Introduction
Osteosarcoma is the most frequent primary bone
malignancy affecting the metaphysis of the long bones in children
and adolescents. Perioperative chemotherapy combined with surgical
treatment has markedly improved five-year disease-free survival and
overall survival rates of 59 and 65% (1); however, treatment-resistant and
recurrent cases, with metastases and poor prognosis, show a mean
survival time of <1 year (2,3).
Driver-gene mutations directly involved in tumorigenesis and growth
have been discovered in many cancers, leading to the development of
molecularly targeted drugs with high efficacy and response rates
(4). Although several molecularly
targeted drugs for osteosarcoma have been tested in clinical
trials, their effectiveness was observed to be insufficient,
probably due to the large number of subtypes and non-unified
genetic abnormalities of the disease (5,6).
Therefore, the mechanisms of tumorigenesis in osteosarcoma must be
determined to improve treatment outcomes.
Survivin, a member of the inhibitor of apoptosis
protein family, inhibits apoptosis by suppressing caspase activity
(7). While it is not expressed in
normal tissues, it is overexpressed in almost all types of cancer,
including osteosarcoma (8). In
addition, patients with high expression of survivin are reported to
have a poor prognosis in various cancers (9–15).
The present authors previously reported that increased expression
of survivin is a poor prognostic factor in osteosarcoma (16).
YM155 is a molecularly targeted drug developed as an
inhibitor of survivin expression, leading to the suppression of
cell proliferation and exhibiting anti-tumor effects by improving
drug sensitivity (17). YM155 is a
potential therapeutic target undergoing clinical testing for lung
cancer and melanoma; however, no difference in patient survival was
reported between treatment with docetaxel alone or in combination
with YM155 in a phase II study of breast cancer (18). Thus, one-to-one drugs have
limitations and therapeutic agents that act more broadly to target
multiple genes may be needed.
Micro (mi)RNAs are small non-coding RNAs of 18–25
nucleotides that regulate gene expression by binding to the
3′-untranslated region of target mRNAs, thereby regulating various
processes such as metabolism, immunity and digestion. Additionally,
miRNAs are involved in the progression of numerous diseases,
including cancer, making them potential therapeutic targets
(19,20).
Of the numerous miRNAs that reportedly target
survivin (Table I) (21), the present study focused on
miR-218, as it targets genes acting on the NF-κB and the
Wnt/β-catenin pathways, which play crucial roles in osteosarcoma
tumorigenesis (Table II)
(22). miR-218 suppresses survivin
expression and exerts antitumor effects in glioblastoma and
cervical cancer (23,24). In addition, it improves drug
sensitivity and induces apoptosis in lung cancer (25). In osteosarcoma, the mitochondrial
apoptotic pathway is thought to be regulated by B-cell lymphoma 2
(Bcl-2) family proteins, including the anti-apoptotic proteins
Bcl-2 and Bcl-xl and pro-apoptotic proteins Bax, Bak and Bad
(26). Although miR-218 is known
to arrest the cell cycle through E2F2 as a tumor suppressor
(27), little is known about its
role in other processes, such as apoptosis. In addition, the
regulation of survivin expression by miR-218 in osteosarcoma has
not been evaluated in vivo.
 | Table I.miRNAs that regulate survivin. |
Table I.
miRNAs that regulate survivin.
| miRNA | Type of cancer | Target gene | Effect |
|---|
| −494 | Acute lymphoblastic
leukemia | TEL-AML1 | Apoptosis |
| −16 | Colorectal
cancers | Direct
survivin | Apoptosis |
| −320a | Acute lymphoblastic
leukemia | TEL-AML1 | Apoptosis |
| −542-3p | Lung cell carcinoma
cell line A549 | Direct
survivin | Cell cycle
arrest |
| −708 | Renal cell
carcinoma | Direct
survivin | Activation
caspase-3, −9 |
| −218 | Nasopharyngeal
carcinoma | Direct
survivin | Apoptosis |
| −143 | Breast cancer cell
line MCF-7 | Direct
survivin | Inhibited
E2-induced cell proliferation |
| −203 | Prostate
cancer | Direct
survivin | Cell death |
|
| Laryngeal
cancer | Direct
survivin | G1 phase cell cycle
arrest |
|
| Hepatocellular
carcinoma | Direct
survivin | Suppressed
proliferation |
|
| Lung cancer | Direct
survivin | Inhibits
proliferation and invasion |
|
| Pancreatic cancer
cells | Direct
survivin | Apoptosis, cell
cycle arrest |
|
| Osteosarcoma | Direct
survivin | Apoptosis, cell
cycle arrest |
| −150 | Burkitt's lymphoma
cell line DG75 | Direct
survivin | Apoptosis |
| −34a | Non-small cell lung
cancer | Notch-1 | Apoptosis |
|
| Neuroblastoma | MYCN | Apoptosis |
|
| Laryngeal squamous
cell carcinoma | Direct
survivin | Cell cycle
arrest |
|
| Gastric cancer | PI3K/AKT/survivin
pathway | Improve the
sensitivity of cisplatin |
|
| Melanoma cell
lines | Direct
survivin | Apoptosis |
|
| Osteosarcoma | Direct
survivin | Apoptosis, cell
cycle arrest |
 | Table II.Target genes of micro-RNA 218. |
Table II.
Target genes of micro-RNA 218.
| Pathway | Gene name | Official name | Functions |
|---|
| RTK pathway | RTKs | Receptor tyrosine
kinase | Cell surface
receptors for growth factors, cytokines and hormones |
|
| PLCy1 | Phosphoinositide
phospholipase C-gamma-1 | Intracellular
transduction of receptor |
|
| ARAF | A-Raf
proto-oncogene | Involvement of cell
growth and development |
| AKT/mTOR
pathway | PIK3C2A | Phosphoinositide
3-kinase-C2-alpha | Cell proliferation,
migration and intracellular protein trafficking |
|
| RICTOR | RPTOR independent
companion of MTOR complex 2 | Embryonic growth
and development |
| Focal adhesion
pathway | Laminin 5 | - | High-molecular
weight proteins of the extracellular matrix |
|
| Integrin | - | Transmembrane
receptors to facilitate cell-extracellular matrix adhesion |
|
| CAV2 | Caveolin 2 | Main component of
the inner surface of caveolae |
|
| ACTN1 |
Alpha-actinin-1 | Spectrin gene
superfamily |
|
| PXN | Paxillin | Actin-membrane
attachment of cell adhesion to ECM |
|
| LASP1 | LIM and SH3 domain
protein 1 | cAMP and cGMP
dependent signaling protein |
|
| SH3GL1 | SH3 domain
containing GRB2 like 1 | Endocytosis and
cell cycle |
| wnt/β-catenin
pathway | FZ | frizzled | Receptors in the
Wnt signaling pathway |
|
| LEF1 | Lymphoid
enhancer-binding factor 1 | Forming a complex
with β-catenin and promotes transcriptional activity |
|
| Survivin | Baculoviral
inhibitor of apoptosis repeat-containing 5 | Inhibitor of
apoptosis (IAP) family |
|
| HMGB1 | High mobility group
box 1 protein | Cell
differentiation and tumor cell migration |
| NF-κB pathway | ECOP | EGFR-coamplified
and overexpressed protein | Increasing NF-κB
transcriptional activity |
|
| IKKβ | Inhibitor of
nuclear factor kappa-B kinase subunit beta | Controlling
activation of NF-kappa-B |
| Cell cycle | CCND1 | Cyclin D1 | Regulation of the
G1/S phase transition |
|
| TET2 | Tet methylcytosine
dioxygenase 2 | Involvement of
myelopoiesis |
|
| BMI1 | B lymphoma Mo-MLV
insertion region 1 homolog | Oncogene by
regulating p16 and p19 |
|
| CDK6 | Cyclin dependent
kinase 6 | G1 phase
progression and G1/S transition |
| Slit-robo
pathway | Robo1 | Roundabout homolog
1 | Regulation of cell
migration and cell adhesion |
| Epigenetic
pathway | HOXB3 | Homeobox B3 | Involvement of
development and biologic subset of AML |
A single miRNA simultaneously targets multiple genes
and thus, may normalize an entire failed biological network;
however, the therapeutic efficacy of miRNAs compared with that of
molecularly targeted drugs remains to be elucidated. The present
study evaluated the in vitro and in vivo anti-tumor
efficacy of miR-218 in osteosarcoma and compared the effects with
those of YM155 to provide a foundation for the development of
therapeutic strategies.
Materials and methods
Osteosarcoma and normal osteoblast
cell lines
The human osteosarcoma cell lines MG63 and HOS were
purchased from the Health Science Research Resource Bank of Japan.
