Introduction
Head and neck cancer represents the sixth most
common malignancy worldwide, with approximately 500,000 new cases
diagnosed each year; it remains one of the leading causes of
cancer-related deaths (1). Head
and neck squamous cell carcinoma (HNSCC) comprises the majority of
head and neck cancers and represents a heterogeneous group of
tumors that arise from the squamous epithelium of the oral cavity,
oropharynx, larynx, and hypopharynx (2). HNSCC metastasizes to lymph nodes,
resulting in poor prognosis (3).
However, few oncogenes have been identified in HNSCC that can be
exploited with targeted agents (4). Therefore, the identification of new
biomarkers and drug targets is necessary for effective treatment of
HNSCC. Many HNSCC patients present with locally advanced disease,
often with prominent involvement of lymph nodes (2). Lymph node metastasis is affected by
various factors in HNSCC. Recent studies have demonstrated that
tumor cells express functionally active chemokine receptors and
that expression of these receptors appears to regulate cellular
functions associated with metastasis (5–7). The
involvement of chemokines and their receptors in both normal and
abnormal physiological behaviors, such as inflammation, immunity,
chemotaxis, and metastasis of tumor cells, is under active
investigation (8–10). CXC chemokine receptor type 7
(CXCR7) is an important marker of lymph node metastasis in HNSCC
(11).
CXCR7 is a G-protein-coupled chemokine receptor; its
ligands are CXCL12 and CXCL11. CXCR7 is upregulated in cancer cells
and mediates a broad range of cellular activities, including
proliferation, survival, and metastasis, by binding CXCL12
(12–14). Elevated levels of CXCR7 were found
to be correlated with tumor aggressiveness and grade in various
types of cancers, including non-small cell lung cancer (15), bladder cancer (16), breast cancer (17), and oral squamous cell carcinoma
(18). In addition, CXCR7
expression is strongly associated with tumor size, lymph node
metastasis, disease recurrence, and poor disease-specific survival,
suggesting that CXCR7 expression results in more biologically
aggressive tumors (19).
Therefore, it will be important to study the role of CXCR7 in HNSCC
cells to reveal potential relevant therapeutic targets to inhibit
tumor progression.
Decursin, a pyranocoumarin compound isolated from
the dried root of the Korean medicinal herb Angelica gigas
Nakai, has been reported to exhibit antitumor activity in breast
and gastric cancer (20–22). Pharmacological effects of this
compound have also been reported, and include antibacterial and
antileukemic activities, as well as antiangiogenic effects
(23,24). Decursin was found to inhibit tumor
growth and to promote apoptosis in several cancer cell types, and
exerts anticancer effects by inducing cell cycle arrest,
particularly in prostate carcinomas (25). However, the relationship between
decursin and its antitumor activity mediated by chemokine receptor
signaling, especially CXCR7, is not fully understood in HNSCC. In
the present study, the role of CXCR7 was investigated in HNSCC
cells and whether decursin exerts antitumor activity by regulating
CXCR7 and its downstream factors was further evaluated.
Materials and methods
Preparation of decursin and
treatments
Decursin was synthesized using decursinol isolated
from the dried roots of Korean angelica (A. gigas).
Decusin was prepared by Professor Song's laboratory at the College
of Pharmacy, Chungnam National University, Deajeon, as previously
described (26). Briefly, in order
to synthesize decursin, decursinol was dissolved in dry methylene
chloride and cooled down in an ice bath. Dicyclohexylcarbodiimide,
3,3-dimethylacrylic acid, and 4-dimethylaminopyridine were then
added with stirring. The reaction mixture was stirred at room
temperature for 24 h and filtered. The filtrate was evaporated
under reduced pressure and purified by flash column chromatography
to obtain decursin as a white powder. The structure of decursin was
confirmed by comparing nuclear magnetic resonance and mass
spectrometry spectra. Decursin was dissolved in dimethyl sulfoxide
(DMSO); in all experiments the concentration of DMSO was limited to
0.1%. The dose of decursin 50 or 100 µM was chosen based on
previous studies (27,28).
Cell culture and stable cell line
establishment
The human HNSCC cell lines SNU1041 (KCBL No. 01041)
and SNU1076 (KCBL No. 01076) were purchased from the Korean Cell
Line Bank (Seoul, Korea). SNU1041 and SNU1076 cells were grown in
RPMI medium (Welgene) supplemented with 10% fetal bovine serum
(FBS) and 1X penicillin/streptomycin. Cells were cultured at 37°C
under 5% CO2 and 95% relative humidity. CXCR7
overexpression in HNSCC cell lines was conducted as previously
described (22). Overexpression of
CXCR7 in HNSCC cells was produced by lentivirus-mediated
transduction of full-length human CXCR7 sub-cloned into a
pLVX-EF1α-IRES-Puro lentiviral vector (Clontech Laboratories, Inc.)
and mock transfected as a negative control. To generate stable
transfectants, the acquired lentiviral vector was co-transfected
into 293T cells with virus packaging mix (Sigma Aldrich; Merck
KGaA) using Lipofectamine 3000 (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's instructions. The
virus was harvested from the supernatant and concentrated with
lenti-X-concentrator (Clontech Laboratories, Inc.), and then added
to SNU1041 and SNU1076 cells along with 5 µg/ml polybrene (Santa
Cruz Biotechnology, Inc.). Puromycin-resistant cells were selected
by culture for 2 weeks in the presence of puromycin. CXCR7
expression levels were analyzed by flow cytometry (Beckman
Coulter), reverse transcriptase PCR (RT-PCR), and western blot
analysis.
Reagents and antibodies
Antibodies against CXCR7 (dilution 1:1,000, cat. no.
ab38089; Abcam), GAPDH (dilution 1:2,000, cat. no. sc-25308; Santa
Cruz Biotechnology, Inc.), cyclin A (dilution 1:1,000, cat. no.
4565; Cell Signaling Technology, Inc.), cyclin E (dilution 1:1,000,
cat. no. 4129; Cell Signaling Technology, Inc.), CDK2 (dilution
1:1,000, cat. no. 2546; Cell Signaling Technology, Inc.),
phosphorylated-STAT3 (p-STAT3; dilution 1:1,000, cat. no. 9134;
Cell Signaling Technology, Inc.), STAT3 (dilution 1:1,000, cat. no.
9132; Cell Signaling Technology, Inc.), c-MYC (dilution 1:1,000,
cat. no. sc-40; Santa Cruz Biotechnology, Inc.) were used in the
western blot analysis and immunofluorescence. Interleukin 6 (IL-6;
cat. no. 200-06; Peprotech, Inc.) and stromal cell-derived factor
1α (SDF-1α or CXCL12; cat. no. 300-28A; Peprotech, Inc.) were
resuspended in medium and used to treat the cells.
Small interfering RNA (siRNA)
treatment and transfection
We obtained CXCR7 siRNA antisense
(5′-CGCUCUCCUUCAUUUACA-3′), sense (5′-UGUAAAUGAAGGAGAGCG-3′)
(Bioneer Corp.), and non-targeting control siRNA (cat. no.
sc-37007; Santa Cruz Biotechnology, Inc.). Cells were transfected
using Lipofectamine RNAi MAX (Invitrogen; Thermo Fisher Scientific,
Inc.).
Reverse transcriptase (RT)-PCR
analysis
Total RNA was isolated using TRIzol reagent (Thermo
Fisher Scientific, Inc.) according to the manufacturer's
instructions. First-strand cDNA was prepared from an RNA template
(2 µg) using the cDNA qPCR RT Master Mix (Toyobo Life
Science). RT-RCR was performed using the EmeraldAmp Master Mix
(Takara Bio). The PCR program included 40 cycles of 95°C for 15
sec, 60°C for 1 min, and 72°C for 1 min. The primers and
oligonucleotide sequences are described in Table SI. The gel was visualized and
analyzed under the GelDoc Xr system (Bio-Rad Laboratories) where
band sizing and molecular weight were determined by Image Lab
analysis software version 4.1 (Bio-Rad Laboratories) in relation to
100 bp DNA ladder (Bioneer).
