Introduction
Lymphomas represent the vast majority of tumors of the lymphoid system and 4.3% of all new cancer cases in the United States (1). Resistance of B cell Non-Hodgkin lymphomas to chemoimmunotherapy still represents a serious health issue. The aggressive subtypes especially pose unmet therapeutic challenges (2). Methylation has gained attention in the treatment of high-risk lymphomas since aberrant methylation may represent a mechanism for chemoresistance (3).
We previously described a novel panel of genes that are differentially methylated in human B lymphocytes purified from follicular lymphoma (FL) and diffuse large B cell lymphoma (DLBCL) (4). In the present work, the layer of regulation represented by gene promoter methylation of Krüppel-like factor 4 (KLF4), death associated protein 1 (DAPK1) and spastic paraplegia 20 (SPG20; protein product, Spartin) in DLBCL-derived cell lines were investigated. These three targets were previously demonstrated to be hypermethylated in primary B lymphocytes of FL and DLBCL (4). By contrast, the present study assessed the methylation status of these loci in peripheral blood mononuclear cells (PBMCs) from healthy donors, which, to the best of our knowledge, has not yet been investigated.
KLF4 is a member of the KLF transcription factor family containing zinc-fingers and is regulated by transcriptional and post-transcriptional mechanisms (5). Promoter hypermethylation has been reported in lymphomas of both B and T cell origin (6,7). KLF4 hypermethylation-mediated inactivation has been associated with pediatric T cell acute lymphoblastic leukemia leading to the pathological activation of the MAP2K7 kinase pathway (7). Hypermethylated KLF4 has also been associated with the poor prognosis of patients with splenic marginal zone lymphomas (SMZL) and SMZL cell lines exposed to hypomethylating agents show a reduced proliferation (6,7).
DAPK1 plays a role in lymphomas, as recently demonstrated by a meta-analysis where hypermethylation of the promoter was found to be associated with the 5-year survival rate of patients (8,9). Overall, DAPK1 hypermethylation corresponds to a significantly lower survival and represents an independent, negative prognostic biomarker (8).
SPG20, which codes for a protein linked to reduced mitochondrial cytochrome c oxidase activity (10,11), was one of the observed novel methylated targets. This gene is involved in Troyer syndrome, which is a form of autosomal recessive hereditary spastic paraplegia caused by deleterious mutations in SPG20 that is associated with the loss of the protein in the skeletal muscle (10,12).
The present study aimed to elucidate the profile of promoter methylation of these selected target genes upon treatment with the two widely used hypomethylating agents 5′-aza-cytidine (5AZA) and 5′-aza-2-deoxycitidine (decitabine) (13). These two epigenetic drugs determine a global hypomethylating effect on the genome, but selectively affect only specific promoters, thus underlying the importance of specific methylation patterns during cell proliferation. DNA methyltransferases (DNMTs) are nuclear enzymes targeted by hypomethylating drugs (13,14). DNMT1 is a well-characterized methyltransferase responsible for maintenance of the methylation status during DNA replication. DNMT3a is considered a de novo DNMT and has been characterized mainly during embryonal development. DNMT3a emerged as a tumor suppressor in several hematological malignancies, including acute myeloid leukemia, myelodysplastic syndrome and T cell lymphoma, and it is now accepted that DNMT3a also contributes to the maintenance and dynamic remodeling of methylation in differentiated cells (15–17).
Thus, the current study investigated DNMT1 and DNMT3a in response to 5AZA or decitabine and by assessing the effects of their specific silencing. The results were aimed at evaluating the effects on promoter methylation of KLF4, DAPK1 and SPG20.
Materials and methods
Cell culture and proliferation assays
The Toledo cell line (ATCC-CRL-2631) was purchased from American Type Culture Collection. NU-DUL-1 cells (ACC 579) were purchased from GSMZ-German Collection of Microorganisms and Cell Cultures GmbH. The GNE-587170 cell line was supplied by Dr Mark Minden's laboratory (Ontario Cancer Institute, Toronto, Canada). Non-Hodgkin lymphoma-derived cell lines were cultured in RPMI-1640 medium (Euroclone SpA) with penicillin-streptomycin at 10 U/ml (Euroclone SpA) and 10% FBS (Gibco; Thermo Fisher Scientific, Inc.). Cells were routinely tested for mycoplasma contamination using the MycoAlert™ Mycoplasma Detection Kit (Lonza Group, Ltd.). Cell lines were authenticated by PCR-single-locus-technology (Eurofins Scientific).
Hypomethylating drugs were used at a concentration ranging between 0.05–10 µM, which is specified in each figure. The drugs were dissolved in water and the control is represented by untreated cells. Cells were incubated in a humidified incubator at 37°C with 5% CO2 for the indicated time each experiment.
Viability assays were performed by using a Cell Counting Kit-8 (CCK-8; Boster Biological Technology). Briefly, 20,000 cells were seeded in a 96-well plate and, after 48 h of incubation, 10 µl/well tetrazolium salt was added. Spectrophotometric reading at 450 nm with a 620 nm filter was performed after 1 h of incubation.
Caspase-3/7 activation assay
Time-course apoptosis assays following either 5AZA or decitabine treatment were performed using an IncuCyte® S3 (Essen Bioscience). Cells were seeded at a density of 20,000 cells/well. The reagents used to label the cells were IncuCyte® Nuclight Rapid Red and Cell Health Reagent Caspase-3/7 (both used at a 1:2,000 final dilution in the wells; Essen Bioscience). Cells were incubated in a humidified incubator at 37°C with 5% CO2. Each condition was performed in triplicate.
Immunocytochemistry
The cytologic specimen was prepared using Cytospin™ 4 (Thermo Fisher Scientific, Inc.) and the glass was fixed with 95% ethanol for 20 min at room temperature. Immunocytochemistry was performed with Monoclonal Mouse Anti-Human Ki-67 Antigen Clone MIB-1 (1:100; cat. no. M724001-2; Dako; Agilent Technologies, Inc.) for 40 min at room temperature. The slides were pre-treated with heat antigen retrieval in EDTA buffer for 64 min. The immunocytochemical staining was developed on Ventana BenchMark ULTRA platform using the pre-diluted UltraView DAB Detection Kit (cat. no. 05269806001; Ventana Medical Systems, Inc.; Roche Diagnostics) for 8 min at 36°C. Slides were counterstained with hematoxylin for 3 min at room temperature. Slides were observed on a Nikon Eclipse 80i light microscope (magnification, ×40; Nikon Corporation).
Analysis of gene-specific, global chromatin and genome-wide methylation
Quantitative gene-specific methylation was performed via EpiTect Methyl II qPCR (Qiagen S.r.l.) as previously described (4).
Global chromatin methylation was performed using the MethylFlash Methylated DNA Quantification kit (EpiGentek Group, Inc.) with 100 ng DNA/sample, according to the manufacturer's instructions. Methylation-specific PCR (MS-PCR) was performed with 50 ng bisulfite-converted genomic DNA. Primers and PCR conditions are presented in Tables SI and SII, respectively. Bisulfite conversion and cleanup was performed with the EpiTect Bisulfite kit (Qiagen S.r.l.). Genome-wide methylation analysis was performed on an Infinium Human Methylation 450K BeadChip array (Illumina, Inc.). DNA samples underwent bisulfite treatment with the EZ DNA methylation kit (Zymo Research Corp.) and were then hybridized on the Infinium 450K BeadChip arrays, following the manufacturer's protocols. Signal intensities and β-values were exported from Beadstudio 2.0 software (Illumina, Inc.), applying default settings. All bioinformatics processing was performed with R version 3.2.0 (https://cran.r-project.org/bin/windows/base/old/3.2.0/). Raw intensity signals were imported and processed using the minfi package (https://www.bioconductor.org/help/course-materials/2015/BioC2015/methylation450k.html). All samples had an average detection P<0.001, indicating good quality data. For quality control (QC), histograms and boxplots were plotted for signal intensities and β-values. All samples were quantile normalized, β-values and M-values were also calculated. The probes containing SNPs were removed as they can have important consequences on the downstream analysis. For clustering and statistical tests, probes mapped on sex chromosomes were excluded. Unsupervised analysis was performed on all the retained probes using hierarchical clustering with Euclidean distance and Ward linkage on the β-values. Supervised analysis of differential methylation between groups was performed using Limma package (https://bioconductor.org/packages/release/bioc/html/limma.html). The Benjamini-Hochberg multiple test correction was applied to control for false positives. Probes showing an unadjusted FDR <0.05 were considered differentially methylated.