The normal osteoblast cell line hFOB1.19 was purchased from ATCC.
The cells were cultured in Dulbecco's modified Eagle's medium
(DMEM; Nacalai Tesque, Inc.) supplemented with 10% fetal bovine
serum (Nichirei Biosciences, Inc.), 0.1 mM non-essential amino
acids (Thermo Fisher Scientific, Waltham, MA, USA) and 100 IU/ml
penicillin-streptomycin-glutamine (Thermo Fisher Scientific, Inc.)
and incubated at 37°C with 5% CO2.
Transfection of miRNA-218 mimics,
YM155 and controls
Osteosarcoma cell lines (1×105 cells)
were treated with a has-miR-218-5p mimic (sequence,
5′-UUGUGCUUGAUCUAACCAUGU-3′; miR-218 group), designed and purchased
from Ambion and negative control miRNA (cat. no. 4464058; Thermo
Fisher Scientific, Inc.; NC-miR group), adjusted to a final
concentration of 50 nM, using Lipofectamine® 2000
Transfection Reagent (Invitrogen; Thermo Fisher Scientific, Inc.)
at 37°C for 48 h. Cells were alternately administered YM155 (YM155
group), purchased from Cayman Chemical Company and adjusted to a
final concentration of 5 nM. Phosphate-buffered saline (PBS; NC
group) was used as a control.
Reverse transcription-quantitative
(RT-q) PCR
At 48 h post-treatment (transfection with miRNA-218
or administration of YM155), total RNA was extracted from each cell
type (1×105 cells) using the RNeasy Mini kit (Qiagen
GmbH). The extracted total RNA was reverse transcribed using Prime
Script RT Master Mix (Takara Bio Inc.) to prepare complementary DNA
(cDNA) at a concentration of 500 ng/µl according to the
manufacturer's recommendations. RT-qPCR for miR-218 was performed
using Takara Prime Script and TaqMan Probe (Takara Bio, Inc.)
according to the manufacturer's instructions. The PCR reactions
were carried out with an initial denaturation for 30 sec at 95°C
followed by 40 cycles of 95°C for 5 sec and 60°C for 30 sec. RNU44
(Thermo Fisher Scientific, Inc.) was used as an endogenous control.
The synthesized cDNA was used to analyze survivin expression levels
using SYBR Premix Ex Taq II (Takara Bio, Inc.). The PCR reactions
were carried out with an initial denaturation for 30 sec at 95°C
followed by 40 cycles of 95°C for 5 sec, 55°C for 10 sec and 72°C
for 30 sec. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was
used as an endogenous control to correct the expression levels with
the 2−ΔΔCq method (28). The experiment was repeated three
times. The specific primers used were as follows: miR-218 (assay ID
000521; Thermo Fisher Scientific, Inc.); RNU44 (assay ID 001094;
Thermo Fisher Scientific, Inc.); survivin-forward,
5′-ACCGCATCTCTACATTCAAG-3′; survivin-reverse,
5′-CAAGTCTGGCTCGTTCTC-3′; GAPDH-forward,
5′-TCACCAGGGCTGCTTTTAAC-3′; GAPDH-reverse,
5′-TGACGGTGCCATGGAATTTG-3′.
Western blotting
The cells were lysed using Pierce RIPA buffer
(Thermo Fisher Scientific, Inc.) containing a protease inhibitor
(Roche Applied Science, Basel, Switzerland). After sonication, the
cells were centrifuged and the supernatant was collected for
protein extraction. The protein concentration was measured using
Bio-Rad DC kits (Bio-Rad Laboratories, Inc.). Cell lysates (20 µg
of protein) were electroblotted with 4–12% NuPAGE Bis-Tris gel
(Thermo Fisher Scientific, Inc.) at 30 mA. The primary antibodies
rabbit anti-survivin (cat. no. 2808; 1:1,000), anti-MMP-2 (cat. no.
13132; 1:1,000) and anti-MMP-9 (cat. no. 13667; 1:1,000; all from
Cell Signaling Technology, Inc.) were transferred to difluoride
membranes by the semi-dry method and blocked at 4°C for 1 h with
Blocking One (Nacalai Tesque, Inc.) to block non-specific binding.
Each primary antibody was diluted with Can Get Signal
Immunoreaction Enhancer Solution 1 (Cytiva) and incubated with the
membrane for 12 h at 4°C. The anti-GAPDH rabbit polyclonal antibody
(cat. no. ab9485; Abcam,) was used as an endogenous control. After
primary antibody treatment, the membranes were washed with 0.1%
Tween-containing PBS and incubated with secondary horseradish
peroxidase-conjugated anti-rabbit IgG antibodies (cat. no. NA934;
Cytiva), each diluted in 0.1% Tween-containing PBS at 1:2,000, for
1 h at 25°C. Chemiluminescence was induced using Chemi-Lumi One
Super assay kit (Nacalai Tesque, Inc.) before bioimaging. Data were
visualized and photographed using the analyzer LAS-4000 (Fujifilm).
Protein expression was semi-quantified using ImageJ software (ver.
1.53; National Institute of Health) and quantified by correcting
for the band concentration of GAPDH for each of the visualized
proteins.
Water-soluble tetrazolium salts 8 cell
proliferation assay
Post-treatment (transfection with miRNA-218 or
administration of YM155), the cells were seeded into 96-well plates
and incubated for 24, 48, or 72 h. Next, the Cell Count Reagent SF
(Nacalai Tesque, Inc.) was added to the culture medium and
incubated at 37°C for 1 h. Absorbance was measured using Wallac
1420 ARVO MX (PerkinElmer, Inc.).
Fluorescence-activated cell sorting
analysis
Following transfection with miRNA-218 or NC-miRNA or
administration of YM155 or PBS, the cells were seeded into 6-well
plates, incubated for 48 h, treated with trypsin/EDTA and
centrifuged at 300 × g for 3 min at 4°C. The pellet was dissolved
in 500 µl of 1X binding buffer, transferred to a
fluorescence-activated cell sorting (FACS) tube, Cells were stained
with Annexin V-FITC and propidium iodide using Annexin V-FITC
Apoptosis Detection kit (cat. no. K101-25, Abcam) according to the
manufacturer's protocols. The cells were sorted by FL1-H (Annexin V
channel) and FL2-H (PI channel) on a FACSCalibur (BD Biosciences)
flow cytometer and analyzed by CellQuest (v. 3.3; BD Biosciences).
The apoptotic rate was calculated as the percentage of early and
late apoptotic cells.
Animal model
A total of 24 eight-week-old male nude mice
(26.4±1.4 g, BALB/cAJcl-nu/nu) were purchased from CLEA Japan, Inc.
and kept in an environment of 22°C and 60% humidity under a 12-h
dark/light cycle, with free access to a food and water.
Osteosarcoma cells were lysed in 100 µl Matrigel and
5.0×106 cells were injected subcutaneously into the
mice. Tumor size was calculated as the long diameter × short
diameter × height/2 and experiments were begun when the tumor
volume was >120 mm3. AteloGene containing miRNAs,
miR-218, or NC-miRNAs was injected subcutaneously at the
recommended concentration of 25 µM, whereas YM155 (3 mg/kg) and PBS
were injected intratumorally (n=6 per group). Each treatment was
applied three times at 0, 7 and 14 days. Tumor volumes were
measured every 2–3 days until tumor resection at 18 days. Animals
were sacrificed with intraperitoneal administration of sodium
pentobarbital (100 mg/kg). The tumor tissue was subjected to
survivin immunostaining according to the manufacturer's protocols
(cat. no. 2808, 1:200; Cell Signaling Technology, Inc.) and the
immunohistochemistry score was calculated as the area × density
(0–15 points), where the area is the percentage of tumor cells per
slide (0, 0%; 1, <5%; 2, 5–20%; 3, 21–50%; 4, 51–75%; 5,
>75%) and density can be absent, weak, moderate, or strong,
ranging from 0–3, respectively. Each slide was observed once by
three individuals. The animal study was approved by the Animal
Experiment Committee of Nihon University School of Medicine, Japan
(approval number AP16M012) and humane endpoints were also in
accordance with this approval.
Wound healing assay
The osteosarcoma cells were seeded into 24-well
plates at 1.0×105 cells/200 µl/well and a gap was
created by scraping the centre of the bottom of the dish 24 h
post-incubation using a cell scratcher scratch stick (AGC Techno
Glass). Following treatment with miRNA-218, NC-miRNA, YM155, or
PBS, medium containing 10% FBS) and mitomycin C (MMC, 0.25 µg/ml)
was added to eliminate the growth effect. The pore area at 0, 24,
48 and 72 h was imaged by phase-contrast microscope Leica DM IL
(Leica Microsystems GmbH), calculated using ImageJ software as
follows: migration rate=[(pore area at 0 h)-(pore area at 24, 48,
or 72 h)]/(pore area at 0 h) ×100.