Flow cytometry
Surface expression of CXCR7 was evaluated by flow
cytometry. Cells were harvested with phosphate-buffered saline
(PBS) containing 0.1% bovine serum albumin (BSA). After cells were
incubated with 10 µl of conjugated PE-CXCR7 (R&D Systems) or
mouse PE-IgG2A antibodies (negative control) by shaking for 1 h at
room temperature. Next, after washing three times with PBS
containing 0.1% BSA, expression levels were measured using flow
cytometry (Beckman Coulter) and the Kaluza analysis program
(version 1.2; Beckman Coulter).
Western blot analysis
Western blot analysis was performed as previously
described (11). Briefly, cells
were washed in PBS and lysed in ProEX™ CETi Lysis buffer (Translab)
containing a protease inhibitor cocktail. Cell lysates were
separated using sodium dodecyl sulfate polyacrylamide gel
electrophoresis (SDS-PAGE) and transferred to polyvinylidene
fluoride membranes (EMD Millipore). Membranes were blocked with the
ProNA™ General-block solution (TLP-115.1P; Translab) for 1 h and
incubated with the indicated primary antibodies at 4°C for
overnight. After membranes had been washed, they were incubated
with the corresponding horseradish peroxidase-conjugated
anti-rabbit IgG (dilution 1:5,000, cat. no. 7074; Cell Signaling
Technology, Inc.) and anti-mouse IgG (dilution 1:5,000, cat. no.
7076; Cell Signaling Technology, Inc.) antibodies diluted in
blocking solution. The immunoreactive polypeptides were detected
using an enhanced chemiluminescence substrate (Thermo Fisher
Scientific, Inc.).
In vitro cell proliferation assay
Medium containing 50 or 100 µM decursin was added to
cells in a 96-well plate. Cell proliferation was measured using the
Cell Counting Kit-8 (CCK-8; Dojindo Molecular Technologies). Cell
proliferation was determined every 24 h for 4 days and absorbance
was measured at 450 nm (Molecular Devices). For the
anchorage-dependent growth assay, cells were seeded in 6-well
plates. After cell attachment, 50 or 100 µM decursin was added to
the wells for 48 h. The cells were then cultured with fresh medium
for 2 weeks. The colonies were fixed in 10% formalin and stained
with 0.1% crystal violet (Sigma Aldrich; Merck KGaA) at room
temperature for 1 h. After stained cells were dissolved in 70%
ethanol, absorbance at 595 nm was measured using a
spectrophotometer.
Gap closure assay
A total of 5×104 cells were seeded on
each side of the chamber (Ibidi, cat. no. 81176; Gräfelfing) with
culture inserts for live-cell analysis. After growth for 24 h, the
chamber inserts were removed, and the two cell islands were washed
with PBS to remove debris and maintained in serum-free medium with
different concentrations of 50 or 100 µM decursin. Cells were
observed at regular intervals and imaged using phase contrast
microscopy.
Migration and invasion assays
The migration and invasion assay were performed
using 8-µm-pore transwell chambers (cat. no. 353097; Corning,
Inc.). The lower chamber was coated with 0.1% gelatin (Sigma
Aldrich; Merck KGaA) for migration, and the upper chamber was
coated with 25 µg/ml Matrigel for the invasion assay (BD
Biosciences). Serum-free medium containing 50 or 100 µM decursin
was added to the upper chamber and 1×105 cells were
seeded in each well. Culture medium containing 10% serum was added
to the lower chamber and the chamber was incubated at 37°C for 24 h
(migration) or 48 h (invasion). After the chambers were removed
from the upper surface of the transwell membrane with a cotton
swab, cells were fixed with 10% formalin and stained with 0.1%
crystal violet. The number of cells passing through the chamber was
counted under a microscope (Olympus IX71). Five randomly chosen
fields were counted for each assay.
Immunofluorescence staining
Cells were seeded in an 8-well chamber slide (Nunc™
Lab-Tek II™; Thermo Fisher Scientific, Inc.) at a concentration of
1×103 cells and treated with 100 µM decursin. After
incubation, the cells were fixed with 10% formalin for 20 min at
room temperature and then washed with PBS. Cell membranes were
permeabilized with 0.3% Triton X-100 (Sigma Aldrich; Merck KGaA)
for 20 min and maintained in blocking buffer (4% BSA in PBS) for 1
h at room temperature. After blocking, the 8-well chamber slide was
reacted with anti-CXCR7 (Abcam) antibody overnight at 4°C. The
slides were washed with PBS and incubated with Alexa 488-conjugated
secondary antibodies (cat. no. A11001; Invitrogen; Thermo Fisher
Scientific, Inc.) in PBS for 1 h at room temperature. For analysis
of the actin cytoskeleton, cells were stained with
rhodamine-conjugated phalloidin (Sigma Aldrich; Merck KGaA) diluted
in PBS for 40 min at room temperature followed by three washes in
PBS. Nuclei were counterstained with DAPI (Vector
Laboratories).
Cell cycle analysis
For cell cycle analysis through propidium iodide
(PI) staining, 1×105 cells were seeded in 6-well plate
and treated with 50 or 100 µM decursin or siRNA for 48 h.
Afterwards, the cells were fixed in 70% ice-cold ethanol overnight
at −20°C and incubated with FxCycle™ PI/RNase Staining Solution
(cat. no. F10797; Invitrogen; Thermo Fisher Scientific, Inc.) for
30 min at room temperature. Fluorescence was analyzed using flow
cytometry (Beckman Coulter) and the Kaluza analysis program
(version 1.2; Beckman Coulter).
GEPIA Database
The Gene Expression Profiling Interactive Analysis
(GEPIA) database (http://gepia.cancer-pku.cn/), a newly developed
web-based tool, provides key interactive and customizable functions
including tumor vs. normal differential expression analysis,
profiling plotting in accordance with cancer types or different
pathological stages, correlation analysis, patient survival
analysis, similar gene detection, and dimensionality reduction
analysis based on the Cancer Genome Atlas (TCGA) and the
genotype-tissue expression data (29). P-values are represented by
asterisks in the GEPIA results.
Statistical analysis
Each experimental condition was performed in
triplicate to evaluate the mean ± standard deviation (SD). All
statistical analysis of the data was performed using Prism
(GraphPad Software, v 5.01; GraphPad Software, Inc.). To determine
the difference between two groups, the unpaired two-tailed
Student's t-test was applied. One-way ANOVA followed by Dunnett's
test was applied to compare multiple data. In all cases, the
minimum statistical significance was set at P<0.05.