Reverse transcription-quantitative PCR (RT-qPCR), immunoblotting and gene silencing
Total RNA was extracted using TRIzol (cat. no. 15596026; Invitrogen; Thermo Fisher Scientific, Inc.) with the RNA isolation protocol (W.M. Keck Foundation Biotechnology microarray Research Laboratory at Yale University) and RT was performed with the iScript™ cDNA Synthesis kit (Bio-Rad Laboratories, Inc.) according to the manufacturer's protocol. Gene expression was performed via qPCR with the SsoFast™ EvaGreen® Supermix (Bio-Rad Laboratories, Inc.) following the manufacturer's protocols. The Cq value of 37 cycles was chosen as the lower threshold (cutoff) for gene expression using the 2−∆∆Cq method of quantification (18). Ribosomal protein S7 was used as the reference gene in RT-qPCR. Primers and amplification conditions are reported in Tables SI and SII, respectively.
Immunoblotting was performed as previously described (19). Briefly, 2×106 cells were lysed in RIPA buffer (150 mM NaCl, 5 mM EDTA pH 8.0, 50 mM Tris pH 8.0, 1% NP-40, 0.5% Na-deoxycholate, 0.1% SDS in water). Total lysates were quantified by spectrophotometric reading at 750 nm [FLUOstar® Omega (BMG Labtech GmbH)] with the Lowry method (DC™ Protein Assay Kit; Bio-Rad Laboratories, Inc.) and 20 µg protein/sample were separated on Bolt Bis-Tris Plus precast 4–12% polyacrylamide gels (Thermo Fisher Scientific, Inc.) and blotted onto nitrocellulose membranes. Blocking was performed with 4% milk in PBS with 0.1% Tween-20 for 30 min at room temperature. Rabbit primary antibodies against DNMT1 (cat. no. 5032), DNMT3a (clone D23G1; cat. no. 3598), SPG20 (cat. no. 13520S), KLF4 (cat. no. 12173) and poly (ADP-ribose) polymerase (PARP; cat. no. 9532) were purchased from Cell Signaling Technology, Inc. Anti-DAPK1 (cat. no. MA1-24696) was purchased from Thermo Fisher Scientific, Inc. All antibodies were used at a final dilution of 1:2,000. Mouse anti-β-actin antibody (clone AC-15; cat. no. A1978) was purchased from Sigma-Aldrich (Merck KGaA) and used at a final dilution of 1:20,000. The incubation with the primary antibodies was performed overnight at 4°C. The membrane was then incubated for 1 h at room temperature with anti-rabbit (cat. no. NA934V; Amersham; Cytiva) and anti-mouse (cat. no. NXA931V; Amersham; Cytiva) secondary antibodies at dilutions of 1:10,000. For visualization ECL detection reagent was used (Amersham; Cytiva) and the chemiluminescence signal was acquired through ChemiDoc MP (Bio-Rad Laboratories, Inc.) and analyzed by ImageLab version 5.2 (Bio-Rad Laboratories, Inc.).
Gene silencing was performed through 4D-Nucleofector™ Core Unit (Lonza Group, Ltd.). The buffer used was the Amaxa™ SF cell line 4D Nucleofector™ X kit (cat. no. V4XC-2024; Lonza Group, Ltd.). Small interfering (si)RNAs were purchased from Ambion (Thermo Fisher Scientific, Inc.), diluted in molecular biology grade water and used at a final concentration of 100 nM. The cells were either transfected with siRNAs against DNMT1 (cat. nos. 4390824, s4216 and s4217; Ambion; Thermo Fisher Scientific, Inc.), DNMT3a (cat. nos. 4392420 and s200426; Ambion; Thermo Fisher Scientific, Inc.) or both. Negative control #2 siRNA (cat. no. 4390846) was used as the control (scramble). Sequences are listed in Table SI. Cells were incubated in a humidified incubator at 37°C with 5% CO2 for 48 h before being collected.
Human PBMCs
PBMCs were purified from the peripheral blood using Histopaque®−1077 (Sigma-Aldrich; Merck KGaA) according to the manufacturer's instructions. Blood samples from healthy donors were collected at the Azienda Unità Sanitaria Locale - IRCCS di Reggio Emilia (Reggio Emilia, Italy), between October 2017 and June 2018. The age range was 19–60 years (five female patients and 22 male patients). The use of PBMCs upon written informed consent was previously approved by the Local Ethics Committee of Reggio Emilia (Comitato Etico Provinciale; approval no. 2016/0025053).
Data mining
IncuCyte® S3 Software 2020B (Essen Bioscience) was used to analyze and graph the time-course fluorescence data. The probes specific to SPG20 locus were sorted from the Infinium Human Methylation 450K BeadChip array (Illumina, Inc.) and the HumanMethylation 27 BeadChip (Illumina, Inc., available through the repository ‘ArrayExpress’ https://www.ebi.ac.uk/arrayexpress/; accession no. E-MTAB-417), and subsequently mapped onto NCBI's genome browser for humans (https://www.ncbi.nlm.nih.gov/genome/gdv/; GRCh38.p13; accession no. GCA_000001405.28). Heatmaps of the β-values were generated through Displayr (https://www.displayr.com/).
Statistical analysis
Data analysis was performed with GraphPad Prism 5.0 (GraphPad Software, Inc.). One-way ANOVA followed by Bonferroni's post hoc test (multiple comparisons) were performed. Data are represented as the mean ± standard error of the mean. Figures were prepared with Adobe Illustrator CS4 (Adobe Systems, Inc.). P<0.05 was considered to indicate a statistically significant difference.
Results
Characteristic profile of promoter methylation on lymphoma cell lines and effects of hypomethylating drugs
It was observed that lymphoma cell lines displayed a characteristic profile of promoter methylation on specific targets, including tumor suppressors. A panel of nine target promoters were previously selected and assessed on a series of primary B lymphocytes sorted from healthy and tumor lymph nodes (4). Among the tested target promoters the three targets that were found to be highly methylated in the tumor samples were chosen. The promoters of the KLF4, DAPK1 and SPG20 genes were fully methylated in Toledo and NU-DUL-1 cell lines (germinal center- and activated B cell like derived, respectively; Fig. 1A). This was consistent with the tumor and metastasis suppressor role reported for KLF4 and DAPK1 (20,21). The positions of the CpG islands investigated by EpiTect Methyl II qPCR are reported in Table SIII.
