Introduction
A high incidence of esophageal cancer (EC) has been
reported in China (1,2). Recent surveys have demonstrated that
there are ~450,000 new cases of EC each year worldwide, with more
than half of these cases originating from China, ranking sixth
among the list malignant tumors in China (3,4). EC
is an invasive from of cancer, with the two main pathological types
being esophageal squamous cell carcinoma (ESCC) and esophageal
adenocarcinoma (EA) (5). ESCC has
been reported as the main histological type of EC in China,
accounting for >95% of reported cases (6,7). The
5-year survival rate of patients with EC is only ~10%, and the
recurrence and mortality rates remain high (8). Radiotherapy, chemotherapy and
targeted therapy have been developed in recent years for the
treatment of EC. However, the clinical efficacy and prognosis of
patients have not been satisfactory (9,10).
This is due to the lack of an in-depth understanding of the
pathogenesis of EC (11).
Therefore, a more detailed elucidation of the pathogenesis of EC at
the molecular level may lead to the discovery of an ideal molecular
target or other effective therapeutic drugs for EC.
The transcriptional co-repressor C-terminal binding
protein (CtBP) has been originally named thus, since it binds to
the five amino acid domains (PLDLS) at the C-terminus of the
adenovirus early region 1A (E1A) protein (12–14).
The CtBP protein, including the CtBP1 and CtBP2 protein isoforms,
is an evolutionarily conserved transcriptional repressor that has
been reported to specifically bind to DNA or protein targets and
function as a bridge molecule between DNA-binding proteins and
transcriptional repressors, thereby inhibiting gene transcription
(15). In addition, CtBP has been
revealed to promote epithelial-mesenchymal transition (EMT) by
inhibiting E-cadherin expression (14,16).
Furthermore, it may function as a transcriptional co-repressor,
negatively regulating certain tumor suppressors, thereby promoting
the occurrence and development of tumors (17,18).
Recent studies have revealed that the CtBP family plays a crucial
role in the occurrence and development of breast, colon, ovarian
and prostate cancer (19–21).
A previous study by the authors found that the
expression of CtBP2 was significantly increased in ESCC tissues,
and was positively associated with the tumor histological grade,
and negatively associated with p16 tumor suppressor gene expression
(22). In addition, CtBP2 has been
demonstrated to promote ECA109 cell proliferation and migration,
and reduce cell susceptibility to cisplatin (23). Moreover, cyclin H/cyclin-dependent
kinase 7 (CCNH/CDK7) has been previously reported to competitively
bind to homeodomain-interacting protein kinase 2 (HIPK2) and CtBP2,
thereby inhibiting the phosphorylation and dimerization of CtBP2,
ultimately regulating its stability in breast cancer cells
(24). CtBP2 has been also
revealed to promote the proliferation and migration, as well as
inhibit the apoptosis of ESCC cells, through the regulation of its
downstream target molecule, basic fibroblast growth factor (FGF2)
(25). These results suggest that
CtBP2 is involved in the occurrence and development of ESCC;
however, the underlying mechanisms remain unknown. Therefore, it
was hypothesized that CtBP2, as a transcriptional co-repressor, may
also interact with other proteins and participate in the
development of ESCC.
The transforming growth-interacting factor (TGIF)
gene is located on chromosome 18p11 and encodes a nuclear protein
composed of 272 amino acid residues with a molecular weight of ~30
kDa (26). It belongs to the
family of three amino acid loop extensions (TALEs), which are
expressed in various cells and tissues (27,28).
TGIF has been reported to be involved in a number of cellular
signal transduction pathways, particularly the TGF-β pathway
(29). It has been demonstrated
that TGIF can bind to Smad2 and Smad3, possibly changing chromatin
structure from loose to dense through the recruitment of histone
deacetylase (HDAC), thereby inhibiting the transcription of target
genes mediated by TGF-β (29).
TGIF can also inhibit the TGF-β signaling pathway through a
Smad-independent mechanism, by recruiting HDAC (30). Previous research has confirmed that
TGIF expression is increased in various tumors and may be related
to the occurrence and development of tumors (26,31,32).
In addition, it was revealed in a previously published study that
high levels of TGIF were associated with high levels of Wnt
signaling pathway components (Axin1, Axin2 and β-catenin) and a
poor survival rate of patients with triple-negative breast cancer
(33). Therefore, it was
hypothesized that an in-depth study of the regulatory mechanisms
between CtBP2 and TGIF may be of utmost significance, in order to
clarify the roles of CtBP2 and TGIF in the occurrence and
development of ESCC.
Materials and methods
Patients and tissue samples
A total of 108 patients with ESCC were identified
and enrolled from 2015 to 2019 at the Affiliated Hospital of
Nantong University (Nantong, China). None of the patients had
previously received radiotherapy, chemotherapy, or immunotherapy
prior to surgery. All fresh tissues (ESCC tissues and matched
adjacent tissues) were collected following surgical resection, were
immediately washed with sterile phosphate-buffered saline (PBS) and
immediately fixed in 4% paraformaldehyde (PFA) for 12 h before
being embedded in paraffin or stored at −80°C. Patient written
informed consent was obtained before the commencement of the study,
according to the guidelines of the Ethics Committee of the
Affiliated Hospital of Nantong University, and ethics approval was
also provided from the respective ethics committee (2015 L132).
Cells and cell culture
The human ESCC cell lines, ECA109 (CC-Y1150), TE-1
(TCHu 89) and KYSE-150 (TCHu236), and the human normal esophageal
epithelial cell line, HEEC (CL0420), were provided by the Cell Bank
of Type Culture Collection of the Chinese Academy of Sciences and
were cultured in high-glucose DMEM (Gibco; Thermo Fisher
Scientific, Inc.) supplemented with 10% FCS (Shanghai Shuangru
Biotechnology Co., Ltd.) and 1% penicillin/streptomycin antibiotic
solution (Beijing Solarbio Science & Technology Co., Ltd.). All
cell lines were cultured at 37°C with 5% CO2.
Lentiviral transduction
To knockdown or overexpress CtBP2 and TGIF,
recombinant lentiviral vectors (sh-CtBP2, sh-TGIF, LV-CtBP2 and
LV-TGIF, respectively) were constructed (Vigen Biotechnology)
(23). The coding sequence of
CtBP2 or TGIF was cloned as an overexpression vector, into a GV492
vector (Vigen Biotechnology). The shRNA of CtBP2 or TGIF, whose
target sequence was 5′-GCGCCTTGGTCAGTAATAG-3′ or
5′-AGCTTCTAGTGGATGTTGC-3′, was cloned into a GV248 vector,
respectively. The lentiviral particles were obtained from Vigen
Biotechnology Co. Ltd. Briefly, 293T packaging cells (The Cell Bank
of Type Culture Collection of the Chinese Academy of Sciences) were
co-transfected with shutter plasmids and packaging vectors using
polyethyleneimine (PEI; Shanghai life ilab Biotechnology;
http://life-ilab.com/) and incubated in 5%
CO2, 37°C incubator for overnight. The following day,
the 293T cells were supplemented with fresh medium. The supernatant
was collected at 48 and 72 h and post-transfection filtered through
0.45-µm filter, and then concentrated with lentivirus concentration
solution. Briefly, the supernatant from 293T cells co-transfected
with pMD2G, psPAX2 and shutter plasmids was collected and
centrifuged at 2,000 × g for 10 min and then filtered through
0.45-µm filters to remove cells and debris. In total, four volumes
of clarified supernatant were mixed with one volume of
concentration reagent. The mixture was incubated at 4°C overnight
followed by centrifugation at 1,500 × g for 45 min at 4°C.
Following centrifugation, the off-white pellet was re-suspended by
PBS. The ECA109 cells were then transfected with recombinant
lentiviral vectors and negative control viruses at a multiplicity
of infection (MOI) of 5. The following formula was used to
calculate to volume of virus to be added: Virus volume = MOI × cell
number/virus titer. In addition, 1 µg/ml polybrene was added, to
improve the transduction efficiency. The mixture was centrifuged at
800 × g for 50 min at 32°C. Following centrifugation, the cells
were seeded into 6-well culture dish and incubated at 37°C
overnight. Transfected ECA109 cells were selected using 2.5 µg/ml
puromycin for 1 week. The efficiency of lentivirus-mediated
knockdown or overexpression of CtBP2 or TGIF was verified using
reverse transcription-quantitative PCR (RT-qPCR) and western blot
analysis.
RT-qPCR
Total RNA extraction and reverse transcription were
performed as previously described (23). Total RNA from cells or tumor
tissues was extracted using TRIzol® LS reagent
(Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's instructions and was quantified using the NanoDrop
ND-1000 spectrophotometer (Thermo Fisher Scientific, Inc.). cDNA
was synthesized by reverse transcription using the PrimeScript RT
reagent kit (Takara Bio, Inc.). qPCR was performed using SYBR
Premix Ex Taq II (Tli RNaseH Plus; Takara Bio Inc.) on a
LightCycler 96 system (Roche Diagnostics). Primers were synthesized
by Sangon Biotech Co., Ltd. The thermocycling conditions were as
follows: Firstly 95°C for 10 min, followed by 95°C for 10 sec, 60°C
for 15 sec and 72°C for 20 sec, for 40 cycles. Data were analyzed
using the 2−ΔΔCq method with GAPDH as the internal
reference control (34). All
results are expressed as the mean ± standard deviation of three
independent experiments. The following primer sequences were used:
GAPDH sense, 5′-GACCTGACCTGCCGTCTA-3′ and antisense,
5′-AGGAGTGGGTGTCGCTGT-3′; CtBP2 sense, 5′-CTGAGTTCCTGGCCTTTCTG-3′
and antisense, 5′-GACTTGATATCCGCGTCCTC-3′; TGIF sense,
5′-GGATGAGGACAGCATGGACA-3′ and antisense,
5′-AGGCATTGTAACGGTGCTCA-3′.