Matrigel invasion assay
Invasion chambers (Corning, Inc.) containing
miR-218, YM155, or NC-miRNA were used to study the invasive
capacity of the cells, which is the ability to degrade and migrate
extracellular substrates beyond the basement membrane. Osteosarcoma
cells (5×105 cells/well) were suspended in minimum
essential medium (MEM) or DMEM, seeded into the Matrigel-coated
upper chamber and incubated with MMC (0.25 µg/ml) in the presence
of 10% fetal bovine serum. After 72 h of treatment, cells that had
passed to the bottom of the chamber were fixed in 4%
paraformaldehyde for 15 min, stained with Giemsa (24°C, 2 min) and
counted under phase-contrast microscope Leica DM IL (Leica
Microsystems GmbH) using ×20 objective and measurements were taken
in 10 fields of view per chamber.
Statistical analysis
Each experiment was performed at least three times.
Data are presented as the means ± standard deviation or median and
range. Student's t-test and Mann-Whitney U test were used for
comparing two groups and one-way ANOVA or mixed repeated measures
ANOVA was used for comparing multiple groups where the Bonferroni's
correction was used to compare differences between groups.
Statistical analysis was performed using GraphPad Prism 5 software
(GraphPad, Inc.). P<0.05 was considered to indicate a
statistically significant difference.
Results
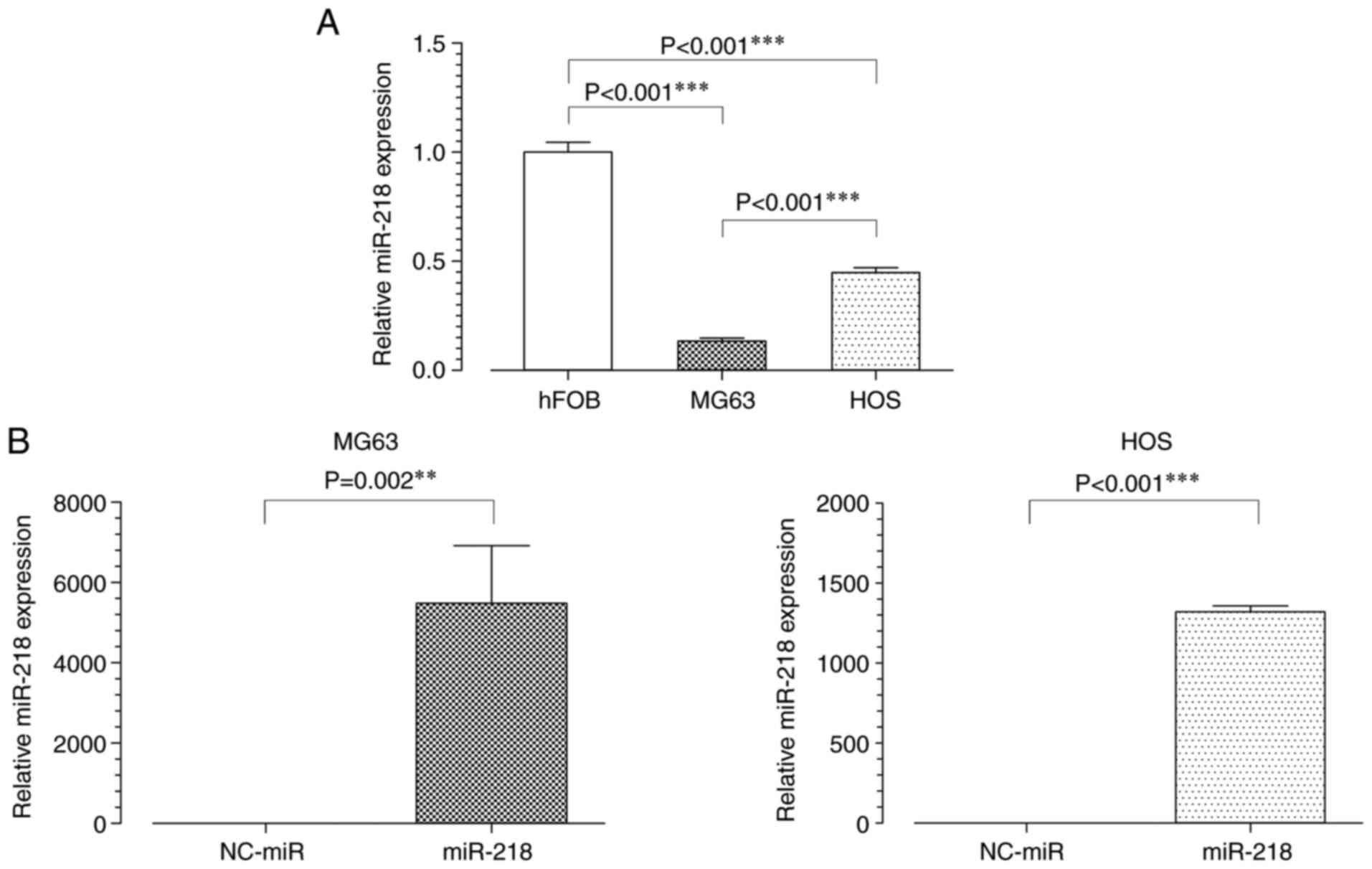
miR-218 expression is downregulated in
osteosarcoma cell lines compared with normal cells
To analyze the function of miR-218 in osteosarcoma,
its expression levels in osteosarcoma and normal osteoblast cells
was first assessed using RT-qPCR. miR-218 expression levels were
significantly lower in osteosarcoma cell lines, MG63 and HOS,
compared with normal osteoblast cells, hFOB (Fig. 1A). After transfection with miR-218,
MG63 and HOS cells overexpressed miR-218 (Fig. 1B). Although differences in
transfection efficiency between the two osteosarcoma cell lines
were observed, the amount of original miR-218 were significantly
lower in MG63 cells than in HOS cells. Therefore, MG63 cells were
used in subsequent experiments as it was easier to evaluate the
effects of miR-218.
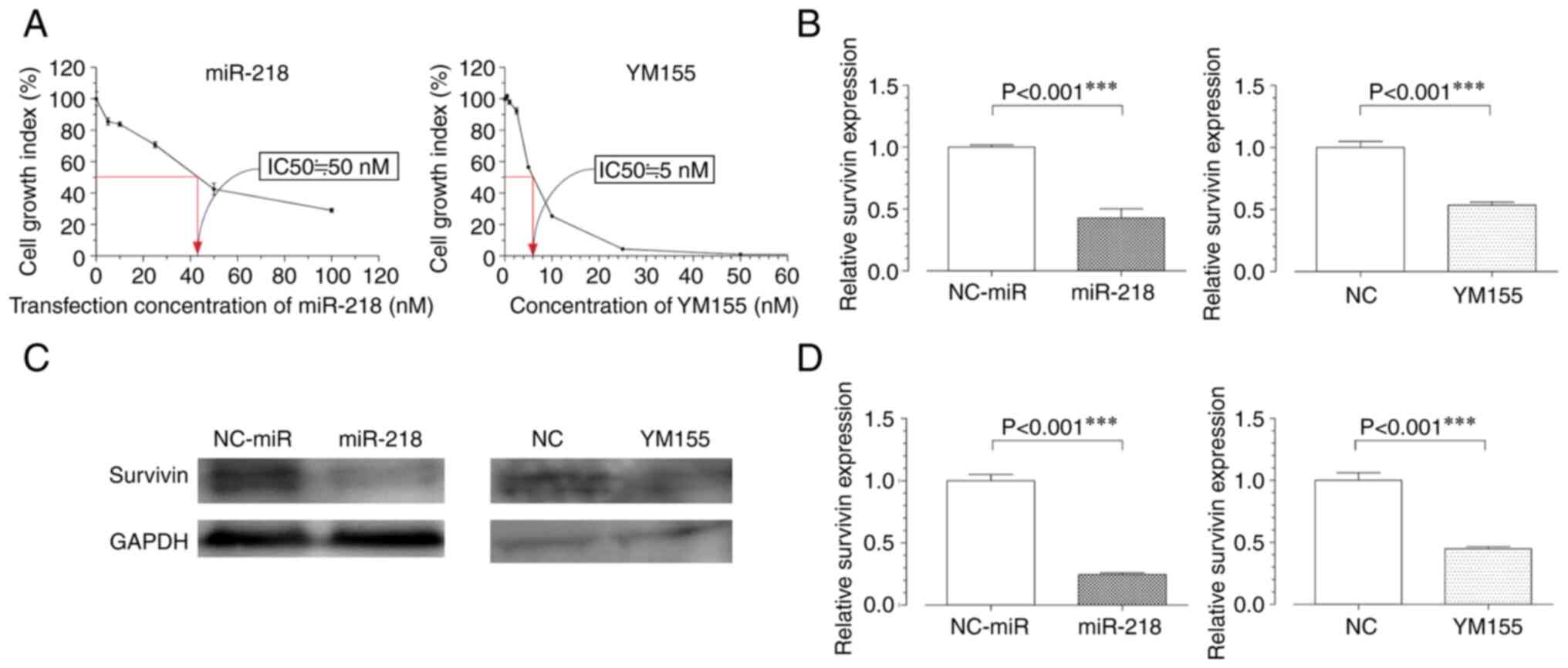
miR-218 overexpression and YM155
administration suppresses survivin expression
The half-maximal inhibitory concentration
(IC50) was measured in a water-soluble tetrazolium salts
8 (WST 8) cell proliferation assay 48 h post-treatment with miR-218
or YM155. The IC50 of miR-218 was 50 nM, whereas that of
YM155 was 5 nM; these concentrations were used in further
experiments involving MG63 cells (Fig.