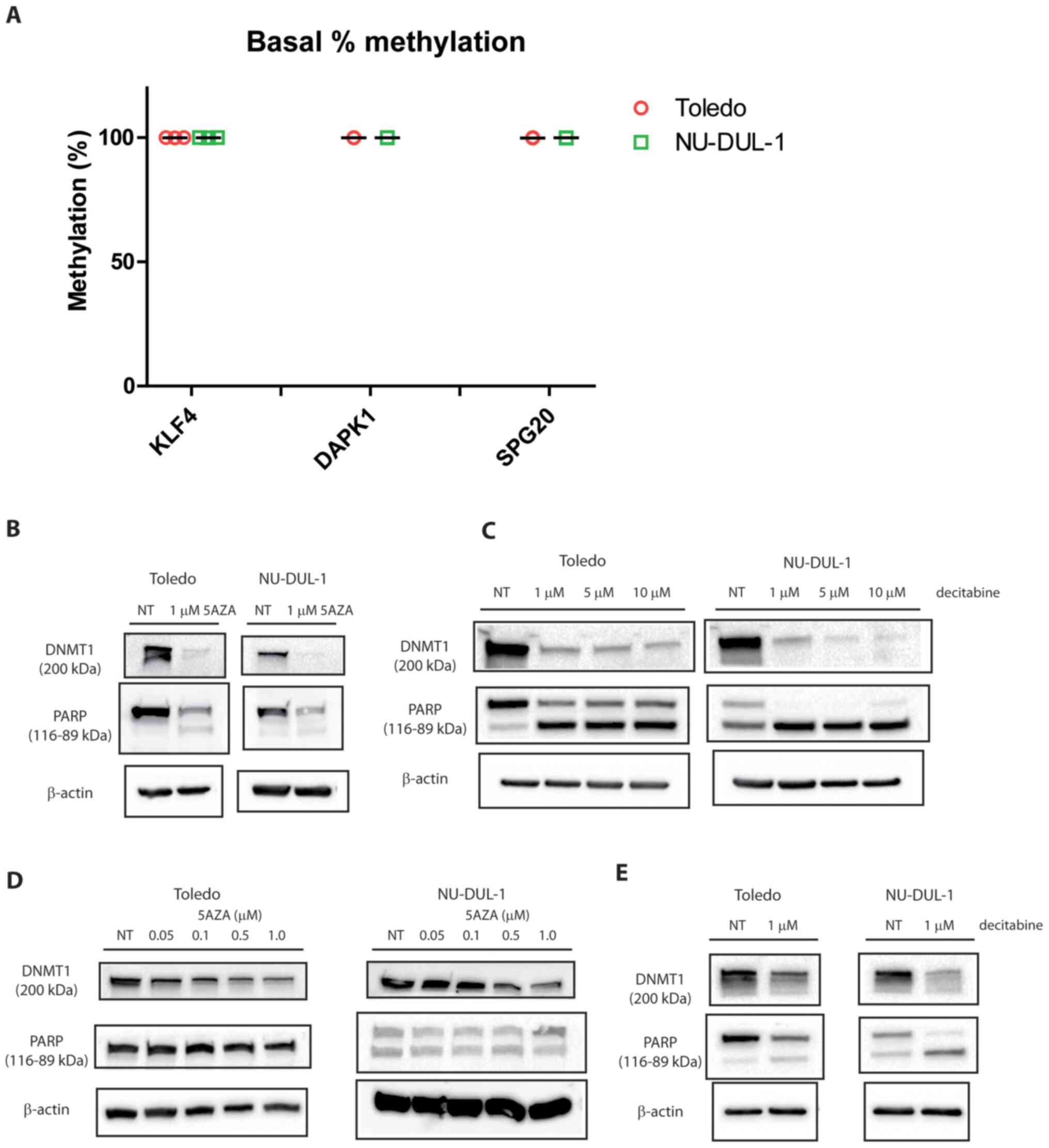 |
Figure 1.
Quantitative promoter methylation in two diffuse large B cell lymphoma cell lines and effects of hypomethylating agents. (A) Toledo and NU-DUL-1 cell lines were analyzed via EpiTect Methyl II qPCR. Data are reported as % promoter methylation and error bars represent SD (n=3). (B) Immunoblotting showing DNMT1, PARP and β-actin after 5AZA treatment for 48 h. (C) Immunoblotting showing DNMT1 and PARP and β-actin after decitabine treatment for 48 h. (D) Immunoblotting showing DNMT1, PARP and β-actin after 5AZA treatments for 7 days. (E) Immunoblotting showing DNMT1 and PARP and β-actin after decitabine treatments for 7 days. n=2. DNMT1, DNA methyltransferase 1; PARP, poly (ADP-ribose) polymerase; NT, not treated; KLF4, Krüppel-like factor 4; DAPK1, death associated protein 1; SPG20, spastic paraplegia 20; 5AZA, 5′-aza-cytidine; decitabine, 5′-aza-2-deoxycitidine.
|
The epigenetic regulation of these targets was initially evaluated through the use of hypomethylating agents. Apoptosis was triggered by 5AZA and decitabine, as demonstrated by caspase-3/7 activation after 24 h (Fig. S1).
DNMT1 was downregulated and PARP was cleaved upon treatment with either 5AZA or decitabine (Fig. 1B and C), which was consistent with the reported mechanism of action for these nucleoside analogs (13). PARP was partially activated by 5AZA and completely activated by decitabine in NU-DUL-1 cells, while Toledo cells only presented a partial cleavage at the highest dose of decitabine. After one week from the day of administration of the hypomethylating drug, DNMT1 was still downregulated and PARP was notably cleaved with both drugs (Fig. 1D and E). Doses of decitabine >1 µM for one week were lethal and therefore data were not analyzable (data not shown).
5AZA- and decitabine-induced global demethylation does not affect KLF4, DAPK1 and SPG20
To assess the specific activity of hypomethylating agents in this study, global chromatin methylation was measured via a fluorescence immunoenzymatic assay with anti-5mC antibody. As expected, chromatin was demethylated upon treatment with 5AZA in both cell lines after 48 h and up to 7 days later (Fig. 2A), which was consistent with the observed DNMT1 downregulation and caspase-3/7 activation (Figs. 1B-E and S1). Next, it was investigated whether the previously selected KLF4, DAPK1 and SPG20 promoters could be modulated by these treatments. These were not affected by 5AZA treatment at any of the tested time points (Fig. 2B, left panel).
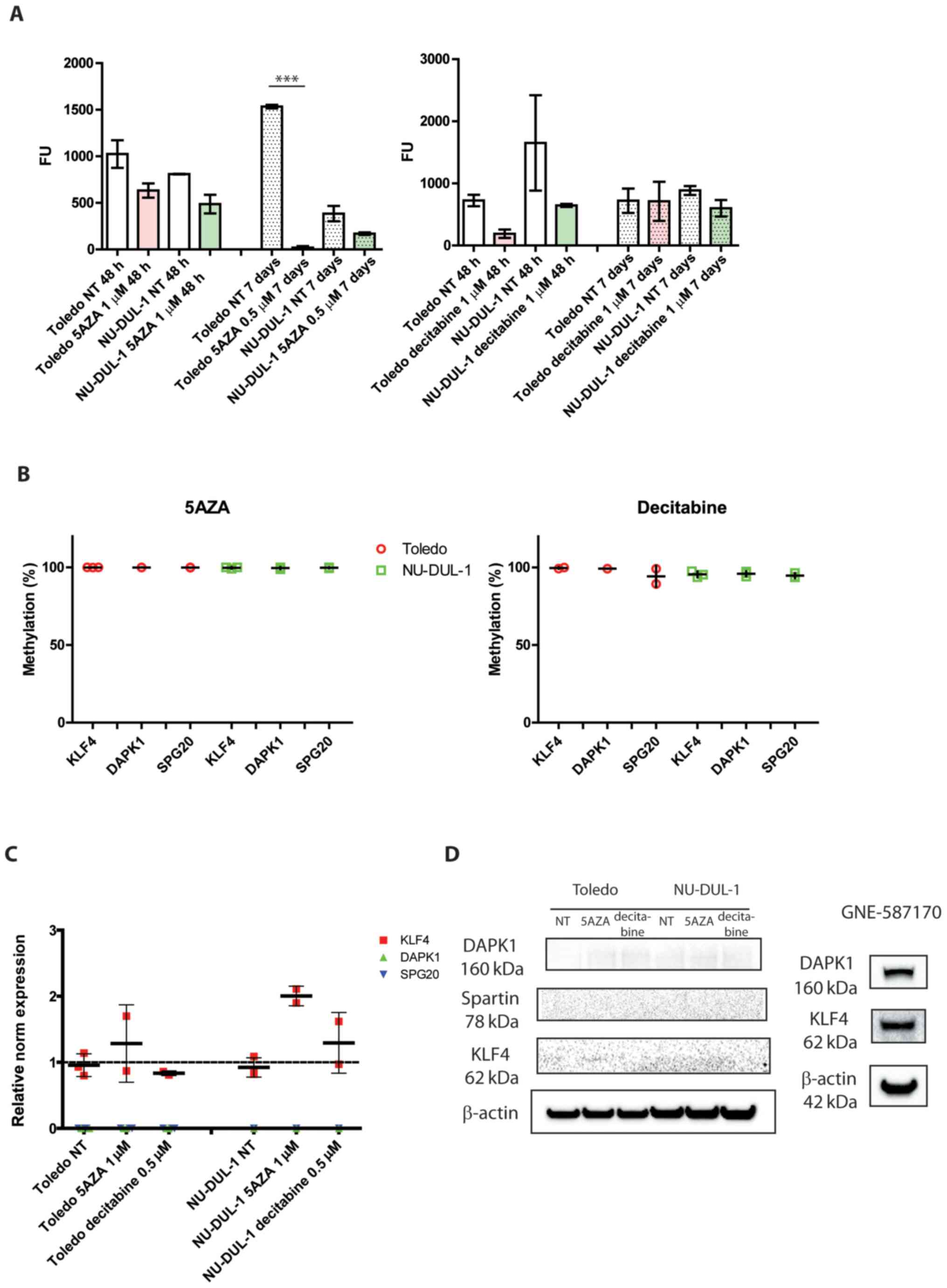 |
Figure 2.