Protein extraction and western blot
analysis
Cells or tissues were lysed with ice-cold lysis
buffer (Beyotime Institute of Biotechnology), as previously
described (23). The protein
concentration of each sample was determined by BCA assay (Thermo
Scientific, Inc.) according to the manufacturer's instructions.
Proteins were separated via 10% SDS-PAGE (50 µg protein/lane), and
then blotted onto PVDF membranes (Bio-Rad, Hercules, CA). The
membranes were blocked with Tris-buffered saline and 0.1% Tween-20
(TBST) supplemented with 5% non-fat milk for 2 h at room
temperature, and the PVDF membranes were incubated with primary
antibodies including, rabbit anti-TGIF (ab52955; 1:1,000; Abcam),
mouse anti-CtBP2 (sc-17759; 1:200; Santa Cruz Biotechnology, Inc.)
and rabbit anti-β-catenin (ab68183; 1:1,000; Abcam) overnight at
4°C. Rabbit anti-β-actin (ab8227; 1:1,000; Abcam) was used as an
internal control. The membranes were then incubated with
HRP-conjugated goat anti-rabbit IgG (A8919; 1:1,000) or rabbit
anti-mouse IgG (A9044; 1:1000) (Sigma-Aldrich; Merck KGaA)
secondary antibodies at room temperature for 2 h. Finally, western
blot images were visualized by incubation with enhanced
chemiluminescence detection reagent (SuperSignal™ West Pico PLUS
Chemiluminescent Substrate, 34577) (Thermo Fisher Scientific, Inc.)
at room temperature for 5 min, and the bands were quantified using
ImageJ (v1.48) software (National Institutes of Health).
Immunohistochemistry (IHC)
Immunohistochemical staining was performed as
previously described (22).
Briefly, 4% PFA-fixed ESCC tissue or normal tissue sections were
deparaffinized in xylene, rehydrated in a graded alcohol series,
and finally washed three times with 0.1 M PBS. The slides were
submerged in Tris-EDTA buffer (100°C, 20 min) for antigen retrieval
and cooled naturally at room temperature. The IHC kit was purchased
from Zhongshan Golden Bridge Biotechnology Co., Ltd. (cat. no.
SP-9000). The sections were blocked with 10% goat serum (containing
0.1% triton-100, reagent 2) for 30 min at 37°C following treatment
with reagent 1 (endogenous peroxidase blockers) for 5 min, followed
by incubation with CtBP2 (sc-17759; 1:200; Santa Cruz
Biotechnology, Inc.) or TGIF (ab52955; 1:1,000; Abcam) antibody
overnight at 4°C. After washing three times with 0.1 M PBS, the
sections were incubated with reagent 3 [HRP-conjugated goat
anti-rabbit IgG (A8919; 1:1,000) or HRP-conjugated rabbit
anti-mouse IgG (A9044; 1:1000, Sigma-Aldrich; Merck KGaA) secondary
antibodies] for 1 h at 37°C, and subsequently incubated with
reagent 4 at 37°C for 1 h. Subsequently, hematoxylin (Beyotime
Institute of Biotechnology) was used for re-staining at room
temperature for 2 h, and finally the sections were dehydrated in
graded alcohol until they were transparent in xylene. The scoring
criteria (semi-quantitative method) were comprehensive and
determined by the staining intensity and proportion of positively
stained cells. The staining intensity score was as follows: 0
points for no staining, 1 point for weak staining (light yellow), 2
points for moderate staining (yellowish-brown), and 3 points for
strong staining (brown). The score for the proportion of positive
cells was as follows: 0 for ≤5%, 1 for 5–25%, 2 for 25–50%, 3 for
50–75%, 4 for >75%. A final score was obtained by multiplying
the two scores, 0–4 indicated negative, 4–8 was weakly positive,
and >8 was strongly positive. CtBP2 and TGIF expression in ESCC
tissues and normal tissues of patients were detected according to
the aforementioned method and score. Low expression was observed in
the negative and weak positive groups, whereas high expression was
observed in the strongly positive group. After staining, five
fields were randomly selected in each section (magnification, ×40)
under a microscope (Axio Imager 2; Carl Zeiss AG). The digitized
images of immunohistochemistry were quantitatively analyzed using
Image-Pro Plus 6.0 software (IPP 6.0; Media Cybernetics, Inc.).
H&E staining
H&E staining was used to distinguish tissue
morphology as per the manufacturer's instructions (Beyotime
Institute of Biotechnology). ESCC tissue or normal tissue sections
(4% PFA-fixed) were deparaffinized in xylene, then rehydrated in a
graded alcohol series. Nuclear staining was performed with
hematoxylin at room temperature for 10 min following by flushing
with running water to yield the color blue. The sections were then
differentiated with 1% hydrochloric acid ethanol for 3 sec and then
washing with running water was continued. The cytoplasm was stained
with eosin at room temperature for 30 sec, followed by 95% ethanol
twice for 5 min, 100% ethanol twice for 5 min, 100% ethanol +
xylene for 5 min (1:1), xylene for 5 min twice, and finally sealed
with neutral gum. H&E staining was observed under a microscope
(Axio Imager 2; Carl Zeiss AG).
Immunofluorescence
Staining reagents included CtBP2 (sc-17759; 1:200;
Santa Cruz Biotechnology, Inc.) and TGIF (ab52955; 1:1,000; Abcam)
primary antibodies, anti-mouse (SAB3701092) or anti-rabbit (F4890)
secondary antibodies (MilliporeSigma), and Hoechst 33342 (Beyotime
Institute of Biotechnology). The cells were digested with trypsin
and pipetted vigorously to make a single-cell suspension. The cells
were placed on glass coverslips (24-well plates), incubated at 37°C
and fixed with 4% PFA for 20 min at 15–25°C. The glass coverslips
were then washed three times with PBS for 10 min each and blocked
with 10% bovine serum albumin (BSA) for 2 h at 15–25°C.
Subsequently, all cells were incubated with primary antibody
(1:200) overnight at 4°C. The following day, the cells were
incubated with secondary antibody (1:2,000) for 2 h at 15–25°C.
Finally, the cells were stained with Hoechst 33258 (Beyotime
Institute of Biotechnology) at room temperature for 10 min, fixed
with anti-fade solution and imaged under a fluorescence microscope
(Zeiss AG).
Co-immunoprecipitation (Co-IP)
assay
The ECA109 cells were transfected with lentiviruses
expressing CtBP2 or TGIF. At 1 week post-transfection, the cells
were subjected to co-immunoprecipitation assay using a commercial
kit (Prod#26149; Thermo Fisher Scientific, Inc.). Briefly, firstly,
for antibody immobilization, ultrapure water, 20X Coupling Buffer
and 10 µg affinity-purified antibody were added directly to the
agarose (50% protein A/G agarose with ratio of 100 µl for a 1 ml
sample) in the spin column. Secondly, for the lysis of the cell
cultures, the culture medium was carefully removed from the cells.
The cells were then washed once with 1X modified Dulbecco's PBS.
This was followed by the addition of 400 µl per well ice-cold IP
lysis/wash buffer (2X 50 ml, 0.025 M Tris, 0.15 M NaCl, 0.001 M
EDTA, 1% NP-40, 5% glycerol; pH 7.4) to the cells cultured in a
6-well plate. The cells were then incubated on ice for 5 min with
periodic mixing. The lysate was then transferred to a
microcentrifuge tube and centrifuged at ~13,000 × g at 4°C for 10
min to pellet the cell debris. The agarose and cell proteins were
then mixed and appropriate experimental controls were prepared,
followed by rocking overnight at 4°C. The column was then
centrifuged at 1,000 × g at 4°C for 1 min. The protocol uses the IP
Lysis/Wash Buffer (1M NaCl) for coupling and washing the immune
complex. The spin column was then placed in a new collection tube.
This was followed by the addition of 50 µl elution buffer and
centrifugation. The tube was centrifuged at 1,000 × g at 4°C for 1
min and the flow-through was collected and analyze for protein. The
resulting immuno-complex was analyzed using western blot analysis,
with an anti-TGIF (ab52955; 1:1,000; Abcam) or anti-CtBP2
(sc-17759; 1:200; Santa Cruz Biotechnology, Inc.) antibody. To
eliminate heavy chain signals, a light chain-specific secondary
antibody (cat. no. 58802; Cell Signaling Technology, Inc.) was
used.
CCK-8 assay
Cell viability was measured using CCK-8 assay
(Dojindo Molecular Technologies, Inc.) according to the
manufacturer's instructions in three independent experiments. The
transfected ECA109 cells at a density of 5×103
cells/well were seeded in 96-well plates with 100 µl 10% FBS. After
culturing the cells for 24, 48 and 72 h with (10 and 100 nM, and 1,
10 and 100 µM) XAV939 (Wnt signaling pathway inhibitor; Merck KGaA)
at 37°C, 10 µl CCK-8 reagent were added to each well and incubated
at 37°C for a further 4 h. Cell viability was then measured based
on the absorbance at 450 nm wavelength (OD450) using a microplate
reader (BioTek Instruments, Inc.).
5-Ethynyl-2′-deoxyuridine (EdU)
staining
Cell proliferation of the transfected ECA109 cells
upon XAV939 treatment was investigated using EdU staining assay.
Briefly, 1×105 transfected ECA109 cells were resuspended
in 200 µl DMEM, and then seeded into 24-well plates. Following
incubation for 48 h with 10 µM XAV939, the cells were stained with
EdU (Beyotime Institute of Biotechnology) at room temperature for
30 min against exposure to light. The cells were re-stained with
Hoechst 33342 for 10 min in the dark, followed by washing with PBS.