Results
CXCR7 promotes proliferation,
migration, and invasion of HNSCC cells
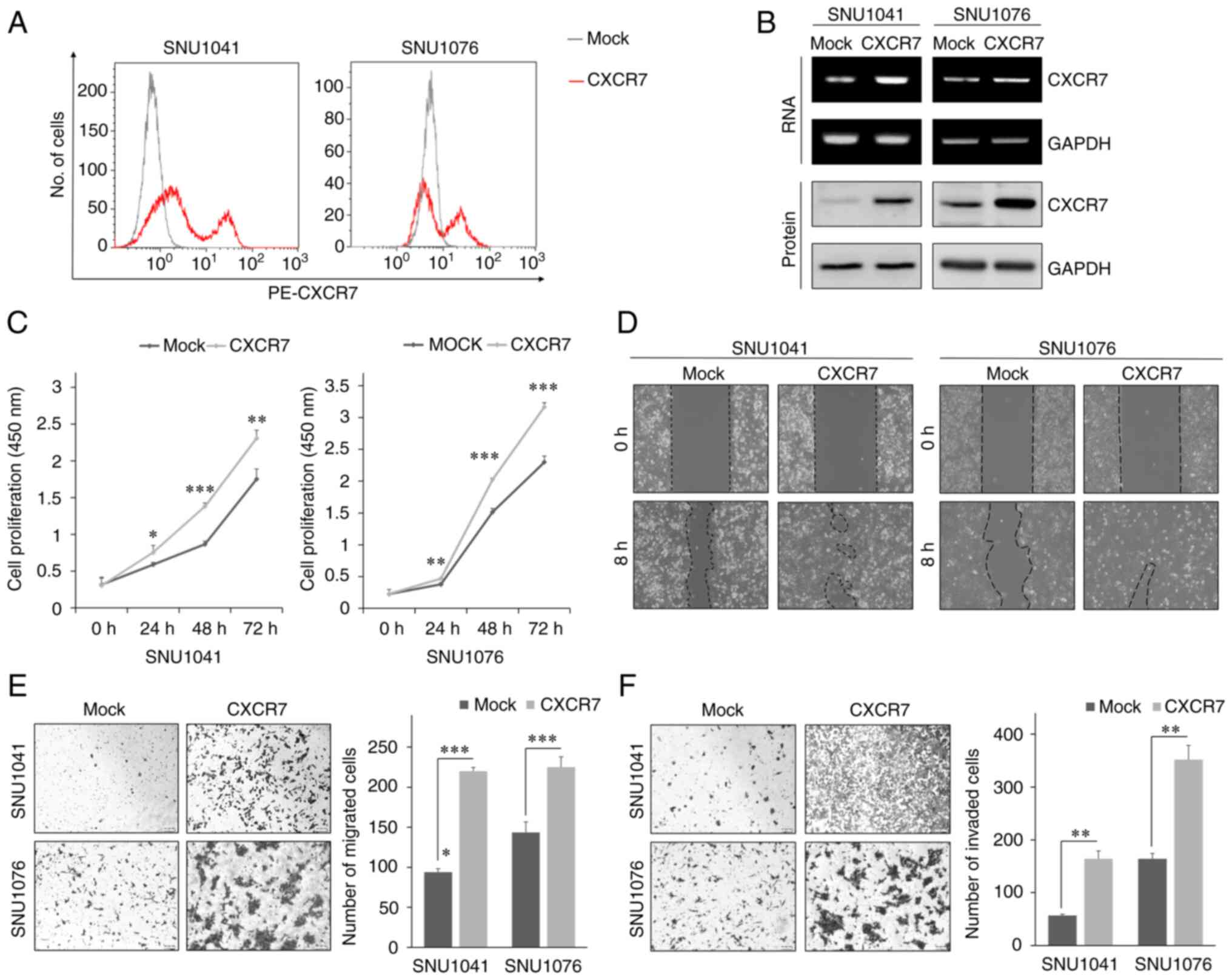
To investigate how CXCR7 regulates tumor progression
in HNSCC, we established stable cell lines overexpressing CXCR7
(SNU1041-CXCR7 and SNU1076-CXCR7) using lentivirus-mediated
transduction of an expression vector for full-length human CXCR7.
Overexpression of CXCR7 was confirmed in the HNSCC SNU1041 and
SNU1076 cell lines using flow cytometry, RT-PCR, western blot
analysis, and immunofluorescence (Fig.
1A and B and S1A).
Overexpression of CXCR7 significantly increased the proliferation
of the HNSCC cell lines (Fig. 1C).
CXCR7 overexpression also significantly increased cell motility in
a gap closure assay. CXCR7-overexpressing cells nearly closed the
wide gaps when compared to the mock cells (Fig. 1D). In addition,
CXCR7-overexpressing cells showed significantly increased migration
and invasion compared with the mock cells (Fig. 1E and F). Furthermore, SDF-1α
treatment increased the cell migration and invasion of the
CXCR7-overexpressed HNSCC cells (Fig.
S1B). These findings indicate that CXCR7 promotes
proliferation, migration, and invasion of HNSCC cells.
Decursin reduces expression of CXCR7
and inhibits proliferation of HNSCC cells
To confirm the effect of decursin on CXCR7, changes
in CXCR7 expression in decursin-treated HNSCC cells were
investigated. Flow cytometry, western blot analysis, and
immunofluorescence confirmed that decursin treatment decreased
expression of CXCR7 in a dose-dependent manner (Fig. 2A-C). Next, decursin treatment
inhibited proliferation of CXCR7-overexpressing cells in a
dose-dependent manner (Fig. 2D).
Decursin-mediated inhibition of proliferation was dose dependent in
bright field imaging (Fig. S2).
In addition, decursin suppressed cell growth in a dose-dependent
manner, as determined by the number of colonies in an
anchorage-dependent growth assay (Fig.
2E). These results suggest that decursin reduces cell
proliferation via downregulation of CXCR7.
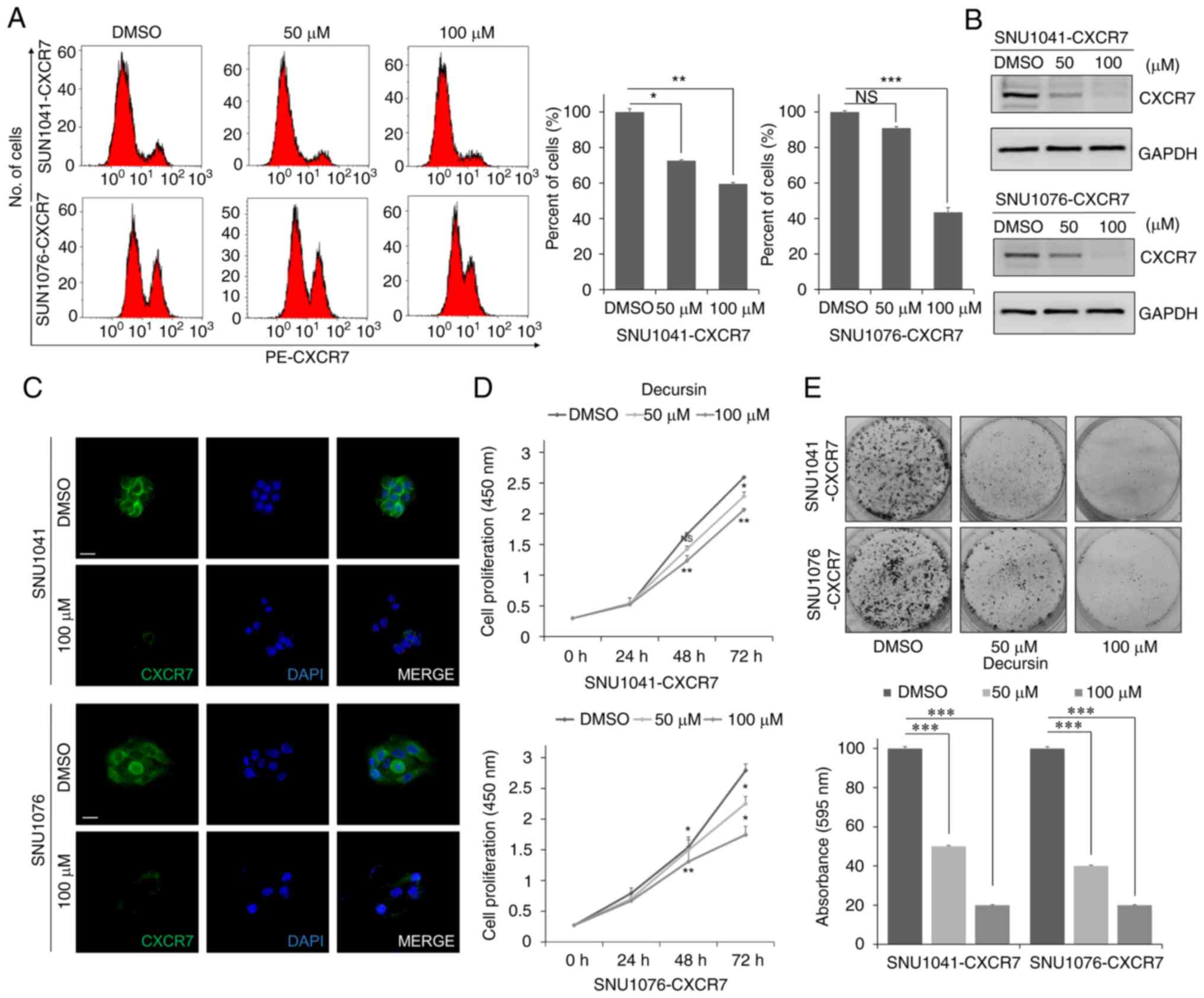 | Figure 2.Decursin reduces CXCR7 expression and
cell proliferation. (A and B) Expression of CXCR7 in SNU1041-CXCR7
and SNU1076-CXCR7 HNSCC cells treated with 50 or 100 µM decursin
for 48 h by flow cytometry and western blot analysis. Three
independent experiments were carried out in triplicate. Blots are
representative of 3 independent experiments. Histograms of the
protein expression levels are presented in Fig. S5B. (C) Representative
immunofluorescence images of CXCR7 in cells treated with 100 µM
decursin for 48 h. Scale bar, 100 µm. (D) Cells were seeded in
96-well plates and treated with 50 or 100 µM decursin for 24, 48,
and 72 h. Following treatment with decursin, cell viability was
measured with the CCK-8 assay. Three independent experiments were
carried out in triplicate. (E) Decursin suppressed cell growth, as
determined by an anchorage-dependent growth assay. SNU1041-CXCR7
and SNU1076-CXCR7 cells were treated with 50 or 100 µM decursin for
48 h. Three independent experiments were carried out in triplicate.