Global chromatin methylation, gene-specific quantitative methylation and gene expression after 5AZA and decitabine treatment. (A) Chromatin methylation after 48 h or 7 days from a single administration of the indicated drug. ***P<0.0001 (n=3; one-way ANOVA). (B) Quantitative promoter methylation of KLF4, DAPK1 and SPG20 after the administration of either 1 µM 5AZA or decitabine 0.5 µM for 48 h. (C) Gene expression of the selected targets upon treatment with hypomethylating drugs (cutoff, 37 cycles). n=2/3. (D) KLF4, DAPK1 and SPG20 protein expression levels after treatment with hypomethylating drugs for 48 h. n=3. The blots of GNE-587170 protein extracts are presented as positive controls. NT, not treated; KLF4, Krüppel-like factor 4; DAPK1, death associated protein 1; SPG20, spastic paraplegia 20; 5AZA, 5′-aza-cytidine; decitabine, 5′-aza-2-deoxycitidine.
|
The effect of decitabine treatment was also evaluated. Global chromatin methylation decreased after 48 h in the Toledo and NU-DUL-1 cell lines compared with the NT groups, but the levels of treated samples returned to the same levels observed in the NT groups after 7 days (Fig. 2A, right panel), which differed from the results observed for 5AZA. Quantitative methylation of the selected targets demonstrated that KLF4, DAPK1 and SPG20 promoters were unaffected (Fig. 2B, right panel).
The gene expression of the three genes of interest was assessed by RT-qPCR. Only KLF4 was found to be partially upregulated by 5AZA treatment in both cell lines, while DAPK1 and SPG20 were still undetectable upon both 5AZA and decitabine treatments (Fig. 2C; mean Cq of 37 was chosen as a cutoff). The immunoblotting confirmed these observations since these three targets were almost undetectable in all the conditions tested. The cell line GNE-587170 was reported as a positive control for KLF4 and DAPK1 proteins (Fig. 2D).
These data collectively suggested that 5AZA and decitabine selectively affected only some regions of the genome, which was consistent with previous findings on colon cancer cells (14). However, unlike in these previous findings, the drugs did not preferentially affect the highly methylated CpG dinucleotides in our targets, which remained unaffected.
DNMT1 is not the only mediator of methylation maintenance in lymphoid cells
To investigate the role of specific DNMTs in the maintenance of methylation at the level of the selected targets, DNMT1 and DNMT3a were individually silenced and promoter methylation was assessed.
DNMT1 and DNMT3a were consistently and specifically downregulated at the RNA and protein levels 48 h after transfection with siDNMT1 and siDNMT3a compared with the scramble siRNA group (Fig. 3A and B). DNMT3a RNA levels were low in both cell lines (Table SIV) and the corresponding protein was undetectable by immunoblotting. This was consistent with the known role played by this enzyme in embryonic development (22).
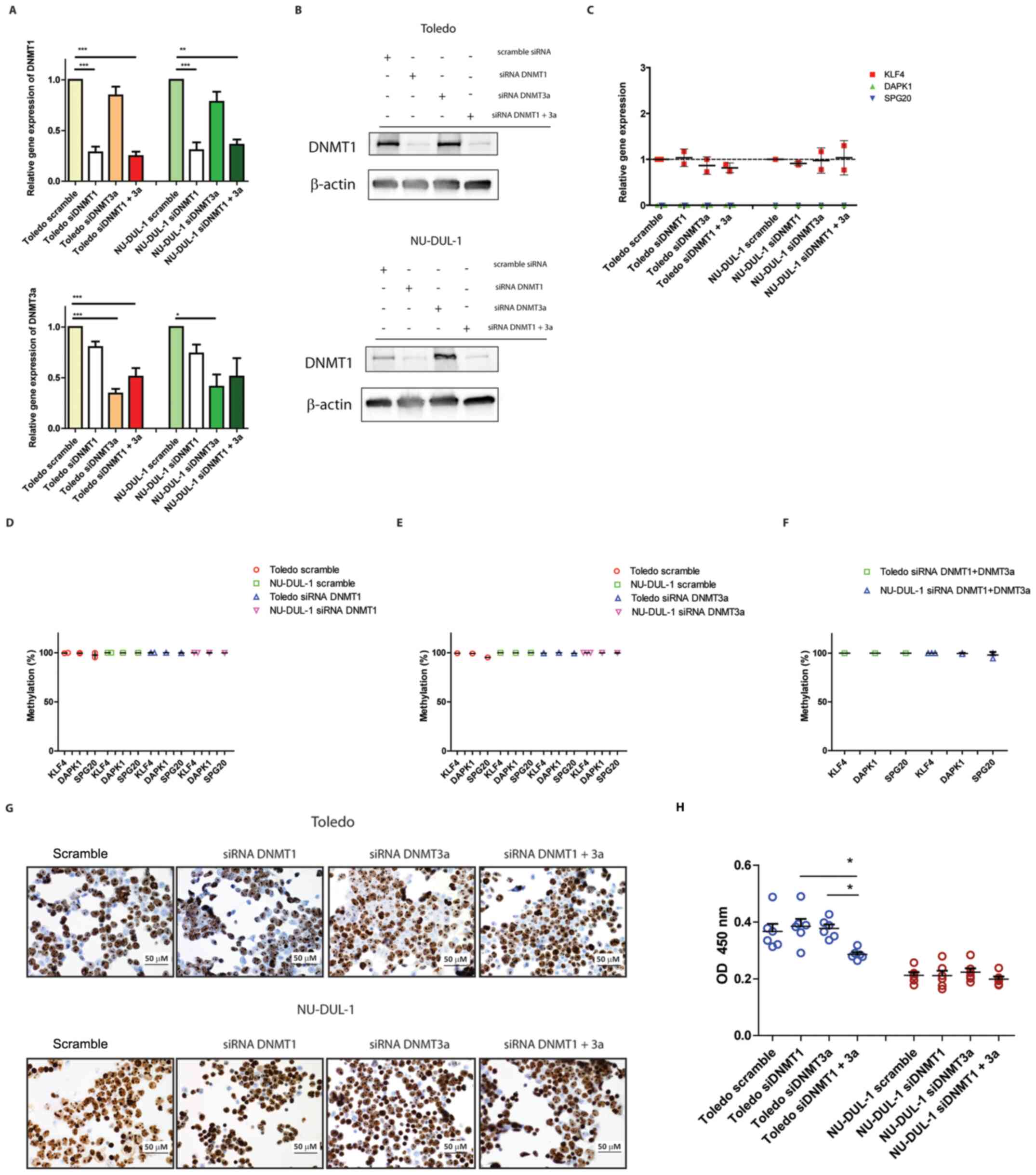 |
Figure 3.