The number of EdU-positive cells was photographed, and five
randomly selected fields were counted, using a fluorescence
microscope (Carl Zeiss AG) (magnification, ×200). The experiments
were performed in triplicate.
Wound healing assay
The migration of the transfected ECA109 cells upon
XAV939 treatment was investigated using a wound healing assay, as
previously described (35). To
ensure the consistency of each initial scratch, a scratch chamber
was used to carry out the experiment. Briefly, 5×104
transfected ECA109 cells were seeded into the scratch chamber (70
µl cell suspension for each side of the scratch chamber), and then
cultured overnight to adhere, followed by the removal of the
scratch chamber. The cells were then washed twice with PBS, in
order to remove any non-adherent cells. Before scratching, the
cells were cultured in high-glucose DMEM containing 10% FBS and
supplemented with 1% penicillin/streptomycin mixture (36). To ensure the consistency of each
initial scratch, ibidi Culture-Inserts (ibidi GmbH) were used to
carry out the experiment. Briefly, 5×104 transfected
ECA109 cells were seeded into the ibidi Culture-Inserts (70 µl cell
suspension for each side of the scratch chamber), and then cultured
overnight to adhere, followed by the removal of the ibidi
Culture-Inserts. Following the removal of the ibidi
Culture-Inserts, the scratch wound was created. After scratching
was complete, the cells were cultured in high-glucose DMEM
containing 2% FBS and supplemented with 1% penicillin/streptomycin
mixture. The migration (wound closure) of the indicated cells was
monitored and photographed randomly at 0, 24 and 48 h. The wound
healing rate (%) = (x h scratch area - 0 h scratch area)/0 h
scratch area ×100. Independent experiments were performed at least
three times.
Transwell assay
The cell migratory and invasive abilities were
examined using a Transwell assay, as previously described (35). Briefly, for the migration assay,
1×105 transfected ECA109 cells were resuspended in 200
µl DMEM, and then seeded into the upper chamber of the Transwell (8
µm pore size; Corning, Inc.) in 24-well plates. For the invasion
assay, the upper chambers were coated with Matrigel matrix (50 µl,
BD Biosciences) before seeding the cells, and 500 µl of 10% FBS
were added to the lower chamber. Following incubation at 37°C for
48 h with 10 µM XAV939, the non-migrating cells were removed using
a cotton swab. However, the migrated or invaded cells on the
underside were fixed with 4% PFA and stained with 0.1% crystal
violet (Beyotime Institute of Biotechnology) at room temperature
for ~40 min. The number of migrating and invading cells was
photographed, and cells in five randomly selected fields were
counted using a phase contrast microscope (Leica DM IL LED)
(magnification, ×200). All experiments were performed in
triplicate.
Bioinformatics analysis
The interactions between CtBP2 and predictive
proteins were analyzed using the STRING database (https://string-db.org). Gene Expression Profiling
Interactive Analysis (GEPIA) (http://gepia.cancer-pku.cn) was used to analyze the
correlation between the expression of two interesting genes in a
given tissue (37).
Statistical analysis
Statistical analysis was performed using GraphPad
Prism version 8.0 (GraphPad Software, Inc.) or SPSS 23.0 (IBM
Corp.). χ2 tests were performed to assess the clinical
association between CtBP2 and TGIF expression and other tumor
characteristics. Survival analysis was performed using the
Kaplan-Meier method and the log-rank test. A Cox proportional
hazards regression model was established to assess the factors
independently associated with patient survival. Differences between
two groups were compared using an unpaired or paired Student's
t-test. To compare more than two groups, one-way ANOVA with post
hoc Holm-Sidak correction for multiple comparisons was performed.
All experimental data are presented as the mean ± SD. P<0.05 was
considered to indicate a statistically significant difference and
all tests performed were two-sided.
Results
Interaction between CtBP2 and
TGIF
The STRING database was used for the detection of
known protein-protein interactions and predictive protein-protein
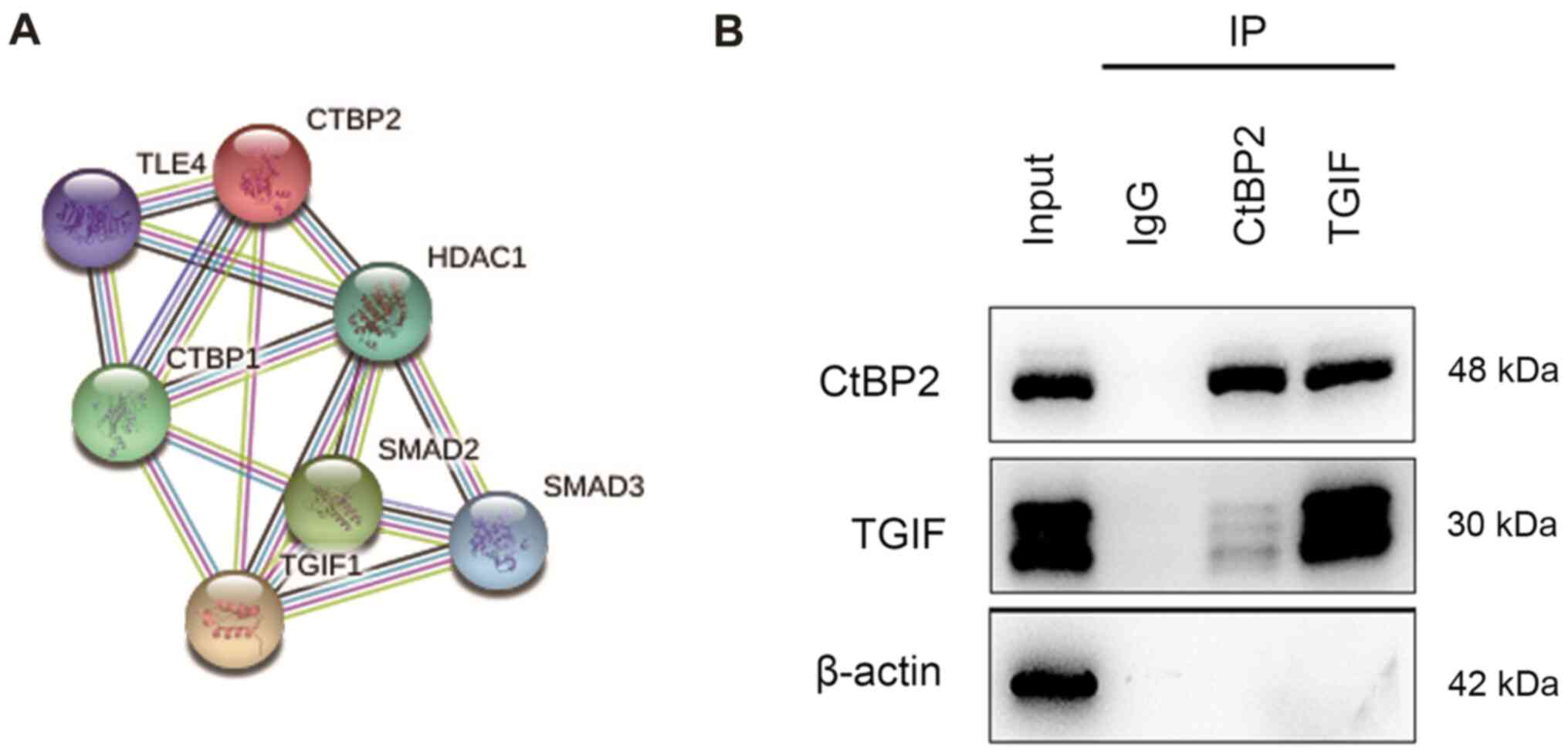
interactions. The CtBP2 protein network is illustrated in Fig. 1A. As a predictive result, an
interaction was detected between CtBP2 and TGIF.
Co-IP was performed to validate the interaction
between CtBP2 and TGIF. Following ECA109 cell transfection with the
with the CtBP2 or TGIF lentiviral transduction vector, the cells
were subjected to Co-IP assay with anti-CtBP2 or anti-TGIF
antibody. Immuno-complexes of CtBP2 and TGIF were observed using
western blot analysis (Fig.
1B).
Expression levels of CtBP2 and TGIF in
ESCC tissues and cells
RT-qPCR and western blot analysis were performed to
examine the expression levels of CtBP2 and TGIF in ESCC tissues and
cells. The mRNA expression levels of CtBP2 and TGIF were robustly
increased simultaneously in the tumor tissues of representative
patients with ESCC (Fig. 2A).
Western blot analysis and IHC were performed to further confirm the
CtBP2 and TGIF expression levels in ESCC tissues. Compared with the
adjacent normal tissues, the expression levels of CtBP2 and TGIF
were significantly upregulated simultaneously in ESCC tissues
(Fig. 2B-E). RT-qPCR and western
blot analysis were performed to evaluate the mRNA and protein
expression levels of CtBP2 and TGIF in the ESCC cell lines. In
comparison with the human normal esophageal epithelial cell line,
HEEC, the mRNA and protein expression levels of both CtBP2 and TGIF
were increased simultaneously in the ESCC cell lines. In ECA109
cells, the levels of CtBP2 and TGIF were the highest (Fig. 2F-H). These results revealed that
CtBP2 and TGIF expression levels were significantly simultaneously
increased in ESCC tissues and cells.