2A). The RT-qPCR results showed that survivin transcription
levels were decreased in both experimental groups compared with
those in the negative control groups (Fig. 2B). Western blot analysis confirmed
these results, revealing decreased protein expression levels of
survivin in the miR-218 and YM155 groups (Fig. 2C and D).
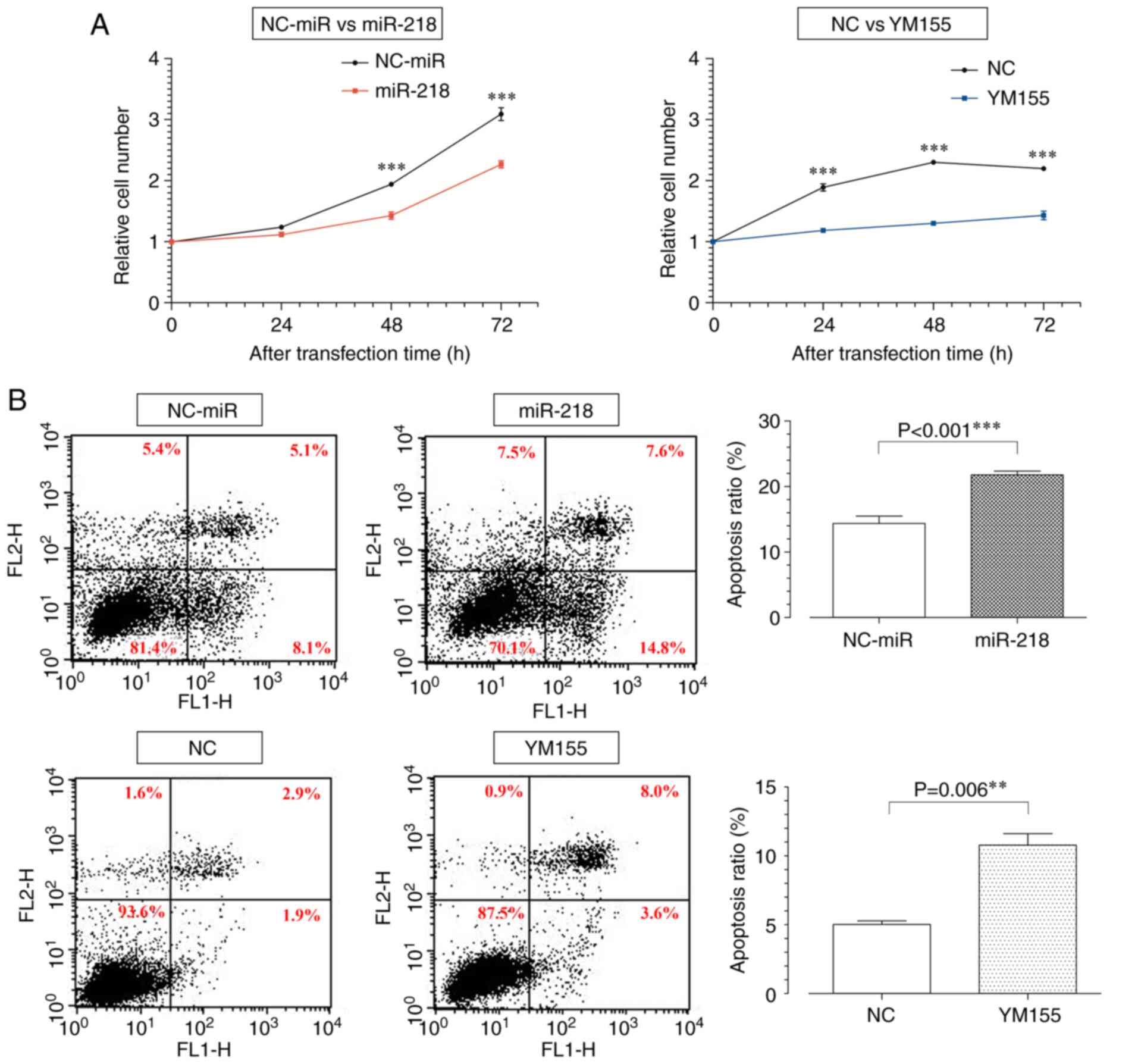
miR-218 overexpression and YM155
administration inhibits cell proliferation
To investigate the effects of miR-218 overexpression
and YM155 administration on cell proliferation in osteosarcoma, a
WST 8 assay and FACS analysis were performed. The WST 8 assay
showed that the miR-218 and YM155 groups had reduced proliferative
capacity compared with the control groups 72 h post-treatment
(Fig. 3A).
Lipofectamine® 2000 (Thermo Fisher Scientific, Inc.),
which was used in gene transfer, has been reported as toxic
(29) and probably caused a
decrease in the cell number in the NC-miR group. For the same
reason, the miR-218 group failed to show a 50% suppression in the
cell number compared with the NC-miR group 48 h post-treatment,
even though the concentration was set to the IC50.
However, the NC group was not affected by gene transfer and reached
the cell number limit 48 h post-treatment. To evaluate apoptosis 48
h post-treatment, Annexin V-PI staining was performed. FACS
analysis showed an increase in the rightward migration of each cell
in the miR-218 and YM155 groups, indicating early and late
apoptosis (Fig. 3B).
miR-218 and YM155 inhibits tumor
volume growth
The effect of miR-218 overexpression on
tumorigenesis in osteosarcoma was evaluated in vivo. In nude
mice, miR-218 and YM155 significantly inhibited tumor growth
compared with that in the NC-miR and NC groups 18 days
post-treatment (Fig. 4A and B).
Survivin immunostaining was performed on tumor specimens and both,
the miR-218 and YM155 groups, showed decreased immunohistochemistry
scores (Fig. 4C).