Magnification, ×200. NS, not significant. *P<0.05, **P<0.01,
and ***P<0.001. HNSCC, head and neck squamous cell carcinoma;
CXCR7, CXC chemokine receptor type 7. |
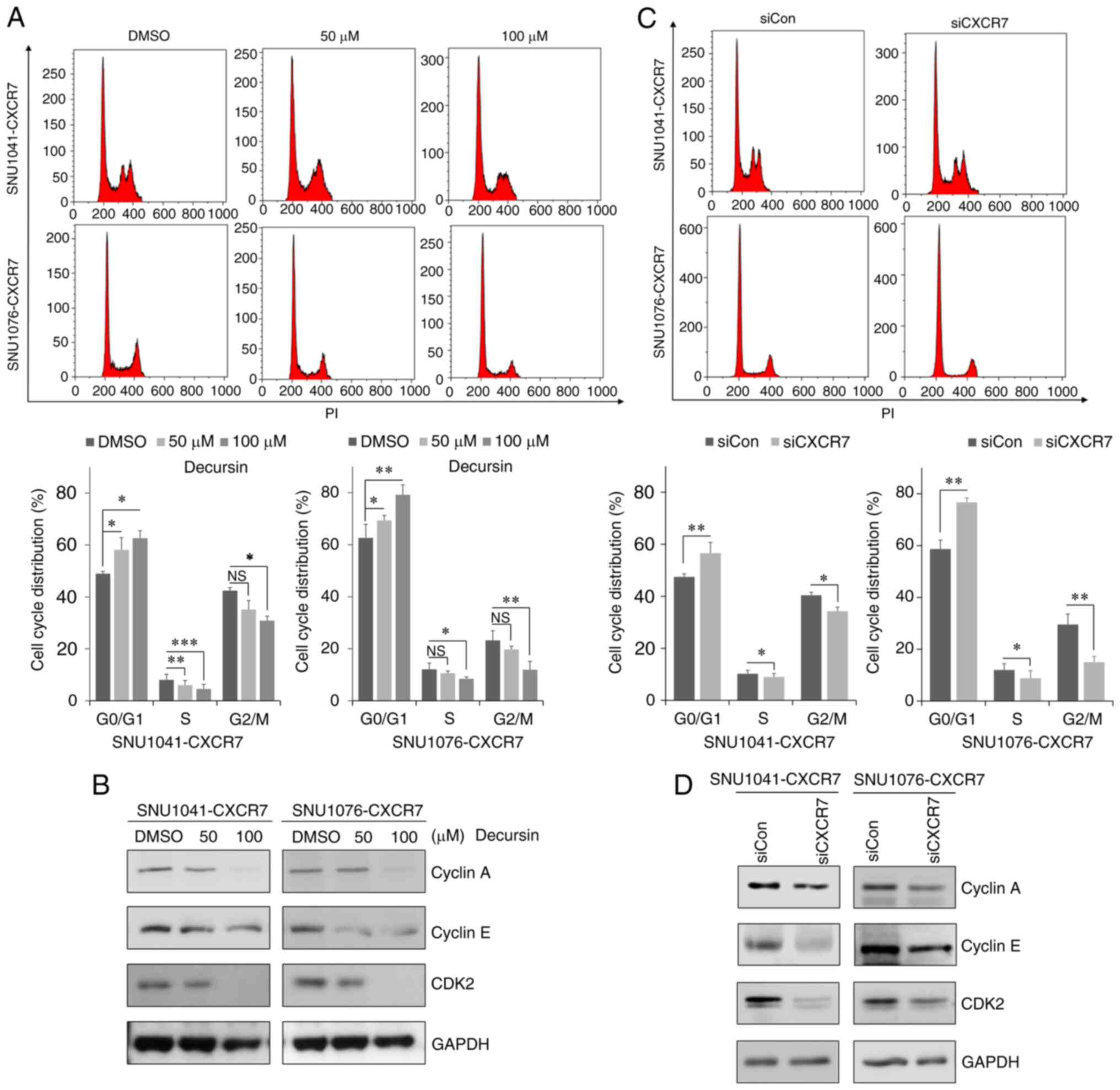
Decursin induces G0/G1 cell cycle
arrest by downregulating CXCR7 expression
To determine whether decursin-mediated inhibition of
proliferation was associated with the cell cycle, the effects of
decursin on the cell cycle distribution of CXCR7-overexpressing
cells were confirmed. Treatment with decursin significantly
increased the percentage of cells in the G0/G1 phase in a
dose-dependent manner compared with the control, whereas the
percentage of cells in the S phase and G2/M phase was decreased in
both CXCR7-overexpressing cell lines (Fig. 3A). In addition, cell cycle
regulatory proteins cyclin A, cyclin E, and CDK2 were downregulated
in a dose-dependent manner following treatment with decursin in the
CXCR7-overexpressing cells (Fig.
3B and S3A). To confirm
whether decursin-induced cell cycle arrest was due to a decrease in
CXCR7, we knocked down CXCR7 expression using siRNA in
CXCR7-overexpressing cells (Fig.
S3B). Knockdown of CXCR7 significantly increased the proportion
of cells in the G0/G1 phase compared with the control, whereas it
decreased the proportion of cells in the S phase and G2/M phase
(Fig. 3C). Knockdown of CXCR7
decreased cyclin A, cyclin E, and CDK2 expression (Fig. 3D). In addition, knockdown of CXCR7
also suppressed cell migration and invasion of CXCR7-overexpressing
cells (Fig. S3C). These results
demonstrated that decursin induced cell cycle arrest in G0/G1 phase
and inhibited cell motility and invasiveness by downregulating
CXCR7 expression.