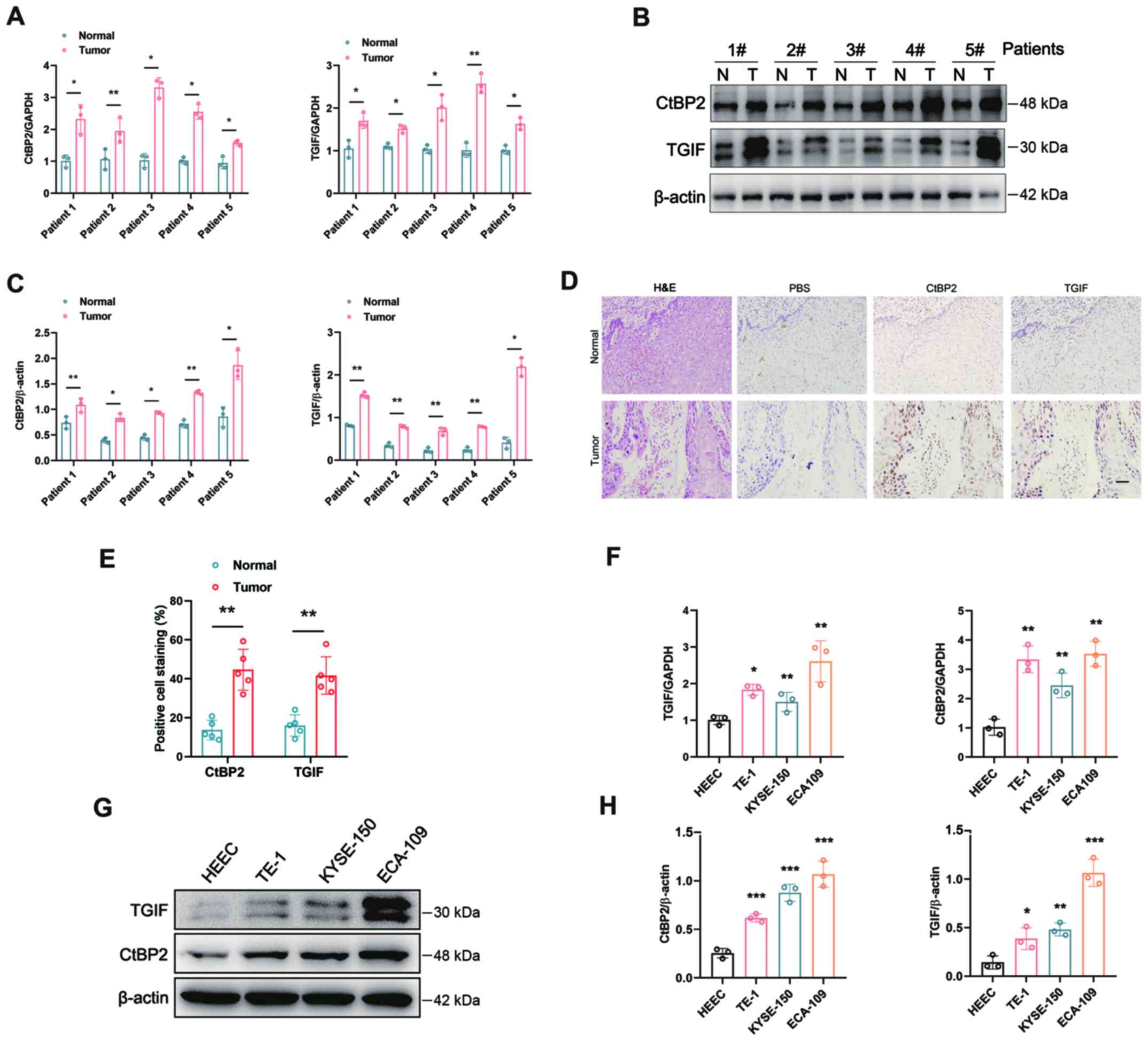 | Figure 2.Expression levels of CtBP2 and TGIF
in tissues or cells of ESCC. (A) mRNA expression levels of CtBP2
and TGIF in ESCC tissues were examined by RT-qPCR. (B) Expression
levels of CtBP2 and TGIF in ESCC tissues were tested by western
blot analysis. N, normal tissues; T, tumor tissues. (C) Statistical
analysis of western blot analysis results. (D) Expression levels of
CtBP2 and TGIF in ESCC tissues were tested by IHC. PBS was used as
the negative control. Scale bar=200 µm. (E) Statistical analysis of
IHC results. (F) Expression levels of CtBP2 and TGIF in the human
normal esophageal epithelial cell line, HEEC, and the human ESCC
cell lines, ECA109, TE-1 and KYSE-150, were examined using western
blot analysis. (G) mRNA expression levels of CtBP2 and TGIF in HEEC
and ECA109, TE-1 and KYSE-150 cells were examined using RT-qPCR.
(H) Statistical analysis of the western blots in panel F. Data are
presented as the mean ± SD, and the Student's t-test and one-way
ANOVA were performed. *P<0.05, **P<0.01 and ***P<0.001.
CtBP2, C-terminal-binding protein 2; TGIF, transforming growth
interacting factor; ESCC, esophageal squamous cell carcinoma; IHC,
immunohistochemistry; RT-qPCR, reverse transcription quantitative
-PCR. |
Association between CtBP2 and TGIF
expression and clinicopathologic characteristics of patients with
ESCC
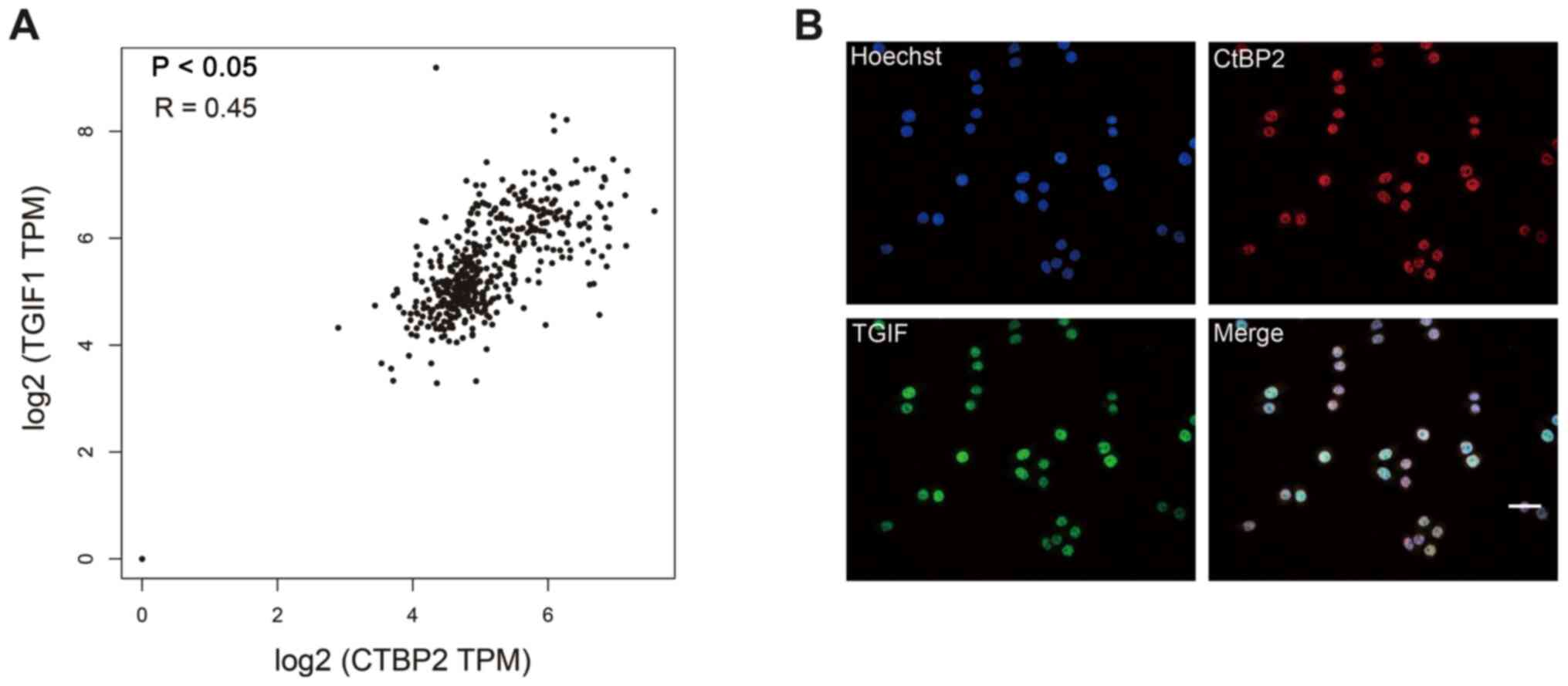
The correlation between CtBP2 and TGIF expression
levels in ESCC based on GEPIA by Pearson's correlation analysis is
depicted in Fig. 3A. There was a
direct correlation between the CtBP2 and TGIF expression levels in
ESCC tissues, with a correlation coefficient of R=0.45
(P<0.05).
Subsequently, since there was a correlation between
CtBP2 and TGIF expression levels in ESCC, their co-localization in
the ESCC cell line, ECA109, was examined. For this purpose,
immunofluorescence staining was performed to evaluate CtBP2 and
TGIF expression co-localization in ECA109 cells. The results
demonstrated that CtBP2 and TGIF expression was co-localized in the
nucleus of ECA109 cells (Fig.
3B).
A total of 108 patients with ESCC was enrolled, and
the association between CtBP2 and TGIF expression and the patient
clinicopathological characteristics was analyzed (Table I). The CtBP2 and TGIF expression
levels were significantly associated with metastasis and survival
(P<0.05).
 | Table I.Association between CtBP2 and TGIF
expression and the clinicopathological characteristics of patients
with ESCC. |
Table I.