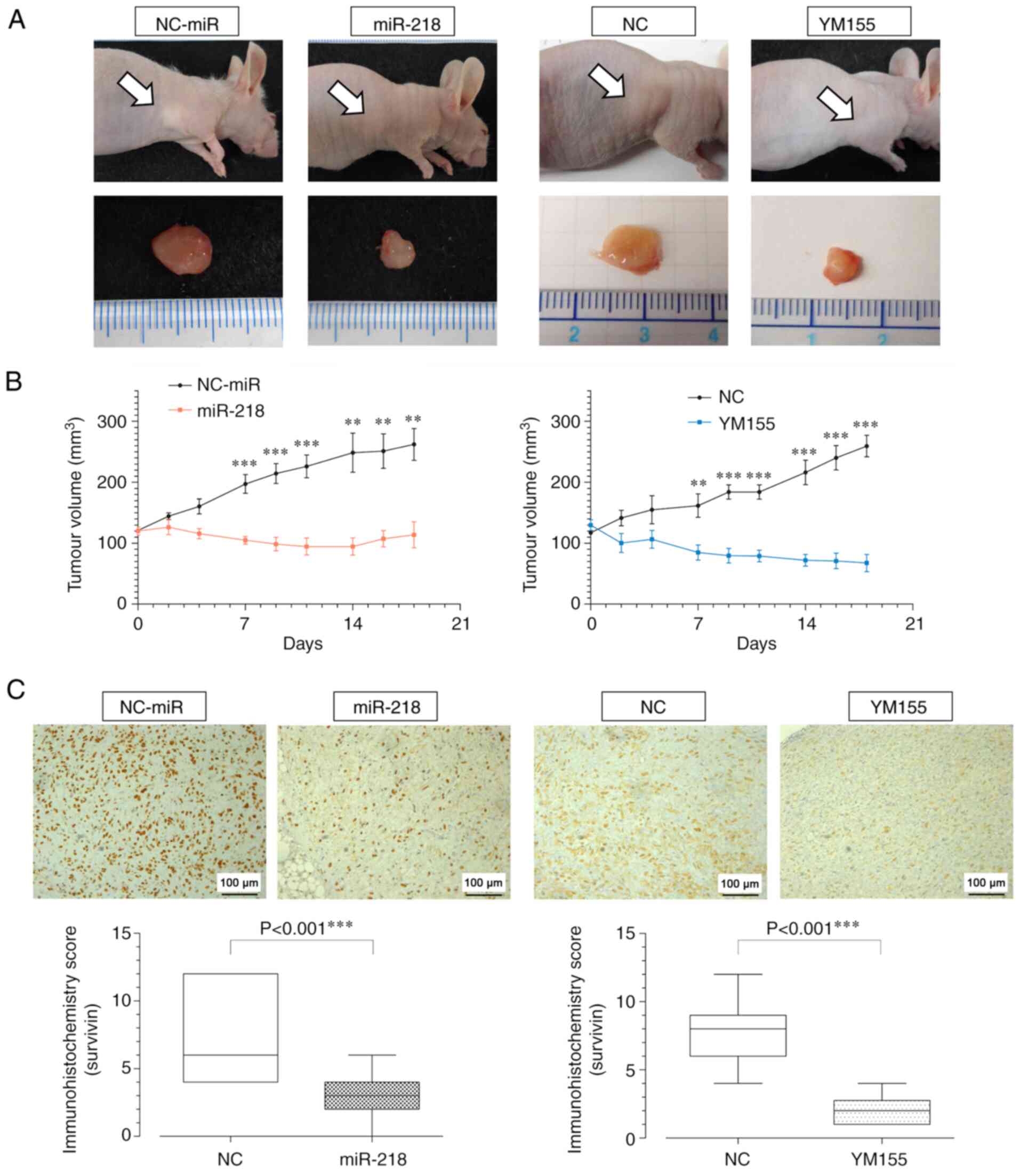 | Figure 4.Tumor volume and survivin expression
following miR-218 overexpression and YM155 treatment (MG63) in an
immunodeficient mouse model. (A) Gross tumor at 18 days
post-treatment. (B) Changes in tumor volume over time. Mixed
repeated measures ANOVA with post hoc Bonferroni-corrected t-test.
(miR-218: 7, 9, 11 days, P<0.001, 14 days, P=0.002, 16 days,
P=0.002, 18 days, P=0.002, days; YM155: 7 days, P=0.009, 9, 11, 14,
16, 18 days, P<0.001) (C) Survivin staining of tumor tissues and
immunohistochemistry score. (miR-218: P<0.001, YM155:
P<0.001). Scale bar=100 µm. **P<0.01, ***P<0.001. miR,
microRNA. |
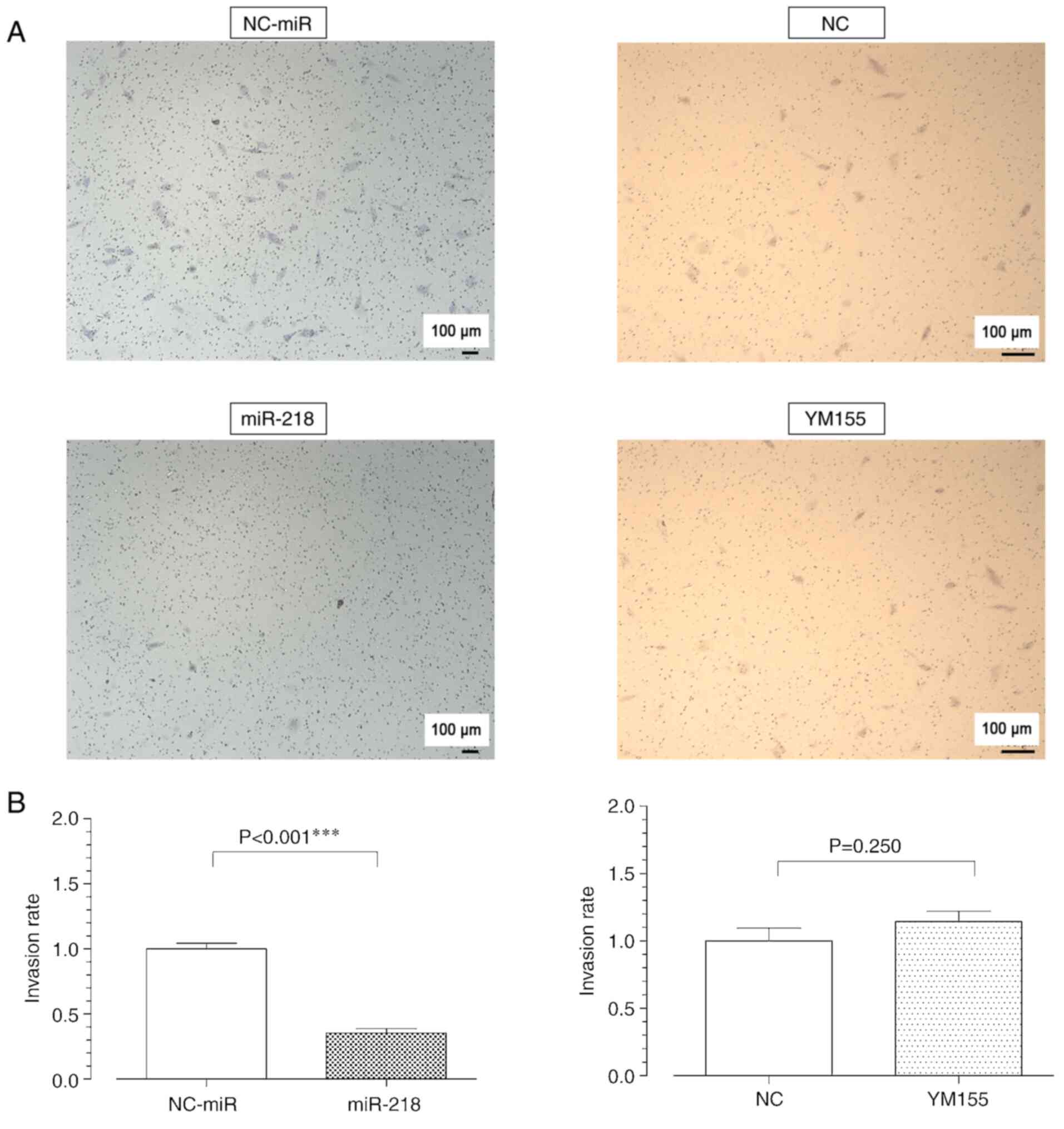
Overexpression of miR-218, but not
YM155, inhibits cell migration and invasion capacity
Effect of the overexpression of miR-218 in
osteosarcoma was assessed using wound healing and migration assays.
The former assay showed a decrease in the pore area of the miR-218
group and consequently in its migration capacity; in contrast, no
significant difference was observed between the YM155 and NC groups
(Fig. 5). Consistently, the
migration assay showed inhibition of the invasive capacity in the
miR-218 group, but no significant difference was observed between
the YM155 and NC groups (Fig. 6).