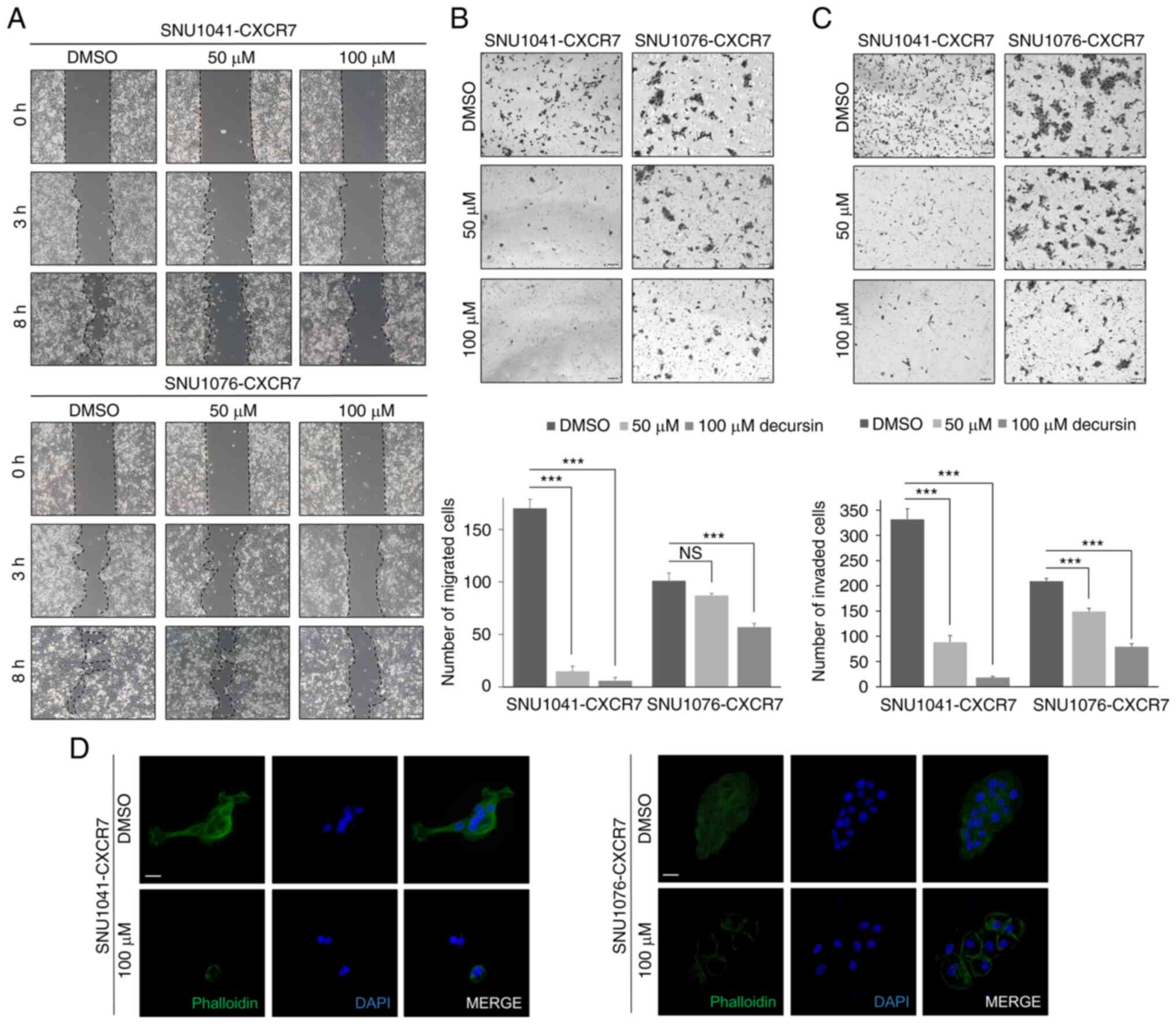
Decursin suppresses cell motility,
migration, and invasion in HNSCC cells
To determine the effect of decursin on motility and
invasiveness, cell motility was assessed using the gap closure and
transwell assays. Decursin significantly impaired the motility of
CXCR7-overexpressing cells (Fig.
4A). In addition, cell migration and invasion were
significantly reduced in CXCR7-overexpressing cells treated with
decursin (Fig. 4B and C). Using
phalloidin staining, it was observed that stress fiber formation
was suppressed in decursin-treated cells (Fig. 4D). Taken together, these findings
indicate that decursin inhibits the motility, migration, and
invasion of CXCR7-overexpressing cells.
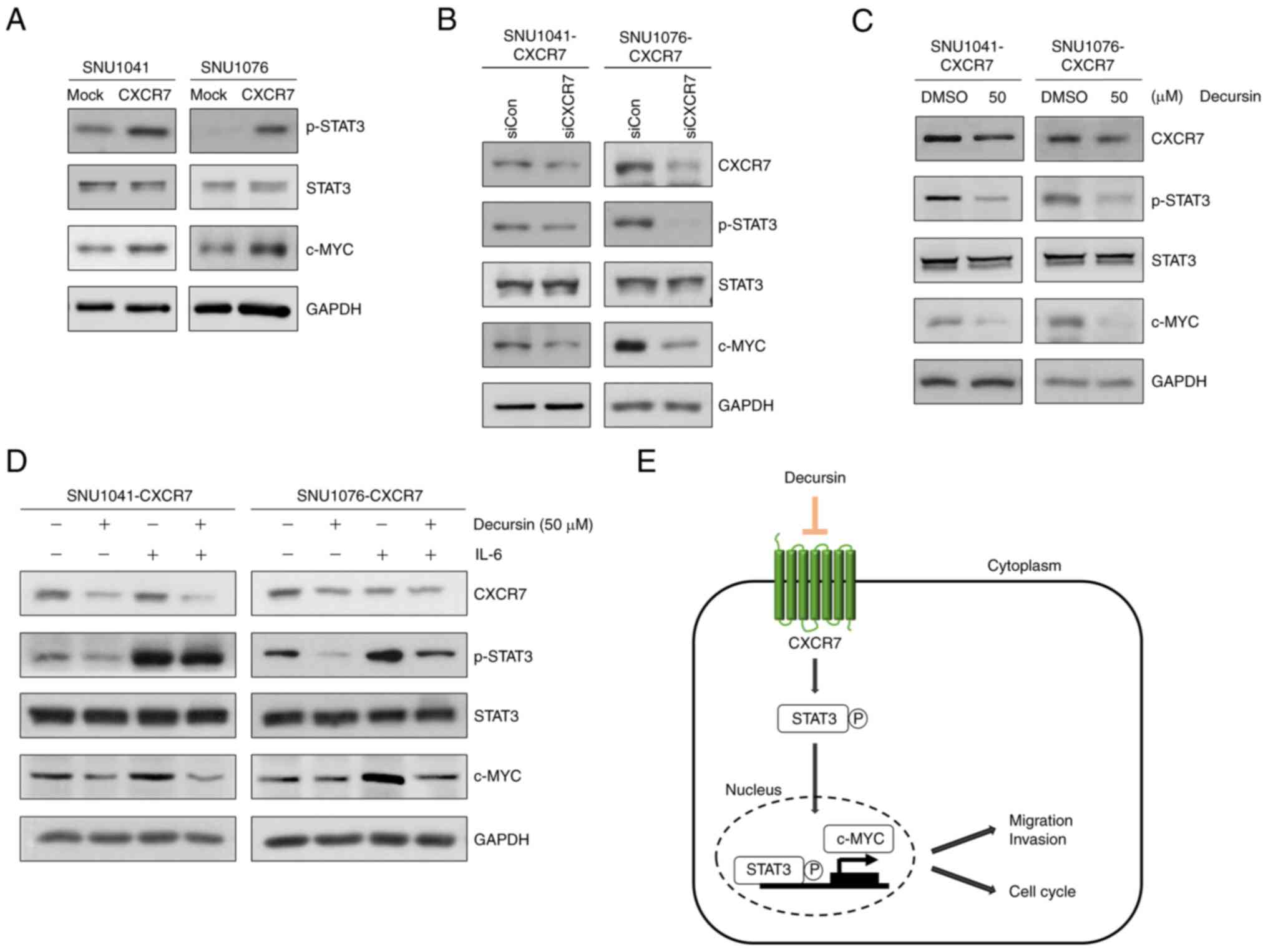
Decursin downregulates c-MYC
expression by inhibiting CXCR7-mediated STAT3 signaling
Next, the mechanism by which decursin inhibits the
proliferation, migration, and invasion of HNSCC cells was
investigated. We screened several signaling pathways and found that
overexpression of CXCR7 increased STAT3 phosphorylation (p-STAT3)
in both CXCR7-overexpressing cell lines (Fig. S4A and Table SII). Furthermore, the GEPIA
dataset showed increased expression of CXCR7 and STAT3 in HNSCC
tissues (Fig. S4B). There was a
correlation between CXCR7 and STAT3 expression (Fig. S4C). Overexpression of CXCR7 also
increased c-MYC and p-STAT3 (Fig.