Association between CtBP2 and TGIF
expression and the clinicopathological characteristics of patients
with ESCC.
|
| CtBP2 |
| TGIF |
|
|---|
|
|
|
|
|
|
|---|
| Characteristic | Low | High | P-value | Low | High | P-value |
|---|
| Age (years) |
|
| 0.912 |
|
| 0.742 |
|
<60 | 10 | 24 |
| 14 | 20 |
|
|
≥60 | 21 | 53 |
| 28 | 46 |
|
| Sex |
|
| 0.154 |
|
| 0.55 |
|
Male | 21 | 62 |
| 31 | 52 |
|
|
Female | 10 | 15 |
| 11 | 14 |
|
| Clinical stage |
|
| 0.147 |
|
| 0.175 |
| I | 3 | 6 |
| 6 | 3 |
|
| II | 24 | 47 |
| 27 | 44 |
|
|
III | 4 | 24 |
| 9 | 19 |
|
| Histological
differentiation |
|
| 0.053 |
|
| 0.379 |
|
Well | 5 | 15 |
| 6 | 14 |
|
|
Good | 12 | 45 |
| 21 | 36 |
|
|
Poor | 14 | 17 |
| 15 | 16 |
|
| Tumor diameter
(cm) |
|
| 0.078 |
|
| 0.097 |
|
<3 | 12 | 17 |
| 15 | 14 |
|
| ≥3 | 19 | 60 |
| 27 | 52 |
|
| T
classification |
|
| 0.695 |
|
| 0.312 |
| T1 | 3 | 11 |
| 6 | 8 |
|
| T2 | 11 | 22 |
| 16 | 17 |
|
| T3 | 17 | 44 |
| 20 | 41 |
|
| N
classification |
|
| 0.108 |
|
| 0.375 |
| N0 | 20 | 47 |
| 27 | 40 |
|
| N1 | 11 | 18 |
| 13 | 16 |
|
| N2 | 0 | 11 |
| 2 | 9 |
|
| N3 | 0 | 1 |
| 0 | 1 |
|
| Metastasis |
|
| 0.015a |
|
| 0.014a |
| No | 17 | 23 |
| 22 | 18 |
|
|
Yes | 14 | 54 |
| 20 | 48 |
|
| Depth |
|
| 0.493 |
|
| 0.345 |
| t0 | 1 | 1 |
| 0 | 2 |
|
| t1 | 2 | 12 |
| 6 | 8 |
|
| t2 | 11 | 19 |
| 14 | 16 |
|
| t3 | 15 | 36 |
| 16 | 35 |
|
| t4 | 2 | 9 |
| 6 | 5 |
|
| Outcome |
|
| 0.003a |
|
| 0.001a |
|
Mortality | 18 | 21 |
| 28 | 11 |
|
|
Survival | 13 | 56 |
| 14 | 55 |
|
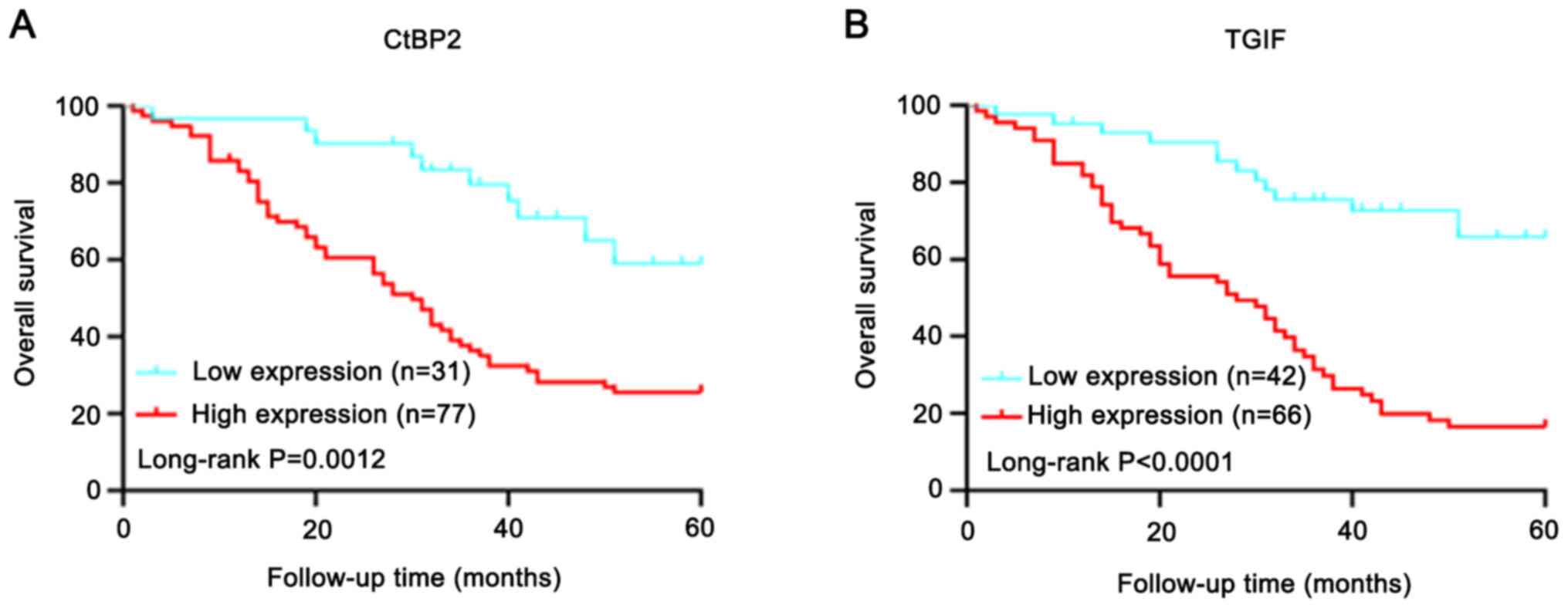
Prognostic value of CtBP2 and TGIF
expression
Kaplan-Meier analysis was performed to examine the
association between CtBP2 or TGIF expression and the survival of
patients with ESCC. A total of 108 patients with follow-up data
were evaluated and IHC staining followed by Kaplan-Meier survival
analysis was performed. The high vs. low expression of CtBP2
(Fig. 4A) or TGIF (Fig. 4B) differed significantly in the
survival curves. Patients with a low expression of CtBP2 or TGIF
had a longer cumulative survival. Univariate analysis, performed
using the Cox proportional hazards regression model, revealed that
CtBP2 (P=0.005), TGIF (P=0.001), clinical stage (P=0.001), tumor
diameter (P=0.005), T classification (P=0.026) and N classification
(P=0.001) were independent prognostic indicators of overall
survival (Table II).
 | Table II.Univariate analyses of various
prognostic parameters in patients with ESCC using Cox regression
analysis. |
Table II.
Univariate analyses of various
prognostic parameters in patients with ESCC using Cox regression
analysis.
|
| Univariate
analysis |
|
|---|
|
|
|
|
|---|
| Parameter | Hazard ratio | 95% confidence
interval | P-value |
|---|
| Age | 1.473 | 0.867–2.503 | 0.152 |
| Sex | 0.763 | 0.43–1.354 | 0.355 |
| Clinical stage | 2.088 | 1.349–3.232 | 0.001a |
| Histological
differentiation | 0.959 | 0.683–1.346 | 0.807 |
| Tumor diameter | 2.381 | 1.299–4.364 | 0.005a |
| T
classification | 1.497 | 1.049–2.137 | 0.026a |
| N
classification | 1.684 | 1.240–2.287 | 0.001a |
| Metastasis | 1.240 | 0.747–2.058 | 0.405 |
| Depth | 1.239 | 0.952–1.613 | 0.112 |
| CtBP2 | 2.361 | 1.288–4.329 | 0.005a |
| TGIF | 4.194 | 2.317–7.590 | 0.001a |
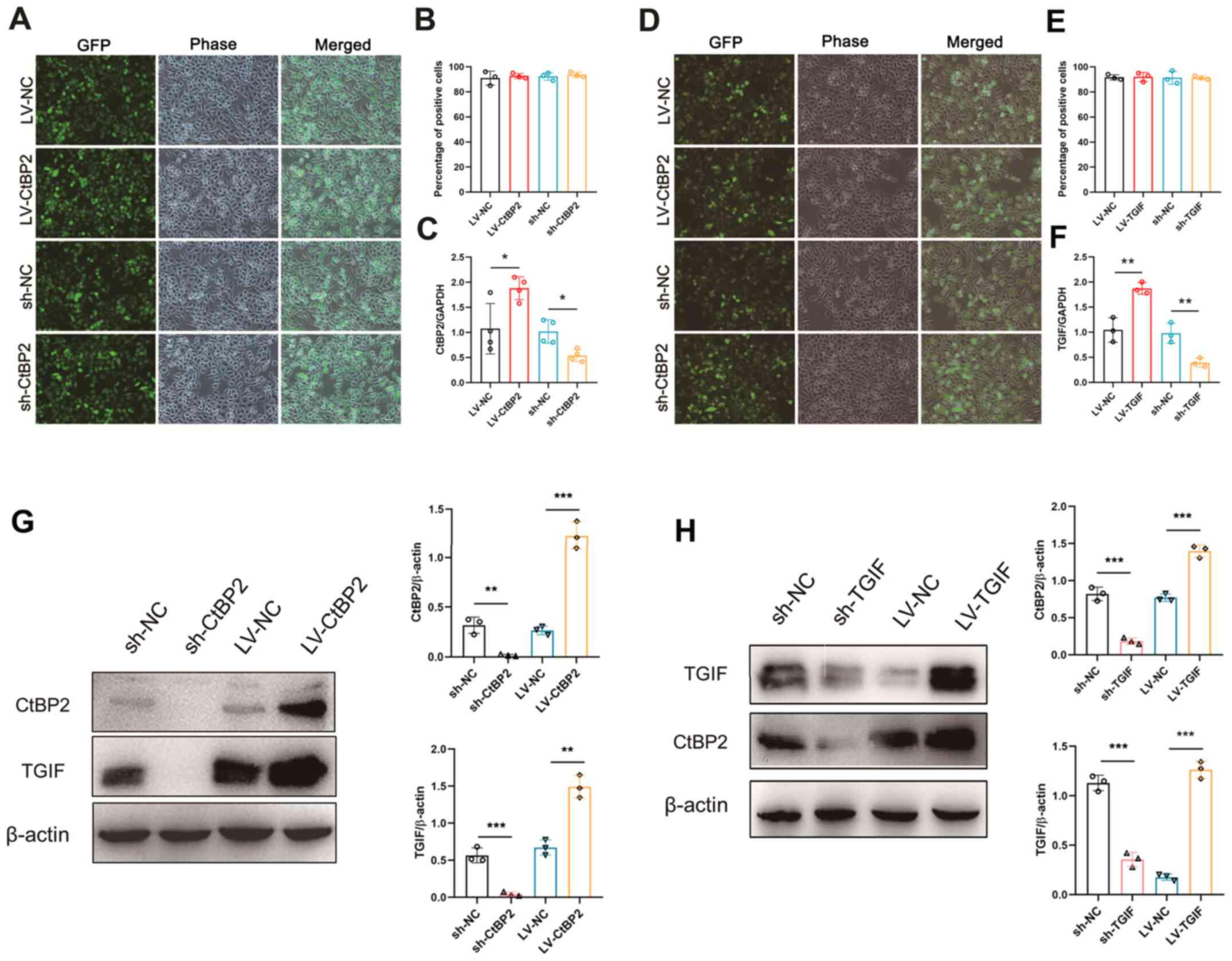
Signaling pathway involved in the of
biological activity of CtBP2 in ECA109 cells
To explore the biological activity of CtBP2 and TGIF
in ECA109 cells, recombinant lentiviral vectors overexpressing
CtBP2 and TGIF (LV-CtBP2 and LV-TGIF) or carrying shRNAs to
knockdown (sh-CtBP2 and sh-TGIF) CtBP2 and TGIF expression were
constructed. Since the recombinant lentiviral vectors contained the
gene encoding the green fluorescent protein (GFP), the transfection
rate could be evaluated directly under a fluorescence microscope.