Effects of DNMT1- and DNMT3a-selective silencing on gene-specific quantitative methylation and expression. (A) Silencing efficiency after 48 h from transfection in Toledo and NU-DUL-1 cell lines as determined by RT-qPCR. *P<0.05, **P<0.01, ***P<0.0001 (n=3; one-way ANOVA). (B) Silencing efficiency 48 h after transfection in Toledo and NU-DUL-1 cell lines as determined by immunoblotting (n=2). (C) Gene expression levels of KLF4, DAPK1 and SPG20 as determined by RT-qPCR (n=2). (D) Quantitative promoter methylation in the panel of target loci 48 h after DNMT1 silencing. (E) Quantitative promoter methylation in the panel of target loci 48 h after DNMT3a silencing. (F) Quantitative promoter methylation in the panel of target loci 48 h after DNMT1 + DNMT3a silencing (n=2/3). (G) Ki67 staining of the transfected cells after 48 h incubation as determined via immunocytochemistry (magnification, ×40). (H) Cell Counting Kit-8 viability assay of the transfected cells after 48 h incubation. *P<0.05 (one-way ANOVA). DNMT, DNA methyltransferase; RT-qPCR, reverse transcription-quantitative PCR; KLF4, Krüppel-like factor 4; DAPK1, death associated protein 1; SPG20, spastic paraplegia 20; si, small interfering RNA.
|
Gene expression analysis showed that none of the KLF4, DAPK1 and SPG20 targets were affected by any of the silencing conditions (Fig. 3C). This was consistent with the finding that the percentage of methylation was not affected after DNMT1 silencing, nor after simultaneous DNMT1- and DNMT3a-silencing (Fig. 3D-F).
To assess whether proliferation was affected by the knockdown of DNMT, Ki67 expression was determined by immunocytochemistry. The majority of the nuclei were Ki67-positive in all the conditions tested after 48 h of incubation (Fig. 3G). In addition to this, viability measured by CCK-8 was not affected after either DNMT1- or DNMT3a-silencing in the two cell lines (Fig. 3H). Therefore, the methylation stability observed in the different conditions was the result of an active process maintaining methylation marks during DNA replication even when DNMTs were deficient.
It was demonstrated that genomic DNA was strongly demethylated by 5AZA and decitabine (Fig. 2A). DNMT1-specific silencing did not lead to hypomethylation at the promoter-specific levels of KLF4, DAPK1 and SPG20 (Fig. 3D-F). Thus, these data collectively demonstrated that DNMT1 downregulation alone is not responsible for chromatin demethylation, even when lymphocytes are actively proliferating.
Genome-wide methylation analysis indicates hypermethylation clusters in SPG20 locus
The data collected thus far demonstrated that the SPG20 promoter was hypermethylated in all the tested cell lines by qPCR and was not affected by hypomethylating drugs nor by the silencing of DNMT1. At present, this locus is poorly characterized in B cell lymphomas, and therefore deserves further investigation.
To investigate the whole SPG20 locus, the data from an Infinium Human Methylation 450K BeadChip array was analyzed. The β-values for 28 DLBCL cell lines were filtered for the SPG20 locus and the results were assembled in a heatmap (Fig. 4A). The β-value distribution showed a hypomethylated region (light blue, bottom) and a hypermethylated region (dark blue, top) in all the tested cell lines. A heterogeneous region (mid-upper) is common to 24 out of 28 cell lines. These three regions were quite clearly clustered and corresponded to exact positions in the SPG20 locus. Looking at the precise mapping of the probes spanning the gene locus, the highest and more homogeneous β-values were clustered in the open sea regions (away from the islands), upstream of the SPG20 transcription starting site (TSS; Fig. 4B). The mean β-values for the nine probes located upstream the TSS were in the 0.60–0.85 range, indicating a hypermethylated status (Fig. 4D). These data suggested a transcriptional silencing, mediated by the hypermethylation of the promoter region.
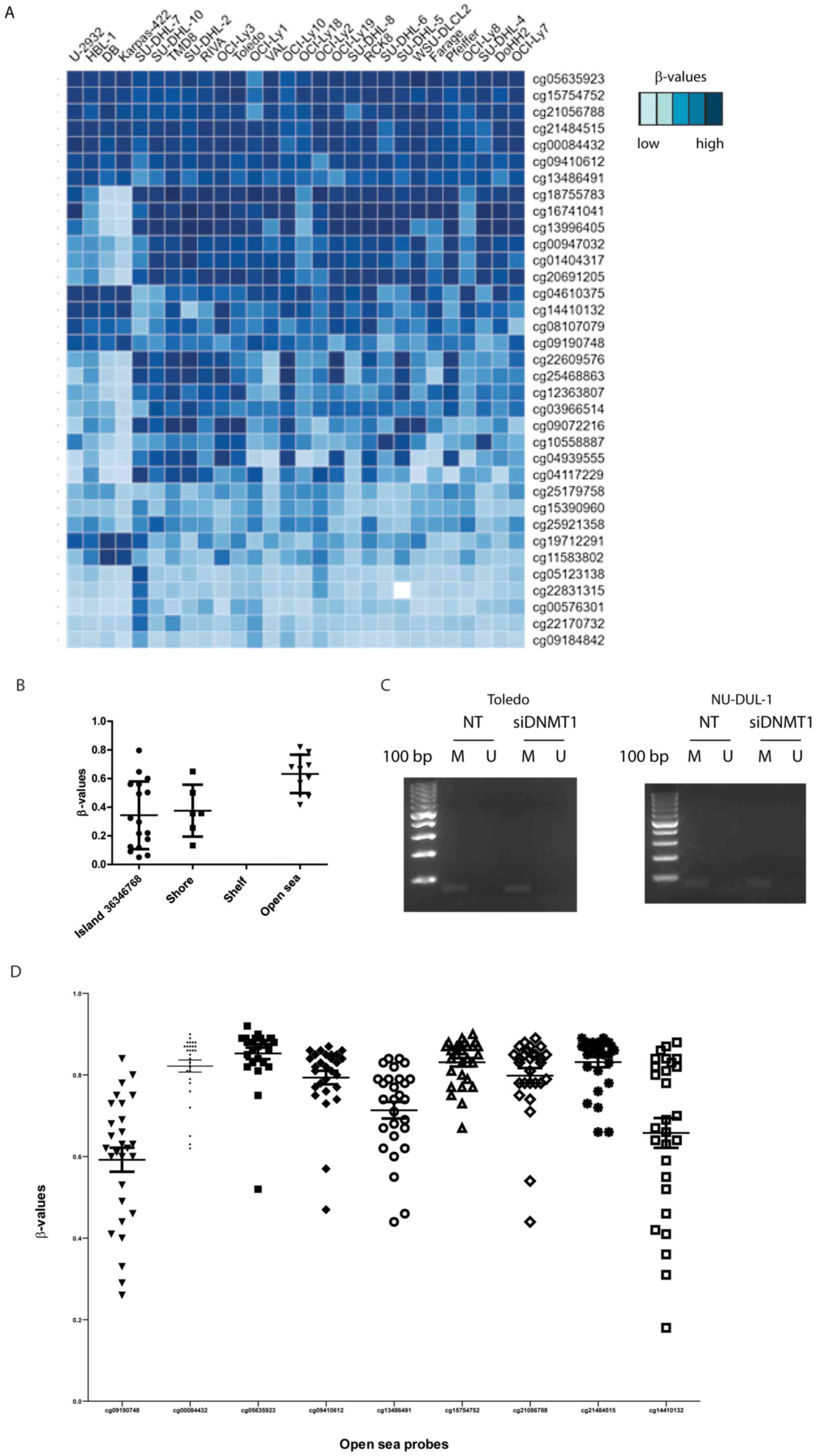 |
Figure 4.
Genome-wide methylation analysis by Infinium Human Methylation 450K BeadChip array and MS-PCR. (A) Heatmap of the β-values of SPG20 locus in 28 diffuse large B cell lymphoma cell lines. (B) Bioinformatics localization of the 35 probes identifying SPG20 locus. (C) MS-PCR on cg15754752 probe with M- or U-specific primers. (D) β-value distribution of the nine probes mapping upstream or on the SPG20 TSS. M, methylated; U, unmethylated; MS-PCR, methylation-specific PCR; SPG20, spastic paraplegia 20; TSS, transcription starting site; DNMT1, DNA methyltransferase 1; NT, not treated; si, small interfering RNA.
|
To confirm the methylation status of this specific open sea region, primers were designed for MS-PCR on the probe cg15754752, located upstream of the TSS (Fig. 4B and D). The selected region was methylated regardless of the silencing of DNMT1, thus confirming the results of the Infinium Human Methylation 450K BeadChip array (Fig. 4C).
It is noteworthy to underline that the data obtained with an older type of array (Illumina 27K) in a colon cancer cell line and available through the repository ‘ArrayExpress’ confirm our findings (14). The probes specific for SPG20 displayed high β-values even after treatment with 5AZA or decitabine (Fig. S2).