5A). In addition, STAT3 phosphorylation and c-MYC expression
were decreased following knockdown of CXCR7 in the
CXCR7-overexpressing cells (Fig.
5B); STAT3 phosphorylation and c-MYC expression were also
reduced in the decursin-treated CXCR7-overexpressing cells
(Fig. 5C). Following treatment of
CXCR7-overexpressing cells with IL-6, a growth factor and inducer
of STAT3 phosphorylation, STAT3 phosphorylation and c-MYC
expression were increased, whereas decursin markedly inhibited
p-STAT3, c-MYC, and CXCR7 expression. Decursin in combination with
IL-6 reduced p-STAT3 and c-MYC to a lesser degree than treatment
with decursin alone (Fig. 5D).
These results demonstrate that decursin suppresses the
CXCR7/STAT3/c-MYC signaling axis and decursin exhibits anticancer
activity in HNSCC (Fig. 5E).
Discussion
The development of effective target therapies for
head and neck squamous cell carcinoma (HNSCC) requires in-depth
knowledge of the underlying mechanisms of the complex signaling
networks related to cancer development and progression. In the
present study, it was demonstrated that decursin, a pyranocoumarin
compound isolated from Angelica gigas Nakai, exhibited
anticancer activity in HNSCC by downregulating the
CXCR7/STAT3/c-MYC signaling pathway and inducing G0/G1 cell cycle
arrest. Moreover, it was shown that decursin inhibited cell
motility, migration, and invasion in vitro. Our findings
showed that CXCR7 promotes cancer progression in HNSCC through the
STAT3/c-MYC pathway and that decursin may be a useful treatment by
which to target CXCR7 in HNSCC.
Chemotherapy, in which cytostatic and cytotoxic
agents target the cellular mechanisms involved in the control of
cell growth and division, is frequently used in cancer treatment
(30). However, chemotherapy alone
does not achieve satisfactory therapeutic results, and resistance
to cytotoxic chemotherapy is a major cause of mortality in
patients. Therefore, the use of natural products is a recent and
innovative strategy aimed at increasing the cytotoxic efficiency of
anticancer drugs, limiting their toxic side effects and delaying
the appearance of acquired chemoresistance (31). Decursin is derived from a natural
product and has shown antitumor effects in several types of
cancers, including gastric (22)
and cervical cancer (32).
However, the effect of this compound on HNSCC, characterized by
high migration and invasion with poor clinical outcome, remains
unclear. In the present study, decursin reduced cell growth,
migration, and invasion in HNSCC by decreasing the expression of
CXCR7. These results suggest that decursin could be an anticancer
agent that exhibits antitumor activity by targeting CXCR7 in
HNSCC.
Chemokine receptor 7 (CXCR7) promotes migration and
invasion in HNSCC and is highly expressed in tumor cells in HNSCC
patients (11). Moreover, several
studies have indicated that CXCR7 regulates the PI3K/Akt, ERK, and
MAPK signaling pathways in several cancer cell types (33,34).
CXCR7 plays an important role in cell division, induces
differentiation-related cell cycle, and promotes proliferation
(35). Consistent with these
roles, decreased expression of CXCR7 has been correlated with the
inhibition of cell cycle progression and marked changes in key
proteins involved in cell cycle regulation (36). A recent study showed that chemokine
pathways have become an important area of investigation for cancer
therapy (37). An emerging
chemokine target for cancer therapy is CXCL12, also known as the
stromal cell-derived factor 1α (SDF-1α), which binds its cognate
receptor CXCR4 and initiates downstream signaling (38,39).
SDF-1α also binds to CXCR7 and has been involved in tumor
development and progression (40).
In our study, CXCR7 overexpression alone increased cell growth via
cell cycle progression and promoted cell motility, migration, and
invasion, independently of SDF-1α, as previously reported by Kim
et al (11). SDF-1α
treatment also increased cell motility and growth.
Signal transducer and activator of transcription 3
(STAT3) is constitutively activated in many cancers and plays a
pivotal role in tumor growth and metastasis (41), including colon cancer (42), gastric cancer (43), cervical cancer (44), and hepatocellular carcinoma
(45). STAT3 is an essential
transcription factor that activates a cascade of survival and
proliferation signaling programs in cells. Expression of
phosphorylated (p)-STAT3 is an important factor related to tumor
invasion and poor prognosis (46,47).
Furthermore, CXCR7-induced STAT3-mediated pathways may enhance
tumor growth by regulating cell proliferative pathways; STAT3
signaling is associated with the upregulation of cyclin and c-MYC
expression, contributing to accelerated cell cycle progression
(17,48). STAT3 is frequently overexpressed in
tumor cells and the importance of STAT3 as a potential target for
cancer therapy has been highlighted (49). STAT3 activity is regulated by
posttranslational modifications, such as phosphorylation,
acetylation, oxidation, and ubiquitination (50). Our results indicate that CXCR7
activates the STAT3/c-MYC signaling axis in HNSCC. Decursin
treatment decreased the expression of CXCR7 and STAT3/c-MYC,
suggesting that decursin may be a useful anticancer drug as it
suppresses CXCR7/STAT3 signaling in HNSCC cells.
c-MYC is an important downstream effector of STAT3
signaling involved in cell growth and transformation (51). MYC is a transcription factor that
responds to and integrates signals into broad changes in gene
expression, supporting cell growth and proliferation (52). STAT3 and c-MYC are activated by
interleukin (IL)-6, which regulates cell growth, differentiation,
and cell survival (53,54). The IL-6/STAT3 pathway is an
attractive pharmacological target in oncology, and different
approaches to target this pathway, such as direct targeting of
STAT3 phosphorylation and activation or downregulation of STAT3
expression, are being pursued pre-clinically and clinically
(55,56). In the present study, we showed that
STAT3 phosphorylation and c-MYC expression were increased by CXCR7,
while decursin treatment decreased STAT3 phosphorylation and
expression of c-MYC and CXCR7. Treatment with IL-6 increased STAT3
phosphorylation and c-MYC expression with no change in CXCR7.
Co-treatment of decursin and IL-6 suppressed STAT3 phosphorylation
and c-MYC expression less than treatment with decursin alone.
Moreover, decursin inhibited cell growth, motility, and
invasiveness. Collectively, these results indicate that decursin
inhibits tumor progression via CXCR7/STAT3/c-MYC signaling in
HNSCC.
To date, there is no established ideal experimental
model for cancers including HNSCC because both in vitro and
in vivo tumor models have advantages and disadvantages
(57). In preclinical stage, most
of cancer research is first investigated in in vitro models
and later in in vivo systems (57). However, this study was performed
using an in vitro model and thus have some limitations which
should be considered and need to be overcome in future research.
For example, cells growing in vitro do not exactly replicate
their in vivo counterparts. Malignant tumors consist of both
cancer cells and tumor microenvironment which makes the tumors a
three-dimensional status with dynamic interaction between them
(57,58). Furthermore, the precise mechanism
by which decursin downregulates expression of CXCR7 is not yet
clear. Thus, we are undergoing further study to elucidate the
molecular mechanism.
In summary, the present study showed the
pro-tumorigenic effect of CXCR7 in HNSCC and revealed that CXCR7
induces p-STAT3 and downstream factors. Decursin suppressed tumor
progression through the inhibition of p-STAT3 and c-MYC by
decreasing CXCR7 expression. Thus, decursin may be a natural
therapeutic drug useful in the treatment of HNSCC.
Supplementary Material
Supporting Data
Acknowledgements
The abstract was presented at the 94th Annual
Meeting of the American Association for Cancer Research Apr 10-May
21, 2021 in Philadelphia, IL and published as abstract no. 2412 in
Cancer Res 81(13_Suppl), 2021: Abstract no. 2412.