Following transfection, the fluorescence images demonstrated that
the percentage of GFP-positive cells was >90% (Fig. 5A, B, D and E). RT-qPCR was
performed to further examine CtBP2 and TGIF mRNA expression in the
ECA109 cells transfected with the vectors. In comparison with the
negative control (NC), the CtBP2 and TGIF mRNA expression levels
were significantly increased or decreased (P<0.05) following
transfection in the ECA109 cells (Fig.
5C and F). In addition, western blot analysis was performed to
examine the CtBP2 and TGIF expression levels in the ECA109 cells
transfected with the vectors. As was anticipated, the CtBP2 and
TGIF expression levels increased or decreased following
transfection in ECA109 cells, as compared with the NC. Of note,
TGIF expression was robustly and simultaneously increased when
CtBP2 expression was significantly upregulated in the cells
transfected with LV-CtBP2 and simultaneously decreased when CtBP2
expression was significantly downregulated in the ECA109 cells
transfected with sh-CtBP2 (Fig. 5G and
H).
XAV939 is an inhibitor of the Wnt signaling pathway
(38), and TGIF plays a role in
tumorigenesis through Wnt signaling (33). Therefore, it was hypothesized that
CtBP2 may interact with TGIF to promote the proliferation and
migration of ECA109 cells through the Wnt/β-catenin signaling
pathway. First, the effects of XAV939 on cell viability and
β-catenin expression were examined in the ECA109 cells. The results
of CCK-8 assay revealed that cell viability was significantly
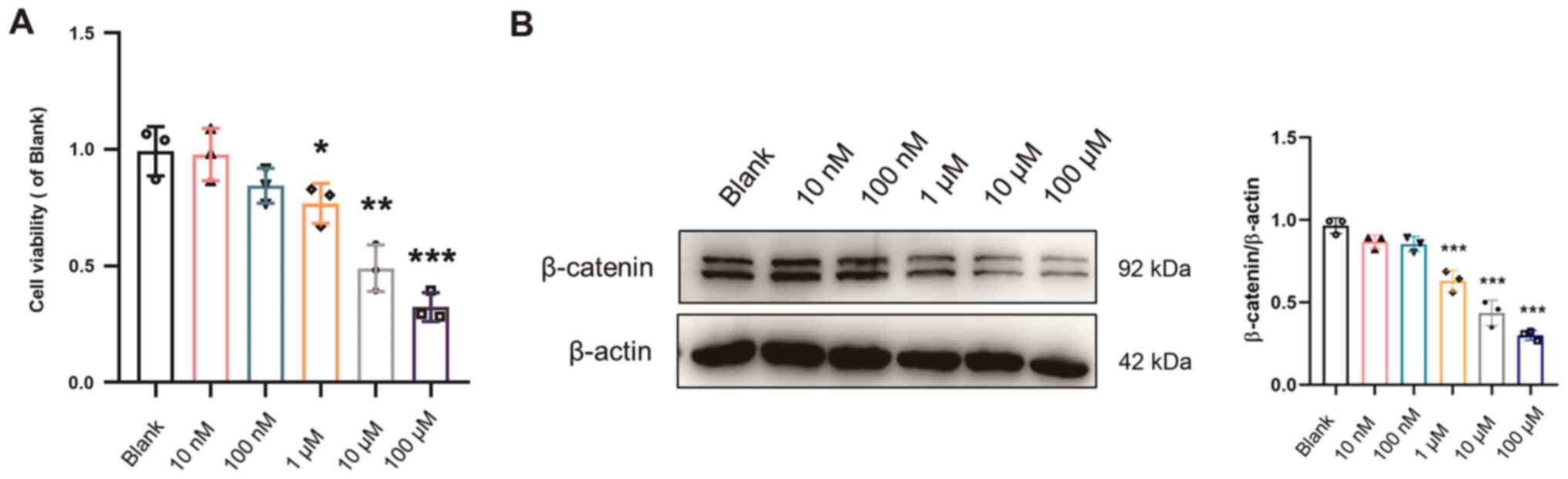
decreased (P<0.05) in the ECA109 cells treated with XAV939 (10
and 100 nM, and 1, 10 and 100 µM) for 48 h (Fig. 6A). The expression of β-catenin was
also significantly downregulated (P<0.001) in the ECA109 cells
treated with XAV939 (10 nM, 100 nM, 1, 10 and 100 µM) for 48 h
(Fig. 6B). Therefore, the optimal
concentration of XAV939 that was used in subsequent experiments was
10 µM.
Furthermore, CCK-8 assay was performed in order to
measure the viability of the ECA109 cells transfected with LV-CtBP2
and treated with 10 µM XAV939 for 24, 48 and 72 h. In comparison
with the negative control (LV-NC), cell viability was markedly
increased in the LV-CtBP2-transfected group; however, it was
markedly decreased (P<0.05) in the XAV939 group, compared with
the LV-NC-transfected group (Fig.
7A). These results indicated that XAV939 inhibited the
CtBP2-mediated viability of ECA109 cells.
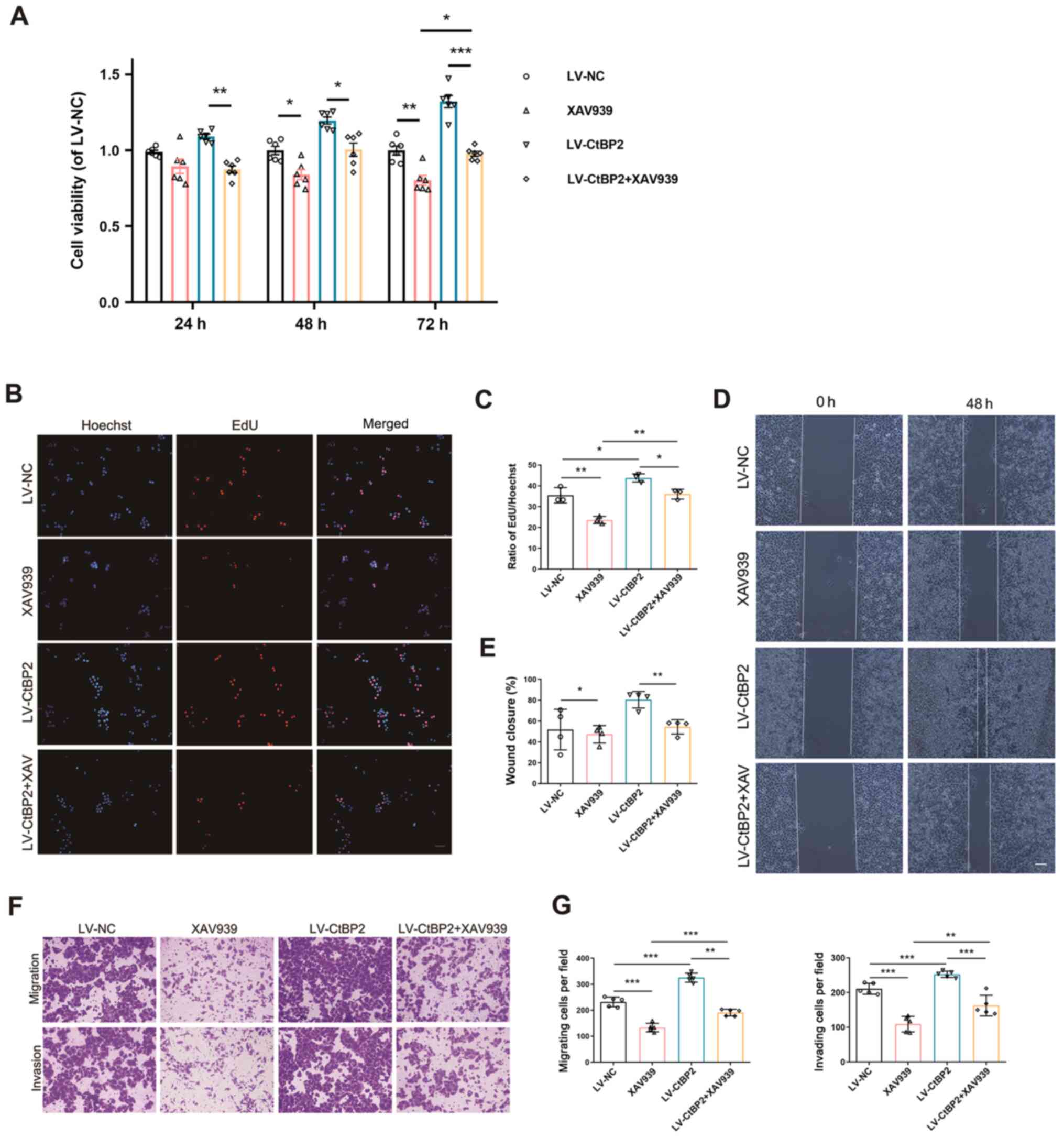 | Figure 7.Signaling pathway involved in the
biological activity of CtBP2 in ECA109 cells. (A) Viability of
ECA109 cells transfected with LV-CtBP2 and treated with 10 µM
XAV939 for 24, 48 and 72 h, was measured using CCK-8 assay. (B) The
proliferation of ECA109 cells transfected with LV-CtBP2 and treated
with 10 µM XAV939 for 48 h was measured using EdU staining. Scale
bar, 50 µm. (C) Statistical analysis of EdU staining results. (D)
The migration of ECA109 cells transfected with LV-CtBP2 and treated
with 10 µM XAV939 for 48 h was measured using wound healing assay.