SPG20 is epigenetically regulated in human PBMCs
Once it had been demonstrated that the SPG20 promoter was fully methylated in the tested DLBCL cell lines (Fig. 4), it was next assessed whether PBMCs from healthy donors displayed a different methylation profile. PBMCs from healthy donors displayed the same methylation pattern as the one shown by non-tumor B lymphocytes immunologically sorted from follicular hyperplasia (Fig. 5A) (4). KLF4, DAPK1 and SPG20 were almost completely unmethylated in the tested PBMCs (Fig. 5A and B), which differed from the results observed in the NHL cell lines (Figs. 2B and 3D-F). This led us to assess Spartin protein levels in healthy, circulating mononuclear cells. Spartin was expressed in all the tested PBMCs (representative of a total of 27 samples; Fig. 5C), demonstrating the epigenetic regulation of this gene through methylation of the promoter. DNMT1 expression levels were highly heterogeneous among different subjects, which suggested that SPG20 methylation status did not rely on this DNMT (Fig. 5C). These data were also consistent with the fact that DNMT1 silencing did not affect SPG20 methylation in Toledo and NU-DUL-1 cell lines (Figs. 3 and 4).
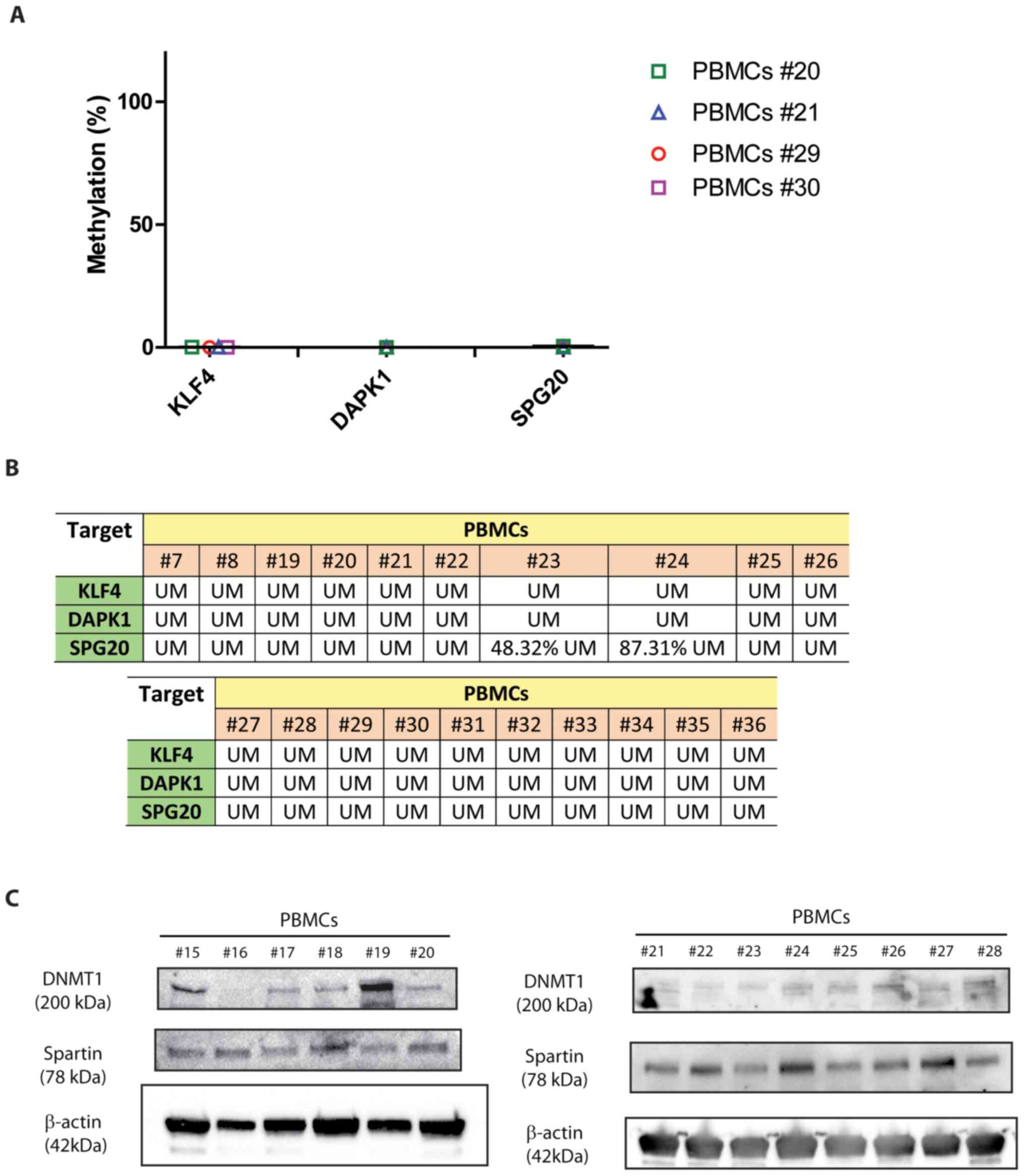 |
Figure 5.
SPG20 is epigenetically regulated in human PBMCs. (A) Methylation profile of the selected targets in four representative PBMCs from healthy donors. (B) Overall methylation profiles of 20 PBMCs. UM corresponds to a % of demethylation ≥91%; lower percentages are specified in each box. (C) SPG20 and DNMT1 protein levels in 20 representative PBMCs from healthy donors (data are representative of a total of 27 independent PBMCs samples). SPG20, spastic paraplegia 20; PBMCs, peripheral blood mononuclear cells; UM, unmethylated; DNMT1, DNA methyltransferase 1; KLF4, Krüppel-like factor 4; DAPK1, death associated protein 1.
|
Discussion
Hypomethylating drugs exhibit antitumoral activity in hematological cancers (3). 5AZA and decitabine are approved for the therapy of myelodysplastic syndromes and acute myeloid leukemias in Europe and the United States (13). They are also under investigation for the therapy of mature B cell neoplasms, namely DLBCL and FL (3,23,24). The rationale for the use of hypomethylating agents is based on the fact that numerous types of cancers display aberrant DNA hypermethylation allowing tumor cells to repress the expression of the genes involved in tumor suppressive pathways (25). However, it is now known that DNA hypermethylation can also positively affect transcription. The oncogene and transcriptional repressor BCL6 plays a pivotal role in mediating the development of B cell lymphomas (26,27). The hypermethylation of intragenic CpG islands within the first intron of the gene associates to high amounts of BCL6 mRNA (28). The underlying mechanism of this phenomenon relies on CCCTC-binding factor (CTCF), which binds DNA in a methylation-sensitive manner (29). DNA methylation of the CTCF-consensus sequences can, indirectly, promote gene transcription by preventing CTCF binding and, thus, abolishing an enhancer block (28). Furthermore, recent evidence has demonstrated how methylation plasticity of tumor cells may impact the expression of extended areas of chromatin in a somehow unpredictable manner. For instance, the translocation of DNMT3a between different compartments of chromatin may lead to a paradoxical hypomethylation of the chromatin under TET deficiency (30).
These previous studies indicate how gene and protein expression can be influenced by the methylation of CpG islands in a complex and context-dependent manner. The data presented in the present study are no exception to this since it demonstrated that the mechanism of action of DNMTs upon silencing are not entirely predictable and some specific targets associated with lymphoma proliferation are differentially and stably methylated compared with circulating, healthy PBMCs.
First of all, the observed dose-dependent downregulation of DNMT1 upon treatment with hypomethylating agents did not affect the methylation status of the key KLF4, DAPK1 and SPG20 promoters, which remained hypermethylated regardless of the treatments. These data were independent of the global chromatin demethylation. 5AZA treatment led to a modest KLF4, but not DAPK1 and SPG20 upregulation, suggesting that silencing of specific targets is maintained by tumor cells through an active mechanism, which is able to overcome the activity of hypomethylating agents. This is strengthened by the fact that the cells were Ki67 positive even upon DNMT1- or DNMT3a-silencing. It has to be specified that the currently used hypomethylating drugs mainly target DNMT1, leaving the other DNMTs mostly unaffected (13,31). To the best of our knowledge these data are the first to specifically describe the effects of these drugs on DLBCL cell lines at the levels of KLF4, DAPK1 and SPG20.