Funding
This study was supported by grants from the National Research
Foundation of Korea (NRF-2017R1D1A1B04034638, NRF-2017R1A5A2015385,
2017R1D1A1B04034379) and the Korea Health Technology R&D
Project through the Korea Health Industry Development Institute
(KHIDI), funded by the Ministry of Health & Welfare, Republic
of Korea (HR20C0025).
Availability of data and materials
The data presented in this study are contained
within the article and the Supplementary Materials.
Authors' contributions
Conceptualization of the study design was carried
out by MJ, JMK, and HJL. Data curation was performed by MJ, YA, SK,
NK and HJJ. Investigation and methodology were the responsibility
of MJ, JBH, and GYS. Formal analysis of the results was carried out
by MJ, JBH, HJJ, and JMK. Funding acquisition was obtained by JMK
and HJL. Writing, review, and/or revision of the manuscript was
carried out by MJ, YA, NK, GYS, JMK, and HJL. Project
administration, resources and software were the responsibility of
HJJ and JMK. Study supervision and data validation were the
responsibility of JMK and HJL. All authors have read and agreed to
the manuscript for publication.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2018. CA Cancer J Clin. 68:7–30. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Solomon B, Young RJ and Rischin D: Head
and neck squamous cell carcinoma: Genomics and emerging biomarkers
for immunomodulatory cancer treatments. Semin Cancer Biol.
52:228–240. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Goesswein D, Habtemichael N, Gerhold-Ay A,
Mazur J, Wünsch D, Knauer SK, Künzel J, Matthias C, Strieth S and
Stauber RH: Expressional analysis of disease-relevant
signalling-pathways in primary tumours and metastasis of head and
neck cancers. Sci Rep. 8:73262018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Isaacsson Velho PH, Castro G Jr and Chung
CH: Novel targeted agents in head and neck squamous cell carcinoma.
Hematol Oncol Clin North Am. 29:993–1009. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Müller A, Homey B, Soto H, Ge N, Catron D,
Buchanan ME, McClanahan T, Murphy E, Yuan W, Wagner SN, et al:
Involvement of chemokine receptors in breast cancer metastasis.
Nature. 410:50–56. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Liotta LA: An attractive force in
metastasis. Nature. 410:24–25. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zlotnik A: Chemokines in neoplastic
progression. Semin Cancer Biol. 14:181–185. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zlotnik A, Morales J and Hedrick JA:
Recent advances in chemokines and chemokine receptors. Crit Rev
Immunol. 19:1–47. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Salazar N, Castellan M, Shirodkar SS and
Lokeshwar BL: Chemokines and chemokine receptors as promoters of
prostate cancer growth and progression. Crit Rev Eukaryot Gene
Expr. 23:77–91. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Pierce KL, Premont RT and Lefkowitz RJ:
Seven-transmembrane receptors. Nat Rev Mol Cell Biol. 3:639–650.
2002. View
Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kim N, Ryu H, Kim S, Joo M, Jeon HJ, Lee
MW, Song IC, Kim MN, Kim JM and Lee HJ: CXCR7 promotes migration
and invasion in head and neck squamous cell carcinoma by
upregulating TGF-β1/Smad2/3 signaling. Sci Rep. 9:181002019.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Liberman J, Sartelet H, Flahaut M,
Mühlethaler-Mottet A, Coulon A, Nyalendo C, Vassal G, Joseph JM and
Gross N: Involvement of the CXCR7/CXCR4/CXCL12 axis in the
malignant progression of human neuroblastoma. PLoS One.
7:e436652012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wang J, Shiozawa Y, Wang J, Wang Y, Jung
Y, Pienta KJ, Mehra R, Loberg R and Taichman RS: The role of
CXCR7/RDC1 as a chemokine receptor for CXCL12/SDF-1 in prostate
cancer. J Biol Chem. 283:4283–4294. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Liu TJ, Guo JL and Xu X: CXC chemokine-7
inhibits growth and migration of oral tongue squamous cell
carcinoma cells, mediated by the epithelial-mesenchymal transition
signaling pathway. Mol Med Rep. 16:6896–6903. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Iwakiri S, Mino N, Takahashi T, Sonobe M,
Nagai S, Okubo K, Wada H, Date H and Miyahara R: Higher expression
of chemokine receptor CXCR7 is linked to early and metastatic
recurrence in pathological stage I nonsmall cell lung cancer.
Cancer. 115:2580–2593. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yates TJ, Knapp J, Gosalbez M, Lokeshwar
SD, Gomez CS, Benitez A, Ekwenna OO, Young EE, Manoharan M and
Lokeshwar VB: C-X-C chemokine receptor 7: A functionally associated
molecular marker for bladder cancer. Cancer. 119:61–71. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wani N, Nasser MW, Ahirwar DK, Zhao H,
Miao Z, Shilo K and Ganju RK: C-X-C motif chemokine 12/C-X-C
chemokine receptor type 7 signaling regulates breast cancer growth
and metastasis by modulating the tumor microenvironment. Breast
Cancer Res. 16:R542014. View
Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yanagiya M, Dawood RI, Maishi N, Hida Y,
Torii C, Annan DA, Kikuchi H, Yanagawa Matsuda A, Kitamura T, Ohiro
Y, et al: Correlation between endothelial CXCR7 expression and
clinicopathological factors in oral squamous cell carcinoma. Pathol
Int. 71:383–391. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Schrevel M, Karim R, ter Haar NT, van der
Burg SH, Trimbos JB, Fleuren GJ, Gorter A and Jordanova ES: CXCR7
expression is associated with disease-free and disease-specific
survival in cervical cancer patients. Br J Cancer. 106:1520–1525.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Jiang C, Guo J, Wang Z, Xiao B, Lee HJ,
Lee EO, Kim SH and Lu J: Decursin and decursinol angelate inhibit
estrogen-stimulated and estrogen-independent growth and survival of
breast cancer cells. Breast Cancer Res. 9:R772007. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kim SH, Lee SW, Park HJ, Lee SH, Im WK,
Kim YD, Kim KH, Park SJ, Hong S and Jeon SH: Anti-cancer activity
of Angelica gigas by increasing immune response and
stimulating natural killer and natural killer T cells. BMC
Complement Altern Med. 18:2182018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kim S, Kim JE, Kim N, Joo M, Lee MW, Jeon
HJ, Ryu H, Song IC, Song GY and Lee HJ: Decursin inhibits tumor
growth, migration, and invasion in gastric cancer by
down-regulating CXCR7 expression. Am J Cancer Res. 9:2007–2018.
2019.PubMed/NCBI
|
|
23
|
Lee S, Shin DS, Kim JS, Oh KB and Kang SS:
Antibacterial coumarins from Angelica gigas roots. Arch
Pharm Res. 26:449–452. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Son SH, Kim MJ, Chung WY, Son JA, Kim YS,
Kim YC, Kang SS, Lee SK and Park KK: Decursin and decursinol
inhibit VEGF-induced angiogenesis by blocking the activation of
extracellular signal-regulated kinase and c-Jun N-terminal kinase.
Cancer Lett. 280:86–92. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yim D, Singh RP, Agarwal C, Lee S, Chi H
and Agarwal R: A novel anticancer agent, decursin, induces G1
arrest and apoptosis in human prostate carcinoma cells. Cancer Res.
65:1035–1044. 2005.PubMed/NCBI
|
|
26
|
Lee JH, Kim MA, Park S, Cho SH, Yun E, O
YS, Kim J, Goo JI, Yun MY, Choi Y, et al: Synthesis and evaluation
of (+)-decursin derivatives as inhibitors of the Wnt/β-catenin
pathway. Bioorg Med Chem Lett. 26:3529–3532. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kim WJ, Lee SJ, Choi YD and Moon SK:
Decursin inhibits growth of human bladder and colon cancer cells
via apoptosis, G1-phase cell cycle arrest and extracellular
signal-regulated kinase activation. Int J Mol Med. 25:635–641.