Scale bar, 100 µm. (E) Statistical analysis of the wound healing
assay results. (F) The migration and invasion of ECA109 cells
transfected with LV-CtBP2 and treated with 10 µM XAV939 for 48 h
was measured using Transwell assay. Scale bar, 100 µm. (G)
Statistical analysis of the Transwell assay results. Data are
presented as the mean ± SD, and the Student's t-test and one-way
ANOVA were performed. *P<0.05, **P<0.01 and ***P<0.001.
CtBP2, C-terminal-binding protein 2; LV, lentiviral; EdU,
5-ethynyl-2′-deoxyuridine. |
EdU staining was then employed to examine the
effects of CtBP2 and XAV939 in ECA109 cells. The results
demonstrated that the number of EdU-positive cells was
significantly decreased (P<0.05) in the LV-CtBP2 + XAV939 group,
compared with the LV-CtBP2-transfected group (Fig. 7B and C). This demonstrated that
XAV939 inhibited the CtBP2-mediated proliferation of ECA109 cells.
The results of wound healing and Transwell assays also revealed
that XAV939 inhibited the CtBP2-mediated migration and invasion of
ECA109 cells (Fig. 7D-G). These
results indicate that CtBP2 exerts its biological activity through
the Wnt/β-catenin pathway in ECA109 cells.
Discussion
The extremely high incidence of EC in certain
regions of China has prompted a number of researchers to
investigate the disease pathogenesis and develop effective
treatment strategies for this disease in China. Due to
comprehensive tumorigenesis and the development of EC involving
complex regulation of oncogenes and tumor suppressor genes, the
in-depth elucidation of the pathogenesis of EC at the molecular
level in order to identify an ideal molecular target or effective
therapeutic drugs for EC is a promising therapeutic approach
(10,39). In the present study, the proteins
interacting with CtBP2 were identified and the mechanisms of the
biological activity of CtBP2 in ESCC were investigated.
CtBP2, a transcriptional co-repressor, acts as a
bridge molecule between DNA-binding proteins and transcriptional
repressors, in order to inhibit gene transcription by specifically
binding to DNA-binding proteins and transcriptional repressors
(12). Therefore, proteins
interacting with CtBP2 were first determined in order to
investigate the underlying mechanisms of CtBP2 biological activity
in ESCC. The STRING database has been used for the detection of
known protein-protein interactions and the prediction of
protein-protein interactions (40). The results obtained from the STRING
database indicated that there was an interaction between CtBP2 and
TGIF and the subsequently performed Co-IP analysis indicated that
these two proteins co-localized in the nucleus.
TGIF, a transcriptional repressor, is involved in a
number of cellular signal transduction pathways, particularly the
TGF-β pathway (41). As an
interaction between CtBP2 and TGIF was observed, the expression of
TGIF and the correlation between CtBP2 and TGIF in ESCC was then
examined. A previous study by the authors revealed that the
expression of CtBP2 was significantly increased in ESCC tissues and
was positively associated with the histological grade of the tumor
(22). As was anticipated, the
expression levels of CtBP2 and TGIF were significantly
simultaneously increased in ESCC tissues and cells. GEPIA was used
to further analyze the correlation between the expression of CtBP2
and TGIF in ESCC. There was a direct correlation between CtBP2 and
TGIF expression in ESCC tissues, with a correlation coefficient of
R=0.45. These results indicated that the interaction of CtBP2 with
TGIF plays a crucial role in ESCC.
The 5-year survival rate of patients with EC has
been reported to be ~10%, and the recurrence and mortality rates
remain high (8). Therefore, there
is an urgent need for the identification of novel prognostic
biomarkers for ESCC. In the present stuyd, Kaplan-Meier analysis
was performed to examine the association between CtBP2 or TGIF
expression and the survival of patients with ESCC. The results
indicated that both CtBP2 and TGIF expression levels were
significantly associated with metastasis and survival. Patients
with a low expression of CtBP2 or TGIF had a longer cumulative
survival. Therefore, CtBP2 and TGIF expression may serve as
prognostic indicators of the clinical outcome of patients with
ESCC.
XAV939 is an inhibitor of the Wnt signaling pathway
(38), and high levels of TGIF are
associated with high levels of the Wnt signaling pathway and a poor
survival rate (33). Wnt exerts
its effects through three signaling pathways, the most classic
being the Wnt/β-catenin signaling pathway, mediated by β-catenin
(42). It has been reported that
the abnormal activation of the Wnt/β-catenin signaling pathway is
involved in a number of diseases, including cancer, genetic
diseases and organ fibrosis, as well as in the occurrence of EMT
(43). Therefore, it was
hypothesized that CtBP2, as a transcriptional co-repressor, may
interact with TGIF and participate in the development of ESCC
through the Wnt/β-catenin signaling pathway. TGIF expression was
robustly increased or decreased simultaneously when CtBP2
expression was significantly upregulated or downregulated, and vice
versa. The results of CCK-8, EdU staining and Transwell assays
indicated that CtBP2 promoted the proliferation, migration and
invasion of ECA109 cells through the Wnt/β-catenin pathway. In
future studies, the authors aim to perform TCF reporter assay, in
order to further verify the signaling pathway involved.
In conclusion, the present study demonstrates the
existence of an interaction between CtBP2 and TGIF expression in
ESCC, and these two proteins were co-localized in the nucleus. The
CtBP2 and TGIF expression levels were robustly increased
simultaneously in ESCC tissues and cell lines, and their expression
was significantly associated with metastasis and survival. CtBP2
interacted with TGIF and promoted the progression of ESCC through
the Wnt/β-catenin pathway.
Acknowledgements
Not applicable.
Funding
The present study was supported by the China Postdoctoral
Science Foundation (2018M632337), Natural Science Foundation of
Shanghai (21ZR1449800), and the Natural Science Research Project of
Nantong Science and Technology Bureau (XG202006-4).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
QJ, MJ and HS contributed to the conception and
design of the study. QJ and HS provided administrative support. QJ,
MJ, WH, QY, ZL and HS provided all the study materials or patient
clinicopathological data and samples. QJ, MJ, WH, QY and HS
performed data collection and assembly. QJ, MJ, WH and HS
contributed to data analysis and interpretation. QJ and HS
confirmed the authenticity of all the raw data. QJ, MJ and HS wrote
the manuscript. All authors have read and approved the final
manuscript.
Ethics approval and consent to
participate
Patient written informed consent was obtained before
the study, according to the guidelines of the Ethics Committee of
the Affiliated Hospital of Nantong University, and ethics approval
has been also provided from the respective ethics committee (2015
L132).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Lin Y, Totsuka Y, Shan B, Wang C, Wei W,
Qiao Y, Kikuchi S, Inoue M, Tanaka H and He Y: Esophageal cancer in
high-risk areas of China: Research progress and challenges. Ann
Epidemiol. 27:215–221. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Yu C, Tang H, Guo Y, Bian Z, Yang L, Chen
Y, Tang A, Zhou X, Yang X, Chen J, et al China Kadoorie Biobank
Collaborative Group, : Hot tea consumption and its interactions
with alcohol and tobacco use on the risk for esophageal cancer: a
population-based cohort study. Ann Intern Med. 168:489–497. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lin Y, Totsuka Y, He Y, Kikuchi S, Qiao Y,
Ueda J, Wei W, Inoue M and Tanaka H: Epidemiology of esophageal
cancer in Japan and China. J Epidemiol. 23:233–242. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Yang S, Lin S, Li N, Deng Y, Wang M, Xiang
D, Xiang G, Wang S, Ye X, Zheng Y, et al: Burden, trends, and risk
factors of esophageal cancer in China from 1990 to 2017: An
up-to-date overview and comparison with those in Japan and South
Korea. J Hematol Oncol. 13:1462020. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Anandavadivelan P and Lagergren P:
Cachexia in patients with oesophageal cancer. Nat Rev Clin Oncol.
13:185–198. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Gao QY and Fang JY: Early esophageal
cancer screening in China. Best Pract Res Clin Gastroenterol.
