI1, a novel gene induced by epithelial-stromal interaction in human breast cancer. Genomics. 79:703–710. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Tran-Thanh D and Done SJ: The role of stromal factors in breast tumorigenicity. Am J Pathol. 176:1072–1074. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
de Neergaard M, Kim J, Villadsen R, Fridriksdottir AJ, Rank F, Timmermans-Wielenga V, Langerød A, Børresen-Dale AL and Petersen OW: Epithelial-stromal interaction 1 (EPSTI1) substitutes for peritumoral fibroblasts in the tumor microenvironment. Am J Pathol. 176:1229–1240. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kim YH, Lee JR and Hahn MJ: Regulation of inflammatory gene expression in macrophages by epithelial-stromal interaction 1 (Epsti1). Biochem Biophys Res Commun. 496:778–783. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kim SC, Hong CW, Jang SG, Kim YA, Yoo BC, Shin YK, Jeong SY, Ku JL and Park JG: Establishment and characterization of paired primary and peritoneal seeding human colorectal cancer cell lines: Identification of genes that mediate metastatic potential. Transl Oncol. 11:1232–1243. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hastie E, Cataldi M, Moerdyk-Schauwecker MJ, Felt SA, Steuerwald N and Grdzelishvili VZ: Novel biomarkers of resistance of pancreatic cancer cells to oncolytic vesicular stomatitis virus. Oncotarget. 7:61601–61618. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li T, Lu H, Shen C, Lahiri SK, Wason MS, Mukherjee D, Yu L and Zhao J: Identification of epithelial stromal interaction 1 as a novel effector downstream of Kruppel-like factor 8 in breast cancer invasion and metastasis. Oncogene. 33:4746–4755. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Capdevila-Busquets E, Badiola N, Arroyo R, Alcalde V, Soler-Lopez M and Aloy P: Breast cancer genes PSMC3IP and EPSTI1 play a role in apoptosis regulation. PLoS One. 10:e01153522015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Fijak M and Meinhardt A: The testis in immune privilege. Immunol Rev. 213:66–81. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kosova G, Scott NM, Niederberger C, Prins GS and Ober C: Genome-wide association study identifies candidate genes for male fertility traits in humans. Am J Human Genet. 90:950–961. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ishii T, Onda H, Tanigawa A, Ohshima S, Fujiwara H, Mima T, Katada Y, Deguchi H, Suemura M, Miyake T, et al: Isolation and expression profiling of genes upregulated in the peripheral blood cells of systemic lupus erythematosus patients. DNA Res. 12:429–439. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Sun JL, Zhang HZ, Liu SY, Lian CF, Chen ZL, Shao TH, Zhang S, Zhao LL, He CM, Wang M, et al: Elevated EPSTI1 promote B cell hyperactivation through NF-kappaB signalling in patients with primary Sjogren's syndrome. Ann Rheum Dis. 79:518–524. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Meng X, Yang D, Yu R and Zhu H: EPSTI1 is involved in IL-28A-mediated inhibition of HCV infection. Mediators Inflamm. 2015:7163152015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yamano Y, Uzawa K, Shinozuka K, Fushimi K, Ishigami T, Nomura H, Ogawara K, Shiiba M, Yokoe H and Tanzawa H: Hyaluronan-mediated motility: A target in oral squamous cell carcinoma. Int J Oncol. 32:1001–1009. 2008.PubMed/NCBI
|
|
26
|
Peng CH, Liao CT, Peng SC, Chen YJ, Cheng AJ, Juang JL, Tsai CY, Chen TC, Chuang YJ, Tang CY, et al: A novel molecular signature identified by systems genetics approach predicts prognosis in oral squamous cell carcinoma. PLoS One. 6:e234522011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Hou J, Aerts J, den Hamer B, van Ijcken W, den Bakker M, Riegman P, van der Leest C, van der Spek P, Foekens JA, Hoogsteden HC, et al: Gene expression-based classification of non-small cell lung carcinomas and survival prediction. PLoS One. 5:e103122010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Douglas NC and Papaioannou VE: The T-box transcription factors TBX2 and TBX3 in mammary gland development and breast cancer. J Mammary Gland Biol Neoplasia. 18:143–147. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Du WL, Fang Q, Chen Y, Teng JW, Xiao YS, Xie P, Jin B and Wang JQ: Effect of silencing the TBox transcription factor TBX2 in prostate cancer PC3 and LNCaP cells. Mol Med Rep. 16:6050–6058. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Khalil AA, Sivakumar S, Lucas FAS, McDowell T, Lang W, Tabata K, Fujimoto J, Yatabe Y, Spira A, Scheet P, et al: TBX2 subfamily suppression in lung cancer pathogenesis: A high-potential marker for early detection. Oncotarget. 8:68230–68241. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ohtaki Y, Shimizu K, Kawabata-Iwakawa R, Gombodorj N, Altan B, Rokudai S, Yamane A, Kaira K, Yokobori T, Nagashima T, et al: Carbonic anhydrase 9 expression is associated with poor prognosis, tumor proliferation, and radiosensitivity of thymic carcinomas. Oncotarget. 10:1306–1319. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Ding M, He SJ and Yang J: MCP-1/CCL2 mediated by autocrine loop of PDGF-BB promotes invasion of lung cancer cell by recruitment of macrophages via CCL2-CCR2 axis. J Interferon Cytokine Res. 39:224–232. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
De la Fuente Lopez M, Landskron G, Parada D, Dubois-Camacho K, Simian D, Martinez M, Romero D, Roa JC, Chahuán I, Gutiérrez R, et al: The relationship between chemokines CCL2, CCL3, and CCL4 with the tumor microenvironment and tumor-associated macrophage markers in colorectal cancer. Tumour Biol. 40:10104283188100592018. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Kitamura T, Qian BZ, Soong D, Cassetta L, Noy R, Sugano G, Kato Y, Li J and Pollard JW: CCL2-induced chemokine cascade promotes breast cancer metastasis by enhancing retention of metastasis-associated macrophages. J Exp Med. 212:1043–1059. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Ling Z, Yang X, Chen X, Xia J, Cheng B and Tao X: CCL2 promotes cell migration by inducing epithelial-mesenchymal transition in oral squamous cell carcinoma. J Oral Pathol Med. 48:477–482. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Liu Y, Liu WB, Liu KJ, Ao L, Cao J, Zhong JL and Liu JY: Overexpression of miR-26b-5p regulates the cell cycle by targeting CCND2 in GC-2 cells under exposure to extremely low frequency electromagnetic fields. Cell Cycle. 15:357–367. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Lee JY, Tokumoto M, Hwang GW, Lee MY and Satoh M: Identification of ARNT-regulated BIRC3 as the target factor in cadmium renal toxicity. Sci Rep. 7:172872017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Watanabe J, Takiyama Y, Honjyo J, Makino Y, Fujita Y, Tateno M and Haneda M: Role of IGFBP7 in diabetic nephropathy: TGF-β1 induces IGFBP7 via Smad2/4 in human renal proximal tubular epithelial cells. PLoS One. 11:e01508972016. View Article : Google Scholar : PubMed/NCBI
|



