2010.PubMed/NCBI
|
|
28
|
Oh ST, Lee S, Hua C, Koo BS, Pak SC, Kim
DI, Jeon S and Shin BA: Decursin induces apoptosis in glioblastoma
cells, but not in glial cells via a mitochondria-related caspase
pathway. Korean J Physiol Pharmacol. 23:29–35. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Tang Z, Li C, Kang B, Gao G, Li C and
Zhang Z: GEPIA: A web server for cancer and normal gene expression
profiling and interactive analyses. Nucleic Acids Res. 45:W98–W102.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Longley DB, Harkin DP and Johnston PG:
5-fluorouracil: Mechanisms of action and clinical strategies. Nat
Rev Cancer. 3:330–338. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
de Oliveira Júnior RG, Christiane Adrielly
AF, da Silva Almeida JR, Grougnet R, Thiéry V and Picot L:
Sensitization of tumor cells to chemotherapy by natural products: A
systematic review of preclinical data and molecular mechanisms.
Fitoterapia. 129:383–400. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Yim NH, Lee JH, Cho WK, Yang MC, Kwak DH
and Ma JY: Decursin and decursinol angelate from Angelica
gigas Nakai induce apoptosis via induction of TRAIL expression
on cervical cancer cells. Eur J Integr Med. 3:e299–e307. 2011.
View Article : Google Scholar
|
|
33
|
Liao YX, Zhou CH, Zeng H, Zuo DQ, Wang ZY,
Yin F, Hua YQ and Cai ZD: The role of the CXCL12-CXCR4/CXCR7 axis
in the progression and metastasis of bone sarcomas (Review). Int J
Mol Med. 32:1239–1246. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Lin L, Han MM, Wang F, Xu LL, Yu HX and
Yang PY: CXCR7 stimulates MAPK signaling to regulate hepatocellular
carcinoma progression. Cell Death Dis. 5:e14882014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wang Y, Xu P, Qiu L, Zhang M, Huang Y and
Zheng JC: CXCR7 participates in CXCL12-mediated cell cycle and
proliferation regulation in mouse neural progenitor cells. Curr Mol
Med. 16:738–746. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Salazar N, Muñoz D, Kallifatidis G, Singh
RK, Jordà M and Lokeshwar BL: The chemokine receptor CXCR7
interacts with EGFR to promote breast cancer cell proliferation.
Mol Cancer. 13:1982014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Duda DG, Kozin SV, Kirkpatrick ND, Xu L,
Fukumura D and Jain RK: CXCL12 (SDF1α)-CXCR4/CXCR7 pathway
inhibition: An emerging sensitizer for anticancer therapies? Clin
Cancer Res. 17:2074–2080. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Sun X, Cheng G, Hao M, Zheng J, Zhou X,
Zhang J, Taichman RS, Pienta KJ and Wang J: CXCL12/CXCR4/CXCR7
chemokine axis and cancer progression. Cancer Metastasis Rev.
29:709–722. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Kojima Y, Acar A, Eaton EN, Mellody KT,
Scheel C, Ben-Porath I, Onder TT, Wang ZC and Richardson AL:
Autocrine TGF-beta and stromal cell-derived factor-1 (SDF-1)
signaling drives the evolution of tumor-promoting mammary stromal
myofibroblasts. Proc Natl Acad Sci USA. 107:20009–20014. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Barbieri F, Bajetto A, Pattarozzi A, Gatti
M, Würth R, Porcile C, Thellung S, Corsaro A, Villa V, Nizzari M
and Florio T: The Chemokine SDF1/CXCL12: A novel
autocrine/paracrine factor involved in pituitary adenoma
development. The Open Neuroendocrinol J. 4:64–76. 2011. View Article : Google Scholar
|
|
41
|
Kamran MZ, Patil P and Gude RP: Role of
STAT3 in cancer metastasis and translational advances. Biomed Res
Int. 2013:4218212013. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Corvinus FM, Orth C, Moriggl R, Tsareva
SA, Wagner S, Pfitzner EB, Baus D, Kaufmann R, Huber LA, Zatloukal
K, et al: Persistent STAT3 activation in colon cancer is associated
with enhanced cell proliferation and tumor growth. Neoplasia.
7:545–555. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Kim DY, Cha ST, Ahn DH, Kang HY, Kwon CI,
Ko KH, Hwang SG, Park PW, Rim KS and Hong SP: STAT3 expression in
gastric cancer indicates a poor prognosis. J Gastroenterol Hepatol.
24:646–651. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Morgan EL and Macdonald A: Autocrine STAT3
activation in HPV positive cervical cancer through a virus-driven
Rac1-NFκB-IL-6 signalling axis. PLoS Pathog. 15:e10078352019.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Lee C and Cheung ST: STAT3: An emerging
therapeutic target for hepatocellular carcinoma. Cancers (Basel).
11:2019. View Article : Google Scholar
|
|
46
|
Sukanya Dimri S and De A: Approaching
non-canonical STAT3 signaling to redefine cancer therapeutic
strategy. Integr Mol Med. 4:2017.
|
|
47
|
Kusaba T, Nakayama T, Yamazumi K, Yakata
Y, Yoshizaki A, Inoue K, Nagayasu T and Sekine I: Activation of
STAT3 is a marker of poor prognosis in human colorectal cancer.
Oncol Rep. 15:1445–1451. 2006.PubMed/NCBI
|
|
48
|
Bartek J and Lukas J: Mammalian G1- and
S-phase checkpoints in response to DNA damage. Curr Opin Cell Biol.
13:738–747. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Chai EZ, Shanmugam MK, Arfuso F,
Dharmarajan A, Wang C, Kumar AP, Samy RP, Lim LH, Wang L, Goh BC,
et al: Targeting transcription factor STAT3 for cancer prevention
and therapy. Pharmacol Ther. 162:86–97. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Im JY, Kim BK, Lee KW, Chun SY, Kang MJ
and Won M: DDIAS promotes STAT3 activation by preventing STAT3
recruitment to PTPRM in lung cancer cells. Oncogenesis. 9:12020.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Tolomeo M and Cascio A: The multifaced
role of STAT3 in cancer and its implication for anticancer therapy.
Int J Mol Sci. 22:6032021. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Schaub FX, Dhankani V, Berger AC, Trivedi
M, Richardson AB, Shaw R, Zhao W, Zhang X, Ventura A, Liu Y, et al:
Pan-cancer alterations of the MYC oncogene and its proximal network
across the cancer genome atlas. Cell Syst. 6:282–300.e282. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Yu CY, Wang L, Khaletskiy A, Farrar WL,
Larner A, Colburn NH and Li JJ: STAT3 activation is required for
interleukin-6 induced transformation in tumor-promotion sensitive
mouse skin epithelial cells. Oncogene. 21:3949–3960. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Wang SW and Sun YM: The IL-6/JAK/STAT3
pathway: Potential therapeutic strategies in treating colorectal
cancer (Review). Int J Oncol. 44:1032–1040. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Aryappalli P, Al-Qubaisi SS, Attoub S,
George JA, Arafat K, Ramadi KB, Mohamed YA, Al-Dhaheri MM, Al-Sbiei
A, Fernandez-Cabezudo MJ and Al-Ramadi BK: The IL-6/STAT3 signaling
pathway is an early target of manuka honey-induced suppression of
human breast cancer cells. Front Oncol. 7:1672017. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Zou S, Tong Q, Liu B, Huang W, Tian Y and
Fu X: Targeting STAT3 in cancer immunotherapy. Mol Cancer.
19:1452020. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Arantes-Rodrigues R, Colaço A, Pinto-Leite
R and Oliveira PA: In vitro and in vivo experimental models as
tools to investigate the efficacy of antineoplastic drugs on
urinary bladder cancer. Anticancer Res. 33:1273–1296.
2013.PubMed/NCBI
|
|
58
|
Varley CL and Southgate J: Organotypic and
3D reconstructed cultures of the human bladder and urinary tract.
Methods Mol Biol. 695:197–211. 2011. View Article : Google Scholar : PubMed/NCBI
|