29:885–893. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Chen W, Li H, Zheng R, Ren J, Shi J, Cao
M, Sun D, Sun X, Cao X, Zhou J, et al: An initial screening
strategy based on epidemiologic information in esophageal cancer
screening: A prospective evaluation in a community-based cancer
screening cohort in rural China. Gastrointest Endosc.
93:110–118.e2. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Chen W, Li H, Ren J, Zheng R, Shi J, Li J,
Cao M, Sun D, He S, Sun X, et al: Selection of high-risk
individuals for esophageal cancer screening: A prediction model of
esophageal squamous cell carcinoma based on a multicenter screening
cohort in rural China. Int J Cancer. 148:329–339. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Liu M, He Z, Guo C, Xu R, Li F, Ning T,
Pan Y, Li Y, Ding H, Zheng L, et al: Effectiveness of intensive
endoscopic screening for esophageal cancer in China: a
community-based study. Am J Epidemiol. 188:776–784. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kelly RJ: Emerging multimodality
approaches to treat localized esophageal cancer. J Natl Compr Canc
Netw. 17:1009–1014. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Borggreve AS, Kingma BF, Domrachev SA,
Koshkin MA, Ruurda JP, van Hillegersberg R, Takeda FR and Goense L:
Surgical treatment of esophageal cancer in the era of multimodality
management. Ann N Y Acad Sci. 1434:192–209. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chen L, Wang L, Qin J and Wei DS: CtBP2
interacts with ZBTB18 to promote malignancy of glioblastoma. Life
Sci. 262:1184772020. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wang H, Xiao Z, Zheng J, Wu J, Hu XL, Yang
X and Shen Q: ZEB1 represses neural differentiation and cooperates
with CTBP2 to dynamically regulate cell migration during neocortex
development. Cell Rep. 27:2335–2353.e6. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhao LJ, Subramanian T, Vijayalingam S and
Chinnadurai G: PLDLS-dependent interaction of E1A with CtBP:
Regulation of CtBP nuclear localization and transcriptional
functions. Oncogene. 26:7544–7551. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Jecrois AM, Dcona MM, Deng X,
Bandyopadhyay D, Grossman SR, Schiffer CA and Royer WE Jr: Cryo-EM
structure of CtBP2 confirms tetrameric architecture. Structure.
29:310–319.e5. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ma Y, Sekiya M, Kainoh K, Matsuda T,
Iwasaki H, Osaki Y, Sugano Y, Suzuki H, Takeuchi Y, Miyamoto T, et
al: Transcriptional co-repressor CtBP2 orchestrates
epithelial-mesenchymal transition through a novel transcriptional
holocomplex with OCT1. Biochem Biophys Res Commun. 523:354–360.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang DP, Gu LL, Xue Q, Chen H and Mao GX:
CtBP2 promotes proliferation and reduces drug sensitivity in
non-small cell lung cancer via the Wnt/β-catenin pathway.
Neoplasma. 65:888–897. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Thio SS, Bonventre JV and Hsu SI: The
CtBP2 co-repressor is regulated by NADH-dependent dimerization and
possesses a novel N-terminal repression domain. Nucleic Acids Res.
32:1836–1847. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhao Z, Hao D, Wang L, Li J, Meng Y, Li P,
Wang Y, Zhang C, Zhou H, Gardner K, et al: CtBP promotes metastasis
of breast cancer through repressing cholesterol and activating
TGF-β signaling. Oncogene. 38:2076–2091. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Dcona MM, Damle PK, Zarate-Perez F, Morris
BL, Nawaz Z, Dennis MJ, Deng X, Korwar S, Singh SJ, Ellis KC, et
al: Active-site tryptophan, the target of antineoplastic C-terminal
binding protein inhibitors, mediates inhibitor disruption of CtBP
oligomerization and transcription coregulatory activities. Mol
Pharmacol. 96:99–108. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Thomas G, Jacobs KB, Yeager M, Kraft P,
Wacholder S, Orr N, Yu K, Chatterjee N, Welch R, Hutchinson A, et
al: Multiple loci identified in a genome-wide association study of
prostate cancer. Nat Genet. 40:310–315. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Guan C, Shi H, Wang H, Zhang J, Ni W, Chen
B, Hou S, Yang X, Shen A and Ni R: CtBP2 contributes to malignant
development of human esophageal squamous cell carcinoma by
regulation of p16INK4A. J Cell Biochem. 114:1343–1354. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Shi H, Mao Y, Ju Q, Wu Y, Bai W, Wang P,
Zhang Y and Jiang M: C-terminal binding protein-2 mediates
cisplatin chemoresistance in esophageal cancer cells via the
inhibition of apoptosis. Int J Oncol. 53:167–176. 2018.PubMed/NCBI
|
|
24
|
Wang Y, Liu F, Mao F, Hang Q, Huang X, He
S, Wang Y, Cheng C, Wang H, Xu G, et al: Interaction with cyclin
H/cyclin-dependent kinase 7 (CCNH/CDK7) stabilizes C-terminal
binding protein 2 (CtBP2) and promotes cancer cell migration. J
Biol Chem. 288:9028–9034. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Shi H, Xu J, Zhao R, Wu H, Gu L and Chen
Y: FGF2 regulates proliferation, migration, and invasion of ECA109
cells through PI3K/Akt signalling pathway in vitro. Cell Biol Int.
40:524–533. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Shah A, Melhuish TA, Fox TE, Frierson HF
Jr and Wotton D: TGIF transcription factors repress acetyl CoA
metabolic gene expression and promote intestinal tumor growth.
Genes Dev. 33:388–402. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wang Y, Shi L, Li J, Li L, Wang H and Yang
H: Long-term cadmium exposure promoted breast cancer cell migration
and invasion by up-regulating TGIF. Ecotoxicol Environ Saf.
175:110–117. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wotton D and Taniguchi K: Functions of
TGIF homeodomain proteins and their roles in normal brain
development and holoprosencephaly. Am J Med Genet C Semin Med
Genet. 178:128–139. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Nakashima H, Tsujimura K, Irie K, Ishizu
M, Pan M, Kameda T and Nakashima K: Canonical TGF-β signaling
negatively regulates neuronal morphogenesis through TGIF/Smad
complex-mediated CRMP2 suppression. J Neurosci. 38:4791–4810. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Sharma A, Sinha NR, Siddiqui S and Mohan
RR: Role of 5′TG3′-interacting factors (TGIFs) in Vorinostat (HDAC
inhibitor)-mediated Corneal Fibrosis Inhibition. Mol Vis.
21:974–984. 2015.PubMed/NCBI
|
|
31
|
Du R, Shen W, Liu Y, Gao W, Zhou W, Li J,
Zhao S, Chen C, Chen Y, Liu Y, et al: TGIF2 promotes the
progression of lung adenocarcinoma by bridging EGFR/RAS/ERK
signaling to cancer cell stemness. Signal Transduct Target Ther.
4:602019. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Liu ZM, Tseng HY, Tsai HW, Su FC and Huang
HS: Transforming growth factor β-interacting factor-induced
malignant progression of hepatocellular carcinoma cells depends on
superoxide production from Nox4. Free Radic Biol Med. 84:54–64.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhang MZ, Ferrigno O, Wang Z, Ohnishi M,
Prunier C, Levy L, Razzaque M, Horne WC, Romero D, Tzivion G, et
al: TGIF governs a feed-forward network that empowers Wnt signaling
to drive mammary tumorigenesis. Cancer Cell. 27:547–560. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Shi H, Shi J, Zhang Y, Guan C, Zhu J, Wang
F, Xu M, Ju Q, Fang S and Jiang M: Long non-coding RNA DANCR
promotes cell proliferation, migration, invasion and resistance to
apoptosis in esophageal cancer. J Thorac Dis. 10:2573–2582. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Mo Y, Wang Y, Zhang S, Xiong F, Yan Q,
Jiang X, Deng X, Wang Y, Fan C, Tang L, et al: Circular RNA
circRNF13 inhibits proliferation and metastasis of nasopharyngeal
carcinoma via SUMO2. Mol Cancer. 20:1122021. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Tang Z, Li C, Kang B, Gao G, Li C and
Zhang Z: GEPIA: A web server for cancer and normal gene expression
profiling and interactive analyses. Nucleic Acids Res. 45((W1)):
W98–W102. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Shetti D, Zhang B, Fan C, Mo C, Lee BH and
Wei K: Low dose of paclitaxel combined with XAV939 attenuates
metastasis, angiogenesis and growth in breast cancer by suppressing
Wnt signaling. Cells. 8:82019. View Article : Google Scholar
|
|
39
|
Huang X, Zhou X, Hu Q, Sun B, Deng M, Qi X
and Lü M: Advances in esophageal cancer: A new perspective on
pathogenesis associated with long non-coding RNAs. Cancer Lett.
413:94–101. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Szklarczyk D, Morris JH, Cook H, Kuhn M,
Wyder S, Simonovic M, Santos A, Doncheva NT, Roth A, Bork P, et al:
The STRING database in 2017: Quality-controlled protein-protein
association networks, made broadly accessible. Nucleic Acids Res.
45(D1): D362–D368. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Wang Y, Shi L, Li J, Wang H and Yang H:
The roles of TG-interacting factor in cadmium exposure-promoted
invasion and migration of lung cancer cells. Toxicol In Vitro.
61:1046302019. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Zhang LN, Zhao L, Yan XL and Huang YH:
Loss of G3BP1 suppresses proliferation, migration, and invasion of
esophageal cancer cells via Wnt/β-catenin and PI3K/AKT signaling
pathways. J Cell Physiol. 234:20469–20484. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Yu F, Yu C, Li F, Zuo Y, Wang Y, Yao L, Wu
C, Wang C and Ye L: Wnt/β-catenin signaling in cancers and targeted
therapies. Signal Transduct Target Ther. 6:3072021. View Article : Google Scholar : PubMed/NCBI
|