In the model of the current study, silencing of either DNMT1, DNMT3a or both did not affect the levels of methylation nor the expression levels of KLF4, DAPK1 and SPG20, as demonstrated by qPCR and MS-PCR.
The present study focused on SPG20, a gene that, when mutated, leads to Troyer syndrome. This syndrome is an autosomal recessive form of paraplegia caused by deleterious mutations in SPG20 associated with the loss of the protein in the skeletal muscle (10,11). SPG20 promoter can be methylated in gastric and colorectal cancers (32,33).
Here, it was demonstrated, for the first time, that lymphoma cell lines displayed stable SPG20 promoter hypermethylation, which was insensitive to the activity of the two widely used drugs 5AZA and decitabine. It was also shown that neither DNMT1 (the main enzyme responsible for methylation maintenance in the nuclei) nor DNMT3a were required to preserve SPG20 hypermethylation, which corresponded to the absence of mRNA and protein. A further in-depth analysis based on the data of the 450K Infinium methylation array led to the identification of a group of nine probes mapping in an open sea region located upstream of the TSS. This region was hypermethylated in most of the 28 DLBCL cell lines, suggesting a conserved mechanism of gene expression regulation.
Most importantly, the present study demonstrated that human PBMCs expressed Spartin protein (the product of SPG20 gene) and that this feature paralleled the unmethylated status of the promoter. These data demonstrated the epigenetic regulation of SPG20 expression in lymphoid cells and primary circulating lymphocytes. Very limited information is available on this protein in lymphomas, thus further experiments are needed to clarify the exact role of Spartin, for instance gain of function experiments aimed at overexpressing this target gene in lymphoma cells. These will provide insights into the exact role of this protein in lymphoid tumors.
A meta-analysis recently published on patients with lymphoma evidenced how the hypermethylation of DAPK1 tumor suppressor represents an independent prognostic biomarker for the 5-year survival of patients (8). DAPK1 promoter methylation has been shown to be methylated in a significant proportion of lymphomas including FL, mucosa-associated lymphoid tissue and DLBCL (4,34). A significant amount of data have demonstrated the prognostic value of DAPK1 promoter methylation in B cell lymphomas (35–39).
KLF4 is an apoptosis inducer and inhibitor of tumor progression in T-acute lymphoblastic leukemia and its methylation is associated with the poor prognosis of patients with splenic marginal zone lymphoma (6,7). However, recent evidence has reported on the dual role of this factor that has been found to also have a pro-oncogenic role in B cell lymphomas (40).
The present data showed that KLF4 and DAPK1 were two homogeneously hypermethylated targets and they were not expressed by the cell lines used in this study. Only 5AZA treatment seemed to have a modest effect on KLF4 expression levels, while DNMT1 silencing did not have any effects. Thus, further investigations are warranted to elucidate their role in DLBCL.
To conclude, the current data indicated that three loci, KLF4, DAPK1 and SPG20, were consistently hypermethylated and not expressed in two DLBCL cell lines. By contrast, normal PBMCs displayed a demethylated profile that reflected the profile of non-tumor B lymphocytes and differed significantly from lymphoma cell lines. Among the tested promoters, SPG20 displayed a hypermethylated status in a significant number of DLBCL cell lines investigated by genome-wide methylation array. Despite the marked pro-apoptotic and hypomethylating effects, 5AZA and decitabine did not affect KLF4, DAPK1 and SPG20 promoters. Indicating that neither DNMT1 nor DNMT3a were necessary to maintain the hypermethylated status of these targets in lymphoma cells. These observations suggested that other mechanisms, such as chromatin accessibility and permissive vs. non permissive conditions, are involved during the regulation of these loci. SPG20 protein levels were measurable in healthy PBMCs where the promoter was demethylated, suggesting that this locus was epigenetically regulated and silenced in DLBCL. Thus, these experiments contributed to the knowledge of the mechanism of action of hypomethylating drugs currently in phase I clinical trial. Future experiments aimed at elucidating the exact role of SPG20 in lymphomas are now warranted.
Supplementary Material
Supporting Data
Acknowledgements
The authors would like to thank Ms. Marina Grassi, Ms. Tonia de Simone (Laboratory of Translational Research, Azienda Unità Sanitaria Locale IRCCS, Reggio Emilia, Italy) and Dr Ione Tamagnini (Pathology Unit, Azienda Unità Sanitaria Locale IRCCS, Reggio Emilia, Italy) for their technical support. They are also thankful to Dr Minden's laboratory (Ontario Cancer Institute, Canada) for the gift of GNE-587170 cells.
Funding
This work was supported by a grant from the Italian Ministry of Health (Ministero della Salute; grant no. GR‑2010‑2320280).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request. The datasets generated and/or analyzed during the current study are available in the Gene Expression Omnibus repository, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE103934 (accession no. GSE103934).
Authors' contributions
RF conceived the present study, performed most of the experiments and wrote the manuscript. VYC and MP performed the experiments. LaC provided the blood samples and performed some of the experiments. LuC obtained the genome-wide methylation data. FB and FM supervised the manuscript and contributed to data interpretation. All authors have read and approved the final manuscript. RF and VYC confirm the authenticity of all the raw data.
Ethics approval and consent to participate
The use of PBMCs from healthy donors upon written informed consent was approved by the Local Ethics Committee of Reggio Emilia (Comitato Etico Provinciale; approval no. 2016/0025053).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
|
1
|
Howlader NNA, Krapcho M, Miller D, Bishop K, Kosary CL, Yu M, Ruhl J, Tatalovich Z, Mariotto A, Lewis DR, et al; Cronin KA: SEER Cancer Statistics Review, 1975–2014. National Cancer Institute; Bethesda, MD: 2017, https://seer.cancer.gov/archive/csr/1975_2014/April 2–2018
|
|
2
|
Novo M, Castellino A, Nicolosi M, Santambrogio E, Vassallo F, Chiappella A and Vitolo U: High-grade B-cell lymphoma: How to diagnose and treat. Expert Rev Hematol. 12:497–506. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Martin P, Bartlett NL, Chavez JC, Reagan JL, Smith SM, LaCasce AS, Jones JA, Drew J, Wu C, Mulvey E, et al: Phase 1 study of oral azacitidine (CC-486) plus R-CHOP in previously untreated intermediate- to high-risk DLBCL. Blood blood. 2021011679. 2021. View Article : Google Scholar
|
|
4
|
Frazzi R, Zanetti E, Pistoni M, Tamagnini I, Valli R, Braglia L and Merli F: Methylation changes of SIRT1, KLF4, DAPK1 and SPG20 in B-lymphocytes derived from follicular and diffuse large B-cell lymphoma. Leuk Res. 57:89–96. 2017. View Article : Google Scholar
|
|
5
|