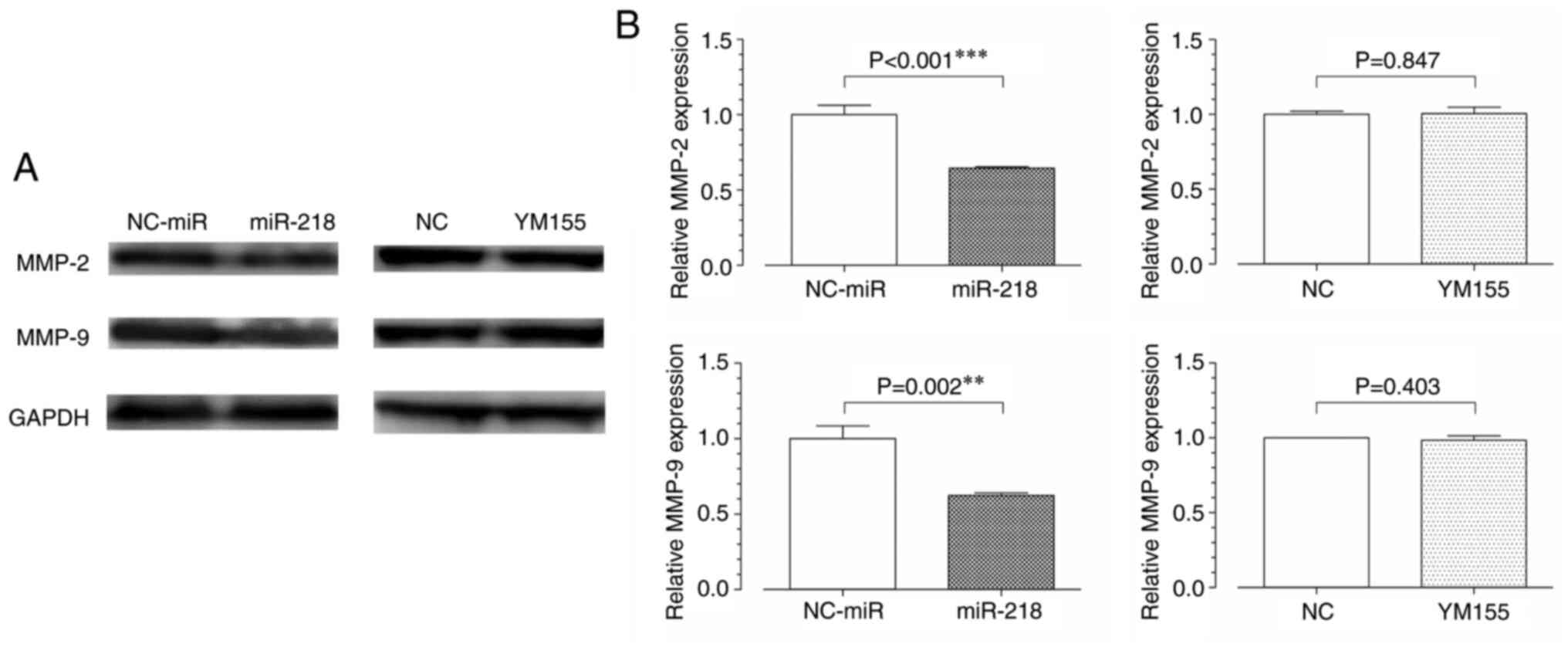
The expression level of MMP-2/9, which is involved in the migration
and invasion ability of cells was further assessed. In western
blotting analysis, MMP-2/9 levels were found to be decreased in the
miR-218 group but showed no significant difference between the
YM155 and NC groups (Fig. 7).
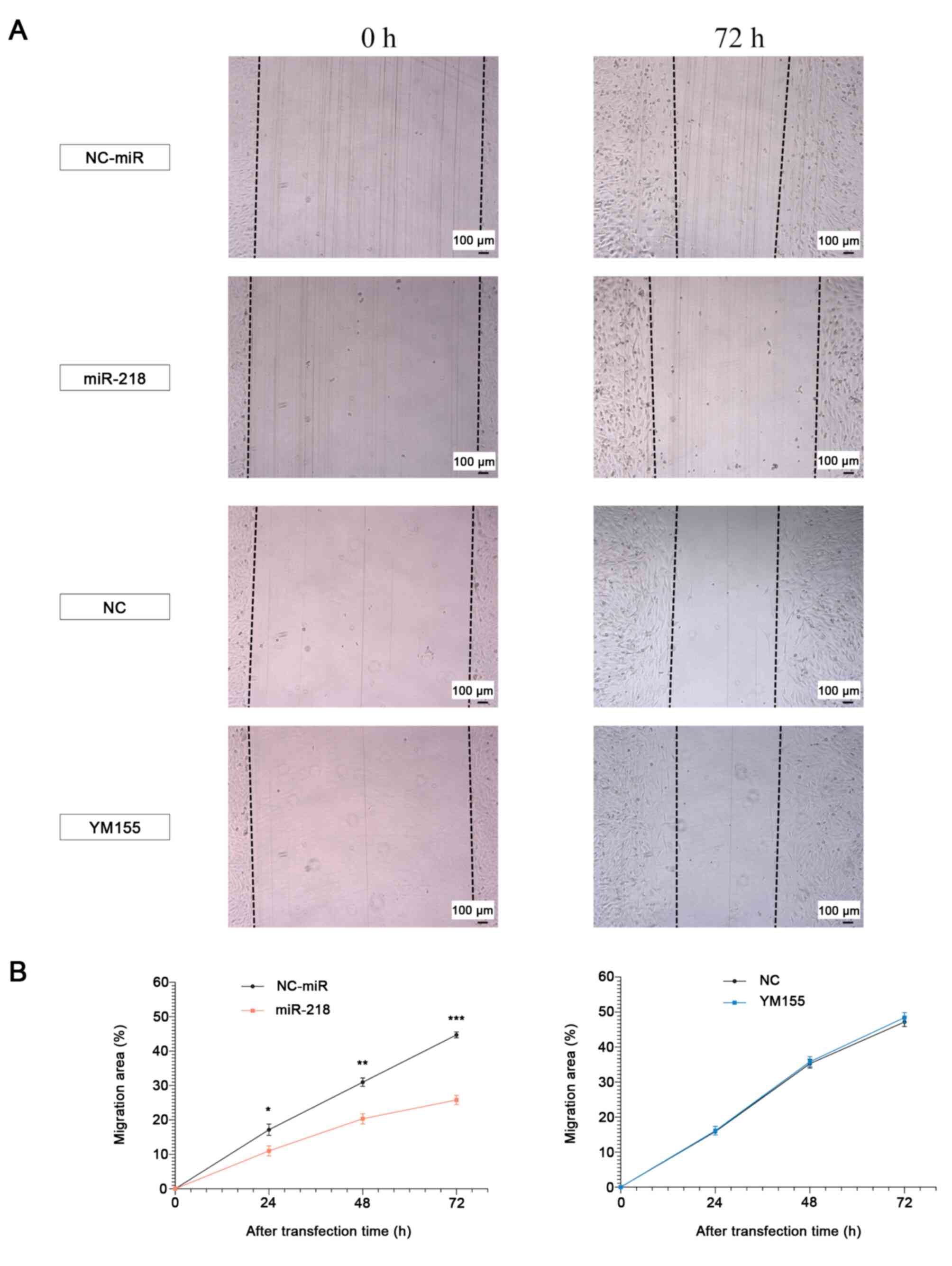 | Figure 5.Migration ability following miR-218
overexpression and YM155 treatment (MG63). (A and B) Changes in
migration capacity 24, 48 and 72 h post-treatment. Mixed repeated
measures ANOVA with post hoc Bonferroni-corrected t-test. (miR-218:
24 h, P=0.049, 48 h, P=0.005, 72 h, P<0.001, YM155: 24 h,
P=0.835, 48 h, P=0.774, 72 h, P=0.547) Scale bar: 100 µm.
*P<0.05, **P<0.01, ***P<0.001. miR, microRNA. |
Discussion
The present study showed that in osteosarcoma,
miR-218 overexpression and YM155 treatment both led to apoptosis
and inhibited the cell proliferative capacity in vivo and
in vitro. However, miR-218 inhibited cell invasion and
migration, whereas YM155 did not. There is no direct comparison in
the literature between the multi-target miR-218 and single-target
molecularly targeted drugs. In osteosarcoma, which shows a
non-unified genetic abnormality, it was demonstrated that miR-218
has a broader effect and is more effective than single-target
drugs.
Cancer is a disease caused by genetic abnormalities;
however, the mechanism of cancer development remains unclear.
Mutations in driver genes directly involved in tumorigenesis and
growth have been discovered in certain types of cancer (such as
breakpoint cluster region-Abelson tyrosine kinase in chronic
myeloid leukemia and genetic mutations of epidermal growth factor
receptor in lung cancer) (30,31).
Molecularly targeted drugs (such as gefitinib used in lung cancer)
are highly efficient against cancers caused by driver-gene
mutations and increase the median survival of patients (32,33).
Clinical trials of various molecularly targeted agents have been
conducted in osteosarcoma. Although antitumor activity of one such
agent, cabozantinib, has been reported, its long-term action and
effect on metastasis remain unclear, probably due to the
non-unified genetic abnormality inherent to osteosarcoma (5,34).
Clinical trials with miRNAs for treating malignant
mesothelioma and hepatitis C showed positive outcomes (35,36);
however, the use of miRNAs for treating osteosarcoma has not been
evaluated. The present study focused on survivin, a therapeutic
target gene for anti-tumor drugs, which is not expressed in normal
tissues but is overexpressed in tumor cells. In osteosarcoma,
miR-34a and miR-203 suppress the survivin gene, causing cell cycle
arrest in the G2-M phase and inducing apoptosis and
improving sensitivity to cisplatin (37). However, their effects on the cell
invasive and migratory capacity, as well as their in vivo
anti-tumor potency are unknown.
miR-218 is a molecular switch that, in addition to
its other actions shown in Table
II, suppresses survivin and the Wnt/β-catenin pathway. Lu et
al (22) and Fernández et
al (38) show that lymphoid
enhancer-binding factor (lef) 1, β-catenin and T-cell factor
(tcf)/lef are involved in survivin expression. Therefore, miR-218
may directly and indirectly regulate survivin, which is highly
expressed in osteosarcoma. Bmi-1, a constituent gene of the NF-κB
pathway, is associated with the development, extension and
prognosis of osteosarcoma. The Bmi-1 antibody, AbBmi-1, suppresses
NF-κB and MMP-9 expression and reduces the proliferative and
migratory potential of osteosarcoma cells; thus, it may be useful
as a novel therapeutic target (39). In addition, epithelial to
mesenchymal transition has been extensively studied for the
migratory and invasive potential of tumor cells. It has been
reported that miR-218 targets and suppresses the expression of
roundabout homolog 1 and epidermal growth factor
receptor-coamplified and overexpressed protein, thereby decreasing
NF-κB transcriptional activity and suppressing epithelial to
mesenchymal transition (40).
Wnt/β-catenin-mediated complexes with transcription factors, such
as tcf and lef, as well as transcriptional coactivators, are
intimately involved in osteosarcoma tumorigenesis, resulting in
transcriptional activity of multiple downstream target oncogenes
(41). In summary, the present
study focused on miR-218 because it is closely involved in
osteosarcoma tumorigenesis and has many targets in the NF-κB and
Wnt/β-catenin pathways, along with affecting the expression of
survivin.
The present study investigated the underlying cause
of the suppression of invasion and migration in the miR-218 group,
but not in the YM155 group, and focused on MMPs, which serve a
central role in cancer metastasis, particularly MMP-2/9, which are
target genes of miR-218. miR-218 and not YM155, suppressed the
expression of MMP-2/9, which is in contrast with a previous study
suggesting that migration and invasion were suppressed by YM155
treatment (42,43). This discrepancy may be attributed
to the considerably higher concentrations of YM155 used in previous
studies (42). Typically, the
anti-tumor effect is proportional to the concentration and duration
of administration of a drug. In clinical practice, high drug
concentrations that show anti-tumor potency often cause strong side
effects. Additionally, MMCs were not administered to prevent
proliferation in previous studies in the assessment of migration
capacity (43). Addition of MMCs
can eliminate the impact of the proliferative capacity, allowing
accurate assessment of migration and invasion capacity (44). The present study revealed a
significant difference between the miR-218 and YM155 groups in the
IC50 for the migration and invasion potentials at the
same degree of proliferative capacity inhibition migration and
invasion ability. Furthermore, MMC administration minimized the
effect of the proliferative capacity, demonstrating further
efficacy and potential of miRNA-targeted treatment methods.
One limitation of the present study is that it used
only MG63 cells to compare the antitumor effects of miR-218 and
YM155. MG63 cells form highly malignant tumors. It has been
reported that osteosarcoma cell lines show characteristic labelling
profiles, producing extracellular matrices with different
compositions and that differences in tumor malignancy exist among
different cell lines (45). Among
them, MG63 cells show a higher methylation rate of WW
domain-containing oxidoreductase and higher proliferative and
invasive capacity compared with HOS cells (46). Additionally, MG63 cells showed high
efficiency of miR-218 transfection, which was suitable for the
comparative study with YM155. The amount of original miR-218 in
MG63 cells was significantly lower than that in HOS cells (Fig. 1A) and the expression of miR-218
following transfection was higher in MG63 cells (Fig. 1B). As the purpose of the present
study was to investigate the anti-tumor effect of miR-218
overexpression in comparative analysis with YM155 via target genes
including survivin, the MG63 cell line was chosen for the present
study. However, further studies in other cell lines with varying
levels of malignancy might help to elucidate the role of miR-218 in
the treatment of osteosarcoma. The second limitation is that the
present study was not able to conduct experiments with clinical
specimens of osteosarcoma. In the future, it will be necessary to
verify the antitumor effects of miR-218 and YM155 in clinical
specimens of osteosarcoma.
Overall, miRNAs may have broad clinical applications
as therapeutic molecules, as they re-express originally intrinsic
genes. Due to the non-unified genetic abnormality inherent to
osteosarcoma, the antitumor effect of miR-218, which regulates more
genes than molecularly targeted drugs, may be enhanced in
osteosarcoma compared with other carcinomas.
Acknowledgements
The authors would like to express our deepest
gratitude to the members of the Department of Orthopedic Surgery,
Nihon University School of Medicine.
Funding
The present study was supported by Nihon University School of
Medicine Alumni Association 60th Anniversary Fund Research Grant
(2016) to Dr Eiji Osaka (Department of Orthopedic Surgery, Nihon
University School of Medicine).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
KS, EO, KF, TT, YT and KN proposed the concept and
design of the study. KS and RF made substantial contributions to
the study execution and acquisition of data. KS and EO confirm the
authenticity of all the raw data. All authors critically revised
the manuscript, commented on drafts and approved the final
manuscript. All authors reviewed and approved the final
manuscript.
Ethics approval and consent to
participate
The animal study was approved by the Animal
Experiment Committee of Nihon University School of Medicine, Japan
(approval number AP16M012).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Eaton BR, Schwarz R, Vatner R, Yeh B,
Claude L, Indelicato DJ and Laack N: Osteosarcoma. Pediatr Blood
Cancer. 68 (Suppl 2):e283522021. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bielack SS, Kempf-Bielack B, Delling G,
Exner GU, Flege S, Helmke K, Kotz R, Salzer-Kuntschik M, Werner M,
Winkelmann W, et al: Prognostic factors in high-grade osteosarcoma
of the extremities or trunk: An analysis of 1,702 patients treated
on neoadjuvant cooperative osteosarcoma study group protocols. J
Clin Oncol. 20:776–790. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kempf-Bielack B, Bielack SS, Jürgens H,
Branscheid D, Berdel WE, Exner GU, Göbel U, Helmke K, Jundt G,
Kabisch H, et al: Osteosarcoma relapse after combined modality
therapy: An analysis of unselected patients in the cooperative
osteosarcoma study group (COSS). J Clin Oncol. 23:559–568. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lee YT, Tan YJ and Oon CE: Molecular
targeted therapy: Treating cancer with specificity. Eur J
Pharmacol. 834:188–196. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Szuhai K, Cleton-Jansen AM, Hogendoorn PC
and Bovée JV: Molecular pathology and its diagnostic use in bone
tumors. Cancer Genet. 205:193–204. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Shaikh AB, Li F, Li M, He B, He X, Chen G,
Guo B, Li D, Jiang F, Dang L, et al: Present advances and future
perspectives of molecular targeted therapy for osteosarcoma. Int J
Mol Sci. 17:5062016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Liu Y, Teng Z, Wang Y, Gao P and Chen J:
Prognostic significance of survivin expression in osteosarcoma
patients: A meta-analysis. Med Sci Monit. 21:2877–2885. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Mita AC, Mita MM, Nawrocki ST and Giles
FJ: Survivin: Key regulator of mitosis and apoptosis and novel
target for cancer therapeutics. Clin Cancer Res. 14:5000–5005.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Islam A, Kageyama H, Hashizume K, Kaneko Y
and Nakagawara A: Role of survivin, whose gene is mapped to 17q25,
in human neuroblastoma and identification of a novel
dominant-negative isoform, survivin-beta/2B. Med Pediatr Oncol.
35:550–553. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kami K, Doi R, Koizumi M, Toyoda E, Mori
T, Ito D, Fujimoto K, Wada M, Miyatake S and Imamura M: Survivin
expression is a prognostic marker in pancreatic cancer patients.
Surgery. 136:443–448. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kappler M, Köhler T, Kampf C,
Diestelkötter P, Würl P, Schmitz M, Bartel F, Lautenschläger C,
Rieber EP, Schmidt H, et al: Increased survivin transcript levels:
An independent negative predictor of survival in soft tissue
sarcoma patients. Int J Cancer. 95:360–363. 2001.PubMed/NCBI
|
|
12
|
Miyachi K, Sasaki K, Onodera S, Taguchi T,
Nagamachi M, Kaneko H and Sunagawa M: Correlation between survivin
mRNA expression and lymph node metastasis in gastric cancer.