Ghaleb AM and Yang VW: Krüppel-like factor 4 (KLF4): What we currently know. Gene. 611:27–37. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Arribas AJ, Rinaldi A, Mensah AA, Kwee I, Cascione L, Robles EF, Martinez-Climent JA, Oscier D, Arcaini L, Baldini L, et al: DNA methylation profiling identifies two splenic marginal zone lymphoma subgroups with different clinical and genetic features. Blood. 125:1922–1931. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Shen Y, Park CS, Suppipat K, Mistretta TA, Puppi M, Horton TM, Rabin K, Gray NS, Meijerink JPP and Lacorazza HD: Inactivation of KLF4 promotes T-cell acute lymphoblastic leukemia and activates the MAP2K7 pathway. Leukemia. 31:1314–1324. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wang H, Zhou LY, Guan ZB, Zeng WB, Zhou LL, Liu YN and Pan XY: Prognostic significance of DAPK promoter methylation in lymphoma: A meta-analysis. PLoS One. 14:e02109432019. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Özdemir İ, Pınarlı FG, Pınarlı FA, Aksakal FNB, Okur A, Uyar Göçün P and Karadeniz C: Epigenetic silencing of the tumor suppressor genes SPI1, PRDX2, KLF4, DLEC1, and DAPK1 in childhood and adolescent lymphomas. Pediatr Hematol Oncol. 35:131–144. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Spiegel R, Soiferman D, Shaag A, Shalev S, Elpeleg O and Saada A: Novel Homozygous Missense Mutation in SPG20 Gene Results in Troyer Syndrome Associated with Mitochondrial Cytochrome c Oxidase Deficiency. JIMD Rep. 33:55–60. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Patel H, Cross H, Proukakis C, Hershberger R, Bork P, Ciccarelli FD, Patton MA, McKusick VA and Crosby AH: SPG20 is mutated in Troyer syndrome, an hereditary spastic paraplegia. Nat Genet. 31:347–348. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Tawamie H, Wohlleber E, Uebe S, Schmäl C, Nöthen MM and Abou Jamra R: Recurrent null mutation in SPG20 leads to Troyer syndrome. Mol Cell Probes. 29:315–318. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Diesch J, Zwick A, Garz AK, Palau A, Buschbeck M and Götze KS: A clinical-molecular update on azanucleoside-based therapy for the treatment of hematologic cancers. Clin Epigenetics. 8:712016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hagemann S, Heil O, Lyko F and Brueckner B: Azacytidine and decitabine induce gene-specific and non-random DNA demethylation in human cancer cell lines. PLoS One. 6:e173882011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Duncan CG, Kondilis-Mangum HD, Grimm SA, Bushel PR, Chrysovergis K, Roberts JD, Tyson FL, Merrick BA and Wade PA: Base-Resolution Analysis of DNA Methylation Patterns Downstream of Dnmt3a in Mouse Naïve B Cells. G3 (Bethesda). 8:805–813. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yang L, Rau R and Goodell MA: DNMT3A in haematological malignancies. Nat Rev Cancer. 15:152–165. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lyko F: The DNA methyltransferase family: A versatile toolkit for epigenetic regulation. Nat Rev Genet. 19:81–92. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Frazzi R, Valli R, Tamagnini I, Casali B, Latruffe N and Merli F: Resveratrol-mediated apoptosis of hodgkin lymphoma cells involves SIRT1 inhibition and FOXO3a hyperacetylation. Int J Cancer. 132:1013–1021. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Guan H, Xie L, Leithäuser F, Flossbach L, Möller P, Wirth T and Ushmorov A: KLF4 is a tumor suppressor in B-cell non-Hodgkin lymphoma and in classic Hodgkin lymphoma. Blood. 116:1469–1478. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Küçük C, Hu X, Jiang B, Klinkebiel D, Geng H, Gong Q, Bouska A, Iqbal J, Gaulard P, McKeithan TW, et al: Global promoter methylation analysis reveals novel candidate tumor suppressor genes in natural killer cell lymphoma. Clin Cancer Res. 21:1699–1711. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Mehdipour P, Murphy T and De Carvalho DD: The role of DNA-demethylating agents in cancer therapy. Pharmacol Ther. 205:1074162020. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Pera B, Tang T, Marullo R, Yang SN, Ahn H, Patel J, Elstrom R, Ruan J, Furman R, Leonard J, et al: Combinatorial epigenetic therapy in diffuse large B cell lymphoma pre-clinical models and patients. Clin Epigenetics. 8:792016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Martin P, Bartlett NL, Rivera IIR, Revuelta M, Chavez JC, Reagan JL, Smith SM, LaCasce AS, Zhang L, Zhai M, et al: A Phase I, Open Label, Multicenter Trial of Oral Azacitidine (CC-486) Plus R-CHOP in Patients with High-Risk, Previously Untreated Diffuse Large B-Cell Lymphoma, Grade 3B Follicular Lymphoma, or Transformed Lymphoma. Blood. 130 (Suppl 1):1922017.
|
|
25
|
Cerchietti L and Leonard JP: Targeting the epigenome and other new strategies in diffuse large B-cell lymphoma: Beyond R-CHOP. Hematology (Am Soc Hematol Educ Program). 2013:591–595. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Hatzi K and Melnick A: Breaking bad in the germinal center: How deregulation of BCL6 contributes to lymphomagenesis. Trends Mol Med. 20:343–352. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Basso K and Dalla-Favera R: Roles of BCL6 in normal and transformed germinal center B cells. Immunol Rev. 247:172–183. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Lai AY, Fatemi M, Dhasarathy A, Malone C, Sobol SE, Geigerman C, Jaye DL, Mav D, Shah R, Li L, et al: DNA methylation prevents CTCF-mediated silencing of the oncogene BCL6 in B cell lymphomas. J Exp Med. 207:1939–1950. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Hark AT, Schoenherr CJ, Katz DJ, Ingram RS, Levorse JM and Tilghman SM: CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature. 405:486–489. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
López-Moyado IF, Tsagaratou A, Yuita H, Seo H, Delatte B, Heinz S, Benner C and Rao A: Paradoxical association of TET loss of function with genome-wide DNA hypomethylation. Proc Natl Acad Sci USA. 116:16933–16942. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Liu K, Wang YF, Cantemir C and Muller MT: Endogenous assays of DNA methyltransferases: Evidence for differential activities of DNMT1, DNMT2, and DNMT3 in mammalian cells in vivo. Mol Cell Biol. 23:2709–2719. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhou Z, Wang W, Xie X, Song Y, Dang C and Zhang H: Methylation-induced silencing of SPG20 facilitates gastric cancer cell proliferation by activating the EGFR/MAPK pathway. Biochem Biophys Res Commun. 500:411–417. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Rezvani N, Alibakhshi R, Vaisi-Raygani A, Bashiri H and Saidijam M: Detection of SPG20 gene promoter-methylated DNA, as a novel epigenetic biomarker, in plasma for colorectal cancer diagnosis using the MethyLight method. Oncol Lett. 13:3277–3284. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Rossi D, Capello D, Gloghini A, Franceschetti S, Paulli M, Bhatia K, Saglio G, Vitolo U, Pileri SA, Esteller M, et al: Aberrant promoter methylation of multiple genes throughout the clinico-pathologic spectrum of B-cell neoplasia. Haematologica. 89:154–164. 2004.PubMed/NCBI
|
|
35
|
Kristensen LS, Treppendahl MB, Asmar F, Girkov MS, Nielsen HM, Kjeldsen TE, Ralfkiaer E, Hansen LL and Grønbæk K: Investigation of MGMT and DAPK1 methylation patterns in diffuse large B-cell lymphoma using allelic MSP-pyrosequencing. Sci Rep. 3:27892013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Amara K, Trimeche M, Ziadi S, Laatiri A, Hachana M and Korbi S: Prognostic significance of aberrant promoter hypermethylation of CpG islands in patients with diffuse large B-cell lymphomas. Ann Oncol. 19:1774–1786. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Nakamichi I, Tomita Y, Zhang B, Sugiyama H, Kanakura Y, Fukuhara S, Hino M, Kanamaru A, Ogawa H and Aozasa K: Correlation between promoter hypermethylation of GSTP1 and response to chemotherapy in diffuse large B cell lymphoma. Ann Hematol. 86:557–564. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Giachelia M, Bozzoli V, D'Alò F, Tisi MC, Massini G, Maiolo E, Guidi F, Cupelli E, Martini M, Larocca LM, et al: Quantification of DAPK1 promoter methylation in bone marrow and peripheral blood as a follicular lymphoma biomarker. J Mol Diagn. 16:467–476. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Kristensen LS, Hansen JW, Kristensen SS, Tholstrup D, Harsløf LB, Pedersen OB, De Nully Brown P and Grønbæk K: Aberrant methylation of cell-free circulating DNA in plasma predicts poor outcome in diffuse large B cell lymphoma. Clin Epigenetics. 8:952016. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Morales-Martinez M, Vega GG, Neri N, Nambo MJ, Alvarado I, Cuadra I, Duran-Padilla MA, Huerta-Yepez S and Vega MI: MicroRNA-7 Regulates Migration and Chemoresistance in Non-Hodgkin Lymphoma Cells Through Regulation of KLF4 and YY1. Front Oncol. 10:5888932020. View Article : Google Scholar : PubMed/NCBI
|



