Gastric Cancer. 6:217–224. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Monzó M, Rosell R, Felip E, Astudillo J,
Sánchez JJ, Maestre J, Martín C, Font A, Barnadas A and Abad A: A
novel anti-apoptosis gene: Re-expression of survivin messenger RNA
as a prognosis marker in non-small-cell lung cancers. J Clin Oncol.
17:2100–2104. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Sarela AI, Macadam RC, Farmery SM, Markham
AF and Guillou PJ: Expression of the antiapoptosis gene, survivin,
predicts death from recurrent colorectal carcinoma. Gut.
46:645–650. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wang H, Xi X, Kong X, Huang G and Ge G:
The expression and significance of survivin mRNA in urinary bladder
carcinomas. J Cancer Res Clin Oncol. 130:487–490. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Osaka E, Suzuki T, Osaka S, Yoshida Y,
Sugita H, Asami S, Tabata K, Sugitani M, Nemoto N and Ryu J:
Survivin expression levels as independent predictors of survival
for osteosarcoma patients. J Orthop Res. 25:116–121. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Gao JZ, Chen FH, Wang L, Wei H and Meng
SL: YM155 inhibits tumor growth and enhances chemosensitivity to
cisplatin in osteosarcoma. Eur Rev Med Pharmacol Sci. 19:2062–2069.
2015.PubMed/NCBI
|
|
18
|
Clemens MR, Gladkov OA, Gartner E,
Vladimirov V, Crown J, Steinberg J, Jie F and Keating A: Phase II,
multicenter, open-label, randomized study of YM155 plus docetaxel
as first-line treatment in patients with HER2-negative metastatic
breast cancer. Breast Cancer Res Treat. 149:171–179. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Di Leva G, Garofalo M and Croce CM:
MicroRNAs in cancer. Annu Rev Pathol. 9:287–314. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Tutar L, Tutar E, Özgür A and Tutar Y:
Therapeutic targeting of microRNAs in cancer: Future perspectives.
Drug Dev Res. 76:382–388. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Huang J, Lyu H, Wang J and Liu B: MicroRNA
regulation and therapeutic targeting of survivin in cancer. Am J
Cancer Res. 5:20–31. 2014.PubMed/NCBI
|
|
22
|
Lu YF, Zhang L, Waye MM, Fu WM and Zhang
JF: MiR-218 mediates tumorigenesis and metastasis: Perspectives and
implications. Exp Cell Res. 334:173–182. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Tong X, Yang P, Wang K, Liu Y, Liu X, Shan
X, Huang R, Zhang K and Wang J: Survivin is a prognostic indicator
in glioblastoma and may be a target of microRNA-218. Oncol Lett.
18:359–367. 2019.PubMed/NCBI
|
|
24
|
Kogo R, How C, Chaudary N, Bruce J, Shi W,
Hill RP, Zahedi P, Yip KW and Liu FF: The microRNA-218~survivin
axis regulates migration, invasion and lymph node metastasis in
cervical cancer. Oncotarget. 6:1090–1100. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zarogoulidis P, Petanidis S, Kioseoglou E,
Domvri K, Anestakis D and Zarogoulidis K: MiR-205 and miR-218
expression is associated with carboplatin chemoresistance and
regulation of apoptosis via Mcl-1 and survivin in lung cancer
cells. Cell Signal. 27:1576–1588. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Dai G, Zheng D, Guo W, Yang J and Cheng
AY: Cinobufagin induces apoptosis in osteosarcoma cells via the
mitochondria-mediated apoptotic pathway. Cell Physiol Biochem.
46:1134–1147. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Xuan C, Jin M, Gao Y, Xu S, Wang L, Wang
Y, Han R and An Q: miR-218 suppresses the proliferation of
osteosarcoma through downregulation of E2F2. Oncol Lett.
17:571–577. 2019.PubMed/NCBI
|
|
28
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Deng W, Fu M, Cao Y, Cao X, Wang M, Yang
Y, Qu R, Li J, Xu X and Yu J: Angelica sinensis polysaccharide
nanoparticles as novel non-viral carriers for gene delivery to
mesenchymal stem cells. Nanomedicine. 9:1181–1191. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Waller CF: Imatinib mesylate. Recent
Results Cancer Res. 212:1–27. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Maemondo M, Inoue A, Kobayashi K, Sugawara
S, Oizumi S, Isobe H, Gemma A, Harada M, Yoshizawa H, Kinoshita I,
et al: Gefitinib or chemotherapy for non-small-cell lung cancer
with mutated EGFR. N Engl J Med. 362:2380–2388. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Kataoka K, Osaka E, Shimizu T, Okamura Y,
Yoshida Y and Tokuhashi Y: Lung squamous cell carcinoma with
brachial soft tissue metastasis responsive to gefitinib: Report of
a rare case. Thorac Cancer. 7:676–680. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Pakkala S and Ramalingam SS: Personalized
therapy for lung cancer: Striking a moving target. JCI Insight.
3:1208582018. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Italiano A, Mir O, Mathoulin-Pelissier S,
Penel N, Piperno-Neumann S, Bompas E, Chevreau C, Duffaud F,
Entz-Werlé N, Saada E, et al: Cabozantinib in patients with
advanced Ewing sarcoma or osteosarcoma (CABONE): A multicentre,
single-arm, phase 2 trial. Lancet Oncol. 21:446–455. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
van Zandwijk N, Pavlakis N, Kao SC, Linton
A, Boyer MJ, Clarke S, Huynh Y, Chrzanowska A, Fulham MJ, Bailey
DL, et al: Safety and activity of microRNA-loaded minicells in
patients with recurrent malignant pleural mesothelioma: A
first-in-man, phase 1, open-label, dose-escalation study. Lancet
Oncol. 18:1386–1396. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Janssen HL, Reesink HW, Lawitz EJ, Zeuzem
S, Rodriguez-Torres M, Patel K, van der Meer AJ, Patick AK, Chen A,
Zhou Y, et al: Treatment of HCV infection by targeting microRNA. N
Engl J Med. 368:1685–1694. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Chen X, Chen XG, Hu X, Song T, Ou X and
Zhang C, Zhang W and Zhang C: MiR-34a and miR-203 inhibit survivin
expression to control cell proliferation and survival in human
osteosarcoma cells. J Cancer. 7:1057–1065. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Fernández JG, Rodríguez DA, Valenzuela M,
Calderon C, Urzúa U, Munroe D, Rosas C, Lemus D, Díaz N, Wright MC,
et al: Survivin expression promotes VEGF-induced tumor angiogenesis
via PI3K/Akt enhanced β-catenin/Tcf-Lef dependent transcription.
Mol Cancer. 13:2092014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Liu J, Luo B and Zhao M: Bmi-1-targeting
suppresses osteosarcoma aggressiveness through the NF-κB signaling
pathway. Mol Med Rep. 16:7949–7958. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Li YJ, Zhang W, Xia H, Zhang BS, Chen P,
Zhao YL and Li J: miR-218 suppresses epithelial-to-mesenchymal
transition by targeting Robo1 and Ecop in lung adenocarcinoma
cells. Future Oncol. 13:2571–2582. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Lin CH, Ji T, Chen CF and Hoang BH: Wnt
signaling in osteosarcoma. Adv Exp Med Biol. 804:33–45. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Zhang W, Liu Y, Li YF, Yue Y, Yang X and
Peng L: Targeting of survivin pathways by YM155 inhibits cell death
and invasion in oral squamous cell carcinoma cells. Cell Physiol
Biochem. 38:2426–2437. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Zhang Z, Ma L and Wang J: YM155 exerts a
growth inhibitory effect on human osteosarcoma in vitro and
in vivo. Oncol Rep. 34:1074–1080. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Imai M, Muraki M, Takamatsu K, Saito H,
Seiki M and Takahashi Y: Spontaneous transformation of human
granulosa cell tumours into an aggressive phenotype: A metastasis
model cell line. BMC Cancer. 8:3192008. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Pautke C, Schieker M, Tischer T, Kolk A,
Neth P, Mutschler W and Milz S: Characterization of osteosarcoma
cell lines MG-63, Saos-2 and U-2 OS in comparison to human
osteoblasts. Anticancer Res. 24:3743–3748. 2004.PubMed/NCBI
|
|
46
|
Liu Y, Wang Q, Yu P, Miao W, Liu C, Pu Y
and Zhang C: Methylation of WWOX gene promotes proliferation of
osteosarcoma cells. J BUON. 25:2708–2713. 2020.PubMed/NCBI
|